Ticks and mosquitoes, as well Culicoides, can transmit viruses in the Reoviridae family. With the help of next-generation sequencing (NGS), previously unreported reoviruses such as equine encephalosis virus, Wad Medani virus (WMV), Kammavanpettai virus (KVPTV), and, with this report, KARYV and KUNDV have been discovered and characterized in India. The isolation of KUNDV and KARYV from Hyalomma anatolicum, which is a known vector for zoonotic pathogens, such as Crimean Congo hemorrhagic fever virus, Babesia, Theileria, and Anaplasma species, identifies arboviruses with the potential to transmit to humans. Characterization of KUNDV and KARYV isolated from Hyalomma ticks is critical for the development of specific serological and molecular assays that can be used to determine the association of these viruses with disease in humans and livestock.
KEYWORDS: Coltivirus, Hyalomma tick, India, reovirus, VP1 gene, electron microscopy, next-generation sequencing, phylogenetic tree, tick-borne virus
ABSTRACT
In 2011, ticks were collected from livestock following an outbreak of Crimean Congo hemorrhagic fever (CCHF) in Gujarat state, India. CCHF-negative Hyalomma anatolicum tick pools were passaged for virus isolation, and two virus isolates were obtained, designated Karyana virus (KARYV) and Kundal virus (KUNDV), respectively. Traditional reverse transcription-PCR (RT-PCR) identification of known viruses was unsuccessful, but a next-generation sequencing (NGS) approach identified KARYV and KUNDV as viruses in the Reoviridae family, Orbivirus and Coltivirus genera, respectively. Viral genomes were de novo assembled, yielding 10 complete segments of KARYV and 12 nearly complete segments of KUNDV. The VP1 gene of KARYV shared a most recent common ancestor with Wad Medani virus (WMV), strain Ar495, and based on nucleotide identity we demonstrate that it is a novel WMV strain. The VP1 segment of KUNDV shares a common ancestor with Colorado tick fever virus, Eyach virus, Tai Forest reovirus, and Tarumizu tick virus from the Coltivirus genus. Based on VP1, VP6, VP7, and VP12 nucleotide and amino acid identities, KUNDV is proposed to be a new species of Coltivirus. Electron microscopy supported the classification of KARYV and KUNDV as reoviruses and identified replication morphology consistent with other orbi- and coltiviruses. The identification of novel tick-borne viruses carried by the CCHF vector is an important step in the characterization of their potential role in human and animal pathogenesis.
IMPORTANCE Ticks and mosquitoes, as well Culicoides, can transmit viruses in the Reoviridae family. With the help of next-generation sequencing (NGS), previously unreported reoviruses such as equine encephalosis virus, Wad Medani virus (WMV), Kammavanpettai virus (KVPTV), and, with this report, KARYV and KUNDV have been discovered and characterized in India. The isolation of KUNDV and KARYV from Hyalomma anatolicum, which is a known vector for zoonotic pathogens, such as Crimean Congo hemorrhagic fever virus, Babesia, Theileria, and Anaplasma species, identifies arboviruses with the potential to transmit to humans. Characterization of KUNDV and KARYV isolated from Hyalomma ticks is critical for the development of specific serological and molecular assays that can be used to determine the association of these viruses with disease in humans and livestock.
INTRODUCTION
The emergence of novel diseases usually occurs following exposure to a vector species, zoonotic transmission and disease outbreaks in humans and livestock. Ticks and other arthropods play an important role in zoonotic disease transmission; specifically, tick-borne viral diseases are caused by more than 38 viral species which belong to 6 different viral families. The Indian subcontinent is endemic to a variety of tick populations, and thus, the human population in India is continually exposed to potential arboviral vectors. Tick-borne viruses like Kyasanur Forest disease virus (KFDV) and Crimean Congo hemorrhagic fever virus (CCHFV) cause outbreaks and affect human and animal populations in India (1). However, many tick-borne viruses pose a serious threat to public health worldwide (2, 3). Novel arboviruses are routinely discovered worldwide (4–7), but the true burden of tick-borne viruses is currently unknown and may be underestimated. While the presence of KFDV- and CCHFV-positive tick reservoirs is known in India, little is known about reovirus reservoirs and tick-borne transmission of reoviruses in India.
Reoviruses are double-stranded RNA viruses that are transmitted between hosts through various routes, including respiratory, fecal-oral, and direct contact, as well as through the bite of arthropod vectors. Reoviridae is divided into two subfamilies, viz., Spinareovirinae, consisting of nine genera, and Sedoreovirinae, containing six genera. The genus Coltivirus belongs to subfamily Spinareovirinae and virions contain 12 double-stranded RNA genomic segments encoding 13 putative proteins. Two tick-borne virus species, namely, Colorado tick fever virus (CTFV) and Eyach virus (EYAV), are placed in the Coltivirus genus (8). CTFV causes Colorado tick fever, which occurs in the Rocky Mountains in the Western United States and Canada and was first isolated from a febrile patient in America in 1946 (6–11). It usually causes mild febrile illness; however, severe symptoms of hemorrhagic fever, pericarditis, and myocarditis have been rarely observed, mainly in children (11). Person-to-person and mother-to-child transmissions are also reported from CTFV infections (12, 13). California hare coltivirus (CTFV-Ca) and Salmon River virus (SRV), different serotypes of CTFV, are also included in this genus. EYAV was first isolated from an Ixodes tick in Germany in 1976 and subsequently from France in 1981. It is another tick-borne virus known to cause neurological disease in humans (14). Serological survey of EYAV has revealed its presence in European rabbits, mice, mountain goats, domestic goats, sheep, and deer (15). In late 2017, two novel viruses belonging to the genus Coltivirus were discovered and characterized, Tarumizu tick virus (TarTV) and Tai Forest reovirus (TFRV), from Japan and Côte d’Ivoire, respectively (16). TarTV was isolated from ticks, and TFRV was isolated from the blood of African free-tailed bats (Chaereophon aloysiisabaudiae) in 2017 (17). However, no human pathogenicity has been associated with these viruses. TFRV is, interestingly, the only Coltivirus reported from bats and not from ticks.
The Orbivirus genus belongs to the Sedoreovirinae subfamily, and its virions contain 10 double-stranded RNA genomic segments encoding 11 putative proteins. Orbiviruses are mainly transmitted by ticks, mosquitoes, and Culicoides midges and can cause infection in arthropods and mammals (18). Outer core protein VP7 (T13) is the main serogroup-specific (species-specific) antigen; however, other viral proteins, such as core proteins NS1, NS2, and VP3, are also conserved between species and can be used to define phylogenetic relationships (19) further. Using these criteria, the orbiviruses can be divided into four serogroups (A to D); group B contains Chenuda virus (CNUV), Ieri virus (IERIV), Wad Medani virus (WMV), and Great Island virus (GIV). Since orbiviruses are transmitted by hematophagous arthropods, they can be further classified by the vector; the tick-borne orbivirus species consist of Chenuda virus, Chobar Gorge virus, WMV, and Great Island virus (20).
In 2011, when animal CCHF was first reported in Gujarat (21), a high priority was declared consisting of increased reporting, surveillance, and tick collection. In 2013, a larger outbreak of CCHF occurred in several Gujarat state districts, and a tick survey was initiated by the National Vector Borne Disease Control Programme (NVBDCP) to evaluate the prevalence of CCHFV in the local tick population. Pools of hard tick samples from buffalo and cattle were collected from the CCHFV-affected areas along with nearby regions. These tick pools were then sent to ICMR-National Institute of Virology (NIV)-Pune, where they were first screened for the presence of CCHFV. On testing, a large number of tick pool samples were found to be negative for CCHFV. These CCHFV-negative tick pools were further investigated for the presence of other pathogenic agents. Virus isolation was attempted from the hard ticks, and two specimens demonstrated cytopathic effects (CPE) in Vero CCL-81 cell culture. Conventional reverse transcription-PCRs (RT-PCRs) specific for several diverse viruses were tried on these samples but failed to identify the virus isolates.
The recent advancement of next-generation sequencing (NGS) technology has assisted in the identification of previously uncharacterized and unknown viruses. Recently, we characterized reoviruses that are transmitted through ticks, mosquitoes, and Culicoides isolated from different parts of India using an NGS platform (22, 23). In this study, we identified and characterized two additional tick-borne reoviruses belonging to the Coltivirus and Orbivirus genera. This is the first identification of a novel virus belonging to the Coltivirus genus in India, and a new strain of WMV, derived from ticks, was also characterized.
RESULTS
Isolation and cytopathic effect of novel infectious agents.
During a CCHFV outbreak in 2011 in the Ahmedabad and Amreli districts of Gujarat State, India, Hyalomma ticks were collected from buffalo, sheep, and goats from the household of CCHFV-positive cases (21). One out of 17 tick pools from Ahmedabad and 1/44 tick pools from Amreli districts were found to positive for the presence of CCHFV (21). To evaluate the presence of other pathogenic agents in the CCHFV-negative Hyalomma ticks, subconfluent Vero CCL-81 cells were inoculated with the tick pools. Two tick pools demonstrated CPE on the 2nd (Kundal virus [KUNDV]) and 4th (Karyana virus [KARYV]) days postinfection (dpi) which increased with time. The KARYV isolate exhibited syncytium formation, cell elongation, rounding, and detachment; in contrast, the KUNDV isolate exhibited cell rounding and detachment (Fig. 1). Isolates were named based on the geographical location of their collection. Susceptibility analysis demonstrated that KUNDV could infect and cause CPE in several cell lines (Vero E6, human rhabdomyosarcoma [RD], and porcine stable kidney [PS] cells) except those derived from Pipistrellus bats and mosquitoes (C6/36) (Fig. 2), whereas KARYV could infect several cell lines (Vero E6, human RD, PS, and Pipistrellus bat embryo cells), except mosquito cells (Fig. 3).
FIG 1.
Isolation and characterization of novel infectious agents. Cytopathic effect of Karyana virus-infected Vero CCL-81 cells at 2 days postinfection (top) and Kundal virus-infected Vero CCL-81 cells at 2 days postinfection (bottom) is shown.
FIG 2.
Cytopathic effect of Kundal virus infection in diverse cell types at 2 days postinfection. RD, human rhabdomyosarcoma cells; PS, porcine stable kidney cells; bat embryo, Pipistrellus ceylonicus bat embryo cells; C6/36, Aedes albopictus cells.
FIG 3.
Cytopathic effect of WMV strain Karyana infection in diverse cell types at 2 days postinfection.
Identification of the novel infectious agents.
Initial identification attempts were performed using RT-PCR specific for pan-flaviviruses, pan-phleboviruses, and specific viruses in the Nairoviridae (Ganjam virus/Nairobi sheep disease virus [NSDV], CCHFV, and nairovirus), Flaviviridae (KFDV), Orthomyxoviridae (Thogoto virus and Dhori virus), Peribunyaviridae (Ingwavuama virus), and Phenuiviridae (Bhanja virus and severe fever with thrombocytopenia syndrome virus [SFTSV]) families, as well as tick-borne encephalitis virus (TBEV) and Powassan virus. None of these assays showed any positive amplification for either of the isolates.
Unbiased NGS of infected monolayers and supernatants identified Karyana and Kundal virus isolates as members of the Reoviridae family, Orbivirus and Coltivirus genera, respectively. The majority of NGS reads were identified with homology to the human genome (45 to 94.9%), and 0.64 to 25.5% of the remaining reads from the supernatants and monolayers were specific to KARYV or KUNDV (Table 1). Small contigs generated from the remaining nonviral, nonhuman reads exhibited homology to mixed eukaryotes and extracellular bacteria, but we did not observe a predominance of reads from other organisms in the KARYV and KUNDV data sets. A single exception consisted of reads specific to Mycoplasma, which was at a significantly lower percentage than KUNDV-specifc reads (99,164 reads [25.5%] viral versus 16,308 reads [4.3%] mycoplasmal).
TABLE 1.
Distribution table depicting number of viral, bacterial, and human reads using bowtie software
| Sample | Total reads | No. of mapped virus-specific reads (bowtie2) (%) | No. of human reads (bowtie2) (%) | No. of nonviral, nonhuman reads (%) | No. of reads that map to Mycoplasma arginini (bowtie2) (%) | Contigs with homology to other organisms |
|---|---|---|---|---|---|---|
| Karyana virus, monolayer | 12,15,692 | 34,985 (2.8) | 1,098,815 (90.3) | 81,892 (6.7) | <5 | Small contigs (<0.45 Kb) with homology to mycoplasma (1) and even-toed ungulates (4) |
| Karyana virus, supernatant | 36,20,400 | 23,178 (0.64) | 3,435,499 (94.9) | 161,723 (4.4) | <40 | Small isolated contings (<1.5 kb) to rRNA from various bacteria. Very small contigs (<0.5 kb) from bacteria, mycoplasmas, and fungi. |
| Kundal virus, monolayer | 5,94,102 | 13,040 (2.2) | 391,676 (65.9) | 189,386 (31.8) | 1,965 (0.3) | Small contings (2.9–1.5 kb and <1 kb) with homology to mycoplasma (5). Very small contigs (<450 bp) with homology to bacteria (6), even-toed ungulates (12), and fungi (3). |
| Kundal virus, supernatant | 3,88,638 | 99,164 (25.5) | 175,261 (45.1) | 114,213 (29.3) | 16,308 (4.2) | Small contings (3.0– 0.3 kb) with homology to mycoplasma (45). Very small contigs (<0.3 kb [n = 9]) with homology to various bacteria (7). |
Through de novo assembly we generated 1,283 and 112 contigs greater than 500 bp long from the paired-end reads generated from KUNDV and KARYV, respectively. BLASTX analysis of KARYV contigs identified 10 segments with the highest similarity to WMV (Table 2). The open reading frames of all 10 segments were obtained. The genome sequence of KARYV was 17,760 nucleotides and encoded proteins ranging from 220 to 1,302 amino acids (Table 2). BLASTX analysis of Kundal contigs identified 12 segments with homology to TarTV and TFRV segments (Table 2).The open reading frames of all 12 segments were obtained. The genome sequence of KUNDV was 28,778 nucleotides and encoded proteins ranging from 170 to 1,434 amino acids (2).
TABLE 2.
Genomic characterization of the Kundal virus (coltivirus) isolate from India and the Wad Medani virus (orbivirus) isolate from India
| Virus | Segment details | Segment length retrieved (bp) | Putative function | Protein size (reference size) | GenBank accession no. |
|---|---|---|---|---|---|
| KUNDV | VP1 | 4,382 | RNA-dependent RNA polymerase | 1,434 | MH327935 |
| VP2 | 3,922 | Methyltransferase | 1,213 | MH327936 | |
| VP3 | 3,533 | Guanylyltransferase | 1,035 | MH327937 | |
| VP4 | 3,149 | VP4 | 1,040 | MH327938 | |
| VP5 | 2,608 | VP5 | 843 | MH327939 | |
| VP6 | 1,992 | Nucleotide binding, NTPase | 639 | MH327940 | |
| VP7 | 2,029 | VP7 | 660 | MH327941 | |
| VP8 | 1,908 | VP8 | 613 | MH327942 | |
| VP9 | 1,850 | VP9 | 596 | MH327943 | |
| VP10 | 1,835 | Kinase, helicase | 576 | MH327944 | |
| VP11 | 963 | VP11 | 305 | MH327945 | |
| VP12 | 607 | RNA replication factors | 170 | MH327946 | |
| KARYV | VP1 | 3,915 | RNA-dependent RNA polymerase | >1,302 (1,304) | MH571964 |
| VP3 | 2,803 | Inner core protein | >619 (623) | MH571965 | |
| VP2 | 1,895 | Capping enzyme | 910 | MH571966 | |
| VP4 | 1,805 | Outer capsid protein | 550 | MH571967 | |
| VP5 | 1,666 | Outer capsid protein 2 | 543 | MH571968 | |
| VP6 | 965 | Helicase | 314 | MH571969 | |
| VP7 | 1,135 | Major core protein | 355 | MH571970 | |
| NS1 | 1,727 | Tubule protein | 536 | MH571971 | |
| NS2 | 1,185 | Nonstructural protein NS2 | 378 | MH571972 | |
| NS3 | 664 | NS3 protein | 220 | MH571973 |
Confirmation of KUNDV and KARYV identification by plaque assay, specific real-time RT-PCR, and viral growth curves.
To confirm that plaques were the result of KUNDV and KARYV replication, and not due to replication from an obligate intracellular organism or tissue culture contaminant, we performed plaque assays and virus-specific RT-PCR. Plaque morphology was assessed in two cell types (Vero CCL-81 and BHK-21), and optimum plaques were identified in Vero CCL-81 cells at 4 dpi. Plaques formed by KARYV were round with a clear center and contained a solid border (Fig. 4A and C). Plaques formed by KUNDV were also round with a clear center but contained a diffuse border. Microscopic examination of KUNDV plaques identified a circumferential zone of spindly cells, which contributed to a diffuse plaque border upon macroscopic examination (Fig. 4B and D). Titration by plaque assay identified that KARYV and KUNDV stocks contained 1 × 106.55 PFU per ml and 1 × 105.79 PFU/ml at 4 dpi.
FIG 4.
Plaque assay for KARYV and KUNDV. (A) Plaques formed by KARYV on Vero CCL-81 cells at 10−4 and 10−3 dilutions at 4 dpi. (B) Plaques formed by KUNDV on Vero CCL-81 cells at 10−4 and 10−3 dilutions at 4 dpi. (C) KARYV plaque on Vero CCL-81 cells at 4 dpi, ×10 magnification. (D) KUNDV plaque on Vero CCL-81 cells at 4 dpi, ×10 magnification.
Using the genomes generated in this study, real-time PCR assays specific to the Vp1 segments of KARYV and KUNDV, respectively, were developed. Eight and 10 plaques from KUNDV and KARYV, respectively, were purified and blind passaged twice on naive Vero CCL-81 cells. Tissue culture fluid was positive for KUNDV and KARYV for all passages.
Growth kinetics for KARYV and KUNDV was evaluated using Vero CCL-81 and BHK-21 cell lines. KARYV replicated well in both Vero CCL-81 and BHK-21 cells and a peak in infectious intracellular KARYV was detected at 3 dpi. Infectious extracellular virus was detected at 5 and 7 dpi and KARYV exhibited a larger burst size in BHK-21 cells than Vero CCL-81 cells (Fig. 5A). In contrast, KUNDV replicated more efficiently in BHK-21 cells than in Vero CCL-81 cells and infectious virus was detected at 2 dpi (intracellular) and 3 to 7 dpi (extraceullular) (Fig. 5B).
FIG 5.
Growth kinetics of KARYV and KUNDV. (A) Growth kinetics of KARYV in Vero CCL-81 and BHK-21 cells. (B) Growth kinetics of KUNDV in Vero CCL-81 and BHK-21 cells.
KUNDV and KARYV identification by RNA segment PAGE.
Through NGS and de novo assembly we identified that Kundal virus contained 12 segments with sizes ranging from 0.6 to 4.3 kbp and Karyana virus contained 10 segments with sizes ranging from 0.6 to 3.9 kbp. These in silico results were confirmed through SDS-PAGE of viral RNA (Fig. 6). We observed that KARYV and KUNDV were released into the supernatant and contained 10 and 12 segments, respectively (Fig. 6A). Viral segment sizes in Fig. 6 (KARYV, VP1 > VP3 > VP2/4 [doublet] > NS1 > VP5 > NS2 > VP7 > VP6 > NS3, and KUNDV, VP1 > VP2 > VP3 > VP4 > VP5 > VP7 > VP6 > VP8 > VP9 > VP10 > VP11 > VP12) were nearly identical to in silico size estimates (Table 2). To confirm that these segments were both packaged and virus specific, we subjected cell monolayers to a freeze-thaw cycle and treated released nucleic acids with DNase and RNase (Fig. 6B). Treatment with nucleases eliminated high-molecular-weight nucleic acids but left viral RNA unaffected, demonstrating that viral RNA is a protected state inaccessible to nucleases.
FIG 6.
RNA segment sizes for Karyana and Kundal viruses. (A) RNA was extracted from the supernatant of Karyana virus-infected (3 dpi), Kundal virus-infected (2 or 5 dpi), or mock-infected cells, separated by 10% SDS-PAGE, and silver stained. RNA from rotavirus strain DS-1 was added as a molecular marker. (B) Cells were infected with Karyana virus (3 dpi) or Kundal virus (2 or 5 dpi) or mock infected, and unprotected nucleic acids from cell monolayers were digested with DNase and RNase or left untreated. RNA from rotavirus strain Wa was added as a molecular marker.
Confirmation of KUNDV and KARYV by electron microscopy analysis of virus particles.
Members of the Reoviridae family contain 1 to 3 concentric layers of capsid proteins with an overall spherical single capsid diameter of 50 to 60 nm and double capsid diameter of 60 to 80 nm (19). By electron microscopy, WMV strain KARYV and KUNDV appeared morphologically similar to the reference reovirus, WMV strain Ar495 (Fig. 7A). WMV strain Ar495 and KARYV and KUNDV viral inclusion bodies (VIB) in the cytoplasm consisted of a granular matrix containing the inner capsid of the virion, known as single capsid (SC) particles (Fig. 7A and Fig. 8A and C), which were comprised of an electron dense nucleoid surrounded by a protein shell. Mature intracellular virions, known as double capsid (DC) particles (Fig. 8A and C), exhibited the electron-dense SC particles surrounded by a fuzzy, diffuse outer capsid and were found surrounding the VIB or free in the cytoplasm. Morphogenesis for WMV strain KARYV and KUNDV exhibited distinguishable differences. Similar to WMV stain Ar495, within the VIB for WMV strain KARYV the SC particles (37.8 ± 3.4 nm) could be scattered across the inclusions (Fig. 7A and Fig. 8B), as opposed to KUNDV, for which the SC particles (42.7 ± 3.4 nm) were mostly at the perimeter of the VIB (Fig. 7A and Fig. 8C). DC particle sizes for WMV strain KARYV (58.6 ± 3.1 nm) and KUNDV (62.4 ± 3.5 nm) were consistent with reovirus DC particle size. Within the VIB of WMV strain KARYV were two structures which were similar in diameter to SC particles, namely, empty capsids (43.0 ± 4.3 nm in diameter) and tubules (35.8 ± 3.2 nm in diameter) (Fig. 8B). Additional virally induced structures in WMV strain KARYV-infected cells were observed during replication. Cytoplasmic hollow tubules (Fig. 8B) (46.0 ± 2.8 nm) with a diameter similar to that of the SC particles were found, as has been reported for bluetongue virus (24). Dense, amphorus cytoplasmic structures with associated SC, similar to VIB (Fig. 8B), and electron-dense filaments in the nucleus (51.2 ± 6.5 nm in diameter) (Fig. 7B) were also observed. Similar nuclear filaments have previously been described for CTFV (25). However, to our knowledge, the dense, amphorus cytoplasmic structures with associated SC are a new observation for reoviruses. Budding virions were sporadically observed at the membrane of intracellular vesicles and the cell surface (Fig. 7C), as has been reported for other orbiviruses (8, 16, 26, 27), and an occasional extracellular enveloped particle could be seen (Fig. 7D). In contrast, the VIB in KUNDV-infected cells did not contain empty capsids or tubules, and cytoplasmic tubules and nuclear filaments were also not observed during viral infection (Fig. 8C).
FIG 7.
Morphology of reoviruses by electron microscopy. (A) Viral inclusion bodies of WMV strain Ar495 (ATCC VR-470) (left image; bar, 200 nm), WMV strain Karyana (middle image; bar, 500 nm) and Kundal virus (right image; bar, 800 nm) all exhibit similar morphologies that are consistent with related reoviruses. Arrowhead, tubules within a viral inclusion body (VIB). (B) Virally induced nuclear filaments (arrowheads) are found in a WMV-infected cell. Nu, nucleus; L, lipid droplet. Bar, 1 μm. (C) Virions (arrowheads) observed budding through the plasma membrane to the extracellular environment or into the lumen of an intracellular vesicle. Bar, 500 nm. (D) Extracellular enveloped particles (arrows) are composed of DC particles enclosed within a viral envelope. Bar, 200 nm.
FIG 8.
Morphology of reovirus particles by electron microscopy. (A) Morphology of WMV strain Karyana. (Left image) Single capsid (sc) particles within a viral inclusion body, composed of an inner nucleoid bordered by the capsid protein. Empty particles (arrowhead) are also found. (Right image) Double capsid (dc) particles show a fuzzy layer surrounding single capsid particles, shown here in the cytoplasm. Bars, 200 nm. (B) Morphology of WMV strain Karyana during viral replication. (Left image) WMV inclusion body containing SC particles (arrow), tubules (arrowhead), and empty capsids. Nu, nucleus. Bar, 200 nm. (Right image) WMV VIB located near virally induced cytoplasmic tubules (arrowheads). Bar, 800 nm. (Bottom image) Dense, amorphous material (arrows) can be found associated with the VIBs of WMV strain Karyana-infected cells. Arrowhead, SC particle. Bar, 800 nm. (C) Morphology of Kundal virus during viral replication. (Left image) Kundal virus inclusion body containing electron-dense SC virions located at the periphery. (Right image) Kundal virus replication center containing mostly DC virions. Bars, 200 nm.
Pathogenesis of novel infectious agents in a mouse model.
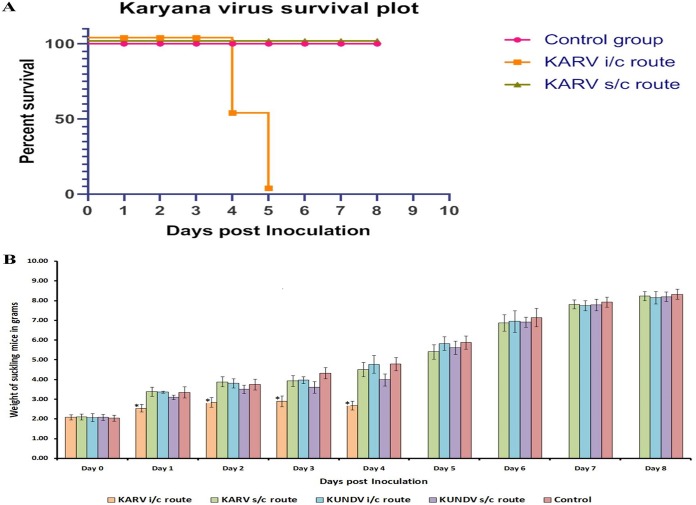
Since KARYV and KUNDV were isolated from Hyalomma ticks (a known vector for other zoonotic diseases) and related coltiviruses (CTFV) can causes mild febrile illness, we evaluated whether they could cause pathogenesis in a mouse model. Morbidity and mortality along with changes in body weight were analyzed in suckling CD1 mice inoculated through the intracranial (i.c.) or subcutaneous (s.c.) route with KUNDV or KARYV isolates. Mice inoculated with KARYV by the i.c. route exhibited reduced feeding, immobility, and hind limb paralysis at 3 dpi. At 1 to 4 dpi, a significant loss of body weight was observed in mice infected subcutaneously (P < 0.0001; 95% confidence interval [CI]). (Fig. 9B). At 4 dpi 50% (4/8) of mice were dead, and mortality increased to 100% at 5 dpi (Fig. 9A). Subcutaneous inoculation of KARYV did not cause any morbidity or mortality, and mice survived until the end of observation (8 dpi) (Fig. 9A). Suckling CD1 mice did not exhibit any signs of sickness after i.c. or s.c. infection with KUNDV. Four additional blind passages of KUNDV were performed in suckling mice, and none of the mice displayed any signs of sickness or mortality.
FIG 9.
Overview of the infectivity of Karyana and Kundal viruses in suckling CD1 mice. (A) Survival plot for suckling mice infected through the intracranial or subcutaneous route with Karyana or Kundal virus. (B) Mouse weight chart for suckling mice infected with Karyana or Kundal virus. Bars represent average weights from eight mice. A significant loss of body weight was observed at 1 to 4 dpi in mice that were infected subcutaneously (P < 0.0001; 95% CI).
Phylogenetics and classification of WMV.
The International Committee on Taxonomy of Viruses (19) defines the assignment of reoviruses to specific genera based on their ability to recombine and reproduce infectious virus progeny and sequence identity greater than 26 to 33% for the RNA-dependent RNA polymerase (RdRp) (VP1). The VP1 segment of KARYV putatively encodes the RdRp, which is supported by the presence of SG (positions 714 and 715) and GDD motifs (positions 765 to 767), as reported for other reoviruses (8). The KARYV RdRp exhibited 13 to 92% amino acid identity with other reoviruses, establishing that it is a member of the Orbivirus genus (Fig. 10). Nucleotide identity greater than 74 to 76% for the major subcore structural protein, VP3, further demarcates orbiviral species (19). The KARYV VP3 gene segment exhibits a maximum of 78.9% nucleotide identity with other WMV isolates, demonstrating that it belongs to the Wad Medani species/serogroup. Within the Wad Medani serogroup, two serotypes exist, namely, WMV and Seletar virus (19). Currently, only 2 complete WMV genomes exist (Seletar virus has not been sequenced). Phylogenetic analysis of the VP1 gene implied that KARYV is the third isolate of WMV (Fig. 11).
FIG 10.
Percent identity and similarity of WMV strain Karyana (GenBank accession no. MH571964). (A) Percent nucleotide identity of the Wad Medani virus to the other reference orbiviruses in the database. (B) Percent amino acid similarity of the Wad Medani virus to the other reference orbiviruses in the database.
FIG 11.
Phylogenetic analysis of members of the Reovirus family using the polymerase (VP1) gene. The sequences belonging to Sedoreovirinae and Spinareovirinae subfamilies are depicted in blue and purple. The new reovirus sequences in the current study are depicted in red.
Phylogenetics and classification of Kundal virus.
The VP1 segment of KUNDV putatively encodes the RNA polymerase which is supported by the presence of motif SG (positions 753 and 754) and GDD motif (positions 814 to 816) reported for other reoviruses (8). The KUNDV RdRp exhibited 56.7 to 62.7% amino acid identity to other reoviruses, establishing that it is a member of the Coltivirus genus. Species demarcation for Coltivirus is defined as nucleotide identity of >89% in the conserved VP12 and amino acid identities of >55%, >57%, and >60%, respectively, in VP6, VP7, and VP12 (the most variable proteins) (19). KUNDV shares a nucleotide identity in the range of 37.2 to 43.6% in the conserved VP12 and maximum amino acid identities of 35.5%, 42.1%, and 31.4% in VP6, VP7, and VP12, respectively (Fig. 12). These data suggest that KUNDV is distinct from the current Coltivirus species and hence forms a tentative new species within the Coltivirus genus. Analysis of the other segments, however, revealed that the KUNDV is more closely related to TarTV in segments 1 to 3, 5 to 7, 10 and 12 (Fig. 12). Phylogenetic analysis of the VP1 gene inferred two separate clusters of Coltivirus species; CTFV and EYAV cluster together, whereas KUNDV forms an ancestral node distinct from the EYAV/CTFV and the TarTV clades (Fig. 11).
FIG 12.
Percent identity and similarity of Kundal virus (GenBank accession no. MH327935). (A) Percent nucleotide identity of the Kundal virus to the other reference coltiviruses in the database. (B) Percent amino acid similarity of the Kundal virus to the other reference coltiviruses in the database.
DISCUSSION
Here we describe the isolation and characterization of unknown infectious agents originally obtained from ticks collected in Gujarat state, India, in 2011 as part of a survey for the presence of CCHFV. However, NGS identified WMV and a novel coltivirus (KUNDV) within the samples when traditional RT-PCR was unable to identify the infectious agents. We established that WMV strain KARYV could cause pathogenesis within mammalian cells lines and mice; in contrast, Kundal virus caused pathogenesis only in tissue culture. Phylogenetic analysis using the RdRp implied the evolutionary history of these reoviruses and showed that WMV strain KARYV is a member of the Sedoreovirinae subfamily, Orbivirus genus, WMV species, whereas KUNDV is a member of the Spinareovirinae subfamily, Coltivirus genus, and represents a novel species. Morphologically, virions from the two isolates were similar to other reoviruses, yet viral inclusion bodies exhibited morphological differences.
Recently, three additional reoviruses (WMV, Kammavanpettai virus [KVPTV], and equine encephalosis virus [EEV]) from India were isolated from different hosts and characterized (22, 23). The tick-borne WMV and mosquito-borne KVPTV have not been associated with human disease, whereas Culicoide-borne EEV had demonstrated pathogenesis in a horse (19). Thus, reoviruses and their disease-causing potential may be underestimated in India and worldwide. In this study, we observed that traditional RT-PCR methods presented limited tools of characterization and NGS methods had revolutionized the discovery of unknown or unidentified viruses (28–30).
Identifying and correlating the presence of infectious organisms with the disease have been enduring challenges to microbiologists. While NGS has revolutionized the field of pathogen discovery, the approach is not without pitfalls. The bioinformatics software can misidentify nucleic acids and build erroneous genomes without appropriate oversight. Despite these drawbacks, we feel confident that the organisms and genomes built in this study are authentic. De novo assembly of WMV strain KARYV generated a genome with significant similarity to preexisting WMV genomes, RT-PCR confirmed the presence of WMV-specific nucleic acids, SDS-PAGE identified RNA segments of the correct predicted sizes, and electron microscopy verified the presence of virions with morphology consistent with reovirus infection (8, 16, 19, 24–27).
In contrast, KUNDV exhibited significant nucleotide and amino acid divergence from preexisting Coltivirus genomes. While the nucleotide homology is so limited that it is beyond the detection of BLASTn, KUNDV exhibits a high degree of amino acid homology with existing Coltivirus genomes, and the conservation of specific residues is consistent with a maintained domain structure in several viral proteins. Additionally, RT-PCR confirmed the presence of KUNDV-specific nucleic acids, SDS-PAGE confirmed RNA segment sizes, and electron microscopy strongly supported the presence of virions with morphology consistent with other reoviruses (19).
Phylogenetic analysis implied that KARYV and KUNDV are related to other reoviruses with worldwide distribution. Specifically, WMV had been isolated from Hyalomma asiaticum ticks in Tajikistan in 1981, Rhipicephalus species in Sudan in 1952, and Hyalomma ticks from India in the 1950s (Fig. 13). Given this broad geographic and temporal distribution, it is surprising that WMV has not been identified in eastern Africa and on the Indian subcontinent since the 1980s. Correspondingly, recent surveillance for novel coltiviruses has similarly identified novel viruses from Dermacentor ticks in the US (2002 to 2013), Ixodes ticks from France (2010), Haemaphysalis ticks in Japan (2013), and Chaereophon bats from Côte d’Ivoire (2016) (Fig. 13). Together, coltiviruses and orbiviruses likely represent an ancient, conserved viral lineage, and increased surveillance will likely identify additional human and animal infections.
FIG 13.
Global prevalence of coltiviruses and orbiviruses. The world map was created using mapchart.net (https://mapchart.net/world.html). The map of India is from ArcGIS Online (https://www.arcgis.com/home) and was edited using Epi Info 7 (version 7.2.1.0; CDC, Atlanta, GA) software.
Considering the wide temporal and geographic distribution of colti- and orbiviruses, the amount of sequence divergence for each viral segment yields new insight into the selective pressures experienced by these viruses. Specifically, we observed that the greatest degree of protein conservation for the RdRp of coltiviruses and VP7 (major core protein) of orbiviruses, consistent with these enzymes experiencing a strong constraint against mutation. Conversely, the outer capsid (VP4), NS2 (NS2), and helicase (VP6) proteins of orbiviruses and VP9, VP8, and nucleotide binding/NTPase (VP6) proteins of coltiviruses exhibit the most diversity, suggesting that these proteins may experience strong selective pressure, potentially leading to protein diversification. Since reoviruses can replicate in both arthropods and mammalian cell lines, proteins required for replication in both hosts may experience less positive selection pressure and represent the existence of a fitness trade-off for replication in both hosts (31). Specifically, we observed that the VP2 (inner core) and methyl transferase (VP2) proteins were conserved in WMV and KUNDV, respectively, indicating that these proteins may be required for replication in two hosts.
Currently unknown is whether WMV and KUNDV cause pathogenesis in humans and livestock. The reoviruses described here were isolated from Hyalomma ticks in India during 2011 CCHF surveillance. Hyalomma anatolicum is the known vector for zoonotic diseases such as CCHFV, Babesia, Theileria, and Anaplasma species, suggesting that it may also transmit other, previously unrecognized pathogens. Human infection with a related coltivirus (CTFV) usually causes mild febrile illness but is known to cause severe symptoms of hemorrhagic fever, pericarditis, and myocarditis, resulting in the hospitalization of approximately 20% of patients (12, 13). Since CTFV usually causes mild febrile illness, the possibility exists that KARYV and KUNDV human infections may mimic other tick-borne infections (such as CCHF, KFD, anaplasmosis, babesiosis, and rickettsioses) and other endemic infections, like malaria. Alternatively, individuals may become coinfected with multiple tick-borne zoonoses following a Hyalomma bite, which may influence disease morbidity and mortality. Specific serological and molecular tests, as well as increased surveillance, are required to further define the prevalence of human and animal reovirus infections and correlate disease with specific epidemiological determinants.
MATERIALS AND METHODS
Ethical statement.
All procedures carried out with mice during this study were approved by the Institutional Animal Ethics Committee (CPCSEA registration no. 43/GO/C/1999/CPCSEA). Suckling CD1 mice were procured from Animal House, ICMR-NIV, Pune.
Cell culture isolation and analysis of susceptibility to novel viruses.
A total of 17 and 44 Hyalomma tick pools were collected from the Ahmedabad and Amreli districts of Gujarat State, India, respectively, from buffalo, sheep, and goats of the household of CCHFV-positive cases (21). CCHFV-negative tick pools were homogenized using a tissue homogenizer (GenoGrinder 2000; BT&C Inc., Lebanon, NJ) in minimum essential medium (MEM). The tick pool homogenates were clarified at 4,000 × g for 2 min at 4°C and 100 μl of supernatant was used to inoculate subconfluent Vero CCL-81 cells in 24-well plates maintained in MEM supplemented with 10% fetal bovine serum (FBS; Gibco), penicillin (100 U/ml), and streptomycin (100 mg/ml). Cells were infected at 37°C and 5% CO2 for 1 h with the intermittent rocking of the plate. After adsorption, the inoculum was removed, and cells were washed using 1× phosphate-buffered saline (PBS) and MEM containing 2% FBS was added to each well. The cells were incubated at 37°C and 5% CO2 and observed daily for any cytopathic effect (CPE) with an inverted light microscope.
Further passage of the tissue culture fluid was done in Vero CCL-81 cells after observation of CPE as described above. Harvesting of the tissue culture fluid (TCF) was performed by three cycles of freeze-thawing of the scraped cells along with the supernatant. The TCF was aliquoted and stored at −80°C until further use. Both of the isolates were tested for the presence of known tick-borne arboviruses, e.g., flavivirus, KFDV, tick-borne encephalitis virus (TBEV), Nairo virus, Ganjam virus (Nairobi sheep disease virus), CCHFV, Thogotovirus, Powssan virus, Ingwavuama virus, Bhanja virus, Dhori virus, severe fever with thrombocytopenia syndrome virus (SFTSV), and phleboviruses (32–39). To evaluate pathogenesis and cell susceptibility toward these infectious agents, a 100-fold dilution of original material was repassaged on African green monkey kidney (Vero E6) cells and human (rhabdomyosarcoma [RD]), porcine (porcine stable [PS] kidney), bat embryo (Pipistrellus ceylonicus), and mosquito (C6/36, Aedes albopictus) continuous cell lines.
Next-generation sequencing of the novel infectious agents.
One milliliter of the cell supernatant and cell debris from both the virus isolates was used for RNA extraction. QIAmp viral RNA extraction (Qiagen) was used for RNA extraction following the manufacturer’s protocol. RNA quantification, depletion to remove rRNA, RNA library preparation, and library quantification were performed as described by Yadav et al. (23).
Bioinformatics analysis.
Next-generation sequencing data from the unknown virus isolate were analyzed using CLC Genomics Workbench version 10 (Qiagen), Geneious (version 10), V-pipe (40), and EDGE (version 2.0.0) (41). De novo assembly of the FASTQ file was performed to generate contigs using CLC. Contigs were classified using the BLASTn and BLASTx programs. The open reading frames (ORFs) and putative functions of the identified segments were assigned using the homology to TarTV.
Confirmation of KUNDV and KARYV identification. (i) Plaque assay and plaque purification.
Five sets of a standard 6-well plate were seeded with 3 ml of growth medium (MEM, 10% FBS) in Vero CCL-81 cells. The growth medium was removed after a monolayer was formed and 200 μl of serially diluted viral stock (KARYV 50% tissue culture infective dose [TCID50], 1 × 105.61/ml, and KUNDV TCID50, 1 × 105.75/ml) was added to each well. The virus stocks were serially diluted with MEM at a ratio of 1:10. This 6-well plate was incubated at 37°C for 1 h with gentle shaking after every 10 min. An overlay of 3 ml of 1% CMC medium per well was added in the 6-well plate and the plate was directly incubated at 37°C and 5% CO2. Each 6-well plate of the set was observed at an interval of 24 h without disturbing the others. Observation was done by gently adding a fixative (3 ml of 0.1% amido black) to each well and incubating it for an hour. Excess stain was removed, cells were further washed twice gently with PBS at room temperature, and plaques were directly observed. Cell monolayers for plaque purification were not stained, and plaques were identified by microscopy. Identified plaques were picked with a syringe (0.7 by 25 mm) manufactured by BD Precision Glide, dissolved in MEM, and used for infecting fresh Vero CCL-81 cells. Blind passages were also performed in Vero CCL-81 cells twice.
(ii) KUNDV- and KARYV-specific real-time RT-PCR.
Real-time RT-PCR primers and probes specific for KARYV and KUNDV were designed using the VP1 sequence retrieved in this study (unpublished data). The primers and probes were standardized and used for detection of plaque-purified virus. All the plaques formed were checked for viral RNA positivity using the test developed for specific viruses.
(iii) Growth kinetics study of the novel viruses.
Tissue culture (24-well) plates were seeded with Vero CCL-81 and BHK-21 cells at a density of 0.5 × 105 and infected at a multiplicity of infection (MOI) of 0.1. These plates were incubated with the inoculum for 1 h, the inoculum was removed, and cells were washed twice with PBS. One milliliter of fresh MEM supplemented with 2% fetal calf serum (FCS) was added to the plate, and the supernatant was harvested immediately (1 h postinfection [hpi]) and thereafter at an interval of 24 h for 7 days. Harvested cells were washed twice with PBS and scraped in 1 ml of MEM supplemented with 2% FCS. Both supernatant and pellets were frozen and stored at −80°C. The amount of infectious virus particles (TCID50) was calculated for both Vero CCL-81 and BHK-21 cells from the supernatants and cell pellets, after controlled thawing, vortexing, and centrifugation.
(iv) RNA segment PAGE.
Low-passage Vero E6 cells at ∼75% confluency in T25 flasks were infected with Karyana or Kundal virus. Virus adsorbed to cells with constant rocking in 500 μl of medium, and after 1 h, Dulbecco modified Eagle medium (DMEM) with 10% FBS was added directly to flasks. Infection proceeded for 2, 3, or 5 days postinfection. After observation of CPE (cell rounding, plaques, and floating cells), cell monolayers were scraped from flasks and cells were pelleted by centrifugation at 1,000 rpm for 5 min. Supernatant and cells were separated, and cells were resuspended in 500 μl of DMEM with 10% FBS and frozen. After a single round of freeze-thawing, unprotected nucleic acids from cell monolayers were treated with 18 U of Baseline-ZERO DNase (Epicentre) and RNase cocktail enzyme mix (12 U of RNase A and 480 U of RNase T1) (Invitrogen) or only 1× Baseline-ZERO buffer for 1 h at 37 C (a single T25 flask was used for each treatment condition). DNase and RNase reaction was terminated by adding 4 volumes of Tripure (Roche). Karyana and Kundal viruses in supernatant were inactivated with 4 volumes of Tripure (2 T25 flasks were used for each condition). RNA was isolated from Tripure using phase separation with 1-bromo-3-chloropropane (Sigma-Aldrich), and RNA was cleaned and concentrated using an RNA Clean and Concentrate-25 column (Zymo Research). In total, viral RNA was concentrated from 14 ml of viral supernatant to 120 μl of RNA or 600 μl of cells to 40 μl of RNA. Viral RNA segments were separated using SDS-PAGE. Viral RNA from supernatants (20 μl) or cell monolayers (10 μl) were mixed with 2× SDS-PAGE loading buffer (Invitrogen) and added to SDS-PAGE gels containing a 4% stacking layer (made with 1.5 M Tris, pH 8.8) and 10% separating layer (made with 1.5 M Tris, pH 8.8). Segments were separated at 90 V for 45 min and 120 V for 3 h (constant voltage) in 1× Novex Tris-glycine SDS running buffer (Invitrogen). Viral RNA was stained using a Silver Stain Plus kit (Bio-Rad). Rotavirus RNA from strains Wa and DS-1 were a kind gift from Mathew Esona and were prepared from rotaviral isolates using RNA Clean and Concentrate-25 columns (Zymo Research). Between 76 and 54 ng of rotaviral RNA was added to the SDS-PAGE gels.
(v) Electron microscopy analysis of reovirus particles.
Ultrapurified virus was prepared by growing Wad Medani virus strains Ar495 (ATCC VR-470) and KARYV (passage 1) and KUNDV (passage 1) in a T150 flask of Vero E6 cells for 6 to 10 days in Eagle’s minimal essential medium (EMEM) supplemented with 10% FBS (Gibco), nonessential amino acids (1×), sodium pyruvate (1 mM), penicillin (100 U/ml), and streptomycin (100 mg/ml). The supernatant was saved, and cell debris was pelleted at 3,200 × g and 4°C for 15 min. Polyethylene glycol 8000 (PEG 8000) was added to viral supernatant to a final concentration of 20% and rotated overnight at 4°C. The next day, the virus was pelleted at 3,200 × g for 30 min at 4°C. Pellet was resuspended in 3.2 ml of EMEM and added to the top of a 20% sucrose cushion (20% sucrose in TNE buffer, i.e., 50 mM Tris [pH 7.5], 100 mM NaCl, and 0.1 mM EDTA). The virus was ultrapurified at 28,000 rpm (average relative centrifugal force [RCF], 74,330) for 2 h at 4°C in an SW-55 rotor. The viral pellet was resuspended in 220 μl of EMEM, and 10 μl was used to infect naive cultures of VeroE6 cells in a 25-cm2 flask for 3 or 6 days. Ultrapurified KUNDV did not generate a high enough titer for electron microscopy (EM) visualization of infected cells; thus, viral supernatant from passage 2 was used to infect naive cells for EM.
To prepare material for EM, cells were scraped from a 25-cm2 flask and pelleted for 5 min at 165 × g. Cells were resuspended in EM buffer (0.1 M sodium phosphate, pH 7.3) and repelleted with 235 × g for 10 min. EM buffer was removed, and cells were fixed with 2.5% glutaraldehyde for 2 h at 4°C. After 2 h, fixative was removed, and the supernatant was replaced with EM buffer. The cell pellet was inactivated with 5 × 106 rads of gamma irradiation and embedded in a mixture of Epon substitute and Araldite resins. Sections were cut, stained with uranyl acetate and lead citrate, and imaged on a Thermo Fisher/FEI Spirit electron microscope.
Mouse susceptibility to KARYV and KUNDV.
The TCF of Vero CCL-81 cells showing CPE was used for virus susceptibility studies with CD1 suckling mice. Eight 0- to 2-day-old suckling CD1 mice were inoculated intracranially (i.c.) as well as subcutaneously (s.c.) with 20 μl of each virus isolate (KARYV TCID50, 1 × 105.61/ml, and KUNDV TCID50, 1 × 105.75/ml). Suckling mice were observed for 8 days for signs of sickness, the weights of individual mice were measured daily, and mortality was recorded. At the onset of symptoms and mortality or after 8 days postinoculation (dpi), irrespective of the presence of sickness, the mice were euthanized using isoflurane overdose; brains were harvested and homogenized to make a 10% suspension in 1.25% bovine serum-supplemented PBS (BAPS). The homogenized 10% suspension was aliquoted and stored at −80°C until further use. Passage of the virus was done using this 10% mouse brain suspension as an inoculum. An unpaired t test was performed to assess whether body weights differed significantly between infection routes.
Phylogenetic analysis.
Phylogenetic analysis of viral sequences was performed using representative Spinareovirinae and Sedoreovirinae reference sequences from GenBank. Alignment of sequences was performed using the ClustalW algorithm as implemented in MEGA software version 7.0 (42). A maximum likelihood evolutionary tree was generated using the general time reversible model + Gamma+ I (GTR+G + I) substitution model. Statistical robustness of the tree was assessed using 1,000 bootstrap replicates. Protein similarity and identity was obtained using the EMBOSS needle program (Blosum62 matrix with a Gap penalty of 10 and an extended penalty of 0.5).
Accession number(s).
The genome sequences have been deposited in GenBank under accession numbers MH571964 to MH571973 for KARYV and MH327935 to MH327946 for KUNDV.
ACKNOWLEDGMENTS
We thank M. D. Gokhale, Scientist-C, ICMR-NIV Pune, for helping with tick identification, and D. H. Joshi, Deputy Director of A.H., FMD, Gujarat state, India, for his contribution to the collection of ticks. We also thank P. T. Joshi, National Vector Borne Disease Control Programme, Gujarat state, for performing a survey for CCHF virus in ticks which led to the identification of these virus isolates. We thank Vijay Aychit for technical support.
Financial support was provided extramurally by the U.S. Centers for Disease Control, Atlanta, and intramurally by Indian Council of Medical Research, New Delhi.
The funders had no role in study design, data collection or interpretation, or the decision to submit the work for publication.
We declare that we have no competing interests. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Mourya DT, Yadav PD, Patil DY. 2014. Highly infectious tick-borne viral diseases: Kyasanur forest disease and Crimean-Congo haemorrhagic fever in India. WHO South East Asia J Public Health 3:8–21. doi: 10.4103/2224-3151.206890. [DOI] [PubMed] [Google Scholar]
- 2.Labuda M, Nuttall PA. 2004. Tick-borne viruses. Parasitology 129:S221–S245. doi: 10.1017/S0031182004005220. [DOI] [PubMed] [Google Scholar]
- 3.Mansfield KL, Johnson N, Phipps LP, Stephenson JR, Fooks AR, Solomon T. 2009. Tick-borne encephalitis virus—a review of an emerging zoonosis. J Gen Virol 90:1781–1794. doi: 10.1099/vir.0.011437-0. [DOI] [PubMed] [Google Scholar]
- 4.Kosoy OI, Lambert AJ, Hawkinson DJ, Pastula DM, Goldsmith CS, Hunt DC, Staples JE. 2015. Novel thogotovirus associated with febrile illness and death, United States, 2014. Emerg Infect Dis 21:760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albariño CG, Zaki SR, Rollin PE, Nicholson WL, Nichol ST. 2012. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- 6.Tokarz R, Sameroff S, Leon MS, Jain K, Lipkin WI. 2014. Genome characterization of Long Island tick rhabdovirus, a new virus identified in Amblyomma americanum ticks. Virol J 11:26. doi: 10.1186/1743-422X-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X-J, Liang M-F, Zhang S-Y, Liu Y, Li J-D, Sun Y-L, Zhang L, Zhang Q-F, Popov VL, Li C, Qu J, Li Q, Zhang Y-P, Hai R, Wu W, Wang Q, Zhan F-X, Wang X-J, Kan B, Wang S-W, Wan K-L, Jing H-Q, Lu J-X, Yin W-W, Zhou H, Guan X-H, Liu J-F, Bi Z-Q, Liu G-H, Ren J, Wang H, Zhao Z, Song J-D, He J-R, Wan T, Zhang J-S, Fu X-P, Sun L-N, Dong X-P, Feng Z-J, Yang W-Z, Hong T, Zhang Y, Walker DH, Wang Y, Li D-X. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attoui H, Mohd Jaafar F, Biagini P, Cantaloube JF, de Micco P, Murphy FA, de Lamballerie X. 2002. Genus Coltivirus (family Reoviridae): genomic and morphologic characterization of Old World and New World viruses. Arch Virol 147:533–561. doi: 10.1007/s007050200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florio L, Miller MS, Mugrage ER. 1950. Colorado tick fever; isolation of the virus from Dermacentar andersoni in nature and a laboratory study of the transmission of the virus in the tick. J Immunol 64:257–263. [PubMed] [Google Scholar]
- 10.Eklund CM, Kohls GM, Brennan JM. 1955. Distribution of Colorado tick fever and virus-carrying ticks. JAMA 157:335–337. doi: 10.1001/jama.1955.02950210031010. [DOI] [PubMed] [Google Scholar]
- 11.Goodpasture HC, Poland JD, Francy DB, Bowen GS, Horn KA. 1978. Colorado tick fever: clinical, epidemiologic, and laboratory aspects of 228 cases in Colorado in 1973-1974. Ann Intern Med 88:303–310. doi: 10.7326/0003-4819-88-3-303. [DOI] [PubMed] [Google Scholar]
- 12.Attoui H, Mohd Jaafar F, de Micco P, de Lamballerie X. 2005. Coltiviruses and seadornaviruses in North America, Europe, and Asia. Emerg Infect Dis 11:1673–1679. doi: 10.3201/eid1111.050868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attoui H, De Micco P, de Lamballerie X. 1997. Complete nucleotide sequence of Colorado tick fever virus segments M6, S1 and S2. J Gen Virol 78:2895–2899. doi: 10.1099/0022-1317-78-11-2895. [DOI] [PubMed] [Google Scholar]
- 14.Moutailler S, Popovici I, Devillers E, Vayssier-Taussat M, Eloit M. 2016. Diversity of viruses in Ixodes ricinus, and characterization of a neurotropic strain of Eyach virus. New Microbes New Infect 11:71–81. doi: 10.1016/j.nmni.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobler G. 1996. Arboviruses causing neurological disorders in the central nervous system. Arch Virol Suppl 11:33–40. [DOI] [PubMed] [Google Scholar]
- 16.Fujita R, Ejiri H, Lim C-K, Noda S, Yamauchi T, Watanabe M, Kobayashi D, Takayama-Ito M, Murota K, Posadas-Herrera G, Minami S, Kuwata R, Yamaguchi Y, Horiya M, Katayama Y, Shimoda H, Saijo M, Maeda K, Mizutani T, Isawa H, Sawabe K. 2017. Isolation and characterization of Tarumizu tick virus: a new coltivirus from Haemaphysalis flava ticks in Japan. Virus Res 242:131–140. doi: 10.1016/j.virusres.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 17.Weiss S, Dabrowski PW, Kurth A, Leendertz SAJ, Leendertz FH. 2017. A novel Coltivirus-related virus isolated from free-tailed bats from Côte d’Ivoire is able to infect human cells in vitro. Virol J 14:181. doi: 10.1186/s12985-017-0843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertens PPC. 1999. Orbiviruses and coltiviruses (Reoviridae). General features, p 1043–1061. In Granoff A, Webster RG, Encyclopedia of virology, 2nd ed Academic Press, London, United Kingdom. [Google Scholar]
- 19.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2011. Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses, p 541–637. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 20.Belaganahalli MN, Maan S, Maan NS, Brownlie J, Tesh R, Attoui H, Mertens P. 2015. Genetic characterization of the tick-borne orbiviruses. Viruses 7:2185–2209. doi: 10.3390/v7052185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mourya DT, Yadav PD, Shete A, Majumdar TD, Kanani A, Kapadia D, Chandra V, Kachhiapatel AJ, Joshi PT, Upadhyay KJ, Dave P, Raval D. 2014. Serosurvey of Crimean-Congo hemorrhagic fever virus in domestic animals, Gujarat, India, 2013. Vector Borne Zoonotic Dis 14:690–692. doi: 10.1089/vbz.2014.1586. [DOI] [PubMed] [Google Scholar]
- 22.Yadav PD, Albariño CG, Nyayanit DA, Guerrero L, Jenks MH, Sarkale P, Nichol ST, Mourya DT. 2018. Equine encephalosis virus in India, 2008. Emerg Infect Dis 24:898–901. doi: 10.3201/eid2405.171844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav PD, Shete AM, Nyayanit DA, Albarino CG, Jain S, Guerrero LW, Kumar S, Patil DY, Nichol ST, Mourya DT. 2018. Identification and characterization of novel mosquito-borne (Kammavanpettai virus) and tick-borne (Wad Medani) reoviruses isolated in India. J Gen Virol 99:991–1000. doi: 10.1099/jgv.0.001102. [DOI] [PubMed] [Google Scholar]
- 24.Murphy FA, Borden EC, Shope RE, Harrison A. 1971. Physicochemical and morphological relationships of some arthropod-borne viruses to bluetongue virus–a new taxonomic group. Electron microscopic studies. J Gen Virol 13:273–288. doi: 10.1099/0022-1317-13-2-273. [DOI] [PubMed] [Google Scholar]
- 25.Murphy FA, Coleman PH, Harrison AK, Gary GW. 1968. Colorado tick fever virus: an electron microscopic study. Virology 35:28–40. doi: 10.1016/0042-6822(68)90302-4. [DOI] [PubMed] [Google Scholar]
- 26.Schnagl RD, Holmes IH. 1976. Characteristics of the genome of human infantile enteritis virus (rotavirus). J Virol 19:267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould AR, Hyatt AD. 1994. The orbivirus genus. Diversity, structure, replication and phylogenetic relationships. Comp Immunol Microbiol Infect Dis 17:163–188. doi: 10.1016/0147-9571(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 28.Capobianchi MR, Giombini E, Rozera G. 2013. Next-generation sequencing technology in clinical virology. Clin Microbiol Infect 19:15–22. doi: 10.1111/1469-0691.12056. [DOI] [PubMed] [Google Scholar]
- 29.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. 2015. Next-generation sequencing for infectious disease diagnosis and management: a report of the Association for Molecular Pathology. J Mol Diagn 17:623–634. doi: 10.1016/j.jmoldx.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Prachayangprecha S, Schapendonk CME, Koopmans MP, Osterhaus A, Schürch AC, Pas SD, van der Eijk AA, Poovorawan Y, Haagmans BL, Smits SL. 2014. Exploring the potential of next-generation sequencing in detection of respiratory viruses. J Clin Microbiol 52:3722–3730. doi: 10.1128/JCM.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes EC. 2003. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol 11:543–546. doi: 10.1016/j.tim.2003.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katargina O, Russakova S, Geller J, Kondrusik M, Zajkowska J, Zygutiene M, Bormane A, Trofimova J, Golovljova I. 2013. Detection and characterization of tick-borne encephalitis virus in Baltic countries and Eastern Poland. PLoS One 8:e61374. doi: 10.1371/journal.pone.0061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshii K, Okamoto N, Nakao R, Klaus Hofstetter R, Yabu T, Masumoto H, Someya A, Kariwa H, Maeda A. 2015. Isolation of the Thogoto virus from a Haemaphysalis longicornis in Kyoto City, Japan. J Gen Virol 96:2099–2103. doi: 10.1099/vir.0.000177. [DOI] [PubMed] [Google Scholar]
- 34.Scaramozzino N, Crance JM, Jouan A, DeBriel DA, Stoll F, Garin D. 2001. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol 39:1922–1927. doi: 10.1128/JCM.39.5.1922-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yadav P, Shete A, Bondre V, Patil D, Kokate P, Chaudhari S, Srivastava S, Jadhav S, Mourya D. 2016. Isolation and characterization of Oya virus a member of Simbu serogroup, family Bunyaviridae, isolated from Karnataka, India. Infect Genet Evol 44:122–126. doi: 10.1016/j.meegid.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 36.Shah KV, Work TH. 1969. Bhanja virus: a new arbovirus from ticks Haemaphysalis intermedia Warburton and Nuttall, 1909, in Orissa, India. Indian J Med Res 57:793–798. [PubMed] [Google Scholar]
- 37.Mourya DT, Yadav PD, Mehla R, Barde PV, Yergolkar PN, Kumar SRP, Thakare JP, Mishra AC. 2012. Diagnosis of Kyasanur forest disease by nested RT-PCR, real-time RT-PCR and IgM capture ELISA. J Virol Methods 186:49–54. doi: 10.1016/j.jviromet.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Atkinson B, Chamberlain J, Logue CH, Cook N, Bruce C, Dowall SD, Hewson R. 2012. Development of a real-time RT-PCR assay for the detection of Crimean-Congo hemorrhagic fever virus. Vector Borne Zoonotic Dis 12:786–793. doi: 10.1089/vbz.2011.0770. [DOI] [PubMed] [Google Scholar]
- 39.Mourya DT, Yadav PD, Shete AM, Gurav YK, Raut CG, Jadi RS, Pawar SD, Nichol ST, Mishra AC. 2012. Detection, isolation and confirmation of Crimean-Congo hemorrhagic fever virus in human, ticks and animals in Ahmadabad, India, 2010–2011. PLoS Negl Trop Dis 6:e1653. doi: 10.1371/journal.pntd.0001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montmayeur AM, Ng TFF, Schmidt A, Zhao K, Magaña L, Iber J, Castro CJ, Chen Q, Henderson E, Ramos E, Shaw J, Tatusov RL, Dybdahl-Sissoko N, Endegue-Zanga MC, Adeniji JA, Oberste MS, Burns CC. 2017. High-throughput next-generation sequencing of polioviruses. J Clin Microbiol 55:606–615. doi: 10.1128/JCM.02121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li P-E, Lo C-C, Anderson JJ, Davenport KW, Bishop-Lilly KA, Xu Y, Ahmed S, Feng S, Mokashi VP, Chain P. 2017. Enabling the democratization of the genomics revolution with a fully integrated web-based bioinformatics platform. Nucleic Acids Res 45:67–80. doi: 10.1093/nar/gkw1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]