The Herpesviridae are structurally complex DNA viruses whose capsids undergo primary envelopment at the inner nuclear membrane and secondary envelopment at organelles in the cytoplasm. In both locations, there is evidence that envelope formation and scission involve the participation of multiple viral proteins and also the cellular ESCRT apparatus.
KEYWORDS: ESCRT, envelopment, Herpesviridae
ABSTRACT
The Herpesviridae are structurally complex DNA viruses whose capsids undergo primary envelopment at the inner nuclear membrane and secondary envelopment at organelles in the cytoplasm. In both locations, there is evidence that envelope formation and scission involve the participation of multiple viral proteins and also the cellular ESCRT apparatus. It nevertheless appears that the best-understood viral strategies for ESCRT recruitment, those adopted by the retroviruses and many other families of enveloped RNA viruses, are not utilized by the Herpesviridae, at least during envelopment in the cytoplasm. Thus, although a large number of herpesvirus proteins have been assigned roles in envelopment, there is a dearth of candidates for the acquisition of the ESCRT complex and the control of envelope scission. This review summarizes our current understanding of ESCRT association by enveloped viruses, examines what is known of herpesvirus ESCRT utilization in the nucleus and cytoplasm, and identifies candidate cellular and viral proteins that could link enveloping herpesviruses to cellular ESCRT components.
INTRODUCTION
The cellular ESCRT (endosomal sorting complex required for transport) apparatus is an ancient and versatile membrane-remodeling machine (1–3) that has been seized by many families of viruses to help construct their envelopes (3–5). As a general strategy, viral structural proteins have evolved binding sites for particular subsets of ESCRT proteins and complexes, recruiting the ESCRT membrane-scission apparatus to pinch off and seal the necks of nascent envelopes. Mechanisms of ESCRT recruitment have been at least partially elucidated for many families of enveloped viruses, including retroviruses, flaviviruses, filoviruses, arenaviruses, and rhabdoviruses, among others (5). For the herpesviruses, the ESCRT apparatus has been implicated in envelopment both at the inner nuclear membrane (INM) and in the cytoplasm, but in each case, our understanding of which ESCRT factors are utilized, and how they are recruited, remains poor. This review summarizes our current understanding of ESCRT function, both in general and in the context of herpesvirus envelopment, with the goal of drawing attention to the major gaps in our knowledge.
ESCRT COMPONENTS AND MEMBRANE REMODELING
The ESCRT apparatus is a membrane-sculpting machine comprising several multisubunit complexes, termed ESCRT-0, -I, -II, and -III, and a number of accessory proteins (Fig. 1) (1, 2). ESCRT components usually drive membrane vesiculation and tubulation “away” from the cytosol, into organellar lumina or outward from the plasma membrane (1), although they can also form membrane tubules with the opposite orientation, projecting into the cytoplasm (6). Various subsets of the core ESCRT machinery and adaptor proteins are used to manipulate membrane bilayers during a diversity of cellular processes, including multivesicular body (MVB) formation, cytokinesis, exosome and microvesicle formation, and repair of plasma and nuclear membranes (1, 2, 4, 7, 8). The “heavy lifting” of ESCRT-mediated membrane deformation is performed by ESCRT-III, a spring-like filament assembled by polymerization of CHMP (charged multivesicular body protein) family members on the surface of a lipid bilayer. Humans express 12 ESCRT-III subunits, including 7 related protein families (CHMP1 to -7), their isoforms, and the CHMP3-like protein IST1 (increased sodium tolerance 1) (2), a diversity that is thought to contribute to the variety of membrane-remodeling events performed by ESCRT-III. The ESCRT-III filament drives the formation of vesicles, tubules, or viral envelopes and then constricts the lipid structure at its neck to catalyze scission, a process accompanied by the disassembly and removal of ESCRT-III via the hexameric AAA ATPase Vps4 (vacuolar protein sorting 4) (1, 2, 6).
FIG 1.
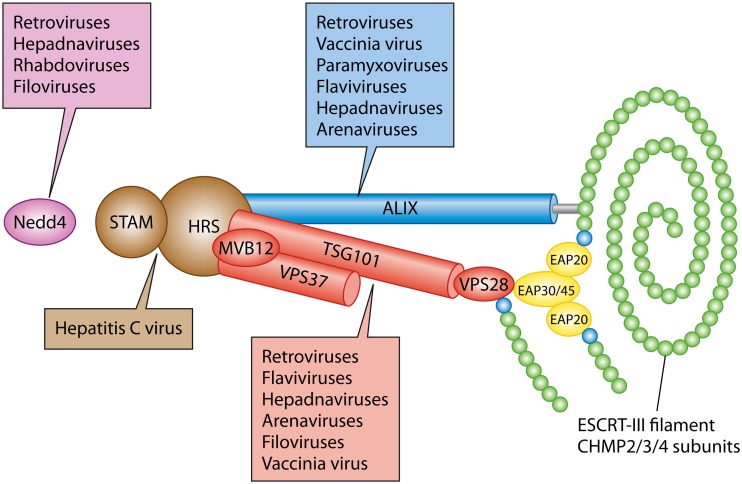
The ESCRT apparatus and its utilization by enveloped viruses. The ESCRT-0 complex (STAM/HRS) is shown in brown, ESCRT-I (TSG101/MVB12/VPS37/VPS28) in red, and ESCRT-II (EAP30/EAP45/EAP202) in yellow. The ESCRT-III filament is shown as a polymer of CHMP2/3/4 subunits (green) capped by nucleating CHMP6 subunits (cyan). ESCRT-II complex EAP20 subunits nucleate filament assembly through the recruitment of CHMP6. ALIX is represented as a blue cylinder making contacts with ESCRT-I and extending its Bro1 domain (gray) to trigger CHMP4 polymerization. Nedd4 family members are shown in purple. Individual viruses or virus families in which one or more members have been reported to utilize Nedd4, ALIX, or an ESCRT complex during envelopment are indicated by boxes of the corresponding color.
It is the responsibility of the ESCRT complexes and associated proteins to control the positioning and polymerization of ESCRT-III filaments, to initiate membrane bending, and, where appropriate, to recruit cargo into assembling vesicles or tubules. How this is accomplished differs from process to process (2), but MVB formation helps to illuminate the general principles (2, 9). This pathway initiates when the ESCRT-0 complex HRS/STAM (hepatocyte growth factor-regulated tyrosine kinase substrate/signal-transducing adaptor molecule) binds to phosphatidylinositol-3-phosphate (PI3P) on the surfaces of endosomes. Multiple ESCRT-0 complexes self-associate, gather ubiquitinated cargo proteins to the nascent budding site for eventual localization to MVBs, and recruit the heterotetrameric complex ESCRT-I via interaction with its subunit TSG101 (tumor susceptibility gene 101). ESCRT-I (like ESCRT-0) binds ubiquitinated cargo but, in addition, can recruit ESCRT-II, a Y-shaped complex with a stem composed of ELL-associated protein (EAP) subunits EAP30 and EAP45 and two arms, each comprising a copy of the protein EAP20. ESCRT-II is capable of binding and activating the ESCRT-III subunit CHMP6. Activated CHMP6, in turn, nucleates ESCRT-III polymerization by binding and activating CHMP4, which, along with CHMP2 and CHMP3, appears to be a major building block of the ESCRT-III fiber (10). CHMP6 can also be recruited via direct interaction with the ESCRT-I subunit VPS28 (1, 2). In addition, Bro1 domain-containing family members, such as Alg-2-interacting protein X (ALIX) (1, 2), join the assembling complexes via association with ESCRT-I. ALIX is a ubiquitin binding protein and cooperates with ESCRT-0 and ESCRT-I in gathering cargo, but its Bro1 domain also binds directly to CHMP4 (11, 12), recruiting it and inducing conformational changes that drive CHMP polymerization (1, 11). ALIX also dimerizes and may favor the polymerization of two ESCRT-III fibers or even double-stranded chains in a manner similar to that of ESCRT-II (1). By such means, early ESCRT components drive the budding of a cargo-laden vesicle into the interior of the endosome and direct the polymerization of an ESCRT-III fiber at the bud neck. Constriction of the ESCRT-III filament results in the scission and “pinching off” of cargo-containing vesicles into the endosomal lumen. Finally, the ATPase Vps4 associates with the ESCRT-III polymer and disassembles it by passing the strand through its central cavity, a process that may be coupled to the scission reaction itself (1, 13–17).
ESCRT recruitment elsewhere in the cell can be triggered by alternative factors to ESCRT-0, for example, by syndecan/syntenin at the surfaces of endosomes to drive the formation of exosomes, by centrosomal protein 55 (CEP55) at the midbody to help initiate ESCRT-mediated abscission at cell division, and by plasma membrane-associated ARRDC1 (arrestin domain-containing 1) to direct the formation and release of microvesicles (1, 3, 4). Nevertheless, in most cases, the overall strategy is to recruit ESCRT-I and ALIX or alternative Bro1 family members and ultimately to trigger ESCRT-III filament deposition at the site of membrane tubulation, vesiculation, and constriction.
STRATEGIES OF ESCRT RECRUITMENT BY ENVELOPED VIRUSES
The majority of enveloped virus families have learned to exploit ESCRT components in order to facilitate envelopment and gain access to ESCRT-III to drive the terminal scission event (3–5). Indeed, subversion of host ESCRT-III/Vps4 for viral budding and replication is a stratagem dating back at least to the Archaea (4, 18). Experimentally, a dominant negative allele of Vps4 termed Vps4-EQ has proven a useful tool for identifying those virus envelopment events (or other cellular processes) that utilize the ESCRT machinery (19). Vps4-EQ carries a glutamate-to-glutamine mutation at position 233 in its ATPase active site, allowing ATP binding but preventing its hydrolysis (20). Vps4 hexamers containing a mixture of wild-type Vps4 and Vps4-EQ subunits associate with ESCRT-III filaments but are unable to drive their disassembly, irreversibly “locking on” to the bud neck and preventing completion of the scission reaction (13, 15, 16, 20). Vps4-EQ expression therefore exerts a dominant negative effect on endogenous Vps4, and its ability to interfere with a membrane-remodeling event strongly implicates the ESCRT apparatus in that process.
In general, viruses acquire ESCRT-III by recruiting one or more early ESCRT components, most commonly ALIX and ESCRT-I via its subunit TSG101 (summarized in Fig. 1). The best-understood example is that of retroviral Gag proteins, where sequence motifs termed late budding domains (L-domains) recruit ALIX via the YPXnL motif and TSG101 via the P(T/S)AP sequence. A third class of L-domain is the PPXY motif, which binds members of the Nedd4 (neural precursor cell-expressed developmentally downregulated gene 4) family of HECT (homologous to the E6AP carboxyl terminus) ubiquitin E3 ligases (5). It is thought that Nedd4 recruitment results in Gag ubiquitination and thus provides an additional pathway by which Gag can recruit ESCRT-I and ALIX, both of which have evolved to bind ubiquitinated proteins as part of their role in MVB formation (described above). L-domains and ubiquitination therefore ensure a multivalent network of Gag/ESCRT-I/ALIX interactions that support envelopment and bud formation prior to ESCRT-III assembly and scission (5, 21). The relative contributions of ESCRT-I, ALIX, and Nedd4 family members to the process of envelopment differ considerably among the retroviruses (4, 5).
A strategy similar to that of retroviral Gag proteins is used by the VP40 matrix proteins of the Filoviridae members Ebola virus and Marburg virus, the Z proteins of the arenaviruses Lassa virus and lymphocytic choriomeningitis virus, and the M (matrix) proteins of the rhabdoviruses vesicular stomatitis virus and rabies virus, all of which possess P(T/S)AP and/or PPXY L-domain-like motifs that may mediate TSG101/ESCRT-I and Nedd4 binding (5). TSG101 and ALIX have also been implicated in the release of enveloped vaccinia virus, and a candidate YPXnL L-domain has been identified in the vaccinia virus F13L protein (22).
Alternative pathways of viral ESCRT recruitment exist but frequently target the same subset of cellular players. Paramyxovirus matrix proteins generally lack YPXnL, P(T/S)AP, and PPXY motifs, but the M proteins of parainfluenza virus 5 (PIV5) and mumps virus recruit the cellular PPXY domain-containing protein AMOTL1 (angiomotin-like 1), using it as a linker to bind Nedd4 family members (5, 23). A similar strategy is utilized by HIV-1 Gag proteins that lack PPXY motifs (24), and TSG101/ESCRT-I may be recruited through novel routes by hepatitis B virus (25), murine leukemia virus (26, 27), and Ebola virus (27, 28). Among the flaviviruses, roles for ALIX have been reported in yellow fever virus (29) and dengue virus (30), and roles for TSG101/ESCRT-I in Japanese encephalitis virus (31). In contrast, hepatitis C virus intercepts the ESCRT machinery very early; polyubiquitination of its nonstructural protein NS2 creates a binding site for the ESCRT-0 component HRS, and additional interactions take place between HRS and NS5A (Fig. 1) (32).
HERPESVIRIDAE AND THE ESCRT APPARATUS: AN OVERVIEW
Herpesvirus capsids are assembled and packaged in the cell nucleus (33–35). They then undergo primary envelopment at the inner nuclear membrane (INM), followed by the delivery of capsids to the cytoplasm via deenvelopment at the outer nuclear membrane (ONM), a pathway resembling that reported for the export of large ribonucleoprotein complexes in Drosophila (36–39). These capsids subsequently undergo a final, secondary envelopment by budding into the lumina of organelles in the cytoplasm (40–48). Primary envelopment at the INM and secondary envelopment in the cytoplasm require the activity of multiple virally encoded proteins, but there is evidence from the three subfamilies of Herpesviridae that the ESCRT apparatus can play a role in both locations.
Among the Alphaherpesvirinae, ESCRT-III and Vps4 appear to be required for the completion of cytoplasmic envelopment by herpes simplex virus 1 (HSV-1) (49, 50) and pseudorabies virus (PRV) (50) and during HSV-1 envelopment at the INM (39). In accord with the pivotal role of Vps4 in HSV-1 envelopment, Vps4 protein and mRNA levels are both suppressed by the interferon response in HSV-1-infected mouse trigeminal ganglia (51). In contrast, the significance of the ESCRT machinery for the replication of Betaherpesvirinae is less well established. In one study, the replication of human cytomegalovirus (HCMV) was reduced by 2 log units upon expression of Vps4-EQ or dominant negative CHMP1A; this reduction was interpreted as due to a block in HCMV cytoplasmic capsid envelopment (52). In contrast, others reported that small interfering RNA (siRNA) knockdown of Vps4 did not inhibit the assembly of enveloped HCMV particles, as measured by particle infectivity and genome number (53). A third study concluded that expression of Vps4-EQ and several dominant negative CHMP alleles had no effect on the envelopment of HCMV capsids but reduced the rate of viral spread, perhaps due to effects on exosome-mediated cell-cell signaling to prepare adjacent cells for infection (54). Among the Gammaherpesvirinae, the ESCRT apparatus has been reported to function during primary envelopment of Epstein-Barr virus (EBV) capsids at the INM (55), but its importance for cytoplasmic envelopment in this subfamily of viruses is unknown.
THE ESCRT APPARATUS AND HERPESVIRUS ENVELOPMENT AT THE INNER NUCLEAR MEMBRANE
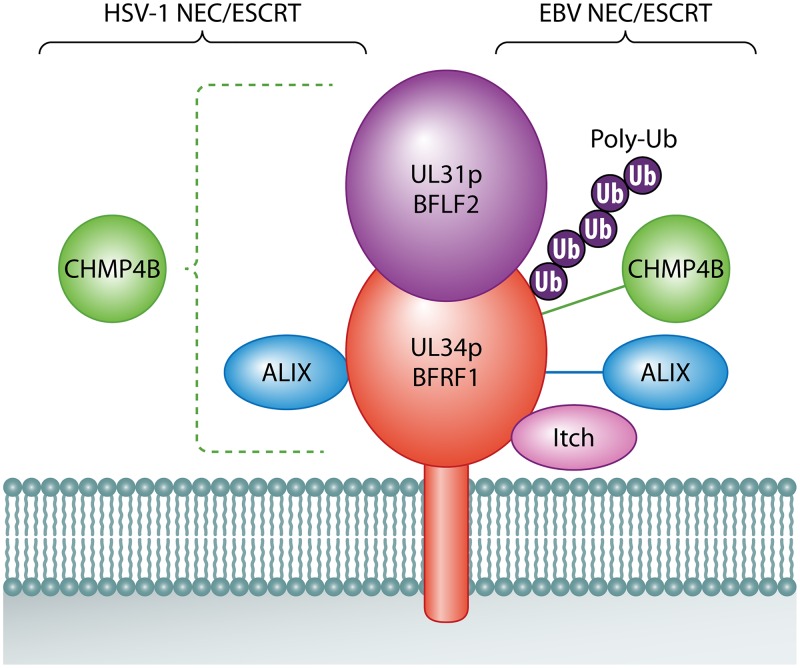
Primary envelopment of herpesviruses at the INM requires the activity of a virally encoded heterodimer termed the nuclear egress complex (NEC) (Fig. 2) (37, 38, 56–60). This complex, found in the INM and ONM, is conserved in all subfamilies of herpesviruses and is composed of a soluble protein (encoded by UL31 in HSV-1) anchored to the nuclear envelope by a type II integral membrane protein (encoded by HSV-1 UL34). In a process remarkably similar to that performed by retroviral Gag proteins at the plasma membrane (61), the UL31p/UL34p NEC forms a curved hexagonal lattice at the INM that drives capsid envelopment and budding into the perinuclear space (36–38, 59, 60, 62) in concert with an array of other viral proteins (63). Strikingly, UL31p/UL34p were necessary and sufficient to localize the ESCRT-III CHMP4 isoform CHMP4B to the nuclear envelope, and loss of the CHMP4 isoforms CHMP4A/B/C or ALIX resulted in an accumulation of virions in the perinuclear space, perhaps trapped because of failure to complete scission at the INM (39). Streptavidin-tagged UL34p coprecipitated with ALIX from HSV-1-infected cells, and glutathione S-transferase (GST) pulldowns indicated a direct interaction between ALIX and UL34p (39). Together, these data suggest that the HSV-1 NEC binds ALIX to mediate ESCRT-III (primarily CHMP4) recruitment to the INM during HSV-1 capsid budding into the perinuclear space. However, expression of the dominant negative Vps4-EQ allele resulted in a modest (2-fold) reduction in the numbers of capsids able to reach the cytoplasm (39). This finding is consistent with earlier published data (detailed below) showing that dominant negative Vsp4-EQ had no effect on the numbers of capsid-associated HSV-1 particles able to reach the cytoplasm (49, 50, 64), implying no nuclear egress defect. It is also consistent with the finding that the NEC alone is sufficient to drive membrane scission in vitro (58). Moreover, a requirement for ALIX in nuclear export is at odds with the observation that ALIX is dispensable for HSV-1 replication (detailed below [65]).
FIG 2.
HSV-1 and EBV NECs and their interactions with ESCRT components. The soluble and membrane-anchored subunits of the herpesvirus NEC are shown in purple and red, respectively, with the lipid bilayer of the INM shown in gray at the bottom. ESCRT components or other relevant molecules that interact with the HSV-1 NEC (UL31p/UL34p) or the EBV NEC (BFLF2/BFRF1) are shown at the left and right of the NEC, respectively. Proteins are represented as touching an NEC subunit in cases where published data imply direct physical contact. Proteins present in a complex with the NEC subunit are connected by a solid line. Dashed lines indicate that the NEC complex influences the distribution of the protein but that no direct or indirect contact has been demonstrated. Purple spheres on EBV BFRF1 (Ub) represent polyubiquitin chains (Poly-Ub) covalently attached to lysine residues.
There are several ways to reconcile these findings. First, although the NEC is capable of driving membrane scission by itself (58), it may nevertheless appropriate ESCRT-III/Vps4 to facilitate HSV-1 budding into the perinuclear space. Thus, the efficiency and rate of traversal of the nuclear envelope may be reduced in the absence of ESCRT function, but not the final yield of cytoplasmic virions. Second, HSV-1 infection in some cell types induces a form of autophagy at the nuclear membranes termed nuclear envelope-derived autophagy (NEDA) (66, 67), and autophagic degradation of inner nuclear membrane lamins is required to facilitate HSV-1 nuclear egress (67, 68). Since the proper closure and sealing of autophagosomes requires ESCRT-III/Vps4 (69), the NEC may be directing ESCRT components to the INM to support autophagic destruction of lamins, clearing away these barriers from the inner membrane bilayer in preparation for NEC-driven capsid budding. Third, NECs might recruit the ESCRT machinery because of its role in helping to maintain the health and integrity of the INM (8, 70), which could be subject to unusual stress and disruption during herpesvirus nuclear egress. However, in the normal repair/resealing pathway, CHMP4B is localized to the INM via the ESCRT-III subunit CHMP7, independently of ALIX (70, 71), neither of which is the case for HSV-1 NEC-mediated CHMP4B INM targeting (39). Finally, the modest effect of Vps4-EQ on HSV-1 capsid export could reflect a role for ESCRT-III that is Vps4 independent, as seen during the engulfment of vesicles by organelles in the poorly understood, unconventional CUPS protein secretion pathway (72).
The NEC of EBV (Fig. 2) has also been reported to interact with ESCRT components during EBV capsid nuclear envelopment (39, 55). The EBV genes BFLF2 and BFRF1 are homologues of the HSV-1 genes UL31 and UL34, respectively (73–75), and the EBV NEC, like that of HSV-1, localizes to the nuclear rim and controls the egress of EBV capsids from the nucleoplasm to the cytoplasm (55, 74, 75). Lytic reactivation of latent EBV is accompanied by redistribution of CHMP4B, CHMP1B, and ALIX to the nucleus (55), and BFRF1 is sufficient to redistribute ALIX and Vps4/Vps4-EQ to the nuclear rim. Furthermore, siRNA-mediated knockdown of ALIX or the expression of Vps4-EQ leads to an accumulation of viral genomes and capsids in the nucleus, suggesting a block in nuclear export (55, 76). These data are consistent with the EBV NEC recruiting ESCRT to facilitate capsid budding at the INM, and indeed, the NEC membrane-anchored subunit BFRF1 coimmunoprecipitates with ALIX, CHMP4B, and Nedd4 ubiquitin E3 ligases, primarily Itch (55, 76). Pulldown experiments suggest that BFRF1 and Itch interact directly (76), but BFRF1 lacks any recognizable L-domain motifs that might mediate ALIX or Nedd4 binding (5). Deletion mapping did not identify a single region of BFRF1 sufficient for ALIX recruitment (55) but suggested that Nedd4 binding involves an ∼150-amino-acid region of BFRF1 adjacent to its membrane-anchoring domain (76). In an interesting parallel with Gag proteins (5), BFRF1 was found to be ubiquitinated at several lysine residues (76), and ubiquitination (or the lysine residues themselves) was important for BFRF1–Nedd4 association. Ablation of these lysines diminished BFRF1-mediated vesiculation of the nuclear membrane (a phenomenon thought to reflect the role of BFRF1 in EBV capsid nuclear export) and reduced the efficiency of secretion of enveloped EBV particles from cells (interpreted as a consequence of reduced delivery of EBV capsids from the nucleus to the cytoplasm) (55, 76). It is not clear whether Nedd4 family members themselves are responsible for the ubiquitination of BFRF1. Like that of HSV-1, the NEC of EBV might recruit ESCRT components to support the scission of primary enveloped virions into the perinuclear space, to promote the degradation of lamins and facilitate access to the INM, or to help maintain the integrity of the nuclear envelope during infection.
THE ESCRT APPARATUS AND HERPESVIRUS ENVELOPMENT IN THE CYTOPLASM
Our understanding of ESCRT function during cytoplasmic herpesvirus envelopment is limited and is mostly derived from study of the Alphaherpesvirinae. Crump and colleagues were the first to demonstrate that Vps4-EQ is a potent inhibitor of HSV-1 cytoplasmic envelopment and virus replication (49). In the presence of Vps4-EQ, no extracellular virus particles were observed, and only unenveloped capsids were present within the cytoplasm. Ultrastructural examination revealed that these capsids were usually associated with membranes but not completely enclosed by them, an observation consistent with a block in sealing and scission. Quantitative electron microscopy confirmed that the numbers of capsids reaching the cytoplasm were similar in the presence of either Vps4 or Vps4-EQ, indicating no apparent defect in nuclear export (49). HSV-1 replication was also reduced by expression of dominant negative alleles of ESCRT-III CHMP family members, with alleles of CHMP1A/1B, CHMP2A/B, CHMP4B, and CHMP5 showing the greatest inhibitory effect (65). Our laboratory subsequently demonstrated that Vps4-EQ inhibited the replication and cytoplasmic envelopment of PRV (50) and that Vps4-EQ expression had no measurable effect on the numbers of HSV-1 or PRV capsids exported from the nucleus but instead inhibited capsid envelopment in the cytoplasm (50, 64). In agreement with transmission electron microscopy studies of Vps4-EQ-arrested virions (49), correlative light and scanning electron microscopy revealed structures in which cytoplasmic HSV-1 capsids sat partially enclosed by Vps4-EQ-bound organelles, resembling an egg sitting in an eggcup (64). The leading edge of the unsealed envelope was decorated by a ring of ∼12-nm-diameter beads (64) that in size and appearance resembled Vps4/Vps4-EQ hexamers seen to align along ESCRT-III polymers in uninfected cells (77). These ultrastructural studies suggest that the terminal HSV-1 phenotype resulting from Vps4-EQ expression is accumulation of partially enveloped cytoplasmic capsids that have failed to undergo “pinching off” into the lumen of a cytoplasmic organelle. Vps4-EQ-mediated arrest of ESCRT-III scission prevents subsequent events in virus egress, blocking the maturation of HSV-1- and PRV-containing organelles into structures that are able to traffic along microtubules in vitro (50).
WHAT IS THE MECHANISM OF ESCRT COMPLEX RECRUITMENT BY CYTOPLASMIC HERPESVIRUSES?
How do cytoplasmic herpesvirus capsids recruit ESCRT-III to support scission during secondary envelopment? Cytoplasmic envelopment requires that capsids be brought together with the full array of tegument and envelope proteins found in the mature viral particle. Figure 3 is a general representation of this process for the Alphaherpesvirinae. Motifs resembling YPXnL and P(T/S)AP Gag L-domains (which mediate ALIX and TSG101 binding, respectively) have been identified by sequence inspection of a number of HSV-1 proteins (65), but expression of dominant negative forms of ALIX and TSG101 had no effect on HSV-1 replication (65). Similarly, siRNA knockdown of ALIX and TSG101 did not diminish HSV-1 titers or growth rates, even when these proteins were depleted simultaneously to exclude the possibility that they act redundantly (65).
FIG 3.
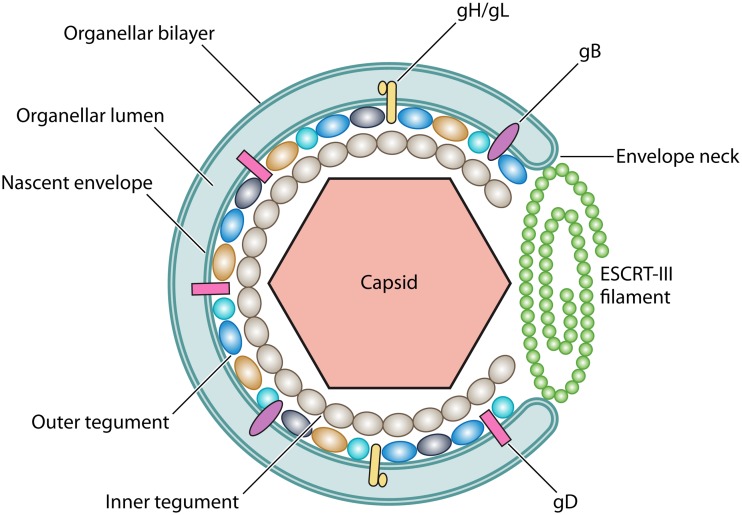
Generalized structure of an alphaherpesvirus cytoplasmic envelopment intermediate. The capsid (pink hexagon) is surrounded by an inner tegument (gray ovals) composed of UL36p/UL37p. This is the foundation for the attachment of the outer tegument subunits (shown in dark blue, cyan, orange, and dark gray), some of which interact with membrane-embedded envelope glycoproteins. There are multiple membrane proteins in the mature alphaherpesvirus envelope, but for clarity, only gB (purple), the gH/gL complex (yellow), and gD (pink) are shown (gD is absent from the alphaherpesvirus VZV). The ESCRT-III filament is assembled from CHMP subunits (green spheres) at the open bud neck. ESCRT-III will constrict to draw the membrane together, sealing the envelope and pinching the enveloped virus into the organellar lumen (light blue space). Constriction and ESCRT-III disassembly are catalyzed by the ATPase Vps4 (not shown).
Several components of the ESCRT machinery are known to bind ubiquitinated proteins as part of their normal cellular function, including (in addition to ALIX and TSG101) the STAM and HRS subunits of ESCRT-0 and the EAP45 subunit of ESCRT-II (2). Furthermore, as detailed above, ubiquitination is known to facilitate ESCRT recruitment to retroviral Gag proteins, to the NS2 protein of hepatitis C virus, and possibly also to the NEC of EBV. Could ubiquitination of herpesvirus proteins be used to recruit the ESCRT apparatus during cytoplasmic envelopment? All three families of herpesviruses encode proteins containing PPXY L-domain-like motifs, and in some cases, these molecules have been demonstrated to bind Nedd4 E3 ligase family members (65, 78). These include the products of the HSV-1 and -2 UL56 genes, ORF0 of varicella-zoster virus (VZV), UL42 of HCMV, and LMP2A of EBV (78). However, there is no evidence that these interactions with Nedd4 play any role in ubiquitin-mediated ESCRT recruitment or viral envelopment. Rather, in most cases, they result in the ubiquitination of Nedd4 itself and reductions in its cellular levels, likely via protein degradation (78–80). Furthermore, loss of the UL56 gene from HSV-2 has only limited consequences for viral replication in culture (79). The Alphaherpesvirinae do encode their own RING finger E3 ubiquitin ligase (termed ICP0 in the case of HSV-1 and -2), which targets multiple cellular proteins for ubiquitination and proteasome-mediated degradation, with consequences for a wide range of cellular processes, including DNA repair, transcription, the cell cycle, and innate and intrinsic antiviral immunity (81, 82). Nevertheless, none of the known ubiquitinated ICP0 substrates (81) are obvious candidates for the recruitment of ESCRT components, and HSV-1 mutants lacking ICP0 exhibit a complex phenotype reflecting altered viral gene expression that differs in severity between cell types (83, 84).
Taken together, the data presented above suggest that HSV (and possibly other herpesviruses) eschews the most commonly used viral strategies for ESCRT-III appropriation. Absent a role for ALIX, ESCRT-I, and Nedd4 binding/ubiquitination, what other mechanisms might be used to recruit ESCRT-III during cytoplasmic envelopment?
(i) ESCRT-II.
Other than ESCRT-I and ALIX, the most obvious remaining cellular candidate to drive ESCRT-III polymerization is ESCRT-II. ESCRT-II does not appear to be a popular choice for this task among enveloped viruses, even those that bind TSG101 and thus are well positioned to recruit ESCRT-II via ESCRT-I. Nevertheless, the apparent dispensability of ESCRT-II in HIV-1 assembly is being reevaluated, and it is possible that ESCRT-II plays a more important role in retroviruses than originally thought (2, 4, 85). Whether ESCRT-II participates in the cytoplasmic envelopment of Herpesviridae remains a major unanswered question. One argument against a role for ESCRT-II is that its primary means of ESCRT-III assembly is via nucleation of CHMP6 polymerization (2, 86), whereas dominant negative CHMP6 had the mildest inhibitory effect on HSV-1 replication of any CHMP family member tested (65). Moreover, if HSV-1 does utilize ESCRT-II, it must have evolved a TSG101/ESCRT-I-independent mechanism to do so. This is not without precedent, since the core protein of hepatitis B virus has been reported to interact directly with ESCRT-II. Core-bound ESCRT-II appears important for pregenomic RNA trafficking, encapsidation, or stabilization of replication-competent hepatitis B virus nucleocapsids (87), but it is not known whether it also plays downstream roles in hepatitis B virus envelopment.
(ii) Cellular Bro1-domain proteins.
Although ALIX is dispensable for HSV-1 envelopment, the human proteome contains four other Bro1-domain proteins with broad tissue expression (88): rhophilin 1 and 2 (RHPN1 and RHPN2), His domain-containing protein tyrosine phosphatase (HD-PTP), and Bro1 domain and CAAX motif containing (BROX) (89). RHPN1 and RHPN2 are Rho-GTP binding proteins associated with the actin cytoskeleton and have not been clearly linked to the ESCRT pathway (89). In contrast, HD-PTP and BROX both participate in ESCRT-III assembly during endosomal cargo sorting and MVB formation (12, 90), positioning them in an appropriate cellular location for use by enveloping HSV-1 capsids (40, 41, 91). HD-PTP recruitment involves several ESCRT components, including ESCRT-0 (2, 92), while the mechanism of BROX assembly is less well defined (2). BROX has an additional interesting property. As mentioned above, of all the CHMP family members, only CHMP4 was thought to be an important target for Bro1 domain-containing proteins (11, 12, 93). However, BROX has been shown to interact with both CHMP4B and CHMP5 and to recruit CHMP5 to endosomes (94). Given that dominant negative alleles of CHMP4B and CHMP5 are two of the most potent inhibitors of HSV-1 replication (65), a role for BROX as a possible cellular regulator of HSV-1 envelopment merits consideration.
(iii) Viral tegument and envelope proteins.
The tegument layer resides between the herpesvirus capsid and the inner surface of the envelope and is a dense complex of virally encoded and host cell proteins (35, 95–97). The “inner” tegument (Fig. 3) is composed principally of the capsid vertex-associated protein UL36p (98) and its binding partner UL37p (99), two large polypeptides conserved among the alpha-, beta-, and gammaherpesviruses (35) that perform a variety of functions, including the recruitment of multiple polypeptides to form the outer tegument (35, 97). UL36p and UL37p are essential for HSV-1 capsid cytoplasmic envelopment and thus for virus replication (35, 100, 101). Loss of UL36p results in the accumulation of aggregates of cytoplasmic capsids (100) and nonenveloped capsids docked to organelles but apparently mislocalized (91, 102), possibly due to the absence of the tethering/targeting function provided by UL37p (103, 104).
The absolute requirement for UL36p in HSV-1 capsid envelopment, and its conservation among the Herpesviridae, make it a reasonable candidate for an ESCRT recruitment molecule. Furthermore, given the role of ubiquitination in ESCRT recruitment (described above), it is interesting that the amino-terminal portion of UL36p contains a conserved cysteine protease activity that functions as a deubiquitinase (DUB) domain, able to remove ubiquitin from posttranslationally modified proteins (105, 106). The HSV-1 and PRV UL36p DUBs regulate the ubiquitination states of several viral proteins during infection, including UL36p itself, and UL36p ubiquitination/deubiquitination controls the ability of PRV to disseminate into the peripheral nervous system (107, 108). Loss of the DUB active-site cysteine reduced PRV titers by 2 log units in tissue culture and led to the accumulation of nonenveloped cytoplasmic capsids (109), but a similar mutation had negligible consequences for HSV-1 replication (110). HSV-1 UL36p has also been reported to modulate the ubiquitination state of TSG101 (111), but given that TSG101 and UL36p DUB activity are both dispensable for HSV-1 replication (65, 110), the significance of this finding for envelopment is unclear.
As a functional test for UL36p in ESCRT recruitment, we reasoned that if UL36p is needed to couple HSV-1 capsids to the ESCRT apparatus, then, in its absence, capsids should no longer become trapped in envelopment complexes by the dominant negative ATPase Vps4-EQ (13, 15, 16, 20). We used correlative light and scanning electron microscopy to confirm the ultrastructures of representative examples of HSV-1 capsid/Vps4-EQ-containing complexes and counted the numbers of capsid/Vps4-EQ complexes in the presence and absence of UL36p (64). The loss of UL36p reduced the numbers of HSV-1 capsid/Vps4-EQ-containing complexes by approximately 70%, suggesting that UL36p plays an important role in the recruitment of capsids to the site of ESCRT-III/Vps4 deposition (64). Nevertheless, UL36p is required for the assembly of multiple outer tegument and envelope proteins into the enveloping particle (99, 104, 112–115), and its role in capsid/ESCRT-III/Vps4 assembly may be indirect (64).
Proteins localized to the outer layers of tegument (Fig. 3) exhibit a network of genetically and biochemically defined interactions with UL36p/UL37p (112–115), with each other, and with the inward-facing (equivalent to cytoplasmically disposed) portions of membrane-embedded envelope proteins (35, 97, 116–119). For HSV-1 and PRV, a number of tegument and envelope proteins have been assigned roles in envelopment and may provide avenues for the recruitment of ESCRT complexes. Six of these (tegument proteins encoded by HSV-1 genes UL7, UL11, UL16, and UL51 and the HSV-1 envelope protein heterodimer gM/gN) are conserved among the alpha-, beta-, and gammaherpesviruses. Another five tegument proteins (encoded by HSV-1 UL21, UL46, UL47, UL48, and UL49), the HSV-1 membrane-anchored tegument protein US9p, and the HSV-1 envelope protein heterodimers gK/UL20p and gE/gI are unique to the Alphaherpesvirinae (35, 120–122). Complicating the interpretation of their roles, some of these outer tegument and envelope proteins are individually required for envelopment, but in other cases, envelopment fails only when several of the genes are deleted simultaneously, suggesting functional redundancy (35). The extent of this redundancy can differ between members of a herpesvirus subfamily and even for a single virus replicating in different cell types. For example, simultaneous deletion of the genes encoding gE/gI and gM resulted in a profound block to PRV capsid envelopment (123, 124), but comparable mutations had a marginal effect on HSV-1 (122). Simultaneous loss of the gE/gI heterodimer and the membrane-anchored tegument protein US9p resulted in an HSV-1 envelopment defect, but one that was specific to neuronal cells (121).
Any tegument and envelope proteins implicated in cytoplasmic envelopment could conceivably play roles in capsid-organelle docking, sequestration of envelope proteins to the assembly site, wrapping of the capsid by membrane, or binding of ESCRT components to support scission. In this regard, the biology of L-particles may provide some insights. L-particles consist of lipid envelope-like structures similar in size to mature virions (though more heterogeneous) and displaying membrane-embedded envelope proteins and tegument but lacking nucleocapsids (125, 126). They have been observed during the tissue culture replication of all alphaherpesviruses examined (126) and, at least for HSV-1, follow an assembly and egress pathway similar to that of infectious particles (35, 125, 127, 128). L-particles may therefore arise when capsids fail to attach to and engage with sites of cytoplasmic tegument deposition, envelope formation, and ESCRT-mediated scission. Consequently, L-particle assembly may not require viral proteins whose primary function is to integrate capsids with the envelopment machinery but presumably would remain dependent on any molecules needed for budding and membrane scission. It is not known whether ESCRT-III/Vps4 are required for the generation of L-particles, but if so, molecules essential for L-particle formation might be good candidates for ESCRT recruitment, as opposed to playing other roles required for the assembly of infectious, capsid-containing virions. Interestingly, bovine herpesvirus 1 (BoHV-1) L-particles contain the same complement of envelope proteins as BoHV-1 virions but lack the inner tegument proteins UL36p and UL37p (129). Thus, if BoHV-1 L-particles require ESCRT-III for their scission, UL36p/UL37p must be dispensable for this process or are subsequently lost from the particle. In contrast, the tegument composition of HSV-1 L-particles resembles that of infectious virions (129).
SUMMARY
The NECs of HSV-1 and EBV provide us with the best available examples of a herpesvirus strategy for ESCRT recruitment (Fig. 2), functioning similarly to retroviral Gag proteins by assembling membrane-deforming hexagonal lattices and (directly or indirectly) binding ALIX (HSV-1) or ALIX/Nedd4/ubiquitin (EBV) at the INM (37, 39, 55, 76). Nevertheless, many unanswered questions remain. The absence of classical L-domains in the NEC suggests that NEC–ALIX/Nedd4 interactions are indirect or utilize yet to be defined binding motifs. It is also unclear whether NECs are directing ESCRT components to support scission at the necks of budding perinuclear virions, in support of autophagy of nuclear lamins, to maintain nuclear envelope health, or to serve other functions. An even greater unknown is herpesvirus envelopment in the cytoplasm—a process of apparently bewildering complexity, encompassing a web of interactions between capsid subunits, inner and outer tegument proteins, membrane proteins in the nascent envelope, and the cellular ESCRT complexes. There is a gap in our understanding of how these two families of proteins, viral and cellular, engage and dance together to coordinate envelopment and scission. Given the apparent dispensability of ALIX and ESCRT-I for HSV-1 envelopment, it remains a possibility that unrecognized mechanisms for the recruitment of ESCRT-II, binding of Bro1-domain proteins other than ALIX, or activation of CHMP subunit polymerization lie buried in the rich proteome of the tegument and envelope.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 AI125244 (to D.W.W.). J.B. acknowledges support from institutional AIDS training grant T32 AI007501, “Training in HIV/AIDS Pathogenesis; Basic and Translational Research.”
REFERENCES
- 1.McCullough J, Frost A, Sundquist WI. 2018. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu Rev Cell Dev Biol 34:85–109. doi: 10.1146/annurev-cellbio-100616-060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. 2017. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem Sci 42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Hurley JH. 2015. ESCRTs are everywhere. EMBO J 34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scourfield EJ, Martin-Serrano J. 2017. Growing functions of the ESCRT machinery in cell biology and viral replication. Biochem Soc Trans 45:613–634. doi: 10.1042/BST20160479. [DOI] [PubMed] [Google Scholar]
- 5.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, Hanson PI, Frost A. 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350:1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alonso Y Adell M, Migliano SM, Teis D. 2016. ESCRT-III and Vps4: a dynamic multipurpose tool for membrane budding and scission. FEBS J 283:3288–3302. doi: 10.1111/febs.13688. [DOI] [PubMed] [Google Scholar]
- 8.Olmos Y, Carlton JG. 2016. The ESCRT machinery: new roles at new holes. Curr Opin Cell Biol 38:1–11. doi: 10.1016/j.ceb.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frankel EB, Audhya A. 2018. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin Cell Dev Biol 74:4–10. doi: 10.1016/j.semcdb.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teis D, Saksena S, Emr SD. 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell 15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 11.McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci U S A 105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichioka F, Kobayashi R, Katoh K, Shibata H, Maki M. 2008. Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4). FEBS J 275:682–692. doi: 10.1111/j.1742-4658.2007.06230.x. [DOI] [PubMed] [Google Scholar]
- 13.Han H, Hill CP. 2019. Structure and mechanism of the ESCRT pathway AAA+ ATPase Vps4. Biochem Soc Trans 47:37–45. doi: 10.1042/BST20180260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoneberg J, Pavlin MR, Yan S, Righini M, Lee IH, Carlson LA, Bahrami AH, Goldman DH, Ren X, Hummer G, Bustamante C, Hurley JH. 2018. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 362:1423–1428. doi: 10.1126/science.aat1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han H, Monroe N, Votteler J, Shakya B, Sundquist WI, Hill CP. 2015. Binding of substrates to the central pore of the Vps4 ATPase is autoinhibited by the microtubule interacting and trafficking (MIT) domain and activated by MIT interacting motifs (MIMs). J Biol Chem 290:13490–13499. doi: 10.1074/jbc.M115.642355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies BA, Azmi IF, Payne J, Shestakova A, Horazdovsky BF, Babst M, Katzmann DJ. 2010. Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol Biol Cell 21:3396–3408. doi: 10.1091/mbc.E10-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mierzwa BE, Chiaruttini N, Redondo-Morata L, von Filseck JM, Konig J, Larios J, Poser I, Muller-Reichert T, Scheuring S, Roux A, Gerlich DW. 2017. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol 19:787–798. doi: 10.1038/ncb3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder JC, Samson RY, Brumfield SK, Bell SD, Young MJ. 2013. Functional interplay between a virus and the ESCRT machinery in Archaea. Proc Natl Acad Sci U S A 110:10783–10787. doi: 10.1073/pnas.1301605110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor GM, Hanson PI, Kielian M. 2007. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J Virol 81:13631–13639. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun S, Li L, Yang F, Wang X, Fan F, Yang M, Chen C, Li X, Wang HW, Sui SF. 2017. Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution. Nat Commun 8:16064. doi: 10.1038/ncomms16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson DS, Bleck M, Simon SM. 2018. Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. Elife 7:e36221. doi: 10.7554/eLife.36221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honeychurch KM, Yang G, Jordan R, Hruby DE. 2007. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J Virol 81:7310–7315. doi: 10.1128/JVI.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray G, Schmitt PT, Schmitt AP. 2019. Angiomotin-like 1 links paramyxovirus M proteins to NEDD4 family ubiquitin ligases. Viruses 11:128. doi: 10.3390/v11020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mercenne G, Alam SL, Arii J, Lalonde MS, Sundquist WI. 2015. Angiomotin functions in HIV-1 assembly and budding. Elife 4:e03778. doi: 10.7554/eLife.03778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann J, Boehm C, Himmelsbach K, Donnerhak C, Roettger H, Weiss TS, Ploen D, Hildt E. 2013. Identification of alpha-taxilin as an essential factor for the life cycle of hepatitis B virus. J Hepatol 59:934–941. doi: 10.1016/j.jhep.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 26.Leung J, Yueh A, Appah FS Jr, Yuan B, de los Santos K, Goff SP. 2006. Interaction of Moloney murine leukemia virus matrix protein with IQGAP. EMBO J 25:2155–2166. doi: 10.1038/sj.emboj.7601097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Qu Y, Liu Y, Jambusaria R, Han Z, Ruthel G, Freedman BD, Harty RN. 2013. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J Virol 87:7777–7780. doi: 10.1128/JVI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpp LN, Galler R, Bonaldo MC. 2011. Interaction between the yellow fever virus nonstructural protein NS3 and the host protein Alix contributes to the release of infectious particles. Microbes Infect 13:85–95. doi: 10.1016/j.micinf.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Pattanakitsakul SN, Poungsawai J, Kanlaya R, Sinchaikul S, Chen ST, Thongboonkerd V. 2010. Association of Alix with late endosomal lysobisphosphatidic acid is important for dengue virus infection in human endothelial cells. J Proteome Res 9:4640–4648. doi: 10.1021/pr100357f. [DOI] [PubMed] [Google Scholar]
- 31.Chiou CT, Hu CC, Chen PH, Liao CL, Lin YL, Wang JJ. 2003. Association of Japanese encephalitis virus NS3 protein with microtubules and tumour susceptibility gene 101 (TSG101) protein. J Gen Virol 84:2795–2805. doi: 10.1099/vir.0.19201-0. [DOI] [PubMed] [Google Scholar]
- 32.Barouch-Bentov R, Neveu G, Xiao F, Beer M, Bekerman E, Schor S, Campbell J, Boonyaratanakornkit J, Lindenbach B, Lu A, Jacob Y, Einav S. 2016. Hepatitis C virus proteins interact with the endosomal sorting complex required for transport (ESCRT) machinery via ubiquitination to facilitate viral envelopment. mBio 7:e01456-16. doi: 10.1128/mBio.01456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heming JD, Conway JF, Homa FL. 2017. Herpesvirus capsid assembly and DNA packaging. Adv Anat Embryol Cell Biol 223:119–142. doi: 10.1007/978-3-319-53168-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai X, Zhou ZH. 2018. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 360:eaao7298. doi: 10.1126/science.aao7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owen DJ, Crump CM, Graham SC. 2015. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mettenleiter TC. 2016. Vesicular nucleo-cytoplasmic transport—herpesviruses as pioneers in cell biology. Viruses 8:266. doi: 10.3390/v8100266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bigalke JM, Heldwein EE. 2017. Have NEC coat, will travel: structural basis of membrane budding during nuclear egress in herpesviruses. Adv Virus Res 97:107–141. doi: 10.1016/bs.aivir.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bigalke JM, Heldwein EE. 2016. Nuclear exodus: herpesviruses lead the way. Annu Rev Virol 3:387–409. doi: 10.1146/annurev-virology-110615-042215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arii J, Watanabe M, Maeda F, Tokai-Nishizumi N, Chihara T, Miura M, Maruzuru Y, Koyanagi N, Kato A, Kawaguchi Y. 2018. ESCRT-III mediates budding across the inner nuclear membrane and regulates its integrity. Nat Commun 9:3379. doi: 10.1038/s41467-018-05889-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J 31:4204–4220. doi: 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harley CA, Dasgupta A, Wilson DW. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J Virol 75:1236–1251. doi: 10.1128/JVI.75.3.1236-1251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turcotte S, Letellier J, Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alwine JC. 2012. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog 8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Granzow H, Klupp BG, Fuchs W, Veits J, Osterrieder N, Mettenleiter TC. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J Virol 75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henaff D, Radtke K, Lippe R. 2012. Herpesviruses exploit several host compartments for envelopment. Traffic 13:1443–1449. doi: 10.1111/j.1600-0854.2012.01399.x. [DOI] [PubMed] [Google Scholar]
- 46.Hambleton S, Gershon MD, Gershon AA. 2004. The role of the trans-Golgi network in varicella zoster virus biology. Cell Mol Life Sci 61:3047–3056. doi: 10.1007/s00018-004-4269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanbo A, Noda T, Ohba Y. 2018. Epstein-Barr virus acquires its final envelope on intracellular compartments with Golgi markers. Front Microbiol 9:454. doi: 10.3389/fmicb.2018.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cepeda V, Esteban M, Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol 12:386–404. doi: 10.1111/j.1462-5822.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- 49.Crump CM, Yates C, Minson T. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol 81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kharkwal H, Smith CG, Wilson DW. 2014. Blocking ESCRT-mediated envelopment inhibits microtubule-dependent trafficking of alphaherpesviruses in vitro. J Virol 88:14467–14478. doi: 10.1128/JVI.02777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cabrera JR, Manivanh R, North BJ, Leib DA. 2019. The ESCRT-related ATPase Vps4 is modulated by interferon during herpes simplex virus 1 infection. mBio 10:e02567-18. doi: 10.1128/mBio.02567-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tandon R, AuCoin DP, Mocarski ES. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J Virol 83:10797–10807. doi: 10.1128/JVI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraile-Ramos A, Pelchen-Matthews A, Risco C, Rejas MT, Emery VC, Hassan-Walker AF, Esteban M, Marsh M. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol 9:2955–2967. doi: 10.1111/j.1462-5822.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 54.Streck NT, Carmichael J, Buchkovich NJ. 2018. A non-envelopment role for the ESCRT-III complex during HCMV infection. J Virol 92:e02096-17. doi: 10.1128/JVI.02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee CP, Liu PT, Kung HN, Su MT, Chua HH, Chang YH, Chang CW, Tsai CH, Liu FT, Chen MR. 2012. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus–associated membrane for the maturation of Epstein-Barr virus. PLoS Pathog 8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roller RJ, Baines JD. 2017. Herpesvirus nuclear egress. Adv Anat Embryol Cell Biol 223:143–169. doi: 10.1007/978-3-319-53168-7_7. [DOI] [PubMed] [Google Scholar]
- 58.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. 2014. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun 5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bigalke JM, Heldwein EE. 2015. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J 34:2921–2936. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bigalke JM, Heldwein EE. 2015. The great (nuclear) escape: new insights into the role of the nuclear egress complex of herpesviruses. J Virol 89:9150–9153. doi: 10.1128/JVI.02530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lingappa JR, Reed JC, Tanaka M, Chutiraka K, Robinson BA. 2014. How HIV-1 Gag assembles in cells: putting together pieces of the puzzle. Virus Res 193:89–107. doi: 10.1016/j.virusres.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeev-Ben-Mordehai T, Weberruß M, Lorenz M, Cheleski J, Hellberg T, Whittle C, El Omari K, Vasishtan D, Dent KC, Harlos K, Franzke K, Hagen C, Klupp BG, Antonin W, Mettenleiter TC, Grünewald K. 2015. Crystal structure of the herpesvirus nuclear egress complex provides insights into inner nuclear membrane remodeling. Cell Rep 13:2645–2652. doi: 10.1016/j.celrep.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banfield BW. 2019. Beyond the NEC: modulation of herpes simplex virus nuclear egress by viral and cellular components. Curr Clin Microbiol Rep 6:1–9. doi: 10.1007/s40588-019-0112-7. [DOI] [Google Scholar]
- 64.Kharkwal H, Smith CG, Wilson DW. 2016. HSV capsid localization to ESCRT-VPS4 complexes in the presence and absence of the large tegument protein UL36p. J Virol 90:7257–7267. doi: 10.1128/JVI.00857-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pawliczek T, Crump CM. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol 83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, Desjardins M. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Radtke K, English L, Rondeau C, Leib D, Lippe R, Desjardins M. 2013. Inhibition of the host translation shutoff response by herpes simplex virus 1 triggers nuclear envelope-derived autophagy. J Virol 87:3990–3997. doi: 10.1128/JVI.02974-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turan A, Grosche L, Krawczyk A, Muhl-Zurbes P, Drassner C, Duthorn A, Kummer M, Hasenberg M, Voortmann S, Jastrow H, Dorrie J, Schaft N, Kraner M, Dohner K, Sodeik B, Steinkasserer A, Heilingloh CS. 2019. Autophagic degradation of lamins facilitates the nuclear egress of herpes simplex virus type 1. J Cell Biol 218:508–523. doi: 10.1083/jcb.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takahashi Y, He H, Tang Z, Hattori T, Liu Y, Young MM, Serfass JM, Chen L, Gebru M, Chen C, Wills CA, Atkinson JM, Chen H, Abraham T, Wang HG. 2018. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat Commun 9:2855. doi: 10.1038/s41467-018-05254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ventimiglia LN, Martin-Serrano J. 2016. ESCRT machinery: damage control at the nuclear membrane. Cell Res 26:641–642. doi: 10.1038/cr.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522:231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- 72.Curwin AJ, Brouwers N, Alonso Y Adell M, Teis D, Turacchio G, Parashuraman S, Ronchi P, Malhotra V. 2016. ESCRT-III drives the final stages of CUPS maturation for unconventional protein secretion. Elife 5:e16299. doi: 10.7554/eLife.16299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lake CM, Hutt-Fletcher LM. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99–106. doi: 10.1016/j.virol.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 74.Gonnella R, Farina A, Santarelli R, Raffa S, Feederle R, Bei R, Granato M, Modesti A, Frati L, Delecluse HJ, Torrisi MR, Angeloni A, Faggioni A. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J Virol 79:3713–3727. doi: 10.1128/JVI.79.6.3713-3727.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farina A, Feederle R, Raffa S, Gonnella R, Santarelli R, Frati L, Angeloni A, Torrisi MR, Faggioni A, Delecluse HJ. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J Virol 79:3703–3712. doi: 10.1128/JVI.79.6.3703-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee CP, Liu GT, Kung HN, Liu PT, Liao YT, Chow LP, Chang LS, Chang YH, Chang CW, Shu WC, Angers A, Farina A, Lin SF, Tsai CH, Bouamr F, Chen MR. 2016. The ubiquitin ligase Itch and ubiquitination regulate BFRF1-mediated nuclear envelope modification for Epstein-Barr virus maturation. J Virol 90:8994–9007. doi: 10.1128/JVI.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanson PI, Roth R, Lin Y, Heuser JE. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koshizuka T, Kobayashi T, Ishioka K, Suzutani T. 2018. Herpesviruses possess conserved proteins for interaction with Nedd4 family ubiquitin E3 ligases. Sci Rep 8:4447. doi: 10.1038/s41598-018-22682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ushijima Y, Koshizuka T, Goshima F, Kimura H, Nishiyama Y. 2008. Herpes simplex virus type 2 UL56 interacts with the ubiquitin ligase Nedd4 and increases its ubiquitination. J Virol 82:5220–5233. doi: 10.1128/JVI.02515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ushijima Y, Luo C, Kamakura M, Goshima F, Kimura H, Nishiyama Y. 2010. Herpes simplex virus UL56 interacts with and regulates the Nedd4-family ubiquitin ligase Itch. Virol J 7:179–189. doi: 10.1186/1743-422X-7-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boutell C, Everett RD. 2013. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol 94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 82.Lanfranca MP, Mostafa HH, Davido DJ. 2014. HSV-1 ICP0: an E3 ubiquitin ligase that counteracts host intrinsic and innate immunity. Cells 3:438–454. doi: 10.3390/cells3020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Everett RD, Boutell C, Orr A. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J Virol 78:1763–1774. doi: 10.1128/JVI.78.4.1763-1774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yao F, Schaffer PA. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol 69:6249–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meng B, Ip NC, Prestwood LJ, Abbink TE, Lever AM. 2015. Evidence that the endosomal sorting complex required for transport-II (ESCRT-II) is required for efficient human immunodeficiency virus-1 (HIV-1) production. Retrovirology 12:72. doi: 10.1186/s12977-015-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. 2005. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J 387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stieler JT, Prange R. 2014. Involvement of ESCRT-II in hepatitis B virus morphogenesis. PLoS One 9:e91279. doi: 10.1371/journal.pone.0091279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell 8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. 2011. Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding. PLoS One 6:e27466. doi: 10.1371/journal.pone.0027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doyotte A, Mironov A, McKenzie E, Woodman P. 2008. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A 105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kharkwal H, Furgiuele SS, Smith CG, Wilson DW. 2015. Herpes simplex virus capsid-organelle association in the absence of the large tegument protein UL36p. J Virol 89:11372–11382. doi: 10.1128/JVI.01893-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J, Oh KJ, Lee D, Kim BY, Choi JS, Ku B, Kim SJ. 2016. Structural study of the HD-PTP Bro1 domain in a complex with the core region of STAM2, a subunit of ESCRT-0. PLoS One 11:e0149113. doi: 10.1371/journal.pone.0149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ichioka F, Takaya E, Suzuki H, Kajigaya S, Buchman VL, Shibata H, Maki M. 2007. HD-PTP and Alix share some membrane-traffic related proteins that interact with their Bro1 domains or proline-rich regions. Arch Biochem Biophys 457:142–149. doi: 10.1016/j.abb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 94.Mu R, Dussupt V, Jiang J, Sette P, Rudd V, Chuenchor W, Bello NF, Bouamr F, Xiao TS. 2012. Two distinct binding modes define the interaction of Brox with the C-terminal tails of CHMP5 and CHMP4B. Structure 20:887–898. doi: 10.1016/j.str.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 96.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 97.Smith GA. 2017. Assembly and egress of an alphaherpesvirus clockwork. Adv Anat Embryol Cell Biol 223:171–193. doi: 10.1007/978-3-319-53168-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fan WH, Roberts AP, McElwee M, Bhella D, Rixon FJ, Lauder R. 2015. The large tegument protein pUL36 is essential for formation of the capsid vertex-specific component at the capsid-tegument interface of herpes simplex virus 1. J Virol 89:1502–1511. doi: 10.1128/JVI.02887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koenigsberg AL, Heldwein EE. 2018. The dynamic nature of the conserved tegument protein UL37 of herpesviruses. J Biol Chem 293:15827–15839. doi: 10.1074/jbc.RA118.004481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol 74:11608–11618. doi: 10.1128/JVI.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Desai P, Sexton GL, McCaffery JM, Person S. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J Virol 75:10259–10271. doi: 10.1128/JVI.75.21.10259-10271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol 82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. 2014. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol 88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koenigsberg AL, Heldwein EE. 2017. Crystal structure of the N-terminal half of the traffic controller UL37 from herpes simplex virus 1. J Virol 91:e01244-17. doi: 10.1128/JVI.01244-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell 19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 106.Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J Virol 79:15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huffmaster NJ, Sollars PJ, Richards AL, Pickard GE, Smith GA. 2015. Dynamic ubiquitination drives herpesvirus neuroinvasion. Proc Natl Acad Sci U S A 112:12818–12823. doi: 10.1073/pnas.1512559112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee JI, Sollars PJ, Baver SB, Pickard GE, Leelawong M, Smith GA. 2009. A herpesvirus encoded deubiquitinase is a novel neuroinvasive determinant. PLoS Pathog 5:e1000387. doi: 10.1371/journal.ppat.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bottcher S, Maresch C, Granzow H, Klupp BG, Teifke JP, Mettenleiter TC. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J Virol 82:6009–6016. doi: 10.1128/JVI.00280-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bolstad M, Abaitua F, Crump CM, O'Hare P. 2011. Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1-2. J Virol 85:8738–8751. doi: 10.1128/JVI.00798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Calistri A, Munegato D, Toffoletto M, Celestino M, Franchin E, Comin A, Sartori E, Salata C, Parolin C, Palu G. 2015. Functional interaction between the ESCRT-I component TSG101 and the HSV-1 tegument ubiquitin specific protease. J Cell Physiol 230:1794–1806. doi: 10.1002/jcp.24890. [DOI] [PubMed] [Google Scholar]
- 112.Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J Virol 84:1397–1405. doi: 10.1128/JVI.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol 79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mijatov B, Cunningham AL, Diefenbach RJ. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31. doi: 10.1016/j.virol.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 115.Svobodova S, Bell S, Crump CM. 2012. Analysis of the interaction between the essential herpes simplex virus 1 tegument proteins VP16 and VP1/2. J Virol 86:473–483. doi: 10.1128/JVI.05981-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chi JH, Harley CA, Mukhopadhyay A, Wilson DW. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J Gen Virol 86:253–261. doi: 10.1099/vir.0.80444-0. [DOI] [PubMed] [Google Scholar]
- 117.Gross ST, Harley CA, Wilson DW. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1–12. doi: 10.1016/j.virol.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 118.Kamen DE, Gross ST, Girvin ME, Wilson DW. 2005. Structural basis for the physiological temperature dependence of the association of VP16 with the cytoplasmic tail of herpes simplex virus glycoprotein H. J Virol 79:6134–6141. doi: 10.1128/JVI.79.10.6134-6141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Regan KJ, Bucks MA, Murphy MA, Wills JW, Courtney RJ. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358:192–200. doi: 10.1016/j.virol.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 120.Jambunathan N, Chouljenko D, Desai P, Charles AS, Subramanian R, Chouljenko VN, Kousoulas KG. 2014. Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J Virol 88:5927–5935. doi: 10.1128/JVI.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.DuRaine G, Wisner TW, Howard P, Williams M, Johnson DC. 2017. Herpes simplex virus gE/gI and US9 promote both envelopment and sorting of virus particles in the cytoplasm of neurons, two processes that precede anterograde transport in axons. J Virol 91:e00050-17. doi: 10.1128/JVI.00050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Browne H, Bell S, Minson T. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J Virol 78:1039–1041. doi: 10.1128/JVI.78.2.1039-1041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brack AR, Dijkstra JM, Granzow H, Klupp BG, Mettenleiter TC. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol 73:5364–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brack AR, Klupp BG, Granzow H, Tirabassi R, Enquist LW, Mettenleiter TC. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J Virol 74:4004–4016. doi: 10.1128/JVI.74.9.4004-4016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Szilagyi JF, Cunningham C. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol 72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 126.Heilingloh CS, Krawczyk A. 2017. Role of L-particles during herpes simplex virus infection. Front Microbiol 8:2565. doi: 10.3389/fmicb.2017.02565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ibiricu I, Maurer UE, Grunewald K. 2013. Characterization of herpes simplex virus type 1 L-particle assembly and egress in hippocampal neurones by electron cryo-tomography. Cell Microbiol 15:285–291. doi: 10.1111/cmi.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rixon FJ, Addison C, McLauchlan J. 1992. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J Gen Virol 73:277–284. doi: 10.1099/0022-1317-73-2-277. [DOI] [PubMed] [Google Scholar]
- 129.Russell T, Bleasdale B, Hollinshead M, Elliott G. 2018. Qualitative differences in capsidless L-particles released as a by-product of bovine herpesvirus 1 and herpes simplex virus 1 infections. J Virol 92:e01259-18. doi: 10.1128/JVI.01259-18. [DOI] [PMC free article] [PubMed] [Google Scholar]