Chronic viral infections such as with HIV and CMV last a lifetime and can continually antagonize the immune system. Both viruses are associated with higher expression of inflammation markers, and recent evidence suggests that CMV may complicate efforts to deplete HIV reservoirs. Our group and others have shown that CMV shedding is associated with a larger HIV reservoir. Subclinical CMV replication could favor HIV persistence via bystander effects on our immune system. In this study, we collected longitudinal PBMC samples from people starting ART and measured immune changes associated with detectable CMV. We found that when CMV was detectable, CD4+ T cell activation was higher and CD8+ T cell degranulation was lower. Both results may contribute to the slower decay of the size of the reservoir during CMV replication, since activated CD4+ T cells are more vulnerable to HIV infection, while the loss of CD8+ T cell degranulation may impede the proper killing of infected cells.
KEYWORDS: CMV, HIV, T cells, immune dysfunction
ABSTRACT
Most people living with HIV (PLWH) are coinfected with cytomegalovirus (CMV). Subclinical CMV replication is associated with immune dysfunction and with increased HIV DNA in antiretroviral therapy (ART)-naive and -suppressed PLWH. To identify immunological mechanisms by which CMV could favor HIV persistence, we analyzed 181 peripheral blood mononuclear cell (PBMC) samples from 64 PLWH starting ART during early HIV infection with subsequent virologic suppression up to 58 months. In each sample, we measured levels of CMV and Epstein-Barr virus (EBV) DNA by droplet digital PCR (ddPCR). We also measured expression of immunological markers for activation (HLA-DR+ CD38+), cycling (Ki-67+), degranulation (CD107a+), and the immune checkpoint protein PD-1 on CD4+ and CD8+ T cell memory subsets. Significant differences in percentages of lymphocyte markers by CMV/EBV shedding were identified using generalized linear mixed-effects models. Overall, CMV DNA was detected at 60/181 time points. At the time of ART initiation, the presence of detectable CMV DNA was associated with increased CD4+ T cell activation and CD107a expression and with increased CD8+ T cellular cycling and reduced CD107a expression on CD8+ T cells. While some effects disappeared during ART, greater CD4+ T cell activation and reduced CD107a expression on CD8+ T cells persisted when CMV was present (P < 0.01). In contrast, EBV was not associated with any immunological differences. Among the covariates, peak HIV RNA and CD4/CD8 ratio had the most significant effect on the immune system. In conclusion, our study identified immune differences in PLWH with detectable CMV starting early ART, which may represent an additional hurdle for HIV cure efforts.
IMPORTANCE Chronic viral infections such as with HIV and CMV last a lifetime and can continually antagonize the immune system. Both viruses are associated with higher expression of inflammation markers, and recent evidence suggests that CMV may complicate efforts to deplete HIV reservoirs. Our group and others have shown that CMV shedding is associated with a larger HIV reservoir. Subclinical CMV replication could favor HIV persistence via bystander effects on our immune system. In this study, we collected longitudinal PBMC samples from people starting ART and measured immune changes associated with detectable CMV. We found that when CMV was detectable, CD4+ T cell activation was higher and CD8+ T cell degranulation was lower. Both results may contribute to the slower decay of the size of the reservoir during CMV replication, since activated CD4+ T cells are more vulnerable to HIV infection, while the loss of CD8+ T cell degranulation may impede the proper killing of infected cells.
INTRODUCTION
During virally suppressive antiretroviral therapy (ART), HIV persists in a rare and heterogeneous population of CD4+ T cells that can avoid immune surveillance and clinical detection (1). These latently infected T cells are sensitive to antigenic stimulation and cellular activation (2) and can reestablish infection in the absence of ART, thus representing a major obstacle to an HIV cure.
Cytomegalovirus (CMV) is another virus causing chronic infection and has high worldwide prevalence, especially in people living with HIV (PLWH), with seroprevalence estimates ranging from 80% to 100% (3–5). Like other herpesviruses, CMV has a latent phase, during which CMV remains asymptomatic and minimally detectable, and a lytic phase, in which the virus replicates and promotes inflammation (reviewed in reference 6). CMV generally remains asymptomatic in people with healthy immune systems; nevertheless, subclinical replication in blood and at various mucosal sites may contribute to chronic inflammation, especially in PLWH (7–9). Over time, CMV maintains an outsized proportion of CMV-specific T cells, and CMV replication is itself favored by inflammatory stimuli (4, 10, 11). Over millions of years of evolution, CMV has evolved a complex arsenal of proinflammatory and proproliferative mediators (12–16) to manipulate the human immune system for its own advantage and possibly also favoring persistence of other coinfecting viruses, such as HIV (16–18) (reviewed in reference 19). We previously demonstrated in a cross-sectional study of PLWH who started ART during chronic infection that asymptomatic shedding of CMV in the genital tract is frequent and is associated with increased systemic T cell immune activation, cellular cycling, and PD-1 expression, as well as with higher levels of HIV DNA in peripheral CD4+ T cells (20, 21, 28). This study had several limitations which may have confounded the analysis, including the lack of longitudinal data and missing information about timing of HIV infection, ART history, and dynamics of viral suppression.
Subsequently, we analyzed longitudinal samples from 107 men starting ART during the earliest phase of their HIV infections (median, 3 months since estimated date of infection) over a median follow-up period of 19 months, and we observed an association between presence of CMV and Epstein-Barr virus (EBV) DNA in peripheral blood mononuclear cells (PBMCs) and slower decay rate of the HIV DNA reservoirs during ART (22). Given these previous findings (22), we need a better understanding of the immunological mechanisms that help HIV persist during CMV replication. In this study, we developed a statistical model to analyze multiparameter flow cytometry and clinical data in the same longitudinal cohort of people starting ART after recent HIV infection to identify possible immunological mechanisms connecting CMV and HIV persistence. We examined the predicted effects of CMV and EBV DNA, peak HIV RNA, CD4/CD8 T cell ratio, and time on immunological functions such as cellular activation, cycling, degranulation, PD-1 expression, and regulatory T cells.
RESULTS
Cohort description.
Study participants (n = 64) were all men who have sex with men (MSM) diagnosed with recent HIV infection and monitored for up to 58 months (median, 28; interquartile range [IQR], 14 to 45 [Table 1]). Participants were started on ART after a median of 3 months (IQR, 1.3 to 5.9) from estimated date of infection (EDI), and half of the participants achieved viral suppression within 3.5 months (IQR, 1.8 to 5.5) of starting ART. The cohort was primarily white, non-Hispanic (71.9%) men who were infected less than 70 days from EDI (79.7%) at the time of enrollment. At study entry, participants were a median of 34.5 years old (IQR, 29 to 42) and had a median baseline CD4+ T cell count of 690 cells/μl (IQR, 519 to 858), median CD4/CD8 ratio of 0.75 (IQR, 0.58 to 1.07), and median peak HIV RNA of 5.61 log10 copies/ml (IQR, 5.05 to 6.27). Among the 64 participants, 40 (62.5%) had at least one time point with detectable CMV, and 33% (n = 60) of all time points exhibited detectable levels of CMV DNA. We detected EBV in 85.1% of samples, with a median level of EBV DNA of 1.33 log10 copies/106 cells (IQR, 0.85 to 0.89). In contrast to the case with CMV, which was only sporadically detectable, most samples had detectable EBV DNA, so we analyzed EBV as a continuous variable. A median of 4 time points was available for each participant (IQR, 3 to 5).
TABLE 1.
Demographics and baseline clinical metrics of study participantsa
| Characteristic | Value for subjects (n = 64) |
|---|---|
| MSM, no. (%) | 64 (100) |
| Race/ethnicity, no. (%) | |
| White (non-Hispanic) | 46 (71.9) |
| Hispanic | 8 (12.5) |
| Other/mixed | 10 (15.6) |
| Age (yrs), median (IQR) | 34.5 (29–42) |
| Baseline CD4 cell count (cells/μl), median (IQR) | 690 (519–858) |
| Baseline CD4/CD8 ratio, median (IQR) | 0.75 (0.58–1.07) |
| Presence of detectable CMV during follow-up, no. (%) | 60/181 (33.1) |
| EBV DNA (log10 copies/106 PBMCs), median (IQR) | 1.33 (0.85–1.89) |
| Peak HIV RNA (log10 copies/ml), median (IQR) | 5.61 (5.05–6.27) |
| Months from EDI to ART start, median (IQR) | 3.0 (1.3–5.9) |
| Months of follow-up, median (IQR) | 28 (14–45) |
| Months from ART start to viral suppression, median (IQR) | 3.5 (1.8–5.5) |
| ART regimen, no. (%) | |
| RT + PI | 42 (65.6) |
| Triple RT | 28 (43.8) |
| RT + PI + MVC | 6 (9.4) |
| RT + INSTI | 4 (6.2) |
ART, antiretroviral therapy; CMV, cytomegalovirus; EBV, Epstein-Barr virus; MSM, men who have sex with men; EDI, estimated date of infection; HIV, human immunodeficiency virus; IQR, interquartile range; RT, reverse transcriptase inhibitor backbone; PI, protease inhibitor; MVC, maraviroc; INSTI, integrase strand transfer inhibitor.
Immunologic trends among memory compartments.
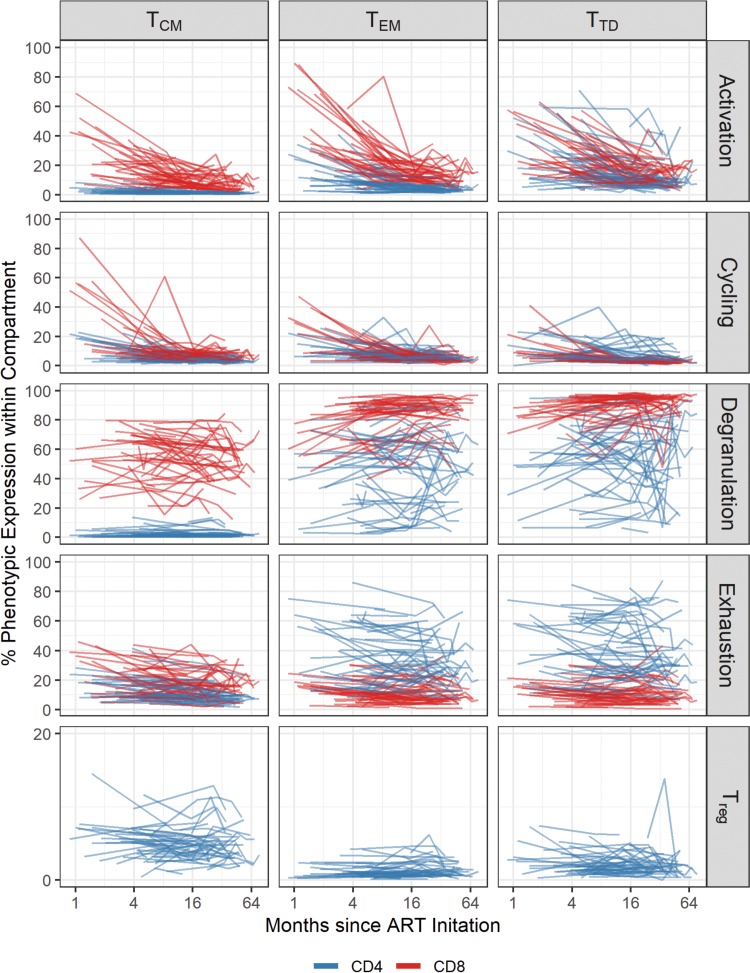
Figure 1 displays the longitudinal analysis of the frequency of the indicated immune markers by memory compartment.
FIG 1.
Longitudinal analysis of the expression of different immunological markers during ART. Levels of each marker in each CD4 and CD8 T cell subset were log transformed and applied to the x axis (time on ART). Each line indicates one subject, and blue and red lines indicate CD4 and CD8 T cells, respectively. Memory subsets analyzed included central memory (TCM), effector memory (TEM), and terminally differentiated (TTD) T cells. We included the longitudinal analysis of levels of activation (HLA-DR+ CD38+), cellular cycling (Ki-67+), degranulation (CD107a+), and PD-1+ in CD4 T cell subsets, as well as the frequency of T regulatory cells (Treg [Fox-P3+ CD25+]).
The level of activation, as measured by dual expression of HLA-DR and CD38, declined over time when all CD4+ memory subsets were combined for analysis. Among individual CD4 T cell subsets, HLA-DR+ CD38+ expression declined most significantly in the central memory (TCM) subset (β = −0.31; P < 0.01). Expression of the cycling marker Ki-67 in CD4+ TCM and effector memory (TEM) cells decreased over time (β = −0.26; P < 0.01); the decay was not significantly different between the two subsets (P = 0.85). In contrast, terminally differentiated (TTD) cells decreased more slowly over time (β = 0.13; P = 0.01) than did TCM cells. Degranulation (expression of CD107a marker) did not change significantly over time in any compartment among CD4+ T cells (P > 0.08 for all). The frequency of cells expressing PD-1 among TCM cells also decreased over time (β = −0.13; P < 0.01). PD-1 expression decreased more slowly in both TEM (β = 0.09; P = 0.01) and TTD (β = 0.10; P < 0.01) cells. While the frequency of T regulatory (Treg) cells with a TCM and TTD phenotype did not significantly change over time (P > 0.4 for both), the frequency of Treg TEM cells increased significantly more than that of Treg TCM cells over time (β = 0.11; P = 0.03).
For CD8+ T cells, the frequency of activated cells, measured as the coexpression of HLA-DR and CD38, decreased over time in TCM cells (β = −0.35; P < 0.01) and declined similarly to that in TEM and TTD cells (P > 0.2 for both). Cellular cycling, measured as the frequency of Ki-67+ cells, declined in TCM cells over time (β = −0.27; P < 0.01), and this decline was similar in TEM and TTD cells (P > 0.2 for both). Degranulation did not significantly change in TCM cells (β = −0.06; P = 0.07), but degranulation increased over time in both TEM (β = 0.18; P < 0.01) and TTD (β = 0.12; P < 0.01) cells. PD-1 expression did not change significantly over time in any compartment among CD8+ T cells (P > 0.1 for all).
Associations between CMV DNA, other covariates, and immunological outcomes.
CMV and relevant covariates had congruent effects among different memory T cells, in that there were no significant interactions between the predictor and subsets. Individual model results for CMV and EBV are presented in Table 2. Individual model results for covariates are presented in the text below, and final adjusted model results are presented in Tables 3 and 4, including significant covariates.
TABLE 2.
Individual model CMV and EBV effect estimates, confidence intervals, and P values for model predictionsa
| Cell type | Phenotype | Estimate (95% confidence interval) |
||
|---|---|---|---|---|
| CMV | CMV × time | EBV | ||
| CD4 | Activation | 0.15 (0.03, 0.26)* | 0.05 (−0.05, 0.15) | |
| Cycling | 0.10 (−0.06, 0.25) | −0.06 (−0.19, 0.06) | ||
| Degranulation | 0.51 (0.11, 0.90)* | −0.13 (−0.22, −0.04)** | 0.07 (−0.03, 0.18) | |
| PD-1 | 0.25 (−0.07, 0.57) | −0.09 (−0.16, −0.02)* | −0.06 (−0.15, 0.03) | |
| Treg | −0.12 (−0.27, 0.03) | −0.07 (−0.20, 0.06) | ||
| CD8 | Activation | 0.04 (−0.10, 0.18) | 0.09 (−0.02, 0.21) | |
| Cycling | 0.98 (0.46, 1.50)** | −0.18 (−0.30, −0.07)** | −0.01 (−0.12, 0.10) | |
| Degranulation | −0.18 (−0.30, −0.06)** | −0.02 (−0.13, 0.08) | ||
| PD-1 | 0.34 (−0.04, 0.72) | −0.11 (−0.20, −0.03)* | −0.05 (−0.15, 0.05) | |
Value for CMV-time interaction (CMV × time) is included if significant. Values for EBV-time interaction were not significant. *, P < 0.05; **, P < 0.01.
TABLE 3.
Adjusted multivariate effects of CMV, EBV, and clinical covariates on CD4+ T cellsa
| Predictor | Multivariate regression coefficient (95% CI) |
||||
|---|---|---|---|---|---|
| Activation | Cycling | Degranulation | Exhaustion | Treg | |
| Detectable CMV | 0.13 (0.02, 0.24)* | 0.51 (0.11, 0.91)* | 0.22 (−0.10, 0.54) | ||
| CMV × time | −0.13 (−0.22, −0.04)** | −0.08 (−0.15, −0.01)* | |||
| EBV DNA (log10 copies/106 cells) | |||||
| Peak HIV RNA (log10 copies/ml) | 0.42 (0.10, 0.74)* | −0.24 (−0.41, −0.07)** | |||
| Peak HIV RNA × time | −0.13 (−0.20, −0.06)** | ||||
| CD4/CD8 ratio | −0.23 (−0.42, −0.04)* | −0.25 (−0.63, 0.13) | 0.38 (0.16, 0.60)** | ||
| CD4/CD8 ratio × time | 0.10 (0.03, 0.17)* | ||||
| Early ART initiation (<3 mo) | 0.39 (0.04, 0.74)* | ||||
| Older age (>45 yrs) | |||||
| Time on ART (log2 mo) | −0.29 (−0.39, −0.19)** | −0.26 (−0.36, −0.16)** | 0.05 (−0.01, 0.11) | −0.09 (−0.16, −0.02)* | −0.02 (−0.08, 0.04) |
| Memory T cell compartment | |||||
| TCM | |||||
| TTM | NA | NA | NA | 0.08 (−0.26, 0.42) | NA |
| TEM | 0.28 (−0.25, 0.81) | −0.03 (−0.51, 0.45) | −0.35 (−0.69, −0.01)* | ||
| TTD | 0.01 (−0.52, 0.54) | −0.51 (−0.99, −0.03)* | −0.35 (−0.69, −0.01)* | ||
| TSCM | −0.73 (−1.26, −0.20)** | NA | NA | NA | NA |
| Compartment × time | |||||
| TCM | |||||
| TTM | NA | NA | NA | −0.02 (−0.09, 0.05) | NA |
| TEM | −0.08 (−0.19, 0.03) | 0.01 (−0.09, 0.11) | 0.08 (0.01, 0.15)* | ||
| TTD | 0.01 (−0.10, 0.12) | 0.14 (0.04, 0.24)** | 0.09 (0.02, 0.16)* | ||
| TSCM | 0.20 (0.09, 0.31)** | NA | NA | NA | NA |
CI, confidence interval; TSCM, stem cell memory T cells; TCM, central memory T cells; TTM, transitional memory T cells; TEM, effector memory T cells; TTD, terminally differentiated T cells; NA, not available. TSCM cells were gated only in the activation panel, and TTM CD4 T cells were gated only in the exhaustion panel. TCM is used as a reference category for memory compartment data in our model. *, P < 0.05; **, P < 0.01.
TABLE 4.
Adjusted multivariate effects of CMV, EBV, and clinical covariates on CD8+ T cellsa
| Predictor | Multivariate regression coefficient (95% CI) |
|||
|---|---|---|---|---|
| Activation | Cycling | Degranulation | Exhaustion | |
| Detectable CMV | 0.86 (0.34, 1.38)** | −0.18 (−0.30, −0.06)** | 0.24 (−0.12, 0.60) | |
| CMV × time | −0.16 (−0.28, −0.04)** | −0.09 (−0.17, −0.01)* | ||
| EBV DNA (log10 copies/106 cells) | ||||
| Peak HIV RNA (log10 copies/ml) | −0.22 (−0.43, −0.01)* | |||
| Peak HIV RNA × time | ||||
| CD4/CD8 ratio | −2.00 (−2.49, −1.51)** | −1.32 (−1.90, −0.74)** | −0.61 (−1.02, −0.20)** | |
| Ratio × time | 0.35 (0.25, 0.45)** | 0.31 (0.18, 0.44)** | 0.20 (0.12, 0.28)** | |
| Early ART initiation (<3 mo) | ||||
| Older age (>45 yrs) | 0.34 (0.06, 0.62)* | |||
| Time on ART (log2 mo) | −0.22 (−0.30, −0.14)** | −0.19 (−0.30, −0.08)** | −0.07 (−0.14, 0.00) | −0.02 (−0.09, 0.05) |
| Memory compartment | ||||
| TCM | ||||
| TEM | −0.66 (−0.96, −0.36)** | |||
| TTD | −0.48 (−0.78, −0.18)** | |||
| Memory × time | ||||
| TCM | ||||
| TEM | 0.18 (0.11, 0.25)** | |||
| TTD | 0.12 (0.05, 0.19)** | |||
TCM is used as a reference category for memory compartment data in our model. *, P < 0.05; **, P < 0.01.
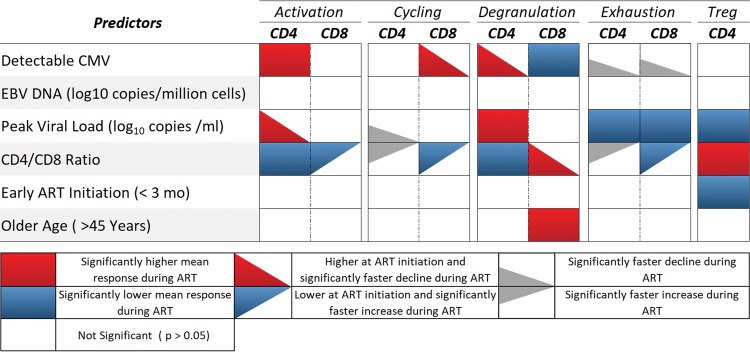
(i) Activation (HLA-DR+ CD38+). In the individual longitudinal models of CD4+ activation by predictor, CD4+ T cells expressed significantly higher levels of activation markers in the presence of detectable CMV before and during ART (β = 0.15; P = 0.01 [Table 2 and Fig. 2]). Further, participants with higher peak HIV RNA before ART had significantly higher levels of CD4+ T cell activation at the time of ART initiation (β = 0.55; P < 0.01), but the frequency of activated cells decreased significantly faster during ART (β = −0.13; P < 0.01). Higher CD4/CD8 ratios were associated with lower frequencies of CD4+ T cell activation during ART (β = −0.29; P < 0.01). We did not detect significant differences in the levels of CD4+ T cell activation based on levels of EBV DNA (P = 0.30), early ART initiation (P = 0.06), or age (P = 0.06). In the multivariate model, all significant effects persisted, along with early ART initiation (β = 0.39; P = 0.04; early ART not significantly different in the individual model) (Table 3).
FIG 2.
Summary of individual longitudinal models’ effects of CMV on immune responses during ART. CMV, cytomegalovirus; activation, HLA-DR+ CD38+; cellular cycling, Ki-67+; degranulation, CD107a+, immune checkpoint protein PD-1+, and Treg (Fox-P3+ CD25+).
For the individual longitudinal models of CD8+ activation, only the CD4/CD8 ratio was significantly associated with the levels of activation. Specifically, higher CD4/CD8 ratios were significantly associated with lower frequencies of activation markers in CD8+ T cells at ART initiation (β = −2.00; P < 0.01) but had a positive effect on slope of activation compared to lower ratios (β = 0.35; P < 0.01). CD8+ T cells did not express significantly different levels of activation in the presence of detectable CMV (P = 0.55), levels of EBV DNA (P = 0.11), peak HIV RNA (P = 0.59), early ART initiation (P = 0.19), or older age (P = 0.55).
(ii) Cycling (Ki-67+). CD4+ T cells did not express significantly different levels of Ki-67 in the presence of detectable CMV DNA (P = 0.21) or EBV DNA (P = 0.33), early ART initiation (P = 0.80), or age (P = 0.57). Similarly, the levels of CD4+ cycling were not significantly different at ART initiation in participants with higher viral loads (β = 0.36; P = 0.06) or CD4/CD8 ratios (β = −0.44; P = 0.14). During ART, the frequency of cycling CD4 declined faster among participants with higher levels of peak HIV RNA (β = −0.08; P = 0.04) but slower among participants with higher CD4/CD8 ratios at presentation (β = 0.13; P = 0.04). In the multivariate model, none of the covariates were found to be significant when included with memory compartment and memory compartment by time interactions.
For the individual longitudinal models of CD8+ T cell cycling, CD8+ T cells expressed significantly higher levels of cycling at ART initiation in the presence of detectable CMV DNA (β = 0.98; P < 0.01), but levels of cycling declined more quickly over time (β = −0.18; P < 0.01) (Table 2). CD8 cycling was not found to be significantly associated with the presence of EBV DNA (P = 0.85) or peak HIV RNA (P = 0.44). Higher CD4/CD8 ratios were significantly associated with lower levels of cycling at ART initiation (β = −1.52; P < 0.01) and slower decline in cycling during ART (β = 0.35; P < 0.01). Early ART initiation (P = 0.76) and age (P = 0.60) had no significant association with cycling. All significant effects persisted in the multivariate models (Table 4).
(iii) Degranulation (CD107a+). CD4+ T cells expressed higher levels of degranulation at ART initiation in the presence of CMV DNA (β = 0.51; P = 0.01), but these increased levels of degranulation declined significantly faster over time in the presence of CMV (β = −0.13; P < 0.01). There was no significant difference in degranulation expression in the presence of EBV DNA (P = 0.18). Participants with higher levels of peak HIV RNA (β = −0.21; P = 0.03) and CD4/CD8 ratios at presentation (β = −0.21; P = 0.05) were more likely to exhibit lower levels of CD4 degranulation during ART, whereas there was no difference in the frequency of CD4 cell expression of the degranulation marker CD107a for those who started ART early (P = 0.65) or were older at presentation (P = 0.07). In the multivariate model, the effect of CMV persisted (Table 3).
In contrast, CD8+ T cells expressed lower levels of degranulation in the presence of detectable CMV (β = −0.18; P < 0.01). The expression levels of CD107a were not found to have a significant association with EBV DNA (P = 0.65) or peak HIV RNA (P = 0.30). Participants with higher CD4/CD8 ratios at presentation were more likely to demonstrate higher levels of CD8 degranulation at ART initiation (β = 0.57; P = 0.01), and their levels of degranulation declined more quickly during ART than in participants with lower CD4/CD8 ratios (β = −0.10; P = 0.02). Early ART initiation was not found to be significantly associated with the frequency of CD8 cells with degranulation markers (P = 0.48), but older participants were found to exhibit higher levels of degranulation during ART (β = 0.35; P = 0.01). In the multivariate model, the effects of CMV and age were also significant (Table 4).
(iv) PD-1. Samples with detectable levels of CMV DNA did not exhibit levels of PD-1 expression in CD4+ T cells significantly different from those with undetectable levels (β = 0.25; P = 0.13), but importantly, the presence of detectable CMV coincided with faster decline in the expression of PD-1 (β = −0.09; P = 0.01). Expression of PD-1 by CD4 T cells was not significantly associated with EBV (P = 0.18). Participants with higher levels of peak HIV RNA demonstrated lower mean levels of PD-1 expression during ART (β = −0.20; P = 0.04), and those with higher CD4/CD8 ratios at presentation displayed no difference in PD-1 expression in CD4 T cells at ART initiation (β = −0.23; P = 0.22), but PD-1 expression declined more slowly (β = 0.10; P < 0.01). CD4+ T cell expression of PD-1 was not associated with either early ART initiation (P = 0.21) or age (P = 0.45). The effects of CMV and the CD4/CD8 ratios persisted in the multivariate model (Table 3).
Similarly, the presence of detectable CMV did not have a significant effect on the mean level of PD-1 expression by CD8+ T cells at ART initiation (β = 0.34; P = 0.08), but PD-1 expression declined more quickly in the presence of CMV (β = −0.11; P = 0.01). Participants with higher levels of peak HIV RNA displayed significantly lower levels of PD-1 expression in CD8+ T cells during ART (β = −0.22; P = 0.02). Participants with higher CD4/CD8 ratios were more likely to have lower expression of PD-1 on CD8+ T cells at ART initiation (β = −0.57; P < 0.01) but displayed significant increases in PD-1 during ART (β = 0.21; P < 0.01). Levels of EBV, early ART initiation, and age did not have a significant effect on PD-1 expression by CD8+ T cells (P = 0.32, P = 0.16, and P = 0.45, respectively). In the multivariate model, all the significant effects present in the individual models endured (Table 4).
(v) Treg (CD4+ Fox-P3+ CD25+). The frequency of the Treg cells was not significantly associated with the presence of detectable CMV (P = 0.12), EBV DNA (P = 0.28), or age (P = 0.34). Participants with higher levels of peak HIV RNA before ART exhibited lower mean levels of CD4+ Treg cells during ART (β = −0.23; P = 0.01), and those who presented with higher CD4/CD8 ratios had higher frequencies of Treg cells during ART (β = 0.39; P < 0.01). The levels of Treg cells were found to be lower in participants with early ART initiation (P = 0.05). In the multivariate model, the effects of peak HIV RNA and CD4/CD8 ratio persisted (Table 3).
DISCUSSION
Several studies demonstrate links between reservoir size, chronic inflammation, and earlier onset of age-related diseases, despite long-term, successful viral suppression (reviewed in reference 23). Similarly, persistent replication by the chronic viral infection CMV, a herpesvirus, is associated with inflammation, age-related disease, and, in PLWH, larger HIV reservoirs (7, 22, 24, 25, 28).
Most PLWH have persistent, subclinical CMV replication (9), and this may antagonize the immune system in ways that favor HIV persistence (19). To identify potential immunological mechanisms by which CMV could favor HIV persistence, in this study, we measured cell-associated CMV DNA in a prospective longitudinal clinical cohort. We analyzed changes in immunological phenotypes of memory T cells over time and identified several immune changes that were associated with detectable CMV DNA. We included an analysis of cell-associated EBV DNA, because similarly to CMV, it is a herpesvirus with high prevalence among PLWH, with frequent reactivation (26). Our inclusion of EBV helps distinguish between general immune effects of a chronic viral infection and those that may be unique to CMV.
In contrast to CMV, our models did not detect any immune changes associated with the detection of cell-associated EBV DNA. This suggests that the higher CD4+ T cell activation and lower CD8+ T cell degranulation observed in the presence of detectable CMV DNA might be attributable to CMV-specific factors. Our comparison of CMV with a similarly endemic herpesvirus supports a model in which CMV-related inflammation mediates HIV persistence, but ultimately, clinical trials of specific CMV drugs will be required to determine the specific impact of CMV replication on immune activation and T cell phenotypes.
Overall, most immunological markers declined over time after initiation of early ART, with the exceptions of the degranulation marker CD107a and the frequency of Treg cells, which were more static (Fig. 1). This may be due to the drop in HIV load accompanying ART initiation, which presumably removes much of the antigen burden driving high levels of activated, cycling, or PD-1-expressing T cells. Consistent with this explanation, HIV load was initially associated with increased CD4+ T cell activation, but over time the effect of the viral load diminished, as samples reached undetectable viral loads at later time points.
This study identified several immunological differences associated with the presence of detectable CMV DNA during early ART. First, expression of the activation markers HLA-DR and CD38 was consistently higher on CD4+ T cells when cell-associated CMV DNA was detected in PBMCs. While this has been previously shown in people starting ART during chronic HIV infection (24), our findings suggest that starting ART early does not prevent CMV-associated immune activation in the CD4+ T cell compartment. Activated CD4+ T cells are more susceptible to HIV infection and may therefore be related to the slower decay of HIV DNA-containing cells observed in people with detectable CMV DNA (22). Second, we found that CD8+ T cells had impaired expression of the early degranulation marker CD107a when CMV DNA was present. In contrast, higher age was associated with higher CD8+ T cell degranulation. CD8+ T cells play an indispensable role in immunity against virus-infected cells (including HIV), by degranulating and thereby killing target cells (27). Although it is unclear whether this was a cause or a consequence of CMV, the potential consequences of impaired CD8 CD107a expression on the HIV reservoir size merit further investigation with more sophisticated functional studies on virus-specific CD8+ T cells.
Third, we found that detectable CMV DNA was associated with increased cycling of CD8+ T cells at ART start, as measured by expression of Ki-67. However, in samples with detectable CMV there was a more rapid decay of Ki-67 expression over time, such that the predicted effects of CMV on CD8+ T cell cycling waned over time. Fourth, we found that expression of the immune checkpoint protein PD-1 decayed significantly faster in both CD4+ and CD8+ T cells when CMV DNA was detected. Although our previous study with virally suppressed PLWH showed that PD-1 expression was increased on CD4+ T cells during seminal CMV shedding (28), that was a cross-sectional study that did not consider the longitudinal effects of ART. The differences in PD-1 expression between these two studies might also reflect the different methodologies used to measure CMV replication. Seminal CMV shedding and the associated mucosal inflammation might elicit a more robust immune response, for which PD-1-expressing memory T cells are more abundant. Interestingly, peak HIV RNA was negatively associated with expression of PD-1, which suggests that PD-1 might reflect acute cellular activation rather than exhaustion during early HIV infection.
In addition to the effects of CMV and EBV discussed above, our analysis also revealed immune changes associated with several other covariates. Specifically, a high CD4/CD8 ratio—a potentially important indicator of overall immune system integrity—was associated with higher frequencies of Treg cells. Regulatory T cells act as a check on our immune system by stifling activation and inflammation through a variety of mechanisms (reviewed in reference 29). Consistent with this paradigm, a higher CD4/CD8 ratio was also associated with lower CD4+ T cell activation and cycling and lower CD8+ T cell expression of PD-1. Conversely, high peak viral load was negatively associated with the frequencies of Treg cells and positively associated with CD4+ T cell degranulation. At ART start, peak viral load was also associated with increased CD4+ T cell activation, but the effect diminished over time. In addition, we analyzed whether ART initiation within 3 months of EDI (median) affected our immune outcomes. Interestingly, earlier ART initiation was associated with higher CD4+ T cell activation and lower Treg cell frequencies. Our study was not randomized, so it is possible that people with more highly activated immune systems are more symptomatic and present earlier.
While some of the immune changes persisted during ART, many of the effects of CMV or covariates waned over time, likely reflecting the restoration of immune homeostasis accompanying the suppression of HIV replication in early-treated individuals. Alternatively, CMV replication may become hindered during the immune restoration that accompanies viral suppression of HIV, thus minimizing CMV’s effect on immune phenotypes.
While we are confident in our model predictions, our study has several limitations. First, our cohort did not have any participants who were CMV or HIV seronegative as a control. Our focus was on immunological changes associated with ongoing subclinical CMV replication during early ART, and people with HIV but no CMV coinfection are rare, especially among MSM (30). For this reason, we compared immune phenotypes within and between participants depending on whether cellular CMV DNA was detectable. Second, this was an observational study, so inferences about the causal relationships between CMV replication should be made with caution. Future interventional studies will be required to determine whether CMV replication is a cause or a consequence of our observed immune phenotypes. Third, our participants were all MSM recruited from one location; findings may differ in people from other regions or with other transmission routes or risk factors. Fourth, measuring cell-associated CMV DNA in PBMCs is an imperfect way to evaluate whether CMV replication is ongoing. The presence of CMV DNA in circulating blood cells (usually monocytes) may reflect low-level viral replication but may also simply reflect latent CMV infection and does not necessarily reflect CMV replication in tissues (e.g., salivary glands and the genital tract). Unfortunately, mucosal samples were not available for this study. Future studies should address whether similar associations are seen with CMV shedding at mucosal sites, particularly in the gut, which is an important site of HIV persistence. Fifth, our cohort of people starting ART has a median age under 35 years old, and for this reason we urge caution in extrapolating our findings to older populations. Our data support other studies showing that ART can dramatically diminish immune dysregulation, and we would expect that increased immune burden of CMV accompanying age would exacerbate our observed phenotypes. Sixth, we did not address the antigen specificity of our PBMC samples, so it is unclear whether the immune changes we observed were due to a global defect on all T cell populations or enrichment for specific antigen-specific populations (i.e., HIV-specific or CMV-specific T cells). Lastly, participants in this study were on a variety of different ART regimens and occasionally changed ART regimens throughout the period of sample collection. However, this could be considered a strength of these findings, because the studied population more closely mirrors real-life populations of people living with HIV and yet still revealed the associations we described between CMV status and immune activation markers.
Despite these limitations, this is a large study investigating in detail the immunological effects of CMV in PLWH who are starting ART. Although virologic suppression of HIV with ART can greatly reduce inflammation, several biomarkers of inflammation can remain elevated (31), and the interaction between reservoir size and increased immune activation remains poorly defined. Based on our findings, CMV replication should be considered in future studies dissecting the relationship between HIV reservoir size and immune activation. For example, in a recent longitudinal study of long-term virally suppressed PLWH, correlations between HIV reservoir size and T cell activation were not detected (32). However, immune activation associated with subclinical CMV replication may have confounded the results.
In summary, CMV likely plays a role in shaping the HIV reservoir, and the data presented here help clarify the interconnectedness between ongoing viral replication and chronic immune dysfunction. Our findings support a model in which CMV can promote HIV reservoir size by increasing the vulnerability of CD4+ T cells via cellular activation and by limiting the antiviral effector functions of CD8+ T cells. Our work underscores the need for future research into the effects of latency-reversing agents on coinfecting viruses, to improve clearance of the HIV reservoir. In examining the pathways by which CMV promotes HIV persistence, insight can be gained into the natural processes that maintain the HIV reservoir and ways to block them.
MATERIALS AND METHODS
Study participants.
Men who have sex with men (MSM) starting ART after recent HIV infection were recruited from the San Diego Primary Infection Consortium. A total of 107 people enrolled in a previous reservoir study, providing a total of 515 longitudinal blood samples (22). Of the 107 participants, we selected 64 who started ART within 1 year of estimated date of infection (EDI), had the longest follow-up, and had enough frozen PBMCs available at baseline and after ART initiation for further analysis by flow cytometry. A summary of participant characteristics is shown in Table 1. There were no significant differences in CD4+ T cell count, peak HIV RNA, or age between the original cohort and the 64 selected for this study. All study participants were at least 18 years of age and provided written informed consent. Our study was approved by The University of California, San Diego Office of Human Research Protections Program.
Sample storage.
PBMCs were isolated from whole blood using a density gradient medium (Lymphoprep; Stemcell Technologies) per the manufacturer’s instructions and cryopreserved at −150°C in 95% fetal bovine serum (FBS) plus 5% dimethyl sulfoxide (DMSO) within 24 h of blood collection.
Detection of EBV and CMV.
DNA was extracted from 5 million PBMCs for each time point using an AllPrep DNA/RNA minikit (Qiagen, CA). Total CMV and EBV DNAs were quantified by droplet digital PCR (ddPCR) as previously described (22, 33). Copy numbers were calculated as the means of replicate PCR measurements and normalized to cell input, determined by RPP30 (34).
Flow cytometry.
Frozen cells were thawed in a 37°C water bath, washed, resuspended in phosphate-buffered saline (PBS; Corning), and counted using a BD Accuri C6 Plus (BD Biosciences). Live cells were then stained using LIVE/DEAD aqua (Invitrogen) in PBS, per the manufacturer’s instructions. Next, the cells were washed and stained for extracellular markers for 30 min at 4°C in the dark in PBS plus 2% FBS plus 0.09% sodium azide. All antibodies, fluorophores, and concentrations used for flow cytometry are listed in Table 5. After extracellular staining, cells were fixed and permeabilized (BD fixation/permeabilization kit; BD Biosciences) for staining of intracellular markers (Table 5). Cells were analyzed on a BD FACS Canto analyzer (BD Biosciences). Compensation and gating were performed using FlowJo (version 10). Representative plots and gating for memory subsets and phenotypic markers are shown in Table 6 and Fig. 3.
TABLE 5.
Flow cytometry antibodies used in this study
| Antibody | Vendor | Catalog no. | Clone | Fluorochromea |
|---|---|---|---|---|
| Ki-67 | BD Biosciences | 556026 | B56 | FITC |
| CD45RA | BD Biosciences | 555488 | HI100 | FITC |
| HLA-DR | BD Biosciences | 555811 | G46-6 | FITC |
| CD57 | BD Biosciences | 555619 | NK-1 | FITC |
| CD45RA | BD Biosciences | 555489 | HI100 | PE |
| CD28 | BD Biosciences | 348047 | L293 | PE |
| CD107a | BD Biosciences | 562628 | H4A3 | PE-CF594 |
| CCR7 (CD197) | BD Biosciences | 562381 | 150503 | PE CF594 |
| CD4 | BD Biosciences | 341654 | SK3 (Leu3a) | PerCP-Cy5.5 |
| CD4 | BD Biosciences | 562659 | RPA-T4 | BV605 |
| CD27 | BD Biosciences | 337169 | L128 | APC |
| CD45RA | BD Biosciences | 560673 | HI100 | Alexa Fuor 700 |
| CD3 | BD Biosciences | 641397 | SK7 | APC-H7 |
| CD25 | BD Biosciences | 560225 | M-A251 | APC-H7 |
| CD8 | BD Biosciences | 560347 | RPA-T8 | V450 |
| CD8 | BD Biosciences | 560774 | RPA-T8 | V500 |
| FoxP3 | BD Biosciences | 560047 | 259D/C7 | Alexa Fuor 488 |
| CD95 | Biolegend | 305612 | DX2 | APC |
| CD38 | Biolegend | 303532 | HIT2 | BV605 |
| PD-1 (CD279) | Biolegend | 329924 | EH12.2H7 | BV605 |
| FoxP3 | eBiosciences | 12-4777-42 | 236A/E7 | PE |
| CD3 | eBiosciences | 45-0036-42 | SK7 | PerCP-Cy5.5 |
| LIVE/DEAD fixable dye | Life Technologies | L34957 | NA | AmCyam/V500 |
FITC, fluorescein isothiocyanate; PE, phycoerythrin; PerCP, peridinin chlorophyll protein; APC, allophycocyanin.
TABLE 6.
Memory subset definitions
| Flow cytometry panel | Memory subset | Definition |
|---|---|---|
| Activation | ||
| Naive | CD27+ CCR7+ CD95− CD45RA+ | |
| Stem cell memory | CD27+ CCR7+ CD95+ CD45RA+ | |
| Central memory | CD27+ CCR7+ CD95+ CD45RA− | |
| Effector memory | CD27± CCR7− CD95+ CD45RA− | |
| Terminally differentiated | CD27− CCR7− CD95+ CD45RA+ | |
| Cycling | ||
| Naive | CD27+ CD45RA+ | |
| Central memory | CD27+ CD45RA− | |
| Effector memory | CD27− CD45RA− | |
| Terminally differentiated | CD27− CD45RA+ | |
| Immune checkpoint | ||
| Naive | CD27+ CD28+ CD45RA+ CCR7+ CD57− | |
| Central memory | CD27+ CD28+ CD45RA− CCR7+ CD57− | |
| Transitional memory | CD27+ CD28+ CD45RA− CCR7− | |
| Effector memory | CD27± CD28− CD45RA− CCR7− | |
| Terminally differentiated | CD27− CD28− CD45RA+ CCR7− CD57+ | |
| Treg/degranulation | ||
| Naive | CD27+ CD45RA+ | |
| Central memory | CD27+ CD45RA− | |
| Effector memory | CD27− CD45RA− | |
| Terminally differentiated | CD27− CD45RA+ |
FIG 3.
Gating strategy for flow cytometry analysis. Representative plots of live CD3+ T cells are shown.
Statistical analysis.
To investigate the effect of CMV DNA on the expression of immunological markers of activation (HLA-DR+ CD38+), cellular cycling (Ki-67+), degranulation (CD107a+), Treg cells (Fox-P3+ CD25+), and PD-1 (an immune checkpoint protein expressed by recently activated or exhausted cells), we developed regression models for the percent expression of each phenotype. These markers were measured in five T-cell memory subsets that were treated as independent variables in the models: stem cell memory (TSCM), central memory (TCM), transitional memory (TTM), effector memory (TEM), and terminally differentiated (TTD). The markers we use to define each subset in on each flow cytometry panel are shown in Table 6. Naive cells were excluded from the analysis, as they had mostly static and low expression of our markers of interest. These outcomes were measured at baseline and up to 5 times after ART initiation, and we used a base 2 log transformation of time on ART to accommodate the model fit. Immunological expression was classified as percent expression of phenotypic markers within memory subsets. To account for inherent differences in expressions between memory subsets, these percentages were normalized within subset after applying a square root transformation to improve the kurtosis. We then modeled the normalized percent expression of the markers with linear mixed-effects models with random intercept and slope for subject and an additional nested random effect for memory compartment within subject. Since CMV DNA was undetectable at 67% of time points, CMV was dichotomized as detectable or undetectable at each time point. Other predictors included in the models were base 10 log-transformed values of EBV DNA and peak HIV RNA, time from EDI to ART initiation, age, and CD4/CD8 ratio.
First, individual models for each panel were built using each predictor, time on ART, and the interaction of the predictor and time. If predictors were found to have a significant interaction with time in the individual models, they were included in the multivariate regression along with their interaction term. We also tested the interaction between the predictor, the memory compartment, and time, but since none were significant, they were excluded from the models. All predictors with P values of <0.2 were considered for the multivariate regression. Since each percent response was normalized within compartment, there is no mean difference by compartment. As a result, memory compartment was included in the multivariate models only if it was found to have a significant interaction with time, adjusting the normalized values to account for difference in slope by compartment. The effect of memory compartment in the model does imply difference in phenotypic response by compartment but adjusts the starting point to compare the difference in slope by compartment. In the multivariate models, predictors were iteratively removed or added based on combination of likelihood ratio test, Akaike information criterion (AIC) (35), and variance inflation factor (VIF) (36). Simulated residuals and QQ plots were constructed to ensure proper model fit and that model assumptions were not violated. Due to the exploratory nature of our study, we did not eliminate effects of multiple comparisons.
ACKNOWLEDGMENTS
We are grateful to Davey Smith for his helpful suggestions and guidance throughout the study design and analysis.
A.C.-Q., M.M., C.S., and S.G. participated in the study design and wrote the preliminary version of the manuscript. A.C.-Q., R.S., S.A.R., M.M., and M.V.-M. optimized and performed flow cytometry staining. C.S., S.A.R., S.G., and R.S. were involved with data interpretation and analysis. S.A.R. collected and summarized the clinical data. A.F., M.N., and C.A. designed the GLMM model and helped edit the manuscript.
We have no conflicts of interests to declare.
This work was supported primarily by a grant from the National Institutes of Health, University of California, San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research, P30-AI027763 (CNIHR), and a California HIV Research Program Ideal award to Sara Gianella, by the department of Veterans Affairs, by the James B. Pendleton Charitable Trust, and by additional grants from the National Institutes of Health: AI100665, MH100974, MH097520, DA034978, AI007384, AI027763, AI106039, AI43638, AI074621, AI036214, MH101012, UL1TR000100, CARE U19 AI096113, and AI068636-09.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Siliciano JM, Siliciano RF. 2015. The remarkable stability of the latent reservoir for HIV-1 in resting memory CD4+ T cells. J Infect Dis 212:1345–1347. doi: 10.1093/infdis/jiv219. [DOI] [PubMed] [Google Scholar]
- 2.Murray AJ, Kwon KJ, Farber DL, Siliciano RF. 2016. The latent reservoir for HIV-1: how immunologic memory and clonal expansion contribute to HIV-1 persistence. J Immunol 197:407–417. doi: 10.4049/jimmunol.1600343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durier N, Ananworanich J, Apornpong T, Ubolyam S, Kerr SJ, Mahanontharit A, Ferradini L, Ruxrungtham K, Avihingsanon A. 2013. Cytomegalovirus viremia in Thai HIV-infected patients on antiretroviral therapy: prevalence and associated mortality. Clin Infect Dis 57:147–155. doi: 10.1093/cid/cit173. [DOI] [PubMed] [Google Scholar]
- 4.Gianella S, Massanella M, Wertheim JO, Smith DM. 2015. The sordid affair between human herpesvirus and HIV. J Infect Dis 212:845–852. doi: 10.1093/infdis/jiv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bate SL, Dollard SC, Cannon MJ. 2010. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin Infect Dis 50:1439–1447. doi: 10.1086/652438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupont L, Reeves MB. 2016. Cytomegalovirus latency and reactivation: recent insights into an age old problem. Rev Med Virol 26:75–89. doi: 10.1002/rmv.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman ML, Lederman MM, Gianella S. 2016. Partners in crime: the role of CMV in immune dysregulation and clinical outcome during HIV infection. Curr HIV/AIDS Rep 13:10–19. doi: 10.1007/s11904-016-0297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arama V, Mihailescu R, Radulescu M, Arama SS, Streinu-Cercel A, Youle M, CMV-HIV Study Group. 2014. Clinical relevance of the plasma load of cytomegalovirus in patients infected with HIV—a survival analysis. J Med Virol 86:1821–1827. doi: 10.1002/jmv.24027. [DOI] [PubMed] [Google Scholar]
- 9.Morris SR, Zhao M, Smith DR, Vargas MV, Little SJ, Gianella S. 2016. Longitudinal viral dynamics in semen during early HIV infection. Clin Infect Dis 64:428–434. doi: 10.1093/cid/ciw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soderberg-Naucler C. 2014. Treatment of cytomegalovirus infections beyond acute disease to improve human health. Expert Rev Anti Infect Ther 12:211–222. doi: 10.1586/14787210.2014.870472. [DOI] [PubMed] [Google Scholar]
- 12.Lisco A, Vanpouille C, Margolis L. 2009. War and peace between microbes: HIV-1 interactions with coinfecting viruses. Cell Host Microbe 6:403–408. doi: 10.1016/j.chom.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Almeida GD, Porada CD, St Jeor S, Ascensao JL. 1994. Human cytomegalovirus alters interleukin-6 production by endothelial cells. Blood 83:370–376. [PubMed] [Google Scholar]
- 14.Suni MA, Ghanekar SA, Houck DW, Maecker HT, Wormsley SB, Picker LJ, Moss RB, Maino VC. 2001. CD4(+)CD8(dim) T lymphocytes exhibit enhanced cytokine expression, proliferation and cytotoxic activity in response to HCMV and HIV-1 antigens. Eur J Immunol 31:2512–2520. doi:. [DOI] [PubMed] [Google Scholar]
- 15.Iwamoto GK, Monick MM, Clark BD, Auron PE, Stinski MF, Hunninghake GW. 1990. Modulation of interleukin 1 beta gene expression by the immediate early genes of human cytomegalovirus. J Clin Invest 85:1853–1857. doi: 10.1172/JCI114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fawaz LM, Sharif-Askari E, Menezes J. 1999. Up-regulation of NK cytotoxic activity via IL-15 induction by different viruses: a comparative study. J Immunol 163:4473–4480. [PubMed] [Google Scholar]
- 17.Saghafian-Hedengren S, Sohlberg E, Theorell J, Carvalho-Queiroz C, Nagy N, Persson JO, Nilsson C, Bryceson YT, Sverremark-Ekstrom E. 2013. Epstein-Barr virus coinfection in children boosts cytomegalovirus-induced differentiation of natural killer cells. J Virol 87:13446–13455. doi: 10.1128/JVI.02382-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overwijk WW, Schluns KS. 2009. Functions of gammaC cytokines in immune homeostasis: current and potential clinical applications. Clin Immunol 132:153–165. doi: 10.1016/j.clim.2009.03.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen-Quick A, Vanpouille C, Lisco A, Gianella S. 2017. Cytomegalovirus and HIV persistence: pouring gas on the fire. AIDS Res Hum Retroviruses 33:S23–S30. doi: 10.1089/aid.2017.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianella S, Anderson CM, Vargas MV, Richman DD, Little SJ, Morris SR, Smith DM. 2013. CMV DNA in semen and blood is associated with higher levels of proviral HIV DNA. J Infect Dis 207:898–902. doi: 10.1093/infdis/jis777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gianella S, Strain MC, Rought SE, Vargas MV, Little SJ, Richman DD, Spina CA, Smith DM. 2012. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J Virol 86:1307–1315. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gianella S, Anderson CM, Var SR, Oliveira MF, Lada SM, Vargas MV, Massanella M, Little SJ, Richman DD, Strain MC, Perez-Santiago J, Smith DM. 2016. Replication of human herpesviruses is associated with higher HIV DNA levels during antiretroviral therapy started at early phases of HIV infection. J Virol 90:3944–3952. doi: 10.1128/JVI.02638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasi M, De Biasi S, Gibellini L, Bianchini E, Pecorini S, Bacca V, Guaraldi G, Mussini C, Pinti M, Cossarizza A. 2017. Ageing and inflammation in patients with HIV infection. Clin Exp Immunol 187:44–52. doi: 10.1111/cei.12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gianella S, Massanella M, Richman DD, Little SJ, Spina CA, Vargas MV, Lada SM, Daar ES, Dube MP, Haubrich RH, Morris SR, Smith DM, California Collaborative Treatment Group 592 Team. 2014. Cytomegalovirus replication in semen is associated with higher levels of proviral HIV DNA and CD4+ T cell activation during antiretroviral treatment. J Virol 88:7818–7827. doi: 10.1128/JVI.00831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith DM, Nakazawa M, Freeman ML, Anderson CM, Oliveira MF, Little SJ, Gianella S. 2016. Asymptomatic CMV replication during early human immunodeficiency virus (HIV) infection is associated with lower CD4/CD8 ratio during HIV treatment. Clin Infect Dis 63(11):1517–1524. doi: 10.1093/cid/ciw612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisco A, Munawwar A, Introini A, Vanpouille C, Saba E, Feng X, Grivel JC, Singh S, Margolis L. 2012. Semen of HIV-1-infected individuals: local shedding of herpesviruses and reprogrammed cytokine network. J Infect Dis 205:97–105. doi: 10.1093/infdis/jir700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee A, Park SP, Park CH, Kang BH, Park SH, Ha SJ, Jung KC. 2015. IL-4 induced innate CD8+ T cells control persistent viral infection. PLoS Pathog 11:e1005193. doi: 10.1371/journal.ppat.1005193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dan JM, Massanella M, Smith DM, Spina CA, Schrier R, Daar ES, Dube MP, Morris SR, Gianella S. 2016. Brief report: effect of CMV and HIV transcription on CD57 and PD-1 T-cell expression during suppressive ART. J Acquir Immune Defic Syndr 72:133–137. doi: 10.1097/QAI.0000000000000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chevalier MF, Weiss L. 2013. The split personality of regulatory T cells in HIV infection. Blood 121:29–37. doi: 10.1182/blood-2012-07-409755. [DOI] [PubMed] [Google Scholar]
- 30.Remis RS, Liu J, Loutfy MR, Tharao W, Rebbapragada A, Huibner S, Kesler M, Halpenny R, Grennan T, Brunetta J, Smith G, Reko T, Kaul R. 2016. Prevalence of sexually transmitted viral and bacterial infections in HIV-positive and HIV-negative men who have sex with men in Toronto. PLoS One 11:e0158090. doi: 10.1371/journal.pone.0158090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenorio AR, Zheng Y, Bosch RJ, Krishnan S, Rodriguez B, Hunt PW, Plants J, Seth A, Wilson CC, Deeks SG, Lederman MM, Landay AL. 2014. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis 210:1248–1259. doi: 10.1093/infdis/jiu254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gandhi RT, McMahon DK, Bosch RJ, Lalama CM, Cyktor JC, Macatangay BJ, Rinaldo CR, Riddler SA, Hogg E, Godfrey C, Collier AC, Eron JJ, Mellors JW, ACTG A5321 Team. 2017. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 13:e1006285. doi: 10.1371/journal.ppat.1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massanella M, Gianella S, Lada SM, Richman DD, Strain MC. 2015. Quantification of total and 2-LTR (long terminal repeat) HIV DNA, HIV RNA and herpesvirus DNA in PBMCs. Bio Protoc 5:e1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akaike H. 1998. Information theory and an extension of the maximum likelihood principle, p 199–213. In Parzen E, Tanabe K, Kitagawa G (ed). Selected papers of Hirotugu Akaike. Springer Series in Statistics (Perspectives in Statistics). Springer, New York, NY. [Google Scholar]
- 36.Rawlings JO, Pantula SG, Dickey DA. 1998. Applied regression analysis : a research tool (2nd ed), p 372–373. Springer, New York, NY. [Google Scholar]