Abstract
Histone deacetylases (HDACs) are known to be key enzymes in cancer development and progression through their modulation of chromatin structure and the expression and post-translational modification of numerous proteins. Aggressive dedifferentiated tumors, like glioblastoma, frequently overexpress HDACs, while HDAC inhibition can lead to cell cycle arrest, promote cellular differentiation and induce apoptosis. Although multiple HDAC inhibitors, such as quisinostat, are of interest in oncology due to their potent in vitro efficacy, their failure in the clinic as monotherapies against solid tumors has been attributed to poor delivery. Thus, we were motivated to develop quisinostat loaded poly(D,L-lactide)-b-methoxy poly(ethylene glycol) nanoparticles (NPs) to test their ability to treat orthotopic glioblastoma. In developing our NP formulation, we identified a novel, pH-driven approach for achieving over 9% (w/w) quisinostat loading. We show quisinostat-loaded NPs maintain drug potency in vitro and effectively slow tumor growth in vivo, leading to a prolonged survival compared to control mice.
Keywords: Glioblastoma, Nanoparticle, HDAC, Quisinostat (JNJ-26481585), PLA-PEG, pH
1. Introduction
Recent advances have highlighted the role of epigenetic aberrations in the development and progression of many cancer types, including glioblastoma (GBM) [1–6]. Histone deacetylases (HDACs) are a class of enzymes capable of producing epigenetic modification of cellular behavior. HDACs are responsible for the deacetylation of lysine residues on histones to regulate chromatin structure, transcription factor binding sites and gene expression, and their overexpression has been observed in dedifferentiated, aggressively proliferating tumors [7–11]. Importantly, molecules that inhibit HDACs (HDAC inhibitors, HDIs) are capable of producing apoptosis and cell cycle arrest, and they also sensitize cells to conventional DNA damaging treatments [12–16]. Currently, three first-generation HDIs are clinically approved for cutaneous T-cell lymphoma [17]; however, despite promising preclinical efficacy of first generation HDIs both in vitro and in vivo, clinical trials of HDIs have failed to show treatment benefits in solid tumors. It has been proposed that inadequate delivery and short biological half-life of most HDIs contribute to their underwhelming in vivo efficacy [18,19]. Second generation HDIs, like quisinostat, were designed and shown to be significantly more selective and potent against class I HDACs with a longer duration of action compared to first generation HDIs, but these agents still failed to show significant efficacy as a monotherapy against solid tumors, presumably due to poor tumor delivery [18,20].
We have shown in previous work polymeric nanoparticles (NPs) can effectively encapsulate poorly water soluble small molecules to improve their tolerability in vivo and delivery to intracranial GL261 GBM tumors, which enables effective treatment of tumors after intravenous administration [21]. Importantly, Wang et al. showed the encapsulation of quisinostat within PLGA-lecithin-PEG core-shell NPs potentiated the effects of radiation in subcutaneous PC3 tumors more effectively than free drug [22]. Thus, the goal of this work was to develop a formulation process that would effectively encapsulate quisinostat in NPs composed of PLA-PEG and to test whether encapsulated quisinostat would be capable of treating orthotopic GBM. Through the process of developing this drug carrier, we identified a novel, pH-driven approach for achieving high quisinostat loading. In contrast to traditional methods that improve drug encapsulation by decreasing the aqueous solubility of the drug to drive it into the polymer core, our novel method achieves high loading by improving the solubility of quisinostat in the aqueous phase prior to solvent evaporation.
2. Materials and methods
2.1. Materials
Quisinostat (JNJ-26481585) was obtained from APExBio (Houston, TX USA). Poly(d,l-lactide)-b-methoxy poly(ethylene glycol) (PLA-PEG, Mw ~5:16 kDa) was purchased from PolySciTech (West Lafayette, IN USA). Endotoxin free (<0.0050 EU/ml) water from G-Biosciences (St. Louis, MO USA) was used throughout nanoparticle fabrication. Dimethyl sulfoxide (DMSO), dichloromethane (DCM), sodium cholate, 1× phosphate buffered saline (PBS), hydrochloric acid (HCl, 0.1001 M) and sodium hydroxide (NaOH, 0.1001 M) were all purchased from Sigma-Aldrich (St. Louis, MO USA). Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS), 0.25% trypsin-EDTA and geneticin selective antibiotic (G-418) were purchased from Gibco Invitrogen (Carlsbad, CA, USA). Greiner T25 tissue culture flasks with filter cap and Costar 96-well assay plates were purchased from VWR International (Radnor, PA, USA). Beetle luciferin (potassium salt) and CellTiter-Glo Luminescent Cell Viability Assay were purchased from Promega (Madison, WI, UAS).
2.2. Nanoparticle fabrication
Nanoparticles were produced by a modified single emulsion-solvent evaporation approach, as previously reported [21,23,24]. 50 mg PLA-PEG dissolved in 2 ml DCM was added dropwise into 4 ml of 1% (w/v) sodium cholate while vortexing, then probe sonicated (Fisher Scientific Model 705 Sonic Dismembrator, Waltham, MA USA) on ice in 3, 10-s bursts at 40% amplitude. The resulting emulsion was added to an evaporation phase consisting of 20 ml of 0.3% (w/v) sodium cholate and allowed to stir for 3 h to evaporate the DCM. Drug loaded nanoparticles were produced by adding 5 mg quisinostat, dissolved in 300 μl DMSO, dropwise into the organic phase or the evaporation phase, as specified for each formulation in Table 1. For nanoparticles made under basic or acidic conditions, the pH of the 0.3% sodium cholate evaporation phase was adjusted to the specified pH by adding dilute (0.1 M) NaOH or HCl. After the 3 h, nanoparticles were washed and concentrated through Amicon Ultra-15 Centrifugal Filters (100 kDa cut-off) for 4, 20 min spins at 5000 RCF. Aliquots were frozen and lyophilized to determine nanoparticle concentration and drug loading. The rest of the nanoparticles were frozen and stored at −80 °C.
Table 1.
Summary of QNP formulation parameters and characterization.
| Formulation | Quisinostat added (mg) | O/Ea | pHb | % Loading (w/w) | Diameter (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|---|---|---|---|
| BNP | 0 | – | 7 | – | 96.3 ± 2.08 | 0.1 ± 0.01 | –13 ± 2.0 |
| QNP-1 | 5 | O | 7 | 1.3 ± 0.71 | 101 ± 2.52 | 0.1 ± 0.01 | −4.9 ± 2.3 |
| QNP-2 | 5 | O | 2 | 0.47 ± 0.25 | 103 ± 2.08 | 0.1 ± 0.01 | −6.2 ± 1.7 |
| QNP-3 | 5 | O | 10 | 5.0 ± 0.51 | 113 ± 10.0 | 0.1 ± 0.01 | −8.2 ± 0.53 |
| QNP-4 | 5 | E | 10 | 9.3 ± 0.29 | 128 ± 8.50 | 0.1 ± 0.02 | −6.0 ± 1.0 |
| QNP-5 | 5 | E | 7 | 2.7 ± 0.15 | 112 ± 2.52 | 0.1 ± 0.01 | −5.2 ± 1.7 |
| QNP-6 | 7.5 | E | 10 | 9.9 ± 0.21 | 129 ± 4.51 | 0.1 ± 0.02 | −9.8 ± 1.1 |
| QNP-7 | 10 | E | 10 | 7.7 ± 0.35 | 121 ± 2.31 | 0.1 ± 0.02 | −9.4 ± 2.1 |
| QNP-8 | 5 | E | 9 | 5.3 ± 0.12 | 115 ± 5.29 | 0.1 ± 0.01 | −8.2 ± 1.4 |
| QNP-9 | 7.5 | E | 11 | 8.9 ± 0.60 | 126 ± 3.21 | 0.1 ± 0.01 | −7.8 ± 1.6 |
(O)rganic/(E)vaporation specifies to which phase the quisinostat was added.
pH of the evaporation phase. Mean ± SD reported for 3 batches of each formulation.
2.3. Nanoparticle characterization
2.3.1. Drug loading
Drug loading was quantified by absorbance (300 nm) on a Tecan plate reader. Lyophilized nanoparticles were dissolved at 5 mg/ml in DMSO. Samples were plated in triplicate (40 μl nanoparticles and 10 μl DMSO per well) in a clear, flat bottom 96-well assay plate. A control curve was constructed in technical triplicate by adding 40 μl blank nanoparticles per well and spiking with 10 μl of known drug concentrations in DMSO. Quisinostat loading was calculated as mass quisinostat/mass polymer (w/w%).
2.3.2. Size and zeta potential
Nanoparticle hydrodynamic diameter and zeta potential were measured using the NanoBrook 90Plus Zeta (Brookhaven Instruments, Holtsville, NY USA). All measurements were done at a nanoparticle concentration of 0.1 mg/ml in triple filtered 1 mM KCl.
2.3.3. Transmission electron microscopy
Transmission electron microscopy (TEM) measurements were measured on the Phillips CM 12 operated at an accelerated voltage of 120 kV using 400 mesh formvar-coated copper grids FCF400-Cu-SB (Electron Microscopy Sciences, PA, USA). Copper-grids were first glow-discharged to increase hydrophilicity on the surface. Samples were then diluted with DI water (final concentration 4 mg/ml). Samples were prepared by pipetting 3 μl of diluted solution to the glow-discharged grids followed by ambient drying using Whatman Filter Paper (Sigma Aldrich, USA).
2.3.4. Controlled release
Quisinostat release from nanoparticles was determined using a protocol adapted from Wang et al. [22]. Nanoparticles were diluted to 20 mg/ml in PBS (pH 7) and 400 μl was transferred to a 3.5 k MWCO Slide-A-Lyzer Dialysis cassette (Thermo Fisher Scientific, Waltham, MA USA) in triplicate. Each cassette was immersed in 2 l PBS (pH 7, replaced at each time point) at 37 °C with gentle stirring (100 rpm). At each time point, 30 μl nanoparticles was removed from the cassette and dissolved in 150 μl DMSO. 60 μl dissolved nanoparticles was added in triplicate to a clear, flat bottom, 96-well plate, and the amount of drug remaining was quantified by absorbance as described in section 2.3.1. A free quisinostat control at the equivalent concentration was included to measure quisinostat movement across the membrane using the same protocol.
2.4. Cell culture
GL261-LucNeo cells were generated by retroviral transduction of parent GL261 cells. The LucNeo construct (obtained from Andrew Kung laboratory, Dana-Farber Cancer Institute) is described in Rubin et al. [25]. Cells were maintained under normal adherent culture conditions supplemented with G-418 as a selection pressure. Cells were grown in T25 flasks in DMEM containing glucose, L-glutamine and 10% FBS at 37 °C and 5% CO2. 0.25% trypsin-EDTA was applied to collect cells and a Cellometer mini (Nexcelom Bioscience, Lawrence, MA USA) was used to count cells prior to all in vitro and in vivo experiments.
2.5. In vitro nanoparticle efficacy
GL261 cells were seeded in 96-well flat, white walled, clear bottom plates at a density of 3k cells/well in 100 μl media and allowed to attach for 4 h prior to adding treatments. Each plate was treated with 10 μl/well of 19 serial dilutions (1:2) ranging from 10 to 0 μM in PBS of either free drug or nanoparticles. After 72 h, cell viability was assessed using CellTiter-Glo, and an IC50 value was calculated in GraphPad Prism (San Diego, CA USA) by a nonlinear fit of the log (inhibitor) vs. response function.
2.6. In vivo tumor treatment
All procedures and animal care practices were performed in accordance with the Barrow Neurological Institute’s Institutional Animal Care and Use Committee.
2.6.1. Tumor induction
Orthotopic GL261-LucNeo tumors were induced in C57BL/6 albino mice (Harlan Laboratories, Indianapolis, IN, USA) as previously reported [23,21]. Briefly, mice were anesthetized with an intraperitoneal injection of ketamine/xylazine (100/10 mg/kg) and mounted in a stereotaxic frame (Kopf Instruments, Tujunga, CA, USA) on top of an infrared heating pad to maintain animal temperature. The animal’s head was shaved and sterilized with three alternating passes each of betadine and ethanol. A 1 cm incision was made over midline, and a burr hole was drilled 2 mm lateral, 0.1 mm posterior of bregma. A hamiltion syringe (29 gauge needle) containing 75k GL261-LucNeo cells in 2 μl DMEM was inserted into the hole to a depth of 2.8 mm and the cells were injected over 2 min. The needle was left in place for 1 min to reduce backflow before the wound was closed with staples. All animals received a subcutaneous (SQ) injection of Buprenorphine SR prior to surgery, and ibuprofen was provided in their water ad lib for 1 week for pain.
2.6.2. Tumor growth
Bioluminescence was used to monitor and measure tumor growth as previously described [21,23]. Imaging was done on the Xenogen IVIS Spectrum in vivo imaging system every 3–4 days starting at day 6 after tumor implantation. Mice received a SQ injection of luciferin (150 mg/kg) and were imaged 25 min post injection under 2% isoflurane. The Living Image software was used to draw an ROI around the tumor signal and measure the size of each tumor (total flux, photons/sec).
2.6.3. Tumor treatment
Quisinostat-loaded nanoparticles were tested in vivo in mice bearing orthotopic GL261 tumors. After the first imaging, mice were randomly assigned to a treatment group. For the free drug study, this included saline control (100 ul) or free quisinostat (10 mg/kg IP, solubilized in 20% hydroxy-propyl-β-cyclodextrin, pH 8.7). For the nanoparticle drug study, this included saline (100 μl), blank nanoparticles (BNP, 1000 mg/kg polymer), or quisinostat-loaded nanoparticles (QNP, 50 mg/kg quisinostat). One mouse in the nanoparticle study was excluded for lack of a tumor signal at the initial imaging. Mice were treated by intravenous injection (lateral tail vein) on days 11, 12, 18, and 19 post tumor induction. Treatment efficacy was measured by tumor growth, and median survival. Mice were monitored daily and euthanized at the sign of symptoms (lack of grooming, abnormal gait, hunched posture etc.) or greater than 15% weight loss.
2.7. Statistics
All statistical tests were performed using the GraphPad Prism 5 software. Tumor growth for each treatment was compared by fitting the average growth with an exponential curve fit and comparing treatments using a one-way ANOVA. Survival differences were compared using a Kaplan-Meier curve and the Mantel-Cox test.
3. Results
3.1. Nanoparticle loading and characterization
NPs produced from amphiphilic polymers such as PLA-PEG possess a hydrophobic core, which is utilized as a favorable environment for the encapsulation of water-insoluble small molecules [26,27]. Our initial attempts to encapsulate quisinostat in PLA-PEG NPs followed a standard single emulsion-solvent evaporation technique under neutral conditions. Quisinostat loaded NPs (QNPs) formed effectively; however, a relatively poor loading of 1.3% (Table 1, QNP-1) was achieved, which is comparable with prior reports of 2.3% (w/w) quisinostat encapsulation within PLGA-lecithin-PEG core-shell NPs [22]. We attempted to improve loading by varying a number of traditional formulation parameters known to affect drug loading (solvent mixtures (acetonitrile, dimethylformamide, acetone, DMSO, DCM, ethyl acetate), nanoprecipitation, feed ratios and temperature) [28,29]; however, none of these changes brought quisinostat loading above 2% (data not shown).
In an emulsion based approach to NP formation, a hydrophobic drug is typically dissolved with the polymer in an organic solvent to aid in the encapsulation of the drug during NP formation, followed by evaporation of the solvent. The final loading of drug within the NP is thought to be determined by diffusion of drug out of the polymer core after NP formation, which is directly related to the solubility of the drug in the aqueous phase. Thus, one approach for improving loading of drug within NPs formed by emulsion is to fabricate particles under conditions that reduce drug solubility in the water phase, which is believed to drive partitioning of drug into the particle core. [28,30,31] Because quisinostat exhibits increased water solubility at a basic pH, we hypothesized that acidifying the evaporation phase to pH 2 would increase quisinostat loading. However, we observed drug loading under acidic conditions significantly decreased compared to NPs produced under neutral conditions to 0.47% (QNP-2). As a negative control, we also tested the effect of raising the evaporation phase pH to 10. Interestingly, a basic evaporation pH resulted in significantly higher loading compared to NP produced at pH 2 or 7, achieving a loading of 5.0% (QNP-3).
The observation that loading improves when quisinostat’s aqueous solubility is increased suggests a loading mechanism that does not rely solely on hydrophobic interactions. Under basic conditions, quisinostat is expected to possess a negative charge due to deprotonation of the hydroxamic acid group, suggesting an ionic mediated loading mechanism. Since we were only altering the pH after NP formation, the ionization could either enable quisinostat retention within the core of the solid NP and/or increase the stability of quisinostat at the water-polymer interface. To test whether quisinostat could be associating with the surface of the NP (as opposed to the core), we generated blank (no drug) NPs in the primary emulsion and added quisinostat directly to the evaporation phase under basic conditions (pH 10). This formulation condition nearly doubled the effective drug incorporation over our prior attempts, achieving a quisinostat loading of 9.3% (QNP-4). Further increases to the mass of quisinostat added to the aqueous phase, from 5 mg to 7.5 or 10 mg, did not result in increased loading (QNP-6 and QNP-7) even at a higher pH (pH 11, QNP −9), supporting a saturable association of drug with the surface of the NP. Formulations at a pH 7 or pH 9, while following an identical post-loading procedure, NP loading dropped to 2.7% (QNP-5) and 5.3% (QNP-8), respectively. When we pre-evaporated the organic phase (DCM) prior to addition of quisinostat, with or without pH change, NP loading dropped to <3% (data not shown). Thus, the highest effective loading of quisinostat (QNP-4) requires the deprotonation of quisinostat at a pH above 10 and can be achieved after NPs are formed but only in the presence of organic solvent. The increase in quisinostat loading as pH increases up to pH 10 with no increase seen at pH 11 supports an ionic association with the full ionization of quisinostat occurring between pH 9 and 10.
One experimental concern is whether the loading measured in these experiments could reflect drug precipitates instead of NP-associated drug; we offer three pieces of evidence that contradict this possibility. First, the optical quality of the emulsion is characteristic of ultra-small polymeric nanoparticles, possessing a translucent/blue hue that is not observed when drug precipitates [32]. Second, TEM characterization does not show drug precipitates (Fig. S1). Third, when PLA-PEG was excluded but post-loading fabrication conditions otherwise maintained, only 10ug of quisinostat was recovered.
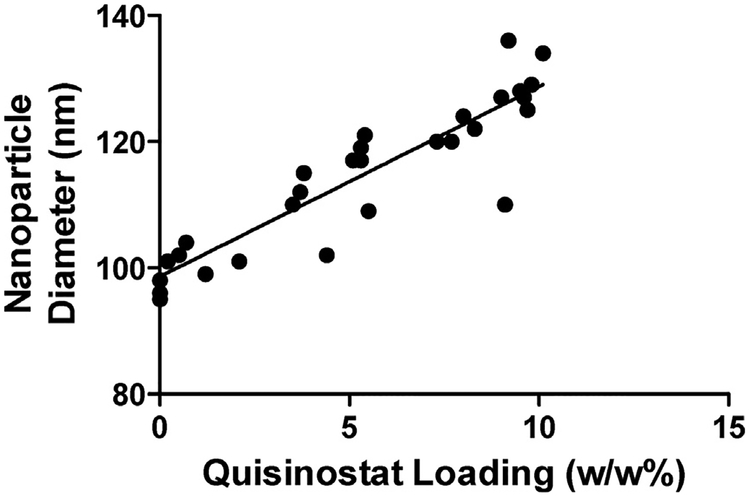
Each NP formulation was also characterized by DLS to measure size and zeta potential. BNPs formed by our standard technique (neutral pH evaporation phase) possessed an average diameter of 96 nm and a zeta potential of −13 mV (Table 1). Alterations to the evaporation phase pH did not significantly alter the biophysical properties of BNPs (data not shown). The presence of quisinostat resulted in NPs with a slightly more neutral surface charge compared to NPs lacking quisinostat, but the amount of quisinostat loaded did not significantly affect the surface charge across QNP formulations. In contrast, the measured NP diameters positively correlated with quisinostat loading, with the average diameter increasing to 129 nm for the formulation with the highest loading (Fig. 1). This phenomenon is consistent with previous reports showing increased NP diameter when drugs are loaded onto the surface of polymeric NPs [33,34]. These observations further support the drug loading measured represents NP-associated quisinostat, as opposed to precipitated drug.
Fig. 1. Nanoparticle size to quisinostat loading correlation.
Nanoparticle’ hydrodynamic diameter, as measured by DLS, positively correlated (Pearson coefficient = 0.9108, p < 0.0001) with the quisinostat loading for each batch. Each data point represents an individual batch.
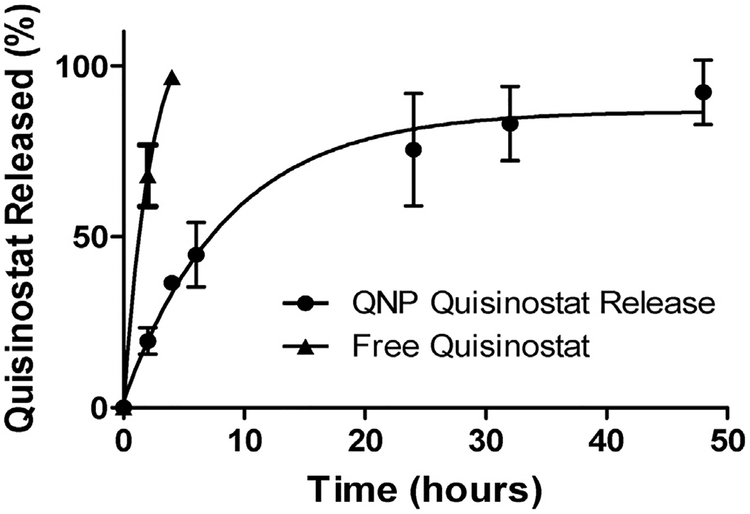
Quisinostat release from QNPs or as free drug at 37 °C in PBS was measured by absorbance after 1, 2, 4, 6, 24, 32 and 48 h. Free quisinostat was completely released from the dialysis cassette by 4 h, whereas only 50% of quisinostat was released from NPs after 6 h, and complete NP release achieved by 48 h (Fig. 2). The fast rate of release from PLA-PEG NPs is in contrast to the 5 days of sustained release previously reported for quisinostat encapsulated within the core of PLGA-lipid hybrid NPs [22]. A rapid burst release supports surface loading of quisinostat [34,35], and the subsequent phase of sustained release is presumably due to electrostatic interactions with the particle, which have previously been demonstrated to enable the sustained release of proteins from PLGA NPs, even in absence of encapsulation [36]. It remains to be determined whether quisinostat resides within the hydrated PEG layer or is within the PLA polymer phase and merely close to the surface. It is not immediately clear that the burst release we observed is a problem for quisinostat drug delivery, since NPs typically distribute and clear over similar time frames to the release kinetics observed here [37,38].
Fig. 2. In vitro quisinostat release from QNPs.
QNPs released quisinostat into PBS at 37 °C over 48 h, with nearly 50% release occurring in the first 6 h. Free quisinostat was completely released from the cassette within 4 h. Points and error bars represent the mean ± SD of 3 samples read in triplicate at each time point.
Our data demonstrate that the pH of the aqueous phase is a major force driving quisinostat loading into or onto PLA-PEG NPs formed by emulsion, and we suggest that the mechanism is charge-mediated. We propose the deprotonation of quisinostat under basic conditions increases NP loading due to electrostatic interactions; presumably, the presence of the organic solvent is required to achieve this because it enhances overall solubility of the drug to enable this interaction. Previous works have described the loading of drugs and proteins onto the surface of inorganic [39,40] and polymeric [34–36] NPs; these effects have been reported to be a function of charge interactions, [34,35,36,37] and their pH-dependency supports ionization as a primary mechanism [34,36,39,40]. Additionally, a charge-dependent loading of proteins onto the surface of PLGA has been demonstrated in a post-fabrication scheme [36]. However, to our knowledge, similar approaches have not yet been demonstrated for loading small molecules on PLA-PEG, and have also not been reported for HDIs.
3.2. QNP activity and efficacy
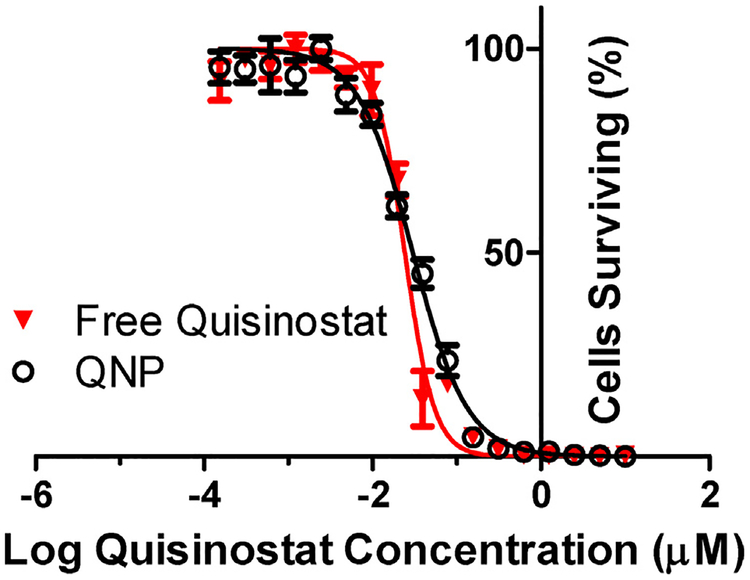
To test whether quisinostat potency is maintained after NP loading, we evaluated growth inhibition produced by free versus NP quisinostat in vitro, in GL261 cultures. Both free and NP-loaded quisinostat effectively inhibited the growth of GL261 cells with IC50 values of 24 and 30 nM, respectively (Fig. 3). We found no significant changes in quisinostat potency due to the NP loading process, and the low nanomolar IC50 is consistent with reported quisinostat IC50 values against other glioblastoma cell lines [19].
Fig. 3. In vitro QNP efficacy against GL261.
QNP and free quisinostat exhibited equipotent growth inhibition against GL261 murine glioma cells in vitro with IC50 s of 30 and 24 nM, respectively. Points and error bars represent the mean ± SD of 3 samples read in triplicate at each dilution.
Multiple investigators have identified HDAC inhibitors as drugs of interest for treating cancer, including GBM [1,8,17,41–43]. While in vitro results have been promising, little success has been observed in vivo [20,44,45]. As a monotherapy, quisinostat and other HDIs have shown the greatest in vivo efficacy against hematological cancers [17,19,46]. Against solid tumors, HDIs are most commonly utilized as a combination therapy to achieve efficacy [6,20,22,47]. Although the mechanism for the in vivo failure of quisinostat or other HDIs as a monotherapy is unknown, it has been suggested that poor delivery may be a factor. NPs have the potential to improve in vivo efficacy of systemically administered agents through a variety of mechanisms, including improved solubility (enabling a higher dose to be delivered), enhanced permeation and retention (EPR) in leaky tumor vasculature, and/or alteration to pharmacokinetic profile of free drug. For example, in previous work, we utilized a NP encapsulation strategy to deliver the otherwise ineffective drug camptothecin (CPT) to intracranial GBM [21]. CPT is a potent drug in cell culture but is very poorly water soluble, inactivated at physiological pH, and cleared rapidly following systemic administration. Encapsulation of CPT within poly(lactic-co-glycolic acid) (PLGA) NPs improved drug tolerability dramatically, which produced a robust slowing of tumor growth and prolongation of survival in mice bearing intracranial tumors. Based on this previous work, we predicted that NP encapsulation would provide a similar benefit to the action of quisinostat.
Prior works using hydroxy-propyl-β-cyclodextrin and/or man-nitol to solubilize quisinostat for injection report the maximum tolerated dose to be in the range of 35–70 mg/kg/week when administered by IP or SQ injection [18,19,46]. In our studies, mice did not show significant weight loss at QNP doses up to 100 mg quisinostat/kg/week IV (Fig. S2), suggesting an improvement in quisinostat tolerability after NP encapsulation. Quisinostat has previously shown efficacy against subcutaneous GBM xenografts [19], which confirms quisinostat demonstrates expected activity against GBM but does not address delivery barriers related to orthotopic tumors. Treatment of orthotopic GBM is significantly hindered by the blood-brain barrier (BBB), which presents both active and passive barriers to restrict the entry of chemotherapies [48,49]. Nearly all drugs of interest for GBM fail to achieve adequate tumor concentrations at a safe dose [50]. Thus, the inability of subcutaneous tumors to recapitulate these unique drug delivery challenges makes intracranial GBM models necessary for evaluating treatment efficacy.
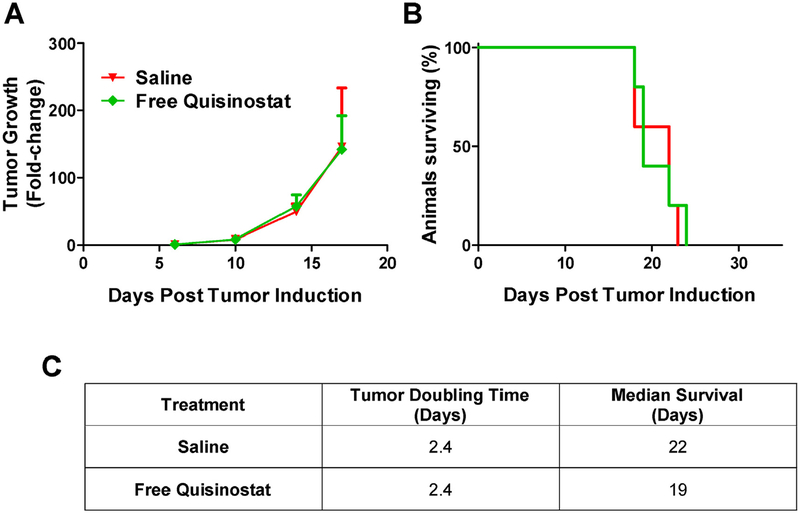
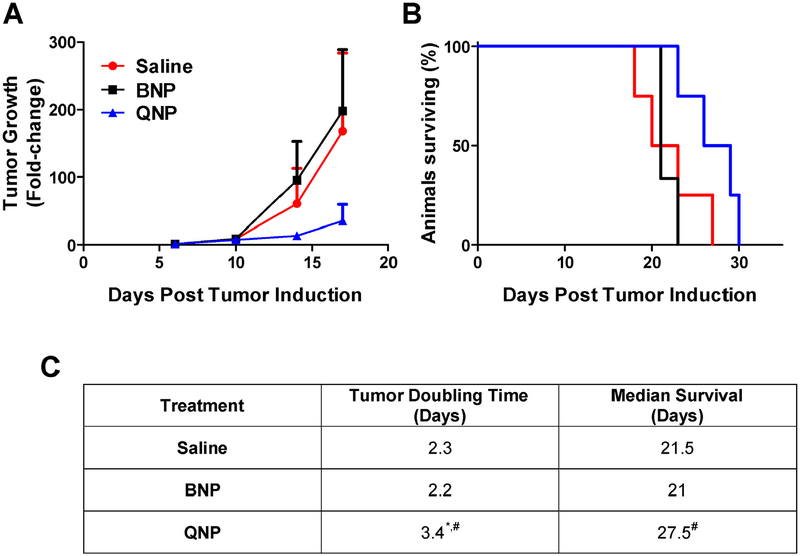
To test whether free quisinostat could treat an orthotopic tumor, intracranial GL261-LucNeo tumors were induced in 10 C57BL/6 albino mice and treated with either saline or free quisinostat (n = 5/group) by IP injection on days 11, 12, 18 and 19. Free quisinostat failed to provide any treatment benefit with a tumor doubling time of 2.4 days for both treatment and a median survival of 22 and 19 days for saline and quisinostat, respectively (Fig. 4). In a separate cohort of 12 mice bearing intracranial tumors, the subjects were divided into 3 treatment groups (saline, BNPs or QNPs) and treatments were administered IV by lateral tail vein injection on days 11, 12, 18 and 19. Tumor growth was exponential in both saline and BNP treated mice with an average tumor doubling time of 2.3 and 2.2 days, respectively, while QNPs significantly (p < 0.05) slowed the tumor doubling time, to 3.4 days (Fig. 5). This delay in tumor growth resulted in a significant increase in median survival to 27.5 days for QNP treated compared to 21 days for those treated with BNPs (p = 0.03) and tended to prolong survival compared to the 21.5 days for saline treated mice (p = 0.10). Although a modest improvement in survival, these data show NP encapsulation of quisinostat can improve its tolerability and efficacy over free drug to effectively slow intracranial GBM growth as a monotherapy.
Fig. 4. Free quisinostat treatment efficacy in mice bearing orthotopic GL261 tumors.
(A) Tumor growth was determined by the change in tumor size (mean ± SD) from day 6, as measured by bioluminescence. (B) Survival is shown on the Kaplan-Meier plot. (C) Saline (n = 5) and Free Quisinostat (n = 5) treated tumors both doubled in size every 2.4 days and had median survival times of 22 and 19 days, respectively.
Fig. 5. QNP in vivo treatment efficacy in mice bearing orthotopic GL261 tumors.
(A) Tumor growth determined by the change in tumor size (mean ± SD) from day 6 as measured by bioluminescence. (B) Survival is shown on the Kaplan-Meier plot. (C) Saline (n = 4) and BNP (n = 3) treated tumors grew exponentially and had median survival times of 21.5 and 21 days, respectively. QNP (n = 4) treatment significantly slowed tumor doubling compared to both controls, leading to a significantly prolonged survival of 27.5 days compared to BNP treatment. # designates significance (p < 0.05) compared to BNP. * designates significance (p < 0.05) compared to saline. Statistical testing on tumor doubling time was performed with a one-way ANOVA followed by Tukey post-hoc testing. Statistical testing on survival was performed by the Mantel-Cox test.
4. Conclusions
This study presents a novel pH driven approach for achieving high quisinostat loading of PLA-PEG NPs, ~9% (w/w), after NP formation. In contrast to the typical approach of reducing drug solubility in the aqueous phase to drive partitioning of drug into the NP core, our data show that quisinostat loading increases as its aqueous solubility increases, which we suggest is due to a charge-mediated association of drug with the nanoparticle surface. QNPs produced by these methods effectively release drug over 48 h and possess equivalent activity to free drug in vitro. Additionally, QNPs were found to robustly slow orthotopic GL261 tumor growth and prolong survival compared to control treated mice. These data support a novel mechanism for loading NPs with quisinostat and further the development of HDIs for the treatment of orthotopic glioblastoma.
Supplementary Material
Acknowledgements
We gratefully acknowledge funding from the Barrow Neurological Foundation, the Rick Oehme Foundation, the ARCS Foundation and the NIH (NINDS RO1NS088448 and NICHD DP2HD084067). The authors would like to thank the LeRoy Eyring Center for Solid State Science at Arizona State University for use of TEM facilities and Jon Yamaguchi for his help in generating the GL261-LucNeo cells.
Footnotes
Conflict of interest
The authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.colsurfb.2018.02.048.
References
- [1].Damaskos C, Valsami S, Kontos M, Spartalis E, Kalampokas T, Kalampokas E, Athanasiou A, Moris D, Daskalopoulou A, Davakis S, Tsourouflis G, Kontzoglou K, Perrea D, Nikiteas N, Dimitroulis D, Histone deacetylase inhibitors: an attractive therapeutic strategy against Breast cancer, Anticancer Res 37 (2017) 35–46. [DOI] [PubMed] [Google Scholar]
- [2].Lane AA, Chabner BA, Histone deacetylase inhibitors in cancer therapy, J. Clin. Oncol 27 (2009) 5459–5468, 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- [3].Lopez CA, Feng FY, Herman JM, Nyati MK, Lawrence TS, Ljungman M, Phenylbutyrate sensitizes human glioblastoma cells lacking wild-type p53 function to ionizing radiation, Int. J. Radiat. Oncol. Biol. Phys 69 (2007) 214–220, 10.1016/j.ijrobp.2007.04.069. [DOI] [PubMed] [Google Scholar]
- [4].Entin-Meer M, Yang X, VandenBerg SR, Lamborn KR, Nudelman A, Rephaeli A, Haas-Kogan DA, In vivo efficacy of a novel histone deacetylase inhibitor in combination with radiation for the treatment of gliomas, Neuro-oncology 9 (2007) 82–88, 10.1215/15228517-2006-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ugur HC, Ramakrishna N, Bello L, Menon LG, Kim S-K, Black PM, Carroll RS, Continuous intracranial administration of suberoylanilide hydroxamic acid (SAHA) inhibits tumor growth in an orthotopic glioma model, J. Neurooncol 83 (2007) 267–275, 10.1007/s11060-007-9337-z. [DOI] [PubMed] [Google Scholar]
- [6].Zagni C, Floresta G, Monciino G, Rescifina A, The search for potent, small-molecule HDACIs in cancer treatment: a decade after vorinostat, Med. Res. Rev (2017), 10.1002/med.21437. [DOI] [PubMed] [Google Scholar]
- [7].Campos B, Bermejo JL, Han L, Felsberg J, Ahmadi R, Grabe N, Reifenberger G, Unterberg A, Herold-Mende C, Expression of nuclear receptor corepressors and class I histone deacetylases in astrocytic gliomas, Cancer Sci 102 (2011) 387–392, 10.1111/j.1349-7006.2010.01792.x. [DOI] [PubMed] [Google Scholar]
- [8].Ceccacci E, Minucci S, Inhibition of histone deacetylases in cancer therapy: lessons from leukaemia, Br. J. Cancer 114 (2016) 605–611, 10.1038/bjc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dvorakova M, Vanek T, Histone deacetylase inhibitors for the treatment of cancer stem cells, Med. Chem. Commun 7 (2016) 2217–2231, 10.1039/C6MD00297H. [DOI] [Google Scholar]
- [10].Giudice FS, P DS Jr., Nör JE, Squarize CH, Castilho RM, Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer, PLoS One 8 (2013) e58672, 10.1371/journal.pone.0058672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cornago M, Garcia-Alberich C, Blasco-Angulo N, Vall-llaura N, Nager M, Herreros J, Comella JX, Sanchis D, Llovera M, Histone deacetylase inhibitors promote glioma cell death by G2 checkpoint abrogation leading to mitotic catastrophe, Cell. Death. Dis 5 (2014) e1435, 10.1038/cddis.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhu L, Wu K, Ma S, Zhang S, HDAC inhibitors: a new radiosensitizer for non-small-cell lung cancer, Tumori 101 (2015) 257–262, 10.5301/tj.5000347. [DOI] [PubMed] [Google Scholar]
- [13].Imai Y, Maru Y, Tanaka J, Action mechanisms of histone deacetylase inhibitors in the treatment of hematological malignancies, Cancer Sci 107 (2016) 1543–1549, 10.1111/cas.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shieh J-M, Tang Y-A, Hu F-H, Huang W-J, Wang Y-J, Jen J, Liao S-Y, Lu Y-H, Yeh Y-L, Wang T-W, Lin P, Wang Y-C, A histone deacetylase inhibitor enhances expression of genes inhibiting Wnt pathway and augments activity of DNA demethylation reagent against nonsmall-cell lung cancer, Int. J. Cancer 140 (2017) 2375–2386, 10.1002/ijc.30664. [DOI] [PubMed] [Google Scholar]
- [15].Newbold A, Falkenberg KJ, Prince HM, Johnstone RW, How do tumor cells respond to HDAC inhibition? FEBS J 283 (2016) 4032–4046, 10.1111/febs.13746. [DOI] [PubMed] [Google Scholar]
- [16].Rugo HS, Vidula N, Ma C, Improving response to hormone therapy in Breast cancer: new targets, new therapeutic options, Am. Soc. Clin. Oncol. Educ. Book Am. Soc. Clin. Oncol. Meet 35 (2016) e40–54, 10.14694/EDBK159198. [DOI] [PubMed] [Google Scholar]
- [17].Mottamal M, Zheng S, Huang TL, Wang G, Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents, Mol. Basel Switz 20 (2015) 3898–3941, 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arts J, King P, Mariën A, Floren W, Beliën A, Janssen L, Pilatte I, Roux B, Decrane L, Gilissen R, Hickson I, Vreys V, Cox E, Bol K, Talloen W, Goris I, Andries L, Jardin MD, Janicot M, Page M, van Emelen K, Angibaud P, JNJ-26481585, a novel second-generation oral histone deacetylase inhibitor shows broad-spectrum preclinical antitumoral activity, Am. Assoc. Cancer Res 15 (2009) 6841–6851, 10.1158/1078-0432.CCR-09-0547. [DOI] [PubMed] [Google Scholar]
- [19].Carol H, Gorlick R, Kolb EA, Morton CL, Manesh DM, Keir ST, Reynolds CP, Kang MH, Maris JM, Wozniak A, Hickson I, Lyalin D, Kurmasheva RT, Houghton PJ, Smith MA, Lock R, Initial testing (Stage 1) of the histone deacetylase inhibitor, quisinostat (JNJ-26481585), by the pediatric preclinical testing program, pediatr, Blood Cancer 61 (2014) 245–252, 10.1002/pbc.24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Venugopal B, Baird R, Kristeleit RS, Plummer R, Cowan R, Stewart A, Fourneau N, Hellemans P, Elsayed Y, Mcclue S, Smit JW, Forslund A, Phelps C, Camm J, Evans TRJ, de Bono JS, Banerji U, A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors, Clin. Cancer Res 19 (2013) 4262–4272, 10.1158/1078-0432.CCR-13-0312. [DOI] [PubMed] [Google Scholar]
- [21].Householder KT, DiPerna DM, Chung EP, Wohlleb GM, Dhruv HD, Berens ME, Sirianni RW, Intravenous delivery of camptothecin-loaded PLGA nanoparticles for the treatment of intracranial glioma, Int. J. Pharm 479 (2015) 374–380, 10.1016/j.ijpharm.2015.01.002. [DOI] [PubMed] [Google Scholar]
- [22].Wang EC, Min Y, Palm RC, Fiordalisi JJ, Wagner KT, Hyder N, Cox AD, Caster J, Tian X, Wang AZ, Nanoparticle formulations of histone deacetylase inhibitors for effective chemoradiotherapy in solid tumors, Biomaterials 51 (2015) 208–215, 10.1016/j.biomaterials.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cook RL, Householder KT, Chung EP, Prakapenka AV, DiPerna DM, Sirianni RW, A critical evaluation of drug delivery from ligand modified nanoparticles: confounding small molecule distribution and efficacy in the central nervous system, J. Control. Release 220 (2015) 89–97, 10.1016/j.jconrel.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deng Y, Saucier-Sawyer JK, Hoimes CJ, Zhang J, Seo Y-E, Andrejecsk JW, Saltzman WM, The effect of hyperbranched polyglycerol coatings on drug delivery using degradable polymer nanoparticles, Biomaterials 35 (2014) 6595–6602, 10.1016/j.biomaterials.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rubin JB, Kung AL, Klein RS, Chan JA, Sun Y, Schmidt K, Kieran MW, Luster AD, Segal RA, A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors, Proc. Natl. Acad. Sci 100 (2003) 13513–13518, 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Letchford K, Burt H, A review of the formation and classification of amphiphilic block copolymer nanoparticulate structures: micelles, nanospheres, nanocapsules and polymersomes, Eur, J. Pharm. Biopharm 65 (2007) 259–269, 10.1016/j.ejpb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- [27].Avgoustakis K, Pegylated poly(lactide) and poly(lactide-co-glycolide) nanoparticles: preparation, properties and possible applications in drug delivery, Curr. Drug Deliv 1 (2004) 321–333. [DOI] [PubMed] [Google Scholar]
- [28].Zhang Y, Ren T, Gou J, Zhang L, Tao X, Tian B, Tian P, Yu D, Song J, Liu X, Chao Y, Xiao W, Tang X, Strategies for improving the payload of small molecular drugs in polymeric micelles, J. Control. Release 261 (2017) 352–366, 10.1016/j.jconrel.2017.01.047. [DOI] [PubMed] [Google Scholar]
- [29].Adesina SK, Ezeonyebuchi U, Akala EO, The effect of formulation variables on drug loading of antitubercular drugs in nanoparticle formulations, Mater. Res. Express 2 (2015), 10.1088/2053-1591/2/9/095403,095403. [DOI] [Google Scholar]
- [30].Peltonen L, Aitta J, Hyvönen S, Karjalainen M, Hirvonen J, Improved entrapment efficiency of hydrophilic drug substance during nanoprecipitation of poly(I)lactide nanoparticles, AAPS PharmSciTech 5 (2004), 10.1208/pt050116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang Y, Li P, Kong L, Peng Z, Luo Y, Formulation optimization for high drug loading colonic drug delivery carrier, 2010 3rd Int Conf. Biomed. Eng. Inform (2010) 1686–1689, 10.1109/BMEI.2010.5640547. [DOI] [Google Scholar]
- [32].McCall RL, Sirianni RW, PLGA nanoparticles formed by single- or double-emulsion with vitamin E-TPGS, J. Vis. Exp 51015 (2013), 10.3791/51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Govender T, Stolnik S, Garnett MC, Illum L, Davis SS, PLGA nanoparticles prepared by nanoprecipitation: drug loading and release studies of a water soluble drug, J. Control. Release 57 (1999) 171–185. [DOI] [PubMed] [Google Scholar]
- [34].Lozano-Pérez AA, Rivero HC, del M, Pérez Hernández C, Pagán A, Montalbán MG, Víllora G, Cénis JL, Silk fibroin nanoparticles: efficient vehicles for the natural antioxidant quercetin, Int. J. Pharm 518 (2017) 11–19, 10.1016/j.ijpharm.2016.12.046. [DOI] [PubMed] [Google Scholar]
- [35].Brasseur N, Brault D, Couvreur P, Adsorption of hematoporphyrin onto polyalkylcyanoacrylate nanoparticles: carrier capacity and drug release, Int. J. Pharm 70 (1991) 129–135, 10.1016/0378-5173(91)90172-K. [DOI] [Google Scholar]
- [36].Pakulska MM, Elliott Donaghue I, Obermeyer JM, Tuladhar A, McLaughlin CK, Shendruk TN, Shoichet MS, Encapsulation-free controlled release: electrostatic adsorption eliminates the need for protein encapsulation in PLGA nanoparticles, Sci. Adv 2 (2016), 10.1126/sciadv.1600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Crawford L, Higgins J, Putnam D, A simple and sensitive method to quantify biodegradable nanoparticle biodistribution using europium chelates, Sci. Rep 5 (2015), 10.1038/srep13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rafiei P, Haddadi A, Docetaxel-loaded PLGA and PLGA-PEG nanoparticles for intravenous application: pharmacokinetics and biodistribution profile, Int. J. Nanomed 12 (2017) 935–947, 10.2147/IJN.S121881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mejías R, Costo R, Roca AG, Arias CF, Veintemillas-Verdaguer S, González-Carreño T, del Puerto Morales M, Serna CJ, Mañes S, Barber DF, Cytokine adsorption/release on uniform magnetic nanoparticles for localized drug delivery, J. Controlled Release 130 (2008) 168–174, 10.1016/j.jconrel.2008.05.028. [DOI] [PubMed] [Google Scholar]
- [40].Curry D, Cameron A, MacDonald B, Nganou C, Scheller H, Marsh J, Beale S, Lu M, Shan Z, Kaliaperumal R, Xu H, Servos M, Bennett C, MacQuarrie S, Oakes KD, Mkandawire M, Zhang X, Adsorption of doxorubicin on citrate-capped gold nanoparticles: insights into engineering potent chemotherapeutic delivery systems, Nanoscale 7 (2015) 19611–19619, 10.1039/C5NR05826K. [DOI] [PubMed] [Google Scholar]
- [41].Adamopoulou E, Naumann U, HDAC inhibitors and their potential applications to glioblastoma therapy, Oncoimmunology 2 (2013), 10.4161/onci.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gryder BE, Sodji QH, Oyelere AK, Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed, Future Med. Chem 4 (2012) 505–524, 10.4155/fmc.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hornig E, Heppt MV, Graf SA, Ruzicka T, Berking C, Inhibition of histone deacetylases in melanoma—a perspective from bench to bedside, Exp. Dermatol 25 (2016) 831–838, 10.1111/exd.13089. [DOI] [PubMed] [Google Scholar]
- [44].El Bahhaj F, Denis I, Pichavant L, Delatouche R, Collette F, Linot C, Pouliquen D, Grégoire M, Héroguez V, Blanquart C, Bertrand P, Histone deacetylase inhibitors delivery using nanoparticles with intrinsic passive tumor targeting properties for tumor therapy, Theranostics 6 (2016) 795–807, 10.7150/thno.13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gilardini Montani MS, Granato M, Santoni C, Del Porto P, Merendino N, D’Orazi G, Faggioni A, Cirone M, Histone deacetylase inhibitors VPA and TSA induce apoptosis and autophagy in pancreatic cancer cells, Cell. Oncol. Dordr 40 (2017) 167–180, 10.1007/s13402-017-0314-z. [DOI] [PubMed] [Google Scholar]
- [46].Deleu S, Lemaire M, Arts J, Menu E, Van Valckenborgh E, King P, Vande Broek I, De Raeve H, Van Camp B, Croucher P, Vanderkerken K, The effects of JNJ-26481585, a novel hydroxamate-based histone deacetylase inhibitor, on the development of multiple myeloma in the 5T2MM and 5T33MM murine models, Leukemia 23 (2009) 1894–1903, 10.1038/leu.2009.121. [DOI] [PubMed] [Google Scholar]
- [47].Deleu S, Lemaire M, Arts J, Menu E, Valckenborgh EV, Broek IV, Raeve HD, Coulton L, Camp BV, Croucher P, Vanderkerken K, Bortezomib alone or in combination with the histone deacetylase inhibitor JNJ-26481585: effect on myeloma bone disease in the 5T2MM murine model of myeloma, Cancer Res 69 (2009) 5307–5311, 10.1158/0008-5472.CAN-08-4472. [DOI] [PubMed] [Google Scholar]
- [48].Bhowmik A, Khan R, Ghosh MK, Blood brain barrier: a challenge for effectual therapy of brain tumors, BioMed Res. Int 2015 (2015), 10.1155/2015/320941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lesniak MS, Brem H, Targeted therapy for brain tumours, Nat. Rev. Drug Discov 3 (2004) 499–508, 10.1038/nrd1414. [DOI] [PubMed] [Google Scholar]
- [50].Mason WP, Blood-brain barrier-associated efflux transporters: a significant but underappreciated obstacle to drug development in glioblastoma, Neuro-oncology 17 (2015) 1181–1182, 10.1093/neuonc/nov122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.