Since acyclovir and its derivatives were launched for herpesviruses control almost four decades ago, the search for novel antivirals has waned. However, as human life expectancy has increased, so has the number of immunocompromised individuals who receive prolonged treatment for HSV recurrences. This has led to an increase in unresponsive patients due to acquired viral drug resistance. Thus, novel treatments need to be explored. Here we explored the HSV-1 ICP0 E3 ligase as a potential antiviral target because (i) ICP0 is expressed before virus replication, (ii) it is essential for infection in vivo, (iii) it is required for efficient reactivation of the virus from latency, (iv) inhibition of its E3 ligase activity would sustain host immune responses, and (v) it is shared by other herpesviruses. We report a compound that inhibits HSV-1 infection in an ICP0-dependent manner by inhibiting ICP0 E3 ligase activity.
KEYWORDS: E3 ubiquitin ligase, HSV, ICP0, high-throughput screening assay, small-molecule inhibitors
ABSTRACT
Herpes simplex virus 1 (HSV-1) has infected more than 80% of the population. Reactivation of the virus causes diseases ranging in severity from benign cold sores to fatal encephalitis. Current treatments involve viral DNA replication inhibitors, but the emergence of drug-resistant mutants is observed frequently, highlighting the need for novel antiviral therapies. Infected cell protein 0 (ICP0) of HSV-1 is encoded by an immediate early gene and plays a fundamental role during infection, because it enables viral gene expression and blocks antiviral responses. One mechanism by which ICP0 functions is through an E3 ubiquitin ligase activity that induces the degradation of targeted proteins. A ΔICP0 virus or mutants with deficiencies in E3 ligase activity cannot counteract beta interferon (IFN-β)-induced restriction of viral infection, are highly immunogenic, are avirulent, and fail to spread. Thus, small molecules interfering with essential and conserved ICP0 functions are expected to compromise HSV-1 infection. We have developed a high-throughput screening assay, based on the autoubiquitination properties of ICP0, to identify small-molecule inhibitors of ICP0 E3 ubiquitin ligase activity. Through a pilot screening procedure, we identified nine compounds that displayed dose-dependent inhibitory effects on ICP0 but not on Mdm2, a control E3 ubiquitin ligase. Following validation, one compound displayed ICP0-dependent inhibition of HSV-1 infection. This compound appeared to bind ICP0 in a cellular thermal shift assay, it blocked ICP0 self-elimination, and it blocked wild-type but not ICP0-null virus gene expression. This scaffold displays specificity and could be used to develop optimized ICP0 E3 ligase inhibitors.
IMPORTANCE Since acyclovir and its derivatives were launched for herpesviruses control almost four decades ago, the search for novel antivirals has waned. However, as human life expectancy has increased, so has the number of immunocompromised individuals who receive prolonged treatment for HSV recurrences. This has led to an increase in unresponsive patients due to acquired viral drug resistance. Thus, novel treatments need to be explored. Here we explored the HSV-1 ICP0 E3 ligase as a potential antiviral target because (i) ICP0 is expressed before virus replication, (ii) it is essential for infection in vivo, (iii) it is required for efficient reactivation of the virus from latency, (iv) inhibition of its E3 ligase activity would sustain host immune responses, and (v) it is shared by other herpesviruses. We report a compound that inhibits HSV-1 infection in an ICP0-dependent manner by inhibiting ICP0 E3 ligase activity.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) has infected more than 80% of the population (1). Primary infection is usually asymptomatic and leads to a lifelong latent infection in sensory neurons (1). Periodically, the virus is reactivated in these neurons due to weakened immune responses, stress, hormonal changes, or UV exposure. The most common manifestation of HSV-1 reactivation is the formation of vesicular lesions at the initial site of infection (1). While HSV-1 can cause genital herpes, most cases are caused by HSV-2, another alphaherpesvirus that is closely related to HSV-1. HSV-2 infection of the genitalia recurs 4 or 5 times per year, while HSV-1 genital reactivation occurs 0 or 1 time, with a tendency to decrease over time. One in five adults in the United States has a genital HSV infection, with over 1 million new cases annually and over 20 million people visiting the hospital due to recurrent genital lesions (1). Transmission from mother to newborn can lead to disseminated infection, with severe consequences for the newborn (3,000 cases/year in the United States) (1). HSV-1 is also associated with more severe diseases such as herpes encephalitis (5,000 adult cases/year in the United States), which has a 70% morbidity rate if left untreated, and HSV-1-induced keratitis, which is a major cause of infectious blindness worldwide (500,000 cases/year in the United States) (1). There is no vaccine for HSV, and current treatments involve nucleoside analogues such as acyclovir (ACV), famciclovir, and valaciclovir, pyrophosphate analogues such as foscarnet, and nucleotide analogues such as cidofovir, all of which inhibit viral DNA replication (2, 3). However, the rates of viral resistance to nucleoside analogues in immunocompromised patients range from 4% to 14% (mainly because of mutations in the UL23 gene encoding the viral thymidine kinase, which phosphorylates ACV before incorporation into the replicating viral genome), and the high toxicity of foscarnet and cidofovir highlight the need for novel treatments (4, 5). The rates of resistance to ACV are expected to increase as people live longer and suffer conditions that compromise their immune systems, e.g., cancer patients and transplant recipients who receive prolonged antiviral treatment (6).
Docosanol is a marketed HSV drug that inhibits viral fusion to the host cell (7–10). Docosanol does not target the virus per se but modifies the host membrane and interferes with the fusion of many viruses; therefore, it is administered only in a topical form, mainly to treat herpes cold sores (7–10).
Given the need for novel herpes treatments, new targets, new types of molecules, and new antiviral mechanisms are being explored, including targeting of the viral DNA helicase and viral glycoproteins and modulation of the immune system (3, 11, 12). In this study, we chose to target the HSV-1 infected cell protein 0 (ICP0) E3 ubiquitin ligase, based on the following rationale. ACV and its derivatives are used as a first line of HSV-1 treatment because they inhibit elongation of the replicating viral genome and they are virus specific. Following the acquisition of viral drug resistance, particularly in patients with underlying immunodeficiency, generic treatments that elicit adverse effects are utilized (3, 13–15). None of these drugs interferes with the ability of the virus to establish latency or to reactivate. ICP0 is expressed following viral DNA release in the nucleus and is required for initiation of viral gene transcription, inhibition of host antiviral responses, and efficient reactivation of the virus from latency (16–18). E3 ligase activity is essential for these ICP0 functions; therefore, E3 ligase inhibitors are expected to affect both the lytic stage and the latent stage of the virus. This is a major advantage over other available drugs against HSV, because they do not affect the ability of the virus to reactivate.
ICP0 is a promiscuous transactivator that enables viral gene expression by disrupting DNA repressor complexes and blocking antiviral responses (16, 17, 19–25). A cysteine-rich region of ICP0 constitutes a C3HC4 zinc-binding really interesting new gene (RING) finger (RF) motif that is essential for these ICP0 functions, as it constitutes an E3 ubiquitin ligase. ICP0 E3 ubiquitin ligase degrades host factors involved in innate immune responses, including the Toll-like receptor (TLR) adaptors Myd88 and Mal, components of ND10 bodies such as promyelocytic leukemia (PML) and SP100 nuclear bodies, the deubiquitination enzyme ubiquitin-specific-processing protease 7 (USP7), and others, resulting in suppression of antiviral responses (26–31). ICP0 is autoubiquitinated both in vivo and in vitro, leading to its degradation by proteasomes. A ΔICP0 virus or mutants with deficiencies in E3 ligase activity cannot counteract the beta interferon (IFN-β)-induced restriction of viral infection, are highly immunogenic, are avirulent, and fail to spread (32–37). In vivo, ICP0 is essential for efficient reactivation of the virus from latency (32–37). Genes coding for ICP0 are present in the genomes of simplex viruses and varicelloviruses, but they are absent from viruses of the Mardivirus genus (38–42). The proteins encoded by these genes show high sequence homology to ICP0, especially within the RF domain. Orthologues of ICP0 are also present in lymphocryptoviruses (e.g., Epstein-Barr virus [EBV]) and cytomegalovirus (CMV) (39, 40, 43). These ICP0 orthologues can rescue a ΔICP0 virus to some extent, by derepressing the viral genome and inducing degradation of components of the ND10 bodies (39–42, 44).
To identify compounds that block the ICP0 E3 ligase activity, we took advantage of the fact that ICP0 is autoubiquitinated and this reaction can be recapitulated in vitro using purified, bacterially expressed exon II of ICP0 (amino acids 20 to 241) (45, 46). Based on these ICP0 properties, we developed a high-throughput screening (HTS) assay that we used to screen a small-compound library composed of 5,160 diversity scaffolds synthesized by the Chemical Methodologies and Library Development Center (CMLD) at the University of Kansas (KU). The KU-CMLD library includes multiple scaffolds that are likely to have pharmacological activity, based on sound drug design principles, and are likely to have drug-like characteristics. Through this pilot study, we identified one hit, a 3,4,5-aryl-substituted isoxazole, that appears to inhibit HSV-1 infection by inhibiting the ICP0 E3 ligase. This is the first study that describes a small-molecule inhibitor for the ICP0 E3 ligase with potential antiviral function.
RESULTS
Development of an HTS assay to monitor the HSV-1 ICP0 E3 ligase activity.
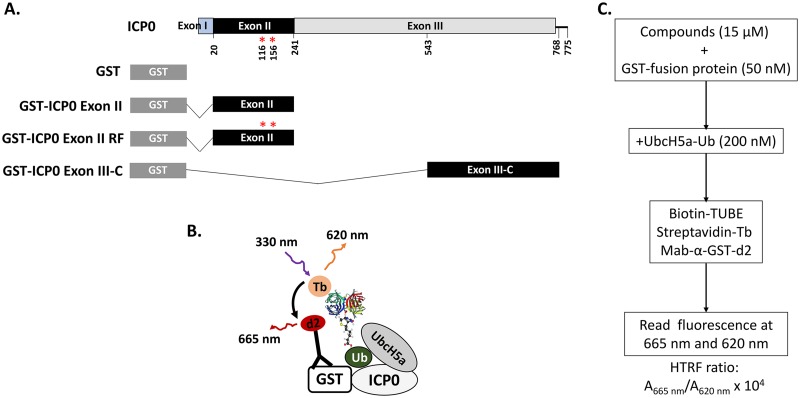
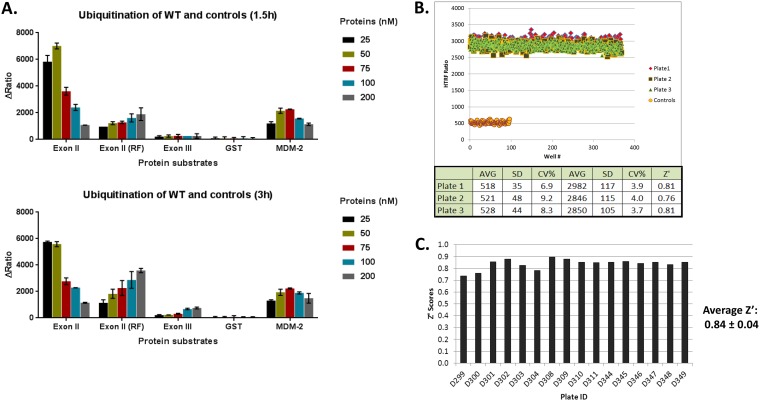
A homogeneous time-resolved fluorescence (HTRF) assay was developed for the E3 ligase encoded by exon II of ICP0 (Fig. 1A) (16, 45, 46). The E3 ligase is essential for ICP0 functions and, to identify inhibitors, we took advantage of the fact that ICP0 is autoubiquitinated in vitro even when purified, bacterially expressed exon II of ICP0 is used in mixtures with the E2 enzyme UbcH5a and ubiquitin. The assay principle is based on recognition of polyubiquitin residues on glutathione S-transferase (GST)-exon II of ICP0 by biotinylated polyubiquitin tandem ubiquitin-binding entities (TUBEs) (Fig. 1B). A fluorescence resonance energy transfer (FRET) is facilitated when streptavidin-terbium (fluorophore energy donor), binding to biotinylated TUBEs, is brought close to d2-labeled (second-generation fluorophore energy recipient) anti-GST antibody, which recognizes the GST tag on ICP0 exon II protein (Fig. 1B). The same form of ICP0 with two amino acid substitutions in the RF domain (C116A and C156A) that are known to compromise ICP0 E3 ligase activity served as a negative control (47). Additional negative controls included the GST protein alone and a portion of exon III of ICP0 (amino acids 543 to 768). We also included GST-Mdm2, an unrelated E3 ubiquitin ligase that is also autoubiquitinated utilizing UbcH5a. All proteins were purified using Escherichia coli strain BL21 (Fig. 1A). A schematic of the HTS assay is depicted in Fig. 1B and C. In a series of control experiments, the HTRF assay was optimized for exon II or control protein concentrations, incubation time, and UbcH5a and MgCl2 concentrations. The optimum conditions were found to be 50 nM for the fusion proteins, 200 nM for UbcH5a (preloaded with ubiquitin), and 2 mM for MgCl2, and the data were collected 90 min after initiation of the reaction. FRET signals due to proximity of ICP0 to the ubiquitin moiety were measured when exon II of ICP0 or the control (Mdm-2) was present but were absent when exon III-C of ICP0 or GST alone was utilized (Fig. 2A). The ICP0 exon II RF domain displayed significantly reduced activity. The overall ubiquitination signal was found to be significantly higher when exon II was used, compared with the GST-Mdm-2 positive control, suggesting that the conditions were favorable for the viral E3 ligase. The assay displayed the desired specificity. Several multiwell plates were tested, and the assay was found to be statistically acceptable, consistently giving a Z′ score of 0.84 ± 0.04, which indicated good separation of positive and negative controls on assay plates (Fig. 2B and C).
FIG 1.
ICP0 HTS assay. (A) GST-ICP0 fusion proteins used for the HTS assay. ICP0 exon II spans amino acids 20 to 241 of ICP0 from HSV-1(F), while the portion of exon III of ICP0 that was used as a control spans amino acids 543 to 768 (ICP0 exon III-C). (B and C) HTS assay measuring the proximity of the ubiquitin (Ub) moiety to ICP0.
FIG 2.
ICP0 ubiquitination assay optimization. The HTRF based ubiquitination assay was performed as discussed in Materials and Methods. (A) Specificity and time dependence of ubiquitination. (Upper) Ubiquitination was found to be specific for the WT GST-ICP0 exon II protein. The RF mutant protein, GST-ICP0 exon II (RF), showed more than 50% reduction in ubiquitination, whereas little or no ubiquitination was observed with GST-ICP0 exon III-C or with the GST protein alone. GST-Mdm2, a control protein, was ubiquitinated to much lower extent than the exon II protein, indicating that the assay parameters were highly specific for ubiquitination of the exon II protein. (Lower) Only a slight increase (1.5-fold) in ubiquitination activity of exon II (RF) was observed after 3 h of incubation, whereas ubiquitination of other proteins remained unchanged over time. ΔRatio was obtained after reads from ICP0 control reactions and UbcH5a control reactions were subtracted from the reads of reactions containing both ICP0 and UbcH5a. (B) Signal uniformity. The uniformity of the HTRF signal was tested in three independent experiments, using low-volume 384-well plates. A window of 5.5 ± 0.2 between complete ubiquitination reactions and controls (lacking exon II) was observed across the three experiments, with an average coefficient of variation of 3.9% and an average Z′ score of 0.8 ± 0.02. (C) Distribution of Z′ scores across all of the plates screened using the optimized assay (∼5,000 compounds). The assay gave an average Z′ score of 0.84 ± 0.04, indicating good separation between the positive and negative controls on each plate.
Identification of HTS hits.
To identify a small-molecule inhibitor for the ICP0 E3 ligase, the desired activity profile was as follows: (i) inhibiting only ICP0 E3 ligase activity, with a significantly lower 50% inhibitory concentration (IC50) than the control (Mdm2); (ii) preventing the degradation of ICP0 substrates; (iii) causing a delay in viral gene expression similar to that observed with an ICP0 RF mutant virus in nonpermissive cell lines; (iv) displaying its effects only on wild-type (WT) cells and not on ΔICP0 virus-infected cells; (v) preventing the self-elimination of ICP0 and increasing its half-life; (vi) binding to ICP0; and (vii) displaying no cellular toxicity. An ICP0 E3 ligase inhibitor should display all of these characteristics.
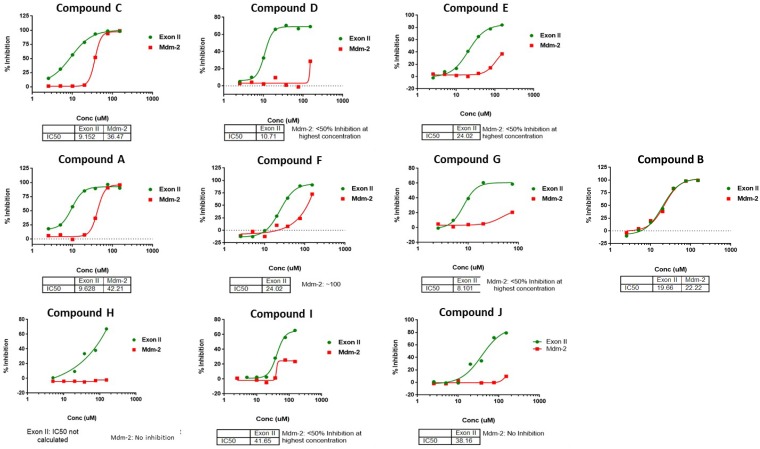
The HTRF assay was used to screen a library of 5,126 drug-like molecules synthesized through the KU-CMLD program. Twenty-seven compounds (0.5% hit rate) were found to inhibit the reaction and were then retested in 5-concentration dose-response experiments, in which 2 did not show a dose-dependent response. Of the 27 hit molecules, we advanced the 10 most potent compounds that were dose responsive for their selectivity against UbcH5a and Mdm2, an unrelated E3 ligase that also utilizes UbcH5a (Fig. 3). Only compound B showed similar inhibition for ICP0 and Mdm2, probably due to inhibition of the E2 ubiquitin conjugation enzyme UbcH5a. Two of the hits (compounds A and C) were resynthesized (>95% purity by liquid chromatography-mass spectrometry [LC-MS] analysis) and retested for activity in the ICP0 ubiquitination assay prior to being used in other downstream cell-based assays (data not shown). These data indicate that we successfully established an in vitro HTS assay to monitor ICP0 E3 ligase activity and identified 9 potential hits capable of inhibiting the ICP0 E3 ligase activity.
FIG 3.
Selectivity of the hits for GST-ICP0 exon II ubiquitination. The hits identified from the primary screen were picked from HTS library stocks, and their activities were confirmed in HTRF assays containing either GST-ICP0 exon II or GST-Mdm2 protein. Except for compound B, all of the hits showed greater activity against ICP0 exon II than against Mdm-2.
Effects of HTS hits on the stability of ICP0 E3 ligase substrates and on viral protein expression.
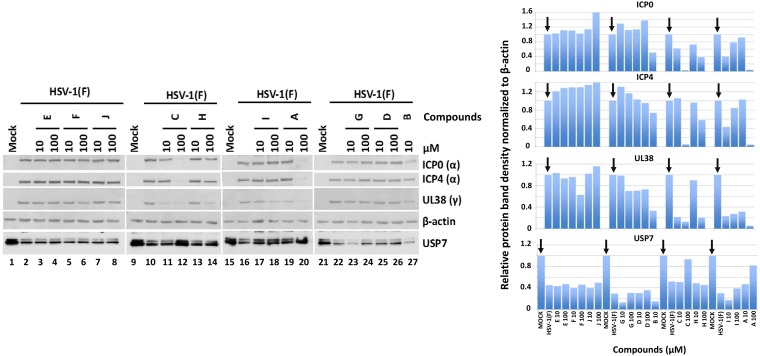
To initially validate the HTS hits, we assessed the ability of the compounds to protect USP7 from ICP0-mediated degradation, as well as their effects on viral protein expression. ICP0 targets several host proteins for degradation, including USP7 (30, 31, 48). ICP0 is also involved in the transactivation of all classes of viral genes (19, 23). Therefore, HEp-2 cells were exposed to HSV-1(F) (5 PFU/cell) and the hit compounds were added to the cultures at the time of infection (10 or 100 μM). The cells were harvested at 8 h postinfection, and the accumulation of USP7 was assessed by immunoblot analysis. HSV-1 causes degradation of USP7, and significant decreases in the amounts of USP7 were observed, compared to uninfected cells (Fig. 4, compare lanes 1, 9, 15, and 21 to lanes 2, 10, 16, and 22). Compounds E, F, J, and G did not restore USP7 accumulation, and they not affect viral protein expression. Compound H did not restore the ICP0 substrate but it caused a delay in viral protein expression at the highest concentration. Treatment with compounds I and D partially restored the levels of USP7, especially at the highest concentration, without altering viral protein accumulation (Fig. 4, compare lanes 25 and 26 to lanes 21 and 22 and lanes 17 and 18 to lanes 15 and 16). Compounds A and C fully restored the levels of USP7 and caused substantial delays in all classes of viral genes, especially at the highest concentration (Fig. 4, compare lanes 19 and 20 to lane 16 and lanes 11 and 12 to lane 10). Finally, compound B was toxic at 100 μM, presumably due to its ability to block the widely used enzyme UbcH5a (Fig. 3), and was discarded. Quantification of the protein bands is depicted in Fig. 4, right. We concluded that at least compound A displayed characteristics of a potential ICP0 E3 ligase inhibitor.
FIG 4.
Effects of compounds on accumulation of ICP0 substrates and on viral gene expression. HEp-2 cells were infected with HSV-1(F) (5 PFU/cell). The compounds were added to the cultures at 10 or 100 μM at the time of infection, and the cells were harvested at 8 h postinfection. Equal amounts of proteins from total cell lysates were analyzed by immunoblot analysis using antibodies against ICP0, ICP4, UL38, β-actin, and USP7 (left). Uninfected cells (lanes 1, 9, 15, and 21) and infected but untreated cells (lanes 2, 10, 16, and 22) served as controls. Quantification of the protein bands was performed using Image J software (right).
HSV-1 growth is impaired by compounds A and C.
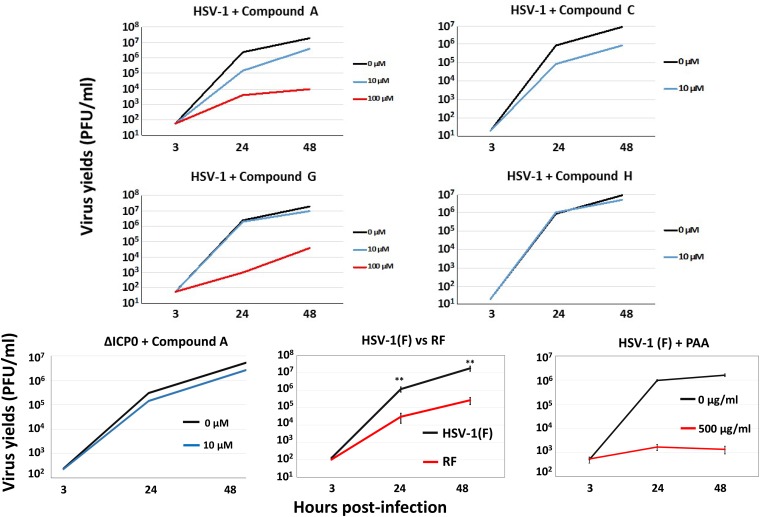
To further characterize the effects of the HTS hits on infection, we monitored the impacts of the compounds on progeny virus production. HEL cells were infected with either the WT virus or a ΔICP0 virus (0.01 PFU/cell), in the presence or absence of the compounds (10 or 100 μM), and intracellular virus was titered at 3, 24, and 48 h postinfection. Compound G strongly inhibited HSV-1 growth at 100 μM but not at 10 μM, despite the lack of an effect on viral protein expression (Fig. 5). Compounds D, F, and J displayed no effects on viral growth at 10 μM (data not shown). Compound A inhibited viral growth in a dose-dependent manner, with 10-fold and 1,000-fold decreases in HSV-1 yields at 10 μM and 100 μM, respectively (Fig. 5). Due to a lethal effect of compound C on viral infection at 100 μM, we tested its ability to inhibit HSV-1(F) at 10 μM. Treatment with compound C reduced viral yields 10-fold (Fig. 5). Compound H was available in only a small quantity and that restricted all of our experiments to a concentration of 10 μM, at which we did not observe any effect on viral growth (Fig. 5). Infection with an ICP0 RF mutant virus (0.01 PFU/cell) and treatment with phosphonoacetic acid (PAA) (500 μg/ml), a DNA polymerase inhibitor, were used as positive controls. As an additional control, we treated ΔICP0 virus-infected cells with compound A (10 μM), and we noticed only a minor decrease in virus growth. At concentrations higher than their IC50 values, all compounds are expected to have off-target effects; therefore, the results obtained with treatment at 10 μM are more relevant. We concluded that some of the hits, including the most potent ICP0 E3 ligase inhibitor, compound A, negatively affected HSV-1 yields.
FIG 5.
Effects of selected hits on HSV-1 growth. HEL cells were infected with HSV-1(F) (0.01 PFU/cell) or ΔICP0 virus (0.01 PFU/cell) and either not treated or treated with compound A, C, G, or H at 10 or 100 μM, added to the cultures at the time of infection. PAA was added at 500 μg/ml at the time of infection. Infection with the ICP0 RF mutant was performed at 0.01 PFU/cell. Cells were harvested at 3, 24, and 48 h postinfection, and titration of progeny viruses was performed in Vero cells. **, P ≤ 0.01.
Compound A inhibits HSV-1(F) but not ΔICP0 virus gene transcription.
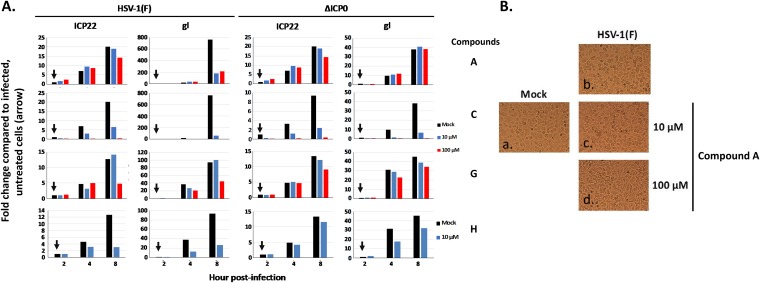
To further test the specificity of the compounds, we infected U2OS cells with HSV-1(F) or a ΔICP0 virus (0.1 PFU/cell), the compounds were added the time of infection, and viral gene expression was monitored by quantitative PCR (qPCR) analysis (Fig. 6A). Ideally, compounds targeting the ICP0 E3 ligase should not affect ΔICP0 virus gene transcription. ICP22 expression is independent of ICP0, but glycoprotein I (gI) expression requires virus replication. We found that compound C inhibited ICP22 and gI expression by both viruses; therefore, it did not fulfill the criteria of an ICP0 E3 ligase inhibitor. Compound G slightly inhibited ICP22 and gI expression in WT virus-infected cells but not in ΔICP0 virus-infected cells, and compound H behaved similarly at 10 μM. The most potent inhibitor, compound A, did not inhibit ΔICP0 virus gene expression or ICP22 expression in WT virus-infected cells but inhibited gI expression even at 10 μM in WT virus-infected cells. Compound A also prevented the cytopathic effects observed during HSV-1 infection (Fig. 6B, compare c and d to a and b). We concluded that compound A displayed features of a potent ICP0 inhibitor and inhibited HSV-1(F) infection in an ICP0-dependent manner.
FIG 6.
Effects of selected hits on HSV-1 and ΔICP0 virus gene expression. (A) U2OS cells were infected with HSV-1(F) or ΔICP0 virus (0.1 PFU/cell) and either not treated or treated with compound A, C, G, or H at 10 or 100 μM, added to the cultures at the time of infection. The cells were harvested at 2, 4, and 8 h postinfection, and viral gene expression was assessed by qPCR analysis. The 18S rRNA primers were used for normalization. (B) HEp-2 cells were treated as in Fig. 4 and pictures were acquired at 24 h postinfection, using an inverted Nikon Eclipse TE2000-S microscope equipped with a Nikon camera.
Analogues of compound A inhibit HSV-1 gene expression in an ICP0-dependent manner.
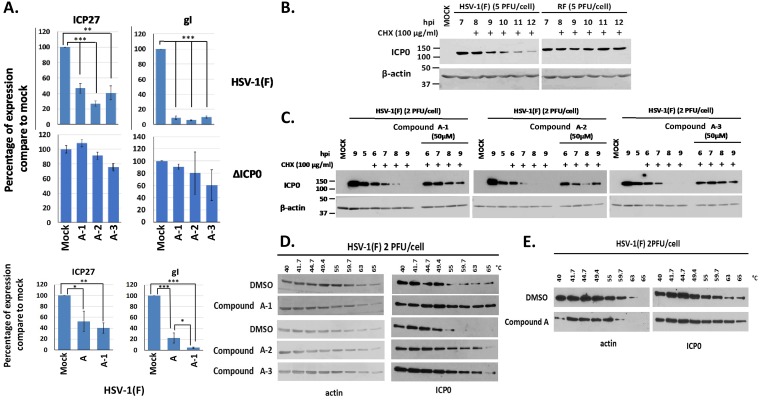
Compound A is an isoxazole with a furan ring in the 2-position. It has been reported that furans tend to undergo oxidative decomposition in air (49, 50). To avoid such undesirable properties, three analogues of compound A in which the furan ring was replaced with a methoxy phenyl group were designed, synthesized, and tested for their ability to block HSV-1 infection. HEL cells were infected with either the WT virus or the ΔICP0 virus (0.1 PFU/cell), and compound A-1, A-2, or A-3 (20 μM) or dimethyl sulfoxide (DMSO) was added to the cultures the time of infection. Cells were harvested at 8 h postinfection, and viral mRNAs were quantified. The amounts of ICP27 and gI transcripts were significantly reduced in WT virus-infected cells after treatment with the compounds, but the amounts were not reduced in ΔICP0 virus-infected cells (Fig. 7A). We also observed that compound A-1 had a slightly greater inhibitory effect on HSV-1 gene transcription than did compound A (Fig. 7A). We concluded that analogues of compound A displayed ICP0-dependent inhibitory effects on HSV-1 infection, like the original compound.
FIG 7.
Effects of the compound A analogues on HSV-1 viral gene expression and the stability of ICP0. (A) HEL cells were infected with HSV-1(F) or ΔICP0 virus (0.1 PFU/cell) and either not treated or treated with compound A, A-1, A-2, or A-3 at 20 μM, added to the cultures at the time of infection. The cells were harvested at 8 h postinfection, and viral gene expression was quantified by qPCR analysis. The 18S rRNA primers were used for normalization. Error bars represent standard deviations. Statistical analysis was performed using unpaired t tests. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B) HEp-2 cells were infected with HSV-1(F) or an ICP0 RF mutant virus (5 PFU/cell). CHX (100 μg/ml) was added to the cultures at 7 h postinfection. The cells were harvested at the time of CHX addition or at 1, 2, 3, 4, and 5 h posttreatment, and ICP0 protein was assessed in equal amounts of cell lysates by immunoblot analysis. β-Actin served as a loading control. (C) HEp-2 cells were infected with HSV-1(F) (2 PFU/cell), compound A-1, A-2, or A-3 (50 μM) was added to the infected cultures at 4.5 h postinfection, and CHX (100 μg/ml) was added to the cultures 0.5 h later. The cells were harvested at 0, 1, 2, 3, and 4 h after the addition of CHX, whereas an infected untreated sample harvested at 9 h after infection served as a control. The amounts of ICP0 protein were assessed in equal amounts of cell lysates by immunoblot analysis. β-Actin served as a loading control. (D) HEp-2 cells were infected with HSV-1(F) (2 PFU/cell) for 4 h and incubated with the indicated compounds (50 μM) for an extra 1 h. Cells were then collected and CETSA was performed. ICP0 temperature stability was assessed in the soluble fraction by immunoblot analysis. (E) The experiment was as in panel D except that HEL cells were used and compound A was added to the cultures at 20 μM for 1 h at 16 h postinfection.
Compounds A-1, A-2, and A-3 prevent ICP0 degradation.
To test the effects of the compound A analogues on ICP0 self-elimination, we infected HEp-2 cells with WT virus (2 PFU/cell) and analyzed the stability of ICP0 after treatment with cycloheximide (CHX) (100 μg/ml), which was added to the cultures at 5 h postinfection, in the presence or absence of the compounds. The degradation of ICP0 was dependent on its RF domain, as protein synthesis inhibition led to RF-domain-dependent degradation of ICP0 (Fig. 7B). Treatment with CHX led to rapid degradation of ICP0, resulting in complete loss of the protein 3 to 4 h posttreatment (Fig. 7C). In contrast, the stability of the ICP0 protein was increased in cells treated with CHX and compound A-1, A-2, or A-3, and ICP0 was still stable at 4 h post-CHX treatment. These results highlight the ability of compounds A-1, A-2, and A-3 to block ICP0 E3 ligase activity during the course of HSV-1 infection.
Compounds A-1, A-2, and A-3 bind to ICP0.
We evaluated the ability of compounds A-1, A-2, and A-3 to directly bind to ICP0 by using the cellular thermal shift assay (CETSA) (51). Briefly, cells were infected with HSV-1(F) for 4 h and either were untreated or were treated with the compounds (50 μM) for an additional 1 h. Cells were harvested, aliquoted, and subjected to transient heating from 40°C to 65°C, followed by lysis and sedimentation to remove cell debris and denatured and precipitated material. The temperature stability of ICP0 was evaluated by immunoblot analysis of the soluble fraction. ICP0 was stable up to 49.4°C but was precipitated at temperatures between 49.4 and 55°C (Fig. 7D). The presence of compounds A-1, A-2, and A-3 prevented precipitation of ICP0 up to 65°C, with a greater effect of compound A-1. In the same assay, we analyzed the stability of β-actin and found that it was identical in cells treated with DMSO or the compounds. We repeated the CETSA experiment in HEL cells but used 20 μM compound A, which was added to the cultures for 1 h at 16 h postinfection (2 PFU/cell). As shown in Fig. 7E, compound A increased the temperature stability of ICP0 even under these conditions. These data suggest that compounds A, A-1, A-2, and A-3 bind to ICP0, leading to its stability at higher temperatures.
DISCUSSION
ICP0 is nonessential for HSV-1 infection in cell culture when infections are performed at a high multiplicity of infection, but it is essential at a low multiplicity of infection in nonpermissive cells (16–19, 52). Most ICP0 functions are performed during the early stages of infection and involve the transcriptional activation of viral genes, the inhibition of antiviral responses, and the modification of the surfaceome of infected cells (16–19, 21, 23–25, 28, 45, 52–56). In vivo, ICP0 is essential for the virus counteracting innate and intrinsic immune responses and for efficient reactivation from latency (16–18, 52). One function of ICP0 is to transfer ubiquitin moieties to targeted substrates via its E3 ubiquitin ligase activity, which results in polyubiquitination and involves one of the two E2 enzymes, UbcH5a or UbcH6 (45, 46, 57, 58). The ICP0-mediated ubiquitination has been linked to degradation of its substrates (59). ICP0 is also autoubiquitinated and degraded, resulting in downmodulation of its functions (45, 46, 57, 58). Importantly, autoubiquitination of ICP0 occurs even in vitro using purified, bacterially expressed exon II of ICP0 carrying the RF domain.
Here, we discuss three major findings, i.e., (i) the development of an in vitro HTS assay that can be used to identify ICP0 E3 ligase inhibitors, (ii) the identification of a compound that fits the profile expected of a potent ICP0 E3 ligase inhibitor, and (iii) the determination of a structure-activity relationship from the three analogues synthesized to expand the scaffold class. For the HTS assay, we relied on the autoubiquitination properties of ICP0 and used the FRET principle. To optimize the procedure, we (i) omitted the E1 enzyme to reduce false-positive results and the need for counter-screening to eliminate compounds acting against the E1 enzyme, (ii) subtracted background light that can be generated in the absence of the E2, and (iii) included Mdm2, an irrelevant E3 ligase that is also autoubiquitinated in the presence of UbcH5a. Mdm2 allowed for the elimination of compounds that interfered with the reaction nonspecifically. An interesting compound that was eliminated via this process was compound B, which was found to inhibit both E3 ligases with similar efficiencies, most likely by interfering with UbcH5a. The use of Mdm2 cannot eliminate false-positive results due to compounds that interfere with the ubiquitin acceptor sites of ICP0, interfere with UbcH5a binding, or bind to ICP0 or GST, causing conformational changes of the protein that disrupt its E3 ligase function. We also included exon II of ICP0 carrying the substitutions C116A and C156A, which disrupted the RF domain (ICP0 RF mutant) and significantly reduced the autoubiquitination activity of ICP0. It should be noted that the activity of ICP0 with mutations in the RF domain was comparable to the activity of Mdm2 E3 ligase but significantly lower than the activity of WT ICP0. A possible explanation is that the conditions of the HTS assay were optimal for the ICP0 E3 ligase and not for Mdm2. The two enzymes might display different catalytic properties and kinetics, and the viral E3 ligase might be more potent than the host E3 ligase under these conditions. Another possibility is that substitution of the two cysteine residues that constitute the ICP0 RF domain is not sufficient to abrogate the ICP0 E3 ubiquitin ligase function, but this is purely hypothetical. Nevertheless, the results for the ICP0 RF mutant highlight the specificity of the assay and enhance our confidence that the signal generated is the result of an autoubiquitination reaction.
Through the HTS analysis, we identified nine hits that inhibited ICP0 exon II, but not Mdm2, activity in a dose-dependent manner. Compound A, a 3,4,5-aryl-substituted isoxazole, was found to satisfy all of the criteria for an ICP0-specific inhibitor. For example, the IC50 of this compound was 9.6 μM for ICP0, compared with 42.2 μM for Mdm2. Compound A prevented degradation of the ICP0 substrate USP7 during infection and prevented ICP0 self-elimination. Moreover, compound A caused a delay in viral gene expression only in WT virus-infected cells and not in ΔICP0 virus-infected cells, with concomitant inhibition of virus-induced cytopathic effects. The defect of HSV-1(F) in the presence of compound A, at lower concentrations, was comparable to that of the ICP0 RF mutant virus, further supporting the mode of action of the compound. Another interesting feature of compound A was the binding to ICP0 in a CETSA, which was observed within 1 h after addition of the compound to the cultures. This binding most likely accounts for the increased stability of ICP0 by preventing self-ubiquitination. Validation of compound A was performed in multiple cell lines. Human fibroblasts (HEL cells) were chosen because they have intact innate immunity pathways and ICP0 is essential for the virus to evade the host; therefore, compound A was anticipated to display the greatest impact on infection. In contrast, ICP0 is nonessential for infection in the human osteosarcoma cell line U2OS, and compound A was expected to have the least effect (60). Importantly, at this stage we cannot exclude off-target effects of compound A or potential effects on ΔICP0 virus infection, particularly at higher doses of the compound. Overall, compound A displays several desired features and could be further explored for optimization, using medicinal chemistry and structural biology approaches, to improve its biological activity and pharmacokinetic properties.
Prior to optimization, small-molecule inhibitors are expected to have limitations. Among the limitations we noticed for compound A, one was reduced solubility, which was estimated to be around 200 μM in phosphate-buffered saline (PBS) supplemented with 8% DMSO-d6. Also, we noticed a small delay in cell proliferation that appeared to be cell type dependent and did not cause morphological changes to the cells or death up to 48 h posttreatment. Thus, the effects on infection observed at concentrations higher than 20 μM most likely represent both specific and off-target effects. Another potential issue is the furan group present in compound A, which can cause metabolic liabilities. To optimize compound A in this respect, three analogues (compounds A-1, A-2, and A-3) were synthesized to potentially increase its stability. These three analogues inhibited viral gene expression like compound A; they could bind to ICP0 during infection and prevented its degradation following protein synthesis inhibition. Additional modifications are needed to further improve the biological activity and pharmacokinetic properties of compound A.
Overall, in this proof-of-concept study, we reported that we have developed a consistent and sensitive assay that can identify potential inhibitors of ICP0 E3 ligase activity with relatively high specificity. We also demonstrated that we have developed a protocol to identify the hits that physically associate with ICP0 and inhibit its function in a biologically relevant manner. Some of the scaffolds identified could be used to develop more potent inhibitors of the E3 ligase. E3 ligases do not contain a canonical active site, and developing highly selective and specific inhibitors is expected to be challenging. This is the first study that describes a small-molecule inhibitor of the HSV-1 ICP0 E3 ligase activity with potential antiviral function.
MATERIALS AND METHODS
Protein purification.
The procedures were described previously (61, 62). The pRB4994 (GST-ICP0 exon II) and pRB4995 (GST-ICP0 exon III-C) plasmids were described previously (63). GST-ICP0 exon II RF was developed from pRB4994 by site-directed mutagenesis to convert cysteine 116 and cysteine 156 to alanine. For protein purification, we used the E. coli BL21 strain, following procedures described elsewhere (53, 58). Eluted proteins were dialyzed against the buffer used for the HTS assay (50 mM Tris-HCl [pH 7.5], 2.5 mM MgCl2, 0.5 mM dithiothreitol, and 20% glycerol) and stored at −80°C until use.
HTS ICP0 autoubiquitination assay.
A CisBio HTRF-based assay was optimized in white, low-volume, 384-well microtiter plates to study autopolyubiquitination of GST-ICP0 exon II. The assay principle is based on recognition of polyubiquitin residues on GST-exon II by biotinylated polyubiquitin TUBEs. A FRET is facilitated when streptavidin-terbium, binding to biotinylated TUBEs, is brought close to d2-labeled anti-GST antibody, which recognizes the GST tag on the ICP0 exon II protein. The assay was developed using ubiquitin-conjugated UbcH5a enzyme and GST-exon II proteins. The enzymatically generated UbcH5a-ubiquitin thioester complex was purified to remove E1 ubiquitin-activating enzyme, uncharged UbcH5a, free ubiquitin, and Mg2+-ATP. Precharged UbcH5a was added directly to in vitro reactions containing ICP0 exon II in the absence of ATP, E1 enzyme, or extra ubiquitin. The optimal concentrations of UbcH5a-ubiquitin and exon II, as well as the detection reagents, were determined from titration of various concentrations of all reactants and reagents. The time of incubation, temperature, and DMSO sensitivity were also optimized. The optimized assay was performed by incubating GST-exon II protein (50 nM) and UbcH5a-ubiquitin protein (200 nM) in buffer A, containing 25 mM Tris-HCl (pH 7.5), 2 mM MgCl2, and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), for up to 90 min. The autoubiquitination of ICP0 was detected by addition of 1.0 nM biotinylated TUBE1 (LifeSensors Inc.) and HTRF detection reagents, 1 nM streptavidin-terbium (donor), and 40 nM anti-GST-d2 (acceptor) monoclonal antibody in detection buffer, in a final volume of 20 μl. After incubation for 1 h at room temperature, the plates were read at 665 and 620 nm with a BioTek Synergy Neo system. The HTRF ratio was calculated as 10,000 × (A665/A620). Percent inhibition was calculated and normalized to DMSO controls with all reaction components and reactions without GST-exon II.
HTS analysis.
The compounds (15 μM) were transferred acoustically to low-volume, 384-well plates (Corning) using an Echo 555 liquid handler (Labcyte Inc.). GST-exon II (50 nM) was preincubated with the compounds for 30 min at room temperature in buffer A (see above), followed by addition of UbcH5a-ubiquitin protein (200 nM). After the ubiquitination reaction at 30°C for 1 h, detection reagents containing biotin-TUBE, streptavidin-terbium, and anti-GST-d2 monoclonal antibody were added to the plates and incubated for 1 h at room temperature. The plates were read at 665 and 620 nm with a BioTek Synergy Neo system. The HTRF ratio and percent inhibition were calculated and normalized to values for DMSO controls with all reaction components and reactions without GST-exon II.
Cells and viruses.
HEL cells (immortalized human embryonic lung fibroblasts) were described previously (66). HEp-2 (human epithelial), U2OS (human osteosarcoma), and Vero (green monkey kidney epithelial) cells were cultured according to the supplier’s instructions (ATCC). HSV-1(F) is a limited-passage isolate described previously (64). The properties of the ΔICP0 virus and the ICP0 RF mutant virus were described elsewhere (47, 65).
Immunoblotting.
HEp-2 cells were infected with HSV-1(F) (5 PFU/cell) in the presence or absence of compounds. Cells were harvested at 8 h postinfection and analyzed by Western blotting as before (25, 61). The ICP0 mouse monoclonal antibody (Santa Cruz) was used at a 1:2,000 dilution. The USP7 mouse monoclonal antibody (Santa Cruz) was used at a 1:1,000 dilution. The ICP4 mouse monoclonal antibody and the UL38 rabbit polyclonal antibody were used at 1:5,000 dilutions and were kindly provided by Bernard Roizman (University of Chicago). The β-actin mouse monoclonal antibody (Sigma) was used at a 1:2,000 dilution. Protein bands were visualized with 5-bromo-4-chloro-3-indolyl phosphate (BCIP)-nitroblue tetrazolium (NBT) (Denville Scientific) or with enhanced chemiluminescence (ECL) Western blotting detection reagents (Amersham Biosciences).
RNA extraction and RT-qPCR analysis.
Cells were harvested in TRIzol reagent, and total RNA was extracted using phenol-chloroform. DNase treatment was performed using Turbo DNase enzyme (Ambion), according to the manufacturer’s instructions. cDNA synthesis was performed using an equal amount of RNA with the SuperScript III first-strand kit (Invitrogen), according to the supplier’s instructions. Reverse transcription (RT)-qPCR analyses were performed using SYBR green reagent (Invitrogen). The 18S rRNA primers (Ambion) were used for normalization. The primers to monitor ICP22, ICP27, and gI expression were described previously (66).
CETSA.
HEp-2 cells were infected with HSV-1(F) (2 PFU/cell) for 4 h, treated with the hits for 1 h, and harvested in PBS supplemented with protease inhibitor cocktail. Cells were aliquoted, subjected to transient heating from 40°C to 65°C for 3 min, and lysed in RIPA buffer (51). Samples were subjected to three freeze-thaw cycles using liquid nitrogen. Samples were centrifuged at 16,000 × g for 20 min at 4°C, and equal amounts of proteins in the supernatants were analyzed by SDS-PAGE (51). For HEL cells, compound A (20 μM) was added to the cultures for 1 h at 16 h postinfection.
Statistical analysis.
Prism 7 software (GraphPad) was used for statistical analysis of the qPCR data. P values were calculated using standard unpaired Student's t tests, and P values of ≤0.05 were considered significant. All statistical analyses were performed using biological replicates.
ACKNOWLEDGMENTS
We thank Bernard Roizman (University of Chicago) for kindly sharing the R7910 virus, the RF mutant virus, and the UL38 and ICP4 antibodies.
Research reported here was supported by the National Institute of General Medical Sciences of the National Institutes of Health (grant P20GM113117). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Antoine TE, Park PJ, Shukla D. 2013. Glycoprotein targeted therapeutics: a new era of anti-herpes simplex virus-1 therapeutics. Rev Med Virol 23:194–208. doi: 10.1002/rmv.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang YC, Feng H, Lin YC, Guo XR. 2016. New strategies against drug resistance to herpes simplex virus. Int J Oral Sci 8:1–6. doi: 10.1038/ijos.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frobert E, Ooka T, Cortay JC, Lina B, Thouvenot D, Morfin F. 2007. Resistance of herpes simplex virus type 1 to acyclovir: thymidine kinase gene mutagenesis study. Antiviral Res 73:147–150. doi: 10.1016/j.antiviral.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Frobert E, Cortay JC, Ooka T, Najioullah F, Thouvenot D, Lina B, Morfin F. 2008. Genotypic detection of acyclovir-resistant HSV-1: characterization of 67 ACV-sensitive and 14 ACV-resistant viruses. Antiviral Res 79:28–36. doi: 10.1016/j.antiviral.2008.01.153. [DOI] [PubMed] [Google Scholar]
- 6.Piret J, Boivin G. 2011. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob Agents Chemother 55:459–472. doi: 10.1128/AAC.00615-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz DH, Marcelletti JF, Khalil MH, Pope LE, Katz LR. 1991. Antiviral activity of 1-docosanol, an inhibitor of lipid-enveloped viruses including herpes simplex. Proc Natl Acad Sci U S A 88:10825–10829. doi: 10.1073/pnas.88.23.10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katz DH, Marcelletti JF, Pope LE, Khalil MH, Katz LR, McFadden R. 1994. n-Docosanol: broad spectrum anti-viral activity against lipid-enveloped viruses. Ann N Y Acad Sci 724:472–488. doi: 10.1111/j.1749-6632.1994.tb38949.x. [DOI] [PubMed] [Google Scholar]
- 9.Leung DT, Sacks SL. 2004. Docosanol: a topical antiviral for herpes labialis. Expert Opin Pharmacother 5:2567–2571. doi: 10.1517/14656566.5.12.2567. [DOI] [PubMed] [Google Scholar]
- 10.Pope LE, Marcelletti JF, Katz LR, Lin JY, Katz DH, Parish ML, Spear PG. 1998. The anti-herpes simplex virus activity of n-docosanol includes inhibition of the viral entry process. Antiviral Res 40:85–94. doi: 10.1016/S0166-3542(98)00048-5. [DOI] [PubMed] [Google Scholar]
- 11.Wilson SS, Fakioglu E, Herold BC. 2009. Novel approaches in fighting herpes simplex virus infections. Expert Rev Anti Infect Ther 7:559–568. doi: 10.1586/eri.09.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleymann G. 2003. Novel agents and strategies to treat herpes simplex virus infections. Expert Opinion Invest Drugs 12:165–183. doi: 10.1517/eoid.12.2.165.21401. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher C, Bean B. 1985. Evaluation of oral acyclovir therapy. Drug Intell Clin Pharm 19:518–524. doi: 10.1177/106002808501900703. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson MA. 1992. Review of the toxicities of foscarnet. J Acquir Immune Defic Syndr 5(Suppl 1):S11–S17. [PubMed] [Google Scholar]
- 15.Levin MJ, Bacon TH, Leary JJ. 2004. Resistance of herpes simplex virus infections to nucleoside analogues in HIV-infected patients. Clin Infect Dis 39(Suppl 5):S248–S257. doi: 10.1086/422364. [DOI] [PubMed] [Google Scholar]
- 16.Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol 78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roizman B. 2011. The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex. J Virol 85:7474–7482. doi: 10.1128/JVI.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roizman B, Whitley RJ. 2013. An inquiry into the molecular basis of HSV latency and reactivation. Annu Rev Microbiol 67:355–374. doi: 10.1146/annurev-micro-092412-155654. [DOI] [PubMed] [Google Scholar]
- 19.Cai W, Schaffer PA. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol 66:2904–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferenczy MW, Ranayhossaini DJ, DeLuca NA. 2011. Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J Virol 85:4993–5002. doi: 10.1128/JVI.02265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu H, Roizman B. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci U S A 104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalamvoki M, Roizman B. 2008. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc Natl Acad Sci U S A 105:20488–20493. doi: 10.1073/pnas.0810879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalamvoki M, Roizman B. 2010. Role of herpes simplex virus ICP0 in the transactivation of genes introduced by infection or transfection: a reappraisal. J Virol 84:4222–4228. doi: 10.1128/JVI.02585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalamvoki M, Roizman B. 2009. ICP0 enables and monitors the function of D cyclins in herpes simplex virus 1 infected cells. Proc Natl Acad Sci U S A 106:14576–14580. doi: 10.1073/pnas.0906905106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalamvoki M, Roizman B. 2010. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A 107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canning M, Boutell C, Parkinson J, Everett RD. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J Biol Chem 279:38160–38168. doi: 10.1074/jbc.M402885200. [DOI] [PubMed] [Google Scholar]
- 27.van Lint AL, Murawski MR, Goodbody RE, Severa M, Fitzgerald KA, Finberg RW, Knipe DM, Kurt-Jones EA. 2010. Herpes simplex virus immediate-early ICP0 protein inhibits Toll-like receptor 2-dependent inflammatory responses and NF-κB signaling. J Virol 84:10802–10811. doi: 10.1128/JVI.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu H, Roizman B. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci U S A 100:8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Wang K, Wang S, Zheng C. 2013. Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-κB activation by interacting with p65/RelA and p50/NF-κB1. J Virol 87:12935–12948. doi: 10.1128/JVI.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boutell C, Canning M, Orr A, Everett RD. 2005. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J Virol 79:12342–12354. doi: 10.1128/JVI.79.19.12342-12354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalamvoki M, Gu H, Roizman B. 2012. Overexpression of the ubiquitin-specific protease 7 resulting from transfection or mutations in the ICP0 binding site accelerates rather than depresses herpes simplex virus 1 gene expression. J Virol 86:12871–12878. doi: 10.1128/JVI.01981-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halford WP, Schaffer PA. 2000. Optimized viral dose and transient immunosuppression enable herpes simplex virus ICP0-null mutants to establish wild-type levels of latency in vivo. J Virol 74:5957–5967. doi: 10.1128/JVI.74.13.5957-5967.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halford WP, Kemp CD, Isler JA, Davido DJ, Schaffer PA. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J Virol 75:6143–6153. doi: 10.1128/JVI.75.13.6143-6153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halford WP, Puschel R, Rakowski B. 2010. Herpes simplex virus 2 ICP0 mutant viruses are avirulent and immunogenic: implications for a genital herpes vaccine. PLoS One 5:e12251. doi: 10.1371/journal.pone.0012251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Sant C, Kawaguchi Y, Roizman B. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci U S A 96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilcox CL, Smith RL, Everett RD, Mysofski D. 1997. The herpes simplex virus type 1 immediate-early protein ICP0 is necessary for the efficient establishment of latent infection. J Virol 71:6777–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khrustalev VV, Barkovsky EV. 2008. An in-silico study of alphaherpesviruses ICP0 genes: positive selection or strong mutational GC-pressure? IUBMB Life 60:456–460. doi: 10.1002/iub.55. [DOI] [PubMed] [Google Scholar]
- 39.Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol 74:10006–10017. doi: 10.1128/JVI.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preston CM, Nicholl MJ. 2005. Human cytomegalovirus tegument protein pp71 directs long-term gene expression from quiescent herpes simplex virus genomes. J Virol 79:525–535. doi: 10.1128/JVI.79.1.525-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Everett RD, Boutell C, McNair C, Grant L, Orr A. 2010. Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J Virol 84:3476–3487. doi: 10.1128/JVI.02544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everett RD, Bell AJ, Lu Y, Orr A. 2013. The replication defect of ICP0-null mutant herpes simplex virus 1 can be largely complemented by the combined activities of human cytomegalovirus proteins IE1 and pp71. J Virol 87:978–990. doi: 10.1128/JVI.01103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adamson AL, Kenney S. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol 75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walters MS, Kyratsous CA, Silverstein SJ. 2010. The RING finger domain of varicella-zoster virus ORF61p has E3 ubiquitin ligase activity that is essential for efficient autoubiquitination and dispersion of Sp100-containing nuclear bodies. J Virol 84:6861–6865. doi: 10.1128/JVI.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boutell C, Sadis S, Everett RD. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol 76:841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagglund R, Van Sant C, Lopez P, Roizman B. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A 99:631–636. doi: 10.1073/pnas.022531599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lium EK, Silverstein S. 1997. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J Virol 71:8602–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol 70:7471–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prisinzano TE. 2015. The importance of molecular design principles in delivering high quality pharmaceutical candidates, p 177–191. In Templeton A, Byrn S, Haskell R, Prisinzano T (ed), Discovering and developing molecules with optimal drug-like properties. Springer, New York, NY. [Google Scholar]
- 50.Olson ME, Abate-Pella D, Perkins AL, Li M, Carpenter MA, Rathore A, Harris RS, Harki DA. 2015. Oxidative reactivities of 2-furylquinolines: ubiquitous scaffolds in common high-throughput screening libraries. J Med Chem 58:7419–7430. doi: 10.1021/acs.jmedchem.5b00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, Martinez MD. 2014. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 52.Roizman B, Taddeo B. 2007. The strategy of herpes simplex virus replication and takeover of the host cell, p 163–176. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, UK. [PubMed] [Google Scholar]
- 53.Cai W, Astor TL, Liptak LM, Cho C, Coen DM, Schaffer PA. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol 67:7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grant K, Grant L, Tong L, Boutell C. 2012. Depletion of intracellular zinc inhibits the ubiquitin ligase activity of viral regulatory protein ICP0 and restricts herpes simplex virus 1 replication in cell culture. J Virol 86:4029–4033. doi: 10.1128/JVI.06962-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu H, Roizman B. 2009. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J Virol 83:181–187. doi: 10.1128/JVI.01940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deschamps T, Dogrammatzis C, Mullick R, Kalamvoki M. 2017. Cbl E3 ligase mediates the removal of nectin-1 from the surface of herpes simplex virus 1-infected cells. J Virol 91:e00393-17. doi: 10.1128/JVI.00393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boutell C, Davido DJ. 2015. A quantitative assay to monitor HSV-1 ICP0 ubiquitin ligase activity in vitro. Methods 90:3–7. doi: 10.1016/j.ymeth.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanni E, Gatherer D, Tong L, Everett RD, Boutell C. 2012. Functional characterization of residues required for the herpes simplex virus 1 E3 ubiquitin ligase ICP0 to interact with the cellular E2 ubiquitin-conjugating enzyme UBE2D1 (UbcH5a). J Virol 86:6323–6333. doi: 10.1128/JVI.07210-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu H, Poon AP, Roizman B. 2009. During its nuclear phase the multifunctional regulatory protein ICP0 undergoes proteolytic cleavage characteristic of polyproteins. Proc Natl Acad Sci U S A 106:19132–19137. doi: 10.1073/pnas.0910920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deschamps T, Kalamvoki M. 2017. Impaired STING pathway in human osteosarcoma U2OS cells contributes to the growth of ICP0-null mutant herpes simplex virus. J Virol 91:e00006-17. doi: 10.1128/JVI.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalamvoki M, Roizman B. 2010. Interwoven roles of cyclin D3 and cdk4 recruited by ICP0 and ICP4 in the expression of herpes simplex virus genes. J Virol 84:9709–9717. doi: 10.1128/JVI.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalamvoki M, Roizman B. 2011. The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J Virol 85:9472–9477. doi: 10.1128/JVI.00876-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawaguchi Y, Bruni R, Roizman B. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol 71:1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 65.Kawaguchi Y, Van Sant C, Roizman B. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol 71:7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deschamps T, Kalamvoki M. 2017. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J Virol 91:e00535-17. doi: 10.1128/JVI.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]