Abstract
Background
Brain tumor surgery requires careful balance between maximizing tumor excision and preserving eloquent cortex. In some cases, the surgeon may opt to perform an awake craniotomy including intraoperative mapping of brain function by direct cortical stimulation (DCS) to assist in surgical decision-making. Preoperatively, functional magnetic resonance imaging (fMRI) facilitates planning by identification of eloquent brain areas, helping to guide DCS and other aspects of the surgical plan. However, brain deformation (shift) limits the usefulness of preoperative fMRI during surgery. To address this, an integrated visualization method for fMRI and DCS results is developed that is intuitive for the surgeon.
Methods
An image registration pipeline was constructed to display preoperative fMRI data corrected for brain shift overlaid on images of the exposed cortical surface at the beginning and completion of DCS mapping. Preoperative fMRI and DCS data were registered for a range of misalignments, and the residual registration errors were calculated. The pipeline was validated on imaging data from five brain tumor patients who underwent awake craniotomy.
Results
Registration errors were well under 5 mm (the approximate spatial resolution of DCS) for misalignments of up to 25 mm and approximately 10–15°. For rotational misalignments up to 20°, the success rate was 95% for an error tolerance of 5 mm. Failures were negligible for rotational misalignments up to 10°. Good quality registrations were observed for all five patients.
Conclusions
A proof-of-concept image registration pipeline is presented with acceptable accuracy for intraoperative use, providing multimodality visualization with potential benefits for intraoperative brain mapping.
Key words: Awake craniotomy, Brain mapping, Brain tumor resection, Electric stimulation, Functional mapping, Multimodal imaging, Surgical planning
Abbreviations and Acronyms: 2D, 2-dimensional; 3D, 3-Dimensional; CT, Computed tomography; DCS, Direct cortical stimulation; FOV, Field of view; fMRI, Functional magnetic resonance imaging; MRI, Magnetic resonance imaging; RE, Registration error; TE, Echo time; TR, Repetition time
Introduction
Functional magnetic resonance imaging (fMRI) has become a useful clinical tool for the surgical management of brain tumor patients, especially in awake craniotomy cases.1 Brain activity maps generated by fMRI allow identification of high-risk eloquent areas engaged in sensorimotor or language processing located proximal to the tumor. Such maps help to inform various aspects of presurgical planning including the optimal amount of brain exposure, the safest entry point, and the necessity and extent of intraoperative mapping, thereby assisting the surgeon in maximizing the benefit-to-risk ratio of the surgery.1, 2, 3, 4 When intraoperative mapping is performed by the gold standard method of direct cortical stimulation (DCS), fMRI activity maps may be visualized on a separate display or mentally recalled to guide the selection of stimulation points or to determine concordance between DCS and fMRI results. However, it can be somewhat challenging to use fMRI data in this manner due to the required mental image transformation, which is further complicated by tissue deformation, or “brain shift” that occurs once the skull bone is removed to reach underlying brain tissue.5, 6, 7
The amount of brain shift depends on multiple factors that include cerebrospinal fluid drainage, gravitational force effects, the use of drugs and surgical tools, tissue loss from tumor resection, and often tumor type, craniotomy size, and head orientation.7, 8, 9, 10 Although the surgeon may try to monitor and estimate the shift visually and mentally, the shift varies considerably (1–12 mm on average to a maximum of 50 mm8) and can include nonlinear spatial deformations across the brain surface.
Methods to tackle brain shift can be categorized into image-based approaches that use intraoperative MRI,11, 12 computed tomography (CT),13 or ultrasound14, 15 with image registration techniques or model-driven approaches that predict brain shift using finite element analysis models.16, 17 Previous work has typically focused on brain anatomy with limited attention toward brain shift correction of functional data, such as that provided by fMRI.15, 18 Despite the interest in using preoperative fMRI to guide DCS procedures, no methods presently exist to covisualize fMRI and DCS data intraoperatively. All DCS and fMRI concordance studies use postoperative analyses.19, 20 Therefore the goal of this work is to enhance the usefulness of preoperative fMRI data during awake craniotomies. A proof-of-concept image registration pipeline is developed for displaying brain shift−corrected fMRI data on the images of exposed cortical surface before and after DCS. As DCS results are conventionally documented simply by taking photographs, the pipeline has been implemented to provide similar 2-dimensional (2D) visualizations to the surgeon with the information of primary concern: the spatial relationship between preoperative fMRI results and intraoperative mapping results on the surface of the exposed brain. The pipeline is validated in silico using imaging data from 5 patients who underwent awake craniotomies.
Methods
Preoperative Data Acquisition
Preoperative functional and anatomic MRI were undertaken on 3T MRI systems at St. Michael's Hospital (Magnetom Skyra, software VD13A, Siemens, Erlangen, Germany) and Sunnybrook Health Sciences Centre (Magnetom Prisma, software VE11C, Siemens, Erlangen, Germany). The protocol included MP-RAGE (magnetization-prepared rapid gradient-echo) T1-weighted anatomic imaging (Magnetom Skyra: [GRAPPA factor 2; repetition time (TR)/echo time (TE)/inversion time/Ɵ = 2300 ms/2.26 ms/900 ms/9o; matrix = 256 × 256; FOV = 256 × 256 mm, 192 sagittal slices; slice thickness = 1 mm] and Magnetom Prisma: [GRAPPA factor 2; TR/TE/inversion time/Ɵ = 1800 ms/2.12 ms/904 ms/10 o; matrix = 256 × 256; FOV = 256 × 256 mm, 176 sagittal slices; slice thickness = 1 mm]), followed by fMRI acquisition using T2*-weighted echo planar imaging using the same protocol on both MRI systems (GRAPPA factor 2; TR/TE/Ɵ = 2000 ms/30 ms/40o; matrix = 64 × 64; FOV = 200 × 200 mm; 35 axial slices; slice thickness = 4 mm) with synchronous recording of pulse and respiratory waveforms. Using an MRI-compatible tablet system, up to 7 previously validated fMRI tasks were administered: phonemic fluency, rhyming, semantic decision, hand clenching, number counting, tongue movement, and foot flexing.21, 22, 23
Surface-Rendered Functional Maps
Functional MRI data were analyzed using Analysis of Functional Neuroimages freeware.24 Data preprocessing included outlier censoring and interpolation (3dDespike), correction of cardiac and respiratory effects (3dretroicor), slice timing correction (3dTshift), rigid-body motion correction (3dvolreg), and alignment with the anatomic images by affine transformation (align_epi_anat.py). Functional data were spatially smoothed with a 6-mm Gaussian filter (3dmerge) and fitted with detrending and autocorrelation corrections (3dREMLfit) to estimate brain activity using a general linear model. Correction for multiple comparisons was achieved using a liberal initial false discovery rate threshold of q ≤ .1 and a small, arbitrary cluster size threshold customized patient to patient to suit the data quality for surgical planning.
The 3D cortical surface was generated from the segmentation of T1-weighted images using FreeSurfer software.25 To create surface rendered fMRI maps, an average value of activations was projected on the cortical surface using the Analysis of Functional Neuroimages surface mapping function (SUMA). A grayscale shading effect from bright for gyri to dark for sulci was created using the “ambient occlusion” filter in MeshLab.26 The cortical surfaces without activations (“MRI”) and with activations (“fMRI”) were visualized in orthographic view such that the region around the tumor was visible. In this orientation, 2D projection images of both “MRI” and “fMRI” cortical surfaces were subsequently screen-captured and used as inputs to the registration pipeline.
Intraoperative Data Collection
Patients were anesthetized using dexmedetomidine and a bupivacaine-based scalp nerve block, providing optimal operative and behavioral testing conditions.27 Each patient's head was held in a Mayfield Skull Clamp (Integra LifeSciences, Plainsboro, New Jersey, USA) to provide rigid fixation. Brain mapping was performed via DCS while the patient performed motor and/or language tasks personalized on the basis of tumor location. Tasks were administered by the surgeon while an Ojemann cortical stimulator (OCS2, Integra LifeSciences) delivered 2- to 6-mA current at 5-mm increments for 1–2 seconds, producing inhibitory or excitatory behavioral responses. No sites were stimulated twice consecutively. If the response occurred at least 3 times, a sterile chip (9 × 4 × 1.5 mm) was placed marking the area as eloquent. The DCS was performed for approximately 10–20 minutes until eloquent areas were identified to the surgeon's satisfaction.
The exposed cortical surface was imaged from the approximate viewing perspective of the surgeon using a 3-dimensional (3D) optical scanner (SCANIFY, Fuel 3D Technologies Limited, Chinnor, UK) at 2 time points for brain shift correction: 1) before brain mapping (“preDCS”) and 2) after brain mapping (“postDCS”). The camera was brought into the surgical field mounted on a Mayo stand and adjusted manually with an attempt to maintain consistency between preDCS and postDCS acquisitions. Although this scanner provides 3D data, only the 2D color image was used for prototype pipeline development to reduce computational burden.
Image Registration Pipeline
Image registration is typically described mathematically as an optimization problem, in which a source image is spatially adjusted (“transformed”) to align to a target image in an iterative procedure that maximizes a similarity metric. These algorithms usually require customization of the overall data processing pipeline to achieve satisfactory results in a given application. In the present case, image registration was implemented using the mutual information similarity metric, as suitable for multimodality input data,28 and affine transformation to account for the nonrigid nature of brain shift.
The pipeline was prototyped in MATLAB (The Mathworks, Inc., Natick, Massachusetts, USA) for operating on 2D input images that provide fast registration compared with potentially time-consuming 3D volumetric registration. The spatial transform was estimated in a 3-level multiresolution framework (‘imregtform’) using the best performing optimizer (“one plus one evolutionary”) parameters (across all patients), and subsequently applied (“imwarp”) to yield the registered output. Before registration, a region of interest corresponding to the craniotomy window was selected.
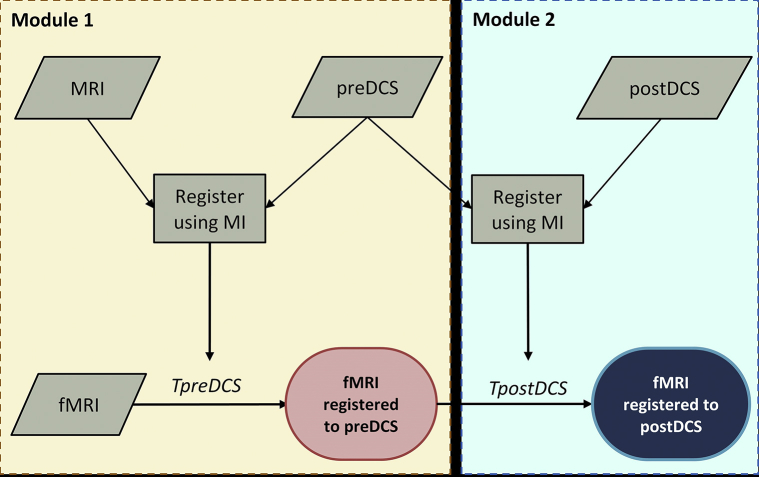
The pipeline was organized in 2 modules according to the time stage when fMRI data were to be visualized intraoperatively on the image of the brain surface (Figure 1). Module 1 performed registration to the preDCS image, accounting for any brain shift that occurred prior to DCS mapping, whereas Module 2 performed registration to the postDCS image, accounting for all brain shift that occurred until DCS completion. Module 1 first registered the anatomical MRI data to the preDCS image. This process accounted for cases where large activation clusters occurred in the craniotomy window, potentially reducing the registration quality. The resulting affine transformation, TpreDCS, was subsequently used to register the fMRI data to the preDCS image, yielding the final output of the module. Module 2 accounted for brain shift that occurred during the DCS procedure by registering the preDCS to the postDCS image. The transform parameters, TpostDCS from this registration were multiplied with TpreDCS, generating a composite transform that was subsequently used to correct fMRI data for the overall brain shift. Thus the final output of Module 2 was fMRI activation data overlaid on the postDCS image, allowing simultaneous visualization of both fMRI and DCS results on the exposed brain tissue. The pipeline was executed on a modest Windows 10−based desktop computer (Intel i5-4590 processor, 8 GB RAM).
Figure 1.
Proposed image registration pipeline using mutual information as the similarity metric for the correction of brain shift.
Experimental Testing in Silico
To validate and quantify the registration accuracy, controlled experimental testing was carried out in silico. For each patient (see “Patient Demographics” later), the ground truth result was generated from an initial registration between the preoperative fMRI and intraoperative optical images. Fifteen landmark points were selected, where the first landmark (x1,y1) was within the region of activations. The fMRI data were then moved out of alignment by translating in x and y directions with a displacement, r between 5 mm and 25 mm and then rotating around x1,y1 by an angle, α between 0 degrees and 20 degrees, sampled over a uniform probability distribution within a preselected range (Figure 2). Each displacement was sampled with 100 random rotations. The misalignments were introduced only in the 2D projection images with a constant projection angle.
Figure 2.
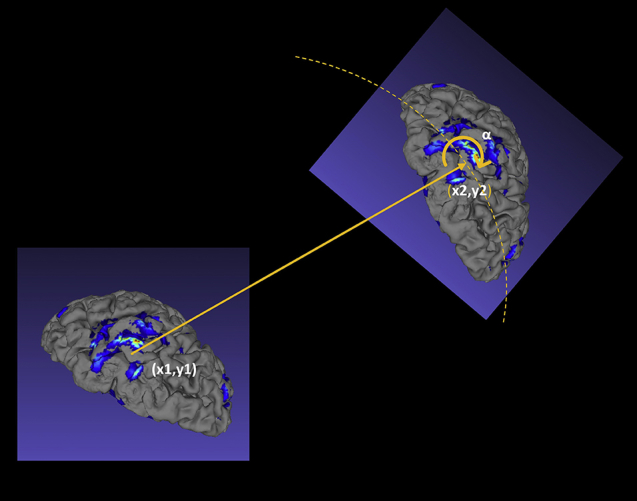
Schematic illustrating the in silico experimental testing procedure for 1 iteration. The landmark point (x1,y1) represents ground truth, whereas point (x2,y2) represents the misalignment produced by translation r and random rotation by angle α. The ability of the image registration algorithm to correct for the misalignment was subsequently quantified. See text for details.
The misaligned fMRI and intraoperative optical images were registered. The mean registration error (RE) in the recovered transformation T was quantified by computing the mean Euclidean distance dist() between N (=15) corresponding landmark points pi of the ground truth and the transformed misaligned image, as in Equation 1.
| (1) |
Patient Demographics
Data were collected for 5 brain tumor patients P1-P5 (mean age 57.4 ± 11.1, Table 1) selected from the ongoing surgical caseload at St. Michael's Hospital that met the following inclusion criteria: selection for awake craniotomy and intraoperative DCS based on possible eloquent cortex adjacent to the tumor and preoperative fMRI showing pertinent brain activations within the planned craniotomy window. Patient recruitment and data collection occurred from November 2016 to December 2018. All patients provided written informed consent for participation in this study, approved by the Institutional Research Ethics Boards at St. Michael's Hospital and Sunnybrook Health Sciences Centre. Patients P1-P4 were imaged at the former institution, and patient P5 was imaged at the latter.
Table 1.
Patient Demographics and Behavioral Testing Response for Intraoperative Brain Mapping
| Patient | Age/Sex | Handedness | Tumor Grade/Pathology | Tumor Location | Direct Cortical Stimulation Mapping Response |
|---|---|---|---|---|---|
| P1 | 39/F | Right | III/Anaplastic oligodendroglioma | L-parietal | Hand movement; face twitching; sensation in hands |
| P2 | 68/F | Right | IV/Glioblastoma | L-frontal | Hand movement |
| P3 | 63/M | Right | IV/Giant cell glioblastoma | L-frontal | Hand movement; foot movement |
| P4 | 60/M | Right | Metastatic adenocarcinoma | L-frontal | Hand movement; foot movement (based on anatomic landmarks) |
| P5 | 57/M | Right | IV/Glioblastoma | L-temporal | Speech arrest; face twitching; reading difficulty; receptive aphasia |
Results
Experimental Testing in Silico: Registration Error and Capture Range
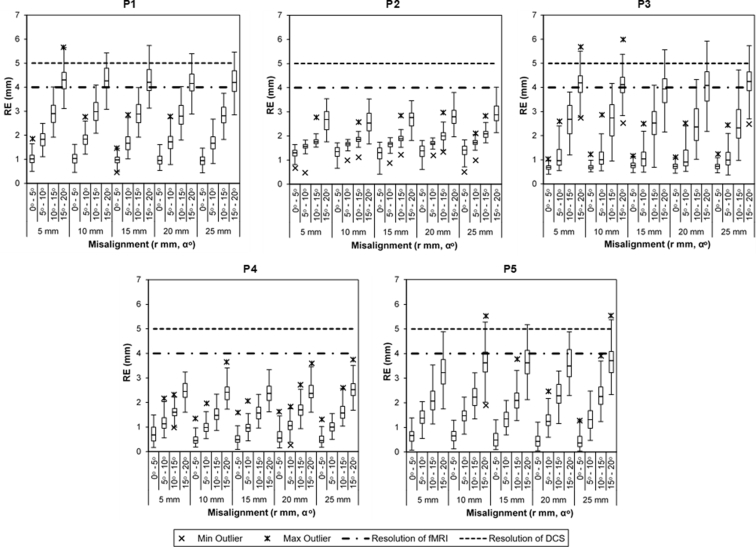
Figure 3 shows box-and-whisker plots illustrating the distribution of RE values for each patient over different combinations of initial misalignments. These plots provide 1) assessment of the capture range—the range of initial translations and rotations that can be recovered by the image registration pipeline and 2) assessment that within this range, RE is within acceptable limits. Both assessments yielded promising results over the patients investigated.
Figure 3.
Distribution of registration error (RE values) for 5 patients (P1-P5) displayed using box-and-whisker plots for each initial misalignment. Each box-and-whisker plot consists of the interquartile range (IQR, box), median (line inside the box), whiskers extending to 1.5*IQR on both ends and minimum and maximum outliers as shown by symbols in the legend.
First, both assessments depend on the RE amplitude that can be tolerated. The horizontal lines (see Figure 3) indicate the spatial resolution limits for fMRI (4 mm, the acquisition slice thickness: dashed-dotted line) and for DCS (nominally 5 mm, the width of the stimulator: dashed line). Ideally, the RE should be well within DCS resolution, as the value for this modality is larger. Initial inspection of Figure 3 shows misalignment ranges that fall well within 5 mm for all patients. Closer inspection shows that the image registration pipeline is robust to simple translation (minimal rotation) misalignment, where the median RE is approximately 1 mm and remains constant up to the maximum translation of 25 mm. When angular misalignments are also considered, larger rotations are associated with larger RE values but no substantial interaction effect between translation and rotation is observed. Patients P1, P3, and P5 show larger RE values with increasing rotational misalignment, whereas the analogous dependency for patients P2 and P4 is weaker. Collectively, the results indicate that for misalignments up to 25 mm and 10–15 degrees, the image registration pipeline produces RE values well within DCS resolution.
To further assess pipeline performance, failure rates were calculated for 2 criteria: RE > 5 mm (Table 2) and RE > 2.5 mm (Table 3) by aggregating experimental testing data across all patients, giving 500 results per misalignment condition. The pipeline performs well under the error tolerance of 5 mm with a maximum failure rate of 5.2%, which increases to 77%–86% when error tolerance is reduced to 2.5 mm for up to 20o misalignments. For both criteria, the failure rates are zero or negligible for misalignments up to 10 degrees.
Table 2.
Registration Failure Rate for Registration Error Tolerance of 5 mm for Maximum Initial Misalignments of 25 mm and 20 Degrees
| Translation (mm) | Rotation (degrees) |
|||
|---|---|---|---|---|
| 0–5 | 5–10 | 10–15 | 15–20 | |
| 5 | 0.0% | 0.0% | 0.0% | 4.6% |
| 10 | 0.0% | 0.0% | 0.0% | 5.2% |
| 15 | 0.0% | 0.0% | 0.0% | 4.8% |
| 20 | 0.0% | 0.0% | 0.0% | 4.6% |
| 25 | 0.0% | 0.0% | 0.0% | 4.8% |
Table 3.
Registration Failure Rate for Registration Error Tolerance of 2.5 mm for Maximum Initial Misalignments of 25 mm and 20 Degrees
| Translation (mm) | Rotation(degrees) |
|||
|---|---|---|---|---|
| 0–5 | 5–10 | 10–15 | 15–20 | |
| 5 | 0.0% | 0.6% | 32.4% | 78.6% |
| 10 | 0.0% | 1.6% | 34.8% | 77.6% |
| 15 | 0.0% | 0.8% | 29.8% | 78.0% |
| 20 | 0.0% | 0.8% | 31.6% | 80.4% |
| 25 | 0.0% | 0.4% | 32.6% | 86.0% |
Registration Visualization
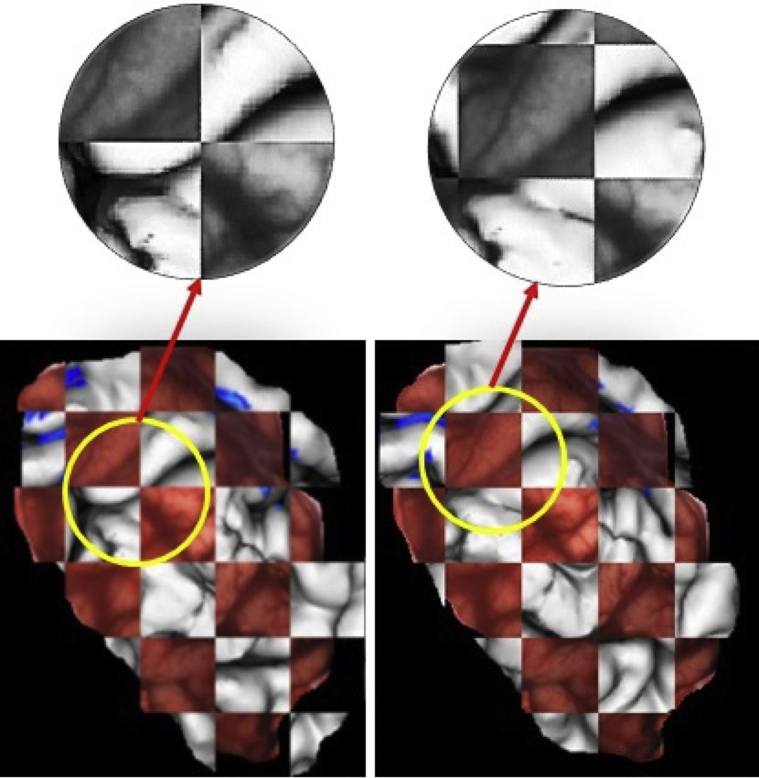
The quality of registration visualized in Figure 4 shows Module 1 output (right) in a checkerboard pattern along with its unregistered counterpart (left) for P2. Before registration, the misalignment between the image pairs can be seen as discontinuities in the sulcal lines, easily observable in the grayscale magnified view. On registration, the misalignment is rectified, shown as the overlap of corresponding sulci and gyri in the registered image pair.
Figure 4.
Checkerboard images of the 2-dimensional projection (gray) of preoperative hand motor fMRI (blue) and intraoperative cortical surface data (red) for patient P2, shown before and after registration for preDCS (pipeline Module 1). The misregistration and its correction following registration can be easily observed in the zoomed-in grayscale view.
The spatial relationship between fMRI and DCS findings is shown in Figure 5, which displays Module 1 (preDCS) and Module 2 (postDCS) outputs for all patients. Preoperative fMRI activations of hand motor tasks are shown for patients P1-P4, whereas for P5 fMRI activations of semantic decision (left cluster) and tongue movement tasks are displayed (right cluster). The preDCS output shows these activations overlaid on the cortical surface for initial guidance of DCS procedures. The postDCS registration output facilitates visual comparison between the sites mapped intraoperatively and the preoperative fMRI activations on the current state of the visible brain surface. The postDCS results indicate that the hand motor activations from DCS mapping (marked as “H”) were proximal to analogous fMRI activations for P1-P4, with partial overlap for patients P3 and P4. In case of P5, DCS mapping identified areas of “facial twitching” (“F”) and “speech arrest” (“SA”) using number counting task near fMRI activations associated with tongue movement and areas of “reading difficulty” (“RD”) and “receptive aphasia” (“RA”) using a reading task, proximal to fMRI activations from a semantic decision task.
Figure 5.
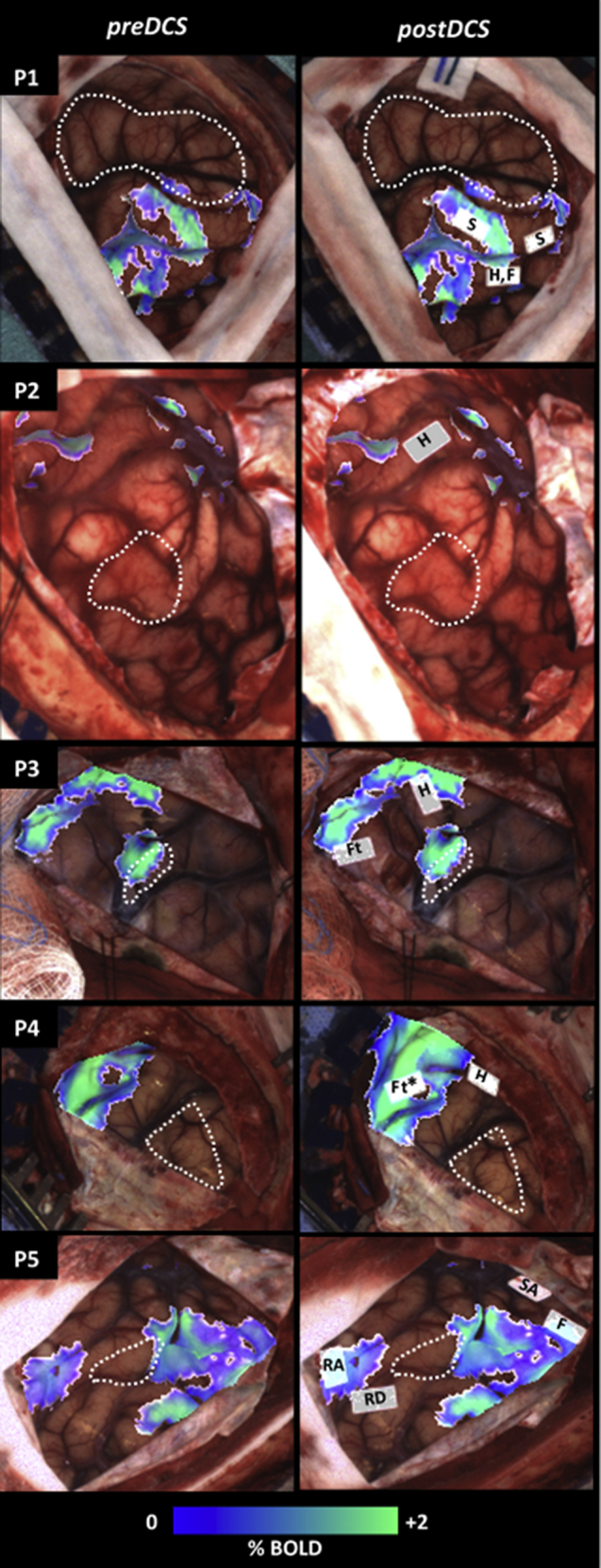
Outputs of the proposed registration pipeline shown for all patients, with additional labeling of DCS sites and delineation of tumor/resection cavity with white dashed line. Each row of images is from the patient indicated on the top left. The first column (preDCS) shows output of pipeline Module 1. For patients P1-P4, fMRI activations of hand motor, and for patient P5 functional magnetic resonance imaging (fMRI) activations of tongue movement (right) and semantic decision tasks (left) are shown on the intraoperative cortical image. The second column (postDCS) shows output of pipeline Module 2, showing fMRI activations overlaid on intraoperative cortical surface with DCS mapping results. The DCS site labels “S,” “H,” “F,” “Ft,” “SA,” “RD,” and “RA” correspond to sites of sensory activation, hand motor, facial twitching, foot motor, speech arrest, reading difficulty, and receptive aphasia, respectively (*marked based on anatomic location).
Discussion
Brain shift−corrected preoperative fMRI data may be helpful to guide DCS mapping efficiently. Lacking a method for visualizing coregistered fMRI and DCS data intraoperatively, we developed a prototype image processing pipeline with such functionality and validated it on 5 brain tumor patients. Overall, the results were promising, showing acceptable accuracy for intraoperative use, for the patients investigated. Over the group, in silico tests showed registration errors well within the spatial sampling resolution of DCS, for misalignments between fMRI and optical image pairs of up to 25 mm (and possibly larger) and approximately 10–15 degrees. For a 5-mm error tolerance, the success rate was 95% for misalignments up to 20 degrees, whereas the analogous rate was 14%−23% for a tolerated value of 2.5 mm. The estimates of capture range are optimistic, however, because in silico tests do not account for the effect of changing the 3D cortical surface's orientation when generating the 2D projection images. As expected, there was some variability in registration accuracy across the patients as well, with patient P3 proving the most challenging and patients P2 and P4 the most robust. Because the patient sample size was small, tests will be necessary in additional patients for a more comprehensive evaluation over a wider range of brain tumor and brain shift presentations.
Nevertheless, this method offers visualizations with potential benefits for intraoperative brain mapping. By using registration to overlay preoperative fMRI data with brain shift correction on the cortical surface image, the surgeon is relieved of transforming these data mentally while using them to streamline DCS workflow and to assess fMRI and DCS concordance intraoperatively.23 This is important because although fMRI has known limitations23 and DCS is regarded as the gold standard for brain mapping, DCS has its own sources of variability (e.g., electrode orientation and amplitude of the stimulation current impact on the activated volume29, 30).
The concordance between fMRI and DCS for all patients is readily assessed qualitatively in Figure 5. It is evident that although both modalities were used to map the motor cortex, a 1-to-1 correspondence was not observed with only partial overlap of the results. Our visualized results are in agreement with the literature and are typical of our ongoing experience with both mapping methods for awake craniotomies involving brain tumor patients.20, 23 Several factors responsible for the imperfect concordance between fMRI and DCS are the different biophysical signals of origin, differences in behavioral task administration and response, and the dependence of fMRI activations on the statistical threshold.20, 23
This work was undertaken within the context of a large body of research concerning brain shift correction methods. Early attempts were focused on accurately establishing the tumor boundary.11, 13 More recent attempts include integrating preoperative functional data into ultrasound-based neuronavigation systems for brain shift correction14, 15 and preliminary work involving intraoperative acquisition of functional data using fMRI31 and near-infrared spectroscopy.32 The present work differs from the existing literature as it provides a quite simple, straightforwardly implemented means of visualizing fMRI data over the exposed brain surface, simultaneously with DCS results, to aid in surgical workflow. Although a similar method exists for epilepsy procedures,33 our study differs in that manual landmark registration is avoided by visually orienting the 3D surface pose according to the 2D intraoperative image and obtaining an analogous region of interest from both input images to constrain the registration search space. In addition, this study validates the use of such simple registration on tumor patient data, which is more challenging to segment and may result in inaccurate surface topology using existing methods. Lastly, here we present the utility of the method for providing guidance both before and after functional mapping in the context of awake brain surgery.
Several limitations are associated with the proof-of-concept pipeline. Brain shift is corrected only at 2 stages: immediately after dural incision and about 10–20 minutes later after DCS mapping. Although brain shift progression during DCS mapping was minimal for the patients investigated, this is not expected to be true in general and intermediate correction steps may be necessary in some cases. The pipeline also does not capture any brain shift in the subsequent stages of the awake craniotomy procedure—when the shift mainly results from tumor excision, causing sagging or sinking of adjacent unsupported tissue due to gravity.7, 34 Because DCS is mainly performed before tumor removal, brain shift correction at further stages of the surgery is not as critical but may still be important in some cases. Furthermore, the pipeline outputs are in 2D, making it impossible to correct for shifts perpendicular to the 2D image plane.
The prototype pipeline used affine transformation to correct for brain shift rapidly. The actual image registration process was completed in seconds for all patients, although some of the manual preprocessing steps took considerably longer. Aspects such as cropping of the craniotomy region can likely be made automatic (and more rapid). In future investigations, cases may be encountered in which affine transformation provides inadequate results. Although more complex transformations are possible, they will introduce additional degrees of freedom that require longer processing times and risk convergence to a suboptimal rather than global optimal result. More pipeline testing are necessary to investigate the interplay among quality of registration, complexity, and execution time.
Lastly, the prototype pipeline needs to be investigated by surgeons to assess the potential for improved workflow during intraoperative brain mapping and its impact on surgical decisions and patient outcome. The present work is a useful starting point for such assessments, which ultimately may lead to a robust visualization tool within the tablet platform previously developed in our laboratory for awake craniotomy procedures.21
Conclusion
An image registration pipeline has been developed that corrects brain shift while integrating preoperative fMRI and intraoperative DCS results for application during awake craniotomy procedures. The pipeline was validated on 5 patients, showing successful registration up to 15 degrees of misalignment. The simple visualization provided by this method has the potential to improve workflow during intraoperative brain mapping and reduce cognitive load on the surgeon.
Footnotes
Conflict of interest statement: The authors declare that the article content was composed in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Petrella J.R., Shah L.M., Harris K.M. Preoperative functional MR imaging localization of language and motor areas: effect on therapeutic decision making in patients with potentially resectable brain tumors. Radiology. 2006;240:793–802. doi: 10.1148/radiol.2403051153. [DOI] [PubMed] [Google Scholar]
- 2.Benson R.R., FitzGerald D.B., LeSueur L.L. Language dominance determined by whole brain functional MRI in patients with brain lesions. Neurology. 1999;52:798–809. doi: 10.1212/wnl.52.4.798. [DOI] [PubMed] [Google Scholar]
- 3.Sakr H.M., Mohamed M.A., Jalalod’din H., Abbas Y.A. Influence of fMRI on operative planning of brain tumors: initial experience in a histopathologically variable subset of tumors. Egypt J Radiol Nucl Med. 2011;42:215–221. [Google Scholar]
- 4.Lee C.C., Ward H.A., Sharbrough F.W. Assessment of functional MR imaging in neurosurgical planning. Am J Neuroradiol. 1999;20:1511–1519. [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts D.W., Hartov A., Kennedy F.E., Miga M.I., Paulsen K.D. Intraoperative brain shift and deformation: a quantitative analysis of cortical displacement in 28 cases. Neurosurgery. 1998;43:749–758. doi: 10.1097/00006123-199810000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Hartkens T., Hill D.L.G., Castellano-Smith A.D. Measurement and analysis of brain deformation during neurosurgery. IEEE Trans Med Imaging. 2003;22:82–92. doi: 10.1109/TMI.2002.806596. [DOI] [PubMed] [Google Scholar]
- 7.Hill D.L.G., Maurer C.R., Maciunas R.J., Barwise J.A., Fitzpatrick J.M., Wang M.Y. Measurement of intraoperative brain surface deformation under a craniotomy. Neurosurgery. 1998;43:514–526. doi: 10.1097/00006123-199809000-00066. [DOI] [PubMed] [Google Scholar]
- 8.Gerard I.J., Kersten-Oertel M., Petrecca K., Sirhan D., Hall J.A., Collins D.L. Brain shift in neuronavigation of brain tumors: a review. Med Image Anal. 2017;35:403–420. doi: 10.1016/j.media.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Nimsky C., Ganslandt O., Cerny S., Hastreiter P., Greiner G., Fahlbusch R. Quantification of, visualization of, and compensation for brain shift using intraoperative magnetic resonance imaging. Neurosurgery. 2000;47:1070–1080. doi: 10.1097/00006123-200011000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Ohue S., Kumon Y., Nagato S. Evaluation of intraoperative brain shift using an ultrasound-linked navigation system for brain tumor surgery. Neurol Med Chir (Tokyo) 2010;50:291–300. doi: 10.2176/nmc.50.291. [DOI] [PubMed] [Google Scholar]
- 11.Clatz O., Delingette H., Talos I.F. Robust nonrigid registration to capture brain shift from intraoperative MRI. IEEE Trans Med Imaging. 2005;24:1417–1427. doi: 10.1109/TMI.2005.856734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimsky C., Ganslandt O., Hastreiter P., Fahlbusch R. Intraoperative compensation for brain shift. Surg Neurol. 2001;56:357–364. doi: 10.1016/s0090-3019(01)00628-0. [DOI] [PubMed] [Google Scholar]
- 13.Yamashita S., Fujisawa M., Kodama K., Ishikawa M., Katagi R. Use of preoperative 3D CT/MR fusion images and intraoperative CT to detect lesions that spread onto the brain surface. Acta Neurochir Suppl. 2013;118:239–244. doi: 10.1007/978-3-7091-1434-6_45. [DOI] [PubMed] [Google Scholar]
- 14.Reinertsen I., Lindseth F., Askeland C., Iversen D.H., Unsgård G. Intra-operative correction of brain-shift. Acta Neurochir (Wien) 2014;156:1301–1310. doi: 10.1007/s00701-014-2052-6. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen I.A., Lindseth F., Rygh O.M. Functional neuronavigation combined with intra-operative 3D ultrasound: initial experiences during surgical resections close to eloquent brain areas and future directions in automatic brain shift compensation of preoperative data. Acta Neurochir (Wien) 2007;149:365–378. doi: 10.1007/s00701-006-1110-0. [DOI] [PubMed] [Google Scholar]
- 16.Škrinjar O., Nabavi A., Duncan J. Model-driven brain shift compensation. Med Image Anal. 2002;6:361–373. doi: 10.1016/s1361-8415(02)00062-2. [DOI] [PubMed] [Google Scholar]
- 17.Sun K., Pheiffer T.S., Simpson A.L., Weis J.A., Thompson R.C., Miga M.I. Near real-time computer assisted surgery for brain shift correction using biomechanical models. IEEE J Transl Eng Heal Med. 2014;2 doi: 10.1109/JTEHM.2014.2327628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Archip N., Clatz O., Whalen S. Non-rigid alignment of pre-operative MRI, fMRI, and DT-MRI with intra-operative MRI for enhanced visualization and navigation in image-guided neurosurgery. Neuroimage. 2007;35:609–624. doi: 10.1016/j.neuroimage.2006.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FitzGerald D.B., Cosgrove G.R., Ronner S. Location of language in the cortex: a comparison between functional MR imaging and electrocortical stimulation. AJNR Am J Neuroradiol. 1997;18:1529–1539. [PMC free article] [PubMed] [Google Scholar]
- 20.Meier M.P., Ilmberger J., Fesl G., Ruge M.I. Validation of functional motor and language MRI with direct cortical stimulation. Acta Neurochir (Wien) 2013;155:675–683. doi: 10.1007/s00701-013-1624-1. [DOI] [PubMed] [Google Scholar]
- 21.Morrison M.A., Tam F., Garavaglia M.M. A novel tablet computer platform for advanced language mapping during awake craniotomy procedures. J Neurosurg. 2016;124:938–944. doi: 10.3171/2015.4.JNS15312. [DOI] [PubMed] [Google Scholar]
- 22.Morrison M.A., Churchill N.W., Cusimano M.D., Schweizer T.A., Das S., Graham S.J. Reliability of task-based fMRI for preoperative planning: a test-retest study in brain tumor patients and healthy controls. PLoS One. 2016;11:e0149547. doi: 10.1371/journal.pone.0149547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison M.A., Tam F., Garavaglia M.M. Sources of variation influencing concordance between functional MRI and direct cortical stimulation in brain tumor surgery. Front Neurosci. 2016;10:1–16. doi: 10.3389/fnins.2016.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 25.Dale A.M., Fischl B., Sereno M.I. Cortical surface-based segmentation. I. segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 26.Cignoni P., Callieri M., Corsini M. MeshLab: an open-source mesh processing tool. Sixth Eurographics Ital Chapter Conf. 2008:129–136. [Google Scholar]
- 27.Garavaglia M.M., Das S., Cusimano M.D. Anesthetic approach to high-risk patients and prolonged awake craniotomy using dexmedetomidine and scalp block. J Neurosurg Anesthesiol. 2014;26:226–233. doi: 10.1097/ANA.0b013e3182a58aba. [DOI] [PubMed] [Google Scholar]
- 28.Maes F., Collignon A., Vandermeulen D., Marchal G., Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 29.Borchers S., Himmelbach M., Logothetis N., Karnath H.-O. Direct electrical stimulation of human cortex—the gold standard for mapping brain functions? Nat Rev Neurosci. 2011;13:63–71. doi: 10.1038/nrn3140. [DOI] [PubMed] [Google Scholar]
- 30.Mandonnet E., Pantz O. The role of electrode direction during axonal bipolar electrical stimulation: a bidomain computational model study. Acta Neurochir (Wien) 2011;153:2351–2355. doi: 10.1007/s00701-011-1151-x. [DOI] [PubMed] [Google Scholar]
- 31.Gasser T., Ganslandt O., Sandalcioglu E., Stolke D., Fahlbusch R., Nimsky C. Intraoperative functional MRI: implementation and preliminary experience. Neuroimage. 2005;26:685–693. doi: 10.1016/j.neuroimage.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh A., Elwell C., Smith M. Cerebral near-infrared spectroscopy in adults. Anesth Analg. 2012;115:1373–1383. doi: 10.1213/ANE.0b013e31826dd6a6. [DOI] [PubMed] [Google Scholar]
- 33.Wang A., Mirsattari S.M., Parrent A.G., Peters T.M. Fusion and visualization of intraoperative cortical images with preoperative models for epilepsy surgical planning and guidance. Comput Aided Surg. 2011;16:149–160. doi: 10.3109/10929088.2011.585805. [DOI] [PubMed] [Google Scholar]
- 34.Ji S., Fan X., Roberts D.W., Hartov A., Paulsen K.D. Cortical surface shift estimation using stereovision and optical flow motion tracking via projection image registration. Med Image Anal. 2014;18:1169–1183. doi: 10.1016/j.media.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]