Abstract
Background
This study aimed to identify the pharmacological targets and mechanisms of action of the traditional Chinese medicine, formononetin, in the treatment of Alzheimer’s disease (AD) using network pharmacological analysis.
Material/Methods
Targets of AD were obtained by using DisGeNET gene discovery platform, the herbal ingredients target (HIT) database, the SuperPred, and the SwissTargetPrediction compound target prediction platforms. Pathogenic and therapeutic targets were imported to the STRING biological database, and Cytoscape network integration software was used to construct component-target and disease-target interaction networks. Core targets were identified by topological analysis and were further tested to identify the biological processes and signaling pathways.
Results
Seven key target genes for formononetin in the treatment of patients with AD were identified, including estrogen receptor alpha (ESR1), peroxisome proliferator-activated receptor gamma (PPARG), tumor protein p53 (TP53), sirtuin 1 (SIRT1), tumor necrosis factor (TNF), cytochrome P450 19A1 (CYP19A1), and nuclear factor (erythroid-derived 2)-like 2 (NFE2L2). The biological processes included hormone metabolism, regulation of nucleoside, nucleotide and nucleic acid metabolism, apoptosis, energy pathways, metabolism, cell communication, and signal transduction. The signaling pathways included histone acetylation and deacetylation (HDAC) class I, regulation of p38-alpha/beta, p38 mitogen-activated protein kinase (MAPK) signaling pathway, bone morphogenetic protein (BMP) receptor signaling, interleukin-1 (IL1) mediated signaling events, the tumor necrosis factor (TNF) receptor signaling pathway, and cytoplasmic and nuclear Smad2/3 signaling.
Conclusions
Pharmacological network analysis was used to identify the gene targets and mechanisms of formononetin treatment in patients with AD.
MeSH Keywords: Alzheimer Disease, Behavior and Behavior Mechanisms, Neuropharmacology
Background
Alzheimer’s disease (AD) is a form of dementia that gradually develops and leads to increased cognitive impairment [1]. Based on reports from the US Centers for Disease Control and Prevention (CDC), it has been estimated that there will be around 14 million cases of AD by 2060 [2]. Although the early symptoms of AD are mild, late-stage symptoms include disorientation, language, mood, and behavioral changes. Also, patients with advanced AD can lose the ability to control bodily functions, and AD can lead to death [3,4]. The etiology of AD is poorly understood. Currently, there are no effective pharmacological treatments for AD. However, some therapeutic approaches can reduce or control the symptoms of AD. Therefore, there remains a need for innovative approaches to identify potential therapeutic targets for AD.
In China, a traditional herb derived from the Astragalus root is used to extract the bioactive component of formononetin, which is used in traditional Chinese medicine. Several studies have shown that formononetin has beneficial therapeutic effects in several diseases, including cancer, diabetes, neurological disease, and cardiovascular disease [5,6]. Recently, the neuroprotective effect of formononetin in patients with AD has been demonstrated in both in vivo and in vitro studies [7,8]. However, the detailed molecular mechanisms for the pharmacological and biological actions of formononetin remain unknown. Following recent developments in bioinformatics, network pharmacology may now be used to identify or predict the biological networks of compound-protein interactions, protein-protein interactions, and biological signaling functions. Also, systematic pharmacology analysis can interpret and integrate these interactive networks using bioinformatics and statistics [9].
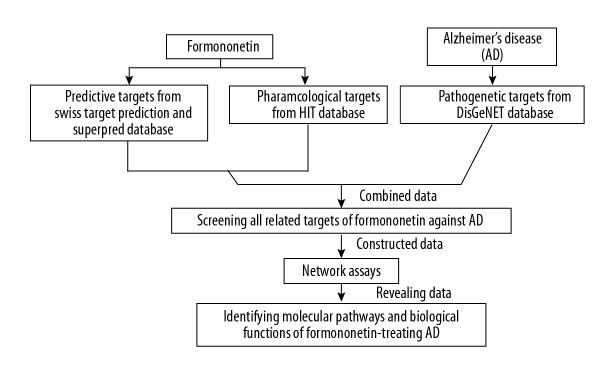
Therefore, this study aimed to identify the pharmacological targets and mechanisms of action of the traditional Chinese medicine, formononetin, in the treatment of AD using network pharmacological analysis. The study design and analytical flowchart is shown in Figure 1.
Figure 1.
The study design and analytical flowchart of the bioinformatics and network pharmacology approach to predicting the hub targets and signaling mechanisms of formononetin in the treatment of Alzheimer’s disease (AD).
Material and Methods
Target data collection for formononetin in the treatment of Alzheimer’s disease (AD)
The web-based Binding Database was used to obtain the verifiable targets of formononetin and to collect the predictive targets of formononetin using the herbal ingredients target (HIT) database, the SuperPred, and the SwissTargetPrediction compound target prediction platforms. The DisGeNET gene discovery platform was used to screen AD-associated genes and to identify the top 200 genes. The overlapping targets from formononetin and AD then allowed identification of gene targets of formononetin in the treatment of AD.
Protein-protein interaction (PPI) network construction and topology analysis
The web-based STRING database was used to analyze the combined targets before obtaining the target function-related protein of formononetin for the treatment of AD. A target-related protein network of formononetin for the treatment of AD was produced through screening the PPIs with scores >0.9 and excluding the duplicated targets. Network-Analyzer in the Cytoscape software was applied to obtain the target-related proteins through assaying topological parameters of the mean degree of freedom and maximum degree of freedom, followed by selection of the core targets according to the value of degree. The upper limit of the screening range was from the maximum degree value in the topological data, and the lower limit was from the two-fold average degree of freedom [10].
Biological function and pathway enrichment analysis
The web-based FunRich functional enrichment software was used to enrich the biological processes and molecular pathways from all core targets of formononetin for the treatment of AD. The histograms of these biological processes and signaling pathways were plotted according to their degree of importance.
Results
Informational presentation of identifiable targets
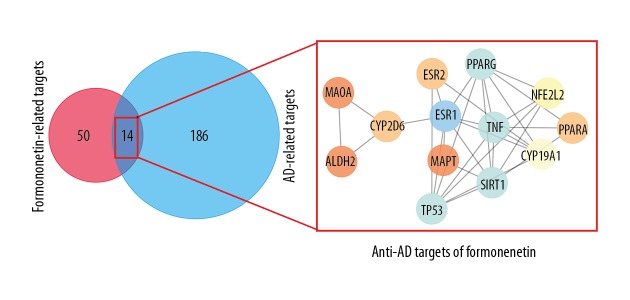
In Alzheimer’s disease (AD), network pharmacological analysis identified 2,244 genes from the DisGeNET database. The top 200 AD genes were screened as investigative targets. Also, 10 and 15 formononetin targets were obtained from the herbal ingredients target (HIT) and the SwissTargetPrediction databases. There were 23 verifiable targets and 20 predictive targets of formononetin that were harvested from the prediction database, and 50 targets were finally obtained by eliminating the duplicates. After mapping, 13 hub targets of formononetin for the treatment of AD were identified, as shown in Figure 2.
Figure 2.
After constructing the targets for formononetin in the treatment of Alzheimer’s disease (AD), a protein-protein interaction (PPI) network was produced to identify the 14 interactive core targets.
The protein-protein interaction (PPI) network of formononetin for the treatment of AD
The STRING database was used to construct the functional PPI network associated with 13 targets. The targets were selected using the data with a confidence score >0.9 before being analyzed by Cytoscape to produce the graphical network of formononetin for the treatment of AD. The protein network had seven nodes and 11 edges that interacted, as shown in Figure 3.
Figure 3.
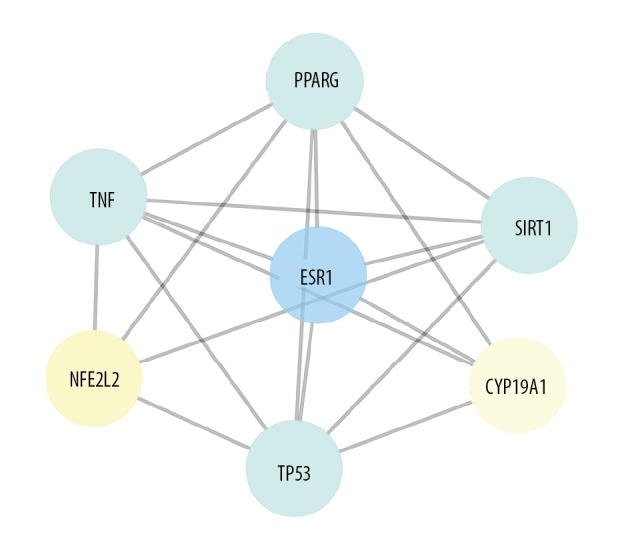
In further assays, the key targets for formononetin in the treatment of Alzheimer’s disease (AD) were obtained. Seven key target genes for formononetin in the treatment of patients with AD were identified, including estrogen receptor alpha (ESR1), peroxisome proliferator-activated receptor gamma (PPARG), tumor protein p53 (TP53), sirtuin 1 (SIRT1), tumor necrosis factor (TNF), cytochrome P450 19A1 (CYP19A1), and nuclear factor (erythroid-derived 2)-like 2 (NFE2L2).
Biological functions and pathway enrichment assays of core targets
The results of biological function enrichment analysis using FunRich functional enrichment software showed that the biological processes of core targets were mainly involved hormone metabolism, regulation of nucleoside, nucleotide and nucleic acid metabolism, apoptosis, energy pathways, metabolism, cell communication, and signal transduction (Figure 4). These data showed that formononetin might exert its therapeutic effects by regulating these biological processes. Also, the data from signaling pathway enrichment analysis by FunRich software showed that the signaling pathways of hub targets mainly involved signaling events mediated by histone acetylation and deacetylation (HDAC) class I, regulation of p38-alpha/beta, p38 mitogen-activated protein kinase (MAPK) signaling pathway, bone morphogenetic protein (BMP) receptor signaling, interleukin-1 (IL-1) mediated signaling events, the tumor necrosis factor (TNF) receptor signaling pathway, and cytoplasmic and nuclear Smad2/3 signaling. (Figure 5).
Figure 4.
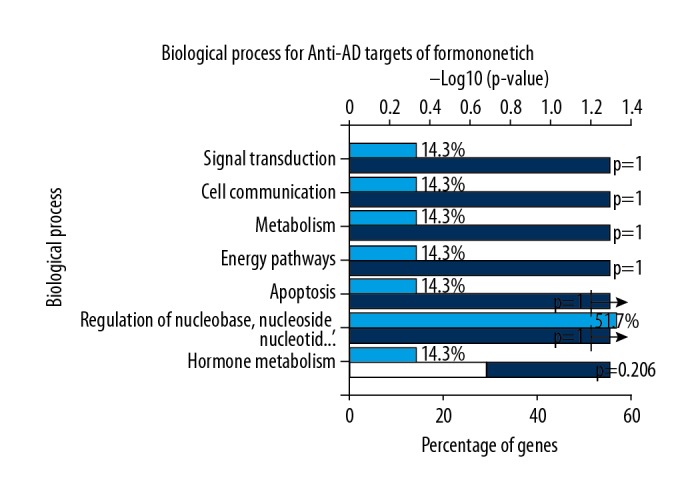
After biological functional analysis of the core targets, the seven main biological processes for formononetin in the treatment of Alzheimer’s disease (AD) are shown. The deep blue and white columns indicate P-values. The light blue columns indicate the percentage of genes (%).
Figure 5.
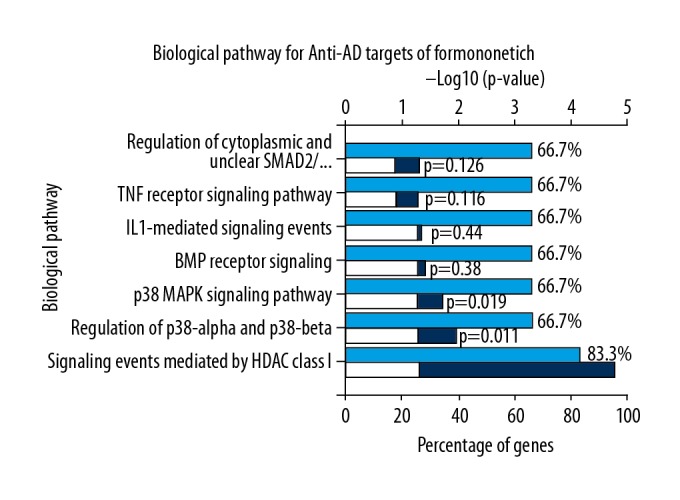
After pathway enrichment analysis of the core targets, the seven main signaling pathways for formononetin in the treatment of Alzheimer’s disease (AD) are shown. The deep blue and white columns indicate P-values. The light blue columns indicate the percentage of genes (%).
Discussion
This study aimed to apply a network pharmacology approach to predict the biological targets of formononetin in the treatment of patients with Alzheimer’s disease (AD) and to identify the biological mechanisms involved. The findings identified seven key target genes for formononetin in the treatment of patients with AD, including estrogen receptor alpha (ESR1), peroxisome proliferator-activated receptor gamma (PPARG), tumor protein p53 (TP53), sirtuin 1 (SIRT1), tumor necrosis factor (TNF), cytochrome P450 19A1 (CYP19A1), and nuclear factor (erythroid-derived 2)-like 2 (NFE2L2).
ESR1 is a key nuclear receptor that is required for neural development. Recent studies have shown that ESR1 has a neuroprotective role through mediating estrogen expression in neurodegenerative disease [11,12]. PPARG is a functional effector in regulating metabolism and has been associated with the pathology of neurodegenerative disorders. In early-stage AD, therapeutic targeting to PPARG may suppress disease progression in the brain [13,14]. TP53 functions as a key tumor suppressor, and when it is activated, TP53 induces neuronal apoptosis, possibly through transcription-associated molecular mechanisms that regulate apoptosis-dependent targets [15]. SIRT1 may have a role as a longevity gene for cellular protection from stress and is a possible therapeutic target for AD as it may modulate the function and survival of hippocampal and cortical neurons [16,17]. Tumor necrosis factor, (TNF) is another therapeutic target for AD through the inhibition of TNF-induced neuroinflammation to promote neuronal survival [18].
Previous studies have shown that the CYP19A1 enzyme functionally controls the biosynthesis of estrogen, and it is indirectly linked to AD, and that CYP19A1 mutation is a risk factor for AD in the Chinese population [19]. NFE2L2 is a transcription factor that is responsible for protecting against oxidative damage due to inflammation and injury, and its cytoprotective effects can suppress AD through antioxidative mechanisms [20]. Therefore, previously published studies support that these key biomolecules may be potential therapeutic targets for patients with AD. Identification of the signaling pathways for the therapeutic effects of formononetin for the treatment of AD was shown in this study, using enrichment analysis. However, these potential pharmacological targets should be validated by conducting future human in vivo and in vitro studies.
Conclusions
This study aimed to identify the pharmacological targets and mechanisms of action of the traditional Chinese medicine, formononetin, in the treatment of Alzheimer’s disease (AD) using network pharmacological analysis. Seven key target genes for formononetin in the treatment of patients with AD were identified, including estrogen receptor alpha (ESR1), peroxisome proliferator-activated receptor gamma (PPARG), tumor protein p53 (TP53), sirtuin 1 (SIRT1), tumor necrosis factor (TNF), cytochrome P450 19A1 (CYP19A1), and nuclear factor (erythroid-derived 2)-like 2 (NFE2L2). The biological processes included hormone metabolism, regulation of nucleoside, nucleotide and nucleic acid metabolism, apoptosis, energy pathways, metabolism, cell communication, and signal transduction. The signaling pathways included histone acetylation and deacetylation (HDAC) class I, regulation of p38-alpha/beta, p38 mitogen-activated protein kinase (MAPK) signaling pathway, bone morphogenetic protein (BMP) receptor signaling, interleukin-1 (IL-1) mediated signaling events, the tumor necrosis factor (TNF) receptor signaling pathway, and cytoplasmic and nuclear Smad2/3 signaling.
Footnotes
Source of support: This study was funded by the National Natural Science Foundation of China (No. 81560134)
Conflict of interest
None.
References
- 1.Lane CA, Hardy J, Schott JM. Alzheimer’s disease. Eur J Neurol. 2018;25:59–70. doi: 10.1111/ene.13439. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer’s Association. 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 2016;12:459–509. doi: 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Eschweiler GW, Leyhe T, Klöppel S, Hüll M. New developments in the diagnosis of dementia. Dtsch Arztebl Int. 2010;107:677–83. doi: 10.3238/arztebl.2010.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheltens P, Blennow K, Breteler MM, et al. Alzheimer’s disease. Lancet. 2016;388:505–17. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 5.Liang K, Ye Y, Wang Y, Zhang J, Li C. Formononetin mediates neuroprotection against cerebral ischemia/reperfusion in rats via downregulation of the Bax/Bcl-2 ratio and upregulation PI3K/Akt signaling pathway. J Neurol Sci. 2014;344:100–4. doi: 10.1016/j.jns.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Liu L, Wang J, Ren B, et al. Formononetin, an isoflavone from Astragalus membranaceus inhibits proliferation and metastasis of ovarian cancer cells. J Ethnopharmacol. 2018;221:91–99. doi: 10.1016/j.jep.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Ou S, Zhou L, et al. Formononetin attenuates Abeta25-35-induced cytotoxicity in HT22 cells via PI3K/Akt signaling and non-amyloidogenic cleavage of APP. Neurosci Lett. 2017;639:36–42. doi: 10.1016/j.neulet.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 8.Fei HX, Zhang YB, Liu T, et al. Neuroprotective effect of formononetin in ameliorating learning and memory impairment in mouse model of Alzheimer’s disease. Biosci Biotechnol Biochem. 2018;82:57–64. doi: 10.1080/09168451.2017.1399788. [DOI] [PubMed] [Google Scholar]
- 9.Su M, Guo C, Liu M, et al. Therapeutic targets of vitamin C on liver injury and associated biological mechanisms: A study of network pharmacology. Int Immunopharmacol. 2019;66:383–87. doi: 10.1016/j.intimp.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 10.Li R, Ma X, Song Y, et al. Anti-colorectal cancer targets of resveratrol and biological molecular mechanism: Analyses of network pharmacology, human and experimental data. J Cell Biochem. :2019. doi: 10.1002/jcb.28404. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Sundermann EE, Maki PM, Bishop JR. A review of estrogen receptor alpha gene (ESR1) polymorphisms, mood, and cognition. Menopause. 2010;17:874–86. doi: 10.1097/gme.0b013e3181df4a19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T. Meta-analysis of PvuII, XbaI variants in ESR1 gene and the risk of Alzheimer’s disease: The regional European difference. Neurosci Lett. 2014;574:41–46. doi: 10.1016/j.neulet.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Helisalmi S, Tarvainen T, Vepsäläinen S, et al. Lack of genetic association between PPARG gene polymorphisms and Finnish late-onset Alzheimer’s disease. Neurosci Lett. 2008;441(2):233–36. doi: 10.1016/j.neulet.2008.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Guan L, Luo D, et al. Gene–gene interaction between PPARG and APOE gene on late-onset Alzheimer’s disease: A case-control study in Chinese Han population. J Nutr Health Aging. 2017;21:397–403. doi: 10.1007/s12603-016-0794-y. [DOI] [PubMed] [Google Scholar]
- 15.Jazvinšćak Jembrek M, Slade N, Hof PR, Šimić G. The interactions of p53 with tau and Aβ as potential therapeutic targets for Alzheimer’s disease. Prog Neurobiol. 2018;168:104–27. doi: 10.1016/j.pneurobio.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Wong SY, Tang BL. SIRT1 as a therapeutic target for Alzheimer’s disease. Rev Neurosci. 2016;27:813–25. doi: 10.1515/revneuro-2016-0023. [DOI] [PubMed] [Google Scholar]
- 17.Rizzi L, Roriz-Cruz M. Sirtuin 1 and Alzheimer’s disease: An up-to-date review. Neuropeptides. 2018;71:54–60. doi: 10.1016/j.npep.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Chang R, Knox J, Chang J, et al. Blood-brain barrier penetrating biologic TNF-α inhibitor for Alzheimer’s disease. Mol Pharm. 2017;14:2340–49. doi: 10.1021/acs.molpharmaceut.7b00200. [DOI] [PubMed] [Google Scholar]
- 19.Zheng J, Yan H, Shi L, et al. The CYP19A1 rs3751592 variant confers susceptibility to Alzheimer disease in the Chinese Han population. Medicine (Baltimore) 2016;95:4742. doi: 10.1097/MD.0000000000004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branca C, Ferreira E, Nguyen TV, et al. Genetic reduction of Nrf2 exacerbates cognitive deficits in a mouse model of Alzheimer’s disease. Hum Mol Genet. 2017;26:4823–35. doi: 10.1093/hmg/ddx361. [DOI] [PMC free article] [PubMed] [Google Scholar]