MLB astroviruses are emerging viruses infecting humans. More studies are required to determine their exact epidemiology, but several reports have already identified them as the cause of unexpected clinical diseases, including severe neurologic diseases. Our study provides the first description of a cell culture system for the propagation of MLB astroviruses, enabling the study of their replicative cycle. Moreover, we demonstrated the unknown capacity of MLB astrovirus to establish persistent infections in cell culture. Whether these persistent infections are also established in vivo remains unknown, but the clinical consequences would be of high interest if persistence was confirmed in vivo. Finally, our analysis of IFN expression provides some trails to understand the mechanism by which MLB astroviruses can cause persistent infections in the assayed cultures.
KEYWORDS: interferon, novel astrovirus, persistence
ABSTRACT
MLB astroviruses were identified 10 years ago in feces from children with gastroenteritis of unknown etiology and have been unexpectedly detected in severe cases of meningitis/encephalitis, febrile illness of unknown etiology, and respiratory syndromes. The aim of this study was to establish a cell culture system supporting MLB astrovirus replication. We used two clinical strains to infect several cell lines, an MLB1 strain from a gastroenteritis case, and an MLB2 strain associated with a neurologic infection. Efforts to propagate the viruses in the Caco-2 cell line were unsuccessful. In contrast, we identified two human nonintestinal cell lines, Huh-7 and A549, permissive for both genotypes. After serial passages in the Huh-7.5 cell line, the adapted strains were able to establish persistent infections in the Huh-7.5, Huh-7AI, and A549 cell lines, with high viral loads (up to 10 log10 genome copies/ml) detected by quantitative reverse transcription-PCR (RT-qPCR) in the culture supernatant. Immunofluorescence assays demonstrated infection in about 10% of cells in persistently infected cultures. Electron microscopy revealed particles of 32 to 33 nm in diameter after negative staining of cell supernatants and capsid arrays in ultrathin sections with a particularly high production in Huh-7.5 cells. Interferon (IFN) expression by infected cells and effect of exogenous IFN varied depending on the type of infection and the cell line. The availability of a cell culture system to propagate MLB astroviruses represents a key step to better understand their replicative cycle, as well as a source of viruses to conduct a wide variety of basic virologic studies.
IMPORTANCE MLB astroviruses are emerging viruses infecting humans. More studies are required to determine their exact epidemiology, but several reports have already identified them as the cause of unexpected clinical diseases, including severe neurologic diseases. Our study provides the first description of a cell culture system for the propagation of MLB astroviruses, enabling the study of their replicative cycle. Moreover, we demonstrated the unknown capacity of MLB astrovirus to establish persistent infections in cell culture. Whether these persistent infections are also established in vivo remains unknown, but the clinical consequences would be of high interest if persistence was confirmed in vivo. Finally, our analysis of IFN expression provides some trails to understand the mechanism by which MLB astroviruses can cause persistent infections in the assayed cultures.
INTRODUCTION
First identified in 1975 in stool samples of children with diarrhea (1), human astroviruses (HAstVs) cause viral gastroenteritis worldwide (2), being the third most common cause in the pediatric population after rotavirus and norovirus. Besides children, HAstV gastroenteritis also frequently occurs in the elderly (3) and in immunocompromised individuals (4–6). HAstVs are small (28 to 41 nm in diameter) nonenveloped single-stranded positive-sense RNA viruses. To date, the family Astroviridae is divided into two genera, Mamastrovirus and Avastrovirus, including viruses infecting mammals and birds, respectively. Their genome codes for three open reading frames (ORFs), with ORF1a and ORF1b encoding the protease and polymerase proteins, respectively, and ORF2 encoding the capsid proteins.
With the advent of next-generation sequencing technologies, two novel groups of highly divergent HAstVs (named MLB and VA/HMO) which are more closely related to certain animal astroviruses than to the classic HAstVs, have been identified in human stools of individuals with diarrhea (7–13) but also in asymptomatic healthy controls (14, 15). To date, no definitive association between novel astroviruses and gastroenteritis has yet been established, but further epidemiologic studies have confirmed the presence of novel HAstVs worldwide (14, 16–21). In addition, novel HAstVs have been recently identified as the cause of unexpected central nervous system infections in (mostly immunocompromised) humans (22–28). Specifically, MLB astroviruses have been involved in one case of acute meningitis in a healthy young adult (28) and in two cases of neurologic infections in immunocompromised patients (27, 28). Both groups of novel HAstVs have been further divided into several genotypes, MLB1 to -3 for MLB astroviruses and VA1 to -5 for VA astroviruses (2, 29).
Novel HAstVs are part of the neurovirulent astroviruses, which also include animal astroviruses (30). Other unexpected clinical manifestations recently associated with human and animal astroviruses include respiratory tract infections (31–37), fever of unknown etiology (38, 39), hepatitis (40, 41), and severe gout in geese (42). Altogether, these findings suggest that there are probably other still unrecognized divergent astroviruses with clinical implications beyond gastroenteritis in humans and animals. The potential for cross-species transmission is high (43, 44), and the increasing number of descriptions of nonenteric severe clinical manifestations in animals, especially neurologic involvement, should prompt us to validate appropriate systems to study the pathogenicity of astroviruses. Among the novel human astroviruses, a cell culture system has been recently described for VA1 (45). The present study aims at describing a cell culture system for the propagation of MLB1 and MLB2 astroviruses, the two MLB genotypes most frequently identified, and providing some clues for understanding their pathogenicity.
RESULTS
MLB astroviruses can be propagated in Huh-7.5 hepatoma cells.
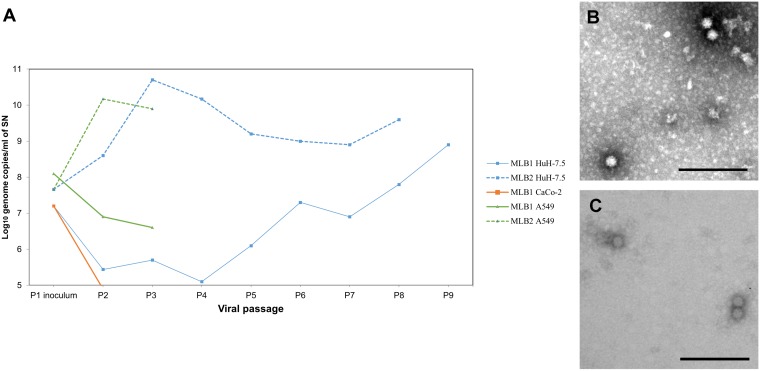
Several MLB clinical specimens were used to infect different cell lines and perform serial viral passages (V-P), following a protocol for an acute infection (see Materials and Methods). Three MLB1 strains were recovered from stool samples of children under 5 years old with symptoms of acute gastroenteritis, and three MLB2 strains were recovered from stool samples and included a neuroinvasive strain identified in an immunocompromised adult patient (28). Among these, only two strains were able to replicate in cell culture (Fig. 1), with one MLB1 strain recovered from a 1-year-old child and the MLB2 neuroinvasive strain recovered from the 37-year-old immunocompromised patient. Attempts to propagate these strains in a Caco-2 cell line resulted in a loss of genome detection after 2 passages (Fig. 2A). Using Huh-7.5 cells, we observed sustained viral genome detection in the culture supernatant (SN) for up to 8 to 9 passages, and some viral passages were also successful in A549 cells (Fig. 1A and 2A). Electron microscopy of the supernatant of acutely infected Huh-7.5 cells confirmed the presence of viral capsids of both genotypes, with a mean size of 33 ± 3 nm for MLB1 and 32 ± 2 nm for MLB2 (Fig. 2B and C).
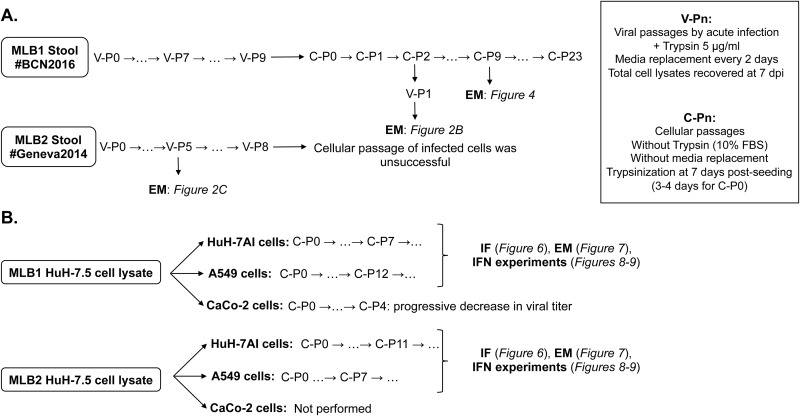
FIG 1.
Description of the viral and cell passages performed with HAstV MLB1 and MLB2 on selected cell lines. (A) Passage history in Huh-7.5 cells. Initially, cells were infected using clinical stool samples as an inoculum. After 7 to 8 viral passages of HAstV MLB1 and MLB2, respectively, infected cells were subcultured. (B) Huh-7AI and A549 cell passages using Huh-7.5 cell lysates as an inoculum to establish persistent infections. V-Pn, viral passages; C-Pn, cellular passages; EM, electron microscopy; IF, immunofluorescence; IFN, interferon.
FIG 2.
Infection of MLB1 and MLB2 in acutely infected cells. (A) Viral genome titers detected in the culture supernatant (SN) by RT-qPCR assays. (B and C) Electron microscopy of SN from Huh-7.5 cells infected with HAstV MLB1, using SN of a persistently infected cell line as inoculum (B) and MLB2 (C). Bars = 200 nm.
Multistep growth curves were performed to define infection kinetics (Fig. 3A and B). While an increase in viral RNA in the cellular fraction showed similar kinetics for the two viruses, with a major increase during the first 2 days after inoculation and a total log10 fold increase from 1 h postinfection (hpi) to 7 days postinfection (dpi) of 3.41 ± 0.37 for MLB1 and 3.22 ± 0.49 for MLB2 (Fig. 3A), the increase in viral RNA in the supernatant fraction was much higher for MLB2 than for MLB1 (5.14 ± 0.03 versus 2.99 ± 0.20, P < 0.05) (Fig. 3B), resulting also in overall higher viral production for MLB2. We also confirmed the occurrence of infectious viruses in the inoculum by treating it for 5 min at 99°C and confirming the lack of viral RNA increase in the supernatant of infected cultures (data not shown).
FIG 3.
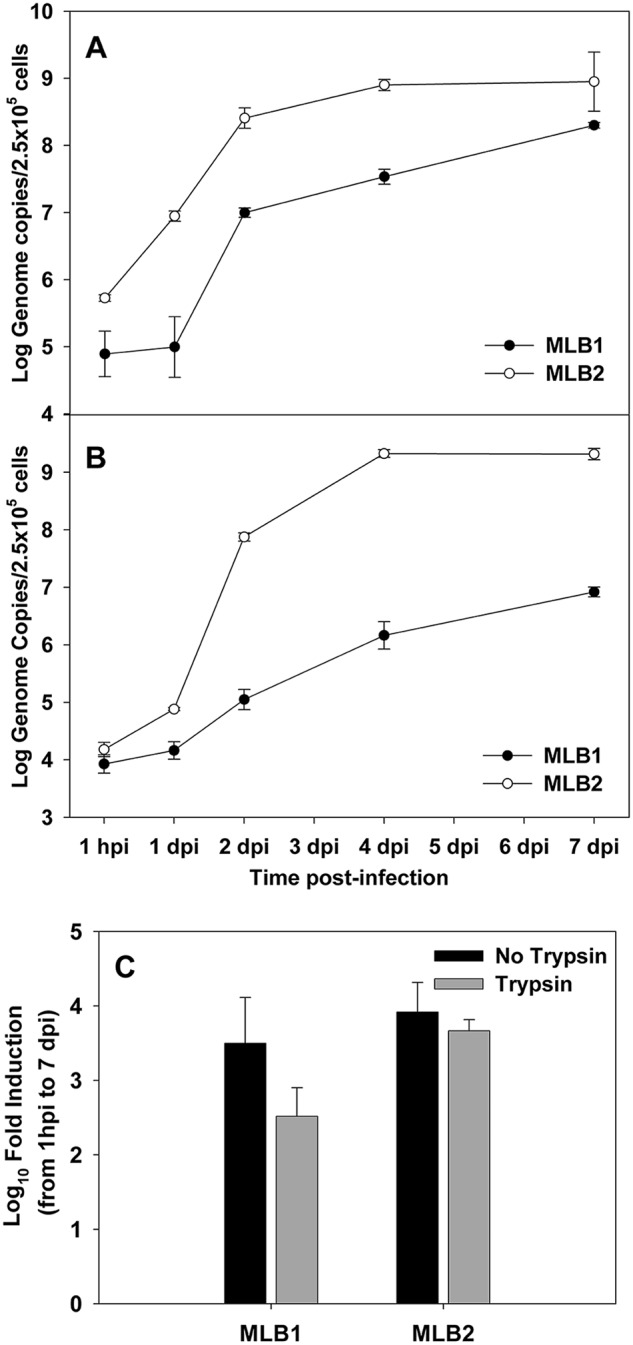
Multistep growth curves of MLB1 and MLB2 on Huh-7.5 cells. Cells were infected using a multiplicity of infection of 20 genome copies/cell, and viral RNA was measured from the cellular (A) and supernatant (B) fractions. Plot shows average values, and error bars indicate one standard deviation from triplicates. (C) Viral replication with or without trypsin for MLB1 and MLB2 on Huh-7.5 cells. Viral replication is expressed as the fold induction of viral genome titers in the SN of infected cells from 1 hpi to 7 dpi. P values comparing fold inductions with and without trypsin for each genotype were not significant (by Mann-Whitney test). Plot shows average values, and error bars indicate one standard deviation. Samples were quantified in duplicate from a single experiment.
A trypsin treatment was initially included, but no significant differences were observed in the efficiency of MLB replication in the presence or absence of trypsin (5 μg/ml) in the postinfection medium (Fig. 3C).
MLB astroviruses can persistently infect cell cultures.
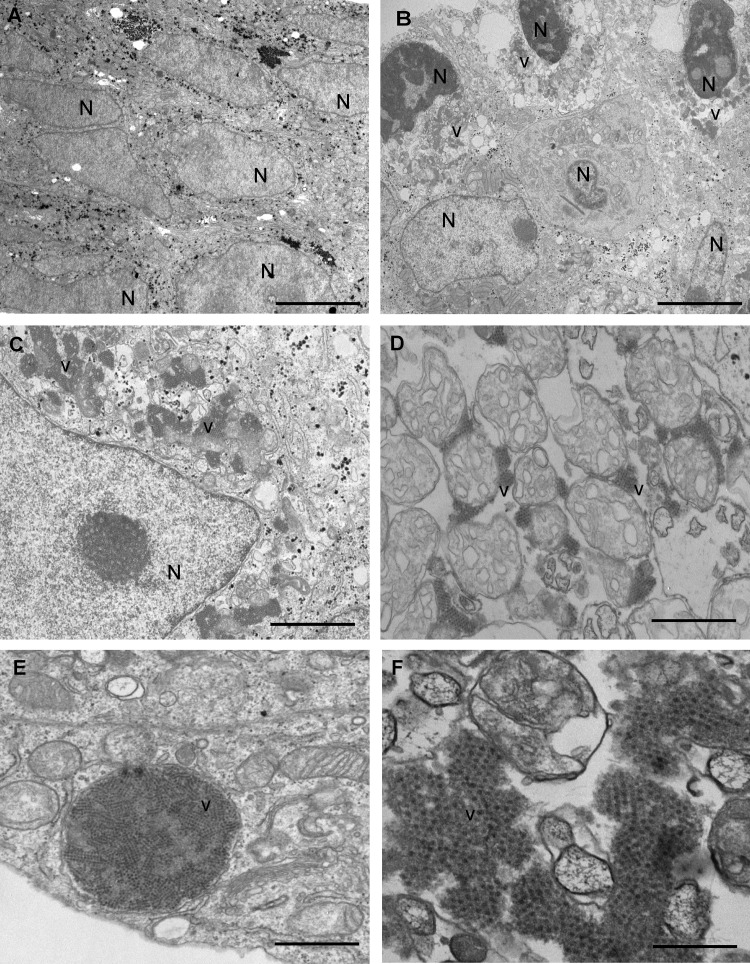
According to the high intracellular viral titer fraction observed for MLB1, we hypothesized that this could reflect a persistent infection. To ascertain whether infected cultures were able to regrow after infection, we used the MLB1 V-P7 cell lysate to establish a persistent infection in the Huh-7.5 cell line. Infected cells were trypsinized at 4 dpi and could be further maintained for up to at least 20 cell passages (C-P) (Fig. 1A). The presence of numerous capsid arrays in persistently infected Huh-7.5 cells, mostly associated with cell membrane vesicles, was observed (Fig. 4). Cells containing capsid arrays showed remarkable cell structure reorganization.
FIG 4.
Electron microscopy analysis of the persistent infections on Huh-7.5 cells. (A to F) Noninfected Huh-7.5 cells (A) and persistently infected Huh-7.5 cells (B to F) showing intracellular capsid arrays of HAstV MLB1 at 4 days postseeding. Aggregates of astrovirus particles (v) accumulated in the cytoplasm of infected cells around the nuclei (N). Bars = 5 μm in panels A and B, 2 μm in panel C, 1 μm in panels D and E, and 200 nm in panel F.
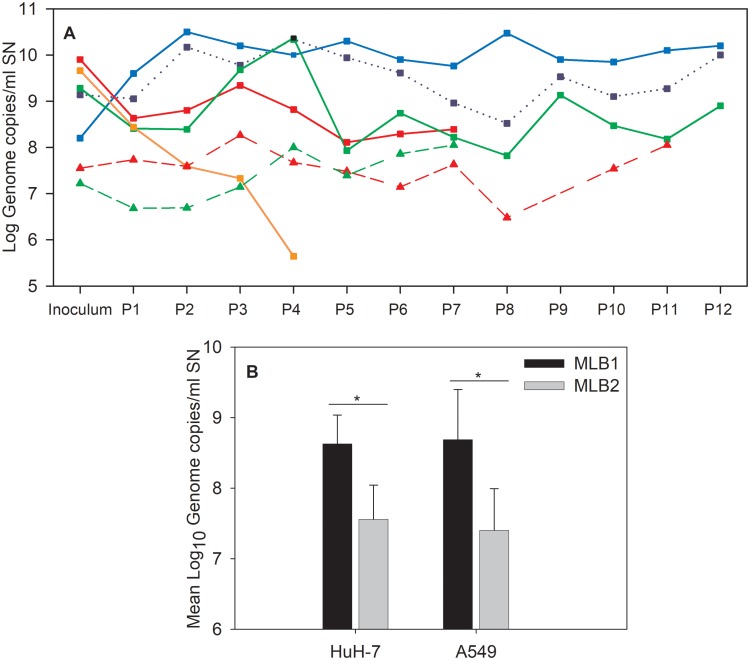
To elucidate if this was due to the described defect in the interferon pathway of the Huh-7.5 cell line (46, 47), we similarly initiated a persistent infection on Huh-7AI cells with both HAstV MLB1 and MLB2 strains recovered from cell lysates, and also on A549 cells, according to the supposed respiratory tropism of novel astroviruses (Fig. 1B). The titers of viral genomes for both strains detected in the supernatant of the two cell lines during passages of persistently infected cultures are shown in Fig. 5A. The mean viral titer for MLB1 was significantly higher than that for MLB2 in the Huh-7AI and A549 cell lines (P < 0.002; Fig. 5B). The MLB1 mean viral titer was also significantly higher in Huh-7.5 cells than in Huh-7AI cells, which could confirm our initial hypothesis that the Huh-7.5 interferon pathway deficiency could promote MLB1 replication (P < 0.001; Fig. 5A).
FIG 5.
Viral genome titers detected in the culture supernatant (SN) by RT-qPCR assays of MLB1 and MLB2 strains in persistently infected cell lines. (A) Blue line refers to the Huh-7.5 cell line, red lines refer to the Huh-7AI cell line, green lines refer to the A549 cell line, and the orange line refers to the Caco-2 cell line. Continuous lines with squares refer to HAstV MLB1 strain, and dotted lines with triangles refer to HAstV MLB2 strain. The dotted blue line with squares corresponds to the MLB1 persistently infected Huh-7.5 cell line, using the original stool sample as an inoculum. (B) Mean viral genome titers of HAstV MLB1 and MLB2 detected by RT-qPCR assays in the SN of persistently infected Huh-7AI and A549 cell lines. The average was calculated based on 7 to 12 numbers, corresponding to the viral genome titers at each cell passage. Error bars indicate one standard deviation. The mean viral titers were significantly different between HAstV MLB1 and HAstV MLB2 in a comparison of the same infected cell line (*, P = 0.0009 in Huh-7AI cells, P = 0.0018 in A549 cells, Mann-Whitney test).
Attempts to establish a MLB2 persistent infection on the Huh-7.5 cell line, however, were unsuccessful, and we decided to pursue the rest of the experiments on Huh-7AI and A549 cells only, in order to be able to compare the results between the two genotypes. Except at 4 days after C-P0, persistently infected cultures did not show cytopathic effect, and cells were morphologically identical to noninfected cells. Of note, attempts to establish a persistent infection on Caco-2 cells showed a progressive decline in the viral titer from one cell passage to another, which is a reason why we did not pursue with this cell culture system either (Fig. 5A).
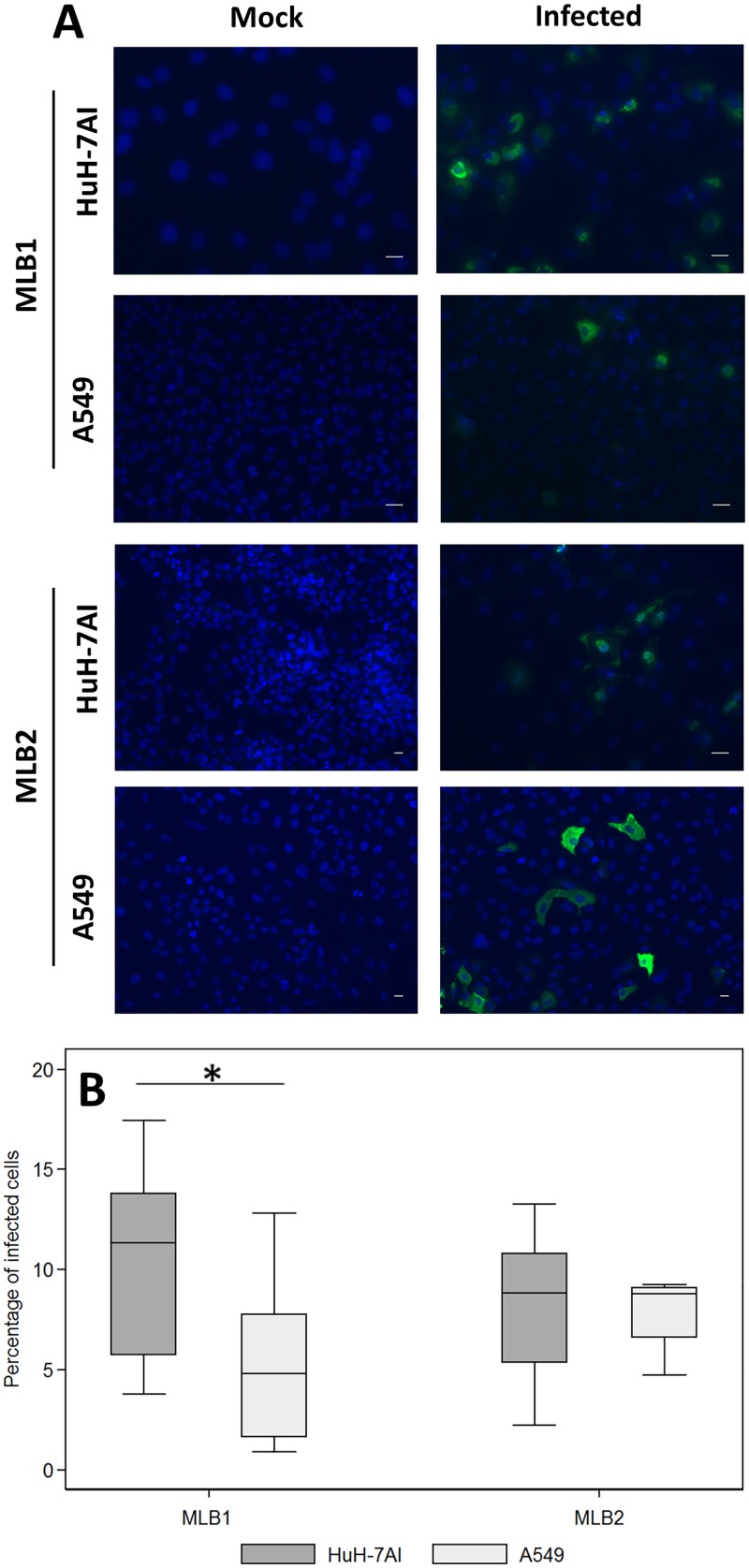
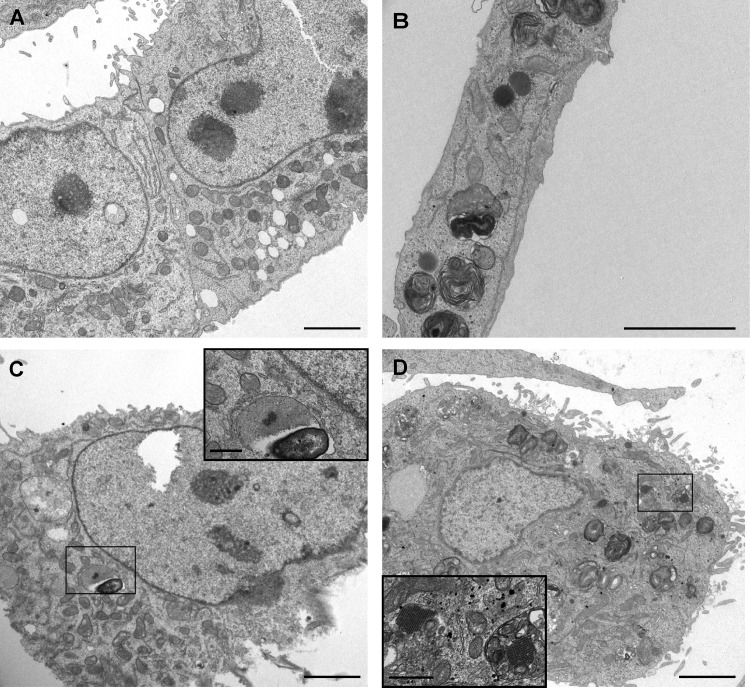
Indirect immunofluorescence assays confirmed the presence of viral capsid proteins in infected cells. Figure 6A shows capsid protein formation in each cell line persistently infected by MLB1 and MLB2 strains. While the fluorescent intensity in each cell line reflects a high production of capsid protein in infected cells, the proportion of cells showing capsid proteins ranged from 1 to 18% (median, 11% and 4.8% for Huh-7AI and A549 persistently infected with MLB1, respectively, and 8.8% for both cell lines persistently infected with MLB2) (Fig. 6B). Overall, for HAstV MLB1, the percentage of capsid-expressing cells was significantly higher in Huh-7AI cells than in A549 cells. No differences were observed for HAstV MLB2. Electron microscopy also confirmed the presence of viral capsid arrays within Huh-7AI- and A549-infected cells (Fig. 7). Table 1 summarizes the results of HAstV MLB1 and MLB2 propagation on selected cell lines.
FIG 6.
Immunofluorescence assay of MLB1 and MLB2 persistently infected cell lines. (A) Infected and noninfected (mock) cultures were fixed at confluence and were incubated with primary (anti-MLB1 and anti-MLB2 antibodies, respectively) and secondary antibodies. Green corresponds to the viral capsid proteins, and blue corresponds to the nuclei. All samples were fixed at 3 to 5 days postseeding (magnification, ×10 to 20). (B) Estimated proportion of persistently infected cells visualized by the immunofluorescence assay. Central line of each box plot represents the median. Each box plot includes data from 7 to 12 fields from 2 different cell passages. For HAstV MLB1, the proportion of Huh-7AI-infected cells was significantly higher than the proportion of A549 cells (*, P = 0.0267, Mann-Whitney test). Bars = 25 μm.
FIG 7.
Visualization of capsid arrays by electron microscopy of persistently infected Huh-7AI and A549 cells. (A) Noninfected Huh-7AI cells. (B) Noninfected A549 cells. (C) Huh-7AI cells persistently infected with HAstV MLB1. (D) A549 cells persistently infected with HAstV MLB2. Bars = 2 μm in main images and 200 nm in enlargements.
TABLE 1.
Summary of results of HAstV MLB1 and MLB2 propagation on selected cell linesa
| Characteristic | HAstV MLB1 | HAstV MLB2 |
|---|---|---|
| Origin of stool sample | 1-yr-old child; Barcelona, Spain | Adult allogeneic stem cell transplant recipient for acute myeloid leukemia; Geneva, Switzerland |
| Patient’s symptoms | Gastroenteritis | Meningoencephalitis; leukemia relapse |
| Viral titer of initial inoculum (genome copies/ml) | 7.9 × 107 | 4.6 × 107 |
| Acute infections | ||
| Infected cell lines | ||
| Caco-2 | − | ND |
| Huh-7.5 | + | + |
| Huh-7AI | + | + |
| A549 | + | + |
| Mean viral titer (genome copies/ml of SN) | ||
| Huh-7.5 | 4.5 × 106 | 2.8 × 109 |
| Persistent infections | ||
| Infected cell lines | ||
| Caco-2 | − | ND |
| Huh-7.5 | + | − |
| Huh-7AI | + | + |
| A549 | + | + |
| Mean viral titer (genome copies/ml of SN) | ||
| Huh-7.5 | 5.6 × 109 | − |
| Huh-7AI | 4.3 × 108 | 3.6 × 107 |
| A549 | 4.9 × 108 | 2.5 × 107 |
| Percentage of infected cells (median) | ||
| Huh-7AI | 11 | 8.8 |
| A549 | 4.8 | 8.8 |
Mean viral titers for Huh-7AI and A549 cell lines acute infections are not provided due to the few number of assays performed. ND, not done; +, successful replication; −, failure to replicate.
Complete genome sequences, using primers detailed in Table 2, were obtained for both strains to analyze whether mutations were occurring during replication compared to viral sequences present in the clinical specimens. The nucleotide sequences of wild-type strains recovered from clinical samples are available at GenBank (accession numbers MK089434 and MK089435). For MLB1, sequences were obtained at V-P6 and C-P2 of acute and persistent Huh-7.5 cell infections, respectively. For MLB2, sequences were obtained at V-P2 on A549 cells and V-P3 in Huh-7.5 cells. No nucleotide changes were detected on the whole genomes throughout the analyzed passages. Nevertheless, we could observe an A-to-C polymorphism at position 1313 and a C-to-T polymorphism at position 5477 of MLB2 at V-P2 in A549 cells, probably reflecting the presence of virus quasispecies. While the latter mutation would be synonymous (and was also present in the wild-type strain), the mutation at position 1313 would result in a substitution of a lysine (K) residue by an arginine (N) in ORF1a. Interestingly, HAstV MLB1 strain directly recovered from the stool sample was also able to establish a persistent infection in the Huh-7.5 cell line, with viral titers comparable to those in the persistent infection with the HAstV MLB1-adapted strain (Fig. 5A), confirming that the ability to establish persistent infections in cell cultures is not dependent on any specific adaptive mutation.
TABLE 2.
Nucleotide sequences of primer pairs used for sequencing of whole genomes of HAstV MLB1 and HAstV MLB2 strains recovered from infected cells
| Segment | Primera | Sequence for strain: |
|
|---|---|---|---|
| MLB1 | MLB2 | ||
| 1 | 1 F | CCAAGAGTGGTGGTATGGC | CCAAGAGTGGTGGTATGGC |
| 1 R | CAGTGCTGTAGACATCCAGAAA | AGCACAAACAACTGATGTAACT | |
| 2 | 2 F | AGAGACCCTGTGTTGCAATAAT | AAAGACCATGTGTTGCCATAAT |
| 2 R | CTAACTTTGGCTTGAGCAACATAA | CTAACTTTAGCCTGGGCCACATAA | |
| 3 | 3 F | CATAGTTACCGCCGCACAT | TATAGTGACAGCAGCACAT |
| 3 R | TTCCCTAGTCAGTCCCTTATCC | TTCTCTGGTTAGGCCTTTATCC | |
| 4 | 4 F | CTGACAGAAGAGGAGTACCAAG | TGGCGCACGTCATAGAA |
| 4 R | CCCATACAGTGGGACCAAA | CCCATACAGTGGGACCAAA | |
| 5 | 5 F | GTACCTTTAGATAGGCCAGTGTATG | GTACCTTTAGATAGGCCAGTGTATG |
| 5 R | CATCAACAAGGTTGGTGGTATTG | CACCCATAAGCGAGAACCGTAAT | |
| 6 | 6 F | GTTGCGCTCCAAAGGTAATAAA | TCCCTTCTTTGGAGGCTTTG |
| 6 R | AGTGAAGCGCCTTGGTAAG | AGTGAAGCGCCTTGGTAAG | |
| 7 | 7 F | CCAGTTGTTGATGGCAAATGA | CCAGTTGTTGATGGCAAATGA |
| 7 R | CCACTCACTAGACGCTGTTT | TTCACAAGGGCCTGAAAC | |
| 8 | 8 F | CTCAACTCATGGTCTGGTCTTG | TTGAATTCATGGTCGGGTC |
| 8 R | CATGTGCCTTGCTGGAAATTG | GGTGGGCAGTACTAGAAATTG | |
| 9 | 9 F | CAGCGGATGTCTATCGTGTTTA | CAGCTGATGTTTACAGAGTTTACAC |
| 9 R | TCCTTAGGTATAGCTGGGTATGT | AATGACCCTGTATGCTGGTATG | |
| 10 | 10 F | GGTCATCAGCACCAGCTAATA | GTTCATCTGCAACATCTGAGA |
| 10 R | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | |
F, forward; R, reverse.
Lack of a strong type I interferon response in MLB-infected cultures.
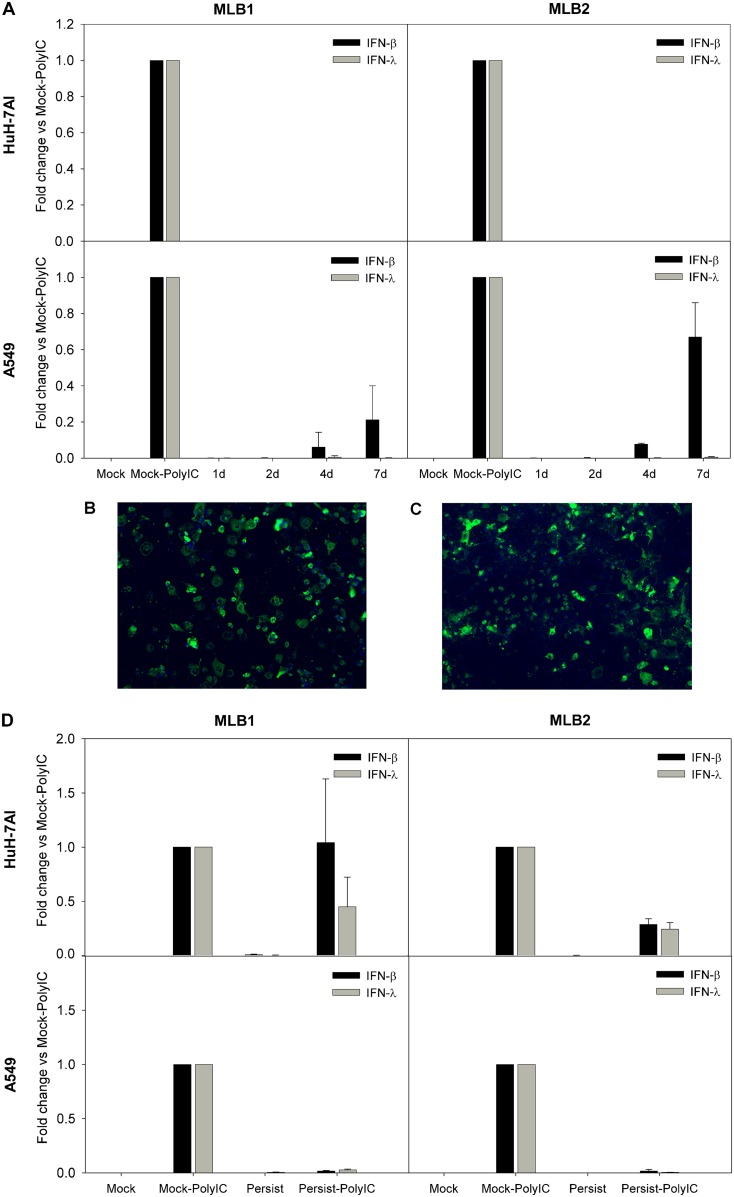
We measured the expression of beta interferon (IFN-β) and IFN-λ1, both known to be implicated in the antiviral innate response. Poly(I·C) transfection was used as a positive control of IFN induction, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were used for normalization. During the acute infection, we could not detect any expression of IFN-λ1 and only a low expression of IFN-β mRNA from 4 to 7 dpi in A549 cells infected with MLB1 and MLB2, compared to those with the poly(I·C) control (Fig. 8A). In order to confirm that the absence of IFN expression in Huh-7AI was not due to the fact that only a small proportion of these cells were infected, we infected them with the highest multiplicity of infection (MOI) possible according to the viral titer of the stocks (MOI, 25,000 genome copies/cell for MLB1 and 680 genome copies/cell for MLB2), which resulted in the infection of >80% of Huh-7AI cells (as visualized by immunofluorescence; Fig. 8B and C), and IFN mRNA remained undetectable. During persistent infections, there was basically no expression of IFN-β nor IFN-λ1 mRNAs in any of the two cell lines infected with MLB1 or MLB2 (Fig. 8D).
FIG 8.
IFN-β and IFN-λ1 expression in infected cell lines. (A) Temporal analysis of IFN-β and IFN-λ1 mRNA expression during acute infection in Huh-7 and A549 cells. Cells were infected with an MOI of 1,000 and 20 genome copies/cell for MLB1 and MLB2, respectively, and poly(I·C)-transfected cells were used as controls. (B and C) Immunofluorescence images correspond to an acute infection using the highest MOI possible (25,000 genome copies/cell for MLB1 and 680 genome copies/cell for MLB2). (D) Analyses of IFN-β and IFN-λ1 expression in persistently infected cultures. Mock, noninfected cells; Mock-PolyIC, noninfected cells transfected with poly(I·C) (positive control); persistent, persistently infected cells; persistent-PolyIC, persistently infected cells that were additionally transfected with poly(I·C); 1d, 2d, 4d, and 7d are the days postinfection (dpi) where IFN expression was measured during acute infections. All samples were quantified at least in duplicate from two distinct experiments.
To understand whether MLB1 and MLB2 replication could block type I IFN expression induced by double-stranded RNA (dsRNA), we analyzed IFN-β and IFN-λ1 mRNA expression after transfecting poly(I·C) in persistently infected cultures. We could observe that IFN-β and IFN-λ1 mRNA expression was almost undetectable in A549 cells for both genotypes compared to the positive control, while in the Huh-7AI cell line, IFN-β and IFN-λ1 induction was only blocked by MLB2. The MLB1 genotype slightly inhibited the expression of IFN-λ1 but not of IFN-β (Fig. 8D). The possibility that A549 persistently infected cultures were refractory to transfection was ruled out by confirming that cells could be efficiently transfected using a green fluorescent protein (GFP)-encoding plasmid (data not shown). Altogether, these results suggest that MLB replication is able to disrupt the innate immune-sensing pathway induced by poly(I·C), although this behavior is cell and genotype (or strain) dependent.
Exogenous IFN inhibits viral replication in a cell-dependent manner.
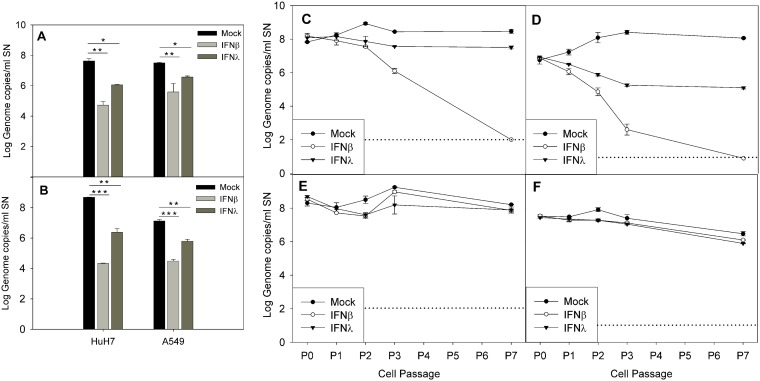
Finally, we tested if the addition of exogenous IFN-β1a and IFN-λ1 at 1,000 U/ml could inhibit viral replication when acutely infected and cure the persistently infected cell lines. During acute infection, pretreatment of cells with both IFN-β1a and IFN-λ1 resulted in a statistically significant reduction in the viral titer compared to the mock-treated controls (P < 0.005 and P < 0.01 for IFN-β1a and IFN-λ1, respectively, during MLB1 infection of both cell lines; P < 0.001 and P < 0.005 for IFN-β1a and IFN-λ1, respectively, during MLB2 infection of both cell lines; Fig. 9A and B). The inhibitory effects of both IFNs were higher in Huh-7AI cells than in A549 cells (P = 0.02). For both genotypes and in both cell lines, the effect of IFN-β1a was approximately 2-fold higher than that of IFN-λ1 (average log10 reductions, 2.91 ± 0.11 versus 1.59 ± 0.11 for MLB1 in Huh-7AI, 1.92 ± 0.52 versus 0.94 ± 0.11 for MLB1 in A549, 4.35 ± 0.03 versus 2.30 ± 0.25 for MLB2 in Huh-7AI, and 2.64 ± 0.20 versus 1.33 ± 0.02 for MLB2 in A549).
FIG 9.
Effect of exogenous IFN during acute infection (A and B) and in persistently infected cultures (C to F). (A and B) Effect of exogenous IFN-β1a and IFN-λ1 on acutely infected Huh-7AI and A549 cells by MLB1 (A) and MLB2 (B). The graphic illustrates the mean viral titer measured in the supernatant at 4 dpi, and error bars show one standard deviation. There was a statistically significant difference in the viral titer between no IFN and IFN-β1a and between no IFN and IFN-λ1 (*, P < 0.01; **, P < 0.005; ***, P < 0.001, ANOVA and Scheffe tests). (C to F) Effect of exogenous IFN-β1a and IFN-λ1 on Huh-7AI cultures persistently infected with MLB1 (C) or MLB2 (D) and on A549 cultures persistently infected with MLB1 (E) and MLB2 (F). Data represent the mean ± standard deviation titer of viral RNA in the supernatant of each passage measured at 4 to 6 days postseeding in the presence or absence of exogenous IFN-β1a or IFN-λ1. All passages were performed in duplicate. P0 corresponds to the viral titer at 1 day after seeding of the first passage with exogenous IFN. Dotted line indicates the limit of detection. P, passage; SN, supernatant.
During persistent infection, the addition of IFN-β1a to the culture medium succeeded in curing the Huh-7AI cell culture from both MLB1 and MLB2 persistent infection after several cell passages. The addition of IFN-λ1 did not eliminate viral replication completely but reduced viral titer 0.96 ± 0.21 and 2.37 ± 0.09 log10 for MLB1 and MLB2, respectively, after 7 passages (Fig. 9C and D). In contrast, in the A549 persistently infected cell cultures, the addition of none of the tested IFNs produced an inhibitory effect on viral replication (Fig. 9E and F). Increase of IFN concentration up to 5,000 U/ml for two additional passages had no effect either (data not shown).
DISCUSSION
We provide the first description of several cell culture systems permissive for the novel HAstV MLB replication. Both Huh-7 and A549 cell lines were infected by viruses present in clinical specimens and sustained viral replication during serial cell passages without the addition of viral inoculum. Expressed viral capsid protein was detected by immunofluorescence within persistently infected cultures, and viral capsids were visualized by electron microscopy both within membrane vesicles and in the cell cytosol. The production of infectious virions released into the supernatant of persistently infected cultures was confirmed by their ability to acutely infect naive cells when used as an inoculum and generate virions that could be observed in the cell culture media after negative staining. In addition, we provide data on the replication of two distinct genotypes, allowing a comparison of the behaviors of HAstV MLB1 and MLB2. Nevertheless, we cannot exclude that the results could be strain dependent, as we only succeeded in propagating one strain of each genotype.
We found that the addition of exogenous trypsin is not required for efficient replication of MLB strains, as previously described for VA1 strains by Janowski et al. (45). The efficient propagation of HAstV MLB in extraintestinal cell lines, hepatic and respiratory, reinforces this information, as these tissues do not secrete as much trypsin as the intestinal tract (48). The capacity of novel HAstVs to infect tissues without the need for capsid activation by trypsin opens the door to a potential wider tissue tropism in vivo, which could explain the diverse clinical manifestations that have been recently described with divergent astrovirus strains, as follows: acute hepatitis, respiratory illnesses, gout, or encephalitis. Nevertheless, our attempts to infect Caco-2 cells were unsuccessful (or at least not as efficient as with Huh-7 and A549 cells, according to the persistent infection assays), which distinguish our results from those of Janowski et al. (45) and those on classical astroviruses. Whether this difference is strain dependent cannot be formally ruled out, but the hypothesis that divergent astroviruses could show an exclusively extraintestinal tropism has also been advanced by other groups (49), who found porcine astrovirus genome in the central nervous system, respiratory tract, and circulatory system of pigs affected by a neurologic syndrome but not in stool samples. Their work suggests that the respiratory tract could be the primary site of astrovirus infection before spreading to the central nervous system. Of note, classical and VA1 astroviruses are also able to infect A549 cells (45, 50).
Apart from providing a cell culture system for HAstV MLB propagation, we have identified the capacity of this clade to establish persistent infection in the studied continuous cell lines. To the best of our knowledge, this is the first description of an experimentally proven persistent infection for astroviruses. RNA viruses make use of several mechanisms for persistence, including the innate immune system evasion, and most persistent infections are asymptomatic (51, 52). Thereby, although continuous cell lines are not the best model to infer issues related to pathology occurring in vivo, it would be very interesting to further study if there is any role for persistent infection in astrovirus diseases, and if so, which are the determinants for the virus to switch from persistence to virulence. Our immunofluorescence assay and structural changes observed by electron microscopy indicate a carrier-state infection, characterized by a small proportion of capsid-expressing cells, associated with a high degree of cell damage and a high production of virus progeny (53) that can infect surrounding noninfected cells. This model of infection is well described for group B coxsackieviruses (54, 55). An alternate hypothesis is that most of the cells in the culture may be resistant to a full cycle of viral replication (precluding capsid visualization by immunofluorescence) and continue to divide. In light of the impossibility of establishing a MLB2 persistent infection in Huh-7.5 cells, it seems that the high rate of replication of MLB2 genotype during the acute infection prevents the survival and regrowth of the infected cells after subculture.
The results of our IFN experiments provide clues to understand the mechanism for HAstV MLB persistence, suggesting that MLB infection does not induce early strong IFN expression, as it has already been described for the classical HAstVs (56). This would avoid a complete clearance of infection by cells and enable persistence. The fact that no IFN expression was observed in any acutely infected cell line before 4 dpi, the time point when infected cultures were subcultivated from C-P0 to study persistence, supports this idea, permitting the virus to continue replicating in permissive cells before the intervention of the innate immune response. Only a certain level of IFN-β mRNA expression was detected in A549 cells during the late course of acute infection. The inhibition of HAstV MLB replication with exogenous IFN during acute infection also reinforces these data; if there was no shutdown of IFN expression, efficient production of IFN by infected cells would inhibit viral replication and thus possibly prevent persistent infections. Of note, sensitivity of astroviruses to IFN when cells are pretreated before infection has also been demonstrated for classical HAstV (56, 57) and VA1 astroviruses (45).
Exogenous IFNs, especially IFN-β1a, were also able to inhibit and even eliminate MLB viruses from persistently infected cultures, but this was only observed for Huh-7AI cells and not for A549 cells. While this difference based on cell type was unexpected, it suggests that persistence may be maintained in both cell lines by dissimilar mechanisms. Indeed, the inhibitory effects of both IFNs on A549 cells were also significantly milder than on Huh-7AI when cells were acutely infected. It is also noteworthy that the effect of MLB replication on IFN mRNA expression induced by poly(I·C) transfection was different in both cell lines. While it could be efficiently blocked on A549 cells, this effect was only partial in Huh-7AI. Our hypothesis is that while MLBs cannot inhibit IFN expression induced by poly(I·C), they may still avoid activation of IFN response by an unknown mechanism, allowing their persistence in the culture unless IFN is added exogenously. On A549 cells, however, MLBs may find the mechanisms to inhibit both arms of the IFN response (induction and action), allowing them to persist in the culture. The fact that efficient counteracting IFN action on A549 cells is only observed when cells are already infected and not when they are acutely infected suggests that a factor expressed at late stages of the replication cycle may be required.
Whether our inability to establish a persistent infection on the Huh-7.5 cell line with the MLB2 strain was due to the activation of other cellular innate responses that would induce the expression of antiviral genes in noninfected neighboring cells or whether it was due to technical factors such as the schedule of culture passaging or to the MOI remains to be elucidated. In addition, we did not measure other types of IFN, such as IFN-α, which could also play a role in the course of HAstV infection. Altogether, we can see a distinct IFN expression and response to exogenous IFN between the persistent versus acute infection, between MLB1 and MLB2, and between different cell lines. These results suggest that there is an actual coevolution between a virus and its host and that many factors (virus strain, cell type, and model of infection) may uniquely influence the course of the infection. Interestingly, Nice et al. recently demonstrated that IFN-λ was able to reduce and to cure persistent infection of murine norovirus in the absence of an adaptive immune response (58, 59) and that the interaction between host IFN-λ response and some viral nonstructural proteins determined viral tropism (59). In addition, these results also suggest that, if persistence was confirmed in vivo, in the case of coinfections, certain MLB HAstVs could potentially promote the replication of other viruses by inhibiting the IFN response.
In summary, we provide a cell culture system for the propagation of the novel HAstV MLB. We have demonstrated that these viruses can establish a carrier-state infection in vitro on extraintestinal human cell lines. IFN expression may be altered by HAstV MLB infections although may vary depending on the strain, the cell line, and the model of infection. Finally, HAstV MLB sensitivity to IFN also depends on the type of infection, the genotype, the cell line, and the type of IFN.
MATERIALS AND METHODS
Cell lines.
Human epithelial colorectal adenocarcinoma (Caco-2 cells; ECACC 86010202), human hepatocyte-derived cellular carcinoma (Huh-7AI cells [60] and Huh-7.5 cells [61]), and adenocarcinoma human alveolar basal epithelial (A549 cells; ATCC CCL-185) cell lines were grown at 37°C with 5% CO2 on minimal essential medium (MEM) with l-glutamine supplemented with 10% fetal bovine serum (FBS; Gibco) and 100 units/ml penicillin and streptomycin (Gibco).
Clinical specimens.
Stool samples positive for MLB1 were collected during a screening of stool samples from children under 5 years old with acute gastroenteritis in Barcelona, Spain. Stool samples positive for MLB2 were identified in a previous study (28). Three samples positive for MLB1 and three samples positive for MLB2 were used for infection, with a viral titer ranging from 1.5 × 106 to 1.2 × 108 genome copies/ml of inoculum for MLB1 and from 2.8 × 104 to 4.6 × 107 genome copies/ml of inoculum for MLB2.
Acute infections.
Stool suspensions of 0.1 g of sample diluted in 900 μl of phosphate-buffered saline (PBS) were filtered through a 0.45-μm filter (Millipore) and then diluted 1:2 with MEM–0% FBS and were used as the initial inoculum. The inoculum was pretreated with trypsin (Gibco), at a final concentration of 10 μg/ml, at 37°C for 30 min. Cells were grown on a 24-well plate to 80 to 100% confluence and washed twice with MEM–0% FBS before infection with 200 μl of the stool inoculum diluted 1:2 after trypsin pretreatment. Cells were incubated for 1 h at 37°C, and the inoculum was then removed and replaced by 500 μl of MEM–0% FBS supplemented with 0.03% kanamycin, penicillin-streptomycin, and 5 μg/ml trypsin. Cells were maintained at 37°C and 5% CO2 for 7 days, and the medium was changed every other day. After 7 days, cells were freeze-thawed three times, and 100 μl of the cell lysate was used for the next viral passage. Subsequent viral passages (V-P) as acute infections were performed without trypsin pretreatment and with the addition of MEM supplemented with 10% FBS without trypsin in the postinfection medium.
Persistent infections.
A cell lysate of the viral passage 7 (V-P7) of the acute viral passages on Huh-7.5 cells was used to persistently infect the Huh-7AI, A549, and Caco-2 cell lines with MLB1 and MLB2, respectively (Fig. 1B). The viral titer in these selected cell lysates was determined as viral genome copies per milliliter by quantitative reverse transcription-PCR (RT-qPCR) assay and as infectious viruses per milliliter by a 50% tissue culture infective dose (TCID50) assay in Huh-7.5 cells. Briefly, for the TCID50 assay, cells were infected with 10-fold serial dilutions of each sample in quadruplicate, as described above for acute infections. After 7 days, nucleic acids were extracted from 50 μl of supernatant from each well, and an RT-qPCR assay was performed for the detection of viral genomes (see below). The TCID50 was calculated using the Spearman-Karber method, with any detection of viral genome in a well being considered infected. The TCID50 corresponded to approximately 1.4 × 104 genomes for MLB1 and to 1.5 × 103 genomes for MLB2. A multiplicity of infection (MOI) of approximately 1,000 genome copies per cell (0.07 infectious viruses per cell) and 20 genome copies per cell (0.01 infectious viruses per cell) was used for MLB1 and MLB2, respectively. For the first passage, cells were grown on 24-well plates to 80 to 100% confluence and washed twice with MEM–0% FBS. Cell lysates were diluted in MEM–0% FBS to a final volume of 200 μl at the desired MOI and were inoculated to the cells. After a 1-h incubation at 37°C, the inoculum was removed and replaced by 500 μl of MEM–10% FBS. There was no pretreatment or addition of trypsin for the persistent infections. Cells were incubated at 37°C for 4 days before being subcultivated by trypsinization at a split ratio of 1:3 for a subsequent passage. Cells were then subcultivated by trypsinization for serial passages without the addition of viral inoculum. Persistently infected cells were maintained in T75 flasks and subcultivated every 7 to 10 days at a split ratio of 1:3 to 1:6. An aliquot of the supernatant before each subculture was collected to monitor viral titer.
Viral RNA extraction and quantitative reverse transcription-PCR assay.
RNA was extracted from the cell culture supernatants using the NucleoSpin RNA virus kit (Macherey-Nagel), following the manufacturer’s instructions, and RT-qPCR-specific assays for MLB astroviruses were performed as previously published (19). Briefly, the following primers and probe for MLB1 were used: forward primer 4320, 5′-GGTCTTGGAGCYCGAATTC-3′; reverse primer 4387, 5′-CGCTGTTTAATGCGCCAAA-3′; and hydrolysis probe 4349, 5′-[FAM]-TAGRGTTGGTTCAAATCT-[MGBNFQ]-3′ (FAM, 6-carboxyfluorescein). The primers and probe used for MLB2 were as follows: forward primer 3762, 5′-CCGAGCTCTTAGTGATGCTAGCT-3′; reverse primer 3832, 5′-CACCCCTCCAAATGTACTCCAA-3′; and hydrolysis probe 3793, 5′-[VIC]-CGCTTCACTCGGAGAC-[MGBNFQ]-3′. Plasmids containing a 125-bp fragment of the MLB1 (spanning nucleotides 4292 to 4416 from the virus with GenBank accession no. FJ222451) and MLB2 (spanning nucleotides 3724 to 3848 from the virus with GenBank accession no. KT224358) genomes were used as controls for quantification, and RT-qPCR was performed using the Kapa Probe Fast universal one-step RT-qPCR master mix (Kapa Biosystems), following the manufacturer’s instructions, on a CFX96 Touch real-time PCR detection system (Bio-Rad). Fifteen microliters of the RT-qPCR master mix was mixed with 5 μl of extracted RNA. The reaction conditions were as follows: 42°C for 15 min, 95°C for 5 min, and then 40 cycles of 95°C for 3 s, 55°C for 20 s, and 72°C for 10 s. Standard curves were constructed based on 10-fold serial dilutions of the corresponding MLB plasmid analyzed in duplicate.
Multistep growth curve.
The multistep growth curve was created using an MOI of 20 genome copies/cell for both MLB1 and MLB2. Infection was performed on 24-well plates according to the protocol used for acute infection (without the use of trypsin nor the change of medium every other day). At each indicated time point, 50 μl of the supernatant as well as the total cells were collected. RNA extraction of the supernatant was performed using the NucleoSpin RNA virus kit (Macherey-Nagel), following the manufacturer’s instructions, and RNA extraction from cells was performed using the GenElute mammalian total RNA miniprep kit (Sigma-Aldrich), as indicated by the manufacturer’s instructions. The RT-qPCR assay was performed as described above. Samples were quantified in triplicate from a single experiment.
Immunofluorescence.
Indirect immunofluorescence assays were performed using rabbit polyclonal MLB1 capsid peptide (DW60) and MLB2 capsid peptide (DW58) antibodies (kindly provided by David Wang, Washington University School of Medicine) (62) as primary antibodies and secondary antibodies labeled with Alexa 488. DAPI (4′,6-diamidino-2-phenylindole) staining was used to detect nuclei. Briefly, cells were rinsed twice with PBS and fixed with 3% paraformaldehyde in PBS for 15 min at room temperature (RT). Permeabilization was performed for 10 min at RT with 0.5% Triton X-100 in 20 mM glycine-PBS. Cells were then blocked for 60 min at RT in 20 mM glycine-PBS containing 10% bovine serum albumin, followed by incubation with primary antibodies at a 1:1,000 dilution for 60 min at 37°C, and then with the secondary antibodies at a 1:500 dilution for another 60 min at 37°C. Incubation with 1 μg/ml DAPI staining was finally performed for 15 min at RT. Cells were washed twice after each step described, except between the blocking step and the incubation with primary antibodies, and were kept in PBS at 4°C until visualization. Negative controls included cells incubated with preimmune serum, primary or secondary antibodies alone, and fixed cells alone (sample autoimmunofluorescence). Nuclei and viral capsids were visualized under a Leica DMIRB/MZFLIII fluorescence microscope.
Electron microscopy.
Cell culture supernatants were analyzed by transmission electron microscopy after negative staining. A 10-μl sample was applied to a carbon-coated 400-mesh copper grid and was stained with 2% phosphotungstic acid at pH 6.4. The grids were examined under a JEOL 1200 electron microscope.
For ultrathin sections, persistently infected cells and noninfected controls were seeded on a 90-mm sterile dish for cell culture until reaching 80 to 90% confluence. After removing the medium, cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (PB) during 60 min. Cells were then scrapped in 1.5 ml of PB and collected. After 10 min of centrifugation at 1,000 × g, the pellet was suspended in PB and washed in agitation at 4°C for 10 min (×4). Cells were then postfixed with 1% osmium tetroxide and 0.8% potassium hexacyanoferrate in 0.1 M PB for 1 to 2 h at 4°C. After extensive washing with Milli-Q water, sample dehydration was performed with a graded series of acetone (50% to 100%) and blocks were prepared in an Eponate 12 kit. Sections of 55 nm were cut with a Leica UC6 ultramicrotome (Leica Microsystems, Inc.). Observation was performed under a JEOL 1200 electron microscope.
Sequence analysis.
Primer pairs were designed to amplify 10 overlapping amplicons covering the complete genomes (Table 2). Reverse transcription was performed using the SuperScript IV reverse transcriptase (Invitrogen), and cDNA amplification was performed using the Pwo DNA polymerase (Roche), under the following reaction conditions: 70°C for 7 min, 50°C for 25 min, and 80°C for 10 min; 94°C for 4 min; and then 40 cycles of 94°C for 30 s, 50 to 55°C for 40 s, 72°C for 2.15 min, and 72°C for 10 min. Amplicons were purified by gel electrophoresis and Sanger sequenced with the ABI Prism BigDye Terminator cycle sequencing ready reaction kit v3.1 on an ABI Prism 3700 automatic sequencer (Applied Biosystems).
IFN expression analysis.
Intracellular RNA was extracted using the GenElute mammalian total RNA miniprep kit (Sigma-Aldrich), as indicated by the manufacturer’s instructions. The resulting eluate was treated with RQ1 RNase-free DNase (Promega) to remove any trace of genomic DNA. Quantitative reverse transcription-PCR (RT-qPCR) was performed using the manufacturer’s instructions for a KiCqStart one-step probe RT-qPCR assay targeting mRNA of GAPDH, IFN-β, and IFN-λ1 on a CFX96 Touch real-time PCR detection system (Bio-Rad). The primer and probe sequences were as follows: GAPDH forward primer, 5′-GAAGGAAATGAATGGGCAGC-3′; GAPDH reverse primer, 5′-TCTAGGAAAAGCATCACCCG-3′; GAPDH probe, 5′-[6FAM]ACTAACCCTGCGCTCCTGCCTCGAT[OQA]-3′; IFN-β forward primer, 5′-CCTCCGAAACTGAAGATC-3′; IFN-β reverse primer, 5′-GCAGTACATTAGCCATCA-3′; IFN-β probe, 5′-[FAM]TAGCCTGTGCCTCTGGGACT[BHQ]-3′; IFN-λ1 forward primer, 5′-CCACCACAACTGGGAAGG-3′; IFN-λ1 reverse primer, 5′-TTGAGTGACTCTTCCAAGGC-3′; and IFN-λ1 probe, 5′-[FAM]AGCGAGCTTCAAGAAGGCCAGGGAC[OQA]-3′. Fifteen microliters of the RT-qPCR master mix was mixed with 5 μl of extracted RNA. The reaction conditions included 50°C for 20 min, 95°C for 1 min, and then 40 cycles of 95°C for 5 s and 60°C for 35 s. GAPDH mRNA titers were used as an endogenous control to normalize all samples versus the number of cells. All samples were quantified at least in duplicate from two distinct experiments. Positive controls were determined by the transfection of the synthetic analog of double-stranded RNA (dsRNA) polyinosine-poly(C) [poly(I·C)] (InvivoGen) at 1 mg/ml on each cell line, using the X-tremeGENE HD transfection reagent (Roche) and Opti-MEM medium (Gibco). Determination by a RT-qPCR assay of the IFN expression 24 h after transfection was then performed as described above. To define a standard curve, 10-fold serial dilutions of the GAPDH, IFN-β, and IFN-λ1 RNA were analyzed for each cell line transfected with poly(I·C).
Inhibition of MLB HAstV replication with the addition of exogenous IFN.
We used human IFN-β1a and IFN-λ1 (PBL Assay Science) to measure inhibition of MLB HAstV replication. For the acute infection, cells were grown on a 24-well plate and were pretreated with IFN-β1a or IFN-λ1 at a concentration of 1,000 U/ml for 24 h before infection. Cells were inoculated with MLB1 and MLB2 at an MOI of 20 genome copies/cell, as described before (without the use of trypsin). IFN-β1a or IFN-λ1 was added in the postinfection medium at a concentration of 1,000 U/ml. Fifty-microliter aliquots were collected from the supernatant at 4 dpi for RNA extraction and RT-qPCR (see above).
For persistent infection, persistently infected cell cultures were subcultured as described before. IFN-β1a or IFN-λ1 was added in the medium postseeding at a concentration of 1,000 U/ml. Fifty microliters of supernatant was collected before the next subculture, between 4 and 6 days postseeding, and viral RNA was extracted and analyzed by RT-qPCR, as described before. All passages were performed in duplicate.
Statistical analyses.
The Mann-Whitney test and analysis of variance (ANOVA), with additional test of Scheffe when appropriate, were used to compare continuous variables. A P value of <0.05 was considered statistically significant. Statistics were performed by Stata/IC 13.1 (StataCorp, College Station, TX, USA).
Data availability.
Complete genome sequences for the MLB1 strain obtained from the stool specimens were deposited in GenBank (accession numbers MK089434 and MK089435).
ACKNOWLEDGMENTS
We thank Laurent Kaiser and Samuel Cordey from Geneva University Hospitals, Switzerland, for providing the clinical stool specimens positive for MLB2 and the RT-qPCR assay sequence primers, probes, and plasmid controls. We are also grateful to David Wang from the Washington University School of Medicine for providing the MLB-specific antibodies.
This work was supported in part by grant 2017-SGR-244 of the Agència de Gestió d’Ajuts Universitaris i de Recerca and the Biotechnology Reference Network (XRB) program of the Generalitat de Catalunya. Diem-Lan Vu was the recipient of a fellowship from Geneva University Hospitals.
REFERENCES
- 1.Appleton H, Higgins PG. 1975. Letter: viruses and gastroenteritis in infants. Lancet i:1297. [DOI] [PubMed] [Google Scholar]
- 2.Bosch A, Pinto RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarchow-Macdonald AA, Halley S, Chandler D, Gunson R, Shepherd SJ, Parcell BJ. 2015. First report of an astrovirus type 5 gastroenteritis outbreak in a residential elderly care home identified by sequencing. J Clin Virol 73:115–119. doi: 10.1016/j.jcv.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Daniel-Wayman S, Fahle G, Palmore T, Green KY, Prevots DR. 2018. Norovirus, astrovirus, and sapovirus among immunocompromised patients at a tertiary care research hospital. Diagn Microbiol Infect Dis 92:143–146. doi: 10.1016/j.diagmicrobio.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallimore CI, Taylor C, Gennery AR, Cant AJ, Galloway A, Lewis D, Gray JJ. 2005. Use of a heminested reverse transcriptase PCR assay for detection of astrovirus in environmental swabs from an outbreak of gastroenteritis in a pediatric primary immunodeficiency unit. J Clin Microbiol 43:3890–3894. doi: 10.1128/JCM.43.8.3890-3894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubitt WD, Mitchell DK, Carter MJ, Willcocks MM, Holzel H. 1999. Application of electronmicroscopy, enzyme immunoassay, and RT-PCR to monitor an outbreak of astrovirus type 1 in a paediatric bone marrow transplant unit. J Med Virol 57:313–321. doi:. [DOI] [PubMed] [Google Scholar]
- 7.Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, Kang G, Wang D. 2009. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol J 6:161. doi: 10.1186/1743-422X-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan TG, Nordgren J, Ouermi D, Simpore J, Nitiema LW, Deng X, Delwart E. 2014. New astrovirus in human feces from Burkina Faso. J Clin Virol 60:161–164. doi: 10.1016/j.jcv.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medici MC, Tummolo F, Calderaro A, Elia G, Banyai K, De Conto F, Arcangeletti MC, Chezzi C, Buonavoglia C, Martella V. 2014. MLB1 astrovirus in children with gastroenteritis, Italy. Emerg Infect Dis 20:169–170. doi: 10.3201/eid2001.131259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitui MT, Bozdayi G, Matsumoto T, Dalgic B, Nishizono A, Ahmed K. 2013. Complete genome sequence of an MLB2 astrovirus from a Turkish child with diarrhea. Genome Announc 1:e00619-13. doi: 10.1128/genomeA.00619-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, Virgin HW, Anderson LJ, Vinje J, Wang D, Tong S. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol 83:10836–10839. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, Zaidi SZ, Delwart E. 2009. Multiple novel astrovirus species in human stool. J Gen Virol 90:2965–2972. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer CT, Bauer IK, Antonio M, Adeyemi M, Saha D, Oundo JO, Ochieng JB, Omore R, Stine OC, Wang D, Holtz LR. 2015. Prevalence of classic, MLB-clade and VA-clade astroviruses in Kenya and The Gambia. Virol J 12:78. doi: 10.1186/s12985-015-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtz LR, Bauer IK, Rajendran P, Kang G, Wang D. 2011. Astrovirus MLB1 is not associated with diarrhea in a cohort of Indian children. PLoS One 6:e28647. doi: 10.1371/journal.pone.0028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumthip K, Khamrin P, Ushijima H, Maneekarn N. 2018. Molecular epidemiology of classic, MLB and VA astroviruses isolated from <5year-old children with gastroenteritis in Thailand, 2011–2016. Infect Genet Evol 65:373–379. doi: 10.1016/j.meegid.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen S, Hohne M, Marques AM, Beslmuller K, Bock CT, Niendorf S. 2018. Co-circulation of classic and novel astrovirus strains in patients with acute gastroenteritis in Germany. J Infect 76:457–464. doi: 10.1016/j.jinf.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Zaraket H, Abou-El-Hassan H, Kreidieh K, Soudani N, Ali Z, Hammadi M, Reslan L, Ghanem S, Hajar F, Inati A, Rajab M, Fakhouri H, Ghanem B, Baasiri G, Melhem NM, Dbaibo G. 2017. Characterization of astrovirus-associated gastroenteritis in hospitalized children under five years of age. Infect Genet Evol 53:94–99. doi: 10.1016/j.meegid.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Cordey S, Vu DL, Zanella MC, Turin L, Mamin A, Kaiser L. 2017. Novel and classical human astroviruses in stool and cerebrospinal fluid: comprehensive screening in a tertiary care hospital, Switzerland. Emerg Microbes Infect 6:e84. doi: 10.1038/emi.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khamrin P, Thongprachum A, Okitsu S, Hayakawa S, Maneekarn N, Ushijima H. 2016. Multiple astrovirus MLB1, MLB2, VA2 clades, and classic human astrovirus in children with acute gastroenteritis in Japan. J Med Virol 88:356–360. doi: 10.1002/jmv.24337. [DOI] [PubMed] [Google Scholar]
- 21.Xavier Mda P, Carvalho Costa FA, Rocha MS, Andrade Jda S, Diniz FK, Andrade TR, Miagostovich MP, Leite JP, Volotão Ede M. 2015. Surveillance of human astrovirus infection in Brazil: the first report of MLB1 astrovirus. PLoS One 10:e0135687. doi: 10.1371/journal.pone.0135687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, Brooks T, Paine SM, Anderson G, Virasami A, Tong CY, Clark DA, Plagnol V, Jacques TS, Qasim W, Hubank M, Breuer J. 2015. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis 60:881–888. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, Grant P, Samayoa E, Federman S, Miller S, Lunn MP, Gant V, Chiu CY. 2015. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frémond ML, Perot P, Muth E, Cros G, Dumarest M, Mahlaoui N, Seilhean D, Desguerre I, Hebert C, Corre-Catelin N, Neven B, Lecuit M, Blanche S, Picard C, Eloit M. 2015. Next-generation sequencing for diagnosis and tailored therapy: a case report of astrovirus-associated progressive encephalitis. J Pediatric Infect Dis Soc 4:e53–e57. doi: 10.1093/jpids/piv040. [DOI] [PubMed] [Google Scholar]
- 26.Lum SH, Turner A, Guiver M, Bonney D, Martland T, Davies E, Newbould M, Brown J, Morfopoulou S, Breuer J, Wynn R. 2016. An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl Infect Dis 18:960–964. doi: 10.1111/tid.12607. [DOI] [PubMed] [Google Scholar]
- 27.Sato M, Kuroda M, Kasai M, Matsui H, Fukuyama T, Katano H, Tanaka-Taya K. 2016. Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol 78:66–70. doi: 10.1016/j.jcv.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Cordey S, Vu DL, Schibler M, L’Huillier AG, Brito F, Docquier M, Posfay-Barbe KM, Petty TJ, Turin L, Zdobnov EM, Kaiser L. 2016. Astrovirus MLB2, a new gastroenteric virus associated with meningitis and disseminated infection. Emerg Infect Dis 22:846–853. doi: 10.3201/eid2205.151807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vu DL, Bosch A, Pinto RM, Guix S. 2017. Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses 9:33. doi: 10.3390/v9020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuter G, Pankovics P, Boros A. 2018. Nonsuppurative (aseptic) meningoencephalomyelitis associated with neurovirulent astrovirus infections in humans and animals. Clin Microbiol Rev 31:e00040-18. doi: 10.1128/CMR.00040-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin HC, Kao CL, Chang LY, Hsieh YC, Shao PL, Lee PI, Lu CY, Lee CY, Huang LM. 2008. Astrovirus gastroenteritis in children in Taipei. J Formos Med Assoc 107:295–303. doi: 10.1016/S0929-6646(08)60090-X. [DOI] [PubMed] [Google Scholar]
- 32.Tseng WC, Wu FT, Hsiung CA, Chang WC, Wu HS, Wu CY, Lin JS, Yang SC, Hwang KP, Huang YC. 2012. Astrovirus gastroenteritis in hospitalized children of less than 5 years of age in Taiwan, 2009. J Microbiol Immunol Infect 45:311–317. doi: 10.1016/j.jmii.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Amaral MS, Estevam GK, Penatti M, Lafontaine R, Lima IC, Spada PK, Gabbay YB, Matos NB. 2015. The prevalence of norovirus, astrovirus and adenovirus infections among hospitalised children with acute gastroenteritis in Porto Velho, state of Rondonia, western Brazilian Amazon. Mem Inst Oswaldo Cruz 110:215–221. doi: 10.1590/0074-02760140381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda Y, Nakaya S, Takagi M, Ushijima H. 1996. Diagnosis and clinical manifestations of diarrheal virus infections in Maizuru area from 1991 to 1994–especially focused on small round structured viruses. Kansenshogaku Zasshi 70:1092–1097. (In Japanese.) doi: 10.11150/kansenshogakuzasshi1970.70.1092. [DOI] [PubMed] [Google Scholar]
- 35.Cordey S, Zanella MC, Wagner N, Turin L, Kaiser L. 2018. Novel human astroviruses in pediatric respiratory samples: a one-year survey in a Swiss tertiary-care hospital. J Med Virol 90:1775–1778. doi: 10.1002/jmv.25246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padmanabhan A, Hause BM. 2016. Detection and characterization of a novel genotype of porcine astrovirus 4 from nasal swabs from pigs with acute respiratory disease. Arch Virol 161:2575–2579. doi: 10.1007/s00705-016-2937-1. [DOI] [PubMed] [Google Scholar]
- 37.Ng TF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. 2015. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol 89:5340–5349. doi: 10.1128/JVI.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. 2012. Sequence analysis of the human virome in febrile and afebrile children. PLoS One 7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordey S, Hartley M-A, Keitel K, Laubscher F, Brito F, Junier T, Kagoro F, Samaka J, Masimba J, Said Z, Temba H, Mlaganile T, Docquier M, Fellay J, Kaiser L, D’Acremont V. 2018. Detection of novel astroviruses MLB1 and MLB2 in the sera of febrile Tanzanian children. Emerg Microbes Infect 7:1. doi: 10.1038/s41426-018-0025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzales-Gustavson E, Timoneda N, Fernandez-Cassi X, Caballero A, Abril JF, Buti M, Rodriguez-Frias F, Girones R. 2017. Identification of sapovirus GV.2, astrovirus VA3 and novel anelloviruses in serum from patients with acute hepatitis of unknown aetiology. PLoS One 12:e0185911. doi: 10.1371/journal.pone.0185911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gough RE, Collins MS, Borland E, Keymer LF. 1984. Astrovirus-like particles associated with hepatitis in ducklings. Vet Rec 114:279. doi: 10.1136/vr.114.11.279-a. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Cao Y, Wang J, Fu G, Sun M, Zhang L, Meng L, Cui G, Huang Y, Hu X, Su J. 2018. Isolation and characterization of an astrovirus causing fatal visceral gout in domestic goslings. Emerg Microbes Infect 7:71. doi: 10.1038/s41426-018-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlsson EA, Small CT, Freiden P, Feeroz MM, Matsen FA IV, San S, Hasan MK, Wang D, Jones-Engel L, Schultz-Cherry S. 2015. Non-human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog 11:e1005225. doi: 10.1371/journal.ppat.1005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boujon CL, Koch MC, Wuthrich D, Werder S, Jakupovic D, Bruggmann R, Seuberlich T. 2017. Indication of cross-species transmission of astrovirus associated with encephalitis in sheep and cattle. Emerg Infect Dis 23:1604–1608. doi: 10.3201/eid2309.170168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janowski AB, Bauer IK, Holtz LR, Wang D. 2017. Propagation of astrovirus VA1, a neurotropic human astrovirus, in cell culture. J Virol 91:e00740-17. doi: 10.1128/JVI.00740-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sumpter R Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koshikawa N, Hasegawa S, Nagashima Y, Mitsuhashi K, Tsubota Y, Miyata S, Miyagi Y, Yasumitsu H, Miyazaki K. 1998. Expression of trypsin by epithelial cells of various tissues, leukocytes, and neurons in human and mouse. Am J Pathol 153:937–944. doi: 10.1016/S0002-9440(10)65635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boros Á, Albert M, Pankovics P, Biro H, Pesavento PA, Phan TG, Delwart E, Reuter G. 2017. Outbreaks of neuroinvasive astrovirus associated with encephalomyelitis, weakness, and paralysis among weaned pigs, Hungary. Emerg Infect Dis 23:1982–1993. doi: 10.3201/eid2312.170804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brinker JP, Blacklow NR, Herrmann JE. 2000. Human astrovirus isolation and propagation in multiple cell lines. Arch Virol 145:1847–1856. doi: 10.1007/s007050070060. [DOI] [PubMed] [Google Scholar]
- 51.Oldstone MB. 2006. Viral persistence: parameters, mechanisms and future predictions. Virology 344:111–118. doi: 10.1016/j.virol.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 52.Randall RE, Griffin DE. 2017. Within host RNA virus persistence: mechanisms and consequences. Curr Opin Virol 23:35–42. doi: 10.1016/j.coviro.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boldogh I, Albrecht T, Porter DD. 1996. Persistent viral infections In Baron S. (ed), Medical microbiology, 4th ed University of Texas Medical Branch at Galveston, Galveston, TX. [PubMed] [Google Scholar]
- 54.Pinkert S, Klingel K, Lindig V, Dorner A, Zeichhardt H, Spiller OB, Fechner H. 2011. Virus-host coevolution in a persistently coxsackievirus B3-infected cardiomyocyte cell line. J Virol 85:13409–13419. doi: 10.1128/JVI.00621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alidjinou EK, Engelmann I, Bossu J, Villenet C, Figeac M, Romond MB, Sane F, Hober D. 2017. Persistence of coxsackievirus B4 in pancreatic ductal-like cells results in cellular and viral changes. Virulence 8:1229–1244. doi: 10.1080/21505594.2017.1284735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guix S, Pérez-Bosque A, Miró L, Moretó M, Bosch A, Pintó RM. 2015. Type I interferon response is delayed in human astrovirus infections. PLoS One 10:e0123087. doi: 10.1371/journal.pone.0123087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marvin SA, Huerta CT, Sharp B, Freiden P, Cline TD, Schultz-Cherry S. 2016. Type I interferon response limits astrovirus replication and protects against increased barrier permeability in vitro and in vivo. J Virol 90:1988–1996. doi: 10.1128/JVI.02367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. 2015. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee S, Wilen CB, Orvedahl A, McCune BT, Kim KW, Orchard RC, Peterson ST, Nice TJ, Baldridge MT, Virgin HW. 2017. Norovirus cell tropism is determined by combinatorial action of a viral non-structural protein and host cytokine. Cell Host Microbe 22:449–459.e4. doi: 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konduru K, Kaplan GG. 2006. Stable growth of wild-type hepatitis A virus in cell culture. J Virol 80:1352–1360. doi: 10.1128/JVI.80.3.1352-1360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holtz LR, Bauer IK, Jiang H, Belshe R, Freiden P, Schultz-Cherry SL, Wang D. 2014. Seroepidemiology of astrovirus MLB1. Clin Vaccine Immunol 21:908–911. doi: 10.1128/CVI.00100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Complete genome sequences for the MLB1 strain obtained from the stool specimens were deposited in GenBank (accession numbers MK089434 and MK089435).