The complex and continuous antigenic evolution of equine influenza viruses (EIVs) remains a major hurdle for vaccine development and the design of effective immunization programs. The present study provides a comprehensive analysis showing the EIV evolutionary dynamics, including the spread and circulation within the Asian continent and its relationship to global EIV populations over a 10-year period. Moreover, we provide a better understanding of EIV molecular evolution in Asian countries and its consequences on the antigenicity. The study underscores the association between the global horse movement and the circulation of EIV in this region. Understanding EIV evolution is imperative in order to mitigate the risk of outbreaks affecting the horse industry and to help with the selection of the viral strains to be included in the formulation of future vaccines.
KEYWORDS: Asia, equine influenza, evolution, H3N8, H7N7, influenza, signature pattern, vaccine
ABSTRACT
Equine influenza virus (EIV) causes severe acute respiratory disease in horses. Currently, the strains belonging to the H3N8 subtype are divided into two clades, Florida clade 1 (FC1) and Florida clade 2 (FC2), which emerged in 2002. Both FC1 and FC2 clades were reported in Asian and Middle East countries in the last decade. In this study, we described the evolution, epidemiology, and molecular characteristic of the EIV lineages, with focus on those detected in Asia from 2007 to 2017. The full genome phylogeny showed that FC1 and FC2 constituted separate and divergent lineages, without evidence of reassortment between the clades. While FC1 evolved as a single lineage, FC2 showed a divergent event around 2004 giving rise to two well-supported and coexisting sublineages, European and Asian. Furthermore, two different spread patterns of EIV in Asian countries were identified. The FC1 outbreaks were caused by independent introductions of EIV from the Americas, with the Asian isolates genetically similar to the contemporary American lineages. On the other hand, the FC2 strains detected in Asian mainland countries conformed to an autochthonous monophyletic group with a common ancestor dated in 2006 and showed evidence of an endemic circulation in a local host. Characteristic aminoacidic signature patterns were detected in all viral proteins in both Asian-FC1 and FC2 populations. Several changes were located at the top of the HA1 protein, inside or near antigenic sites. Further studies are needed to assess the potential impact of these antigenic changes in vaccination programs.
IMPORTANCE The complex and continuous antigenic evolution of equine influenza viruses (EIVs) remains a major hurdle for vaccine development and the design of effective immunization programs. The present study provides a comprehensive analysis showing the EIV evolutionary dynamics, including the spread and circulation within the Asian continent and its relationship to global EIV populations over a 10-year period. Moreover, we provide a better understanding of EIV molecular evolution in Asian countries and its consequences on the antigenicity. The study underscores the association between the global horse movement and the circulation of EIV in this region. Understanding EIV evolution is imperative in order to mitigate the risk of outbreaks affecting the horse industry and to help with the selection of the viral strains to be included in the formulation of future vaccines.
INTRODUCTION
Equine influenza A virus (EIV) infection causes an acute disease with variable morbidity and low mortality in horses and odd-toed ungulates. Equine influenza (EI) is one of the most economically important respiratory diseases of horses in most parts of the world, due to its highly contagious nature and rapid spread among susceptible hosts (1).
The influenza A virus has a genome composed of eight segments of negative-sense single-strand RNA and is subtyped according to the combination of two surface glycoproteins: hemagglutinin (HA) and neuraminidase (NA) (2). To date, 18 HA and 11 NA subtypes have been identified in different susceptible hosts (3). In particular, two EIV subtypes have been reported in equines: H7N7 and H3N8 (1). Whereas the last confirmed report of H7N7 in horses was in 1979 (4), H3N8 viruses have been detected worldwide since 1963 (5). The phylogenetic analysis of the HA gene revealed that equine H3N8 viruses have diverged into distinct evolutionary lineages. The strains that circulate today belong to two clades that emerged in the Americas around 2002, designated Florida clade 1 (FC1) and Florida clade 2 (FC2) (6). FC1 viruses are endemic in North America and FC2 viruses are predominant in Europe. However, both subtypes have also caused outbreaks in Europe, South Africa, South America, and Asia (7).
EIVs of FC1 and FC2 clades were reported in Asian and Middle East countries in recent times. In relation to FC1, several EI outbreaks were described in Japan in local and imported horses from 2007 to 2017 (5, 8, 9). Moreover, FC1 strains closely related to those detected in Japan in 2007 were detected in South Korea in 2011 (10). In 2012, an EI outbreak was reported in the quarantine facilities of Dubai, and the isolated viruses were genetically related to an FC1 outbreak reported in South America in the same year (11, 12). In 2015, FC1 strains were also detected in Malaysia.
FC2 strains have been detected in Asian countries since 2007. Several EI outbreaks occurred in China from 2007 to 2017, and all were caused by strains belonging to the FC2 lineage (13, 14). Besides, EIV was also reported in India (2008 and 2009) and Mongolia (2008, 2012, and 2016). Phylogenetic analyses showed that all Indian and Mongolian strains were similar to the FC2 viruses circulating in China between 2007 and 2008 (9, 15–17). Finally, EIV was also detected in Kazakhstan during 2007 and 2012, and the isolates were closely related to Mongolian and Chinese FC2 strains (18).
Most of the evolutionary and molecular studies in EIV have focused on the HA and NA gene segments, because they code for the proteins traditionally used for typing (2). These genes undergo antigenic drift, i.e., nonsynonymous point mutations are fixed by antibody-mediated selection and can cause significant changes in the antigenicity of the virus. Even when the antigenic drift in HA and NA is the most important mechanism for influenza virus evolution and drives the divergence of lineages, the conservation of viral fitness requires an optimal interaction between the proteins encoded by all the genomic segments (19). However, there are only a few studies in EIV that have analyzed the other genomic segments (6, 10, 12, 20, 21).
The import of competition horses from regions of endemicity (mainly Europe and North America) has resulted in several major outbreaks in susceptible populations in Asia (5, 8–10, 13, 14, 16–18). However, in addition to the sports industry, the breeding of working equines constitutes an important agricultural resource in Central Asia. The large population of horses in this area provides a favorable ecological context for sustained transmission and evolution of EIV in equine hosts (22). Moreover, it has been described that EIV isolated in Central Asia in the last years constitutes a local lineage with a specific genetic signature (13, 18).
In addition to the genealogical reconstruction, the viral genomic data contain valuable information regarding the spatiotemporal spread of virus that can be extracted using phylodynamic approaches (23). The integration of phylogenetical and geographical available data might help to depict the evolutionary history, the spread pattern, and the epidemiological links of the EIV strains in the Asian continent. Moreover, if the HA of Asian lineages has diverged significantly from that of reference strains, the evaluation of the amino acid changes, in particular, those located in or near antigenic sites, allows assessing its potential impact in vaccination programs and the need for vaccine update (24).
The aim of this study was to describe the evolution, epidemiology, and molecular characteristics of the EIV lineages, with focus on those detected in Asian countries from 2007 to 2017. To this end, we performed phylodynamic, genomic, and structural analyses of H3N8 EIV complete genome sequences available in public databases.
RESULTS
EI viruses.
EIV was successfully isolated from three samples from the 2015 Chinese outbreak. The complete genome sequence was obtained from strain A/equine/Urumqi/1/2015 (accession numbers MK215812 to MK215819), and the HA gene sequence was obtained for A/equine/ChengDu/1/2015 and A/equine/WuHan/1/2015 strains (accession numbers MK215821 and MK215822).
Data set quality analyses.
The estimated evolutionary models for data set 1 (DS1) were: PB2 (TVM+I+G), PB1 (TPM1uf+G), PA (TPM1uf+G), HA (HKY+G), NP (HKY+I), NA (TPM1uf+G), MP (HKY+G), and NS (TPM1uf + G). For data set 2 (DS2), the evolutionary model was HKY+G. The phylogenetic network and the pairwise homoplasy index (PHI) test indicated no significant evidence of intrasubtype recombination events in all data sets (P values: 1-PB2, 0.8592; 2-PB1, 0.8736; 3-PA, 0.0815; 4-HA, 0.1078; 5-NP, 0.4808; 6-NA, 0.6498; 7-MP, 0.0567; 8-NS, 0.6407; HA1 DS2, 0.9994). The likelihood mapping showed that all genomic segments contained adequate phylogenetic information. In addition, there was a strong correlation between the genetic distance of each sequence to the root of the phylogeny and the date of strain sampling for all genomic segments (data not shown).
Bayesian phylogeny and global population dynamic of H3N8 EIV.
The phylodynamic analysis of the complete genome, that is, where all genomic fragments were concatenated and analyzed together, had a deficient performance. Multiple combinations of models and priors were tested, but the convergence of relevant parameters was not achieved. Given the low confidence of estimations, the results of these analyses are not presented.
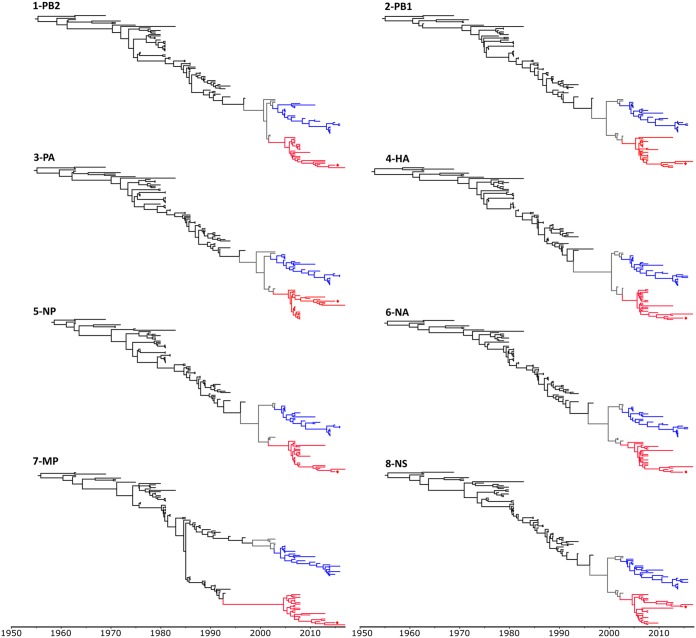
On the other hand, genomic segments were analyzed individually. The topologies of all individual gene trees were similar, except for the MP gene (Fig. 1). Consistent topologies were obtained by other phylogenetical methods (neighbor joining and maximum likelihood) (data not shown). Asian strains from 2007 to 2017 were distributed in both FC1 and FC2 and showed a geographical structure. All Japanese, Korean, Malaysian, and Arabic strains belong to FC1, whereas the isolates from mainland countries (China, India, Kazakhstan, and Mongolia) belong to FC2. Moreover, all Asian FC2 strains were monophyletic.
FIG 1.
Time-scaled phylogeny of EIV H3N8 genes. The analyses were performed under the Bayesian skyline plot (BSP) demographic model and a relaxed clock model (UCLN) calibrated with terminal nodes (time of sampling, in years). FC1 and FC2 lineages are colored in blue and red, respectively. Strain A/equine/Urumqi/1/2015 sequenced in this study is indicated with a diamond.
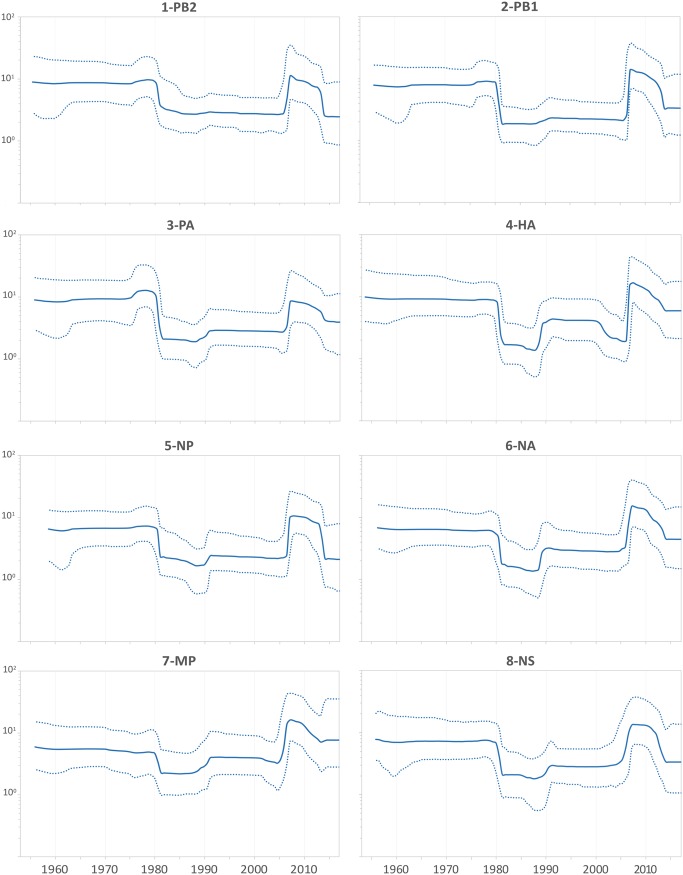
The demographic history of an EIV population was inferred through the effective number of infections through time (Neτ). All genomic segments showed similar skyline profiles (Fig. 2). Our analysis reproduced the behavior described by Murcia et al. (6) when analyzing the sequences that were available until 2008. The recent demographic history showed an exponential increase in viral diversity from 2005 to 2008. This high diversity level showed a slight decrease over the next 5 years. Around 2015, viral diversity markedly decreased, and it has remained at low levels since then.
FIG 2.
Population dynamics of EIV H3N8 genes. The effective numbers of infections through time (Neτ) obtained from the analysis of EIV H3N8 individual genes, under the UCLN-BSP models. Median values are denoted by the solid lines, while dotted lines denote the 95% highest posterior density (HPD) values.
Phylogeographic reconstruction of FC1 and FC2 spread based on HA gene.
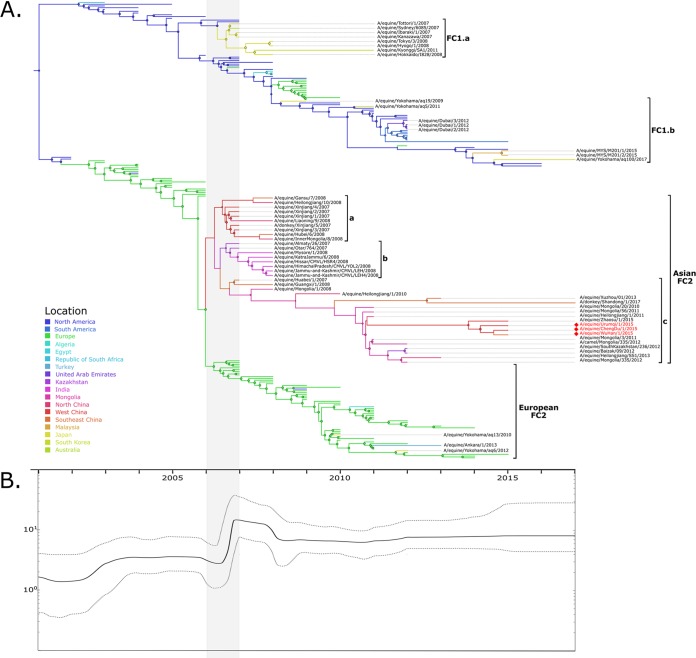
To elucidate the global phylodynamics of FC1 and FC2, a more informative a data set of the HA1 gene (DS2) including sequences isolated from a larger number of countries was compiled. The ancestral location reconstruction indicated that the common ancestor and basal lineages of both clades were in North America, with a high posterior probability. After their divergence, FC1 evolved mainly in North America, whereas FC2 established in Europe in 2002 (Fig. 3).
FIG 3.
Phylodinamics of FC1 and FC2. (A) Time-scaled phylogeny from the discrete phylogeographic analysis. Branches are colored according to the most probable location of the parental node of each lineage (color codes are shown at bottom left). Circles on nodes are sized according to the location posterior probability. The major Asian lineages discussed in the text are shown with brackets. (B) Temporal fluctuation of the effective numbers of infections through time (Neτ,; y axis). Solid line, median Neτ values; dotted lines, 95% HPD.
All nodes in the central trunk of the FC1 phylogeny represented ancestors located in North America with high posterior probability. Lineages from North America had caused outbreaks in other geographical regions. Two groups containing Asian sequences can be highlighted in FC1 (“a” and “b”). The FC1a group included strains from an outbreak detected in 2007 and 2008 in Japan, which affected Australia also; this lineage was detected for the last time in South Korea in 2011. The FC1b group included Asian strains that circulated from 2009 to 2017 in Japan, United Arab Emirates, and Malaysia. These lineages derived directly from ancestral nodes located in North America, although all of them represent independent introductions to Asian countries. It is worth mentioning that the strains detected in 2012 in United Arab Emirates derived from a South American lineage and not directly from North America (Fig. 4).
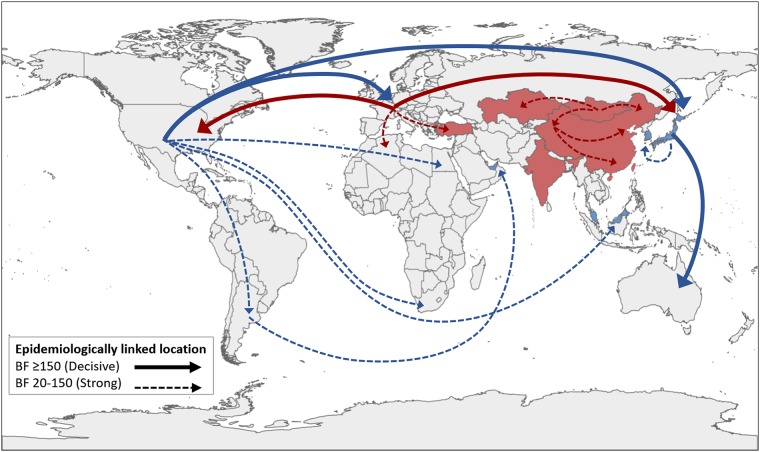
FIG 4.
Epidemiologically linked locations. The FC1 circulation is plotted in blue and that for FC2 in red. The figure shows all the nonzero rates with significant epidemiological links (Bayes factor [BF] ≥ 20). Asian countries with HA1 sequences available and included in the analysis are highlighted in blue and red. The map data were obtained from the National Geomatics Center of China (NGCC) and compiled by using QGIS 3.4 (https://www.qgis.org).
The FC2 basal phylogeny shows short branches that arise from the central trunk, characteristic of a succession of seasonal variants. However, in 2006, they split into two defined groups: the European and Asian sublineages, which evolved separately and circulated in different geographical regions until today. The strains from Japan (Yokohama/aq13/2010 and Yokohama/aq6/2012) and Turkey (Ankara/1/2013) belonged to the European sublineage. On the other hand, all strains from mainland countries (China, India, Mongolia, and Kazakhstan) belonged to a single monophyletic group. This Asian sublineage had a common ancestor located in West China (posterior probability [pp] = 0.42), also in Mongolia with considerable probability (pp = 0.35). At least three clusters can be defined in Asian FC2. The oldest clusters are related to outbreaks that occurred between 2007 and 2009 in China and Kazakhstan (Asian FC2a) and in India (Asian FC2b). After that, a new dispersion from an ancestor located in Mongolia occurred. All the strains that have been detected in recent years in mainland countries belong to this lineage (Asian FC2c). Furthermore, several interspecies transmissions were caused by Asian FC2 strains: horse-to-donkey transmissions were detected in China in 2007 (25) and 2017, and the first horse-to-camel interspecies transmission was detected in Mongolia in 2012 (26). These non-horse strains were genetically related to horse strains isolated in the same area in previous years (Fig. 3).
The main epidemiological linkages between geographic locations were identified using a Bayes factor (BF) test. Among the most significant rates involving the introduction and/or circulation of Florida lineages in Asian countries, those in Japan linked to both North America (related to the introduction of FC1 lineages) and Europe (involving European FC2 sublineage). Other significant links involving migrations of FC1 lineages included those from North America to Malaysia, from South America to the United Arab Emirates, and from Japan to South Korea. In relation to the Asian FC2 sublineage dispersion, that in Mongolia was related to those in all neighbouring locations (Kazakhstan and West and North China), which suggests that it plays a central role in the dispersion of EIV in mainland Asia. Significant links were also detected between internal regions of China, suggesting a west-to-east migratory flow (Fig. 4 and Table 1).
TABLE 1.
Significant epidemiologically linked locations
| From | To | Bayes factor |
|---|---|---|
| North America | Japan | 14161,84 |
| North America | Europe | 12872,92 |
| Europe | North America | 4034,60 |
| Japan | Australia | 465,95 |
| Europe | Japan | 246,26 |
| West China | Southeast China | 130,32 |
| Europe | Algeria | 121,62 |
| North America | Egypt | 114,26 |
| North America | South America | 95,08 |
| South America | UAE | 71,06 |
| Japan | South Korea | 66,23 |
| North America | South Africa | 62,12 |
| West China | North China | 52,13 |
| Mongolia | Kazakhstan | 38,30 |
| North America | Malaysia | 37,15 |
| Mongolia | North China | 32,43 |
| Mongolia | West China | 28,74 |
| Europe | Turkey | 26,51 |
Complete genome signature pattern.
The eight RNA segments of FC1 and FC2 strains were compared to assess specific amino acid residues that characterize the clades. The divergent evolution of FC1 and FC2 was reflected in a signature pattern throughout the eight genomic segments (Table 2). Interestingly, the HA segment showed seven mutations, four in addition to the three characteristic sites described previously (12).
TABLE 2.
Signature patterns observed in FC1 and FC2 EIV
| Gene | FC1 vs FC2a |
|---|---|
| PB2 | N79S, T105A, R251K, H300Q, I570M, R574K |
| PB1 | L94F, V119M, Q329R, D377E, E618D, R621K |
| PA | E64D, M86I, K158R/G, E237K, S259P, S321N, T354I, N409S, V465I, A476T, V505I, K626R, E684G |
| HA | L(-8)F, G7N, A79V, S159N, A372T, G379E, V527I |
| NP | M136I, I183V, I257T, M341L, T359A, N450S |
| NA | M8I, A9T, F12S, A35V, R40E, D42G, Y66H, T70A, S78P, V191I, V209I, D235N, N337S |
| MP | V80I, R95K, R208K |
| NS | S48I, V84I, G96D |
FC1 versus FC2: amino acidic residues in FC1 and FC2 in the respective positions. All segments were numbered starting in the first methionine of the protein with exception of HA, which was codified with the H3 numbering (Burke and Smith [54]).
HA signature pattern.
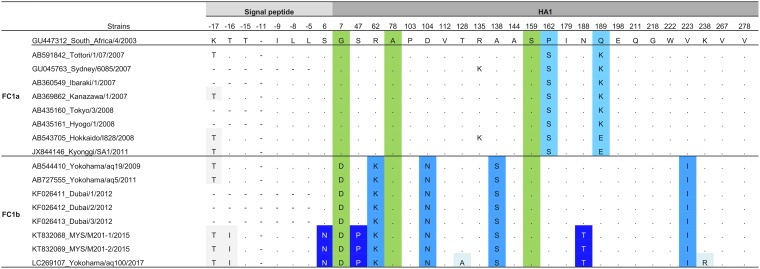
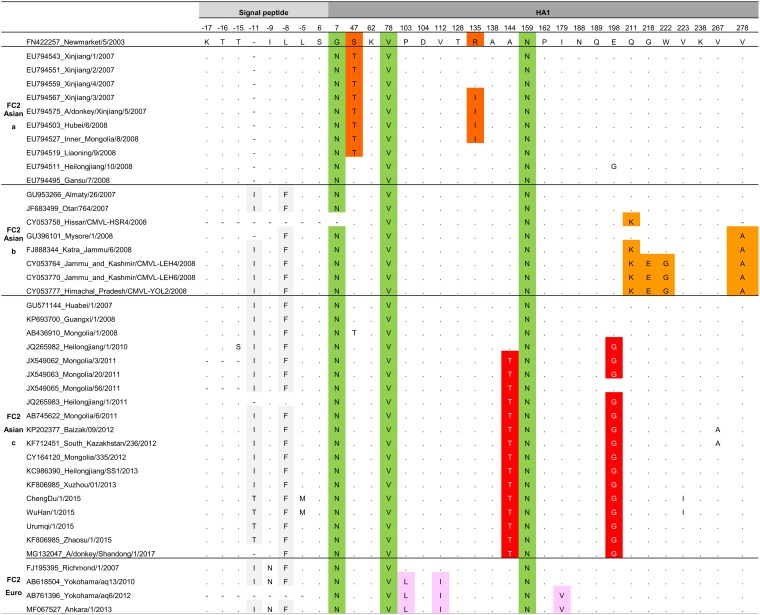
The Asian strains in the HA DS2 contrasted with the FC1 and FC2 EIV reference strains A/equine/SouthAfrica/4/2003 and A/equine/NewMarket/5/2003, respectively. First, all Asian strains showed the characteristic amino acids described in FC1 and FC2 strains worldwide. That is, the FC1 strains possess A78 and S159 and all FC2 strains possess N7, V78, and N159 (12) (Tables 3 and 4, green columns).
TABLE 3.
Signature pattern of FC1 Asian populationa
The colors show the amino acids that characterized the FC1 populations. In gray, the amino acid substitutions present in the signal peptide. In green, the amino acids that characterized the FC1 clade. In light blue, the amino acids that characterized the FC1a population. In medium blue, the amino acids that characterized the FC1b population. In dark blue, the amino acids that characterized the population that evolved from 2015 to 2017. In light blue shadow, the amino acid substitutions present in the 2017 strain.
TABLE 4.
Signature pattern of FC2 Asian populationa
The colors show the amino acids that characterized the FC2 populations. In gray, the amino acid substitutions present in the signal peptide. In green, the amino acids that characterized the FC2 clade. In dark orange, the amino acids that characterized the Asian FC2a population. In orange, the amino acids that characterized the Asian FC2b population. In red, the amino acids that characterized the Asian FC2c population. In pink, the amino acids that characterized the European FC2 population.
In concordance with the HA phylogenetic tree, two patterns were observed in Asian FC1 strains (clades FC1a and FC1b). The FC1a possessed two characteristic amino acid substitutions, P162S and Q189K, whereas FC1b showed four characteristic amino acid substitutions (R62K, D104N, A138S, and V223I). Unlike these mutations that were also reported elsewhere in the world, three additional substitutions (S6N, S47P, and N188T) were found in 2015 and 2017 Asian strains (Tables 3 and 4). Lastly, changes in the signal peptide were also identified; all FC1 strains showed one additional mutation, K−17T, in comparison with the prototype strain. Moreover, FC1b incorporated the T−16I substitution (Tables 3 and 4, gray columns).
On the contrary, the Asian FC2 lineage presented some specific substitutions that were not reported in FC2 worldwide strains. The Asian FC2 displayed three viral lineages (a, b, and c). The Asian FC2a showed the S47T amino acid substitution in all strains except Gansu/7/2008 and Heilongjian/10/2008 (Table 4, dark orange). FC2b possessed the substitutions Q211K and V278A, except Mysore/1/2008 that only had the last one (Table 4, orange). The Asian FC2c showed two amino acid changes: A144T and E198G (Table 4, red). Meanwhile, the European FC2 lineages detected in Asian countries from 2010 to 2013 showed the three characteristic amino acid mutations in positions P103L, V112I, and I179V that were described in European FC2 strains (Table 4, pink). Finally, Asian FC2b, FC2c, and European FC2 shared I−11 and L−8F substitutions in the signal peptide (Tables 3 and 4, gray columns).
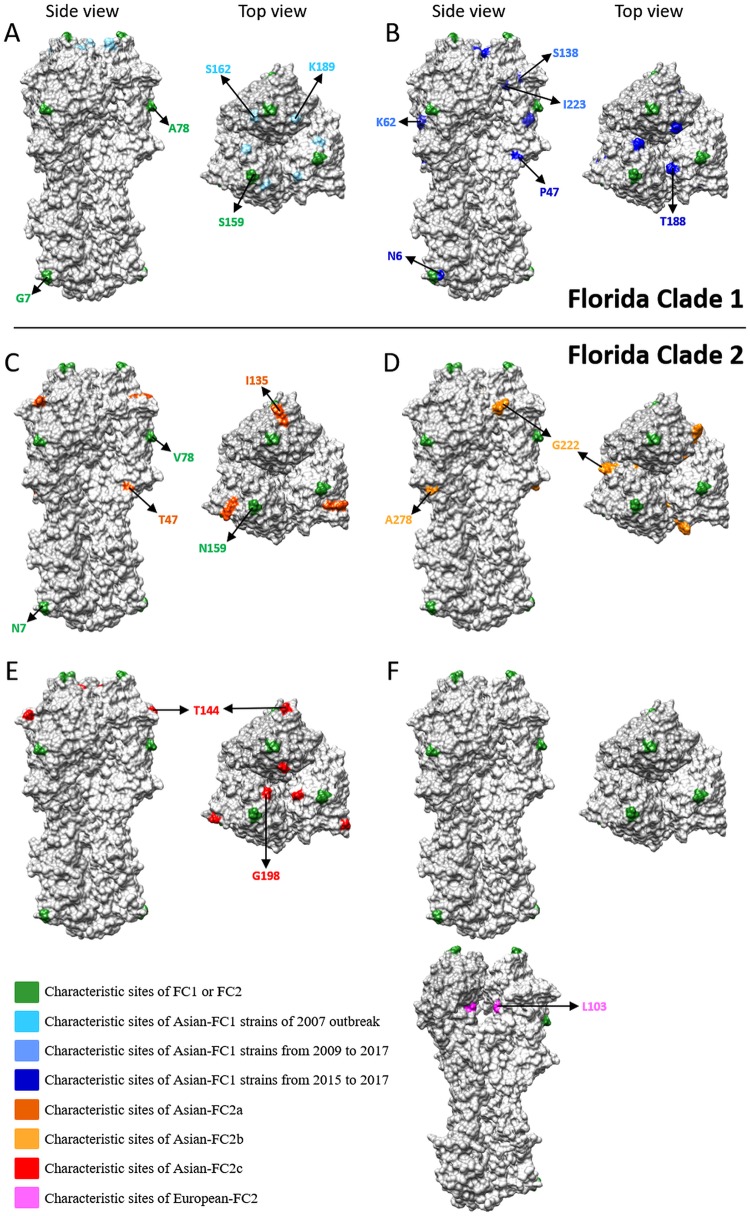
HA1 structural analysis.
Representative strains of the Asian FC1 and FC2 lineages were selected, and their characteristic changes were mapped. In the Asian FC2 sublineages, all the observed substitutions were at the surface of the trimer, whereas in the European FC2 sublineages, the characteristic substitutions were inside the trimer (Fig. 5).
FIG 5.
HA1 characteristic sites in the Asian FC1 and FC2 lineages. Representation of the modeled equine HA trimer representing the viral population described in both Asian FC1 and FC2. The amino acids labeled in green correspond to those characteristic of FC1 (G7, A78, and S159) or FC2 (N7, B78, and N159). (A) FC1a, the characteristic amino acid in light blue; (B) FC1b, the characteristic amino acid in medium blue and the latest amino acid acquisitions in blue; (C) Asian FC2a, the characteristic amino acid substitution in orange-red; (D) Asian-FC2b, the characteristic amino acid substitution in orange; (E) Asian FC2c, the characteristic amino acid substitution in red; (F) European FC2, the inner view of the HA trimer is also showed to highlight the characteristic amino acid substitution (in pink).
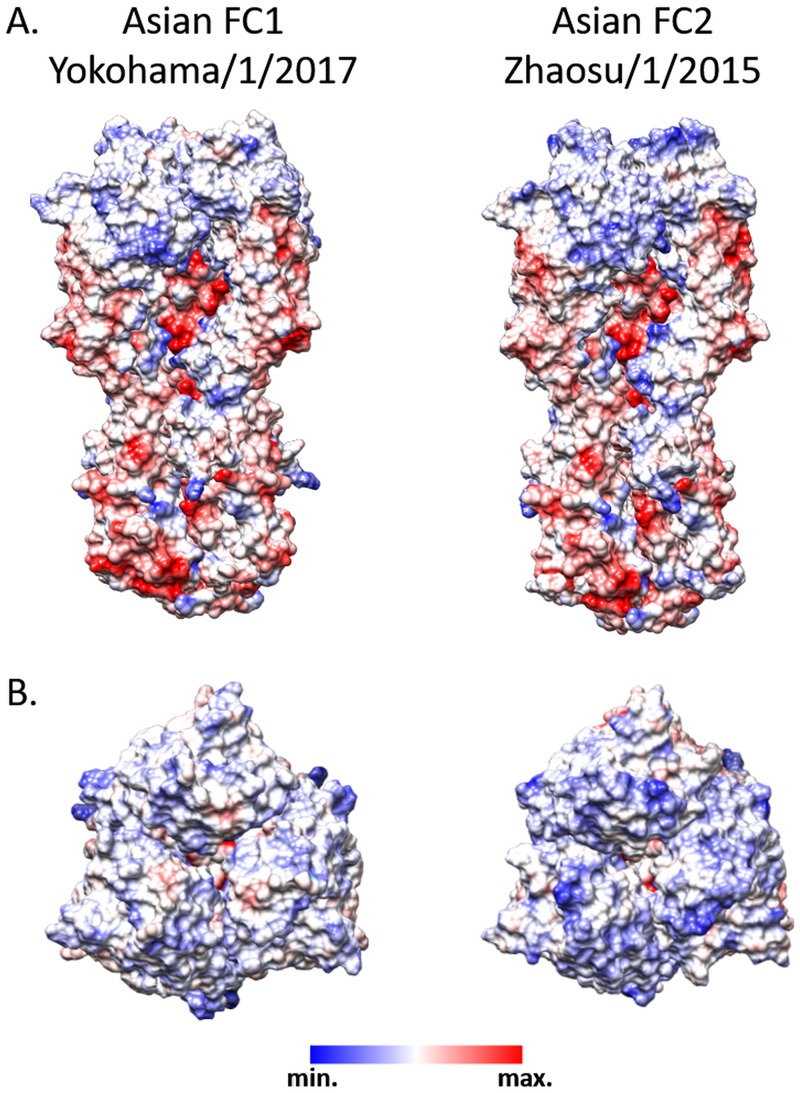
Moreover, the roles of these substitutions in the overall hydrophobicity and the electrostatic potential of the modeled structures were explored. The electrostatic potential in HA1 mostly differed at the top of the trimer: the FC1 strains showed a nonpolar surface, while the Asian FC2 strains showed a basic surface (Fig. 6).
FIG 6.
HA1 hydrophobicity and electrostatic potential. Representation of the modeled equine HA trimers (Asian FC1 and FC2) showing the electrostatic potential of the surfaces. Positive charges are indicated by blue, and negative by red. Scale, ±10 kcal/mol·e.
DISCUSSION
EIV outbreaks are reported every year worldwide, and since 2002, these outbreaks have been caused by strains that belong to Florida sublineage clade 1 and clade 2 (27). Although FC1 and FC2 are endemic in America and Europe, respectively, they have also caused several outbreaks in Asia from 2007 to 2017. In this work, we report the origin, spread, and molecular characteristics of these Asian EIV isolates. The FC1 outbreaks were caused by independent introductions from the American continent; these Asian isolates were genetically similar to the contemporary American lineages. Otherwise, the FC2 strains detected in mainland Asian countries conform to an autochthonous monophyletic group with a common ancestor dating back to 2006. This Asian FC2 sublineage showed evidence of a continual evolution in local hosts and showed several amino acid differences in HA in comparison with prototype strains.
Due to the segmented nature of the viral genome, reassortment is a major mechanism of genetic diversification in influenza viruses. The analysis of EIV H3N8 complete genomes showed that all segments had a similar genealogy since their emergence in equines in the 60s, except for the MP segment. Moreover, the recent phylogeny showed that FC1 and FC2 constituted separate and divergent groups in the eight genomic segments, without evidence of reassortment between clades. This behavior is consistent with that previously described by Murcia et al. (6) and suggests that reassortment has not played a significant role in the evolution of FC1 and FC2 after their emergence in 2002. On the other hand, the MP genealogy showed the Florida clade as a polyphyletic group with a common ancestor dating back to 1985. The MP gene in FC2 was genetically more related to the H3N8 strains that circulated in the late 80s and the beginning of the 90s than to those strains circulating at the time of emergence of the Florida clade. This pattern could be a consequence of an intrasubtype reassortment event. We hypothesize that the pre-FC2 lineage that circulated in early 2000 acquired the MP gene from an “ancient” H3N8 strain. This ancient strain could belong to the 1990s Eurasian lineage, which circulated in nature since then and evolved at a very low rate under a phenomenon of “frozen evolution” (28). Establishment of the origin of the EIV Florida clade was beyond the aims of this study. However, further research should be conducted to show whether the MP genes in the Florida clade have different origins and, moreover, their possible impact on the emergence and diversification of FC1 and FC2 lineages.
Nowadays, the international movement of horses for competitions, breeding, or sales is essential in the horse industry and is one of the most important factors in the spread of EIV (29). EIV H3N8 strains were responsible for outbreaks in Asia, and it is noteworthy that H7N7 sequences were not reported to public databases during the analyzed period (2007 to 2017), supporting the assumption that the last subtype could be extinct in equine populations in nature. Usually, the characterization of circulating strains is based on sequencing of the HA segment and, consequently, there are more HA sequences than complete genomes reported in public databases (4, 27). Therefore, the HA sequences of H3N8 FC1 and FC2 spanning 27 countries were analyzed to depict the FC1 and FC2 phylogeny with particular focus on the origin and phylogenetic relationships of Asian EIV strains. FC1 showed the characteristic stepped pattern of EIV and evolved as a single lineage (30). Conversely, FC2 showed a divergence event around 2004 giving rise to two well-supported sublineages, European and Asian, which coexist today. This divergence event was also reflected by the population dynamics over time. The skyline shows an expansion in 2006 and 2007, and it could be reflecting both: the increase of genetic diversity as a consequence of the FC2 spread into a new ecological niche in Asia and also the increase in the number of infections by the outbreaks reported around the world at that time (5, 8–10, 13, 16–18, 31). After that, the diversity decreased, reaching a baseline in 2009, suggesting that the global EIV population reaches an equilibrium level and returns to a more typical behavior of lineage succession within the successful clades (FC1, Asian FC2, and European FC2).
The spatiotemporal spread pattern and the epidemiological links obtained by phylogeographic methods allow us to know the global EIV dynamics and identify movements to, from, and within the Asian continent. Two different spread patterns of EIV in Asia were detected by the phylogeographic analysis of the HA protein. On one hand, all the Asian FC1 outbreaks were caused by strains introduced from the Americas. The directional epidemiological links from North America to Japan, Japan to Australia, North America to Malaysia, and Argentina to Dubai are well documented in epidemiological reports. In fact, all the FC1 detections in Asian countries were performed in the context of commercial events or horse competitions (5, 8, 10, 13, 16–18, 32). Similarly, the detection of European FC2 strains in Japan and Turkey could be related to their close economic relationships with European countries (9, 33). This global circulation pattern reflects the endemic evolution of EIV H3N8 in America and Europe and highlights the role of these locations as an important primary source of new strains and also the relevance of the international displacements of horses in the worldwide spread of the virus, mainly of the FC1 and European FC2 lineages (12, 34, 35).
Conversely, the continuous local detection and the monophyly of the Asian FC2 strains constitute strong evidence of the establishment of an endemic population. It seems that this lineage emerged in Central Asia around 2006, and at least three different clades have circulated in the region since then. Most of the lineages of the central trunk were located in Mongolia. In addition, Mongolia linked epidemiologically to all neighbouring locations (Kazakhstan and West and North China), suggesting that it could play a central role in the maintenance and dispersion of EIV in mainland Asia. The large population of horses in this area (more than 20 million) and the sanitary and ecological conditions provided an adequate context for the endemic maintenance of EIV. It might be possible that the low vaccination coverage (<10% of the horse population) and the absence of official campaign and control in this region might have favored the spread of EIV in the horse population (36). The massive and free horse movement across international borders for transport, cultural celebrations, and equestrian events has been associated with the occurrence of outbreaks in the area (37, 38). Besides, the report of frequent outbreaks and interspecies transmission events constituted additional evidence of the wide dispersion of EIV (25, 26). These factors point out an extensive and continual circulation of the Asian FC2 lineage and the need for control measures appropriated to local ecology.
The major evolutionary process in influenza virus is the antigenic drift of the HA protein. This process also causes fixation of mutations elsewhere in the viral genome by a hitchhiking effect and thereby contributes to the divergence among EIV lineages in all eight viral gene segments (19). The molecular changes that have arisen during the evolutionary history of FC lineages generated a clade-specific signature pattern, where each segment possesses several typical mutations. The FC1 and FC2 viral populations could be interpreted as two optimal combinations of the eight segments. The reassortment between these two genomic constellations could generate suboptimal combinations, due to incompatibility among proteins and segment mismatch, and then be negatively selected (19). This may be the reason, because reassortant strains between FC1 and FC2 have not been reported yet, despite their cocirculation in several geographic regions.
Vaccination is the most useful strategy for prophylaxis and control of EI. The continuous genetic evolution of the virus, mainly, the substitutions in the HA protein, could impact the vaccine efficacy and the natural immunity, and demands the genetic characterization of currently circulating EIVs for the selection of a candidate vaccine strain (39). The mutations described in FC1 and FC2 older strains were conserved through the lineage evolution and were present even in the current strains (12, 18). The FC1 populations that caused the Asian outbreak during 2007 and 2011 had their particular substitutions, which differed from the current FC1 lineage. The FC1 lineage that circulates today, including those detected in Asia, had several substitutions different from the vaccine strain South Africa/2003 (12). In addition to the residues inherited from ancestral lineages, the current Asian FC2 strains showed many differences in HA in comparison with the reference strains from 2003 (12). The three Asian FC2 lineages observed in the phylogeny have fixed different amino acid substitutions. In particular, the Asian FC2 and the European sublineages that are cocirculating at present showed a different substitution, which should be taken into account in the vaccine formulation (24).
In summary, in this study, we showed that there are two circulation patterns of EIV in Asia. The international movement of horses for competitions or sales remains an important factor in the spread of EIV. Moreover, there has been an endemic circulation of EIV in the Asian mainland since 2006. The autochthonous Asian FC2 sublineage had its own characteristic molecular pattern in the HA protein. Further studies are needed to assess if these lineages remain antigenically related to the strains that are used in current vaccine formulation. This study emphasizes the importance of continuous EIV surveillance in local horses in order to monitor the behavior of the Asian FC2 sublineages and to establish suitable control strategies.
MATERIALS AND METHODS
EIV detection and isolation.
In 2015, three cities of China (Urumqi, Chengdu, and Wuhan) reported EI disease outbreaks. Nasopharyngeal swabs in viral transport medium were collected from the infected animals, and the EIV was confirmed by reverse transcription-PCR (RT-PCR) against M and HA genes. Virus isolation was carried out from all RT-PCR-positive samples according to a protocol described elsewhere (13). RNA extraction, EIV gene amplification, and sequencing were carried out with the Sanger method as described previously (13).
Data set assembly, alignment, and sequence quality control.
Different analytical data sets were assembled to achieve the objectives. (i) Data set 1 (DS1) (see Table S1A in the supplemental material) contains the complete genome sequences newly generated in this work that were combined with H3N8 EIV publicly available sequences obtained from the NCBI Influenza Virus Database in January 2018 (n = 102 for PB1, PB2, HA, NA, NP, MP, and NS; n = 101 for PA); (ii) Data set 2 (DS2) (see Table S1B) contains HA1 sequences belonging to FC1 and FC2 (n = 203) that were selected as follows. H3N8 nucleotide sequences isolated from equines since 2002 were downloaded from the NCBI Influenza Virus Database; identical sequences in an outbreak were represented by the oldest sequence in the group. Maximum likelihood phylogenetic analysis was performed using RAxML (40). Then, sequences that formed a monophyletic clade including the reference sequences A/equine/South Africa/4/2003 (FC1) and A/equine/NewMarket/5/2003 (FC2) were selected.
All data sets were built and edited in BioEdit v7.1.3.0 (41) and aligned with Muscle (42) with default parameters. The molecular evolutionary models were inferred according to the Bayesian information criterion (BIC) statistics obtained with jModelTest v2.1 (43).
Prior to Bayesian or maximum likelihood phylogenetic analyses, the information contained in DS1 and DS2 was assessed. Briefly, recombination was evaluated by PHI test (44) using SplitsTree4 (45). The presence of a phylogenetic signal was assessed by a likelihood mapping method implemented in IQ-Tree (46). The presence of a temporal signal was examined by root-to-tip regression using TempEst v1.5.1 software (47).
Bayesian phylogeny and global population dynamic of H3N8 EIV.
A Bayesian coalescent approach was used to estimate the phylogenetic relationship, divergence times, and the population dynamics of EIV complete genome sequences (DS1). The analysis was carried out in BEAST v1.8.4 package (48) in the CIPRES server (49). First, the EIV complete genome was analyzed, and each segment was introduced as an independent partition with the corresponding molecular evolutionary model. Second, genomic segments were analyzed individually. All the analyses were carried out setting up flexible models, i.e., a Bayesian skyline plot (BSP) demographic model and an uncorrelated lognormal (UCLN) molecular clock, and the temporal calibration was based on the tip dates. Markov chain Monte Carlo (MCMC) sampling was performed in duplicates, and samples were examined with Tracer v1.6 to evaluate the convergence of parameters (effective sample size [ESS] of ≥200, acceptable mixing without tendencies in traces, with a burn-in of 10%). The maximum clade credibility (MCC) tree was summarized using Tree Annotator v1.8.4 and visualized with FigTree v1.4.2 (available at http://tree.bio.ed.ac.uk/software/figtree/).
Phylogeographic reconstruction of FC1 and FC2 spread based on HA gene.
To investigate the global spread of FC1 and FC2 EIV and to test the significant linkages between geographical locations, a phylogeographic analysis was performed in BEAST v1.8.4. Only HA1 (DS2) was considered in order to include a larger number of strains to represent the global EIV epidemiology. DS2 was analyzed under the BSP and UCLN models. A discrete phylogeographical model with an asymmetric substitution matrix over the sampling locations was used, implementing the Bayesian stochastic search variable selection (BSSVS) procedure. The sequence location states span eighteen geographic regions (Table S1B). MCMC samples were inspected in Tracer v1.6, and the MCC tree was summarized as described above. The significant epidemiological links were analyzed and visualized by SpreaD3 (50), and Bayes factor (BF) values >20 were considered well supported, as proposed by Kass and Raftery (51).
Signature pattern.
In order to obtain a complete genome characteristic amino acid (signature pattern), the FC1 and FC2 strains were selected from DS1. The eight segments were compared between them to assess specific amino acid residues that characterize each clade. Additionally, to perform an in-depth analysis of Asian strains, these were selected from HA1 (DS2) and were contrasted with the FC1 and FC2 EIV reference strains A/equine/SouthAfrica/4/2003 and A/equine/NewMarket/5/2003, respectively.
Structural modeling and analyses.
The molecular modeling was carried out using the I-Tasser online server (http://zhanglab.ccmb.med.umich.edu/I-TASSER) (52). Representative strains of Asian lineages were used for HA structural modeling (A/equine/Huabei/1/2007, A/equine/Zhaosu/1/2015, and A/equine/Yokohama/1/2017). The C-scores, which are confidence scores provided by I-Tasser and range from −5 to 2 (where higher values signify higher confidence), were 1.12, 0.93, and 0.93 for the three HA1 models and 0.63 for the HA2 model. Note that modeling was based on the experimentally solved structure from Aichi/68 strains (PDB 3HMG) that present high similarity (≥90%) with the HA1 proteins from the equine strains used in this study (data not shown). The HA trimer was reconstructed using the atomic model of the EIV strain (H3; PDB 3HMG). Molecular visualizations and graphs were obtained using the UCSF Chimera package (53). Electrostatic potentials of the surface were calculated using the coulombic surface coloring option with default settings as implemented in UCSF Chimera. Electric charges for each atom were assigned using AMBER ff14SB. Molecular visualizations and graphs were obtained using UCSF Chimera.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Instituto Nacional de Tecnología Agropecuaria (INTA), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), and Chinese Academy of Agricultural Sciences (CAAS).
We thank Juan Pablo Jaworski for the English revision of the manuscript.
Author contributions were as follows: study design, S.M., L.M., R.C., and X.W.; virus identification and sequence analyses, W.G., H.Z., T.Q., C.D., X.Z., and J.W.; phylogenetic, molecular, and structural analyses, S.M. and L.M.; data analyses and interpretation, S.M. and L.M.; writing of the manuscript, S.M. and L.M.; revision of the manuscript, R.C. and X.W.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00116-19.
REFERENCES
- 1.Landolt GA, Townsend HGG, Lunn DP. 2014. Equine influenza infection, p 141–151. In Sellon DC, Long M (ed), Equine infectious diseases 2nd ed Saunders Elsevier, St. Louis, MO. [Google Scholar]
- 2.Wright PF, Neumann G, Kawaoka Y. 2013. Ortomyxoviruses, p 1186–12443. In Knipe D, Howley P (ed), Fields virology 6th ed Wolters Kluwer-Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Durães-Carvalho R, Salemi M. 2018. In-depth phylodynamics, evolutionary analysis and in silico predictions of universal epitopes of influenza A subtypes and influenza B viruses. Mol Phylogenet Evol 121:174–182. doi: 10.1016/j.ympev.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Webster RG. 1993. Are equine 1 influenza viruses still present in horses? Equine Vet J 25:537–538. doi: 10.1111/j.2042-3306.1993.tb03009.x. [DOI] [PubMed] [Google Scholar]
- 5.Ito M, Nagai M, Hayakawa Y, Komae H, Murakami N, Yotsuya S, Asakura S, Sakoda Y, Kida H. 2008. Genetic analyses of an H3N8 influenza virus isolate, causative strain of the outbreak of equine influenza at the Kanazawa racecourse in Japan in 2007. J Vet Med Sci 70:899–906. doi: 10.1292/jvms.70.899. [DOI] [PubMed] [Google Scholar]
- 6.Murcia PR, Wood JLN, Holmes EC. 2011. Genome-scale evolution and phylodynamics of equine H3N8 influenza A virus. J Virol 85:5312–5322. doi: 10.1128/JVI.02619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullinane A, Newton JR. 2013. Equine influenza—a global perspective. Vet Microbiol 167:205–214. doi: 10.1016/j.vetmic.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Yamanaka T, Niwa H, Tsujimura K, Kondo T, Matsumura T. 2008. Epidemic of equine influenza among vaccinated racehorces in Japan in 2007. J Vet Med Sci 70:623–625. doi: 10.1292/jvms.70.623. [DOI] [PubMed] [Google Scholar]
- 9.Motoshima M, Okamatsu M, Asakura S, Kuribayashi S, Sengee S, Batchuluun D, Ito M, Maeda Y, Eto M, Sakoda Y, Sodnomdarjaa R, Kida H. 2011. Antigenic and genetic analysis of H3N8 influenza viruses isolated from horses in Japan and Mongolia, and imported from Canada and Belgium during 2007–2010. Arch Virol 156:1379–1385. doi: 10.1007/s00705-011-1000-5. [DOI] [PubMed] [Google Scholar]
- 10.Na W, Kang B, Kim H-I, Hong M, Park S-J, Jeoung H-Y, An D-J, Moon H, Kim J-K, Song D. 2014. Isolation and genetic characterization of naturally NS-truncated H3N8 equine influenza virus in South Korea. Epidemiol Infect 142:759–766. doi: 10.1017/S095026881300143X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perglione CO, Gildea S, Rimondi A, Miño S, Vissani A, Carossino M, Cullinane A, Barrandeguy M. 2016. Epidemiological and virological findings during multiple outbreaks of equine influenza in South America in 2012. Influenza Other Respir Viruses 10:37–46. doi: 10.1111/irv.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodward AL, Rash AS, Blinman D, Bowman S, Chambers TM, Daly JM, Damiani A, Joseph S, Lewis N, Mccauley JW, Medcalf L, Mumford J, Newton JR, Tiwari A, Bryant NA, Elton DM. 2014. Development of a surveillance scheme for equine influenza in the UK and characterisation of viruses isolated in Europe, Dubai and the USA from 2010–2012. Vet Microbiol 169:113–127. doi: 10.1016/j.vetmic.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 13.Qi T, Guo W, Huang WQ, Li HM, Zhao L-P, Dai L-L, He N, Hao X-F, Xiang W-H. 2010. Genetic evolution of equine influenza viruses isolated in China. Arch Virol 155:1425–1432. doi: 10.1007/s00705-010-0724-y. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C, Li Q, Guo W, Lu G, Yin X, Qi T, Xiang W, Ran D. 2013. Complete genomic sequences of an H3N8 equine influenza virus strain isolated in China. Genome Announc 1:e00654-13. doi: 10.1128/genomeA.00654-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sack A, Daramragchaa U, Chuluunbaatar M, Gonchigoo B, Bazartseren B, Tsogbadrakh N, Gray G. 2017. Low prevalence of enzootic equine influenza virus among horses in Mongolia. Pathogens 6:61. doi: 10.3390/pathogens6040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yondon M, Heil GL, Burks JP, Zayat B, Waltzek TB, Jamiyan BO, Mckenzie PP, Krueger WS, Friary JA, Gray GC. 2013. Isolation and characterization of H3N8 equine influenza A virus associated with the 2011 epizootic in Mongolia. Influenza Other Respir Viruses 7:659–665. doi: 10.1111/irv.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Virmani N, Bera BC, Singh BK, Shanmugasundaram K, Gulati BR, Barua S, Vaid RK, Gupta AK, Singh RK. 2010. Equine influenza outbreak in India (2008–09): virus isolation, sero-epidemiology and phylogenetic analysis of HA gene. Vet Microbiol 143:224–237. doi: 10.1016/j.vetmic.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Karamendin K, Kydyrmanov A, Kasymbekov Y, Khan E, Daulbayeva K, Asanova S, Zhumatov K, Seidalina A, Sayatov M, Fereidouni SR. 2014. Continuing evolution of equine influenza virus in Central Asia, 2007–2012. Arch Virol 159:2321–2327. doi: 10.1007/s00705-014-2078-3. [DOI] [PubMed] [Google Scholar]
- 19.Lowen AC. 2017. Constraints, drivers, and implications of influenza A virus reassortment. Annu Rev Virol 4:105–121. doi: 10.1146/annurev-virology-101416-041726. [DOI] [PubMed] [Google Scholar]
- 20.Bera BC, Virmani N, Shanmugasundaram K, Vaid RK, Singh BK, Gulati BR, Anand T, Barua S, Malik P, Singh RK. 2013. Genetic analysis of the neuraminidase (NA) gene of equine influenza virus (H3N8) from epizootic of 2008–2009 in India. Indian J Virol 24:256–264. doi: 10.1007/s13337-013-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boukharta M, Azlmat S, Elharrak M, Ennaji MM. 2015. Multiple alignment comparison of the non-structural genes of three strains of equine influenza viruses (H3N8) isolated in Morocco. BMC Res Notes 8:471. doi: 10.1186/s13104-015-1441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.FAO. 2016. FAOStat3-2016 FAOSTAT–FAO statistical database. Food and Agriculture Organization of the United Nations, Rome, Italy. [Google Scholar]
- 23.Dellicour S, Baele G, Dudas G, Faria NR, Pybus OG, Suchard MA, Rambaut A, Lemey P. 2018. Phylodynamic assessment of intervention strategies for the West African Ebola virus outbreak. Nat Commun 9:2222. doi: 10.1038/s41467-018-03763-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamanaka T, Cullinane A, Gildea S, Bannai H, Nemoto M, Tsujimura K, Kondo T, Matsumura T. 2015. The potential impact of a single amino-acid substitution on the efficacy of equine influenza vaccines. Equine Vet J 47:456–462. doi: 10.1111/evj.12290. [DOI] [PubMed] [Google Scholar]
- 25.Qi T, Guo W, Huang W, Dai L, Zhao L, Li H, Li X, Zhang X, Wang Y, Yan Y, He N, Xiang W. 2010. Isolation and genetic characterization of H3N8 equine influenza virus from donkeys in China. Vet Microbiol 144:455–460. doi: 10.1016/j.vetmic.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Yondon M, Zayat B, Nelson MI, Heil GL, Anderson BD, Lin X, Halpin R. a, Mckenzie PP, White SK, Wentworth DE, Gray GC. 2014. Equine influenza A (H3N8) virus isolated from Bactrian camel, Mongolia. Emerg Infect Dis 20:2144–2147. doi: 10.3201/eid2012.140435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.OIE. 2017. OIE expert surveillance panel on equine influenza vaccine composition, OIE headquarters, 22 March 2017. Conclusions and Recommendations. World Organisation for Animal Health, Paris, France. [Google Scholar]
- 28.Endo A, Pecoraro R, Sugita S, Nerome K. 1992. Evolutionary pattern of the H3 haemagglutinin of equine influenza viruses: multiple evolutionary lineages and frozen replication. Arch Med Res 123:73–87. doi: 10.1007/BF01317139. [DOI] [PubMed] [Google Scholar]
- 29.Gildea S, Quinlivan M, Arkins S, Cullinane A. 2012. The molecular epidemiology of equine influenza in Ireland from 2007–2010 and its international significance. Equine Vet J 44:387–392. doi: 10.1111/j.2042-3306.2011.00472.x. [DOI] [PubMed] [Google Scholar]
- 30.Volz EM, Koelle K, Bedford T. 2013. Viral phylodynamics. PLoS Comput Biol 9:e1002947. doi: 10.1371/journal.pcbi.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryant NA, Rash AS, Woodward AL, Medcalf E, Helwegen M, Wohlfender F, Cruz F, Herrmann C, Borchers K, Tiwari A, Chambers TM, Newton JR, Mumford JA, Elton DM. 2011. Isolation and characterisation of equine influenza viruses (H3N8) from Europe and North America from 2008 to 2009. Vet Microbiol 147:19–27. doi: 10.1016/j.vetmic.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 32.Watson J, Daniels P, Kirkland P, Carroll A, Jeggo M. 2011. The 2007 outbreak of equine influenza in Australia: lessons learned for international trade in horses. Rev Sci Tech 30:87–93. doi: 10.20506/rst.30.1.2021. [DOI] [PubMed] [Google Scholar]
- 33.Gahan J, Garvey M, Gildea S, Gür E, Kagankaya A, Cullinane A. 2018. Whole-genome sequencing and antigenic analysis of the first equine influenza virus identified in Turkey. Influenza Other Respir Viruses 12:374–382. doi: 10.1111/irv.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perglione C, Golemba M, Torres C, Barrandeguy M. 2016. Molecular epidemiology and spatio-temporal dynamics of the H3N8 equine influenza virus in South America. Pathogens 5:61. doi: 10.3390/pathogens5040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khadka R. 2010. Thesis Global horse population with respect to breeds and risk status. Swedish University of Agricultural Sciences, Uppsala, Sweden. [Google Scholar]
- 36.Mushtaq MH, Khan A, Ahmad MUD, Khan A, Farooqi SH, Hussain A, Oladunni F. 2018. An epidemiological investigation of associated risk factors with equine influenza (H3N8) epidemic 2015–16 in Pakistan. Acta Trop 186:63–68. doi: 10.1016/j.actatropica.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Khan A, Mushtaq MH, Ahmad MD, Nazir J, Fatima Z, Khan A, Farooqi SH. 2018. The equine influenza outbreak in Pakistan 2016: seroprevalence and geo-temporal epidemiology of a large propagating outbreak. Pak J Zool 50:453–461. [Google Scholar]
- 38.UNESCO. 2010. Fifth session of the intergovernmental committee (5.COM) - Nairobi, Kenya, 15 to 19 November 2010. United Nations Educational, Scientific and Cultural Organization, Paris, France. [Google Scholar]
- 39.White MC, Steel J, Lowen AC. 2017. Heterologous packaging signals on segment 4, but not segment 6 or segment 8, limit influenza A virus reassortment. J Virol 91:e00195-17. doi: 10.1128/JVI.00195-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. 2015. Using RAxML to infer phylogenies. Curr Protoc Bioinformatics 51:6.14.1–6.14.14. doi: 10.1002/0471250953.bi0614s51. [DOI] [PubMed] [Google Scholar]
- 41.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 41:95–98. [Google Scholar]
- 42.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rambaut A, Lam TT, Carvalho LM, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 2010.
- 50.Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. 2016. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol 33:2167–2169. doi: 10.1093/molbev/msw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kass RE, Raftery AE. 1995. Bayes factors. J Am Stat Assoc 90:773–795. doi: 10.2307/2291091. [DOI] [Google Scholar]
- 52.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Burke DF, Smith DJ. 2014. A recommended numbering scheme for influenza A HA subtypes. PLoS One 9(11):e112302. doi: 10.1371/journal.pone.0112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.