The papillomavirus L2 capsid protein plays an essential role in infectious entry, where it directs the successful trafficking of incoming viral genomes to the nucleus. However, nothing is known about how potential posttranslational modifications may affect different aspects of capsid assembly or infectious entry. In this study, we report the first phospho-specific modification of the BPV-1 and HPV-16 L2 capsid proteins. The phospho-acceptor site is very highly conserved across multiple papillomavirus types, indicating a highly conserved function within the L2 protein and the viral capsid. We show that this modification plays an essential role in infectious entry, where it modulates susceptibility of the incoming virus to capsid disassembly. These studies therefore define a completely new means of regulating the papillomavirus L2 proteins, a regulation that optimizes endocytic processing and subsequent completion of the infectious entry pathway.
KEYWORDS: HPV, L2, infection, protein phosphorylation
ABSTRACT
The human papillomavirus (HPV) capsid comprises two viral proteins, L1 and L2, with the L2 component being essential to ensure efficient endocytic transport of incoming viral genomes. Several studies have previously reported that L1 and L2 are posttranslationally modified, but it is uncertain whether these modifications affect HPV infectious entry. Using a proteomic screen, we identified a highly conserved phospho-acceptor site on the HPV-16 and bovine papillomavirus 1 (BPV-1) L2 proteins. The phospho-modification of L2 and its presence in HPV pseudovirions (PsVs) were confirmed using anti-phospho-L2-specific antibodies. Mutation of the phospho-acceptor sites of both HPV-16 and BPV-1 L2 resulted in the production of infectious virus particles, with no differences in efficiencies of packaging the reporter DNA. However, these mutated PsVs showed marked defects in infectious entry. Further analysis revealed a defect in uncoating, characterized by a delay in the exposure of a conformational epitope on L1 that indicates capsid uncoating. This uncoating defect was accompanied by a delay in the proteolysis of both L1 and L2 in mutated HPV-16 PsVs. Taken together, these studies indicate that phosphorylation of L2 during virus assembly plays an important role in optimal uncoating of virions during infection, suggesting that phosphorylation of the viral capsid proteins contributes to infectious entry.
IMPORTANCE The papillomavirus L2 capsid protein plays an essential role in infectious entry, where it directs the successful trafficking of incoming viral genomes to the nucleus. However, nothing is known about how potential posttranslational modifications may affect different aspects of capsid assembly or infectious entry. In this study, we report the first phospho-specific modification of the BPV-1 and HPV-16 L2 capsid proteins. The phospho-acceptor site is very highly conserved across multiple papillomavirus types, indicating a highly conserved function within the L2 protein and the viral capsid. We show that this modification plays an essential role in infectious entry, where it modulates susceptibility of the incoming virus to capsid disassembly. These studies therefore define a completely new means of regulating the papillomavirus L2 proteins, a regulation that optimizes endocytic processing and subsequent completion of the infectious entry pathway.
INTRODUCTION
Human papillomaviruses (HPVs) form a large family of nonenveloped DNA viruses, with more than 200 different types. Most cause only benign epithelial lesions; however, a small subset of HPVs with transforming potential are responsible for almost all cases of cervical cancer and a majority of other anogenital and oropharyngeal cancers (1–3).
HPVs infect epithelial cells, and their life cycle is intimately linked to the differentiation program of the infected epithelium. The virus infects basal keratinocytes through microtraumas in the skin, but new infectious virus particles can only be produced in the upper terminally differentiated layers of the skin. The productive viral life cycle requires the viral oncoproteins E6 and E7, which create an environment favorable for viral DNA replication in the mid-epithelial layers, where DNA replication would normally not be possible. In rare cases, persistent infection and perturbation of the productive viral life cycle may lead to development of cancer (4, 5).
Papillomaviruses (PVs) have an 8-kb DNA genome packaged in an icosahedral capsid comprising 360 copies of the major capsid protein L1 and 12 to 36 copies of the minor capsid protein L2 (6, 7). While both proteins play essential functions in capsid assembly and virus entry, the L2 protein also appears to ensure that the incoming viral genome is trafficked correctly to the nucleus (8, 9).
HPV infection involves multiple sequential steps, not all of which are well understood. After the initial attachment of the virus particle to the extracellular matrix, both capsid proteins undergo conformational changes that allow the virus to bind the target cell and enter by endocytic uptake (10–13). Intracellular trafficking through the endosomal compartment and acidification of the endocytic vesicles are essential for virus uncoating and release of the L2/viral DNA complex. The majority of the L1 protein is sorted into lysosomes where it undergoes degradation (14–16). The L2/viral DNA complex then exits the endocytic compartment (17–19) and is transported to the nucleus through the trans-Golgi network (TGN) and, possibly, through the endoplasmic reticulum (20, 21). The nuclear entry of the L2/viral DNA complex is also critically dependent upon mitosis (22–24).
Interactions of the viral capsid proteins with cellular components play an important role in papillomavirus infectious entry (25–27). Intracellular processing of the viral capsid is dependent on a number of different host factors, such as furin (13), cyclophilin (16), and gamma-secretase (21, 28–30), among others. The cellular cargo-sorting machinery, including the ESCRT complex (31–33), SNX17 (34), SNX27 (35), and components of both the retromer (36, 37) and retriever (38) complexes are also required for efficient virus infection. The sequence of events and the definition of all the cellular components utilized by HPV virions during their transport to the nucleus remain important questions.
Phosphorylation of different viral proteins plays an essential role during the different phases of a virus life cycle, regulating such processes as virion trafficking, uncoating, replication, protein-protein interactions, assembly, and viral egress (39, 40). While such modifications of the E6 and E7 oncoproteins have been well characterized, little is known about the potential roles of posttranslational modifications of the HPV capsid proteins. Indeed, previous studies had shown that HPV-16 L1 and L2 could be phosphorylated when expressed in a baculovirus expression system (41), as well as potentially being modified by sumoylation (42) and ubiquitination (31). What role these potential modifications might have with respect to viral capsid protein function remains completely unknown. Our aim was to identify any potential conserved phosphorylation sites on the HPV-16 and bovine papillomavirus 1 (BPV-1) L2 proteins. We used purified HPV-16 and BPV-1 pseudovirions (PsVs) produced in cultured cells and, for comparison, native purified BPV particles isolated from bovine papillomas. We have identified a highly conserved phospho-acceptor site on the L2 proteins of both HPV-16 and BPV-1, mutation of which greatly reduces virus infection and capsid uncoating.
RESULTS
Identification of a conserved phospho-acceptor site at T62 in HPV-16 L2.
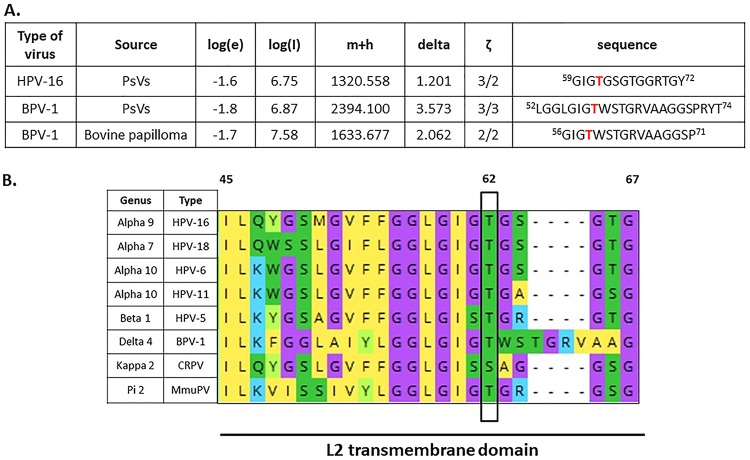
Previous studies had shown that HPV-16 L2 was phosphorylated when it was expressed in insect cells (41); however, it was unknown whether L2 is phosphorylated in the context of intact viral capsids. To determine possible sites of L2 phosphorylation, we performed proteomic analyses on native BPV-1 virions (43) and on HPV-16 and BPV-1 PsVs produced and purified from HEK293TT cells. The mass spectroscopy (MS) spectra for L2 were analyzed for phospho-modifications and compared with the spectra obtained from native BPV-1 virions, as described in Materials and Methods. A number of phospho-modifications on L2 were identified, but among these the threonine residue (T62 in HPV-16 L2 and T59 in BPV-1 L2) was found to be phosphorylated in both PsVs and in native virus (Fig. 1A). T62 is located in the transmembrane (TM) domain of L2 and was the most highly conserved phospho-acceptor site that we identified, being highly conserved between high- and low-risk mucosal alpha-papillomaviruses, as well as in other genera of the Papillomaviridae family, such as betapapillomaviruses (HPV-5), deltapapillomaviruses (BPV-1), pipapillomaviruses (murine papillomavirus, MmuPV) and kappapapillomaviruses (cottontail rabbit papillomavirus 1, CRPV-1) (serine residue), as illustrated in Fig. 1B.
FIG 1.
Identification of the conserved phospho-acceptor site T62 in HPV-16 L2 capsid protein using mass spectrometry analysis. (A) The bicistronic plasmids expressing HPV-16 or BPV-1 L1 and L2 were transfected into HEK293TT cells, together with a luciferase reporter plasmid. After 48 h the cells were harvested, and pseudovirions were purified by cesium chloride gradient and analyzed by mass spectrometry. The spectra for L2 were analyzed for phospho-modifications and compared with those obtained from native BPV-1 virions isolated from a bovine papilloma. The threonine residue (T62 in HPV-16 L2 and T59 in BPV-1 L2) was phosphorylated in both PsVs and native virus. The table shows results from the mass spectrometric analysis: log(e), the base 10 log of the expectation that the assignment is stochastic; log(l), the base 10 log of the sum of the intensities of the fragment ion spectra; m+h, the calculated mass of the protonated parent ion for this sequence assignment; delta, the difference between the measured and calculated protonated parent ion masses; ζ, the ratio of the measured charge of the parent ion to the number of basic sites in the assigned peptide sequence. The sequence of the assigned peptide is also shown. (B) Alignment of L2 transmembrane (TM) domains of selected PVs. Alignment was generated with Mega X software. Conserved threonine/serine phospho-acceptor (T62/S62) sites in the L2 sequences are highlighted in green and boxed. Note that residue T62/S62 is highly conserved across a wide evolutionary spectrum of PV types, ranging from high- and low-risk HPV types to BPV-1, MmuPV, and CRPV-1.
Confirmation of L2 phosphorylation using a phospho-specific HPV-16 L2-pT62 antibody.
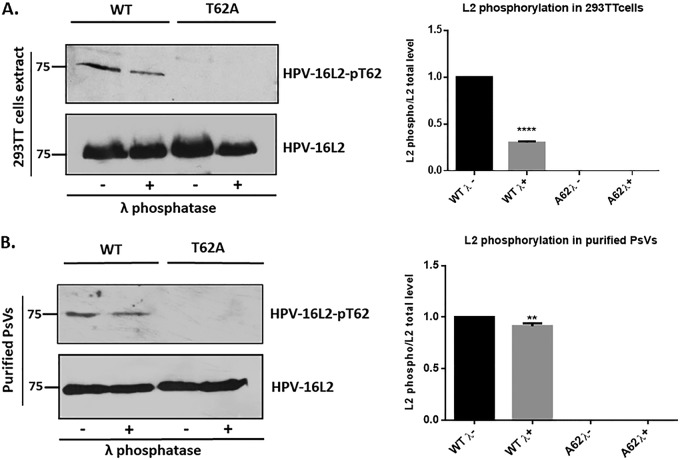
To confirm that the HPV-16 L2 protein was phosphorylated at T62, we generated a rabbit phospho-specific anti-HPV-16 L2 antibody and used it to determine whether L2 was phosphorylated in cell extracts and purified HPV-16 PsVs. To verify specificity of the antibody, we generated the mutant T62A in HPV-16 L2, in the context of the bicistronic plasmid expressing both L1 and L2 of HPV-16. HEK293TT cells were transfected with plasmids expressing the wild-type and the mutant (T62A) L2 and harvested after 24 h; then the cell extracts were incubated either with or without lambda phosphatase for 30 min. Western blotting was then performed for total and phosphorylated L2. As shown in Fig. 2A, there was a clear phosphorylation of HPV-16 L2 in HEK293TT cell extracts, which decreased by 70% following lambda phosphatase treatment and was completely absent with the T62A mutant. To verify that phosphorylated L2 was also incorporated into purified HPV-16 PsVs, a similar analysis was performed on wild-type and T62A mutant-containing HPV-16 PsVs purified by cesium chloride gradient centrifugation and treated with lambda phosphatase for 30 min. The PsVs were then analyzed by Western blotting for total and phosphorylated L2. The results shown in Fig. 2B confirm the presence of phosphorylated L2 T62 in purified viral capsids. Taken together, these results demonstrate that the HPV-16 L2 protein is phosphorylated at T62 and can be detected in both cell extracts and purified PsVs.
FIG 2.
Confirmation of L2 phosphorylation using a phospho-specific HPV-16 L2-pT62 antibody. (A) HEK293TT cells were transfected with plasmids expressing wild-type (WT) and mutant (T62A) L2. After 24 h the cells were harvested and incubated in the presence (+λ) or absence (−λ) of lambda phosphatase for 30 min. Western blotting was then performed for phosphorylated L2 (top panel) and total L2 (bottom panel). Quantification of phosphorylated L2 normalized to the amount of total L2 is shown in the histogram. Note that wild-type HPV-16 L2 is clearly phosphorylated in HEK293TT cell extracts, with a decrease in signal following treatment with lambda phosphatase, and no phosphorylation is seen with the T62A mutant. (B) The cesium chloride gradient-purified wild-type and mutant (T62A) PsVs were also analyzed by Western blotting using the same phospho-specific HPV-16 L2-pT62 antibody. Purified PsVs were incubated with (+λ) or without (−λ) lambda phosphatase and then analyzed by Western blotting using the anti-HPV-16 L2 phospho-specific antibody and the total anti-L2 antibody. Quantification of phosphorylated L2 normalized to the amount of total L2 is shown in the histogram. **, P < 0.01; ****, P < 0.0001. Note that these results again confirm the presence of a phospho-acceptor site at T62 in the context of purified viral capsids. Taken together, these results demonstrate that the HPV-16 L2 protein is phosphorylated at T62 both in cell extracts and in purified PsVs.
The T62 and T59 L2 phospho-acceptor sites are required for efficient infection with HPV-16 and BPV-1 PsVs.
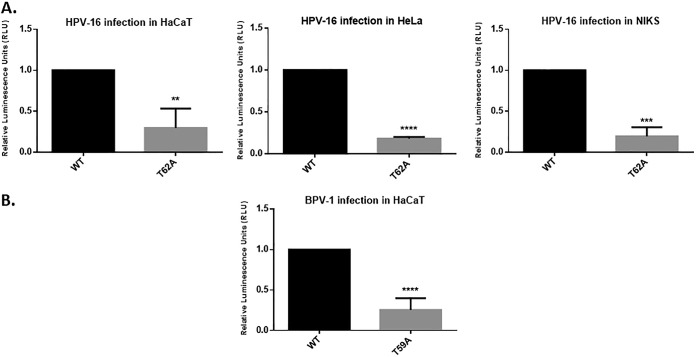
Having found that HPV-16 and BPV-1 L2 proteins were phosphorylated at the same highly conserved amino acid residue, we determined whether this residue was required for virus infection by comparing the abilities of the wild-type PsVs and those of HPV-16 L2 T62A and BPV-1 T59A mutant-containing PsVs to transduce a luciferase reporter construct. Infections were performed in HaCaT, HeLa, and NIKS cell lines, and luciferase activity was measured in cells harvested at 48 h postinfection. As shown in Fig. 3, the T62A substitution in HPV-16 L2 (Fig. 3A) and the T59A substitution in BPV-1 L2 (Fig. 3B) had profound inhibitory effects upon the ability of HPV-16 and BPV-1 PsVs to infect the different cell lines. Taken together, these results suggest that an intact phospho-acceptor at these positions in L2 plays an important role in infection by different PV types.
FIG 3.
The T62 and T59 L2 phospho-acceptor sites are required for efficient infection with HPV-16 and BPV-1 PsVs. (A) HaCaT, HeLa, and NIKS cells were infected with wild-type (WT) and mutant (T62) HPV-16 PsVs carrying equal amounts of encapsidated luciferase reporter plasmid. After 48 h the cells were harvested, and the levels of luciferase activity were measured in triplicate by luminometry. The efficiency of infection of T62A mutant HPV-16 PsVs was calculated by normalizing the values to those of wild-type HPV-16 PsVs. (B) HaCaT cells were infected with the wild-type (WT) and mutant (T59) BPV-1 PsVs carrying equal amounts of encapsidated luciferase reporter plasmid. After 48 h the cells were harvested, and the levels of luciferase activity were measured in triplicate by luminometry. The efficiency of infection of T59A mutant BPV-1 PsVs was calculated by normalizing the values with wild-type BPV-1 PsVs. Throughout the experiments, equal amounts of total cell protein extract were used in the luciferase measurements. Results are expressed as means, with the standard deviations of at least three independent experiments. **, P ≤ 0.01; ***, P ≤ 0.001. ****, P ≤ 0.0001.
Mutation of the L2 phospho-acceptor site does not affect PsV assembly.
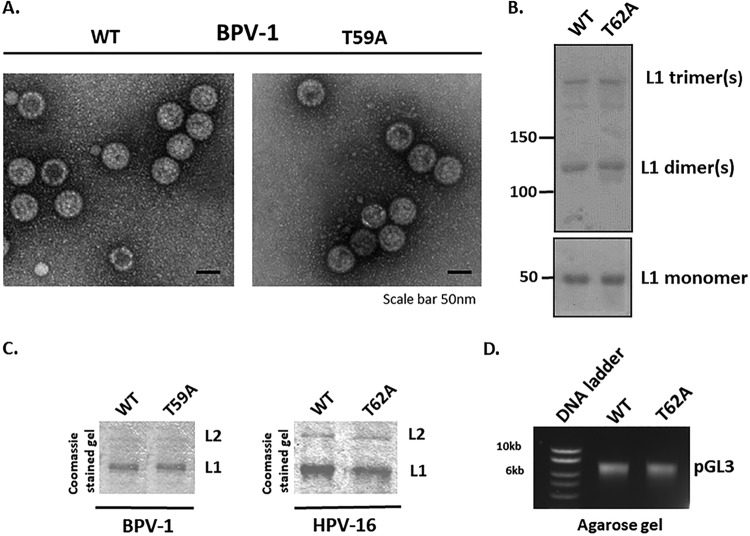
To verify that the difference in infectivities of the wild-type and mutant PsVs was not caused by a defect in PsV assembly, we monitored the integrity of wild-type and mutant (T59A) BPV-1 PsVs by electron microscopy (EM) of the purified PsVs (44). As can be seen in Fig. 4A, no evident structural differences were observed between the wild-type and mutant PsV preparations. It is known that capsid maturation requires intramolecular disulfide bonds that contribute to the stability of papillomavirus capsids (45). To check whether there are any differences in the levels of disulfide cross-linking between wild-type and mutant PsV preparations, we analyzed them on nonreducing denaturing gels. As can be seen in Fig. 4B, the wild-type and HPV-16 mutant virion preparations show the same patterns of L1 dimers and trimers, which correspond to fully mature capsids, as previously described (46). To determine whether wild-type and mutant PsVs have similar ratios of L1 to L2 within the capsid, purified PsVs were resolved by 10% SDS-PAGE. The Coomassie-stained gels (Fig. 4C) showed similar capsid protein levels and proportions in BPV-1 wild-type and T59A mutant PsVs. This was also seen with wild-type and T62A mutant HPV-16 PsVs, indicating that coassembly of L1/L2 proteins into particles was unaffected by the loss of the phospho-acceptor site in either BPV-1 or HPV-16 L2. Furthermore, agarose gel electrophoresis of the pGL3:luciferase vector extracted from purified HPV-16 wild-type and T62A mutant PsVs confirmed that they contained similar levels of the plasmid (Fig. 4D). This result was confirmed by real-time PCR analysis. The average numbers (the mean results from at least three independent experiments) of pseudogenome (pGL3 plasmid) copies incorporated per 1 mg of L1 were similar for the two PsVs preparations (2.08E+11 copies/mg L1 for wild-type PsVs and 1.8E+11 copies/mg L1 for T62A mutant PsVs) and were typical for standard HPV-16 PsV preparations in HEK293TT cells (2.0E+11 copies/mg L1) (47). These results demonstrate that the lack of phosphorylation of the T62 residue of L2 does not affect PsV assembly.
FIG 4.
Mutation of the L2 phospho-acceptor site does not affect PsVs assembly. (A) The integrity of purified wild-type and mutant (T59A) BPV-1 PsVs was monitored by electron microscopic analysis. Note that no evident structural differences were observed between the wild-type and mutant PsV preparations. (B) The patterns of disulfide bonds in the purified wild-type and T62A HPV-16 PsVs were examined by nonreducing denaturing gel analysis and Ponceau S staining (upper panel). The lower panel shows conventional denaturing gel analysis of both PsV preparations. Note that the two PsV preparations show similar levels of disulfide cross-linking, which correspond to those of fully mature capsids. (C) The Coomassie-stained gels of the purified PsVs show similar levels of capsid protein for wild-type and T59A BPV-1 PsVs and for wild-type and T62A HPV-16 PsVs, indicating that coassembly of L1/L2 proteins into particles is not affected by the T62A substitution. (D) The agarose gel shows similar amounts of encapsidated DNA (pGL3 vector carrying the luciferase reporter gene) extracted from the purified wild-type and T62A HPV-16 PsVs.
Mutation of the L2 phospho-acceptor site alters PsVs stability.
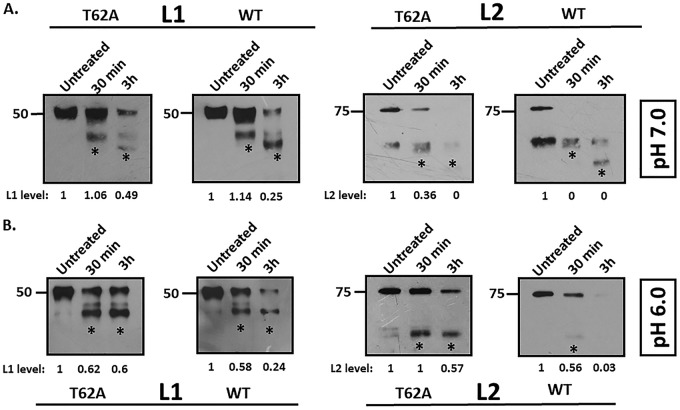
We next investigated whether T62A influences capsid stability. A previous analysis had shown that a brief digestion with trypsin has little effect on mature and correctly folded HPV-16 PsVs (44, 48). To determine whether subtle conformational differences in wild-type and T62A HPV-16 PsVs could be discerned by trypsin sensitivity, we performed trypsin digestion of PsV preparations for either 30 min or 3 h (Fig. 5). Detection of L1, following trypsin digestion at pH 7.0, showed similar tryptic products in the two preparations. However, the extent of L1 digestion was greater in the wild-type HPV-16 PsVs than in the T62A HPV-16 PsVs at both the 30-min and 3-h time points, as indicated by more intense bands of tryptic products (Fig. 5A, left panel). Interestingly, examination of L2 showed a dramatic difference between the two PsV preparations (Fig. 5A, right panel), with more distinct digestion products being evident at 3 h in the wild-type PsVs than in the T62A PsVs. Likewise, the total L2 protein levels were more reduced in the wild-type than in T62A PsVs, with the wild-type L2 protein being almost undetectable after 30 min of trypsin digestion. Previous studies have shown that dissociation of L1 from L2 is most efficient at pH 6.0, a pH closer to that of the environment in the acidifying endosome (16). The assay was therefore repeated at pH 6.0, and the results in Fig. 5B show similar L1 tryptic products in the wild-type and mutant PsV preparations. However, it is also clear that digestion of L1 was much faster in the wild-type PsV preparations than in the T62A PsVs (Fig. 5B, left panel). Similarly for L2, both the total levels and the intensity of tryptic products of wild-type PsVs were significantly lower than those of T62A PsVs after 30 min and 3 h of trypsin digestion (Fig. 5B, right panel). Taken together, these data show that the T62 residue influences virion sensitivity to trypsin, with PsVs lacking the T62 phospho-acceptor site being significantly more resistant to trypsin digestion than the wild-type PsVs. This result may suggest that a possible cause for the defect in infection is related to a defect in capsid uncoating.
FIG 5.
Mutation of the L2 phospho-acceptor site alters PsV stability. (A) Trypsin digestion of HPV-16 PsVs at pH 7.0. Purified T62A and wild-type HPV-16 PsVs were incubated with an equal volume of 0.05% trypsin at pH 7.0 for either 30 min or 3 h and compared with untreated samples by Western blot analysis. Panels show anti-L1 and anti-L2 immunoblots. Note the increased intensity of the lower bands (*, tryptic products) in the trypsin-treated wild-type PsVs. Clearly, the total L2 protein levels are greatly reduced in wild-type preparations in comparison to those in the T62A preparations, with the wild-type L2 protein being almost undetectable after 30 min of trypsin digestion. (B) Trypsin digestion of HPV-16 PsVs at pH 6.0. Purified T62A and wild-type HPV-16 PsVs were incubated with an equal volume of 0.05% trypsin at pH 6.0 for either 30 min or 3 h and compared with untreated samples by Western blot analysis. Panels show anti-L1 and anti-L2 immunoblots. Note that L1 proteins from wild-type and T62A PsVs show similar tryptic products following trypsin treatment at pH 6.0. However, digestion of L1 was much more rapid in the wild-type PsV preparations than in the T62A PsVs. In the case of L2, both the total levels and the tryptic products of wild-type L2 were significantly lower than those of T62A PsVs after 30 min of trypsin digestion at pH 6.0. Taken together, these data show that the T62 residue in the L2 phosphorylation site has a major influence on viral sensitivity to trypsin, indicating that PsVs lacking the T62 phospho-acceptor site are significantly more resistant to trypsin digestion than the wild-type PsVs. For all experiments, total L1 and total L2 protein levels were quantified by measuring the pixel intensity of each band using densitometry (ImageJ) and are shown normalized to levels of their respective (untreated) controls.
The T62A phospho-acceptor site has no effect on virus internalization but does affect intracellular processing.
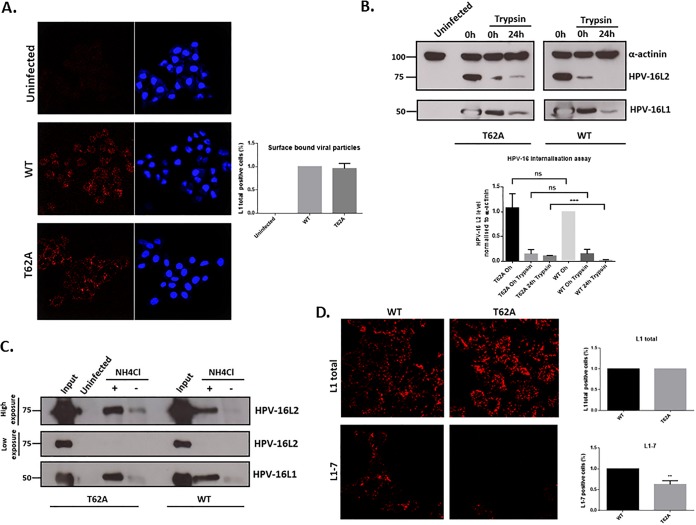
To understand further the potential function of L2 phosphorylation in infectious entry, we investigated how mutation of the T62 residue might affect the intracellular trafficking of HPV-16 PsVs. To assess whether the T62A substitution affects PsV binding to the cell surface, HeLa cells were incubated with wild-type or T62A HPV-16 PsVs for 24 h. The cells were then washed, fixed, and subjected to quantitative immunofluorescence analysis of nonpermeabilized cells using an L1-specific antibody. As shown in Fig. 6A, similar levels of L1 staining were seen with the wild-type and mutant PsVs, indicating that similar amounts of the two PsV were bound to the cells.
FIG 6.
The T62A phospho-acceptor site has no effect on virus internalization but does affect intracellular processing. (A) PsV binding to the cell surface. HeLa cells were exposed to wild-type and T62A HPV-16 PsVs for 24 h. The cells were then washed, fixed, and subjected to quantitative immunofluorescence analysis of nonpermeabilized cells using an L1-specific antibody (red). The nuclei were detected with 4′,6′-diamidino-2-phenylindole (blue). Representative pictures are shown. Quantification of surface-bound viral particles is also shown. Note that similar amounts of wild-type and T62A HPV-16 L1-positive PsVs are observed after L1 staining, confirming that there are no major differences in how both types of PsVs bind to the surface of infected cells. (B) PsV internalization assay. HeLa cells were infected with wild-type and T62A HPV-16 PsVs. At 1 h and 24 h postinfection, the cells were treated with trypsin to remove any noninternalized virus. They were then harvested, and the levels of L1 and L2 protein remaining within the cells were monitored by Western blotting. Quantification of HPV-16 L2 levels, normalized to alpha-actinin, is shown in the histogram. Note that there are similar levels of L1 and L2 in wild-type and T62A PsVs at 1 h postinfection (0 h trypsin), indicating no major differences in levels of PsV entry. Interestingly, the intensity of T62A L2 is much higher than that of wild-type L2 at 24 h postinfection (24 h trypsin), suggesting that the processing of T62A PsVs in HeLa cells is slower than that of the wild-type. (C) Intracellular processing of PsVs. HaCaT cells were exposed to wild-type and T62A PsVs and incubated for 24 h at 37°C. The cells were then removed with 0.25% trypsin, washed twice with PBS, and lysed in RIPA lysis buffer. Clarified lysates were incubated with mouse anti-HPV-16 L1 (H16.V5) antibody followed by protein A-conjugated Sepharose beads. Immunoprecipitated complexes were collected by centrifugation, washed three times, resolved by 10% SDS-PAGE, and analyzed by Western blotting. HPV-16 L1 was detected with the CamVir-1 antibody. For determination of L1-associated L2 protein, mouse anti-HPV-16 L2 (16.D4 64-81) was used. The acidification inhibitor NH4Cl was included as a positive control for L2 retention. Note that in the input capsids, the levels of T62A and wild-type capsid-associated L2 protein were equal. However, in the untreated cells, the T62A HPV-16 PsVs showed a much stronger association between L1 and T62A L2 than with the wild-type L2. When the cells were treated with NH4Cl, increased levels of both T62A and wild-type L2 remained associated with L1 compared to levels with untreated cells; however, there was no major difference in T62A and wild-type L2 levels. These results further indicate a delay in the processing of HPV-16 T62A mutant PsVs compared with that of wild-type virus. (D) Capsid disassembly assay. HeLa cells were infected with wild-type and T62A HPV-16 PsVs. At 7 h postinfection the cells were fixed and stained using mouse anti-L1 antibody (CamVir-1) for total L1 or the uncoating-specific anti-L1 antibody 33L1-7 (red). Representative pictures are shown. Quantification of virus disassembly based on the 33L1-7 staining is also shown. Note that capsid disassembly is approximately 40% lower in the T62A PsV-infected cells than in wild-type-infected cells. These results indicate that the absence of an L2 phospho-acceptor site at T62 increases capsid stability and delays the process of capsid uncoating. **, P < 0.01; ***, P < 0.001.
To determine whether the L2 phospho-acceptor site influenced virus internalization, HeLa cells were infected with wild-type or T62A HPV-16 PsVs. At 1 h and 24 h postinfection, the cells were treated with trypsin to remove any noninternalized virus and then harvested, and the levels of L1 and L2 protein remaining in the cells were determined by Western blotting. The results in Fig. 6B show similar levels of L1 and L2 in wild-type and T62A PsVs at 1 h postinfection (0-h trypsin), indicating no major differences in the entry efficiencies of the PsVs. Interestingly, the intensity of T62A L2 was much higher than that of wild-type L2 at 24 h postinfection (24-h trypsin), suggesting that the processing of T62A PsVs in HeLa cells may be slower than that of the wild type.
Having found an apparent delay in intracellular processing of T62A PsVs, we next evaluated whether this might indicate a reduced efficiency of L2 separation from the capsid (which occurs prior to L2’s egress from the late endosome (LE)/lysosome) (16, 20). We therefore determined the levels of L1-associated T62A or wild-type L2 at 24 h postinfection by immunoprecipitating L1 from the cell extracts using anti-L1 (H16.V5) antibody, followed by Western blotting to detect L1 and L2 proteins (Fig. 6C). The levels of wild-type and T62A capsid-associated L2 proteins were the same for the input capsids, as would be expected from the results shown in Fig. 4. However, by 24 h postinfection, there was essentially no association between L1 and L2 in the wild-type PsV-infected cells, whereas in the T62A PsV-infected cells there was a residual association of L1 and L2. As a further control, cells were treated with the acidification inhibitor NH4Cl, and in this case the levels of L1 and L2 association at 24 h postinfection were the same in the wild-type and T62A mutant PsV-infected cells. These results further indicate a delay in the processing of HPV-16 T62A mutant PsVs compared with that of wild-type virus.
The above results indicate defects in trafficking PsVs that contain the T62A L2 mutation and would predict a delay in capsid uncoating. To investigate whether this was indeed the case, HeLa cells were infected with wild-type and T62A HPV-16 PsVs, and at 7 h postinfection the cells were fixed and analyzed by immunofluorescence using the 33L1-7 antibody that recognizes an epitope on L1 that is a marker of capsid disassembly (49). The results in Fig. 6D show that capsid disassembly is approximately 40% less efficient in the T62A PsV-infected cells than in wild-type-infected cells. These results indicate that the absence of an L2 phospho-acceptor site at T62 increases capsid stability and delays capsid uncoating, suggesting that phosphorylation of L2 at this site in diverse PV types confers a structural alteration that favors capsid disassembly in the acidifying endosome.
DISCUSSION
HPV infectious entry involves a complex series of steps, including the attachment of virus to the extracellular matrix, the structural remodelling of the virion, receptor attachment, endocytic uptake, intracellular trafficking through the endosomal compartment, uncoating, and the recruitment of different components of the cellular transport machinery to ensure delivery of the incoming viral genomes to the nucleus through the trans-Golgi network (25–27). However, no studies have been performed to investigate whether potential phosphorylation of the HPV capsid proteins can play any roles in any of these processes. Indeed, recent studies have shown important roles for phosphorylation of viral capsid proteins from many different viruses during various aspects of virus assembly and infection (39, 40). Since previous studies had suggested that HPV-16 L1 and L2 can be phosphorylated when expressed in insect cells (41), we were particularly interested in investigating whether any potential phospho-modifications could be found in native virus particles and, if so, whether these modifications might influence either virion assembly or the intracellular trafficking of the virus.
To determine whether the papillomavirus capsid proteins were subject to posttranslation modification and, in particular, whether these modifications were found within intact capsids, we performed a proteomic analysis of native BPV-1 capsids compared with BPV-1 and HPV-16 PsVs. This analysis identified a number of potential phosphorylated residues, but the most striking was the T62 site in L2 in HPV-16 PsVs that was also found in BPV-1 native virions and PsVs at the equivalent residue in L2, T59. Furthermore, this putative threonine/serine phospho-acceptor site that is close to residues required for L2 transmembrane spanning (17) is highly conserved across a wide evolutionary spectrum of PV types, ranging from high- and low-risk HPV types, to MmuPV and CRPV-1. This suggests that the T62 phospho-acceptor site may have an evolutionarily conserved function.
We confirmed a phospho-threonine modification at T62/T59 in the proteomic analysis using an anti-phospho-specific L2 antibody. This antibody specifically detects phosphorylation at the T62 residue in wild-type HPV-16 L2, both from L2 in cell extracts and in purified PsV preparations. The mutant T62A L2 that has lost the phospho-acceptor site did not react with the antibody, and, furthermore, treatment with lambda phosphatase resulted in decreased reaction with wild-type HPV-16 PsVs. Interestingly, loss of reactivity in the presence of lambda phosphatase was not complete, indicating that not all phosphorylated L2 residues were accessible by the phosphatase, which suggests that some of the phosphorylated sites may be sterically inaccessible within the capsid. In addition, the stoichiometry of L2 phosphorylation is unknown since in the proteomic analyses both phosphorylated and nonphosphorylated peptides were detected, but our proteomic protocols did not allow any conclusions to be made concerning overall levels of phosphorylation in the PsVs.
Having confirmed that HPV-16 L2 and BPV-1 L2 were subject to phosphorylation at T62 and T59, respectively, we investigated whether an intact phospho-acceptor site was required for virus infection. We found a dramatic decrease in the capacity of mutant PsVs to transduce a luciferase reporter construct, indicating that an intact phospho-acceptor site was important for virus infection. This result might be a consequence of a packaging or assembly defect or, alternatively, a block in the infectious entry. To distinguish between these possibilities, we analyzed the capsid integrity of the wild-type and mutant PsVs using EM, gel electrophoresis, and quantitative PCR (qPCR). In all assays we found no differences between the wild-type PsVs and those containing the mutations in the T62 and T59 phospho-acceptor sites. These results suggest that mutation of these residues has no major effect upon virion assembly and packaging.
We then investigated how mutation of this phospho-acceptor site might affect virus uptake and intracellular trafficking. Analysis of surface-bound particles by immunofluorescence or internalization assays (trypsin digestion of cell surface-bound virions) indicated that cell binding and uptake of the virus were not affected by the loss of the L2 phosphorylation site. However, we did find evidence for a defect in capsid uncoating. Comparing the susceptibilities of wild-type and L2 phospho-site mutant PsVs to trypsin digestion under both neutral and mildly acidic conditions revealed a striking increase in capsid resistance in in vitro trypsin digestion assays with the mutant PsVs. Furthermore, the separation of L2 from L1 during the course of endocytic trafficking was delayed with the mutant PsVs compared to that with the wild type. As a further confirmation of a defect in the capsid disassembly process, we also analyzed the exposure of the 33L1-7 epitope on L1, which is indicative of capsid disassembly (49). Again, there was a dramatic reduction in the 33L1-7-reactive signal in the L2 phospho-site mutants compared with that in the wild-type PsVs. Taken together, these results suggest that this highly conserved phospho-acceptor site in the L2 protein plays a critical role in infectious entry and appears to contribute toward the process of capsid disassembly during the process of endocytic acidification. It should be emphasized that at this stage we do not know whether these defects are due to lack of phosphorylation per se or whether the defects are a direct consequence of the T/A substitution itself. Studies are currently being undertaken to address this by identifying the responsible kinase and then producing virions under conditions in which the kinase activity has been ablated.
These results offer interesting parallels with a number of other viruses such as poliovirus, human immunodeficiency virus type 1 (HIV-1), hepatitis B virus, and human respiratory syncytial virus (HRSV) for which the phosphorylation of the viral capsid proteins has been shown to affect infectious entry and capsid stability and disassembly (50–53). In the case of PVs, it would appear that phosphorylation at a site just downstream of the transmembrane domain of L2 aids capsid disassembly. These results obviously raise a number of important questions concerning the identity of the kinase and why this phosphorylation would be necessary. The identity of the kinase is unknown but highlights potential differences in generation of capsids in raft cultures versus that in HEK293TT cells. Phospho-modifications of the viral capsid might be different under these conditions, which, in turn, might be sufficient to explain some of the reported differences between these different virion preparations (54). Future studies will aim to determine whether similar phospho-modifications are observed in viruses produced in raft cultures although identifying a phospho-acceptor site in virus obtained from a cow wart would seem to be biologically the most relevant source, in the context of this particular study.
More intriguing, however, is the role of this phospho-acceptor site in capsid uncoating. Papillomavirus capsids are extremely stable in the environment (55). However, this stability may create a problem for the virus during infection if efficient uncoating is not achieved prior to L1 entry into the lysosomal compartments (16). Therefore, this phospho-acceptor site does seem to play a significant role in aiding capsid disassembly and separation of L2 from L1 through the very subtle decrease in virion stability resulting from L2 phosphorylation, which is really only apparent once the virion is already partially disassembled by some of the structural alterations that occur following capsid attachment and internalization (10–13). This phosphorylation site is highly conserved through the evolution of papillomaviruses, pointing to a highly conserved function in facilitating capsid disassembly in the acidifying endosomal compartments.
MATERIALS AND METHODS
Cell lines.
HeLa, HaCaT, and HEK293TT cell lines (from ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin (100 U/ml), and glutamine (300 mg/ml). The NIKS cells (normal immortalized keratinocytes) were maintained in F medium (0.66 mM Ca2+), composed of 75% Ham’s F12 medium and 25% DMEM and supplemented with 5% FBS, adenine (24 μg/ml), cholera toxin (8.4 ng/ml), epidermal growth factor (10 ng/ml), hydrocortisone (2.4 μg/ml), and insulin (5 μg/ml) (56).
Plasmids.
The T62A substitution in the L2 phosphorylation site in the context of the pXULL plasmid (used for T62A PsV production) was accomplished using a GeneArt site-directed mutagenesis system (Invitrogen). The mutant plasmids were confirmed by DNA sequencing.
Antibodies.
The phospho-specific HPV-16 L2-pT62 antibody (custom-made by Eurogentec) was raised against the peptide C+GIG-T(PO3H2)-GSGTGGRT in rabbits. For detection of phospho-L2 in Western blotting, the membrane was blocked for 1 h at room temperature using 1% milk and 3% bovine serum albumin (BSA) in Tris-buffered saline (TBS) containing 0.01% Tween 20 (TBST). The blot was then incubated overnight with the phospho-specific HPV-16 L2-pT62 antibody diluted in blocking buffer (1:200). After three washes with TBST, the blot was incubated with anti-rabbit antibody conjugated with horseradish peroxidase (HRP; Dako) and developed using an ECL detection system (Amersham).
The primary antibodies mouse anti-HPV-16 L1 (CamVir-1) and mouse anti-alpha-actinin (H-2) were obtained from Santa Cruz Biotechnology. Other antibodies were kindly provided by Martin Müller (mouse anti-16 L2 [16.D4 64-81]), Martin Sapp (mouse 33L1-7), and Neil Christensen (H16.V5). Secondary antibodies, conjugated to horseradish peroxidase (HRP; Dako) or rhodamine (Molecular Probes), were used as indicated in the text.
PsV production.
Pseudovirions (PsVs) containing a luciferase reporter plasmid (pGL3:luciferase) were generated in HEK293TT cells, as described previously (57). Purity and capsid protein content were determined by SDS-PAGE and Coomassie brilliant blue staining; the encapsidated DNA was analyzed by real-time PCR, and the copy number was quantified using a standard curve of reporter plasmid DNA.
Mass spectrometry analysis.
HPV-16 and BPV-1 PsVs produced in cultured cells and native BPV-1 virions isolated from bovine papillomas were purified by cesium chloride gradient centrifugation.
Samples were then digested by the addition of 100 ng of trypsin or chymotrypsin in 20 μl of 20 mM triethylammonium bicarbonate (pH 8.5) for 16 h at room temperature. Following digestion, the supernatant was passed two time over a stop-and-go extraction (STAGE) tip (58) and eluted with 15 μl of 65% acetonitrile and 0.1% formic acid. The samples were dried and resuspended in 10 μl of 0.1% formic acid and injected onto a 170-mm by 0.075-mm custom-packed column. The column was custom packed using Ascentis Express RPA resin (Sigma-Aldrich). The column was developed over 60 min using a gradient of 0.1% formic acid to 80% acetonitrile. The effluent of the column was sprayed directly into an Amazon ETD mass spectrometer (Bruker Daltonics). Each precursor scan was followed by five fragmentation scans using a dynamic exclusion window of 30 s. Mass spectrometry analysis was triggered by the neutral loss of phosphate in the tandem MS (MS/MS) spectra. The resulting spectra were converted into peak lists using data analysis software (Bruker Daltonics) and analyzed using the X!Tandem search engine (59). The spectra for L2 were analyzed for phospho-modifications and compared with the spectra obtained from the native BPV-1 virions (43).
Infectivity assays.
HaCaT, HeLa, and NIKS cells were seeded in 12-well plates at a density of 0.5 × 105 cells/well. After 24 h cells were exposed to 100 viral genome equivalents (vge)/cell of wild-type and T62A luciferase reporter-positive PsVs. Infection was monitored after 48 h by luminometric analysis of firefly luciferase activity, using a luciferase assay system (Promega). The efficiency of infection of T62A mutant PsVs was calculated by normalizing the values to those of wild-type PsVs. Equal amounts of total cell protein extract were used in the luciferase measurements.
BPV-1 virion preparation.
BPV-1 virions were isolated from cow warts as previously described (45). A 100-ml volume of buffer H (20 mM HEPES, pH 8.0, 100 mM NaCl, 0.1 mM CaCl2) was added to 100 g of minced tissue, and the suspension was homogenized (Brinkmann Kinematica) at setting 4 or 5 for 5 to 10 min. The homogenate was then centrifuged at 14,000 × g for 15 min at 4°C. The supernatant was saved. The pellet was rehomogenized in 50 ml of buffer H with 1 M NaCl and centrifuged as before. The supernatant was saved, and the pellet was rehomogenized with buffer H with 0.05% sodium deoxycholic acid. A 25-ml volume of 1,1,2-trichlorotrifluoroethane (Freon) was added, and the mixture was rehomogenized and centrifuged as described above. All supernatants were combined, sodium deoxycholic acid was added to a final concentration of 0.05%, and the mixture was centrifuged for 30 min at 20,000 × g and 4°C. The clarified supernatant was then centrifuged in an SW28 rotor for 4 h at 25,000 rpm and 20°C. The supernatant was discarded, and the pellets were gently resuspended with 1 ml of buffer R (20 mM HEPES, pH 8.0, 1 mM CaCl2) with agitation at 4°C for 24 to 48 h. CsCl (≈100 mg) was added to each resuspended pellet before Dounce homogenization with a B pestle (Wheaton). The homogenate was brought to a final volume of 10 ml with a stock solution containing 1.33 g of CsCl per ml and 20 mM HEPES (pH 8.0). Samples were then centrifuged in an SW50.1 rotor at 35,000 rpm for 18 h at 20°C. Virion fractions were banded twice in CsCl before use. Final virion preparations were assayed by electron microscopy and SDS-PAGE for purity.
Electron microscopy.
For electron microscopy analysis, purified BPV-1 virions (100 ng) were applied to glow-discharged, carbon-coated, 400-mesh grids and stained with 2% uranyl acetate. The images were photographed with a Philips CM 10 electron microscope at a nominal magnification of 339,000 or 373,000 (45).
Nonreducing denaturing gel analysis.
The pattern of disulfide bonds in the PsV capsids was examined as previously described (46). HPV-16 pseudovirion preparations were alkylated using N-ethyl maleimide (NEM; Sigma). Alkylation was conducted by diluting roughly 1 μg of pseudovirion preparations into 10 mM NaPO4 (pH 6.5) and then adding 10 mM NEM for 10 min at room temperature. Samples were next mixed with an equal volume of nonreducing SDS-PAGE loading buffer (2% SDS, 100 mM Tris, pH 6.8, 10 mM NEM, 5 mM EDTA, 10% glycerol, and 0.01% bromophenol blue) and incubated at room temperature for 10 min and then at 65°C for 10 min prior to electrophoresis on a 7.5% polyacrylamide gel. Samples were then transferred to nitrocellulose membrane and stained using Ponceau S solution (Sigma).
Trypsin digestion of capsids.
The sensitivity of the PsVs to trypsin digestion was determined as previously described (48). Briefly, 10 μl of PsVs (500 ng) was incubated with 10 μl of 0.05% trypsin at 37°C for either 30 min or 3 h. Following the digestion period, 4 μl of the PsV mixture was processed for L1 detection, and 16 μl was processed for L2 detection. Samples were resolved on a 10% SDS-PAGE gel followed by Western blotting; L1 was detected with CamVir-1 and L2 with mouse anti-16 L2 (16.D4 64-81) antibody.
PsV internalization assay.
HeLa cells were grown overnight in six-well dishes at a density of 2.5 × 105 cells/well. Cells were infected with 900 ng/well PsVs (based on L1 protein quantitation). After 1 h at 4°C to allow PsV attachment, cells were immediately lysed in SDS sample buffer to monitor total input virus. A second set was treated with trypsin for 15 min and then lysed in SDS sample buffer to detect the amount of virus internalized during the first hour of incubation. The third set of cells was incubated at 37°C for 24 h and then treated with trypsin for 15 min to remove any noninternalized virus. The cells were extracted in SDS sample buffer, and the levels of L1 and L2 were detected by Western blotting. Alpha-actinin was used as the loading control.
Intracellular processing of PsVs.
PsV intracellular processing was examined as previously described (60). Briefly, HaCaT cells were seeded in six-well plates at 4 × 105 cells/well. After 24 h, cells were exposed to 900 ng/well wild-type and T62A PsVs (based on L1 protein) and incubated for a further 24 h at 37°C. The cells were then removed with 0.25% trypsin, washed twice with phosphate-buffered saline (PBS), and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer containing protease inhibitors. Cellular debris was removed by centrifugation. Clarified lysates were incubated overnight with mouse anti-HPV-16 L1 (H16.V5) antibody and then incubated with protein A-conjugated Sepharose beads for 1 h at room temperature. Immunoprecipitated complexes were collected by centrifugation and washed three times in E1A buffer (25 mM HEPES, pH 7.0, 0.1% NP-40, 150 mM NaCl). The remaining complexes were resolved by 10% SDS-PAGE and analyzed by Western blotting. HPV-16 L1 was detected with the CamVir-1 antibody. For determination of L1-associated L2 protein, mouse anti-HPV-16 L2 (16.D4 64-81) was used. The acidification inhibitor NH4Cl was included as a positive control for L2 retention and was added to a final concentration of 20 mM prior to the infection.
Immunofluorescence.
Papillomavirus binding to the cell surface was analyzed by immunofluorescence. HeLa cells were seeded on coverslips at 1 × 105 cells/well; after 24 h cells were exposed to 500 vge/cell wild-type and T62A HPV-16 PsVs for 1 h at 4°C, washed, and then incubated for a further 24 h at 37°C. The cells were then washed, fixed, and analyzed by quantitative immunofluorescence of the nonpermeabilized cells using anti-L1 antibody (CamVir-1), followed by rhodamine-conjugated rabbit anti-mouse secondary antibody. Samples were visualized using a Zeiss Axiovert 100 M microscope attached to an LSM 510 confocal unit. For quantification, the relative number of total L1-positive cells was determined, based on the total number of cells in each assay. At least 100 cells were analyzed in each experiment.
To determine whether there were defects in PsV capsid disassembly, quantitative immunofluorescence analysis was performed. HeLa cells were seeded on coverslips at 1 × 105 cells/well; after 24 h, cells were infected with wild-type and T62A HPV-16 PsVs at 500 vge/cell. At 7 h postinfection the cells were fixed in 3.7% paraformaldehyde for 15 min and then stained for total L1 using mouse anti-L1 antibody (CamVir-1) or the uncoating-specific anti-L1 antibody 33L1-7, followed by rhodamine-conjugated rabbit anti-mouse secondary antibody. Samples were visualized using a Zeiss Axiovert 100 M microscope attached to an LSM 510 confocal unit. For quantification, the relative numbers of L1-positive cells and 33L1-7-positive cells were determined based on the total number of cells in each group. At least 100 cells were analyzed in each experiment.
Statistics.
All experiments were performed at least three times. Statistical significance was calculated using GraphPad Prism, version 6, software and an unpaired t test.
ACKNOWLEDGMENTS
We are very grateful to Martin Müller for kindly providing the mouse anti-16 L2 (16.D4 64-81) antibody, Martin Sapp for providing mouse 33L1-7 antibody, and Neil Christensen for the anti-L1 neutralizing antibody (H16.V5). We are also grateful to Miranda Thomas for valuable comments on the manuscript.
J.B. gratefully acknowledges support from the Umberto Veronesi Foundation (Post-Doctoral Fellowship, year 2018). This work was supported in part by a grant from the Associazione Italiana per la Ricerca sul Cancro (to L.B.), by a Collaborative Research Programme (CRP) grant from the International Centre for Genetic Engineering and Biotechnology (CRP/SLO13-04RG to M.B.M.), and by a grant from the National Cancer Institute (R01CA37667 to R.L.G.).
REFERENCES
- 1.zur Hausen H. 1996. Papillomavirus infections–a major cause of human cancers. Biochim Biophys Acta 1288:F55–F78. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 3.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V, WHO International Agency for Research on Cancer Monograph Working Group. 2009. A review of human carcinogens–part B: biological agents. Lancet Oncol 10:321–322. doi: 10.1016/S1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 4.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30:F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 5.Ganti K, Broniarczyk J, Manoubi W, Massimi P, Mittal S, Pim D, Szalmas A, Thatte J, Thomas M, Tomaic V, Banks L. 2015. The human papillomavirus E6 PDZ binding motif: from life cycle to malignancy. Viruses 7:3530–3551. doi: 10.3390/v7072785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modis Y, Trus BL, Harrison SC. 2002. Atomic model of the papillomavirus capsid. EMBO J 21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, Trus BL. 2008. Arrangement of L2 within the papillomavirus capsid. J Virol 82:5190–5197. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day PM, Roden RB, Lowy DR, Schiller JT. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol 72:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day PM, Baker CC, Lowy DR, Schiller JT. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc Natl Acad Sci U S A 101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joyce JG, Tung JS, Przysiecki CT, Cook JC, Lehman ED, Sands JA, Jansen KU, Keller PM. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J Biol Chem 274:5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- 11.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang R, Day PM, Yutzy WH IV, Lin KY, Hung CF, Roden RB. 2003. Cell surface-binding motifs of L2 that facilitate papillomavirus infection. J Virol 77:3531–3541. doi: 10.1128/JVI.77.6.3531-3541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day PM, Lowy DR, Schiller JT. 2008. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J Virol 82:12565–12568. doi: 10.1128/JVI.01631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JL, Campos SK, Wandinger-Ness A, Ozbun MA. 2008. Caveolin-1-dependent infectious entry of human papillomavirus type 31 in human keratinocytes proceeds to the endosomal pathway for pH-dependent uncoating. J Virol 82:9505–9512. doi: 10.1128/JVI.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabydeen SA, Meneses PI. 2009. The role of NH4Cl and cysteine proteases in human papillomavirus type 16 infection. Virol J 6:109. doi: 10.1186/1743-422X-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bienkowska-Haba M, Williams C, Kim SM, Garcea RL, Sapp M. 2012. Cyclophilins facilitate dissociation of the human papillomavirus type 16 capsid protein L1 from the L2/DNA complex following virus entry. J Virol 86:9875–9887. doi: 10.1128/JVI.00980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bronnimann MP, Chapman JA, Park CK, Campos SK. 2013. A transmembrane domain and GxxxG motifs within L2 are essential for papillomavirus infection. J Virol 87:464–473. doi: 10.1128/JVI.01539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiGiuseppe S, Keiffer TR, Bienkowska-Haba M, Luszczek W, Guion LGM, Müller M, Sapp M. 2015. Topography of the human papillomavirus minor capsid protein L2 during vesicular trafficking of infectious entry. J Virol 89:10442–10452. doi: 10.1128/JVI.01588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, Monteiro da Silva G, Deatherage C, Burd C, DiMaio D. 2018. Cell-penetrating peptide mediates intracellular membrane passage of human papillomavirus L2 protein to trigger retrograde trafficking. Cell 174:1465–1476.e13. doi: 10.1016/j.cell.2018.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. 2013. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol 87:3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Kazakov T, Popa A, DiMaio D. 2014. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires gamma-secretase activity. mBio 5:e01777. doi: 10.1128/mBio.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. 2009. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog 5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kuhbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A, Schelhaas M. 2014. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog 10:e1004162. doi: 10.1371/journal.ppat.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broniarczyk J, Massimi P, Bergant M, Banks L. 2015. Human papillomavirus infectious entry and trafficking is a rapid process. J Virol 89:8727–8732. doi: 10.1128/JVI.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aksoy P, Gottschalk EY, Meneses PI. 2017. HPV entry into cells. Mutat Res Rev Mutat Res 772:13–22. doi: 10.1016/j.mrrev.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campos SK. 2017. Subcellular trafficking of the papillomavirus genome during initial infection: the remarkable abilities of minor capsid protein L2. Viruses 9:E370. doi: 10.3390/v9120370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddiqa A, Broniarczyk J, Banks L. 2018. Papillomaviruses and endocytic trafficking. Int J Mol Sci 19:E2619. doi: 10.3390/ijms19092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H-S, Buck CB, Lambert PF. 2010. Inhibition of gamma secretase blocks HPV infection. Virology 407:391–396. doi: 10.1016/j.virol.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karanam B, Peng S, Li T, Buck C, Day PM, Roden R. 2010. Papillomavirus infection requires γ secretase. J Virol 84:10661–10670. doi: 10.1128/JVI.01081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue T, Zhang P, Zhang W, Goodner-Bingham K, Dupzyk A, DiMaio D, Tsai B. 2018. γ-Secretase promotes membrane insertion of the human papillomavirus L2 capsid protein during virus infection. J Cell Biol 217:3545–3559. doi: 10.1083/jcb.201804171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broniarczyk J, Bergant M, Gozdzicka-Jozefiak A, Banks L. 2014. Human papillomavirus infection requires the TSG101 component of the ESCRT machinery. Virology 460–461:83–90. doi: 10.1016/j.virol.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Gräßel L, Fast LA, Scheffer KD, Boukhallouk F, Spoden GA, Tenzer S, Boller K, Bago R, Rajesh S, Overduin M, Berditchevski F, Florin L. 2016. The CD63-syntenin-1 complex controls post-endocytic trafficking of oncogenic human papillomaviruses. Sci Rep 6:32337–32337. doi: 10.1038/srep32337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broniarczyk J, Pim D, Massimi P, Bergant M, Goździcka-Józefiak A, Crump C, Banks L. 2017. The VPS4 component of the ESCRT machinery plays an essential role in HPV infectious entry and capsid disassembly. Sci Rep 7:45159. doi: 10.1038/srep45159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergant Marusic M, Ozbun MA, Campos SK, Myers MP, Banks L. 2012. Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17. Traffic 13:455–467. doi: 10.1111/j.1600-0854.2011.01320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pim D, Broniarczyk J, Bergant M, Playford MP, Banks L. 2015. A novel PDZ domain interaction mediates the binding between human papillomavirus 16 L2 and sorting nexin 27 and modulates virion trafficking. J Virol 89:10145–10155. doi: 10.1128/JVI.01499-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie M-A, Poffenberger AC, Nelson CDS, Atwood WJ, DiMaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. 2015. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog 11:e1004699. doi: 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nagele H, Morris LL, Singla A, Overlee BL, Heesom KJ, Sessions R, Banks L, Collins BM, Berger I, Billadeau DD, Burstein E, Cullen PJ. 2017. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol 19:1214–1225. doi: 10.1038/ncb3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keating JA, Striker R. 2012. Phosphorylation events during viral infections provide potential therapeutic targets. Rev Med Virol 22:166–181. doi: 10.1002/rmv.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoover H, Cheng Kao C. 2016. Phosphorylation of the viral coat protein regulates RNA virus infection. Virus Adapt Treat 8:13–20. doi: 10.2147/VAAT.S118440. [DOI] [Google Scholar]
- 41.Xi SZ, Banks LM. 1991. Baculovirus expression of the human papillomavirus type 16 capsid proteins: detection of L1-L2 protein complexes. J Gen Virol 72:2981–2988. doi: 10.1099/0022-1317-72-12-2981. [DOI] [PubMed] [Google Scholar]
- 42.Marusic MB, Mencin N, Licen M, Banks L, Grm HS. 2010. Modification of human papillomavirus minor capsid protein L2 by sumoylation. J Virol 84:11585–11589. doi: 10.1128/JVI.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belnap DM, Olson NH, Cladel NM, Newcomb WW, Brown JC, Kreider JW, Christensen ND, Baker TS. 1996. Conserved features in papillomavirus and polyomavirus capsids. J Mol Biol 259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buck CB, Thompson CD, Pang Y-Y, Lowy DR, Schiller JT. 2005. Maturation of papillomavirus capsids. J Virol 79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Beard P, Estes PA, Lyon MK, Garcea RL. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol 72:2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardone G, Moyer AL, Cheng N, Thompson CD, Dvoretzky I, Lowy DR, Schiller JT, Steven AC, Buck CB, Trus BL. 2014. Maturation of the human papillomavirus 16 capsid. mBio 5:e01104. doi: 10.1128/mBio.01104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerqueira C, Thompson CD, Day PM, Pang Y-Y, Lowy DR, Schiller JT. 2017. Efficient production of papillomavirus gene delivery vectors in defined in vitro reactions. Mol Ther Methods Clin Dev 5:165–179. doi: 10.1016/j.omtm.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day PM, Thompson CD, Pang YY, Lowy DR, Schiller JT. 2015. Involvement of nucleophosmin (NPM1/B23) in assembly of infectious HPV16 capsids. Papillomavirus Res 1:74–89. doi: 10.1016/j.pvr.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sapp M, Kraus U, Volpers C, Snijders PJF, Walboomers JMM, Streeck RE. 1994. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J Gen Virol 75:3375–3383. doi: 10.1099/0022-1317-75-12-3375. [DOI] [PubMed] [Google Scholar]
- 50.Ratka M, Lackmann M, Ueckermann C, Karlins U, Koch G. 1989. Poliovirus-associated protein kinase: destabilization of the virus capsid and stimulation of the phosphorylation reaction by Zn2+. J Virol 63:3954–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wacharapornin P, Lauhakirti D, Auewarakul P. 2007. The effect of capsid mutations on HIV-1 uncoating. Virology 358:48–54. doi: 10.1016/j.virol.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Kang H, Yu J, Jung G. 2008. Phosphorylation of hepatitis B virus core C-terminally truncated protein (Cp149) by PKC increases capsid assembly and stability. Biochem J 416:47–54. doi: 10.1042/BJ20080724. [DOI] [PubMed] [Google Scholar]
- 53.Asenjo A, González-Armas JC, Villanueva N. 2008. Phosphorylation of human respiratory syncytial virus P protein at serine 54 regulates viral uncoating. Virology 380:26–33. doi: 10.1016/j.virol.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 54.Biryukov J, Meyers C. 2015. Papillomavirus infectious pathways: a comparison of systems. Viruses 7:4303–4325. doi: 10.3390/v7082823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Broniarczyk J, Ring N, Massimi P, Giacca M, Banks L. 2018. HPV-16 virions can remain infectious for 2 weeks on senescent cells but require cell cycle re-activation to allow virus entry. Sci Rep 8:811. doi: 10.1038/s41598-017-18809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allen-Hoffmann BL, Schlosser SJ, Ivarie CAR, Meisner LF, O’Connor SL, Sattler CA. 2000. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J Investig Dermatol 114:444–455. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 57.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 119:445–462. doi: 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- 58.Rappsilber J, Friesen WJ, Paushkin S, Dreyfuss G, Mann M. 2003. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal Chem 75:3107–3114. doi: 10.1021/ac026283q. [DOI] [PubMed] [Google Scholar]
- 59.Fenyo D, Beavis RC. 2003. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem 75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 60.Day PM, Thompson CD, Lowy DR, Schiller JT. 2017. Interferon gamma prevents infectious entry of human papillomavirus 16 via an L2-dependent mechanism. J Virol 91:e00168-17. doi: 10.1128/JVI.00168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]