Feline leukemia virus (FeLV) is a member of the genus Gammaretrovirus, which causes malignant diseases in cats. The most prevalent FeLV among cats is FeLV subgroup A (FeLV-A), and specific binding of FeLV-A Env to its viral receptor, thiamine transporter feTHTR1, is the first step of infection. In infected cats, novel variants of FeLV with altered receptor specificity for viral entry have emerged by mutation or recombination of the env gene. A novel FeLV variant arose from a subtle mutation of FeLV-A Env, which altered the specific interaction of the virus with its receptor. RFC, a folate transporter, is a potential receptor for the novel FeLV variant. The perturbation of specific retrovirus-receptor interactions under selective pressure by the host results in the emergence of novel viruses.
KEYWORDS: cats, endogenous retrovirus, feline leukemia virus, retroviruses, viral envelope, viral infection, viral pathogenesis, viral receptor
ABSTRACT
Feline leukemia virus (FeLV) is horizontally transmitted among cats and causes a variety of hematopoietic disorders. Five subgroups of FeLV, A to D and T, each with distinct receptor usages, have been described. Recently, we identified a new FeLV Env (TG35-2) gene from a pseudotyped virus that does not belong to any known subgroup. FeLV-A is the primary virus from which other subgroups have emerged via mutation or recombination of the subgroup A env gene. Retrovirus entry into cells is mediated by the interaction of envelope protein (Env) with specific cell surface receptors. Here, phenotypic screening of a human/hamster radiation hybrid panel identified SLC19A1, a feline reduced folate carrier (RFC) and potential receptor for TG35-2-phenotypic virus. RFC is a multipass transmembrane protein. Feline and human RFC cDNAs conferred susceptibility to TG35-2-pseudotyped virus when introduced into nonpermissive cells but did not render these cells permissive to other FeLV subgroups or feline endogenous retrovirus. Moreover, human cells with genomic deletion of RFC were nonpermissive for TG35-2-pseudotyped virus infection, but the introduction of feline and human cDNAs rendered them permissive. Mutation analysis of FeLV Env demonstrated that amino acid substitutions within variable region A altered the specificity of the Env-receptor interaction. We isolated and reconstructed the full-length infectious TG35-2-phenotypic provirus from a naturally FeLV-infected cat, from which the FeLV Env (TG35-2) gene was previously isolated, and compared the replication of the virus in hematopoietic cell lines with that of FeLV-A 61E by measuring the viral RNA copy numbers. These results provide a tool for further investigation of FeLV infectious disease.
IMPORTANCE Feline leukemia virus (FeLV) is a member of the genus Gammaretrovirus, which causes malignant diseases in cats. The most prevalent FeLV among cats is FeLV subgroup A (FeLV-A), and specific binding of FeLV-A Env to its viral receptor, thiamine transporter feTHTR1, is the first step of infection. In infected cats, novel variants of FeLV with altered receptor specificity for viral entry have emerged by mutation or recombination of the env gene. A novel FeLV variant arose from a subtle mutation of FeLV-A Env, which altered the specific interaction of the virus with its receptor. RFC, a folate transporter, is a potential receptor for the novel FeLV variant. The perturbation of specific retrovirus-receptor interactions under selective pressure by the host results in the emergence of novel viruses.
INTRODUCTION
Retroviral envelope (Env) proteins consist of a trimer of heterodimers formed between the surface subunit (SU) and the transmembrane subunit (TM). Interaction of the retroviral SU with a receptor on the host cell surface is the initial step in viral entry. The specific SU-receptor interaction begins with the fusion of viral and host cell membranes, resulting in viral entry into the host cell. Therefore, viral tropism is determined by whether the target cell expresses a surface receptor protein and can bind to the viral SU protein (1). Infection of the target cell by virus usually prevents successive rounds of infection in the same cell as a result of masking or downregulation of the receptor by the viral Env protein. This phenomenon is known as superinfection interference, and this phenomenon identifies whether the virus uses the same or different receptors (2, 3). Therefore, elucidation of the molecular basis for the retrovirus-receptor interaction contributes to our understanding of viral entry and infection.
Feline leukemia virus (FeLV) belongs to the genus Gammaretrovirus and is transmitted horizontally among domestic cats (Felis silvestris catus) (4). A recent epidemiological survey of FeLV infection in Japan detected FeLV in 12.2% of the 1,770 cats tested (5). This virus is known to induce various diseases in domestic cats, such as lymphoma, myelodysplastic syndrome, anemia, acute myelogenous leukemia, and immune deficiency (6, 7). The mechanisms by which this virus induces the multifarious symptoms of FeLV-associated diseases are still unclear; however, genetic polymorphisms resulting from substitution or recombination have led to changes in FeLV pathogenicity and unexpected symptoms (8–14). The analysis of superinfection interference properties has identified FeLV variants comprising FeLV subgroups A, B, C, D, and T and FeLV variant (TG35-2) (15–20). FeLV-A is the primary virus transmitted among cats (21–23), and FeLV subgroups are thought to be generated in cats infected with FeLV-A. FeLV subgroups B and D arise from recombination between FeLV-A env and the env genes of endogenous FeLV (enFeLV) or endogenous retrovirus of the domestic cat (ERV-DC) (17, 24, 25); subgroups C and T and FeLV TG35-2 possibly arise from mutations in the FeLV-A env gene (8–10, 18). The cellular viral receptors for FeLV subgroups A, B, C, and T have been identified; FeLV-A uses the feline thiamine transporter receptor (feTHTR1) (26), while FeLV-B uses the phosphate transporter receptors (Pit1/2) (27–30). FeLV-C uses a heme transporter (FLVCR-1/2) as its receptor along with THTR1 (31–33). FeLV-T, a T-cytopathic FeLV subgroup, also uses Pit1 as a receptor, but it requires a second host protein known as FeLIX, a truncated envelope protein produced by enFeLV for entry (34).
We previously identified the FeLV env gene, TG35-2, in a 1-year-old castrated male cat, TG35, presenting with a bite injury, stomatitis, loss of appetite, and FeLV infection, although he had been vaccinated with inactivated FeLV. He eventually died without diagnosis (5, 18). TG35-2 Env is not classified to any known interference subgroup of FeLV and shows distinct cell tropism from FeLV-A (18). The env sequences of this clone clustered phylogenetically with those of genotype I/clade I FeLV, found mainly in Japan (5). In this study, we used phenotypic screening of radiation hybrid (RH) cell lines (35) to identify SLC19A1, the feline reduced folate carrier (feRFC) as an entry receptor for FeLV TG35-2-phenotypic virus. Substitution of a few amino acids within variable region A (VRA) in Env altered the specificity of the Env-receptor interaction, including facilitating the occurrence of a dual-tropic virus. Furthermore, we isolated and reconstructed the full-length infectious FeLV TG35-2-phenotypic provirus from a naturally FeLV-infected cat, from which the FeLV Env (TG35-2) gene had previously been isolated. Our results provide tools for further investigation of FeLV infectious disease.
RESULTS
Identification of reduced folate carrier as an entry receptor for FeLV variant.
FeLV TG35-2-phenotypic virus (FeLV 33TGE2), a chimeric infectious virus, infects human but not hamster cells (18), indicating that it might be possible to map the position of the receptor of FeLV TG35-2-phenotypic virus by analyzing the susceptibility of human-hamster RH cell lines to infection by FeLV 33TGE2. We used the G3 panel of human RH cell lines from the Stanford Human Genome Center (SHGC) (36) for phenotypic mapping of the receptor for FeLV TG35-2-phenotypic virus. This panel had been previously genotyped using array comparative genomic hybridization (37, 38).
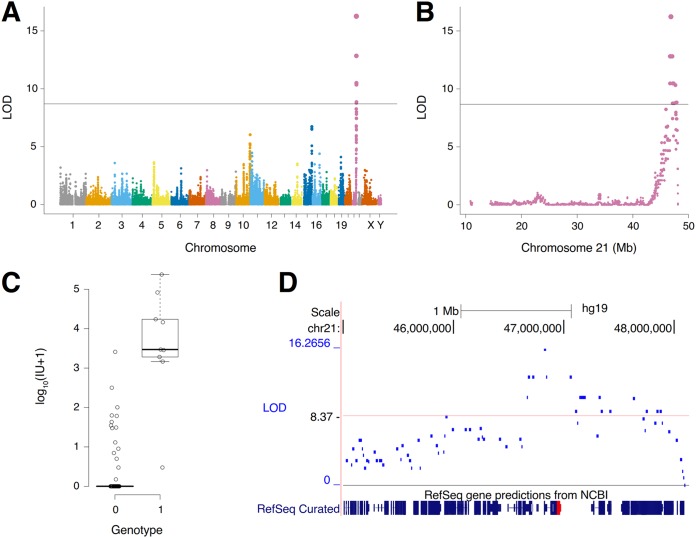
We first confirmed that FeLV 33TGE2 does not infect the recipient A23 hamster cells used in the construction of the G3 panel. We then correlated the genotypes of the RH clones with their susceptibility to FeLV TG35-2-phenotypic virus infection. The combined narrow-sense (additive) heritability, h2, of this phenotype was indistinguishable from 1 (0.99 ± 0.12 standard deviation [SD]), suggesting a simple monogenic architecture (39). Consistent with this observation, we identified a single genome-wide significant locus with a logarithm of the odds (LOD) score of 16.3 on chromosome 21q22.3, with a peak marker at 46,822,915 bp (Fig. 1A and B). The mean log10(IU + 1) (infectious units/milliliter supernatant + 1) was 3.6 ± 0.5 standard error of the mean (SEM) for RH clones with a peak marker and 0.3 ± 0.1 SEM for clones without (Fig. 1C). The additive heritability for the locus was 0.63 ± 0.13 SD, explaining the majority of the overall narrow-sense heritability and showing consistency with a monogenic trait.
FIG 1.
Mapping of a locus for FeLV TG35-2-phenotypic virus infectivity. (A) A genome-significant locus resides on chromosome 21. LOD, logarithm of the odds. (B) The locus is located near the telomere on the long arm of chromosome 21 at 21q22.3. (C) Log10(IU + 1) for RH clones containing the peak marker (genotype = 1) and for clones without (genotype = 0). IU, infectious units per milliliter supernatant. (D) The peak marker is close to the RFC gene (red).
The 2 LOD critical region of the chromosome 21 locus extended from 46,677,060 bp to 47,058,655 bp, or from 146 kb to the left of the peak marker (in the direction of the centromere) to 236 kb to the right (in the direction of the q telomere) (Fig. 1D). Examination of this region of 21q22.3 showed that none of the previously mapped retroviral receptors localized to the same position. Thus, the receptor for FeLV TG35-2-phenotypic virus most likely represents a new FeLV receptor. The gene closest to the peak marker was COL18A1, which was 52.5 kb to the right. The second closest gene was the reduced folate carrier (RFC) gene (SLC19A1), which was 95.2 kb in the same direction.
To determine whether RFC might function as the receptor for FeLV TG35-2-phenotypic virus, we isolated human RFC (huRFC) and feRFC cDNAs from HEK293T cells and feline peripheral blood mononuclear cells (PBMCs), respectively. We generated retroviral expression vectors expressing the cDNAs encoding huRFC or feRFC and introduced them into Mus dunni tail fibroblast (MDTF) cells, because MDTF cells were resistant to infection with Env-pseudotyped FeLV-A and FeLV TG35-2 (18). MDTF cells carrying huRFC (MDTF-huRFC) and feRFC (MDTF-feRFC) were tested for permissiveness to Env-pseudotyped viruses of FeLV-A (FeLV-A clone 33), FeLV-B (FeLV-B Gardner-Arnstein), FeLV-C (FeLV-C Sarma), FeLV-D (FeLV-D Ty26), FeLV TG35-2, ERV-DC10, and amphotropic murine leukemia virus (ampho-MuLV; MuLV 4070A) carrying a LacZ reporter gene, which were prepared in GPLac cells. Ampho-MuLV is known to infect mouse and human cells and was used as a positive control (18).
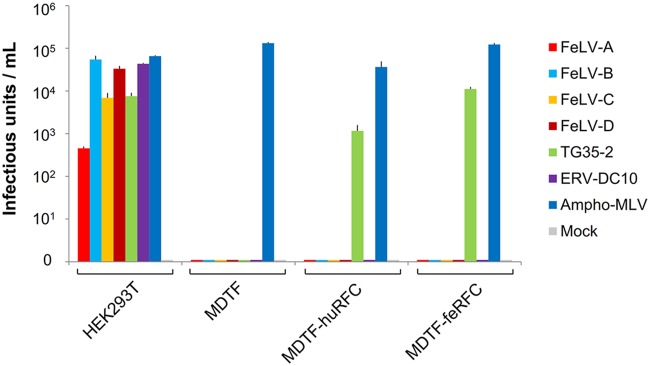
MDTF-huRFC and MDTF-feRFC cells were susceptible to FeLV TG35-2-pseudotyped virus infection with >103 infectious units (Fig. 2). However, MDTF cells carrying an empty vector were not susceptible to FeLV TG35-2-pseudotyped virus infection (Fig. 2). Other feline retroviruses, FeLV-A, FeLV-B, FeLV-C, FeLV-D, and ERV-DC10-pseudotyped viruses, could not infect MDTF-huRFC or MDTF-feRFC cells, whereas FeLV-A, FeLV-B, FeLV-C, FeLV-D, FeLV TG35-2, and ERV-DC10-pseudotyped viruses successfully infected HEK293T cells. These results indicated that huRFC and feRFC conferred susceptibility to FeLV-TG35-2-pseudotyped virus infection.
FIG 2.
Infection of cell lines by LacZ-carrying Env-pseudotyped viruses. Env-pseudotyped viruses of FeLV-A (FeLV-A clone 33), FeLV-B (FeLV-B Gardner-Arnstein), FeLV-C (FeLV-C Sarma), FeLV-D (FeLV-D Ty26), FeLV TG35-2, ERV-DC10, and ampho-MuLV (MuLV 4070A) were tested in the cell lines, HEK293T, MDTF, MDTF expressing human RFC (MDTF-huRFC), and MDTF expressing feline RFC (MDTF-feRFC) shown on the x axis. The y axis indicates the infectious units using log10 of β-galactosidase (LacZ)-positive cells per ml of supernatant. Data were obtained from three independent experiments in triplicates and represent the averages from nine results and the standard deviation.
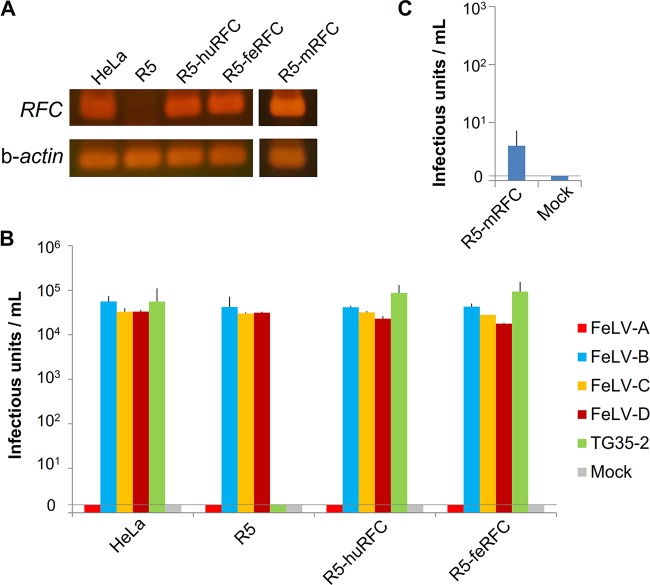
Expression of human or feline RFC renders HeLa-R5 cells susceptible to FeLV TG35-2-pseudotyped virus.
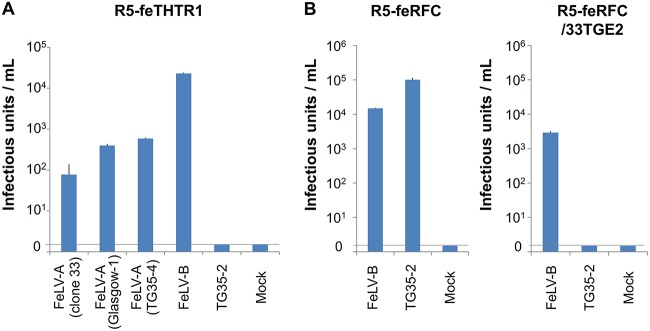
We previously showed that FeLV TG35-2-phenotypic virus (FeLV 33TGE2), a chimeric infectious virus, can infect HeLa cells (18). HeLa-R5 cells, a derivative of HeLa cells, are characterized by the genomic deletion of RFC as a result of exposure to methotrexate (40). As shown in Fig. 3A, we confirmed that human RFC was not expressed in HeLa-R5 cells but was expressed in HeLa cells by reverse transcriptase PCR (RT-PCR). Therefore, we used HeLa-R5 cells to determine susceptibility to FeLV TG35-2-phenotypic virus infection. FeLV-A, FeLV-B, FeLV-C, FeLV-D, and FeLV TG35-2 Env-pseudotyped viruses were prepared in GPLac cells and tested in the cell lines indicated below. As expected, HeLa-R5 cells were nonpermissive for FeLV TG35-2 Env-pseudotyped virus infection, while FeLV TG35-2 Env-pseudotyped virus successfully infected the parent HeLa cells (Fig. 3B). FeLV-B, FeLV-C, and FeLV-D Env-pseudotyped viruses infected HeLa and HeLa-R5 cells, while FeLV-A Env-pseudotyped virus could not infect HeLa cells or HeLa-R5 cells. Next, a retroviral expression vector expressing huRFC or feRFC was introduced into HeLa-R5 cells to generate R5-huRFC and R5-feRFC cells (Fig. 3A), and the cells were tested for infectivity with FeLV TG35-2 Env-pseudotyped virus as well as FeLV-A, FeLV-B, FeLV-C, and FeLV-D Env-pseudotyped viruses. As shown in Fig. 3B, both R5-huRFC and R5-feRFC cells were permissive for FeLV TG35-2 Env-pseudotyped virus infection with >104 infectious units, as well as for FeLV-B, FeLV-C, and FeLV-D Env-pseudotyped virus infection. However, R5-huRFC and R5-feRFC cells were nonpermissive for FeLV-A Env-pseudotyped virus infection. A retroviral expression vector expressing the cDNA encoding mouse RFC was introduced into HeLa-R5 cells to generate R5-mRFC cells (Fig. 3A), and these cells were tested for infectivity with FeLV TG35-2 Env-pseudotyped virus. As shown in Fig. 3C, FeLV TG35-2-pseudotyped virus infected R5-mRFC cells at a much lower titer (approximately 10 infectious units); however, no infection of the mouse MDTF cell line was detected (Fig. 2). These results indicated that transduction of huRFC and feRFC into HeLa-R5 cells rendered them susceptible to viral entry and FeLV TG35-2-phenotypic virus infection. Because HeLa and HeLa-R5 cells were not permissive for FeLV-A infection, we conducted the following experiment. A retroviral expression vector expressing feline THTR1, which was known to be the receptor for FeLV-A (26), was introduced into HeLa-R5 cells, termed R5-feTHTR1, and the cells were tested for FeLV-A Env-pseudotyped viruses from FeLV-A clone 33 (41), FeLV-A Glasgow-1 (42), FeLV-A TG35-4 from a TG35 case (18), and FeLV TG35-2 Env-pseudotyped virus. As shown in Fig. 4A, all FeLV-A Env-pseudotyped viruses infected R5-feTHTR1 cells, but FeLV TG35-2 Env-pseudotyped virus could not. FeLV-B Env-pseudotyped virus was used as a positive control. The results indicated that transduction of feTHTR1 into HeLa-R5 cells could not render cells susceptible to FeLV TG35-2 Env-pseudotyped virus infection. We next conducted an interference assay to determine whether classification of FeLV TG35-2-phenotypic virus depends on the feRFC receptor. R5-feRFC cells preinfected with FeLV 33TGE2 (R5-feRFC/33TGE2 cells) were tested for FeLV TG35-2 Env-pseudotyped virus infection, and the FeLV TG35-2 Env-pseudotyped virus could not infect R5-feRFC/33TGE2 cells but infected R5-feRFC cells (Fig. 4B).
FIG 3.
Infection of cell lines by LacZ-carrying Env-pseudotyped viruses. (A) The expression of RFC was tested in the cell lines HeLa, HeLa-R5 (R5), R5 expressing human RFC (R5-huRFC), R5 expressing feline RFC (R5-feRFC), and R5 expressing mouse RFC (R5-mRFC) by RT-PCR. β-Actin was used as a control. (B) The Env-pseudotyped viruses of FeLV-A (FeLV-A clone 33), FeLV-B (FeLV-B Gardner-Arnstein), FeLV-C (FeLV-C Sarma), FeLV-D (FeLV-D Ty26), and FeLV TG35-2 were tested for infection in the cell lines shown on the x axis. (C) The Env-pseudotyped viruses of FeLV TG35-2 were tested in the cell line R5 expressing mouse RFC (R5-mRFC), shown on the x axis. The y axis indicates the infection units for log10 of β-galactosidase (LacZ)-positive cells per milliliter of supernatant. Data represent the averages from three independent experiments, with the standard deviations shown.
FIG 4.
Infection and interference assay of LacZ-carrying Env-pseudotyped viruses. (A) The Env-pseudotyped viruses of FeLV-A Clone 33, FeLV-A Glasgow-1, FeLV-A (TG35-4), FeLV-B (FeLV-B Gardner-Arnstein), and FeLV TG35-2 were tested for infection in R5 expressing feTHTR1 cells (R5-feTHTR1). (B) The Env-pseudotyped FeLV-B (FeLV-B/Gardner-Arnstein) and FeLV TG35-2 viruses were tested for infection in the R5 cells expressing feline RFC (R5-feRFC) and R5-feRFC cells preinfected with FeLV 33TGE2 (R5-feRFC/33TGE2). The infection units were indicated by log10 of β-galactosidase (LacZ)-positive cells per milliliter of supernatant. Data represent the averages from three independent experiments, with the standard deviations shown.
Taken together, these results indicated that both feline and human RFC are receptors for FeLV TG35-2-phenotypic virus and that viral interference of the FeLV TG35-2-phenotypic virus depends on the RFC receptor.
Isolation of cDNA encoding feline RFC.
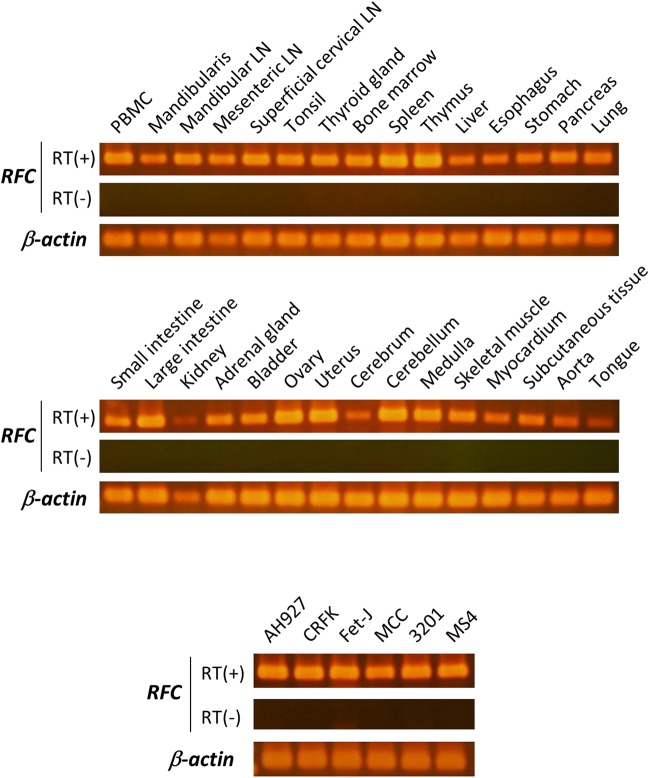
RFC (SLC19A1) transports folates but not thiamine (43). Feline RFC has not been isolated previously. In this study, feline cDNA isolated from feline PBMCs was sequenced and predicted to encode a protein of 522 amino acids. The similarity between feline and human RFC and between feline and mouse RFC was 92.1% and 90.4%, respectively. An alignment of the predicted amino acid sequences of the proteins encoded by the feline and human RFC genes is shown in Fig. 5. The amino acid sequence of huRFC obtained from HEK293T cells was used in this alignment. There were 69 amino acid differences between the feline and human proteins. Phylogenetic analysis of RFC sequences and related sequences, including FeLV receptors, indicated that our clones were likely to be feRFC (Fig. 6). We examined mRNA expression by RT-PCR using total RNA extracted from various feline tissues. Feline RFC was detected in all feline tissues tested (Fig. 7). The feline RFC transcript was detected in the CRFK feline kidney cell line (44), AH927 feline embryo fibroblasts (45), Fet-J feline T-cells, MCC feline large granular lymphoma (46), 3201 feline T-cell lymphoma (47), and MS4 feline B-cell lymphoma (48) (Fig. 7).
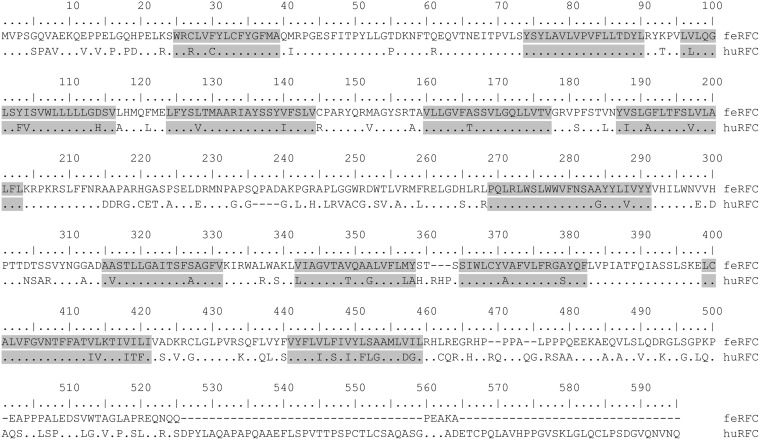
FIG 5.
Alignment of the amino acid sequences of feRFC and huRFC. The alignment in single letter amino acid code was conducted using MUSCLE software (66). Dots indicate conserved amino acid residues, and positions where there are differences between feline and human RFC sequences are shown as letters. Hyphens indicate gaps in the amino acid sequence. Transmembrane domains (gray boxes) based on huRFC were predicted using the constrained consensus topology prediction (74).
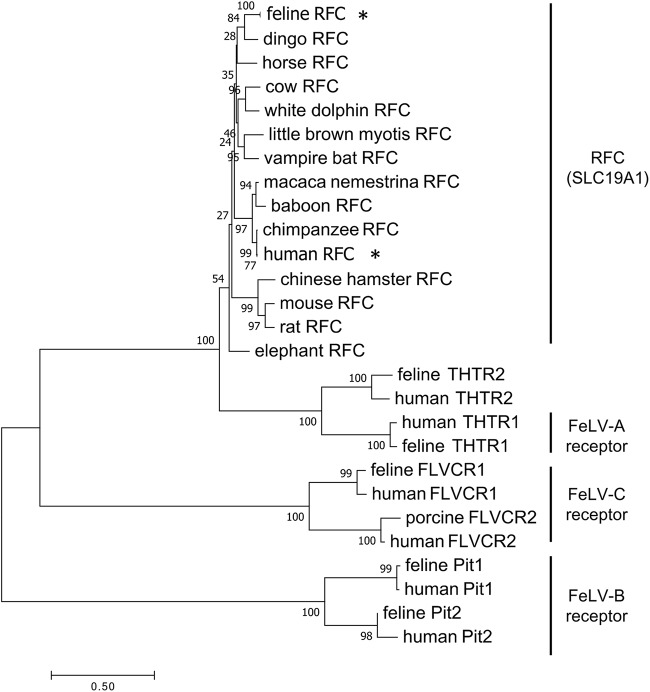
FIG 6.
Phylogenetic analysis of RFC and related proteins with FeLV receptors. A neighbor-joining tree was generated from the amino acid sequences of mammalian RFC, including human RFC (*) and feline RFC (*) with proteins indicated for the FeLV-A, FeLV-B, and FeLV-C receptors. The scale bar indicates evolutionary distance in amino acid substitutions per site.
FIG 7.
RFC expression in feline tissues and feline cell lines. Detection of RFC by RT-PCR using total RNA isolated from the indicated tissues and cell lines (AH927, CRFK, Fet-J, MCC, 3201, and MS4). A representative 2% agarose gel with electrophoresed PCR product (305 bp) is shown. The gels were stained by ethidium bromide. RT(+) and RT(−) controls were included during cDNA synthesis.
Determination of the amino acids in the Env protein that are required for the FeLV TG35-2 phenotype.
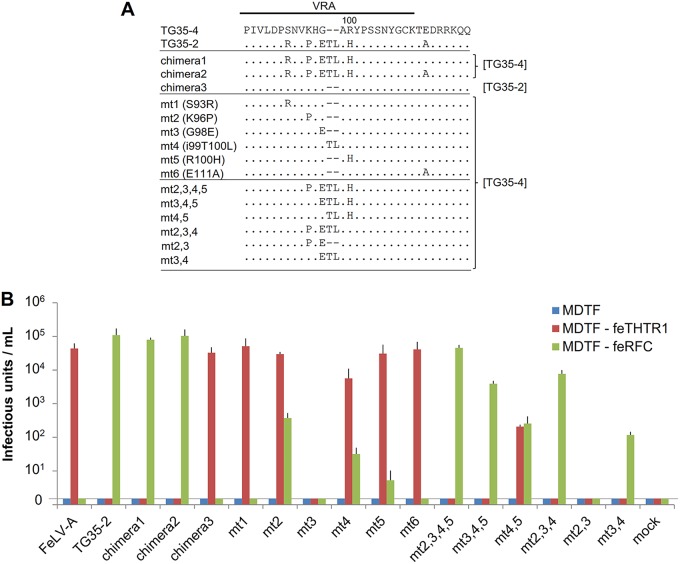
We previously showed that a subtle change in the VRA altered the interference patterns of the FeLV TG35-2 and FeLV-A phenotypes (18). In this study, a series of Env mutants (Fig. 8A) were tested for receptor usage using MDTF-feTHTR1 and MDTF-feRFC cells. FeLV-A TG35-4 isolated in a cat infected with FeLV TG35-2 was used for the construction of mutants. As shown in Fig. 8B, FeLV-A (TG35-4) Env-pseudotyped virus infected MDTF-feTHTR1 cells but not MDTF-feRFC cells. However, FeLV TG35-2 Env-pseudotyped virus infected MDTF-feRFC but not MDTF-feTHTR1 cells. Chimeras 1 and 2, which contained the VRA of TG35-2 and the backbone of TG35-4, infected MDTF-feRFC but not MDTF-feTHTR1 cells, while chimera 3, which comprised the VRA of TG35-4 and the backbone of TG35-2, infected MDTF-feTHTR1 but not MDTF-feRFC cells. These results indicated that the VRA conferred specific receptor usage to FeLV-A and FeLV TG35-2.
FIG 8.
Determination of the amino acids in the Env protein that are required for the FeLV TG35-2 phenotype. (A) The indicated mutant FeLV env genes, constructed using either the TG35-2 or TG35-4 (FeLV-A) env gene, were generated by site-directed mutagenesis or recombination of the VRA (18). The mutant FeLV env gene, mt3,4, was newly generated in this study. The env sequences other than the VRA, derived from TG35-2 or TG35-4 env, are referenced on the right. (B) The indicated Env-pseudotyped viruses were tested for infection in the MDTF, MDTF expressing feline THTR1 (MDTF-feTHTR1), and MDTF expressing feline RFC (MDTF-feRFC) cell lines. The infection units were indicated by log10 of β-galactosidase (LacZ)-positive cells per milliliter of supernatant. Data represent the averages from three independent experiments, with the standard deviations shown.
A further 12 Env mutants with substituted amino acids in the VRA with the TG35-4 backbone were tested for infectivity in MDTF-feTHTR1 and MDTF-feRFC cells. Some mutants [mt2(K96P), mt4(i99T100L), mt5(R100H), and mt4,5] exhibited infectivity in both MDTF-feTHTR1 and MDTF-feRFC cells. The infectious unit measurements of mt2, mt4, and mt5 were higher in MDTF-feTHTR1 than in MDTF-feRFC cells, while the mt4,5 mutant infected MDTF-feTHTR1 and MDTF-feRFC cells to a similar extent. These Env mutants showed a dual-tropic phenotype combining that of FeLV-A and FeLV TG35-2. Thus, one or three amino acid substitutions in the VRA of FeLV-A (mt2 or mt4,5 mutants, respectively) effectively altered the FeLV-A-specific phenotype to a FeLV-A and FeLV TG35-2 dual phenotype. The mt2,3,4,5, mt2,3,4, mt3,4,5, and mt3,4 mutants demonstrated infectivity in MDTF-feRFC cells but not MDTF-feTHTR1, indicating that they were of the FeLV TG35-2 phenotype. The mt3,4 mutant, which was newly constructed in this study, had only three amino acid substitutions in the FeLV-A VRA. These results indicated that subtle mutation of the FeLV VRA alters the specificity of infection via the viral receptor, feTHTR1 or feRFC.
Isolation and construction of infectious FeLV provirus.
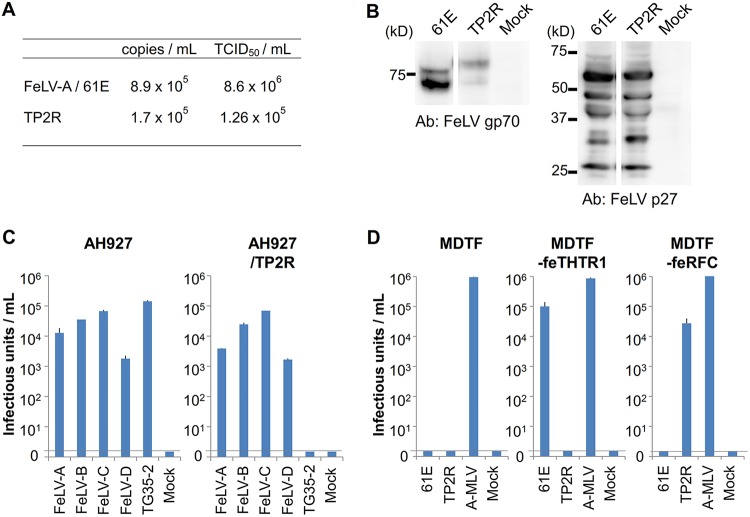
We isolated the FeLV provirus from the genome of a cat TG35, in which the TG35-2 Env clone was detected. PCR primers designed in the U3 region of the 5ʹ long terminal repeat (LTR) and 3ʹ LTR were used for amplification of the provirus. The infectious provirus was reconstructed as described in Materials and Methods and was termed TP2R clone. The amino acid sequence of Env from FeLV TP2R was almost the same as that of TG35-2 Env (Fig. 9). Phylogenetic analysis classified FeLV TP2R as belonging to genotype I/clade 1, which is often observed in Japanese FeLV strains (data not shown) (5). The LTRs of TP2R did not contain tandem repeats in the enhancer. The 293T cells were transfected with FeLV TP2R and FeLV-A 61E (49) plasmids, and the supernatants of the cells were prepared as a virus stock. AH927 cells infected with FeLV TP2R persistently produced the virus at a high titer, as well as FeLV-A 61E when the supernatant from the cells was measured using two methods: quantitative real-time RT-PCR and determination of the 50% tissue culture infectious dose (TCID50) (Fig. 10A). FeLV Env and Gag proteins were detected in AH927 cells infected with FeLV TP2R by Western blot analysis, and the molecular weight of FeLV TP2R Env was slightly higher than that of FeLV-A 61E (Fig. 10B). The results indicated that FeLV TP2R was replication competent. To determine the viral interference group of FeLV TP2R, AH927 cells infected with FeLV TP2R (AH927/TP2R cells) were tested with the FeLV-A, -B, -C, -D, and TG35-2 Env-pseudotyped viruses. FeLV-A, FeLV-B, FeLV-C, and FeLV-D Env-pseudotyped viruses infected AH927/TP2R cells, whereas FeLV TG35-2 Env-pseudotyped virus could not. By contrast, FeLV-A, -B, -C, -D, and TG35-2 Env-pseudotyped viruses infected AH927 cells (Fig. 10C). Next, to determine the receptor of FeLV TP2R, FeLV-A 61E and FeLV TP2R viruses were prepared from 293Lac cells that contained the LacZ-coding retroviral vector, and viral infection of MDTF-feRFC and MDTF-feTHTR1 cells was analyzed. As shown in Fig. 10D, FeLV TP2R infected MDTF-feRFC cells but not MDTF or MDTF-feTHTR1 cells. By contrast, FeLV-A 61E infected MDTF-feTHTR1 cells but not MDTF or MDTF-feRFC cells. These results demonstrated that FeLV TP2R could be classified as the FeLV TG35-2 phenotype and used feRFC as its receptor.
FIG 9.
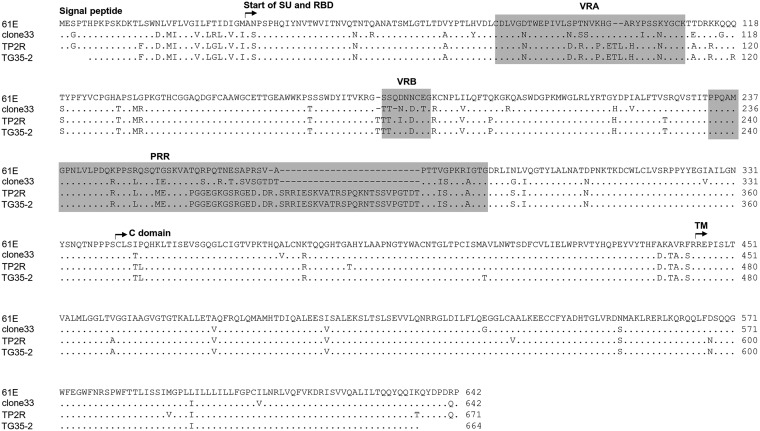
Alignment of the amino acid sequence of FeLV Env. Surface subunit (SU), transmembrane subunit (TM), receptor-binding domain (RBD), proline-rich region (PRR), and C domain of the Env protein are shown for FeLV-A 61E (49), FeLV-A clone 33 (41), and FeLV TG35-2 env clone (18) compared with FeLV TP2R. The variable regions, VRA, VRB, and PRR, are also shown by gray boxes. Dots indicate identical residues, and dashes indicate spaces that were introduced for the amino acid alignment. The Env sequences were aligned with the Genetyx program (Genetyx Corporation, Tokyo, Japan).
FIG 10.
Characterization of FeLV TP2R. (A) Viral copies and titers of FeLV-A 61E and FeLV TP2R harvested from the supernatants of FeLV-infected AH927 cells are shown as copies per milliliter and the 50% tissue culture infectious doses (TCID50) per milliliter. (B) FeLV proteins were detected in AH927 cells infected with FeLV-A 61E (61E) or FeLV TP2R (TP2R) using anti-FeLV gp70 Env and anti-FeLV p27 Gag antibodies by Western blot analysis. (C) Interference assay of FeLV TP2R. Env-pseudotyped viruses of FeLV-A (FeLV-A clone 33), FeLV-B (FeLV-B Gardner-Arnstein), FeLV-C (FeLV-C Sarma), FeLV-D (FeLV-D Ty26), and FeLV TG35-2 were tested for infection of AH927 cells and AH927 cells preinfected with FeLV TP2R (AH927/TP2R). (D) The replication-competent viruses of FeLV-A 61E, FeLV TP2R, and ampho-MuLV carrying LacZ were tested for infection of MDTF, MDTF-feTHTR1, and MDTF-feRFC cells. The infection units are by log10 of β-galactosidase (LacZ)-positive cells per milliliter of supernatant. Data represent the averages from three independent experiments, with the standard deviations shown.
FeLV TP2R and FeLV-A 61E viruses were prepared from AH927 cells, and viral replication was analyzed in different cell lines (CRFK, Fet-J, MCC, 3201, and MS4 cells) by determining the viral copy number at 10 days postinfection. As shown in Fig. 11, FeLV TP2R and FeLV-A 61E viruses exhibited replication with high copy numbers in CRFK cells. In particular, the copy number of FeLV-A 61E was significantly higher than that of FeLV TP2R in CRFK cells (P < 0.01). Both viruses replicated in hematopoietic cells, and FeLV TP2R virus exhibited significantly higher viral copy numbers than FeLV-A/61E in Fet-J feline T-cells, MCC feline large granular lymphoma cells, and MS4 cells (Fig. 11). These results indicated that FeLV TP2R can replicate to a high titer, similarly to FeLV-A 61E, in cultured cell lines.
FIG 11.
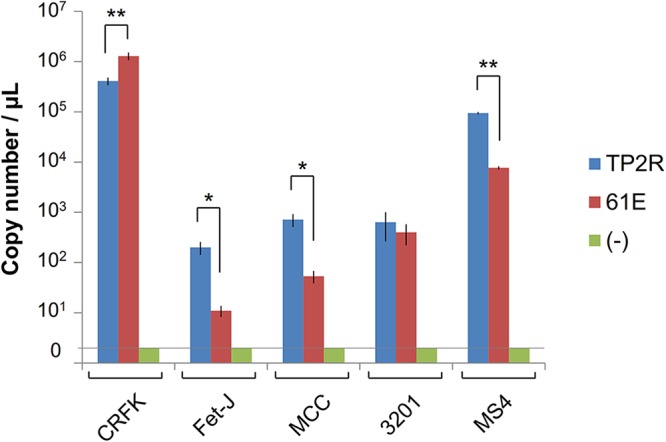
FeLV replication in different cell lines. The cells (CRFK, Fet-J, MCC, 3201, and MS4) were infected with 2 × 103 TCID50 of FeLV TP2R or FeLV-A 61E virus. The viral copy number was measured in the culture supernatants at 10 days postinfection by quantitative real-time RT-PCR. The y axis indicates the viral copy number. **, P < 0.01, *, P < 0.05 (Student’s t test). Data represent the averages from three independent experiments, with the standard deviations shown.
DISCUSSION
FeLV is transmitted among domestic cats at high prevalence in Japan and shows high genetic diversity due to the mutation or recombination of viral genes (5, 50). Mutation of the FeLV Env sequence, especially in the VRA, may lead to a change in the viral receptor interference group. Our previous study identified a novel FeLV interference group of TG35-2 based on FeLV Env (18). In this study, we report that the receptor of FeLV TG35-2-phenotypic virus is RFC, which is classified as SLC19A1. We mapped the receptor for FeLV TG35-2-phenotypic virus to within region q22.3 of chromosome 21 using phenotypic screening of RH cell lines (Fig. 1), and further analyses demonstrated that RFC confers susceptibility to FeLV TG35-2-phenotypic virus infection. Expression of feline and human RFC cDNA in nonpermissive MDTF cells rendered these cells susceptible to FeLV TG35-2-phenotypic virus infection. Sequence similarity and phylogenetic analysis indicated that the feline receptor is an orthologue of huRFC and is most likely a folic acid transport protein. Further functional experiments clarified that it is a folic acid transporter.
Analysis of the amino acid sequence encoded by feRFC revealed gene polymorphisms encoding Ala or Gly at position 249, and both cDNAs functioned as receptors for FeLV TG35-2-phenotypic virus (data not shown). Genetic variants in the huRFC gene locus have been found and are reported to be associated with differences in folate homeostasis (51). Two huRFCs were isolated from HEK293T cells and were found to contain polymorphisms compared with the huRFC sequence with gene accession numbers AAC50180 and NM_194255, and both function as receptors for FeLV TG35-2-phenotypic virus (data not shown). The feRFC sequence revealed high amino acid similarity (more than 90%) with huRFC and mRFC. Forced expression of huRFC rendered cells permissive for FeLV TG35-2-pseudotyped virus infection (Fig. 2). However, despite high amino acid similarity with mRFC, FeLV TG35-2-phenotypic virus could not infect mouse cells (MDTF cells) (Fig. 2 and 10) or NIH 3T3 cells (18), and infected cells ectopically expressed mRFC at a much lower titer (Fig. 3C). This may indicate that additional factors influence viral entry and infection. FeLV subgroup was classified by viral interference and its in vitro host range properties, and we demonstrated here that the FeLV TG35-2-phenotypic virus required RFC (Fig. 4B).
The utilization of transport proteins for cell entry is a common feature of gammaretroviruses, including extinct retroviruses (1, 52, 53). For example, ecotropic murine leukemia virus utilizes mCAT, the cationic amino acid transporter (54). To date, all known receptors for feline and murine gammaretroviruses have been multitransmembrane receptors (4, 55). The discovery of RFC as an entry receptor for FeLV TG35-2-phenotypic virus follows this pattern of multipass membrane transport molecules acting as retroviral receptors. RFC was recently reported to be the receptor for murine endogenous retrovirus (56). Because some gammaretroviruses are known to share a receptor, such as gibbon ape leukemia virus (GaLV), FeLV-B, koala retrovirus A (KoRV-A), and 10A1-MuLV, which all use Pit1 (27–29, 57, 58), it is plausible that other known viruses may also use RFC as a receptor.
FeLV-A is transmitted among domestic cats, and novel subgroups of FeLV are usually generated in vivo. In other words, FeLV-A evolves into the FeLV-B, FeLV-C, FeLV-D, and FeLV-T subgroups in FeLV-A-infected cats, and each of these subgroups display altered tropisms because of their differential receptor use. Furthermore, FeLV TG35-2-phenotypic virus may be generated from the FeLV-A env gene, as can be seen from the similarity of FeLV-A env sequences (18). The RFC transporter belongs to the SLC19 family of reduced folate transporters, of which there are three members: two thiamine transporters, THTR1 and THTR2, and RFC. It is known that huRFC is ubiquitously expressed in tissues (43), and as shown in Fig. 7, feRFC is also ubiquitous in feline tissues, including in the ovaries and uterus. FeLV TG35-2-phenotypic virus, utilizing RFC as its entry receptor, may have the potential to be endogenized in cats, as seen for murine endogenous retrovirus (56).
In experiments using Env mutants, the VRA region of FeLV-A and FeLV TG35-2 determined the specificity of viral receptors. A slight change in the amino acid sequence altered the tropism from the FeLV-A to the FeLV TG35-2 phenotype (Fig. 8). In other words, receptor usage was changed from feTHTR1 to feRFC. The amino acid residues ETL in the VRA partly contribute to this specific shift to FeLV TG35-2 phenotype tropism. Interestingly, some Env mutants with point mutations in the VRA utilized both feTHTR1 and feRFC. This indicates that the structure of the VRA of Env determines the interaction with the receptor, and this interaction leads to the specificity of viral infection. Although Env mutants mt 3,4,5 and mt 4,5 infected MDTF cells expressing the receptor (Fig. 8B), these mutants did not infect AH927 cells (18). Therefore, this also suggests that additional factors may influence viral entry and infection.
Since FeLV TG35-2-phenotypic virus was detected in FeLV-A-vaccinated cats, this may indicate that selection pressure may have led to the emergence of FeLV TG35-2-phenotypic virus. Dual-tropic FeLVs with different receptor usages have been reported (19, 33, 59), and dual-tropic mutants that use both feTHTR1 and feRFC may occur as an intermediate phenotype, but this phenotype may not be sufficiently stable. KoRV is known to use THTR1 and Pit1 as receptors (57, 60, 61), as seen for FeLV (26–30). Therefore, our results suggest that KoRV may have potency in evolving into new viruses that use koala RFC as a receptor, due to a subtle mutation in Env.
Methotrexate is a chemotherapeutic agent and immune system suppressant that is transported by RFC (43, 51). In a similar way, FeLV TG35-2 Env-pseudotyped virus carrying retroviral expression vector can be utilized in transduction via RFC.
The FeLV TP2R provirus isolated from cat case TG35 was reconstructed as infectious provirus. FeLV TP2R was characterized as belonging to the FeLV TG35-2 phenotype (Fig. 10) and was able to preferentially replicate in hematopoietic cells compared with FeLV 61E (Fig. 11), which may be due to FeLV TP2R promoter activity or viral receptor usage.
In this study, we identified an entry receptor for FeLV TG35-2-phenotypic virus and isolated and reconstructed an infectious FeLV TG35-2-phenotypic provirus. However, the prevalence and pathogenicity of FeLV TG35-2-phenotypic virus in cats remains to be determined. Identification of an entry receptor for FeLV TG35-2-phenotypic virus may therefore help establish a strategy to detect FeLV variants in domestic cats. Multiple FeLV subgroups, including FeLV-A, can simultaneously infect one cat, and genetic polymorphisms in FeLV can be generated in FeLV-infected cats (5). The study of FeLV variants such as FeLV TG35-2-phenotypic virus helps to elucidate the relationship between genetic polymorphisms or genetic changes in FeLV and the pathogenicity of FeLV. Therefore, our study provides a tool for further investigations into FeLV-induced diseases.
MATERIALS AND METHODS
Cells.
The CRFK feline kidney cell line (44), AH927 feline embryo fibroblasts (45), 3201 feline T-cell lymphoma cells (47), HEK293T, and Mus dunni tail fibroblasts (MDTF) (62) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1× penicillin/streptomycin. Fet-J feline T-cells (ATCC CRL-11967) and MCC feline large granular lymphoma cells (46) were cultured in RPMI 1640 with 10% FCS and 1× penicillin/streptomycin. MS4 feline B-cell lymphoma cells (48) were cultured in RPMI 1640 with 20% FCS and 1× penicillin/streptomycin. HeLa cells and RFC-null HeLa (R5) cells (40) (kindly provided by I. David Goldman at Albert Einstein College of Medicine) were cultured in DMEM with 10% FCS and 1× penicillin/streptomycin. MDTF and R5 cells expressing feline, human, and mouse RFC, MDTF-feRFC, MDTF-huRFC, R5-feRFC, R5-huRFC, and R5-mRFC, MDTF cells expressing feline THTR1 (MDTF-feTHTR1) (18), and R5 cells expressing feline THTR1 (R5-feTHTR1) were cultured in DMEM with 10% FCS, 1× penicillin/streptomycin, and 0.6 mg/ml G418. PLAT-E and PLAT-A retroviral packaging cells (63), GPLac cells (5), an env-negative packaging cell line containing β-galactosidase (LacZ)-coding pMXs retroviral vector (5, 63), and 293Lac cells (64) containing LacZ-coding pMXs retroviral vector, were cultured in DMEM with 10% FCS and 1× penicillin/streptomycin.
Viruses.
Feline retroviruses were prepared from the supernatants of AH927 cells infected with FeLV-B Gardner-Arnstein (65), FeLV-A 61E (49), and FeLV 33TGE2 (18), a replication-competent virus (33TGE2) containing the TG35-2 env gene and the LTR genes, gag and pol, of FeLV-A clone 33 (41). Ampho-MuLV was prepared from the supernatants of NIH 3T3 cells infected with ampho-MuLV (18).
The retroviral vector pMXs encoding LacZ with FeLV-A 61E, FeLV TP2R, and ampho-MuLV were harvested from 293Lac cells infected with each replication-competent virus. Virus-containing cell supernatants were filtered through 0.22-μm-pore filters and stored as viral stocks at −80°C until use.
Isolation and reconstruction of FeLV TP2R provirus.
Genomic DNA was isolated from the blood of case TG35 (5) using the QIAamp DNA Blood kit (Qiagen, Venlo, Netherlands). The FeLV provirus was amplified by PCR using KOD FX Neo (Toyobo, Osaka, Japan) with a primer pair designed to the FeLV 5ʹ LTR and 3ʹ LTR: Fe-227S (5ʹ-TTACCCAAGTATGTTCCCRTGAGATANAAGGAAGT-3ʹ, nucleotide positions 67 to 101 of FeLV-A 61E; GenBank accession number M18247) and Fe-7R (5ʹ-GTCAACTGGGGAGCCTGGAGAC-3ʹ, nucleotide positions 8174 to 8195 of FeLV-A 61E). The resulting PCR products of 8 to 10 kbp were cloned into pCR-Blunt II-TOPO vectors (Invitrogen, Carlsbad, CA, USA) and sequenced by dye terminator cycle sequencing carried out by Fasmac Co., Ltd., Kanagawa, Japan. Two clones, TG35LL1 and TG35LL2, were isolated and used for further experiments. The 5ʹ LTR U3 and the 3ʹ LTR R-U5 of the TG35LL1 and TG35LL2 clones were repaired based on each LTR sequence using the In-Fusion HD Cloning kit (TaKaRa, Shiga, Japan), and were renamed TP1 and TP2. Clone TP1 was replication competent with an FeLV-A phenotype, but the TP1 virus did not cause persistent infection in the cultured cells and the reason for this was unknown (data not shown). The 1.5-kb restriction fragment, generated by excision at the NruI restriction enzyme site located upstream of the pol gene and the BspT104I restriction site located in the pol gene in clone TP2, was replaced with that of clone TP1. The results indicated that the phenylalanine at amino acid position 384 of Pol was changed to isoleucine, and the isoleucine at amino acid position 485 of Pol was changed to leucine to ensure that the reconstructed provirus, designated TP2R, was replication competent. The isoleucine at amino acid position 384 of Pol was conserved among replication-competent FeLVs such as FeLV-A/61E, FeLV-A/clone 33, and FeLV-A/Glasgow-1. The nucleotide sequence was deposited in the GenBank database under accession number LC462187.
HEK293T cells were seeded in six-well plates 1 day prior to transfection, and the TP2R expression plasmid was transfected using Lipofectamine 3000 (Invitrogen). Two days posttransfection, the supernatant was filtered through 0.22-μm-pore filters and was used to infect AH927 cells in the presence of 10 μg/ml Polybrene (Santa Cruz Biotechnology, Dallas, TX, USA). The cells were cultured for more than 14 days and the virus-containing cell supernatant was filtered through 0.22-μm-pore filters and stored as a viral stock at −80°C until use.
Screening of the G3 RH panel.
The RH cell lines from the human/hamster G3 panel were initially obtained from A. Dusty Miller (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). The cells were maintained in α minimum essential medium with 10% fetal bovine serum (FBS), 1× penicillin/streptomycin (Wako Pure Chemical Industries, Osaka, Japan), and 1× hypoxanthine-aminopterin-thymidine (HAT) (100 μm hypoxanthine, 0.4 μm aminopterin, and 16 μm thymidine; Life Technologies, Carlsbad, CA, USA) (36). A total of 79 clones were available, of which 75 were tested in our experiment. Prior to the infection assay, the cells were weaned from HAT by growing in HAT medium for 1 week, then 2 weeks in HT medium lacking aminopterin, and then 1 week in nonsupplemented medium (38).
The RH cell lines were plated at 104 cells per well in a 24-well plate and exposed to FeLV TG35-2-phenotypic virus carrying LacZ (33TGE2-LacZ) the next day. Two days after infection, the cells were stained with X-Gal (5-bromo-4-chloro-indolyl-β-d-galactopyranoside; Wako Pure Chemical Industries). Viral titers were determined as infectious units (IU) per milliliter by counting the blue-stained nuclei.
The 75 hybrid cell lines were RH1 to RH83, omitting RH36, RH38, RH48, RH49, RH69, RH71, RH76, and RH78. The viral titers for the cell lines (IU/ml) were 0, 0, 0, 0.70, 0, 1.54, 0, 0, 0, 0, 1.48, 0, 0, 4.92, 0.85, 0.95, 0, 1.49, 0, 0, 0, 2.50, 1.11, 0, 0, 1.63, 2.00, 0, 1.79, 0, 0, 0, 0, 0, 0, 0, 4.24, 0, 4.16, 0, 0, 5.37, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1.80, 0, 0, 3.41, 0, 0, 0, 0.48, 0, 0, 3.28, 0, 3.46, 0.48, 3.17, 0, 3.47, 0.18, 0, 0, and 0, respectively. The cell lines had previously been genotyped using 235,789 markers by array comparative genomic hybridization (37, 38).
Human markers were binned into 0 or 1 extra copies (diploid or triploid) using coordinates from the GRCh37/hg19 genome assembly (https://genome.ucsc.edu). Markers were discarded if they possessed the same genotype vector, were present in four or fewer clones, or were present in all clones. A total of 54,560 markers remained. The phenotype used for mapping in each RH clone was log10 of the IU/ml value plus one. The LOD scores were computed and genome-wide significance levels were set by permutation as described previously (38). A 5% family-wise error rate (FWER) was used as the threshold for genome-wide significance.
Isolation of RFC and construction of an RFC expression vector.
Total RNA was isolated from feline PBMCs (66) and HEK293T cells using RNAiso Plus (TaKaRa), and the extracted RNA was treated with recombinant DNase I (RNase-free) (TaKaRa). cDNA was synthesized with a PrimeScript II first-strand cDNA synthesis kit (TaKaRa) using oligo(dT) primers. FeRFC cDNA was amplified by PCR using the primers fRFC-1S (5ʹ-CCGCCCGCCCGCCCGGGTACCTGGGGAG-3ʹ) and fRFC-1R (5ʹ-GCCAGCCCGCAGTGCCCCAGCAGGCAGCGGGAT-3ʹ) and was then cloned into pCR-Blunt II-TOPO vectors (Invitrogen) and sequenced. FeRFC and huRFC were amplified by PCR using the primers fRFCEI (5ʹ-GCGAATTCACAGCAAGCATGGTGCCCTCCGGCCAGGTGGCGG-3ʹ) and fRFCBII (5ʹ-GGAGATCTTCACAGGTCTTCTTCAGAGATCAGTTTCTGTTCGGCTTTGGCCTCGGGCTGCTGGTTCTGTT-3ʹ; underlining indicates the myc tag sequence) for feRFC, and huRFC-S (5ʹ-CGCTCGAGATGGTGCCCTCCAGCCCAGCGGTGGAG-3ʹ) and huRFC-R (5ʹ-CGAGATCTTCACAGGTCTTCTTCAGAGATCAGTTTCTGTTCCTGGTTCACATTCTGAACACCGT-3ʹ; underlining indicates the myc tag sequence) for human RFC.
Mouse RFC (mRFC) cDNA (clone H4025H01) was obtained from Riken BRC (National Research and Development Institute of RIKEN Bioresource Center). Mouse RFC was amplified by PCR using a specific primer pair: mRFC-F (5ʹ-CTGGGCACCATGGTGCCCACTGGCCAGGTGGCAG-3ʹ) and mRFC-R (5ʹ-AGAGATCTAGATCTTCACAGATCCTCTTCTGAGATGAGTTTTTGTTCCTGGTTCACATTCTGAACACCGTCGCTTGGAAGACA-3ʹ; underlining indicates the myc tag sequence).
PCRs were conducted with KOD FX Neo DNA polymerase (Toyobo). PCR products were digested with EcoRI and BglII for feRFC and mRFC, and with XhoI and BglII for huRFC, and then each fragment was inserted into pMSCVneo retroviral vector (TaKaRa).
Generation of cell lines.
The feRFC, huRFC, and feTHTR1 (26) retroviral expression vectors were transfected into PLAT-E (ecotropic MuLV packaging) cells or PLAT-A (amphotropic MuLV packaging) cells using Lipofectamine 3000 (Invitrogen). The pMSCVneo empty vector was used as a control. Two days posttransfection, supernatants were collected and filtered through a 0.22-μm filter, and 1 ml of the filtrate was used to infect MDTF and RFC-null HeLa (R5) cells, which were then seeded into 12-well plates. Polybrene, at a concentration of 8 μg/ml, was added to the infection. Cells were maintained in complete medium containing G418 at a concentration of 0.6 mg/ml.
Preparation of LacZ-carrying Env-pseudotyped viruses.
GPLac cells, an env-negative packaging cell line containing a LacZ-coding retroviral vector, were seeded in 6-well plates 1 day prior to transfection and were transfected with env expression plasmids to produce LacZ-carrying Env-pseudotyped virus. After 48 h, cell culture supernatants were collected, filtered through a 0.22-μm filter, and stored at −80°C. env expression plasmids for pseudotyped virus preparations, namely, pFUΔss clone 33 (FeLV-A clone 33 env), pFUΔss GB (FeLV-B Gardner-Arnstein env), pFUΔss SC (FeLV-C Sarma env), pFUΔss Ty2.0 (FeLV-D Ty26 env), pFUΔss TG35-2 (FeLV TG35-2 env), pFUΔss TG35-4 (FeLV-A TG35-4 env), pFUΔss DC10 (ERV-DC10 env), and pFUΔss 4070A (amphotropic MuLV 4070A env), have been described previously (17, 18).
env expression plasmids for the mutant FeLV env genes, constructed in either TG35-2 or TG35-4 env were chimera 1, chimera 2, chimera 3, mt1, mt2, mt3, mt4, mt5, mt6, mt2,3,4,5, mt2,3,4, mt3,4,5, mt2,3, and mt4,5, as previously described (18). In this experiment, the env expression plasmid, mutant mt3,4, was newly generated by site-directed mutagenesis with Fe-602S (5ʹ-CTAGCAATGTAAAACATGAAACCCTCGCTCGTTATCC-3ʹ) and its complementary sequence in the pFUΔss vector. The sequences of the Env mutants are shown in Fig. 8A.
Viral infection and titration by a LacZ assay.
Target cells seeded in 24-well plates were inoculated with 250 μl of Env-pseudotyped viruses and cultured in medium containing 10 μg/ml of Polybrene. After 48 h, cells were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and single-cycle infectivity was titrated by counting blue-stained nuclei under the microscope.
Detection of RFC by RT-PCR.
Feline tissues were obtained from a specific-pathogen-free cat (Kyoto-SPF1) described in our previous study (66). Total RNA was extracted from the tissues with an RNAiso Plus kit (TaKaRa) and from cell lines using miRNeasy (Qiagen) and recombinant DNase I (TaKaRa). cDNA was synthesized with the PrimeScript II first-strand cDNA synthesis kit (TaKaRa). PCR for detecting RFC in the feline tissues and cell lines was performed using KOD One PCR Master Mix Blue (Toyobo) with primer set Fe-626S (5ʹ-CACCGACTACCTGCGCTACA-3ʹ) and Fe-601R (5ʹ-CGTAGTTGACCGTGGAGAAGG-3ʹ). Thermal cycling conditions were 35 cycles of 98°C for 10 s, 60°C for 5 s, and 68°C for 1 s. PCR for detecting RFC in HeLa, R5, R5-huRFC, and R5-feRFC cells was performed using the KOD One PCR Master Mix Blue (Toyobo) with primer set Fe-649S (5ʹ-AGAGCTTCATCACCCCCTAC-3ʹ) and Fe-625R (5ʹ-GCTGTAGAAGAGCTCCATGA-3ʹ). Thermal cycling conditions were 35 cycles of 98°C for 10 s, 55°C for 5 s, and 68°C for 1 s. PCR for detecting β-actin was performed using the same master mix kit and a previously reported primer set (17). Thermal cycling conditions were 30 cycles of 98°C for 10 s, 52°C for 5 s, and 68°C for 1 s.
Phylogenetic analysis.
A phylogenetic tree was constructed using the following sequences: human RFC (AAC50180 and NM_194255), mouse RFC (XP_006513480), chimpanzee RFC (XP_001157360.2), pigtail monkey RFC (XP_011724100.1), baboon RFC (XP_021790831.1), horse RFC (XP_023486074.1), cow RFC (XP_024848105.1), rat RFC (NP_001030309.1), Chinese hamster RFC (XP_007639501.1), dingo RFC (XP_025317973.1), white dolphin RFC (XP_022452800.1), little brown myotis RFC (XP_014320861.1), vampire bat RFC (XP_014320861.1), elephant RFC (XP_003421994.1), human THTR1 (NP_008927), feline THTR1 (ABD61002.1), human THTR2 (NP_079519), feline THTR2 (AFV75033), human FLVCR1 (NP_054772), feline FLVCR1 (NP_001009302), porcine FLVCR2 (NP_001136312), human FLVCR2 (NP_060261), human Pit1 (NP_005406), feline Pit1 (NP_001009840), human Pit2 (NP_006740), and feline Pit2 (NP_001009839). The MEGA7 program package was used for the phylogenetic analysis (67). The alignments for each phylogenetic tree were conducted using MUSCLE software (68). The phylogenetic tree was constructed using 341 positions, the neighbor-joining method (69), and the JTT matrix-based method (70), and the robustness of each tree was evaluated by the bootstrap method (1,000 times) (71).
Viral titration.
AH927 cells were seeded into 24-well plates 1 day prior to infection. Then, 250 μl of diluted virus stock (10-fold serial dilutions) was added in the presence of 10 μg/ml Polybrene in quadruplicates. Eight hours postinfection, 250 μl of medium was added to each well. Three days postinfection, the cells were fixed with 3.7% formaldehyde solution, permeabilized with 0.2% Triton X-100, and then blocked with 1% bovine serum albumin. The cells were stained overnight at 4°C with a FeLV Gag p27 antibody and then stained for 1 h at room temperature with horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody (Cell Signaling, Danvers, MA, USA). The cells were added with 3,3'-diaminobenzidine (DAB)-peroxidase substrate solution (Nacalai Tesque, Kyoto, Japan), and colonies of brown cells were counted under a microscope. The viral titer was calculated as the 50% tissue culture infectious dose (TCID50) according to the Reed-Muench method (72).
Quantification of viral RNA by quantitative real-time RT-PCR.
Fet-J, MCC, MS4, and 3201 cells were seeded at 4 × 104 cells/well into 24-well plates and infected with 2 × 103 TCID50 of virus stock in the presence of 10 μg/ml Polybrene in a total volume of 500 μl. Twenty-four hours postinfection, the culture medium was changed and the cells were cultured for 10 days. Then the culture supernatants were collected after centrifugation at 300 × g at 4°C for 5 min.
The culture supernatants (200 μl) were treated for 40 min at 37°C with 10 mM MgCl2 and 20 μg/ml of DNase I, and total RNA was extracted using the High Pure Viral RNA kit (Roche, Basel, Switzerland). cDNA was generated from 8 μl of RNA using the PrimeScript II 1st Strand cDNA Synthesis kit (TaKaRa) with random 6-mers in a total volume of 20 μl. For real-time RT-PCR of FeLV 61E, a probe (FeLV_U3-probe) and primers (forward, FeLV_U3-exo-f; reverse, FeLV_U3-exo-r) against FeLV LTR were used as previously reported by Tandon et al. (73). For real-time RT-PCR of FeLV TP2R, a reverse primer was designed (FeLV_U3-exo-r2, 5ʹ-TTATAGCAAAAAGCGCGGG-3ʹ). The probe was labeled at the 5ʹ end with the fluorescent reporter dye FAM (6-carboxyfluorescein) and at the 3ʹ end with the fluorescent quencher dye TAMRA (carboxytetramethylrhodamine). Then, 2 μl of cDNA was amplified in a total volume of 25 μl using Premix Ex Taq (TaKaRa) with a final concentration of 300 nM forward primer, 300 nM reverse primer, and 200 nM probe. Reactions were performed using CFX96 Touch (Bio-Rad, Hercules, CA, USA). Thermal cycling conditions were 95°C for 30 s and then 45 cycles of 95°C for 5 s and 60°C for 30 s. Plasmid p61E (a gift from Edward Hoover), which contains the full-length FeLV-A 61E provirus subcloned into pUC18, and TP2R, were used as standards for PCR quantification. The plasmid standard copy number was calculated from optical density measurements at 260 nm. A 10-fold dilution series of the plasmid standard template DNA was made in 10 mM Tris-Cl (pH 8.5). Quantification of the sample amplicon was achieved by comparing the threshold cycle value of the sample with the standard curve of the coamplified standard template DNA.
Western blot analysis.
Cell lysates were prepared by resuspending the cells in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 1 mM Na3VO4, and 1 μg/ml each of aprotinin and leupeptin) followed by incubation on ice for 20 min. Insoluble components were removed by centrifugation, and the protein concentrations were determined using a protein assay kit (Bio-Rad). Proteins were separated by electrophoresis on 7.5% or 10% to 20% gradient Tris-glycine mini gels (Oriental Instruments, Kanagawa, Japan) under reducing conditions (3.5 × 10−2 M 2-mercaptoethanol) and then transferred electrophoretically to nitrocellulose filters (Invitrogen) for Western blotting using goat anti-FeLV gp70 and goat anti-FeLV p27 primary antibodies (NCI-Frederick), and an HRP-conjugated anti-goat IgG secondary antibody (Cell Signaling). Detected proteins were visualized using 20× LumiGLO (Cell Signaling).
Ethical approval.
Animal studies were conducted according to the guidelines for the Care and Use of Laboratory Animals of the Ministry of Education, Culture, Sports, Science, and Technology, Japan. All experiments were approved by the Genetic Modification Safety Committee of Yamaguchi University, Yamaguchi, Japan.
Data availability.
The nucleotide sequences reported in this study were deposited in the DDBJ, EMBLE, and GenBank databases under accession numbers LC223819, LC223820, and LC462187.
ACKNOWLEDGMENTS
We thank A. Dusty Miller for providing the RH clones. We thank Hajime Tsujimoto for providing the MCC and 3201 cells and the FeLV-A/pFGA5, FeLV-B/pFGB, and FeLV-C/pFSC plasmids, Tsutomu Hohdatsu for providing the Fet-J cells, and Yoshinao Kubo for providing the MDTF cells. We thank I. David Goldman for providing the HeLa and HeLa-R5 cells, Edward A. Hoover for providing the FeLV-A 61E plasmid, and Toshio Kitamura for providing the PLAT-E and PLAT-A cells. We thank Julie Overbaugh for providing the feline THTR1. We also thank Kate Fox from the Edanz Group (Fukuoka, Japan) for editing a draft of the manuscript.
This work was supported by JSPS KAKENHI 15H04602 to K.N. and NIH R21 HG007405 to A.H.K. and D.J.S.
REFERENCES
- 1.Greenwood AD, Ishida Y, O'Brien SP, Roca AL, Eiden MV. 2018. Transmission, evolution, and endogenization: lessons learned from recent retroviral invasions. Microbiol Mol Biol Rev 82:e00044-17. doi: 10.1128/MMBR.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter E. 1997. Viral entry and receptor, p 71–120. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 3.Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu ES, Hoover EA, VandeWoude S. 2018. A retrospective examination of feline leukemia subgroup characterization: viral interference assays to deep sequencing. Viruses 10:29. doi: 10.3390/v10010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe S, Kawamura M, Odahara Y, Anai Y, Ochi H, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2013. Phylogenetic and structural diversity in the feline leukemia virus env gene. PLoS One 8:e61009. doi: 10.1371/journal.pone.0061009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann K. 2012. Clinical aspects of feline retroviruses: a review. Viruses 4:2684–2710. doi: 10.3390/v4112684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hisasue M, Nagashima N, Nishigaki K, Fukuzawa I, Ura S, Katae H, Tsuchiya R, Yamada T, Hasegawa A, Tsujimoto H. 2009. Myelodysplastic syndromes and acute myeloid leukemia in cats infected with feline leukemia virus clone33 containing a unique long terminal repeat. Int J Cancer 124:1133–1141. doi: 10.1002/ijc.24050. [DOI] [PubMed] [Google Scholar]
- 8.Overbaugh J, Donahue PR, Quackenbush SL, Hoover EA, Mullins JI. 1988. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science 239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 9.Borjatsch J, Kristal BS, Viglianti GA, Khiroya R, Hoover EA, Mullins JI. 1992. Feline leukemia virus subgroup C phenotype evolves through distinct alterations near the N terminus of the envelope surface glycoprotein. Proc Natl Acad Sci U S A 89:8457–8461. doi: 10.1073/pnas.89.18.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigby MA, Rojko JL, Stewart MA, Kociba GJ, Cheney CM, Rezanka LJ, Mathes LE, Hartke JR, Jarrett O, Neil JC. 1992. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J Gen Virol 73:2839–2847. doi: 10.1099/0022-1317-73-11-2839. [DOI] [PubMed] [Google Scholar]
- 11.Sheets RL, Pandey R, Jen WC, Roy-Burman P. 1993. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J Virol 67:3118–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsatsanis C, Fulton R, Nishigaki K, Tsujimoto H, Levy L, Terry A, Spandidos D, Onions D, Neil JC. 1994. Genetic determinants of feline leukemia virus-induced lymphoid tumors: patterns of proviral insertion and gene rearrangement. J Virol 68:8296–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishigaki K, Okuda M, Endo Y, Watari T, Tsujimoto H, Hasegawa A. 1997. Structure and function of the long terminal repeats of feline leukemia viruses derived from naturally occurring acute myeloid leukemias in cats. J Virol 71:9823–9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandhasin C, Coan PN, Levy LS. 2005. Subtle mutational changes in the SU protein of a natural feline leukemia virus subgroup A isolate alter disease spectrum. J Virol 79:1351–1360. doi: 10.1128/JVI.79.3.1351-1360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarma PS, Log T. 1971. Viral interference in feline leukemia-sarcoma complex. Virology 44:352–358. doi: 10.1016/0042-6822(71)90266-2. [DOI] [PubMed] [Google Scholar]
- 16.Jarrett O, Laird HM, Hay D. 1973. Determinants of the host range of feline leukaemia viruses. J Gen Virol 20:169–175. doi: 10.1099/0022-1317-20-2-169. [DOI] [PubMed] [Google Scholar]
- 17.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. doi: 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyake A, Watanabe S, Hiratsuka T, Ito J, Ngo MH, Makundi I, Kawasaki J, Endo Y, Tsujimoto H, Nishigaki K. 2016. Novel feline leukemia virus interference group based on the env gene. J Virol 90:4832–4837. doi: 10.1128/JVI.03229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohn JL, Moser MS, Gwynn SR, Baldwin DN, Overbaugh J. 1998. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J Virol 72:2686–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donahue PR, Quackenbush SL, Gallo MV, deNoronha CMC, Overbaugh J, Hoover EA, Mullins JI. 1991. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol 65:4461–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy WD Jr, Hess PW, MacEwen EG, McClelland AJ, Zuckerman EE, Essex M, Cotter SM, Jarrett O. 1976. Biology of feline leukemia virus in the natural environment. Cancer Res 36:582–588. [PubMed] [Google Scholar]
- 22.Jarrett O, Russell PH. 1978. Differential growth and transmission in cats of feline leukemia viruses of subgroups A and B. Int J Cancer 21:466–472. doi: 10.1002/ijc.2910210411. [DOI] [PubMed] [Google Scholar]
- 23.Jarrett O, Hardy WD Jr, Golder MC, Hay D. 1978. The frequency of occurrence of feline leukemia virus subgroups in cats. Int J Cancer 21:334–337. doi: 10.1002/ijc.2910210314. [DOI] [PubMed] [Google Scholar]
- 24.Elder JH, Mullins JI. 1983. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J Virol 46:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheets RL, Pandey R, Klement V, Grant CK, Roy-Burman P. 1992. Biologically selected recombinants between feline leukemia virus (FeLV) subgroup A and an endogenous FeLV element. Virology 190:849–855. doi: 10.1016/0042-6822(92)90924-E. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Hara B, Johann SV, Klinger HP, Blair DG, Rubinson H, Dunn KJ, Sass P, Vitek SM, Robins T. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ 1:119–127. [PubMed] [Google Scholar]
- 28.Johann SV, Gibbons JJ, O'Hara B. 1992. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol 66:1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi Y, Vile RG, Simpson G, O'Hara B, Collins MK, Weiss RA. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol 66:1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson MM, Lauring AS, Robertson S, Dirks C, Overbaugh J. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J Virol 75:10563–10572. doi: 10.1128/JVI.75.22.10563-10572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tailor CS, Willett BJ, Kabat D. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J Virol 73:6500–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093–1099. [PubMed] [Google Scholar]
- 33.Shalev Z, Duffy SP, Adema KW, Prasad R, Hussain N, Willett BJ, Tailor CS. 2009. Identification of a feline leukemia virus variant that can use THTR1, FLVCR1, and FLVCR2 for infection. J Virol 83:6706–6716. doi: 10.1128/JVI.02317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson MM, Lauring AS, Burns CC, Overbaugh J. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 35.Rasko JE, Battini JL, Kruglyak L, Cox DR, Miller AD. 2000. Precise gene localization by phenotypic assay of radiation hybrid cells. Proc Natl Acad Sci U S A 97:7388–7392. doi: 10.1073/pnas.130200097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, Lee R, Maratukulam A, O’Connor K, Perkins S, Piercy M, Qin F, Reif T, Sanders C, She X, Sun W-L, Tabar P, Voyticky S, Cowles S, Fan J-B, Mader C, Quackenbush J, Myers RM, Cox DR. 1997. An STS-based radiation hybrid map of the human genome. Genome Res 7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 37.Wang RT, Ahn S, Park CC, Khan AH, Lange K, Smith DJ. 2011. Effects of genome-wide copy number variation on expression in mammalian cells. BMC Genomics 12:562. doi: 10.1186/1471-2164-12-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan AH, Bloom JS, Faridmoayer E, Smith DJ. 2016. Genetic screening reveals a link between Wnt signaling and antitubulin drugs. Pharmacogenomics J 16:164–172. doi: 10.1038/tpj.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, Keurentjes JJ, van Eeuwijk FA. 2015. Marker-based estimation of heritability in immortal populations. Genetics 199:379–398. doi: 10.1534/genetics.114.167916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao R, Gao F, Hanscom M, Goldman ID. 2004. A prominent low-pH methotrexate transport activity in human solid tumor cells: contribution to the preservation of methotrexate pharmacologic activity in HeLa cells lacking the reduced folate carrier. Clin Cancer Res 10:718–727. doi: 10.1158/1078-0432.CCR-1066-03. [DOI] [PubMed] [Google Scholar]
- 41.Nishigaki K, Hanson C, Thompson D, Yugawa T, Hisasue M, Tsujimoto H, Ruscetti S. 2002. Analysis of the disease potential of a recombinant retrovirus containing Friend murine leukemia virus sequences and a unique long terminal repeat from feline leukemia virus. J Virol 76:1527–1532. doi: 10.1128/JVI.76.3.1527-1532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart MA, Warnock M, Wheeler A, Wilkie N, Mullins JI, Onions DE, Neil JC. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol 58:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao R, Goldman ID. 2013. Folate and thiamine transporters mediated by facilitative carriers (SLC19A1-3 and SLC46A1) and folate receptors. Mol Aspects Med 34:373–385. doi: 10.1016/j.mam.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crandell RA, Fabricant CG, Nelson-Rees WA. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176–185. doi: 10.1007/BF02618435. [DOI] [PubMed] [Google Scholar]
- 45.Rasheed S, Gardner MB. 1980. Characterization of cat cell cultures for expression of retrovirus, FOCMA and endogenous sarc genes, p 393–400. In Hardy WD Jr, Essex M, McClelland AJ (ed), Proceedings of the third international feline leukemia virus meeting. Elsevier/North-Holland Publishing Co., New York, NY. [Google Scholar]
- 46.Cheney CM, Rojko JL, Kociba GJ, Wellman ML, Di Bartola SP, Rezanka LJ, Forman L, Mathes LE. 1990. A feline large granular lymphoma and its derived cell line. In Vitro Cell Dev Biol 26:455–463. doi: 10.1007/BF02624087. [DOI] [PubMed] [Google Scholar]
- 47.Snyder HW, Hardy WD, Zuckerman EE, Fleissner E. 1978. Characterisation of a tumour-specific antigen on the surface of feline lymphosarcoma cells. Nature 275:656–658. doi: 10.1038/275656a0. [DOI] [PubMed] [Google Scholar]
- 48.Mochizuki H, Takahashi M, Nishigaki K, Ide T, Goto-Koshino Y, Watanabe S, Sato H, Sato M, Kotera Y, Fujino Y, Ohno K, Uchida K, Tsujimoto H. 2011. Establishment of a novel feline leukemia virus (FeLV)-negative B-cell cell line from a cat with B-cell lymphoma. Vet Immunol Immunopathol 140:307–311. doi: 10.1016/j.vetimm.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 49.Donahue PR, Hoover EA, Beltz GA, Riedel N, Hirsch VM, Overbaugh J, Mullins JI. 1988. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J Virol 62:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawamura M, Watanabe S, Odahara Y, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2015. Genetic diversity in the feline leukemia virus gag gene. Virus Res 204:74–81. doi: 10.1016/j.virusres.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 51.Matherly LH, Hou Z, Deng Y. 2007. Human reduced folate carrier: translation of basic biology to cancer etiology and therapy. Cancer Metastasis Rev 26:111–128. doi: 10.1007/s10555-007-9046-2. [DOI] [PubMed] [Google Scholar]
- 52.Soll SJ, Neil SJ, Bieniasz PD. 2010. Identification of a receptor for an extinct virus. Proc Natl Acad Sci U S A 107:19496–19501. doi: 10.1073/pnas.1012344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco-Melo D, Gifford RJ, Bieniasz PD. 2017. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. Elife 6:e22519. doi: 10.7554/eLife.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albritton LM, Tseng L, Scadden D, Cunningham JM. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 55.Kozak CA. 2015. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. doi: 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsang J, Ribet D, Heidmann T, Dewannieux M. 2018. Identification of the receptor used by the ecotropic mouse GLN endogenous retrovirus. J Virol 93:e01125-18. doi: 10.1128/JVI.01125-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliveira NM, Farrell KB, Eiden MV. 2006. In vitro characterization of a koala retrovirus. J Virol 80:3104–3107. doi: 10.1128/JVI.80.6.3104-3107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller AD, Wolgamot G. 1997. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J Virol 71:4531–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng HH, Anderson MM, Hankenson FC, Johnston L, Kotwaliwale CV, Overbaugh J. 2006. Envelope determinants for dual-receptor specificity in feline leukemia virus subgroup A and T variants. J Virol 80:1619–1628. doi: 10.1128/JVI.80.4.1619-1628.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu W, Stadler CK, Gorman K, Jensen N, Kim D, Zheng H, Tang S, Switzer WM, Pye GW, Eiden MV. 2013. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc Natl Acad Sci U S A 110:11547–11552. doi: 10.1073/pnas.1304704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shojima T, Yoshikawa R, Hoshino S, Shimode S, Nakagawa S, Ohata T, Nakaoka R, Miyazawa T. 2013. Identification of a novel subgroup of koala retrovirus from koalas in Japanese zoos. J Virol 87:9943–9948. doi: 10.1128/JVI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lander MR, Chattopadhyay SK. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol 52:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 64.Kuse K, Ito J, Miyake A, Kawasaki J, Watanabe S, Makundi I, Ngo MH, Otoi T, Nishigaki K. 2016. Existence of two distinct infectious endogenous retroviruses in domestic cats and their different strategies for adaptation to transcriptional regulation. J Virol 90:9029–9045. doi: 10.1128/JVI.00716-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nunberg JH, Williams ME, Innis MA. 1984. Nucleotide sequences of the envelope genes of two isolates of feline leukemia virus subgroup B. J Virol 49:629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. doi: 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 70.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 71.Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 72.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 73.Tandon R, Cattori V, Gomes-Keller MA, Meli ML, Golder MC, Lutz H, Hofmann-Lehmann R. 2005. Quantitation of feline leukaemia virus viral and proviral loads by TaqMan real-time polymerase chain reaction. J Virol Methods 130:124–132. doi: 10.1016/j.jviromet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 74.Hou Z, Ye J, Haska CL, Matherly LH. 2006. Transmembrane domains 4, 5, 7, 8, and 10 of the human reduced folate carrier are important structural or functional components of the transmembrane channel for folate substrates. J Biol Chem 281:33588–33596. doi: 10.1074/jbc.M607049200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequences reported in this study were deposited in the DDBJ, EMBLE, and GenBank databases under accession numbers LC223819, LC223820, and LC462187.