Human cytomegalovirus (HCMV) is an important human pathogen that causes severe symptoms, especially in immunocompromised hosts. Furthermore, congenital HCMV infection is the leading viral cause of severe birth defects. Development of antiviral drugs to prevent the production of infectious virus progeny is challenging due to a complex and multistep virion morphogenesis. The mechanism of secondary envelopment is still not fully understood; nevertheless, it represents a potential target for antiviral drugs. Our identification of the role of a positively charged motif in the pUL71 C terminus for efficient HCMV secondary envelopment underlines the importance of pUL71 and, especially, its C terminus for this process. It furthermore shows how cell-associated spread and virion release depend on secondary envelopment. Ultrastructural analyses of different stages of envelopment contribute to a better understanding of the mechanisms underlying the process of secondary envelopment. This may bring us closer to the development of novel concepts to treat HCMV infections.
KEYWORDS: HCMV, Human betaherpesvirus 5, TEM analysis, UL71, secondary envelopment, tetralysine motif
ABSTRACT
Human cytomegalovirus (HCMV) secondary envelopment requires the viral tegument protein pUL71. The lack of pUL71 results in a complex ultrastructural phenotype with increased numbers of viral capsids undergoing envelopment at the cytoplasmic virus assembly complex. Here, we report a role of the pUL71 C terminus in secondary envelopment. Mutant viruses expressing C-terminally truncated pUL71 (TB71del327-361 and TB71del348-351) exhibited an impaired secondary envelopment in transmission electron microscopy (TEM) studies. Further mutational analyses of the C terminus revealed a tetralysine motif whose mutation (TB71mutK348-351A) resulted in an envelopment defect that was undistinguishable from the defect caused by truncation of the pUL71 C terminus. Interestingly, not all morphological alterations that define the ultrastructural phenotype of a TB71stop virus were found in cells infected with the C-terminally mutated viruses. This suggests that pUL71 provides additional functions that modulate HCMV morphogenesis and are harbored elsewhere in pUL71. This is also reflected by an intermediate growth defect of the C-terminally mutated viruses compared to the growth of the TB71stop virus. Electron tomography and three-dimensional visualization of different stages of secondary envelopment in TB71mutK348-351A-infected cells showed unambiguously the formation of a bud neck. Furthermore, we provide evidence for progressive tegument formation linked to advancing grades of capsid envelopment, suggesting that tegumentation and envelopment are intertwined processes. Altogether, we identified the importance of the pUL71 C terminus and, specifically, of a positively charged tetralysine motif for HCMV secondary envelopment.
IMPORTANCE Human cytomegalovirus (HCMV) is an important human pathogen that causes severe symptoms, especially in immunocompromised hosts. Furthermore, congenital HCMV infection is the leading viral cause of severe birth defects. Development of antiviral drugs to prevent the production of infectious virus progeny is challenging due to a complex and multistep virion morphogenesis. The mechanism of secondary envelopment is still not fully understood; nevertheless, it represents a potential target for antiviral drugs. Our identification of the role of a positively charged motif in the pUL71 C terminus for efficient HCMV secondary envelopment underlines the importance of pUL71 and, especially, its C terminus for this process. It furthermore shows how cell-associated spread and virion release depend on secondary envelopment. Ultrastructural analyses of different stages of envelopment contribute to a better understanding of the mechanisms underlying the process of secondary envelopment. This may bring us closer to the development of novel concepts to treat HCMV infections.
INTRODUCTION
The Human betaherpesvirus 5, also known as human cytomegalovirus (HCMV), is a globally spread pathogen that causes severe disorders in immunocompromised hosts. HCMV is also the major viral cause of birth sequelae in newborns following congenital infection (1). Mature HCMV virions show the characteristic herpesvirus organization, consisting of four major structural components, which can be unambiguously identified in electron micrographs (2, 3). The icosahedral capsid has an electron-dense core, which contains the tightly packed double-stranded DNA genome. The capsid is surrounded by a layer of mainly viral but also cellular proteins, termed tegument, that connect the capsid with the viral envelope (4). The latter is a host cell-derived lipid bilayer and contains several viral glycoprotein complexes.
Virion morphogenesis is completed in a virally regulated process, referred to as secondary envelopment, in which capsids acquire their envelope. Secondary envelopment takes place in the cytoplasm of infected cells in a perinuclear region, the virus assembly complex (vAC) (5). The vAC is formed around the centrioles during HCMV infection by a virus-induced rearrangement of microtubules and cytoplasmic membranes (6–11). The membranes involved in vAC formation are, in particular, of the Golgi-apparatus and of the secretory and endocytic pathways (6–9, 12).
Because of the high resolution that is needed to visualize secondary envelopment, our current understanding of this process is obtained mainly from studying virus morphogenesis by transmission electron microscopy (TEM). The introduction of advanced sample preparation techniques, such as high-pressure freezing and freeze substitution, has led to improved preservation of viral and cellular structures, especially of membranes, and thus provides high-quality and high-resolution micrographs (3, 13, 14). Previous studies have indicated that HCMV secondary envelopment is a multistep process (3). Consistent with this, different stages of maturation can be observed, three of which can be easily categorized based on their appearance in TEM micrographs. Cytoplasmic capsids at their first stage of maturation are partially tegumented but not associated with membranes. Capsids at later stages are in contact with membranes that wrap around the capsid to form the envelope, a process also referred to as budding. The budding/envelopment process is finished by a membrane scission event, resulting in an enveloped capsid within a vesicle, which is the third stage of maturation. Those intracellular enveloped capsids are morphologically identical to mature extracellular virions. The intracellular enveloped capsids are transported within vesicles to the plasma membrane where they are released into the extracellular space by fusion of the vesicle membrane with the plasma membrane.
The driving forces for membrane curvature during secondary envelopment of HCMV and membrane scission are still unknown. There is some evidence that budding and membrane scission are regulated by viral proteins (14–22). One viral protein with a reported function for HCMV secondary envelopment is the tegument protein pUL71 (14). HCMV pUL71 is conserved in all herpesvirus subfamilies, e.g., pUL51 in the alphaherpesviruses herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PrV) and BSRF1 in the gammaherpesvirus Epstein-Barr virus (EBV). Similarly to HCMV pUL71, pUL51 proteins of HSV-1 and PrV are involved in secondary envelopment, and mutant viruses lacking pUL51 are impaired in virus spread and release (23, 24). In HCMV-infected cells, pUL71 accumulates at the vAC, which is consistent with a function of pUL71 in secondary envelopment (14, 25–27). Its function for HCMV secondary envelopment was identified by quantitative ultrastructural analysis showing that the absence of pUL71 results in an accumulation of budding capsids at the vAC and hence to a decreased ratio of enveloped capsids. The envelopment defect is additionally characterized by the formation of extended membrane compartments associated with multiple budding events. Furthermore, viral capsids accumulated at the membrane and inside enlarged multivesicular bodies (MVBs) in the periphery of the vAC (14). These structural alterations are associated with an impaired virus release and virus spread.
With 361 amino acids, HCMV pUL71 is one of the largest herpesvirus homologs (14, 24, 28–31). Sequence comparison showed that the N terminus is highly conserved, whereas the C terminus exhibits low sequence homology (32). We and others identified important functional domains in the N terminus of pUL71 and its homologs. HSV-1 pUL51 has been shown to be palmitoylated (33). Furthermore, a cell-type-specific function of HSV-1 pUL51 for cell-to-cell spread was identified at the N terminus; however, mutation of the motif had no impact on virus release, suggesting a partial functional uncoupling of replication, release, and cell-to-cell spread of HSV-1 pUL51 (34). HCMV pUL71 harbors a basic leucine zipper-like domain, which is involved in the oligomerization of pUL71. Mutation of this motif did not alter the localization or the levels of pUL71 at the vAC but resulted in defective viral growth and secondary envelopment comparable to that of a virus mutant lacking pUL71 (26). We have recently reported the endocytic trafficking of HCMV pUL71 from the plasma membrane to membranes of the vAC and the requirement of an N-terminal tyrosine-based trafficking motif (25). Disruption of this motif results in impaired viral growth and a defect in secondary envelopment similar to what has been reported for a pUL71-deficient virus mutant.
Here, we investigated the role of the pUL71 C terminus for HCMV secondary envelopment. Two mutant viruses expressing C-terminally truncated versions of pUL71 were generated and analyzed by TEM. Both mutant viruses exhibited impaired secondary envelopment similar to that of a TB71stop virus but lacked other ultrastructural alterations such as enlarged MVBs or extended membrane compartments with multiple budding events. Subsequent targeted mutagenesis at the C terminus identified a positively charged tetralysine motif at position 348 to 351. Mutation of this motif was sufficient to cause the defect in secondary envelopment that was observed in C-terminally truncated mutants. C-terminal mutant viruses exhibited better growth and spread than the TB71stop virus, although all mutant viruses showed similar increases in the ratio of nonenveloped capsids at the vAC. By scanning transmission electron microscopy (STEM) tomography of capsids at various stages of secondary envelopment, we found evidence for progressive tegument formation linked to advancing grades of capsid envelopment, suggesting that these processes are intertwined. Using three-dimensional electron microscopy, we furthermore show for the first time that the envelopment process leads to the formation of a narrow bud neck. In conclusion, we demonstrated the importance of a C-terminal tetralysine motif for the regulation of secondary envelopment and investigated tegumentation and secondary envelopment by three-dimensional electron microscopy.
RESULTS
Generation and characterization of pUL71 C-terminally truncated virus mutants.
Previous studies have shown an important role of the HCMV tegument protein pUL71 for late stages of virion morphogenesis, specifically, for the process of secondary envelopment (14, 26, 27). The ultrastructural phenotype of a UL71-stop virus is quite complex and not restricted to a defect in secondary envelopment alone. Two functional motifs located in the N terminus of pUL71, a basic leucine zipper-like domain required for oligomerization of pUL71 and a tyrosine-based trafficking motif required for localization of pUL71 at the vAC, have already been identified as being crucial for the function of pUL71 in secondary envelopment (Fig. 1A). Among its human herpesvirus homologs, HCMV pUL71 is the largest representative. Interestingly, the C terminus is the least conserved part when comparing sequences of pUL71 homologs (32). Due to the complex ultrastructural phenotype in the absence of pUL71 and the low conservation of the C terminus of pUL71, we hypothesized that the C terminus might contain functional domains that at least partially contribute to the phenotype.
FIG 1.
(A) Sequence alignment of HCMV pUL71 (APG57260.1) and HSV pUL51 (ALM22621.1) by the online tool MUSCLE (68). Asterisks mark two predicted palmitoylation sites of pUL71 (CSS-Palm 4.0). Motifs that have been shown to be functional during virus morphogenesis are the following: 1, palmitoylation site of pUL51 (33); 2 and 3, tyrosine-based trafficking motifs in pUL71 and pUL51 (25, 34); 4, basic leucine zipper-like motif in pUL71 (26); 5, phosphorylation site of pUL51 (69); 6, tetralysine motif of pUL71 (this study). (B) Generation of C-terminal pUL71 mutant viruses. Shown is the genomic organization up- and downstream of the UL71 gene in parental virus TB40-BAC4 and the amino acid sequence of pUL71 in the indicated mutant viruses. TB71del327-361 and TB71del348-361 express C-terminally truncated versions of pUL71. Mutation of the underlined amino acids to the amino acids depicted in bold resulted in the point mutants TB71mutK348-351A and TB71mutL357-358A. The revertant virus TB71revK348-351A was generated by restoring the amino acid sequence of TB71mutK348-351A to the sequence of the parental virus TB40-BAC4.
We tested this hypothesis by generating two recombinant viruses expressing C-terminally truncated forms of pUL71 in the background of the infectious bacterial artificial chromosome (BAC) clone TB40-BAC4 of the endotheliotropic HCMV strain TB40/E (35). One mutant was lacking the last 35 C-terminal amino acids (TB71del327-361), whereas the introduction of a premature stop codon at codon 348 in the second mutant resulted in expression of a truncated pUL71 lacking the last 14 C-terminal amino acids (TB71del348-361) (overview in Fig. 1B). Sequence analysis of the mutated regions confirmed the integrity of the generated mutants. Both mutant viruses were reconstituted by electroporation of the respective BAC DNAs into human fibroblasts.
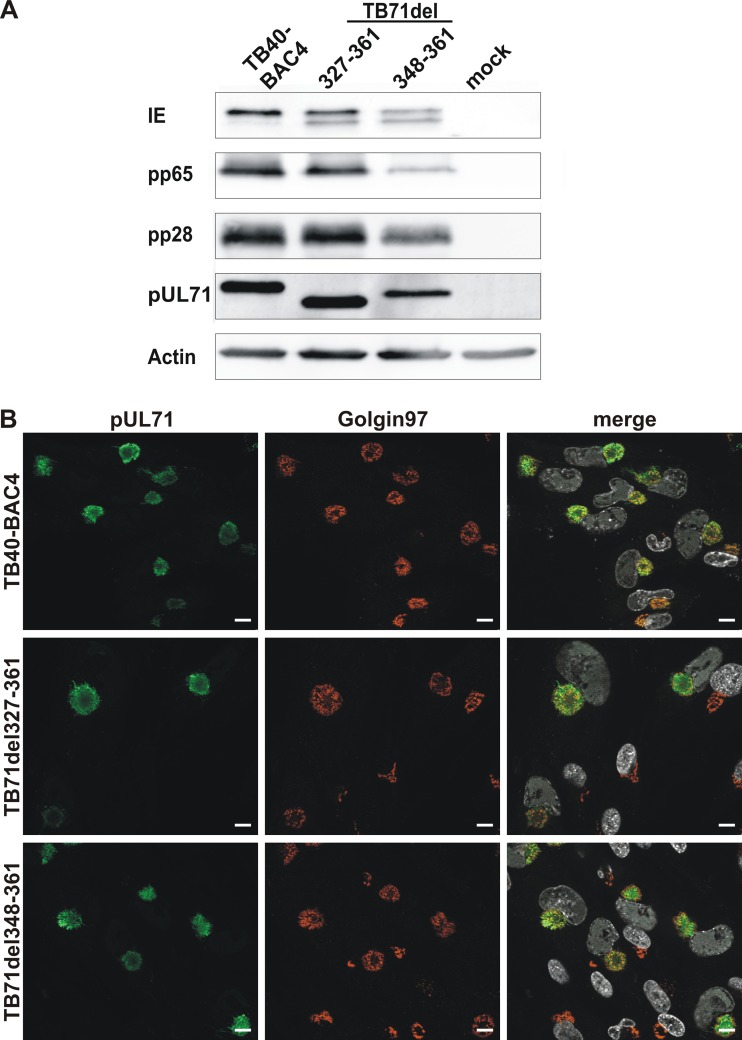
To confirm expression of the truncated versions of pUL71, lysates of mutant- and wild-type-virus-infected human foreskin fibroblasts (HFFs) at 120 h postinfection were examined by Western blot analysis. A polyclonal anti-pUL71 antibody (14) detected specific bands of lower molecular weight in lysates of TB71del327-361- and TB71del348-361-infected cells than of full-length pUL71 in lysates of wild-type-virus-infected cells (Fig. 2A). In addition, comparable band intensities of pUL71 in all virus-infected cell lysates indicated that neither expression nor stability of pUL71 was considerably affected by the C-terminal truncations.
FIG 2.
Characterization of pUL71 C-terminal deletion mutants. (A) Cell lysates of TB40-BAC4-, TB71del327-361-, TB71del348-361-, and mock-infected HFFs (120 h postinfection) were processed for Western blot analysis. The blot was probed with antibodies against viral IE, pp65, pp28, pUL71, and cellular actin. (B) Representative confocal images of the subcellular localization of pUL71 (green) and trans-Golgi network protein golgin-97 (red) in HFFs infected with the indicated viruses (MOI of 1) at 120 h postinfection. Nuclei were stained with DAPI (white). Scale bars, 10 μm.
Next, we investigated whether the subcellular localization of pUL71 is affected by the C-terminal truncations, because trafficking of pUL71 to the vAC is required for its function during infection (25). The intracellular localization of both C-terminally truncated forms of pUL71, 71del327-361 and 71del348-361, was indistinguishable from that of full-length pUL71 at 120 h postinfection and exhibited overlapping signals to cellular Golgi marker protein golgin-97, indicating that the C-terminal truncations had no gross effect on the intracellular trafficking of pUL71 to the vAC and its accumulation at this site (Fig. 2B).
The C terminus of pUL71 is involved in secondary envelopment.
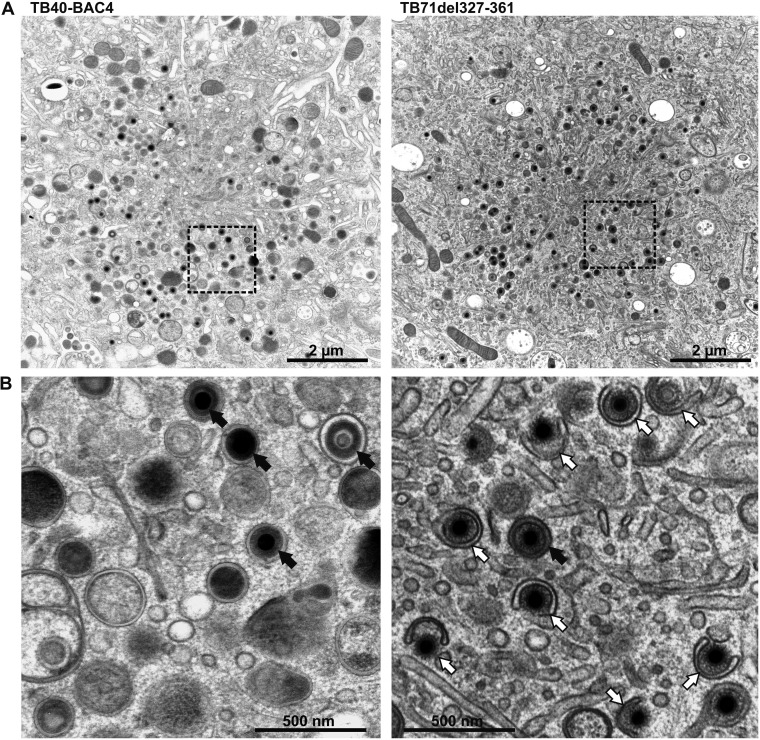
It was previously shown that HCMV virion morphogenesis, particularly, secondary envelopment, is severely impaired in cells infected with a UL71-stop virus (14). Since secondary envelopment can only be visualized by electron microscopy, we performed ultrastructural analyses to investigate the role of the C terminus of pUL71 for HCMV secondary envelopment. HFFs were infected with parental TB40-BAC4, the mutant viruses TB71del327-361, TB71del348-361, and the previously published TB71stop virus (14). Infected cells were prepared for TEM analysis by high-pressure freezing and freeze substitution at 120 h postinfection. This sample preparation protocol preserves cellular membranes close to their native state and provides excellent membrane contrast (13), which is crucial for the quantification of the various envelopment stages of virus particles at the vAC. Virus particles in two-dimensional micrographs of ultrathin sections of the area of the vAC were evaluated regarding their envelopment stage and divided into three categories (14). Category I (naked) comprises nonenveloped cytoplasmic capsids, which are not in contact with cellular membranes. Category II (budding) contains all those capsids, which are in a budding process characterized by the bending of membranes around capsids. Category III (enveloped) contains capsids that are surrounded by a complete envelope.
The quantitative analysis of at least 15 cells for each virus showed that the majority of capsids (83.9%) at the vAC were enveloped in parental TB40-BAC4-virus-infected fibroblasts, while only 15.5% of all capsids underwent budding (Fig. 3, left, and Table 1). In contrast, only approximately 30% of the cytoplasmic capsids of both C-terminally truncated mutants were enveloped, while the majority of capsids (65.5% and 72.1%) were in the process of budding at membranes of small vesicles (Fig. 3, right). These results indicated that the last 14 C-terminal amino acids of pUL71 are required for efficient HCMV secondary envelopment.
FIG 3.
(A) Electron micrographs of the vAC of HCMV TB40-BAC4- (left) and TB71del327-361-infected (right) HFFs at 120 h postinfection. (B) Higher magnification of selected areas from panel A shows fully enveloped virus capsids (black arrows) in TB40-BAC4-infected cells (left), whereas TB71del327-361-virus-infected cells (right) contain a high number of capsids in the process of secondary envelopment (white arrows).
TABLE 1.
Ultrastructural quantification of secondary envelopment of HCMV virus particles in vACs of infected HFFs at 120 h postinfection
| Virus | No. of cells analyzed | No. of capsids total | % particles by type (mean ± SD)a
|
||
|---|---|---|---|---|---|
| Enveloped | Budding | Naked | |||
| TB40-BAC4 | 30 | 1,040 | 83.9 ± 14.8 | 15.5 ± 14.7 | 0.5 ± 1.7 |
| TB71del327-361 | 22 | 211 | 31.5 ± 18.9 | 65.5 ± 20.3 | 2.9 ± 5.3 |
| TB71del348-361 | 25 | 980 | 26.4 ± 12.4 | 72.1 ± 10.9 | 1.5 ± 2.8 |
| TB71mutK348-351A | 15 | 514 | 21.9 ± 14.0 | 76.5 ± 14.3 | 1.7 ± 3.1 |
| TB71revK348-351A | 14 | 418 | 77.1 ± 10.7 | 20.5 ± 6.8 | 2.4 ± 5.4 |
| TB71mutL357-358A | 17 | 599 | 82.5 ± 8.8 | 13.8 ± 8.5 | 3.7 ± 3.2 |
| TB71stop | 30 | 1,564 | 25.9 ± 8.6 | 69.9 ± 9.7 | 3.5 ± 4.3 |
Given are the mean percentages and standard deviations (SD) of enveloped particles, budding particles (nonenveloped particles attached to membranes), and naked particles (nonenveloped particles not attached to membranes) from at least two independent experiments. For each virus, boldface value highlights the stage of secondary envelopment at which the majority of particles were counted.
Another interesting finding of this TEM study was that secondary envelopment of capsids was predominantly found as budding of single capsids into small crescent-shaped vesicles in TB71del327-361- and TB71del348-361-infected cells (Fig. 3, right). Consequently, most enveloped capsids were also found as single capsids within small vesicles in these mutant-virus-infected cells, which is the predominant phenotype of enveloped particles in parental virus-infected cells. This is in contrast to a UL71-stop virus, where multiple budding events into large vesicles are frequently observed (14). We concluded from this that the truncation of the last 14 amino acids of pUL71 leads to a defective secondary envelopment without affecting the overall morphology of vAC membranes.
Identification of functional domains of the pUL71 C terminus.
The defect in secondary envelopment in mutant viruses expressing C-terminally truncated pUL71 indicated that the 14 endmost C-terminal amino acids harbor a function that is involved in the regulation of the secondary envelopment process.
Sequence examination revealed two motifs within the 14 endmost C-terminal amino acids: a dileucine motif (357LL358) and a positively charged tetralysine motif (348KKKK351). Dileucine motifs are known trafficking motifs that regulate sorting of transmembrane proteins to endosomes and lysosomes (36, 37). The function of positively charged amino acids is less clear. They are often part of nuclear localization signals (38, 39) but also mediate membrane association of proteins by their interaction with negatively charged phospholipids (40–43). To clarify whether one of these two motifs has a function in secondary envelopment, two additional recombinant viruses were generated in which either the dileucine motif (TB71mutL357-358A) or the tetralysine motif (TB71mutK348-351A) was mutated to alanine residues (Fig. 1A).
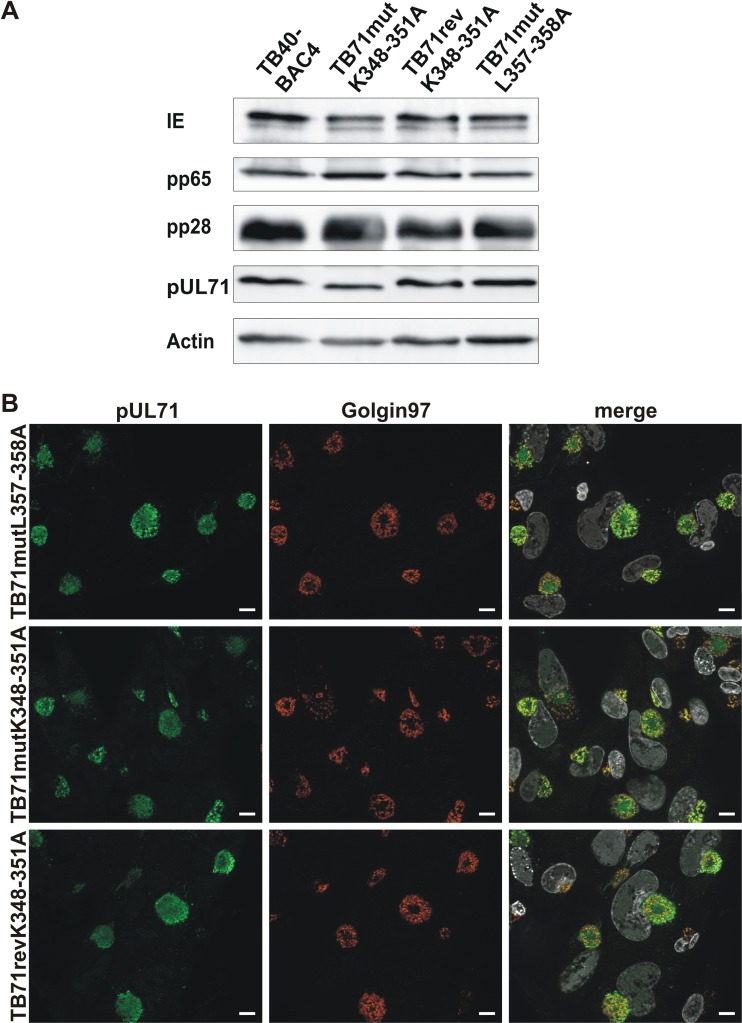
First, we analyzed the expression of the point-mutated versions of pUL71 by Western blot analysis. Protein levels of pUL71 were similar in lysates of TB71mutL357-358A-virus-infected cells and TB40-BAC4-infected cells (Fig. 4A). Interestingly, the mutant protein pUL71mutK348-351A exhibited a slightly faster migration in Western blot analysis than wild-type pUL71, which is likely caused by the exchange of four lysine residues with smaller alanine residues. To exclude unwanted mutations elsewhere in the TB71mutK348-351A mutant, a revertant virus was generated in which the four lysine residues 348 to 351 were reintroduced (TB71revK348-351A). Protein levels and the migration pattern of the repaired pUL71 were similar to those of wild-type pUL71 in Western blot analysis, which indicated that the K348-351A mutation accounts for the observed alterations of pUL71 in TB71mutK348-351A infections.
FIG 4.
Characterization of pUL71 C-terminal point mutants. (A) Western blot analysis of cell lysates of TB40-BAC4, TB71mutL357-358A, TB71mutK348-351A, and TB71revK348-351A (120 h postinfection). The blot was probed with antibodies against IE, pp65, pp28, pUL71, and cellular actin. (B) Representative confocal images of the subcellular localization of pUL71 (green) and trans-Golgi network protein golgin-97 (red) in HFFs infected with the indicated viruses (MOI of 1) at 120 h postinfection. Nuclei were stained with DAPI (white). Scale bars, 10 μm.
As expected, the point mutants TB71mutK348-351A and TB71mutL357-358A exhibited an intracellular localization at the vAC indistinguishable from that of pUL71 in TB40-BAC4 and revertant virus infection (Fig. 4B), which is in line with the C-terminal deletion mutants.
Regulation of secondary envelopment by a C-terminal tetralysine motif.
To analyze the importance of the dileucine (357LL358) and the tetralysine (348KKKK351) motifs for HCMV morphogenesis, HFFs were infected with TB71mutL357-358A and TB71mutK348-351A viruses and subjected to quantitative TEM analysis of secondary envelopment at 120 h postinfection. The majority (82.5%) of all capsids at the area of the vAC were enveloped in TB71mutL357-358A-virus-infected cells (Table 1), indicating that mutation of the C-terminal dileucine motif to two alanines had no adverse effect on secondary envelopment. Thus, the point mutant TB71mutL357-358A was excluded from further examinations.
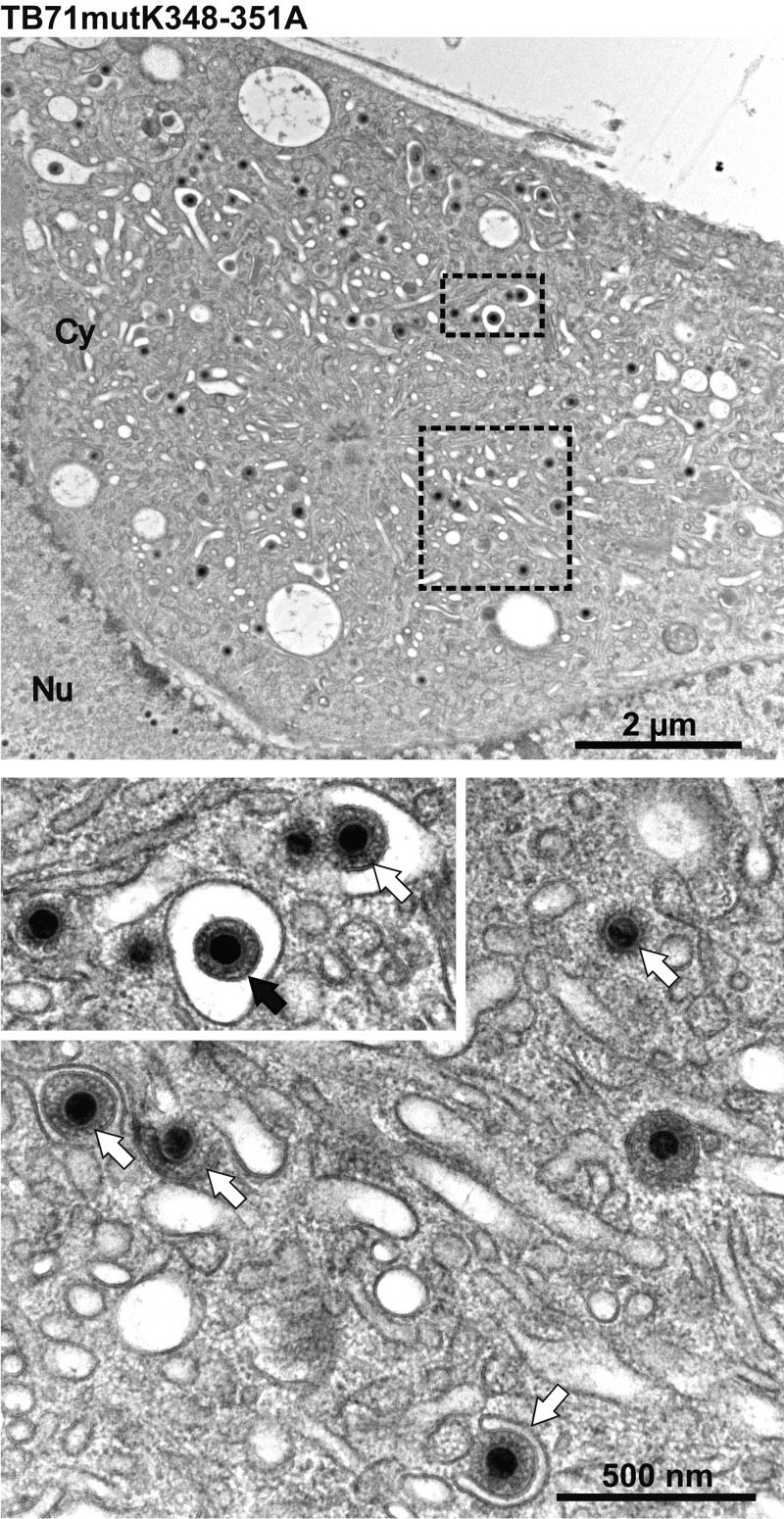
In contrast, only 21.9% of all capsids were enveloped in TB71mutK348-351A-infected cells (Table 1), which is comparable with the findings in TB71del327-361-, TB71del348-361-, and TB71stop-virus-infected cells. Budding of capsids was mostly observed at small vesicles in TB71mutK348-351A-infected cells (Fig. 5), which is consistent with the phenotype of the two C-terminally truncated mutants. This defect in secondary envelopment was reversed in the revertant TB71revK348-351A virus to the phenotype of parental TB40-BAC4-virus-infected cells (Table 1). This indicates that the tetralysine motif (348KKKK351) in the C terminus of pUL71 is required for efficient HCMV secondary envelopment.
FIG 5.
Electron micrographs of the vAC of HCMV TB71mutK348-351A-infected HFFs at 120 h postinfection. Higher magnifications below show many nonenveloped capsids (white arrows) and one enveloped virus particle (black arrow).
Impaired virus spread and release is associated with a defect in secondary envelopment.
The truncation of the pUL71 C terminus or mutation of the C-terminal tetralysine motif resulted in a clear defect in secondary envelopment, which was similar to that of the TB71stop virus. Therefore, we expected to find a similar defect in viral growth in C-terminally mutated-virus-infected cells as it was described for the TB71stop virus (14). However, monitoring of viral growth during reconstitution of the C-terminal mutant viruses suggested a slightly better growth than expected.
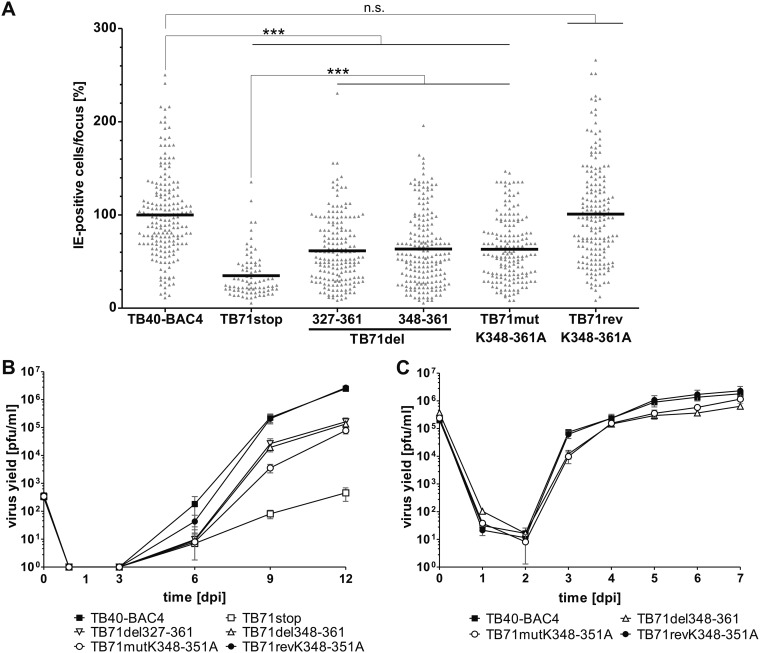
To address this systematically, we first performed a focus expansion assay to compare cell-associated spread of the C-terminal deletion mutants and the TB71mutK348-351A mutant with the spread of TB40-BAC4, TB71revK348-351A, and TB71stop viruses. HFFs were infected and cultured under methylcellulose overlay conditions for 9 days. The size of individual foci was determined by counting the number of immediate early protein (IE)-positive cell nuclei per focus. As shown in Fig. 6A, the relative focus size of both C-terminally truncated viruses, TB71del327-361 and TB71del348-361, as well as of the tetralysine mutant TB71mutK348-351A, reached around 60% of the focus size of the parental TB40-BAC4 and the revertant TB71revK348-351A virus. Foci of the C-terminally mutated viruses were significantly larger than the foci formed by the TB71stop virus, which reached only 35% of the focus size of the parental virus. From this we concluded that the lack of 35 and 14 C-terminal amino acids as well as the mutation of the C-terminal tetralysine motif of pUL71 caused an intermediate spread phenotype compared to the spread defect of a TB71stop virus. This spread defect is most likely the result of impaired secondary envelopment.
FIG 6.
Growth of C-terminal pUL71 mutants. (A) Focus expansion assay of the indicated viruses in HFFs under methylcellulose overlay. HCMV-infected cells were detected by indirect immunofluorescence staining for IE1/2 antigen at 9 days postinfection (dpi). Each data point represents the relative number of IE1/2-positive nuclei per focus. Shown is the mean focus size for each virus (black line) normalized to the mean focus size of TB40-BAC4. At least 80 foci from three independent experiments were determined for each virus. Significance testing was performed by a Kruskal-Wallis test followed by a Dunn’s multiple-comparison test (P < 0.05). Significance is given compared to TB40-BAC4 and TB71stop. ***, P < 0.0001; n.s., not significant. (B) Multistep growth kinetics experiments of the indicated viruses were performed by infecting HFFs with an infection rate of 0.3%. Virus yields in the supernatants of infected cells were determined at the indicated times by titration on HFFs. Growth curves show the mean virus yields and standard deviations from three independent virus supernatants. Virus yields at time zero represent the starting infection rates determined at 24 h postinfection. (C) Single-step growth kinetics experiments of the indicated viruses were performed on HFFs with an MOI of 3. Virus yields in the supernatants of infected cells were determined at the indicated times by titration on HFFs. Growth curves show the mean virus yields and standard deviations from three independent virus supernatants. Virus yields at time zero represent the virus yields of the inocula.
Next, the production and release of infectivity of the C-terminally mutated viruses were assessed by multistep growth kinetics in comparison to that of the parental virus. As shown in Fig. 6B, extracellular virus yields of TB71del327-361, TB71del348-361, and TB71mutK348-351A were reduced 20-fold compared to virus yields of TB40-BAC4 and TB71revK348-351A, whereas the TB71stop virus showed a 5,000-fold reduction in extracellular virus yields at 12 days postinfection.
Taken together, the intermediate growth of the C-terminally mutated viruses and the more severe growth defect of the TB71stop virus indicated that pUL71 possesses additional functions that contribute to virus growth, which are separate from the regulation of secondary envelopment by the pUL71 C terminus.
Now the question arose whether the spread defect alone accounts for the impaired growth in the multistep growth kinetics experiment or whether virus release is also affected by the C-terminal deletion or the mutation of the tetralysine motif. To answer this, we performed a single-step growth kinetics experiment. Since the two deletion mutants behaved the same in the previous experiments, we compared the extracellular virus yields of only TB71del348-361 with TB71mutK348-351A, TB40-BAC4, and TB71revK348-351A (Fig. 6C). Parental and revertant viruses reached their maximum virus yields at day 7 postinfection. At that time, both mutants showed a mild ∼1.5-fold reduction in extracellular virus yields. The maximum reduction of TB71mutK348-351A virus yields compared to that of TB40-BAC4 was approximately 10-fold at 3 days postinfection. Thus, we assumed that not only viral spread is affected by the defect in secondary envelopment but also virus release to a minor extent. However, the mutation of the C-terminal tetralysine motif in pUL71 affected egress of virus progeny to a surprisingly small extent, considering the profound impairment in secondary envelopment.
Three-dimensional visualization of tegumentation and secondary envelopment.
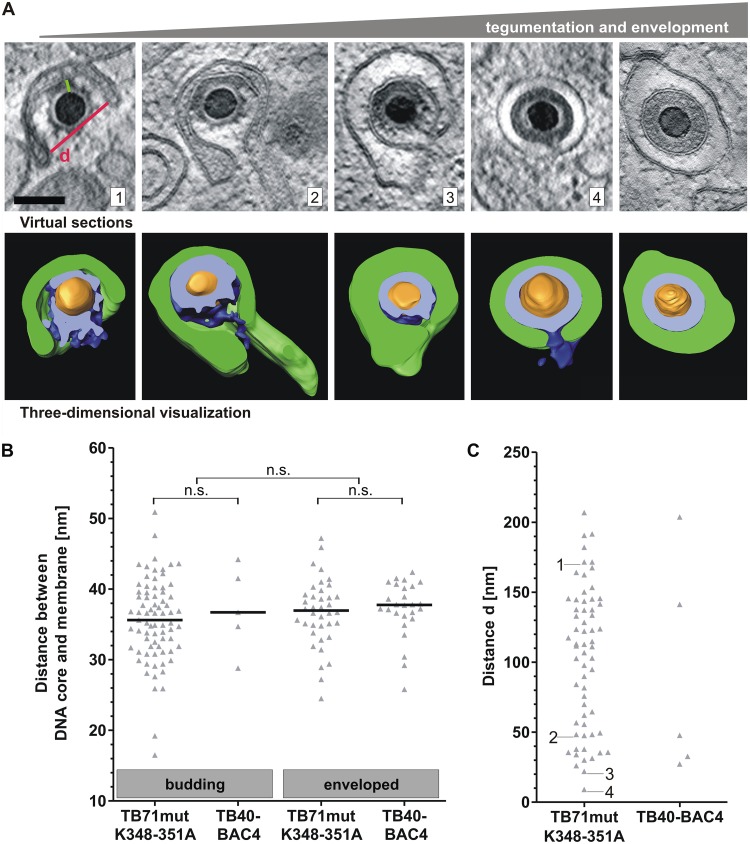
Finally, we sought to investigate the stages of secondary envelopment by three-dimensional electron microscopy to gain more insights into its mechanism. Secondary envelopment is not very frequently observed in wild-type-virus-infected cells. To analyze sufficient numbers of the various stages, we made use of the mutant virus TB71mutK348-351A, which exhibits a morphologically indistinguishable secondary envelopment to wild-type virus (budding of single capsids into small vesicles) with the advantage of being able to observe increased numbers of cytoplasmic capsids undergoing secondary envelopment. Sixty-two capsids at different stages of secondary envelopment were analyzed from 11 tomograms of TB71mutK348-351A-infected cells, whereas only 5 budding capsids were found in 8 tomograms of wild-type-virus-infected cells. As shown in Fig. 7A, envelopment starts with capsids that are already decorated with electron-dense material, which is most likely formed by tegument proteins. This surprisingly regularly spaced material is protruding from capsids and makes contact with the membrane. As the membrane wraps around the capsid, this material appears to keep a specific median distance (approximately 36 nm) to the membrane (Fig. 7B). This distance in TB71mutK348-351A budding capsids is similar to what is measured in budding capsids of the wild-type virus and also similar to the median distance in intracellular enveloped capsids of the mutant as well as of the wild-type virus (Fig. 7B).
FIG 7.
Stages of HCMV secondary envelopment. (A) TB71mutK348-351A virus capsids at different stages of secondary envelopment based on three-dimensional STEM tomography. Images from left to right show representative capsids with increasing amounts of tegument in correlation with different stages of secondary envelopment. In three-dimensional visualizations, membranes are segmented in green, tegument in blue, and capsids in orange. The cross sections through the tegument are shown in bright blue. Scale bar, 100 nm. (B) The distance between the outline of the DNA core and the enwrapping membrane (green line in image A1) of budding and enveloped capsids was measured in tomograms of TB71mutK348-351A- and TB40-BAC4-infected cells. Every triangle represents the distance measurement for a single virus particle. The black lines indicate the median distance. Significance was calculated by using the Kruskal-Wallis test followed by a Dunn’s multiple-comparison test (P < 0.05). (C) Distance d (red line in image A1) was determined for 62 TB71mutK348-351A capsids and 5 TB40-BAC4 capsids. Each data point represents d of a single budding capsid. The numbers 1 to 4 in the diagram indicate the distance of the respective capsids shown in A.
In contrast to the tegument of capsids at the initial phase of envelopment, the tegument in intracellular enveloped virions of mutant and wild-type viruses appears as a uniform electron-dense layer without distinct structures. Numerous stages of envelopment of capsids were recorded from mutant-virus-infected cells. It was interesting to note that the formation of a uniform tegument appears to start at one side of the budding capsid. Capsids in advanced stages of membrane wrapping, defined by the smaller distance between the two arms of the enveloping vesicle, usually exhibit larger portions of uniform tegument, suggesting that the tegument forms along with the process of secondary envelopment. Secondary envelopment is completed by a membrane scission event, which requires the previous formation of a narrow bud neck. Numerous capsids were found at a stage where the two arms of the enveloping membrane were only separated by a small gap in TB71mutK348-351A-infected cells. Quantitative analysis of the three-dimensional data of these virus particles revealed the formation of a narrow bud neck at these vesicles, with a minimal observable neck width of 9.1 nm (Fig. 7C).
Next, we investigated whether the C-terminal mutation in pUL71 has an adverse effect on a specific stage of secondary envelopment that would lead to an accumulation of capsids in that specific stage. The stages of secondary envelopment were defined based on the distance between the two arms of the enveloping vesicle. In STEM tomograms, this distance ranged from 9.1 nm (bud neck) (Fig. 7A, capsid 4) to 206.9 nm (crescent-shaped vesicle) (Fig. 7A, capsid 1). There was no apparent accumulation of capsids at a specific distance, suggesting that the tetralysine motif of pUL71 is not involved in a specific stage of secondary envelopment (Fig. 7C). The distance d in the few observed budding wild-type capsids was distributed in a similar range as in the mutant virus.
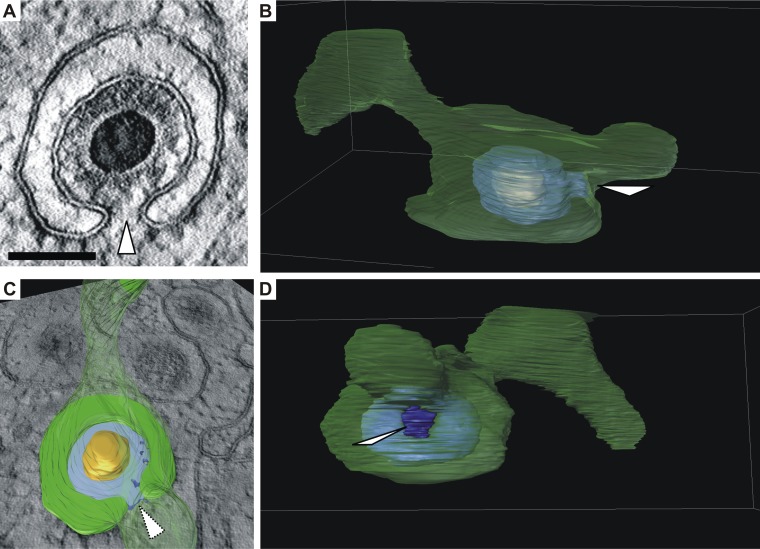
Another interesting observation in STEM tomograms was that many capsids were budding into vesicles that were not merely spherical but exhibited more complex shapes, frequently featuring tubular extensions (Fig. 8 and see also Movie S1 in the supplemental material). These additional three-dimensional membrane features are usually not apparent in two-dimensional micrographs, which are obtained from ultrathin sections. Due to a section thickness of only 70 nm, ultrathin sections contain only a small portion of these vesicles. Thus, it frequently appears in two-dimensional cross sections that capsids bud into small spherical vesicles (crescent-shaped profile in two dimensions), which seem to provide not enough membrane to finish envelopment.
FIG 8.
Formation of a bud neck (white arrowheads). (A) Virtual section of a representative budding HCMV capsid based on three-dimensional STEM tomography showing a bud neck. Scale bar, 100 nm. (B and C) Three-dimensional visualizations of the capsid in panel A which show that the vesicle membrane (green, semitransparent) has a large tubular shape. Orange, capsid; blue, tegument. (C) Cross section through the vesicle as shown in image A. The cross section through the tegument is shown in light blue. (B and D) Views of the vesicle from different angles (see Movie S1 in the supplemental material).
In summary, the detailed analysis of HCMV secondary envelopment in TB71mutK348-351A-infected cells suggests a coordinated process by which the tegument layer is formed during the secondary envelopment process, which culminates in a narrow bud neck that exhibits a critical width prior to the scission event.
DISCUSSION
The mechanism of HCMV secondary envelopment is currently not understood. One of the few viral proteins known to modulate this process is pUL71. It is interesting to note that pUL71 is highly conserved among its herpesvirus homologs and appears to share similar functionality, such as being involved in secondary envelopment (14, 23, 24, 27). HCMV pUL71 is one of the largest proteins among its homologs, whereas its C terminus is the least conserved part. Here, we found that the C terminus of pUL71, specifically a tetralysine motif, is required for efficient HCMV secondary envelopment. This adds another important functional region to the already identified motifs in the N terminus of pUL71. Our new findings underline the role of pUL71 for the process underlying the budding of capsids at membranes, just before the final membrane scission event that is required to complete HCMV secondary envelopment.
Ultrastructural analysis reveals a function of the pUL71 C terminus in secondary envelopment.
The identification of the tetralysine motif for secondary envelopment relied, as in previous analyses of pUL71 mutant viruses, on a detailed quantitative TEM analysis of virus-infected HFFs at late stages of infection. The exceptional structural preservation of biological membranes by the use of high-pressure freezing and freeze substitution for EM sample preparation allowed us to distinguish and quantify the different stages of maturation that cytoplasmic capsids undergo to obtain their envelope. Sample processing, image acquisition, and quantification of envelopment stages in this study were conducted exactly as for other mutant viruses of pUL71 (14, 26), which justifies a comparison of the results of this study with previous quantifications. In addition, we included the well-characterized UL71-deficient mutant virus, TB71stop, in this study for direct comparison. While the impairment of secondary envelopment of the C-terminal deletion mutants as well as of the tetralysine mutant is similar to the defect quantified in TB71stop infections, other ultrastructural characteristics of UL71 mutant viruses were not found, including enlarged MVBs, budding of numerous capsids into MVBs, and multiple budding events of capsids at extended membrane compartments (14, 25, 26). This leads to two major conclusions. First, the C-terminal tetralysine motif plays a role for HCMV secondary envelopment without visibly affecting the morphology of MVBs and other membranes of the vAC. Thus, it seems that the pUL71 C terminus is mechanistically involved in the secondary envelopment process. Second, pUL71 harbors additional functions other than modulating secondary envelopment, which are connected to the organization of membranes of the vAC and to an altered morphology of MVBs. As far as we know, these additional functions of pUL71 are connected to a leucine-zipper like motif involved in oligomerization of pUL71 (26) and a tyrosine-based trafficking motif regulating accumulation of pUL71 at the vAC (25).
Contribution of the pUL71 tetralysine motif to secondary envelopment.
One still unanswered but important question is how the C terminus and the tetralysine motif contribute to secondary envelopment. Due to the lack of solid structural data for pUL71, we can only speculate. However, we can exclude that an altered intracellular localization of pUL71 accounts for the impaired secondary envelopment upon mutation of the tetralysine motif (Fig. 4 and 5; Table 1). We can also exclude that the slightly lower pUL71 protein levels in TB71mutK348-351A-infected cells are responsible because of unaltered pUL71 levels and a similar phenotype of the C-terminally truncated mutants (Fig. 2A, 3, and 6). Mechanistically, pUL71 can fulfill its role in secondary envelopment only when it is able to associate with membranes at the vAC. It was recently shown that pUL71 is localized to the cytoplasmic face of the Golgi membranes (25). Since pUL71 lacks a transmembrane domain, it requires means to associate with membranes. Lysine is a positively charged polar amino acid. By mutating the tetralysine motif, the positively charged polar lysine residues were exchanged with uncharged nonpolar alanine residues. There is good evidence that positively charged amino acids can directly interact with negatively charged components of the membrane, such as phospholipids (40–43). Interestingly, several known lipid-binding domains, including PH and FYVE domains, are dominated by positively charged amino acids, supporting the idea that lysine and arginine interact with lipids (41). Thus, the pUL71 C terminus could be involved in lipid interaction and thereby contribute to secondary envelopment. This hypothesis is difficult to address, because membrane interaction of positively charged amino acids is mostly transient and facilitated by electrostatic interaction with lipids. Our data showing that the mutation of the tetralysine motif to uncharged alanines has no detectable effect on the intracellular localization of pUL71 and its localization at golgin-97-positive membranes at the vAC suggest that membrane association and trafficking of pUL71 are not detectably impaired. This is perhaps not surprising, considering that pUL71 is likely palmitoylated at conserved N-terminal cysteine residues similarly to its homolog pUL51 in HSV-1 and that palmitoylation facilitates membrane association of pUL51 (33). Proteins that are involved in a membrane deformation process, such as envelopment of viral membranes, often require multiple membrane interactions (44). Hence, it is feasible to postulate that the tetralysine motif might, while not being required for association of pUL71 with membranes, interact with the membrane and align the protein in such a way that allows pUL71 to function in membrane deformation and scission. The induction of membrane curvature is very important for the secondary envelopment process, as it facilitates the engulfment of the capsid by cellular membranes. Three mutually nonexclusive ways are postulated by which proteins can induce membrane curvature: (i) the scaffold mechanism, (ii) the local spontaneous curvature mechanism, and (iii) the bilayer-couple mechanism (reviewed in reference 45). It would be interesting to investigate whether pUL71 exerts any of these mechanisms to support secondary envelopment.
It is also possible that the mutation of the tetralysine motif alters protein folding. In this context, positively charged amino acids stabilize protein folding by their interaction with negatively charged regions. It is currently unknown to us whether the C-terminal tetralysine motif is present at the surface or buried in the context of the folded protein.
Alternatively, the C terminus could be involved in an interaction with cellular or viral proteins to either recruit these proteins to the vAC or to form a functional protein complex. Potential interaction partners of pUL71 that were found in a yeast-to-hybrid screen are UL24, UL26, UL89.2, and UL51 (46). The only verified interaction with pUL71, however, is with pUL103 (47, 48). The exact role of the protein complex for virus morphogenesis is currently under investigation.
Another functional aspect of positively charged amino acid motifs for nuclear import is their function as nuclear localization signals (NLSs) (49, 50). For example, the NLS of the simian virus 40 large T-antigen consists of a single cluster of the amino acids PKKKRKV (51, 52). Despite the lack of experimental proof, we think it is unlikely that the tetralysine motif functions as an NLS, because nuclear localization of pUL71 has not been found with either transient expression (25) or infection (27, this paper). In addition, it is difficult to imagine how nuclear localization of pUL71 would contribute to HCMV secondary envelopment.
Thus, the exact function or contribution of the tetralysine motif for the process of secondary envelopment remains elusive. It would be very interesting to solve the structure of pUL71 in future studies, which might help to clarify how the C terminus contributes to HCMV secondary envelopment.
Impact of impaired secondary envelopment on viral growth.
The generation of mutant viruses with a defect in secondary envelopment without affecting the morphology of MVBs and other vAC membranes allowed us to determine the impacts of this defect on viral spread and release. The C-terminal deletion mutants and the tetralysine mutant exhibited impaired cell-associated spread and decreased extracellular virus yields in multistep growth kinetics experiments, consistent with what would be expected when fewer enveloped intracellular particles are generated (Fig. 6A and B). Remarkably, cell-associated growth and release of infectivity were less severely impaired in C-terminal mutant viruses than in a mutant virus that is unable to express pUL71, despite a comparable impairment of secondary envelopment (Table 1 and Fig. 6A and B). Furthermore, the 5-fold reduction of enveloped capsids at the vAC in the C-terminal pUL71 mutants led to a reduction of extracellular virus titers in a single-step growth kinetic of only 1.5-fold (Fig. 6C). This suggests that secondary envelopment is not the rate-determining step for the release of infectious virus particles. This is in line with our previous three-dimensional ultrastructural study showing that the total number of enveloped capsids at one vAC largely exceeds the number of infectious virions released from an infected cell (3, 53).
Considering the ultrastructural alterations in TB71stop-virus-infected cells, the structure and/or origin of membranes and vesicles at which secondary envelopment occurs is also of great importance for the efficiency of viral growth and seems to be at least partially controlled by pUL71.
Electron microscopic analyses of virus morphogenesis show the ultrastructure of infected cells at a given time. The finding of enveloped virus particles at vACs and infectious virus in supernatants indicates that the generation of mature virus particles is not completely blocked in UL71-mutant-virus-infected cells. However, secondary envelopment appears to be impeded, as indicated by an accumulation of capsids in the process of secondary envelopment in the examined UL71 mutants.
Three-dimensional imaging of budding capsids leads to a better understanding of tegumentation and secondary envelopment.
The increased number of capsids undergoing envelopment in TB71mutK348-351A-infected cells and the similar morphology of this process compared to that with wild-type virus infection allowed us to investigate the various stages of secondary envelopment in greater detail with the help of three-dimensional electron microscopy.
Initiation of envelopment seems to be unaffected in UL71 mutant viruses, as the vast majority of capsids in close contact with membranes are at least partially enwrapped by them. STEM tomography of individual capsids at various stages of maturation suggested that the formation of the tegument proceeds with the progression of envelopment. We want to emphasize that the bulk of these data originated from TB71mutK348-351A-infected cells and thus has to be translated carefully regarding envelopment stages in wild-type-virus-infected cells. However, we do find similar stages in wild-type-infected cells albeit with much lower frequency (Table 1 and Fig. 7B and C). The same examination in wild-type-infected cells would have required us to acquire approximately ten times as many tomograms to reach sufficient numbers of budding capsids for quantitative analysis.
With all due caution, we postulate a model of tegument formation in which the majority of tegument proteins is already associated with the capsid or the membrane at the step of envelopment initiation (16) and in which their concentration by protein-protein interactions and further maturation steps (e.g., posttranslational modifications) result in an ultrastructurally uniform tegument layer. This notion is supported by numerous data reporting on the accumulation of tegument proteins at the vAC (16, 19, 25) and is in agreement with the current model of tegumentation (54–57).
By showing that the thickness of the tegument layer was similar in budding and intracellular enveloped particles, we have reason to believe that the thickness of the tegument layer is fully established at the start of secondary envelopment. The regularly spaced tegument material around the capsid, which has also been observed by STEM tomography of wild-type capsids (3), most likely serves as a scaffold for the forming tegument.
The envelopment process culminates in the formation of a narrow bud neck with a minimal distance of 9.1 nm between the membranes. This bud neck appears as a small hole in the enveloping vesicle membrane in three dimensions. This is, to our knowledge, the first report that shows the formation of a narrow bud neck in herpesvirus secondary envelopment using three-dimensional electron microscopy. At least in TB71mutK348-351A-infected cells and the few wild-type capsids, no apparent protein accumulation was visible at the bud neck. This is in contrast to human immunodeficiency virus (HIV) infection (58), where the gap between the opposing membranes is filled with electron densities originating from cellular proteins associated with endosomal sorting complex required for transport (ESCRT)-mediated membrane scission events. However, this has to be interpreted with caution, since our preparation method is different from the method that was used for the study of HIV infection. Nevertheless, it is still unclear how the membrane scission is facilitated in HCMV infection. The lack of electron-dense material at the bud neck supports recent published data that the ESCRT machinery is not required for this process in HCMV (59). Thus, it is feasible to hypothesize that scission in HCMV infection may be mediated by the action of viral proteins, e.g., pUL71, or with the aid of other cellular proteins that still need to be identified. Unravelling the mechanism of secondary envelopment will certainly require a lot more work.
MATERIALS AND METHODS
Cells.
Human foreskin fibroblasts (HFFs) were maintained in minimal essential medium (MEM; Gibco-BRL) and were used before passage 23. Human embryonic lung fibroblasts (MRC-5; European Collection of Cell Cultures) were maintained in Dulbecco’s modified Eagle medium (DMEM; Gibco-BRL) and used between passage 21 and 30 for virus reconstitution. Both MEM and DMEM were supplemented with 2 mM l-glutamine (200 mM; PAA Laboratories), 10% fetal calf serum, 100 U of penicillin, and 100 μg of streptomycin (all three from Gibco-BRL) and 1× nonessential amino acids (Biochrom AG).
Generation of mutant viruses.
The parental virus of all recombinant viruses of this study was reconstituted from the HCMV bacterial artificial chromosome (BAC) clone TB40-BAC4 derived from the endotheliotropic HCMV strain TB/40E (accession number EF999921.1 [35]). Recombinant viruses were generated by using the markerless two-step RED-GAM recombination as described previously (60, 61).
This recombination method uses primers that contain homology to the UL71 sequence upstream and downstream of the sequence to be mutated and homology to the pEPkan-S template plasmid (60). The bacmid of recombinant virus TB71del327-361 was generated with the primers ep_71del35aa_for (5′-CAGCCCCGACGTGCCTCGGGAGGCGGTGATGTCACCGACCTGAACAGCCTGGCACGTTTTAGGATGACGACGATAAGTAGGG-3′) and ep_71del35aa_rev (5′-CGTGATCACGTACGTTTTCCAAAACGTGCCAGGCTGTTCAGGTCGGTGACATCACCGCCTCAACCAATTAACCAATTCTGATTAG-3′), which deleted codons 327 to 361 of the UL71 sequence. The primers ep_71del14aa_for (5′-GATACCCTTTGTGGGTTCCGCGCGTGAACTCAGGGGCGTGTGGAGAAAAAGAAACCCACGGCGAGGATGACGACGATAAGTAGGG-3′) and ep_71del14aa_rev (5′-GCGGAGGACAGCAAGGCCGCCGCCGTGGGTTTCTTTTTCTCCACACACGCCCCTGAGTTCACGCGCAACCAATTAACCAATTCTGATTAG-3′) were used to generate the bacmid of the TB71del348-361 mutant virus. Instead of a deletion, a stop codon was introduced at codon 348 followed by a frameshift, which leads to the expression of a C-terminally truncated pUL71. The bacmid of recombinant virus TB71mutK348-351A was generated with the primers ep-UL71-K348-51A_for (5′-GATACCCTTTGTGGGTTCCGCGCGTGAACTCAGGGGCGTGGCGGCCGCTGCGCCCACGGCGGAGGATGACGACGATAAGTAGGG-3′) and ep-UL71-K348-51A_rev (5′-AGGCTGTTCACGCGGAGGACAGCAAGGCCGCCGCCGTGGGCGCAGCGGCCGCCACGCCCCTGCAACCAATTAACCAATTCTGATTAG-3′). Bacmid TB71mutK348-351A and primers ep-UL71-rescK348_51A_for (5′-GATACCCTTTGTGGGTTCCGCGCGTGAACTCAGGGGCGTGAAGAAGAAGAAACCCACGGCGGAGGATGACGACGATAAGTAGGG-3′) and ep-UL71-rescK348_51A_rev (5′-AGGCTGTTCACGCGGAGGACAGCAAGGCCGCCGCCGTGGGTTTCTTCTTCTTCACGCCCCTGCAACCAATTAACCAATTCTGATTAG-3′) were used to restore the mutation K348-351A back to the parental amino acid sequence in the revertant virus. In order to distinguish TB40-BAC4 and TB71revK348-351A at the DNA level, a silent mutation was introduced in TB71revK348-351A. The bacmid of the virus TB71mutL357-358A was generated with the primers ep-UL71-L357/8A_for (5′-ACTCAGGGGCGTGAAGAAAAAGAAACCCACGGCGGCGGCCGCTGCGTCCTCCGCGTGAACAGCAGGATGACGACGATAAGTAGGG-3′) and ep-UL71-L357/8A_rev (5′-GTACGTTTTCCAAAACGTGCCAGGCTGTTCACGCGGAGGACGCAGCGGCCGCCGCCGTGGGTTCAACCAATTAACCAATTCTGATTAG-3′). Sequencing of the mutated regions confirmed the correct introduction of the mutations. The bacmids of the recombinant HCMV viruses were reconstituted in MRC-5 as described previously (62, 63). HFFs were then used for further virus propagation and generation of virus stocks. The generation of the UL71-deficient virus (TB71stop) is described elsewhere (14).
Antibodies.
HCMV and cellular proteins were detected with monoclonal antibodies (MAb) in indirect immunofluorescence stainings, Western blot analyses, focus expansion assays, and virus titrations. The mouse MAbs were directed against HCMV IE1/2 (MAb 63-27), HCMV major capsid protein (MCP; MAb 28-4), pp65 (pUL83; MAb 65-33, kindly provided by W. Britt, Alabama University, Birmingham), pp28 (clone 5C3; Santa Cruz Biotechnology), cellular golgin-97 (IgG1; Invitrogen), and cellular actin (Sigma). HCMV pUL71 was detected in indirect immunofluorescence experiments and Western blot analyses with a polyclonal antibody raised against the C terminus of pUL71 (amino acids [aa] 162 to 361) (14). In indirect immunofluorescence experiments and focus expansion assays, goat anti-rabbit and goat anti-mouse antibodies conjugated to Alexa Fluor (AF) 488 or AF555 (Invitrogen) were used as secondary antibodies. Goat anti-mouse or goat anti-rabbit antibodies conjugated to StarBright Blue 700 or StarBright Blue 520 (Bio-Rad) were used as secondary antibodies in Western blot analyses.
Indirect immunofluorescence staining.
HFFs were seeded in μ-Slide 8-well chamber slides (Ibidi GmbH; Germany) and synchronized by serum starvation for 48 h prior to infection with the respective viruses at a multiplicity of infection (MOI) of 1. At various times postinfection, cells were washed with phosphate-buffered saline (PBS) and subsequently fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at 4°C. Indirect immunofluorescence stainings were performed as described previously (16, 25). Briefly, cells were permeabilized with 0.1% Triton X-100, and unspecific binding sites were blocked with PBS containing 1% bovine serum albumin and 10% human serum from HCMV-negative individuals. Primary and secondary antibodies were diluted in blocking solution. DNA was stained with 30 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Roche) diluted in PBS. Confocal images were acquired with the 63× lens objective of the Axio-Observer.Z1 fluorescence microscope equipped with an Apotome, and images were processed with the Zen 2.3 software (Zeiss).
Virus growth analyses.
For multistep growth kinetics experiments, confluent HFFs were infected in triplicates with an infection rate of approximately 0.3% for each virus. The titers of the inocula were calculated from the actual infection rate, which was determined 24 h postinfection by detection of IE-positive cells. The first supernatants were harvested at 24 h postinfection by removing the input virus followed by three washings with warm PBS, addition of complete MEM, and an incubation for 10 min at 37°C (day 1). Additional supernatants were harvested at days 3, 6, 9, and 12 postinfection. All supernatants were stored at −80°C until titration.
For the single-step growth kinetics experiments, confluent HFFs were infected in duplicates at an MOI of 3. To control for the same initial infection, virus yields of the inocula were determined on HFFs by titration. Input virus was removed, cells were washed with warm PBS, and fresh MEM was added 90 min postinfection. Supernatants were harvested every 24 h and stored at −80°C. Determination of virus yields in the inocula and the harvested supernatants from the multistep and single-step growth kinetics experiments was performed in triplicates by titration on HFFs as described previously (14).
The ability of cell-associated spread was determined in a focus expansion assay, as described previously (25). Briefly, confluent HFFs in 12-well plates (2 × 105 HFFs/well) were infected with 50, 100, and 150 PFU/well of the respective virus. After 24 h, the cells were washed thoroughly with warm PBS and cultivated for another 24 h in fresh MEM. Cells were kept under a 0.65% methylcellulose overlay medium from 3 days postinfection until the medium was removed, and then the cells were washed with warm PBS and fixed with ice-cold methanol for 10 min at −20°C at day 9 of infection. HCMV-infected cells were detected by staining of IE1/2 antigen. The nuclei were counterstained with DAPI. The experiment was repeated three times, and at least 50 foci of each experiment and virus were acquired in the Axio-Observer.Z1 fluorescence microscope with the 10× lens objective. The number of IE-positive nuclei of each focus was determined using ImageJ software (64) with the cell-counter plugin (https://imagej.nih.gov/ij/plugins/cell-counter.html; Kurt De Vos, University of Sheffield, Academic Neurology). The mean focus size for each virus was calculated and normalized to the mean focus size in TB40-BAC4-infected wells, and statistical analysis was performed by Kruskal-Wallis test (P < 0.05) followed by Dunn’s multiple-comparison testing (P < 0.05).
Electron microscopy.
Sample preparation for transmission electron microscopy (TEM) and quantitative studies of virus morphogenesis by TEM were performed as described previously (3). In short, HFFs were grown on carbon-coated sapphire discs (3-mm diameter, 50-μm thickness; Wohlwend GmbH), infected with the respective viruses at an MOI of 1, and fixed at 120 h postinfection by high-pressure freezing (HPF Compact 01; Wohlwend GmbH) followed by freeze substitution (13) and stepwise embedding in epoxy resin (Fluka). Ultrathin sections of 70-nm thickness of HCMV-infected cells were examined with a JEM-1400 (Jeol) TEM equipped with a charge-coupled-device (CCD) camera at an accelerating voltage of 120 kV. Enveloped, budding, and naked cytoplasmic capsids were quantified in micrographs of regions of vACs of randomly chosen virus-infected cells from at least three independent experiments for each virus.
Fixation and embedding of infected cells for STEM tomography was the same as described for TEM. Subsequent sample preparation steps were conducted as described previously (65, 66). Briefly, sections of infected cells with a thickness between 500 and 700 nm were prepared and mounted on EM grids with 200 parallel grid bars (Plano) which had been pretreated with poly-l-lysine (10% in water). Mounted sections were again treated with poly-l-lysine before attachment of colloidal gold fiducials and finally coated with a 5-nm-thick layer of carbon. Tomogram acquisition was conducted on a STEM JEM-2100F (Jeol) with an accelerating voltage of 200 kV. Tilt series were acquired from at least +68° to −68° with a 1.5° increment using the bright-field detector. Image series were reconstructed to tomograms by weighted back projection with the IMOD software package (67). The distance of the two arms of the enveloping vesicle was measured from virtual sections of tomograms of budding capsids using the IMOD software (Fig. 7). The distances were determined for 62 capsids of the mutant TB71mutK348-351A from 11 tomograms and 5 capsids of the TB40-BAC4 virus from 8 tomograms. Furthermore, the distance between the DNA core and the membrane (thickness of tegument plus thickness of the capsid shell) was measured in virtual sections for 38/75 (enveloped/budding) capsids from 11 tomograms in the mutant virus and 26/9 (enveloped/budding) capsids from 8 tomograms of the wild-type virus. Three-dimensional visualization of budding capsids was performed using Avizo 6.3 software (Visualization Science Group, Burlington, MA, USA) by manual segmentation of the membrane and capsid structure. Threshold segmentation was used for the visualization of tegument densities in these three-dimensional reconstructions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Claudia Ploil, Anke Lüske, Jutta Hegler, Monika Dürre (Institute of Virology, Ulm, Germany), Renate Kunz, and Reinhard Weih (Central Facility of Electron Microscopy, Ulm, Germany) for their technical assistance. We also thank Andrea Dietz, Sascha Suffner, and Daniela Fischer (Institute of Virology, Ulm, Germany) for their experimental help.
This work was partially supported by the Baustein 3.2. program of Ulm University and the MWK Baden-Wuerttemberg through the Junior Professor Program.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02244-18.
REFERENCES
- 1.Griffiths P, Baraniak I, Reeves M. 2015. The pathogenesis of human cytomegalovirus. J Pathol 235:288–297. doi: 10.1002/path.4437. [DOI] [PubMed] [Google Scholar]
- 2.Mocarski E, Shenk T, Pass R. 2006. Cytomegaloviruses, p. 2701–2771. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA (ed), Fields virology 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Schauflinger M, Villinger C, Mertens T, Walther P, von Einem J. 2013. Analysis of human cytomegalovirus secondary envelopment by advanced electron microscopy: HCMV morphogenesis. Cell Microbiol 15:305–314. doi: 10.1111/cmi.12077. [DOI] [PubMed] [Google Scholar]
- 4.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi: 10.1128/JVI.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alwine JC. 2012. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog 8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cepeda V, Esteban M, Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol 12:386–404. doi: 10.1111/j.1462-5822.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- 8.Das S, Pellett PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol 85:5864–5879. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol 81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa-Goto K, Tanaka K, Gibson W, Moriishi E, Miura Y, Kurata T, Irie S, Sata T. 2003. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J Virol 77:8541–8547. doi: 10.1128/JVI.77.15.8541-8547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Procter DJ, Banerjee A, Nukui M, Kruse K, Gaponenko V, Murphy EA, Komarova Y, Walsh D. 2018. The HCMV assembly compartment is a dynamic Golgi-derived MTOC that controls nuclear rotation and virus spread. Dev Cell 45:83.e7–100.e7. doi: 10.1016/j.devcel.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homman-Loudiyi M, Hultenby K, Britt W, Söderberg-Nauclér C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J Virol 77:3191–3203. doi: 10.1128/JVI.77.5.3191-3203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walther P, Ziegler A. 2002. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J Microsc 208:3–10. doi: 10.1046/j.1365-2818.2002.01064.x. [DOI] [PubMed] [Google Scholar]
- 14.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol 85:3821–3832. doi: 10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mach M, Osinski K, Kropff B, Schloetzer-Schrehardt U, Krzyzaniak M, Britt W. 2007. The carboxy-terminal domain of glycoprotein N of human cytomegalovirus is required for virion morphogenesis. J Virol 81:5212–5224. doi: 10.1128/JVI.01463-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cappadona I, Villinger C, Schutzius G, Mertens T, von Einem J. 2015. Human cytomegalovirus pUL47 modulates tegumentation and capsid accumulation at the viral assembly complex. J Virol 89:7314–7328. doi: 10.1128/JVI.00603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips SL, Cygnar D, Thomas A, Bresnahan WA. 2012. Interaction between the human cytomegalovirus tegument proteins UL94 and UL99 is essential for virus replication. J Virol 86:9995–10005. doi: 10.1128/JVI.01078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips SL, Bresnahan WA. 2012. The human cytomegalovirus (HCMV) tegument protein UL94 is essential for secondary envelopment of HCMV virions. J Virol 86:2523–2532. doi: 10.1128/JVI.06548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo J-Y, Britt WJ. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J Virol 80:5611–5626. doi: 10.1128/JVI.02630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo J-Y, Britt WJ. 2007. Cytoplasmic Envelopment of Human Cytomegalovirus Requires the Postlocalization Function of Tegument Protein pp28 within the Assembly Compartment. J Virol 81:6536–6547. doi: 10.1128/JVI.02852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva MC, Yu Q-C, Enquist L, Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol 77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahlqvist J, Mocarski E. 2011. Cytomegalovirus UL103 controls virion and dense body egress. J Virol 85:5125–5135. doi: 10.1128/JVI.01682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp BG, Granzow H, Klopfleisch R, Fuchs W, Kopp M, Lenk M, Mettenleiter TC. 2005. Functional analysis of the pseudorabies virus UL51 protein. J Virol 79:3831–3840. doi: 10.1128/JVI.79.6.3831-3840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nozawa N, Kawaguchi Y, Tanaka M, Kato A, Kato A, Kimura H, Nishiyama Y. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J Virol 79:6947–6956. doi: 10.1128/JVI.79.11.6947-6956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietz AN, Villinger C, Becker S, Frick M, von Einem J. 2017. A tyrosine-based trafficking motif of the tegument protein pUL71 is crucial for human cytomegalovirus secondary envelopment. J Virol 92:e00907-17. doi: 10.1128/JVI.00907-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meissner CS, Suffner S, Schauflinger M, von Einem J, Bogner E. 2012. A leucine zipper motif of a tegument protein triggers final envelopment of human cytomegalovirus. J Virol 86:3370–3382. doi: 10.1128/JVI.06556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Womack A, Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio 1:e00282-10. doi: 10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumeister J, Klupp BG, Mettenleiter TC. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J Virol 69:5560–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenk M, Visser N, Mettenleiter TC. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J Virol 71:5635–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mettenleiter TC. 2004. Budding events in herpesvirus morphogenesis. Virus Res 106:167–180. doi: 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Roller RJ, Fetters R. 2015. The herpes simplex virus 1 UL51 protein interacts with the UL7 protein and plays a role in its recruitment into the virion. J Virol 89:3112–3122. doi: 10.1128/JVI.02799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nozawa N, Daikoku T, Koshizuka T, Yamauchi Y, Yoshikawa T, Nishiyama Y. 2003. subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J Virol 77:3204–3216. doi: 10.1128/JVI.77.5.3204-3216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roller RJ, Haugo AC, Yang K, Baines JD. 2014. The herpes simplex virus 1 UL51 gene product has cell type-specific functions in cell-to-cell spread. J Virol 88:4058–4068. doi: 10.1128/JVI.03707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 36.Kropff B, Koedel Y, Britt W, Mach M. 2010. Optimal replication of human cytomegalovirus correlates with endocytosis of glycoprotein gpUL132. J Virol 84:7039–7052. doi: 10.1128/JVI.01644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 38.Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NFW, Curmi PM, Forwood JK, Bodén M, Kobe B. 2011. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta 1813:1562–1577. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 39.Oka M, Yoneda Y. 2018. Importin α: functions as a nuclear transport factor and beyond. Proc Jpn Acad Ser B Phys Biol Sci 94:259–274. doi: 10.2183/pjab.94.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. 1996. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys J 71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheglmann D, Werner K, Eiselt G, Klinger R. 2002. Role of paired basic residues of protein C-termini in phospholipid binding. Protein Eng 15:521–528. doi: 10.1093/protein/15.6.521. [DOI] [PubMed] [Google Scholar]
- 42.Pelassa I, Fiumara F. 2015. Differential occurrence of interactions and interaction domains in proteins containing homopolymeric amino acid Repeats. Front Genet 6:345. doi: 10.3389/fgene.2015.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan HM, He T, Fuglebakk E, Grauffel C, Yang B, Roberts MF, Gershenson A, Reuter N. 2016. A role for weak electrostatic interactions in peripheral membrane protein binding. Biophys J 110:1367–1378. doi: 10.1016/j.bpj.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madsen KL, Bhatia VK, Gether U, Stamou D. 2010. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett 584:1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 45.Zimmerberg J, Kozlov MM. 2006. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7:9. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 46.To A, Bai Y, Shen A, Gong H, Umamoto S, Lu S, Liu F. 2011. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 6:e17796. doi: 10.1371/journal.pone.0017796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer D. 2012. Dissecting functional motifs of the human cytomegalovirus tegument protein pUL71. Dissertation, Ulm University, Ulm, Germany. [Google Scholar]
- 48.Ortiz DA, Glassbrook JE, Pellett PE. 2016. Protein-protein interactions suggest novel activities of human cytomegalovirus tegument protein pUL103. J Virol 90:7798–7810. doi: 10.1128/JVI.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makkerh JP, Dingwall C, Laskey RA. 1996. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol 6:1025–1027. doi: 10.1016/S0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 50.Nakielny S, Dreyfuss G. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677–690. doi: 10.1016/S0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 51.Kalderon D, Richardson WD, Markham AF, Smith AE. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 52.Lanford RE, Butel JS. 1984. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 37:801–813. doi: 10.1016/0092-8674(84)90415-X. [DOI] [PubMed] [Google Scholar]
- 53.Sampaio KL, Cavignac Y, Stierhof Y-D, Sinzger C. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J Virol 79:2754–2767. doi: 10.1128/JVI.79.5.2754-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mettenleiter TC. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet Microbiol 113:163–169. doi: 10.1016/j.vetmic.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 55.Crump C. 2018. Virus assembly and egress of HSV, p. 23–44. In Kawaguchi Y, Mori Y, Kimura H (ed), Human herpesviruses. Springer, Singapore. [DOI] [PubMed] [Google Scholar]
- 56.Owen DJ, Crump CM, Graham SC. 2015. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Close WL, Anderson AN, Pellett PE. 2018. Betaherpesvirus virion assembly and egress, p. 167–207. In Kawaguchi Y, Mori Y, Kimura H (ed), Human herpesviruses. Springer, Singapore. [DOI] [PubMed] [Google Scholar]
- 58.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Streck NT, Carmichael J, Buchkovich NJ. 2018. Nonenvelopment role for the ESCRT-III complex during human cytomegalovirus infection. J Virol 92:e02096-17. doi: 10.1128/JVI.02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 61.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 62.Brock I, Kruger M, Mertens T, von Einem J. 2013. Nuclear targeting of human cytomegalovirus large tegument protein pUL48 is essential for viral growth. J Virol 87:6005–6019. doi: 10.1128/JVI.03558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chevillotte M, Landwehr S, Linta L, Frascaroli G, Luske A, Buser C, Mertens T, von Einem J. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J Virol 83:2480–2490. doi: 10.1128/JVI.01818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Höhn K, Sailer M, Wang L, Lorenz M, Schneider ME, Walther P. 2011. Preparation of cryofixed cells for improved 3D ultrastructure with scanning transmission electron tomography. Histochem Cell Biol 135:1–9. doi: 10.1007/s00418-010-0765-z. [DOI] [PubMed] [Google Scholar]
- 66.Villinger C, Schauflinger M, Gregorius H, Kranz C, Höhn K, Nafeey S, Walther P. 2014. Three-dimensional imaging of adherent cells using FIB/SEM and STEM. Methods Mol Biol 1117:617–638. doi: 10.1007/978-1-62703-776-1_27. [DOI] [PubMed] [Google Scholar]
- 67.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 68.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato A, Oda S, Watanabe M, Oyama M, Kozuka-Hata H, Koyanagi N, Maruzuru Y, Arii J, Kawaguchi Y. 2018. Roles of the phosphorylation of herpes simplex virus 1 UL51 at a specific site in viral replication and pathogenicity. J Virol 92:e01035-18. doi: 10.1128/JVI.01035-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.