T cells are central to an effective defense against persistent viral infections that can be related to human cytomegalovirus (HCMV) or human herpesvirus 6 (HHV-6). However, knowledge of HHV-6-specific T-cell responses is limited. In order to deepen our knowledge of T-cell responses to HHV-6, we characterized HHV-6A- and HHV-6B-specific CD4+ and CD8+ T-cell responses directly ex vivo from healthy coinfected blood donors. Despite the strong similarity of HHV-6 and HCMV from a virologic point of view, we observed immunological differences, particularly in relation to the frequency and phenotype of effector/memory and regulatory virus-specific T cells. This suggests that different immune factors are solicited in the control of HHV-6 infection than in that of HCMV infection. Our findings may encourage immunomonitoring of patients with viral replication episodes to follow the emergence of effector versus regulatory T cells.
KEYWORDS: HCMV, HHV-6, effector T cells, regulatory T cells
ABSTRACT
Human herpesvirus 6 (HHV-6) infects >90% of the population and establishes a latent infection with asymptomatic episodes of reactivation. However, HHV-6 reactivation is associated with morbidity and sometimes mortality in immunocompromised patients. To date, control of the virus in healthy virus carriers and the failure to control it in patients with disease remain poorly understood. In particular, knowledge of HHV-6-specific T-cell responses is limited. Here, we characterized HHV-6A- and HHV-6B-specific CD4+ and CD8+ T-cell responses from peripheral blood mononuclear cells (PBMCs) of healthy donors. We studied the phenotype of effector HHV-6-specific T cells ex vivo, as well as of induced specific suppressive regulatory CD4+ T cells in vitro poststimulation, in comparison to human cytomegalovirus (HCMV) responses. Compared to that for HCMV, we show that ex vivo T-cell reactivity in peripheral blood is detectable but at very low frequency, both for HHV-6A and -6B viruses. Interestingly, the phenotype of the specific T cells also differs between the viruses. HHV-6A- and HHV-6B-specific CD4+ T lymphocytes are less differentiated than HCMV-specific T cells. Furthermore, we show a higher frequency of HHV-6-specific suppressive regulatory T cells (eTregs) than HCMV-specific eTregs in coinfected individuals. Despite the strong similarity of HHV-6 and HCMV from a virologic point of view, we observed immunological differences, particularly in relation to the frequency and phenotype of effector/memory and regulatory virus-specific T cells. This suggests that different immune factors are solicited in the control of HHV-6 infection than in that of HCMV infection.
IMPORTANCE T cells are central to an effective defense against persistent viral infections that can be related to human cytomegalovirus (HCMV) or human herpesvirus 6 (HHV-6). However, knowledge of HHV-6-specific T-cell responses is limited. In order to deepen our knowledge of T-cell responses to HHV-6, we characterized HHV-6A- and HHV-6B-specific CD4+ and CD8+ T-cell responses directly ex vivo from healthy coinfected blood donors. Despite the strong similarity of HHV-6 and HCMV from a virologic point of view, we observed immunological differences, particularly in relation to the frequency and phenotype of effector/memory and regulatory virus-specific T cells. This suggests that different immune factors are solicited in the control of HHV-6 infection than in that of HCMV infection. Our findings may encourage immunomonitoring of patients with viral replication episodes to follow the emergence of effector versus regulatory T cells.
INTRODUCTION
Human herpesvirus 6 (HHV-6) is a member of the Herpesviridae family, Betaherpesvirinae subfamily, and Roseolovirus genus. HHV-6 is classified as two species, HHV-6A and HHV-6B, with a high level of nucleotide similarity (70% to 95%, depending on the genes considered) (1, 2); these betaherpesviruses are ubiquitous pathogens that infect more than 90% of the population (3). After primary infection, usually occurring during infancy and associated with a self-limiting or benign acute disease (exanthema subitum), the virus establishes latent infection, with asymptomatic episodes of reactivation (4). However, in immunocompromised individuals, HHV-6 reactivation is associated with significant clinical pathologies, such as encephalitis, hematopoiesis disorders, graft versus host disease (GvHD), and cutaneous rash, and can lead to increased mortality in the context of transplantation (5, 6). While the effects of chronic HHV-6 infection are yet to be fully understood, the virus has been implicated as a possible trigger for autoimmune diseases such as multiple sclerosis (MS) (7) and myocarditis (8).
HHV-6 preferentially replicates in activated CD4+ T lymphocytes and uses specific cell receptors that permit virus anchorage to the cell surface. HHV-6A uses CD46, a regulator of complement activation expressed on all nucleated cells, while CD134 (also called OX40), a member of the tumor necrosis factor (TNF) receptor superfamily present only on activated T lymphocytes, functions as a specific entry receptor for HHV-6B. HHV-6 also has important immunomodulatory properties through the induction of lytic infection of CD4+ and/or cytotoxic effector T cells, the impairment of antigen-presenting cell functions, the production of inflammatory and immunosuppressive cytokines and/or chemokines, and the downmodulation of T-cell receptor expression (9). Despite these properties, immune control of the virus in healthy virus carriers is effective. The mechanisms of immune control as well as its failure in patients with diseases nonetheless remain poorly understood.
Interestingly, there is a high degree of homology (66%) between HHV-6 and human cytomegalovirus (HCMV), another persistent herpesvirus frequently carried by healthy individuals that may reactivate in immunocompromised individuals (10). Adaptive immune responses to herpesviruses are critical in controlling the virus (11). It has already been shown that HCMV responses inflate over time (12, 13). The bulk of the HCMV-specific T-cell response arises from the effector memory (TEM) compartment in CD4+ T cells and, to a similar extent, from the TEM and terminally differentiated effector memory (TEMRA) compartments in CD8+ T cells. This accumulation of late-differentiated memory HCMV-specific T cells in older adults has been associated with impaired immunity (14, 15). Because it skews the T-cell receptor repertoire (15, 16), HCMV infection is a major contributor to the immune risk profile (IRP) reported in older humans (17, 18). Nevertheless, in contrast to that for HCMV, knowledge of HHV-6-specific T-cell responses is limited. This caveat may result from the difficulty of detecting HHV-6-specific T cells ex vivo in healthy adults (19–21). Indeed, most studies have performed analyses after in vitro expansion (22, 23).
Increasing evidence suggests that induction of regulatory T cells (Tregs) is another important immune-escape mechanism used by viruses to establish chronic infections or latency. The existence of HHV-6-specific interleukin-10 (IL-10)-producing CD4+ T cells has been demonstrated in HHV-6-infected individuals that exhibited T regulatory activity in vitro (24). After generation of clones, these HHV-6-specific Tregs suppress the proliferation of naive CD4+ T cells, inhibit the function of HHV-6-specific CD4+ effector T cells, and also block dendritic cell maturation and function (24, 25). However, whether HHV-6 infection can directly induce virus-specific Tregs, thus dampening antiviral immunity, is still unknown. In order to deepen our knowledge of T-cell responses to HHV-6, we characterized HHV-6A- and HHV-6B-specific CD4+ and CD8+ T-cell responses directly ex vivo from healthy blood donors.
RESULTS
Ex vivo HHV-6A- and HHV-6B-specific CD4+ and CD8+ T-cell detection.
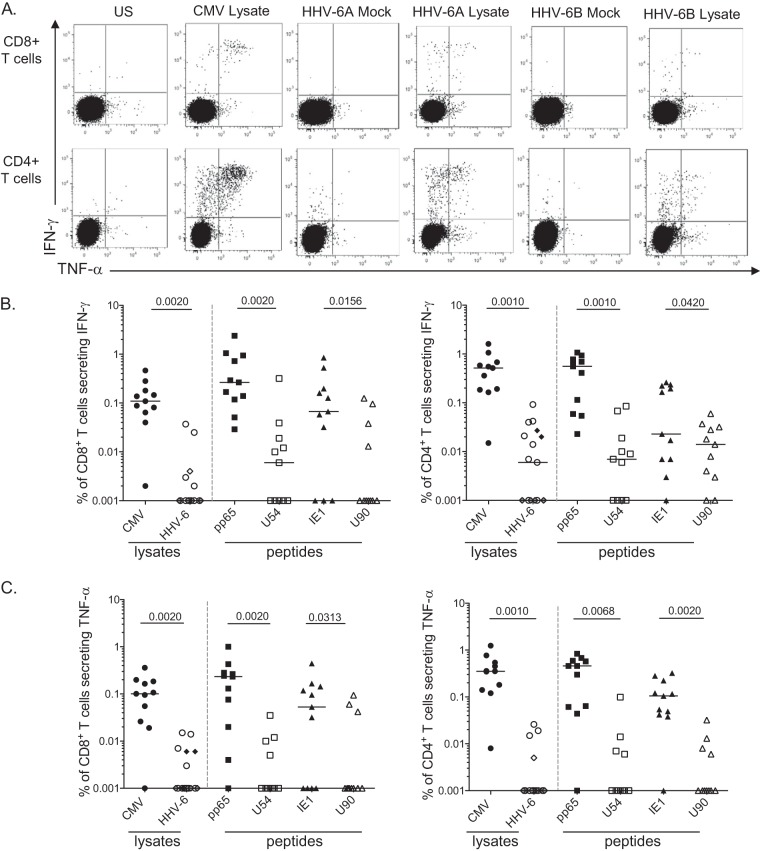
Given the high degree of homology between HHV-6 and HCMV, we set out to determine if T-cell responses could be detected directly ex vivo. We used lysates from HHV-6 (i.e., the HHV-6A SIE strain and the HHV-6B MAR strain) as well as HCMV (i.e., the AD169 strain). We also focused on two HHV-6 antigens from HHV-6, products of U54 and U90, which are positional homologues of the pp65 tegument protein and the nonstructural immediate early protein 1 (IE1) from HCMV, respectively. The frequency of CD4+ and CD8+ T cells (secreting either gamma interferon [IFN-γ] or tumor necrosis factor alpha [TNF-α] in responses to HCMV or HHV-6 antigens) was analyzed after stimulating peripheral blood mononuclear cells (PBMCs) from a panel of 11 dually infected blood donors for adequate comparison. A representative example of the gating strategy employed to analyze HHV-6 antigen-specific CD8+ and CD4+ T cells is shown in Fig. 1A.
FIG 1.
Frequencies of HHV-6-specific CD4+ and CD8+ T cells. (A) Representative flow cytometry dot plots showing the frequencies of HCMV- or HHV-6-specific CD8+ (top) and CD4+ (bottom) IFN-γ- and/or TNF-α-responding T cells after overnight stimulation with the appropriate lysates. Frequencies of IFN-γ-secreting (B) or TNF-α-secreting (C) HCMV- or HHV-6-specific T cells are depicted. Overnight stimulation was performed using either viral lysates (HCMV, •; HHV-6A, ○; HHV-6B, ♦) or overlapping peptides (pp65 and U54 values are denoted by squares, IE1 and U90 values by triangles), and the frequencies of responder T cells are shown for CD8 (left) or CD4 (right). Each symbol represents a donor (coinfected by HHV-6 and HCMV), and horizontal bars indicate the medians. P values were calculated using the Wilcoxon pair test for group comparisons.
Although we detected HHV-6A- or HHV-6B-specific T cells in several patients ex vivo, their frequency was very low compared to that of HCMV-specific cells. CD4+ T-cell responses specific for HHV-6A/B lysates (median, 0.006% IFN-γ and 0.001% TNF-α) were significantly lower than responses to HCMV lysate (0.52% IFN-γ and 0.35% TNF-α; P = 0.001) (Fig. 1B and C). Interestingly, CD4+ T-cell responses specific for HHV-6B were dominated by U90 protein (median, 0.014% CD4+ IFN-γ+ [range, 0% to 0.06%]) over U54 protein (median, 0.007% CD4+ IFN-γ+ [range, 0% to 0.085%]), contrary to what is described for their HCMV positional homologues IE1 and pp65, respectively. However, this immunodominance was not sustained for TNF-α secretion. HHV-6-specific CD8+ T-cell responses were even lower, just above detection limits upon stimulation with lysates (median of detection of 0.001% CD8+ IFN-γ+ or TNF-α) or with HHV-6B overlapping peptides (median of detection of 0.001% CD8+ IFN-γ+ for U90 compared to a median of detection of 0.006% CD8+ IFN-γ+ for U54). Altogether, these results confirm that HHV-6-specific T-cell responses are scarce but functionally detectable ex vivo (when based on IFN-γ or TNF-α secretion as readouts) (Fig. 1).
Ex vivo comparison between HCMV- and HHV-6-specific T cell phenotypes.
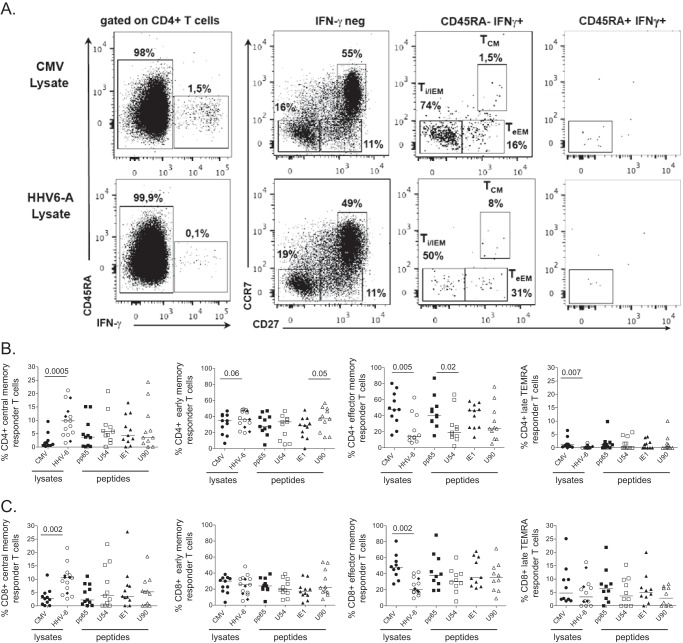
In healthy carriers, the persistence of HCMV may have a major impact on the host through modulation of innate and adaptive immune responses. In particular, this can lead to substantial expansion of HCMV-specific CD8+ memory T cells over time, a process described as memory inflation (26). These HCMV-specific T cells are typically defined as effector memory (TEM) CD27− CD28− CD45RA− or TEMRA CD27− CD28− CD45RA+ cells. We therefore decided to analyze the ex vivo phenotype of HHV-6-specific CD4+ and CD8+ T cells ex vivo, in comparison to that of HCMV-specific T cells. PBMCs from 18 HHV-6-seropositive blood donors (including the 11 HCMV coinfected) were stimulated overnight with the different lysates or with single-tube 15-mer PepMix pools for each HHV-6B or HCMV antigen. The phenotype of the responding CD4+ and CD8+ T cells identified by IFN-γ secretion was studied. Using CCR7 and CD27 within the CD45RA− cells, we distinguished the less differentiated central memory (TCM) and early effector memory (TeEM) cells and the more differentiated intermediate/late effector memory (Ti/lEM) T cells. CCR7− CD27− TEMRA T cells were identified from CD45RA+ IFN-γ+ subsets (Fig. 2A).
FIG 2.
Phenotypes of HCMV- and HHV-6-specific CD4+ and CD8+ T cells. (A) Representative flow cytometry dot plots depicting the gating strategy used to phenotype nonresponder (IFN-γ neg) and responder (CD45RA− IFN-γ+ or CD45RA+ IFN-γ+) T cells. Gating strategy identifies central memory (TCM), early effector memory (TeEM), and intermediate/late effector memory (Ti/lEM) and TEMRA T cells (left), using CCR7 and CD27 within the nonresponder and responder T cells. Differentiation phenotypes of HCMV- or HHV-6-specific CD4+ (B) and CD8+ (C) T cells from coinfected healthy blood donors (HHV-6+ HCMV+) were analyzed by stimulation with either HCMV (HCMV lysate [•], overlapping peptides covering pp65 [■], or IE1 [▲]) or HHV-6 antigens (lysates for HHV-6A [○] or HHV-6B [♦], overlapping peptides U54 [□] or U90 [△]). Each symbol represents a donor, and horizontal bars indicate the medians. P values were calculated using the Wilcoxon pair test for group comparisons.
We observed that the phenotype of the responder T cells differed according to the virus considered. The frequency of the HHV-6-specific CD4+ TCM and TeEM cells was significantly higher than that of HCMV-specific cells after stimulation with the specific lysates (P = 0.0005 and P = 0.06, respectively) (Fig. 2B). Inversely, the frequencies of the HHV-6-specific CD4+ Ti/lEM and TEMRA cells were significantly lower than those of HCMV-specific cells after stimulation with their respective lysates (P = 0.005 and P = 0.007, respectively) and after stimulation with the peptides U54 and U90 (P = 0.02 and P = 0.1, respectively, for CD4+ Ti/lEM). HHV-6-specific CD8+ T cells also exhibited a higher frequency of TCM cells and a lower frequency of Ti/lEM cells (P = 0.006 and P = 0.002, respectively) (Fig. 2C). Overall, these data indicate that HHV-6A- and HHV-6B-specific T lymphocytes are less differentiated than HCMV-specific T cells.
Frequency of virus-specific regulatory T cells.
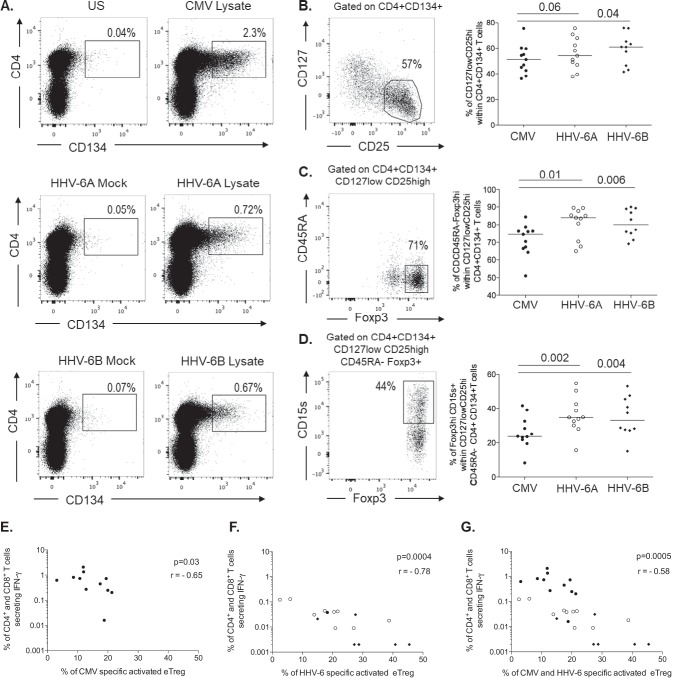
Since we did not observe a high frequency of HHV-6-responding CD4+ or CD8+ T cells from infected blood donors, we speculated whether reduced responses could be due to negative immune regulation. It has been described that HHV-6 infection can directly induce virus-specific Tregs in vitro (25). We therefore decided to address the frequency and phenotype of induced HHV-6-specific Tregs directly ex vivo. Antigen-specific memory CD4+ T cells can be defined using dual expression of CD134 (OX40) and CD25 after 45 h of stimulation with antigen (27, 28). In combination with classical markers such as CD25, CD127, Foxp3, and CD45RA, we used CD15s (sialyl Lewis x), recently described to be specifically expressed by suppression-competent Foxp3+ suppressive effector Tregs (eTregs) but not by nonsuppressive cytokine-secreting Foxp3+ non-Tregs (29).
PBMCs from the 11 dually infected donors were stimulated for 45 h with the appropriate lysates, and the phenotype of the Tregs was analyzed according to the gating strategy shown in Fig. 3A to D. This allowed us to define antivirus-specific Tregs (CD4+ CD134+) based on the expression of either CD127low CD25high alone or combined with CD45RA− Foxp3high or CD15s+. Our data demonstrate that the median frequency of CD134+ CD4+ T cells was higher upon stimulation with the HCMV lysate (1.8%) than with the HHV-6 lysates (0.67% for HHV-6A and 0.62% for HHV-6B). Interestingly, among these cells, a higher frequency of HHV-6-specific cells appeared to be Tregs than of HCMV-specific cells (as exemplified in Fig. 3B and C). In combination with CD127, CD25, CD45RA, and Foxp3, we focused on the expression of CD15s, which has been described to define suppressive effector Tregs (eTregs). Like for the other Treg subsets (CD127low CD25high or CD45RA− Foxp3high) (Fig. 3B and C, respectively), the frequency of HHV-6-specific eTregs was higher (median, 34.8% of HHV-6A-specific eTregs and 33.1% of HHV-6B-specific eTregs) than that of HCMV-specific cells (23.7% of HCMV specific to eTregs) (Fig. 3D). No statistical difference was observed between the frequencies of HHV-6A-specific eTregs and HHV-6B-specific eTregs. Overall, the frequency of HHV-6A- and HHV-6B-specific suppressive eTregs was significantly higher than HCMV-specific eTregs (P = 0.002 and P = 0.004, respectively) (Fig. 3D). Of note, there was a negative correlation between the frequency of specific eTregs and the frequency of specific effector T-cell responses both for HCMV (Fig. 3E) (r = −0.65, P = 0.03) and for HHV-6 (Fig. 3F) (r = −0.78, P = 0.0004), suggesting that virus-specific eTregs may limit specific conventional effector/memory T-cell immune responses through their potent suppressive activity (Fig. 3G) (r = −0.58; P = 0.0005).
FIG 3.
Induced specific Treg phenotypes. (A) Representative flow cytometry dot plots showing the frequencies of activated CD134+ CD4+ T cells observed poststimulation with either HCMV lysate or with HHV-6A/HHV-6B lysates and their respective negative controls (unstimulated or mock lysates). (B to D) Gating strategy used to identify the frequencies of Treg subsets defined with the expression of CD127low CD25high (B), CD45RA− Foxp3high (C), and CD15s+ Foxp3high (D) effector Tregs (eTregs). Their respective frequencies are summed up for HCMV (•) or HHV-6-induced (HHV-6A, ○; HHV-6B, ♦) specific Tregs. Each symbol represents an individual donor, and horizontal bars indicate the medians. (E to G) P values were calculated using the Wilcoxon pair test for group comparisons. Correlations between the frequency of IFN-γ-secreting effector T cells and the frequency of induced eTregs specific for either HCMV (•) (E), HHV-6 (HHV-6A, ○; HHV-6B, ♦) (F), or pooled specificities (HCMV and HHV-6) (G). The correlation coefficients were derived from Spearman analyses.
DISCUSSION
HHV-6 exhibits wide cell tropism in vivo and, like other herpesviruses, induces a lifelong latent infection in humans (2). It is assumed that herpesviruses are permanently under the control of virus-specific T cells, and viral reactivations occur due to therapy-related immunosuppression. In agreement with this concept, adoptive transfer of virus-specific T cells prevents and eliminates Epstein-Barr virus (EBV)-, HCMV-, and even HHV-6-associated diseases after transplantation (30, 31).
However, in contrast to those of other human herpesviruses, HHV-6-specific T cells have been difficult to characterize so far. Indeed, HHV-6-specific T-cell responses were reported to be absent or of really low frequency in peripheral blood (20, 21, 23, 32, 33), particularly in regard to CD8+ T cells (34). It has also been reported that HHV-6A impacts T cell subsets differentially; i.e., in response to HHV-6A stimulation, naive or central memory cells were more prone to apoptosis, whereas proliferation was increased for the late memory compartment (35).
Our work here focused on the characterization of T-cell responses specific for selected HHV-6 proteins: the viral protein U54 and the immediate early transactivator U90. The reason for choosing these antigens was based on their correspondence to immunogenic proteins of human HCMV. However, despite the existence of certain commonalities between recognition patterns of HCMV and HHV-6 antigens, a recent publication showed that the overall composition and diversity of the HHV-6-specific CD8+ T-cell repertoire stands in marked contrast to that of HCMV (34). Therefore, we also characterized global antiviral responses through lysate stimulation and found HHV-6-specific responses directly detectable ex vivo, both for CD8+ and for CD4+ T cells. Contrary to HCMV, HHV-6 elicits very low T-cell responses, suggesting that our knowledge of anti-HCMV immunity is limited in predicting responses to HHV-6 (data not shown). Interestingly, we found that HHV-6 and HCMV, despite their homology, induced different types of T-cell responses in terms of phenotype, HHV-6-specific T cells being in general less differentiated than their HCMV counterparts. This indicates that although HHV-6 and HCMV may be closely related viruses, virus-host equilibrium is different for and specific to each infection.
Our work emphasizes, in fact, that HHV-6 infection induces a high frequency of virus-specific Tregs compared to HCMV-specific T-cell responses. These Tregs can have a broad immunosuppressive capacity and inhibit innate and adaptive immune-cell functions, which may be crucial for HHV-6-mediated latency and survival (24, 25, 36). In line with this possibility, we found that the frequency of HHV-6-specific regulatory T cells was inversely correlated with the frequency of conventional effector/memory T cells. We also found that stimulation with HHV-6 results in a higher level of IL-10 production and a lower level of TNF-α production than stimulation with HCMV (see Fig. S1 in the supplemental material). This suggests that the induction of Tregs over limited effector T cells by HHV-6 may explain the emergence of HHV-6-related pathologies. It seems, therefore, relevant to monitor both subsets in immunocompromised settings (transplantation or autoimmune disorders).
A key difference of HHV-6 from other human herpesviruses is that HHV-6 genomic DNA can be integrated into the subtelomeric region of chromosomes in every nucleated cell of the body, including germ cells, resulting in a condition known as inherited chromosomally integrated HHV-6 (iciHHV-6) that is present in approximately 1% of the general population and is vertically transmitted. Despite the detection of specific immune responses to HHV-6 in this particular condition (37), more detailed investigation needs to be conducted, specifically on the induction of IL-10 in infected cells, which may lead to a severe clinical outcome in the case of reactivation (38).
MATERIALS AND METHODS
Blood donors and serology.
Venous blood was obtained from 18 healthy adult volunteers (11 dually infected with HHV-6 and HCMV), ranging in age from 18 to 55 years (median age, 33 years), from EFS (Etablissement Français du Sang) of the Pitié-Salpêtrière Hospital in Paris. PBMCs were isolated by Ficoll density gradient centrifugation Eurobio AbCys (Les Ulis, France). Diagnosis of HCMV was performed on plasma samples by enzyme-linked immunosorbent assay (ELISA), using an Euroimmun anti-CMV kit (Bussy-Saint-Martin, France) according to the manufacturer’s instructions. Serological status for HHV-6 was determined using immunofluorescence assays (IFAs) to detect HHV-6 IgG (Biotrin International, Dublin, Ireland).
Virus stocks and peptides.
The HHV-6A SIE strain and the HHV-6B MAR strain were previously isolated and propagated in PBMCs from healthy blood donors in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% heat-inactivated fetal calf serum in the presence of 20 mg/ml amikacin and vancomycin and 29 ± 2 mg/ml l-glutamine. Amplification of HHV-6A (SIE) and HHV-6B (MAR) was performed on preactivated PBMCs until 50% to 70% of 500 to 700 million cells were infected. After 1 to 3 weeks of incubation at 37°C in the presence of 5% CO2, infectious particles were recovered by freeze-thaw-centrifugation cycles (39, 40). Clarified viral stocks were concentrated and purified by ultracentrifugation through a solution of 10% iohexol coupled with a 70% cushion (Nycodenz). The presence of infectious viral particles was determined by titration expressed as 50% tissue culture infective dose (TCID50/ml) calculated with the Spearman-Karber formula. Proteins were quantified using the Bradford method and used at 10 μg/ml.
The HCMV lysate strain AD169 from Tebu-Bio (Le Perray-en-Yvelines, France) was aliquoted at 1 mg/ml and used at 1/100 dilution. Single-tube PepMix pools containing 15-mer peptides (overlapping by 11 amino acids) spanning the whole protein for HHV-6B antigens U54 and U90 were obtained from JPT Technologies (Berlin, Germany). Fifteen-amino-acid-long synthetic peptides overlapping by 10 amino acids and spanning both HCMV proteins, pp65 and IE1, were provided by GeneCust (Ellange, Luxembourg). Our lyophilized peptides were reconstituted in phosphate-buffered saline (PBS) containing 0.2% dimethyl sulfoxide (DMSO), as suggested by the manufacturer. Stock concentrations were diluted in PBS for performing experiments at a final concentration of 1 μM. To establish the optimal peptide dose used for stimulation, we performed titration experiments comparing T-cell responses obtained with 3 doses of overlapping peptides (titer A, 1 μM; titer B, 3.3 μM; titer C, 10 μM). Similar ranges of responses, secretion of either IFN-γ or TNF-α, were observed for both CD4 and CD8 (see Fig. S2). Consequently, we used a concentration of 1 μM throughout the sets of experiments.
Detection and phenotypic analyses of antigen-specific T cells.
Directly conjugated antibodies were obtained from the following vendors: CD45RA (V450), CD4 (HV500), CD27 (phycoerythrin [PE]), CCR7 (PE-CF594), TNF-α (PE-Cy7), IFN-γ (AF700), CD8 (allophycocyanin [APC]-Cy7), CD15s (BV510), CD134 (PE), and CD4 (APC-Cy7) from BD Biosciences (San Jose, CA); CD45RA (ECD) from Beckman Coulter (Pasadena, CA); CD3 (BV650), CD25 (BV785), and FOXP3-AF647 from BioLegend (San Diego, CA); CD127 (eF450) from eBioscience (San Diego, CA); and IL-2 (APC) from Miltenyi Biotec (Bergisch Gladbach, Germany). Staining for cell surface markers was performed with a standard method as previously described (12). Cells were analyzed on a Fortessa flow cytometer (Becton, Dickinson). Data were analyzed using FlowJo 9.3.2 (Tree Star, Inc.) and FunkyCells (FunkyCells, Paris, France) software (41). To assess the phenotype and functional capacity of HCMV-specific or HHV-6-specific CD8+ and CD4+ T cells, PBMCs were stimulated with the synthetic peptides spanning either the whole of HCMV proteins pp65 and IE1 (1 μM) or the whole of HHV-6 proteins U54 and U90 (1 μM). After 1 h, the secretion inhibitors brefeldin A (5 μg/ml) and monensin (2.5 μg/ml) (Sigma-Aldrich, St. Louis, MO) were added, and incubation was pursued overnight at 37°C in 5% CO2 atm in RPMI medium supplemented with 10% fetal calf serum (FCS; Thermo Fisher Scientific). Cells were thereafter stained for cell surface markers, including CCR7, CD45RA, CD4, CD27, and CD8, and Cytofix/Cytoperm (BD Biosciences) was used to fix/permeabilize the cells prior to staining for intracellular CD3, IFN-γ, and TNF-α. To assess the phenotype and functional capacity of HCMV-specific or HHV-6-specific CD4+ regulatory T cells, PBMCs were stimulated with synthetic peptides spanning either the whole of HCMV proteins pp65 and IE1 or the whole of HHV-6 proteins U54 and U90 and incubated 40 h at 37°C in 5% CO2 atm in IMDM medium (Life Technologies) with 10% FCS. After the addition of brefeldin A, incubation was pursued for 5 h at 37°C in 5% CO2 atm. Cells were thereafter stained for cell surface markers, including CD127, CD15s, CD25, CD134, CD45RA, and CD4. Cytofix/Cytoperm (BD Biosciences) was used to fix/permeabilize the cells prior to staining for intracellular CD3, Foxp3, IFN-γ, and TNF-α. Functional measures were corrected by subtracting background signals obtained by analyzing the same samples incubated in medium alone or under mock conditions (software from FunkyCells, Paris, France) (41). Data in Table S1 summarize background levels for individual responses, i.e., mock stimulation for HHV-6 lysates and unstimulated condition (medium alone) serving as negative controls for peptide stimulation.
Statistical analysis.
Univariate statistical analysis was performed using GraphPad Prism software. Groups were compared using the nonparametric Mann-Whitney test. The Wilcoxon paired test was used to compare samples of the different viruses from the same dually infected healthy donor. P values of <0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank the healthy volunteers that participated in this study. We also thank Angélique Godet for providing virological tools and Henri Agut for constructive scientific discussions.
S.F. performed experiments, analyzed the data, and wrote the manuscript; C.B. performed experiments; M.L. analyzed the data; P.M. performed HHV-6 serologies; P.B. performed experiments and provided virological tools; N.B. designed research; A.G.-D. performed experiments and provided virological tools; V.A. designed research and wrote the manuscript; D.S. designed research, performed experiments, analyzed the data, and wrote the manuscript.
We declare no financial or commercial conflict of interest.
M.L. and D.S. are inventors of the Polyfunctionality Index (patent number WO2013127904). M.L. is proprietary owner of the Funky Cells ToolBox software.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02321-18.
REFERENCES
- 1.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agut H, Bonnafous P, Gautheret-Dejean A. 2017. Update on infections with human herpesviruses 6A, 6B, and 7. Med Mal Infect 47:83–91. doi: 10.1016/j.medmal.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 3.De Bolle L, Naesens L, De Clercq E. 2005. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev 18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol 73:8040–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Pagter PJ, Schuurman R, Meijer E, van Baarle D, Sanders EA, Boelens JJ. 2008. Human herpesvirus type 6 reactivation after haematopoietic stem cell transplantation. J Clin Virol 43:361–366. doi: 10.1016/j.jcv.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh M, Yoshizawa S, Katagiri S, Suguro T, Asano M, Kitahara T, Akahane D, Okabe S, Tauchi T, Ito Y, Ohyashiki K. 2014. Human herpesvirus 6 reactivation on the 30th day after allogeneic hematopoietic stem cell transplantation can predict grade 2-4 acute graft-versus-host disease. Transpl Infect Dis 16:440–449. doi: 10.1111/tid.12229. [DOI] [PubMed] [Google Scholar]
- 7.Leibovitch EC, Jacobson S. 2014. Evidence linking HHV-6 with multiple sclerosis: an update. Curr Opin Virol 9:127–133. doi: 10.1016/j.coviro.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leveque N, Boulagnon C, Brasselet C, Lesaffre F, Boutolleau D, Metz D, Fornes P, Andreoletti L. 2011. A fatal case of human herpesvirus 6 chronic myocarditis in an immunocompetent adult. J Clin Virol 52:142–145. doi: 10.1016/j.jcv.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 9.Dagna L, Pritchett JC, Lusso P. 2013. Immunomodulation and immunosuppression by human herpesvirus 6A and 6B. Future Virol 8:273–287. doi: 10.2217/fvl.13.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence GL, Chee M, Craxton MA, Gompels UA, Honess RW, Barrell BG. 1990. Human herpesvirus 6 is closely related to human cytomegalovirus. J Virol 64:287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moss P, Rickinson A. 2005. Cellular immunotherapy for viral infection after HSC transplantation. Nat Rev Immunol 5:9–20. doi: 10.1038/nri1526. [DOI] [PubMed] [Google Scholar]
- 12.Appay V, Fastenackels S, Katlama C, Ait-Mohand H, Schneider L, Guihot A, Keller M, Grubeck-Loebenstein B, Simon A, Lambotte O, Hunt PW, Deeks SG, Costagliola D, Autran B, Sauce D. 2011. Old age and anti-cytomegalovirus immunity are associated with altered T-cell reconstitution in HIV-1-infected patients. AIDS 25:1813–1822. doi: 10.1097/QAD.0b013e32834640e6. [DOI] [PubMed] [Google Scholar]
- 13.Sauce D, Larsen M, Fastenackels S, Duperrier A, Keller M, Grubeck-Loebenstein B, Ferrand C, Debre P, Sidi D, Appay V. 2009. Evidence of premature immune aging in patients thymectomized during early childhood. J Clin Invest 119:3070–3078. doi: 10.1172/JCI39269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher JM, Vukmanovic-Stejic M, Dunne PJ, Birch KE, Cook JE, Jackson SE, Salmon M, Rustin MH, Akbar AN. 2005. Cytomegalovirus-specific CD4+ T cells in healthy carriers are continuously driven to replicative exhaustion. J Immunol 175:8218–8225. doi: 10.4049/jimmunol.175.12.8218. [DOI] [PubMed] [Google Scholar]
- 15.Hadrup SR, Strindhall J, Kollgaard T, Seremet T, Johansson B, Pawelec G, Thor Straten P, Wikby A. 2006. Longitudinal studies of clonally expanded CD8 T cells reveal a repertoire shrinkage predicting mortality and an increased number of dysfunctional cytomegalovirus-specific T cells in the very elderly. J Immunol 176:2645–2653. doi: 10.4049/jimmunol.176.4.2645. [DOI] [PubMed] [Google Scholar]
- 16.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PA. 2002. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol 169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 17.Olsson J, Wikby A, Johansson B, Lofgren S, Nilsson BO, Ferguson FG. 2001. Age-related change in peripheral blood T-lymphocyte subpopulations and cytomegalovirus infection in the very old: the Swedish longitudinal OCTO immune study. Mech Ageing Dev 121:187–201. doi: 10.1016/S0047-6374(00)00210-4. [DOI] [PubMed] [Google Scholar]
- 18.Pawelec G, Akbar A, Caruso C, Effros R, Grubeck-Loebenstein B, Wikby A. 2004. Is immunosenescence infectious? Trends Immunol 25:406–410. doi: 10.1016/j.it.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Becerra-Artiles A, Dominguez-Amorocho O, Stern LJ, Calvo-Calle JM. 2015. A simple proteomics-based approach to identification of immunodominant antigens from a complex pathogen: application to the CD4 T cell response against human herpesvirus 6B. PLoS One 10:e0142871. doi: 10.1371/journal.pone.0142871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halawi M, Khan N, Blake N. 2015. Identification of novel CD8+ T cell epitopes in human herpesvirus 6B U11 and U90. Immun Inflamm Dis 3:118–131. doi: 10.1002/iid3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iampietro M, Morissette G, Gravel A, Dubuc I, Rousseau M, Hasan A, O'Reilly RJ, Flamand L. 2014. Human herpesvirus 6B immediate-early I protein contains functional HLA-A*02, HLA-A*03, and HLA-B*07 class I restricted CD8+ T-cell epitopes. Eur J Immunol 44:3573–3584. doi: 10.1002/eji.201444931. [DOI] [PubMed] [Google Scholar]
- 22.Becerra A, Gibson L, Stern LJ, Calvo-Calle JM. 2014. Immune response to HHV-6 and implications for immunotherapy. Curr Opin Virol 9:154–161. doi: 10.1016/j.coviro.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin LK, Schub A, Dillinger S, Moosmann A. 2012. Specific CD8+ T cells recognize human herpesvirus 6B. Eur J Immunol 42:2901–2912. doi: 10.1002/eji.201242439. [DOI] [PubMed] [Google Scholar]
- 24.Wang F, Yao K, Yin QZ, Zhou F, Ding CL, Peng GY, Xu J, Chen Y, Feng DJ, Ma CL, Xu WR. 2006. Human herpesvirus-6-specific interleukin 10-producing CD4+ T cells suppress the CD4+ T-cell response in infected individuals. Microbiol Immunol 50:787–803. doi: 10.1111/j.1348-0421.2006.tb03855.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang F, Chi J, Peng G, Zhou F, Wang J, Li L, Feng D, Xie F, Gu B, Qin J, Chen Y, Yao K. 2014. Development of virus-specific CD4+ and CD8+ regulatory T cells induced by human herpesvirus 6 infection. J Virol 88:1011–1024. doi: 10.1128/JVI.02586-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klenerman P, Oxenius A. 2016. T cell responses to cytomegalovirus. Nat Rev Immunol 16:367–377. doi: 10.1038/nri.2016.38. [DOI] [PubMed] [Google Scholar]
- 27.Seddiki N, Cook L, Hsu DC, Phetsouphanh C, Brown K, Xu Y, Kerr SJ, Cooper DA, Munier CM, Pett S, Ananworanich J, Zaunders J, Kelleher AD. 2014. Human antigen-specific CD4+CD25+CD134+CD39+ T cells are enriched for regulatory T cells and comprise a substantial proportion of recall responses. Eur J Immunol 44:1644–1661. doi: 10.1002/eji.201344102. [DOI] [PubMed] [Google Scholar]
- 28.Zaunders JJ, Munier ML, Seddiki N, Pett S, Ip S, Bailey M, Xu Y, Brown K, Dyer WB, Kim M, de Rose R, Kent SJ, Jiang L, Breit SN, Emery S, Cunningham AL, Cooper DA, Kelleher AD. 2009. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40). J Immunol 183:2827–2836. doi: 10.4049/jimmunol.0803548. [DOI] [PubMed] [Google Scholar]
- 29.Miyara M, Chader D, Sage E, Sugiyama D, Nishikawa H, Bouvry D, Claer L, Hingorani R, Balderas R, Rohrer J, Warner N, Chapelier A, Valeyre D, Kannagi R, Sakaguchi S, Amoura Z, Gorochov G. 2015. Sialyl Lewis x (CD15s) identifies highly differentiated and most suppressive FOXP3high regulatory T cells in humans. Proc Natl Acad Sci U S A 112:7225–7230. doi: 10.1073/pnas.1508224112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naik S, Nicholas SK, Martinez CA, Leen AM, Hanley PJ, Gottschalk SM, Rooney CM, Hanson IC, Krance RA, Shpall EJ, Cruz CR, Amrolia P, Lucchini G, Bunin N, Heimall J, Klein OR, Gennery AR, Slatter MA, Vickers MA, Orange JS, Heslop HE, Bollard CM, Keller MD. 2016. Adoptive immunotherapy for primary immunodeficiency disorders with virus-specific T lymphocytes. J Allergy Clin Immunol 137:1498.e1–1505.e1. doi: 10.1016/j.jaci.2015.12.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papadopoulou A, Gerdemann U, Katari UL, Tzannou I, Liu H, Martinez C, Leung K, Carrum G, Gee AP, Vera JF, Krance RA, Brenner MK, Rooney CM, Heslop HE, Leen AM. 2014. Activity of broad-spectrum T cells as treatment for AdV, EBV, CMV, BKV, and HHV6 infections after HSCT. Sci Transl Med 6:242ra83. doi: 10.1126/scitranslmed.3008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerdemann U, Keukens L, Keirnan JM, Katari UL, Nguyen CT, de Pagter AP, Ramos CA, Kennedy-Nasser A, Gottschalk SM, Heslop HE, Brenner MK, Rooney CM, Leen AM. 2013. Immunotherapeutic strategies to prevent and treat human herpesvirus 6 reactivation after allogeneic stem cell transplantation. Blood 121:207–218. doi: 10.1182/blood-2012-05-430413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nastke MD, Becerra A, Yin L, Dominguez-Amorocho O, Gibson L, Stern LJ, Calvo-Calle JM. 2012. Human CD4+ T cell response to human herpesvirus 6. J Virol 86:4776–4792. doi: 10.1128/JVI.06573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin LK, Hollaus A, Stahuber A, Hubener C, Fraccaroli A, Tischer J, Schub A, Moosmann A. 2018. Cross-sectional analysis of CD8 T cell immunity to human herpesvirus 6B. PLoS Pathog 14:e1006991. doi: 10.1371/journal.ppat.1006991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta S, Agrawal S, Gollapudi S. 2009. Differential effect of human herpesvirus 6A on cell division and apoptosis among naive and central and effector memory CD4+ and CD8+ T-cell subsets. J Virol 83:5442–5450. doi: 10.1128/JVI.00106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustafsson RK, Engdahl EE, Hammarfjord O, Adikari SB, Lourda M, Klingstrom J, Svensson M, Fogdell-Hahn A. 2013. Human herpesvirus 6A partially suppresses functional properties of DC without viral replication. PLoS One 8:e58122. doi: 10.1371/journal.pone.0058122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strenger V, Kayser S, Witte KE, Lassner D, Schwinger W, Jahn G, Urban C, Feuchtinger T. 2016. Individuals with inherited chromosomally integrated human herpes virus 6 (ciHHV-6) have functionally active HHV-6 specific T-cell immunity. Clin Microbiol Infect 22:209.e5–209.e8. doi: 10.1016/j.cmi.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Bonnafous P, Marlet J, Bouvet D, Salame E, Tellier AC, Guyetant S, Goudeau A, Agut H, Gautheret-Dejean A, Gaudy-Graffin C. 2018. Fatal outcome after reactivation of inherited chromosomally integrated HHV-6A (iciHHV-6A) transmitted through liver transplantation. Am J Transplant 18:1548–1551. doi: 10.1111/ajt.14657. [DOI] [PubMed] [Google Scholar]
- 39.Agut H, Guetard D, Collandre H, Dauguet C, Montagnier L, Miclea JM, Baurmann H, Gessain A. 1988. Concomitant infection by human herpesvirus 6, HTLV-I, and HIV-2. Lancet 1:712. [DOI] [PubMed] [Google Scholar]
- 40.Aubin JT, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux JM, Agut H. 1991. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol 29:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen M, Sauce D, Arnaud L, Fastenackels S, Appay V, Gorochov G. 2012. Evaluating cellular polyfunctionality with a novel polyfunctionality index. PLoS One 7:e42403. doi: 10.1371/journal.pone.0042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.