Abstract
During protein synthesis, the mRNA and tRNAs must be moved rapidly through the ribosome while precisely maintaining the translational reading frame. This complex dynamic process is coupled to large- and small-scale conformational rearrangements in the ribosome, mainly in its ribosomal RNA. The free energy of peptide bond formation and GTP hydrolysis are most likely used to impose directionality on these movements. We suggest that this is accomplished by coupling this energy to two pawls, consisting of tRNA and EF-G, which enable two ratchet mechanisms that act separately and sequentially on the two ribosomal subunits.
Introduction
Ribosomes are the ancient ribonucleoprotein complexes responsible for translation, playing a unique role in the complex biological drama that enables expression of genetic information, spanning the divide that separates the two very different worlds of nucleic acids and proteins. Fittingly, the ribosome is itself composed of both RNA and protein, likely reflecting its own descent from an RNA world. We now know that the mechanisms of at least two of the three fundamental steps of protein synthesis - codon recognition by aminoacyl-tRNAs and catalysis of peptide bond formation - are based on ribosomal RNA1–3; i.e., the ribosome is a ribozyme. But what about the third step, arguably the most complex operation of protein synthesis: translocation? It has long been known that the coupled translocation of mRNA and tRNA is catalyzed by the GTPase elongation factor EF-G. But while textbook descriptions have often implied that GTP hydrolysis by EF-G provides some kind of “power stroke”, the role of the ribosome in this process had long been overlooked. This situation has been undergoing rapid change.
How do the mRNA and tRNAs move?
Following formation of each peptide bond, tRNAs must be moved from the A and P sites to the P and E sites, to vacate the A site for the next incoming aminoacyl-tRNA. This process must guard against slippage of the translational reading frame, a catastrophic event that results in rampant misincorporation and termination at out-of-frame stop codons. Research from several groups in recent years has begun to reveal the underlying mechanisms of translocation, which involve precise rearrangements in the structure of the ribosome itself, both large- and small-scale, that mirror the movements of the mRNA and tRNAs. These structural changes have been inferred from cryo-EM and crystallographic snapshots of ribosomes trapped in intermediate states of translocation, and followed directly in real time by approaches such as ensemble and single-molecule FRET. It is now becoming apparent that the tRNAs and mRNA are translocated under the guidance of precise structural rearrangements in the ribosome and EF-G.
Large-Scale Ribosomal Movements During Translocation
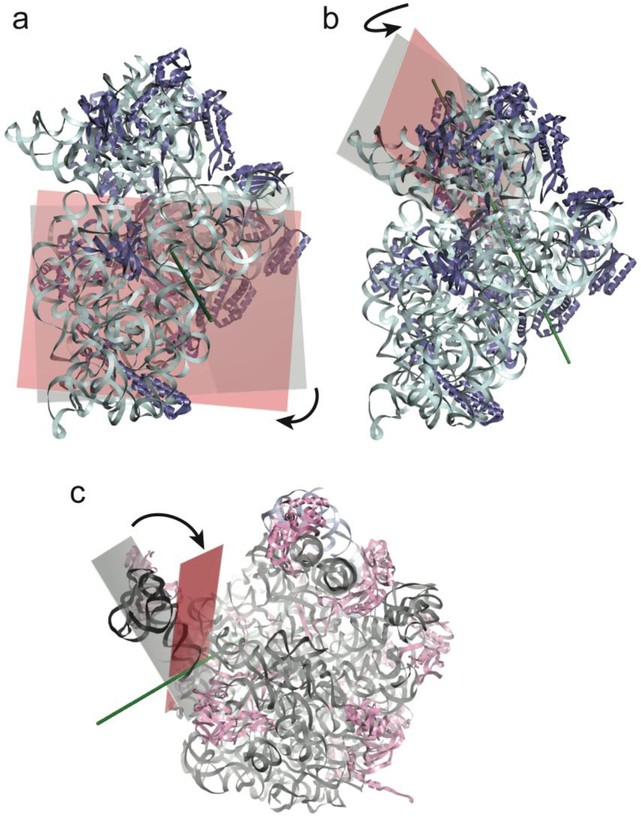
Large-scale conformational changes in the ribosome were first seen in cryo-EM reconstructions by Frank and Agrawal4 showing a ~6° rotational rearrangement of the orientation of the two (30S and 50S) ribosomal subunits in ribosomes bound with EF-G (Fig. 1a,b,e). They proposed that this intersubunit rotation is part of a ratchet mechanism that is involved in translocation of mRNA and tRNA. This proposal was tested by reversibly crosslinking the two subunits together via a disulphide bond between two proteins5. Formation of the crosslink specifically abolished translocation, which was immediately restored upon reduction of the disulfide bond. This experiment provided evidence that some kind of intersubunit movement is essential for translocation.
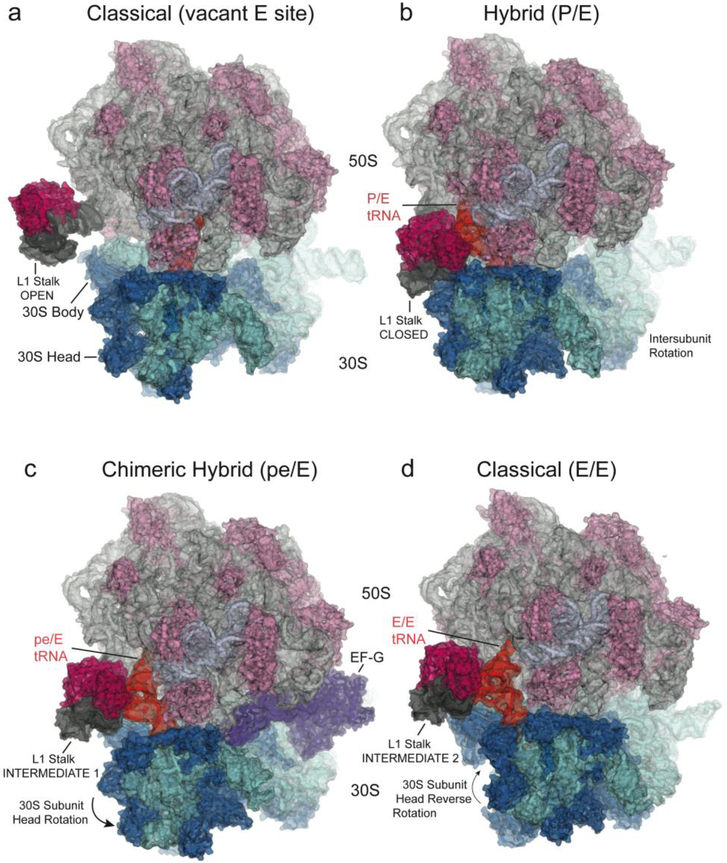
Fig. 1. Intermediate states of ribosomal translocation.
(top) Schematics showing four states of the translocation cycle. (a) Classical (pre-translocation) non-rotated state, with P/P tRNA, vacant E site, and L1 stalk in its open position. (PDB ID: 4GD2)50 =; (b) Hybrid state, with P/E tRNA, ~6° intersubunit rotation, ~6° 30S subunit head rotation and 30° rotation of the L1 stalk to its closed position, where it contacts the elbow of the P/E tRNA and the 30S subunit.55 (PDB ID: 4V9H) (c) Chimeric hybrid state, with pe/E tRNA, 21° 30S subunit head rotation, ~2° intersubunit rotation and L1 stalk in Int 1 position15. (PDB ID: 4V9K); (d) Classical (post-translocation) non-rotated state, with E/E tRNA, L1 stalk in Int 2 position28. (PDB ID: 4V67) Components shown are: 16S rRNA (cyan); 23S rRNA (light grey); 5S rRNA (blue-grey); 30S proteins (dark blue); 50S proteins (light magenta); L1 stalk (dark grey); L1 protein (magenta; docked in panel (a) from Zhou et al. (2013)15); deacylated tRNA (orange). The head of the L1 stalk maintains contact with the elbow of the deacylated tRNA as it moves through states b to d. (e-g) The three large-scale motions are shown by planes bisecting the (e) 30S body domain; (f) 30S head domain and (g) L1 stalk in the structures of the non-rotated (cyan or grey) and rotated (pink) states; Euler-Rodrigues axes20 are represented by grey rods.
Subsequently, it has been found that intersubunit rotation does not result in productive translocation, but instead creates an intermediate state that is likely the hybrid state that was inferred from chemical probing experiments6. In this first translocation step, movement of the acceptor ends of the tRNAs on the large subunit results in rearrangement of the tRNA binding states from their classical A/A and P/P states to the A/P and P/E hybrid states. This step can occur spontaneously, in the absence of EF-G and GTP, and is reversible6,7; at 37°, its spontaneous rate is ~40 sec−1, comparable to the rate-limiting step of translocation8. But completion of translocation in the absence of EF-G, i.e., the steps beyond the hybrid state, is about 4 orders of magnitude slower than the EF-G-catalyzed rate9–11. Thus, intersubunit rotation is coupled to tRNA movement on the large subunit, but not the small subunit.
The question then arose, how are the mRNA and the anticodon stem-loops (ASLs) of the tRNAs translocated? An early clue came from the first all-atom structure of a 70S ribosome, which revealed a 13Å constriction between the head and body domains of the 30S subunit in the path leading from the P site to the E site, presenting a steric block to movement of the P-site ASL12. The authors proposed that this block could be resolved by rotation of the 30S head domain, by analogy with a cryo-EM structure of the yeast 80S ribosome bound with elongation factor eEF2 in the presence of sordarin, in which the head domain of the 40S subunit was found to be rotated13.
Such a mechanism was exactly borne out by the first view of a tRNA with its ASL trapped in an intermediate state of movement, as revealed in a 7.8Å cryo-EM reconstruction of the T. thermophilus ribosome bound with EF-G in the presence of fusidic acid, called the TIPOST state14. This structure showed opening of the head/body constriction that allowed movement of the tRNA on the 30S subunit. But the big surprise was that the tRNA was not just diffusing from the P to the E site, but instead remained attached to the 30S head in an intermediate pe/E state, in which the ASL was bound between the P site of the head domain and the E site of the body domain, while its acceptor end is bound to the 50S E site. Crystal structures of three EF-G-ribosome complexes trapped with fusidic acid or the non-hydrolyzable GTP analogue GDPNP were then solved at resolutions between 3.5Å and 4.1Å, showing movement of the ASL coupled to 18° to 21° rotations of the 30S head domain15(Fig. 1c,f). Precise contact was maintained between the ASL and all P-site elements of the 30S head domain in the rotated states, while contact with the P-site elements of the body domain were disrupted and replaced by contact with E-site elements. As reported for the TIPOST state, the acceptor end of the tRNA was bound to the 50S E site in all three complexes. These structures all pointed to coupling of ASL-mRNA translocation to rotation of the 30S head domain.
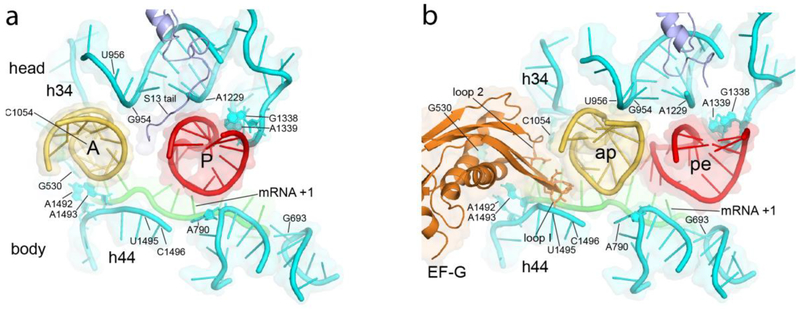
A shortcoming of the latter studies was that the trapped complexes contained only a single tRNA, whereas in vivo two tRNAs are translocated. This was addressed in subsequent cryo-EM16 and x-ray17 structures of authentic translocation intermediates trapped with EF-G, mRNA and two tRNAs. In these complexes, the P-site tRNA again moved into the pe/E chimeric hybrid state, while the A-site tRNA was moved into an ap/ap chimeric state (Fig. 1c; Fig. 2). Movement of the ASLs of the two tRNAs precisely followed the 21° rotation of the 30S head domain. In contrast, the ASL of the ap/ap tRNA moved further than the 30S head, possibly resulting from contact with the mobile domain IV of EF-G (Fig. 2b). The 3.8Å crystal structure of the complex containing EF-G, mRNA and two tRNAs17 provided a detailed description of numerous small-scale structural changes in the trapped complex, as discussed below.
Fig 2. Movement of the tRNA Anticodon Stem-Loop (ASL) on the 30S Subunit.
Crystal structures showing positions of the A- and P-site ASLs in the (a) classical-state ribosome, prior to translocation56, and in the (b) trapped chimeric hybrid-state translocation intermediate17. The P-site ASL moves toward the E site into the pe chimeric state, precisely following rotation of the 30S head domain as shown in (b); the A-site ASL moves further than head rotation, likely following the movement of the tip of domain IV of EF-G into the ap chimeric state. Apart from the tail of protein S13 and domain IV of EF-G, virtually all of the ASL contacts are with 16S ribosomal RNA.
A third large-scale rearrangement involves the L1 stalk, a feature of the 50S subunit comprised of helices H76, H77 and H78 of 23S rRNA and protein L1. As the deacylated tRNA moves into the P/E hybrid state following peptide bond formation, the head domain of the L1 stalk moves through a distance of more than 60Å to create a stacking contact with the elbow of the tRNA (Fig. 1b,g). As the tRNA moves from the P/E to the pe/E and E/E states, the L1 stalk flexes in order to maintain its stacking interaction with the tRNA elbow (Fig. 1). In the P/E hybrid state, the head of the stalk also contacts the 30S subunit head and body, forming a transient intersubunit bridge (Fig. 1b)18,19.
Molecular Mechanics of Large-Scale Conformational Changes
What is the source of the conformational changes that underlie translocation? The two most dramatic movements are the 18°−21° rotation of the 30S head domain and the ~60Å movement of the L1 stalk. Comparative analysis of different trapped intermediate complexes has revealed the structural basis of these dynamic events18,20. In both cases, the movements are based on bending or hinging of ribosomal RNA, showing that translocation, like the other two fundamental steps of protein synthesis, is RNA-based.
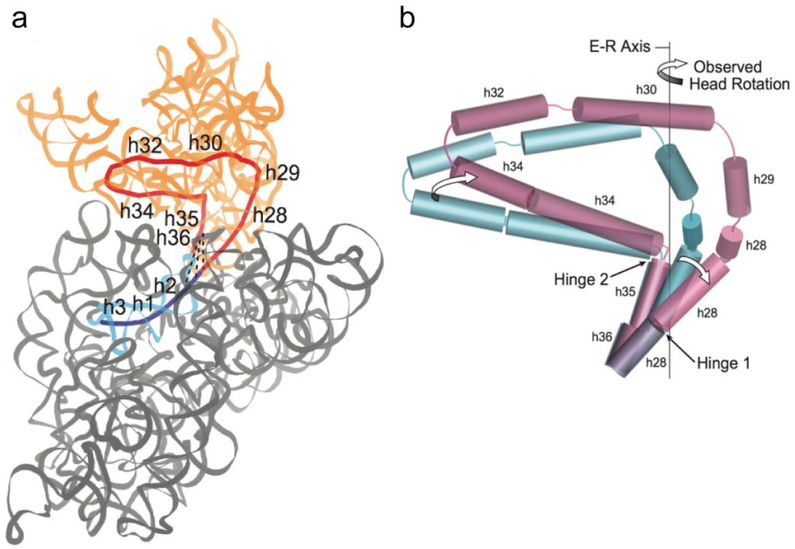
Rotation of the 30S head domain originates at two hinge points located near the two points of connection between the head domain and the 30S body20. Hinge 1 is located in the middle of helix h28 of 16S rRNA, the sole covalent connection between the head and body (Fig. 3a). It is positioned at the bulged G926, which is flanked by two G-U wobble pairs. Hinge 2 is located in a Family A 3-way helical junction21 in the linker between helices h34 and h35, near a non-covalent connection between the head and body mediated by an interaction between the tetraloop of helix h36 and helix h2 (Fig. 3a). Flexing at both hinge points combines to create an overall rotational movement of the head domain (Fig. 3b).
Fig. 3. Origins of 30S head domain rotation.
(a) Coaxial helical core of the 16S rRNA component (red) of the 30S head domain (orange) contains helices h28 to h36 (red). Helices h2, h1 and h3 of the body domain (blue) are coaxial with the core helices of the head. Dotted lines represent conserved A-minor interactions between the h36 tetraloop and helix h2 in the 30S body (grey). (b) Positions of coaxial 16S rRNA helical axes are represented by cylinders for the non-rotated31 (blue) and rotated15 (magenta) states. During head rotation, movement of Hinge 1 is caused by straightening of the kinked helix h28; movement of Hinge 2 is the result of swiveling of helix h34 around the static h35. Rotation of the 30S head around the Euler-Rodrigues (E-R) axis results from combined bending at Hinges 1 and 2.
Rearrangement of the L1 stalk also originates at two main hinge points18,22. An essentially rigid-body hinging motion occurs in another Family A 3-way helical junction, in the linker between 23S rRNA helices H75 and H76, while a distributed bending movement is seen in a G-U-rich segment of helix H76. These two motions combine to allow the head of the L1 stalk to preserve its stacking contact with the elbow of the deacylated tRNA as it moves through its ~60Å trajectory from P/E to pe/E to E/E states and finally to its open state upon release of the tRNA23–25.
Both movements originate at Family A 3-way RNA helical junctions and in G-U-rich helical segments, suggesting that they may represent general strategies for functional movements of RNA. Another common motif is the occurrence of non-canonical base pairs or base-base mismatches at dynamic stacking interfaces, such as the hinge between the D stem and anticodon stem of tRNA15,26–28 and in the 3-way junctions of hinge 2 and of the L1 stalk18,20.
tRNA Constrains the Ribosome
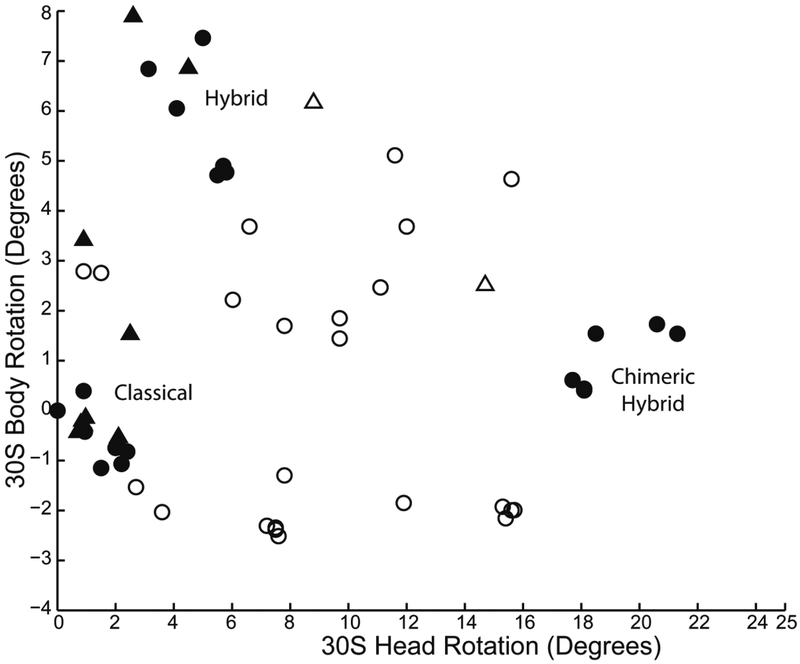
In the absence of tRNA, the three largest-scale motions - 30S head and body rotation and L1 stalk movement - are relatively unconstrained, showing a wide range of observed values (Fig. 4). However, when one or more tRNAs are bound, the conformational freedom of these motions becomes dramatically constrained, clustering around three sets of allowed values (Fig. 4). These correspond to the classical, hybrid and chimeric-hybrid states, which we presume correspond to relative free energy minima.
Fig. 4. tRNA constrains the conformational freedom of the ribosome.
In vacant ribosomes, (empty symbols), head and body rotation angles are distributed across a wide range of possible values. In the presence of tRNA (filled symbols), the ribosomes are dramatically constrained into three clusters, corresponding to the classical, hybrid and chimeric-hybrid states. Data are extracted from 69 X-ray and cryo-EM structures of 8Å or better resolution of bacterial (circles) and eukaryotic (triangles) ribosomes.
How can tRNA constrain the conformational state of the ribosome? Although we still have a limited understanding, there are some clues to stabilization of the classical and hybrid states. The strongest tRNA binding state is for a peptidyl-tRNA bound in the P/P classical state, because of its high affinity for both the 30S and 50S P sites29; the P/P state enforces the minimal intersubunit rotation observed in the classical state30,31. The classical state is also stabilized by peripheral intersubunit bridges, disruption of which lead to accelerated translocation32,33.
When the P-site tRNA becomes deacylated, its 3’-deacylated CCA end is then able to bind to the 50S E site, which likely helps to stabilize the ~6° intersubunit rotation seen in the hybrid state. Intersubunit rotation in the hybrid state is further stabilized (and limited) by contact of the head of the L1 stalk with the 30S subunit when it moves to stack on the P/E tRNA, creating a steric block to rotation of the 30S head domain18,34 (Fig. 1b). This block may act as a “check-point” to ensure that the acceptor end of the deacylated tRNA has entered the 50S E site prior to mRNA and tRNA translocation on the 30S subunit.
Structural Rearrangements of the tRNA Binding Sites
Contacts between the mRNA and tRNAs and their ribosomal binding sites present barriers to movement. Given the critical importance of moving the tRNAs accurately from one binding site to the next, it is not surprising that this process does not proceed by simple diffusion. An important insight from the x-ray structures of different functional complexes is that the tRNA binding sites are not static structures, but rearrange to bind or release the tRNAs, and in some cases actually appear to reach out to escort the tRNAs from one site to the next. The first such example was the flipping of 16S rRNA bases G530, 1492 and 1493 during binding of tRNA to the 30S A site1, a rearrangement that must be reversed during translocation. In another instance, conformational rearrangement of the loop of 16S rRNA helix h31 disrupts the packing of m2G966 against ribose 34 of the P-site tRNA, re-positioning it to pack against ribose 34 of the translocating A-site tRNA as it moves into the P site17. Other elements of the 30S P site, including G1338 and A1339 of 16S rRNA and the C-terminal tails of proteins S9 and S13, remain in contact with the P-site tRNA as it moves with rotation of the 30S head domain toward the E site. Thus, the tRNA ASL appears to maintain contact with tRNA binding-site elements throughout its movement from the 30S A site to the P site and from the P site to the E site15,17.
Most critical is the need to maintain the translational reading frame during coupled movement of mRNA and tRNA between the 30S A and P sites. This presents a problem because, unlike P-site tRNA, the primary contacts with the A-tRNA ASL are with the 30S subunit body rather than the head. Thus, when the head rotates, it does not carry the A-tRNA with it. Moreover, the body interactions involving 16S rRNA nucleotides A1492, A1493 and G530 that stabilize codon-anticodon interaction in the 30S A site must be disrupted, overcoming what may be the main barrier to the rate-limiting step (“unlocking”) of translocation35,36. Both of these issues may be solved by EF-G. Cryo-EM and x-ray structures of chimeric hybrid-state complexes containing EF-G and 2 tRNAs show that the tip of domain IV of EF-G contacts the minor groove of the codon-anticodon helix of the transient A-site tRNA (Fig. 2b)16,17, and it has been proposed that this same interaction disrupts the minor-groove contacts with 16S rRNA in the A site16,37–39. Thus, elongation factor EF-G may play two critical roles in translocation: (i) disrupting binding of the codon-anticodon duplex to the 30S A site to overcome the barrier to the second step of translocation and (ii) stabilizing the otherwise fragile codon-anticodon pairing, to prevent loss of the translational reading frame during translocation.
At their acceptor ends, the conserved CCA tails of the A-site and P-site tRNAs are bound initially to the A and P loops, respectively, of 23S rRNA in the 50S subunit3,40,41. During translocation, the A-tRNA must disrupt its interactions with the A loop (helix H92 of 23S rRNA), and bind to the P loop (helix H80 of 23S rRNA). The x-ray structure of the chimeric hybrid-state complex revealed an unexpected intermediate state in which the CCA end of the A-site tRNA contacts both the A and P loops, the structures of which are distorted, reaching toward each other to contact the tRNA, in what has been named the ap/ap chimeric hybrid state17.
In between the binding sites for the two tRNA extremities lies intersubunit bridge B1a, formed between helix H38 of 23S rRNA (the so-called A-site finger) and the head of the 30S subunit. H38 blocks movement of the elbow of A-site tRNA into the position of the P-site elbow. Intersubunit rotation or 30S head rotation both reposition H38, disrupting bridge B1a, allowing the tRNA elbow to reach its 50S P site position17,42,43. This explains how mutational disruption of bridge B1a results in an increase in the rate of EF-G-independent spontaneous translocation32,33,44. Thus, the movements of the tRNAs are facilitated by numerous conformational changes involving dynamic elements of ribosomal RNA as well as EF-G.
Ratchets and Pawls
Although it has often been implied that translocation is driven by a ‘power stroke’ generated by the energy released upon hydrolysis of GTP by EF-G, even very large-scale movements, including rapid rotation of the entire ~800kD 30S subunit can occur in the complete absence of EF-G or GTP7. Thus, thermal energy alone should, in principle, be sufficient to drive translocation. However, ribosomal translocation must be executed unidirectionally, in contrast to the randomness of Brownian motion. This is the basis of the idea that translocation must be rectified by some sort of ratchet mechanism(s)4,45–47. Thus, external sources of energy are required for constraining thermal motions to impose directionality.
If ribosomal translocation is based on a ratchet-like mechanism or mechanisms, what are the pawls - the elements that impose unidirectional movement? One external source of energy comes from peptide bond formation, which uses the energy of the aminoacyl- and peptidyl-tRNA ester linkages, which in turn originate from the ATP that is used for aminoacylation of tRNA. As Spirin has pointed out48, this energy is used indirectly to impose directionality on movement of tRNA on the 50S subunit. As the chemical identity of theacceptor end of the tRNA changes during peptide bond formation, its specificity for binding to the 50S A, P and E sites changes. Peptidyl tRNA becomes deacylated upon peptide bond formation, enabling its CCA end to bind to the 50S E site, which is sterically unable to bind an acylated tRNA. This creates a vacant 50S P site, which has strong affinity for the acceptor end of the peptidyl-tRNA. These two events result in formation of the P/E and A/P hybrid states, imposing directionality on the movement of the acceptor ends of the tRNAs on the 50S subunit, Thus, the tRNAs effetively act as pawls in rectifying their own movement on the 50S subunit, driven by the energy of peptide bond formation.
On the 30S subunit, it has been proposed that insertion of the tip of domain IV of EF-G into the minor groove of the codon-anticodon duplex disrupts its binding to the A site, overcoming the barrier to the second step of translocation, as described above16,37–39; preservation of this contact during movement to the P site could then act as a kind of door-stop to prevent reversal of tRNA movement. EF-G could thus act as a pawl in a 30S subunit ribosomal ratchet.
According to this view, translocation is constrained to proceed in a forward direction by two ratchet mechanisms - one for each of the two ribosomal subunits, mirroring the fact that translocation occurs independently on the two subunits. According to the second law of thermodynamics, there must be an energetic cost for constraining the direction of these movements. The energy for directional movement of the acceptor ends of the tRNAs through the 50S subunit would originate in the chemistry of peptide bond formation, and the energy to constrain directionality of movement of the anticodon ends of the tRNAs, (along with the mRNA) through the 30S subunit would come from EF-G-catalyzed hydrolysis of GTP. Thus, the two ratchet mechanisms are coupled to the two sources of external (i.e., non-thermal) energy that are in play during translocation. In effect, tRNA and EF-G then represent the pawls of these two ratchets.
Another type of pawl is suggested by the intercalation of two universally conserved bases of 16S rRNA between bases of the mRNA. In chimeric-hybrid state structures, intercalation of A1503 between bases −1 and −2, and C1397 between bases −9 and −10 was observed15. Moreover, these two conserved nucleotides are tethered to each other by the conserved C1399-G1504 tertiary Watson-Crick base pair. The role of these intercalations is not known, but they may be involved in preventing slippage of the mRNA at some point during translocation.
An Emerging Picture of the Mechanics of Translocation
Translocation can be summarized as the series of events that take place between peptide bond formation and the emergence of a vacant A site for the next incoming aminoacyl-tRNA. A view of this process is beginning to emerge that combines structural and functional findings from the last decades of investigation of this complex and intriguing mechanism (Fig. 5). As discussed above, ribosome structural dynamics enable and guide the movements of the tRNAs and mRNA during translocation. Nevertheless, there remain many outstanding questions, and of course not all laboratories are in agreement about the specifics. The following summary attempts to merge structural, functional and kinetic observations to characterize translocation in terms of a series of conformational changes that take place between the three most extensively characterized states of the ribosome.
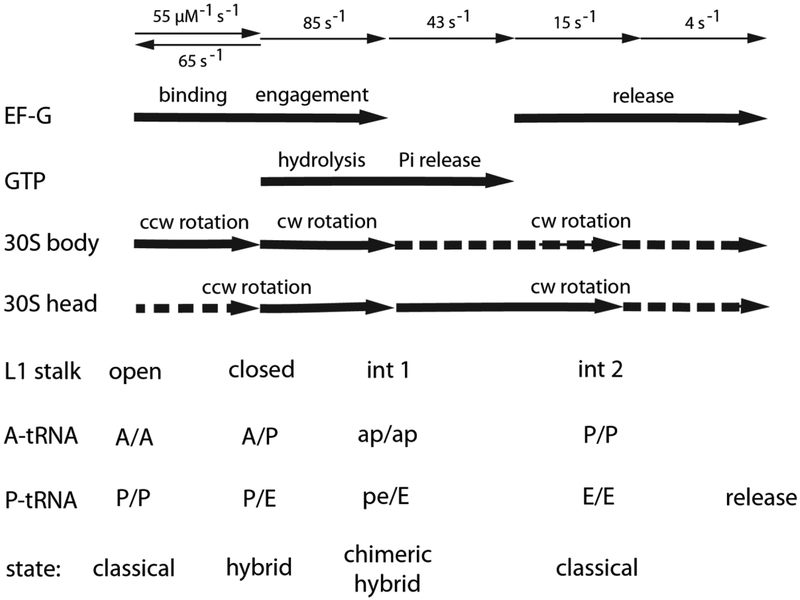
Fig. 5. Merging the results of kinetic and structural studies on translocation.
At the top are shown the rates of individual steps of translocation, corresponding to binding, engagement and release of EF-G; and hydrolysis and release of GTP54. In the middle are the timing of the corresponding large-scale rotational movements of the 30S body and head domains34,54,57,58, and movements of the L1 stalk25. At the bottom are the corresponding binding states of the tRNAs16,17. Dotted arrows indicate smaller-scale rotational movements.
Following peptide bond formation, EF-G·GTP joins the ribosome, binding preferentially to the hybrid-state ribosome34,49. Hybrid-states formation, which can occur independently of EF-G or GTP, involves a ~6–8° rotation of the 30S body around an axis orthogonal to the subunit interface, a ~5° rotation of the 30S head domain and rotation of the L1 stalk to its closed state50. However, it is unclear how EF-G engages with the ribosome in this state. A 7.6 Å cryo-EM structure of a hybrid-state ribosome bound to EF-G in the presence of viomycin shows a 12° hyper-rotation of the body domain of the 30S subunit that allows the elongated form of EF-G to bind to its canonical binding site on the 50S subunit without being sterically blocked by the ASL of the A/P tRNA51. A further possibility is suggested by the crystal structure of a classical-state ribosome bound to EFG-GDP52. In this structure, domain IV of EFG-GDP is dramatically compacted, allowing a ball-like form of EF-G to bind the 50S subunit without sterically clashing with the ASL of the A/A tRNA. Although it is clear that EF-G initially binds the ribosome in its GTP form, smFRET evidence indicates that free EF-G·GTP has a more compact structure in solution than when bound to the ribosome53. This compact conformation of EF-G·GTP may be essential for its initial interaction with the ribosome.
In the next step, the head domain of the 30S subunit rotates by ~18–21° along with a nearly complete reversal of intersubunit rotation14–17. This event appears to be timed with GTP hydrolysis (Fig. 5)54. The tip of domain IV of EF-G contacts the A-site codon-anticodon duplex, likely displacing it from the decoding site and helping to transport it into the ap chimeric state on the 30S subunit. The acceptor end of the peptidyl-tRNA moves into an ap state on the 50S subunit, with its CCA end shared between the A and P loops of 23S rRNA17. The deacylated tRNA moves from the P/E state into the chimeric hybrid pe/E state, as its anticodon end is moved by rotation of the 30S head domain into a state where it is held between P-site elements of the 30S head domain and E-site elements of the 30S body. The L1 stalk moves to maintain its stacking contact with the tRNA elbow. The resulting chimeric hybrid-state ribosome then contains a peptidyl-tRNA in the ap/ap state and a deacylated tRNA in the pe/E state.
Finally, upon Pi release54 the tRNAs move into the classical P/P and E/E states coupled to reverse rotations of the 30S head and body domains. This results in a vacant ribosomal A site, ready to accept an incoming aminoacyl-tRNA. Upon or during completion of these movements, EF-G·GDP is released from the ribosome54. During all of these steps, tRNA itself undergoes extensive conformational flexing, particularly around the junction of its D stem and anticodon stem17. Thus, the mechanism of translocation is deeply rooted in the dynamic properties of RNA, as are most, if not all of the basic functions of the ribosome, reflecting its likely origins in an RNA world.
Fig. 6.
References
- 1.Ogle JM et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Demeshkina N, Jenner L, Westhof E, Yusupov M & Yusupova G A new understanding of the decoding principle on the ribosome. Nature 484, 256–9 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Nissen P, Hansen J, Ban N, Moore PB & Steitz TA The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–30 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Frank J & Agrawal RK A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–22 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Horan LH & Noller HF Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci U S A 104, 4881–5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moazed D & Noller HF Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Cornish PV, Ermolenko DN, Noller HF & Ha T Spontaneous intersubunit rotation in single ribosomes. Mol Cell 30, 578–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma H et al. Kinetics of Spontaneous and EF-G-Accelerated Rotation of Ribosomal Subunits. Cell Rep 16, 2187–96 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM & Spirin AS Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol 101, 537–52. (1976). [DOI] [PubMed] [Google Scholar]
- 10.Gavrilova LP & Spirin AS Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Lett 17, 324–326 (1971). [DOI] [PubMed] [Google Scholar]
- 11.Fredrick K & Noller HF Catalysis of ribosomal translocation by sparsomycin. Science 300, 1159–62 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Schuwirth BS et al. Structures of the bacterial ribosome at 3.5 A resolution. Science 310, 827–34 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Spahn CM et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23, 1008–19 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratje AH et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Lancaster L, Donohue JP & Noller HF Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramrath DJ et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci U S A 110, 20964–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Lancaster L, Donohue JP & Noller HF How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345, 1188–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohan S & Noller HF Recurring RNA structural motifs underlie the mechanics of L1 stalk movement. Nat Commun 8, 14285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bock LV, Blau C, Vaiana AC & Grubmuller H Dynamic contact network between ribosomal subunits enables rapid large-scale rotation during spontaneous translocation. Nucleic Acids Res 43, 6747–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohan S, Donohue JP & Noller HF Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci U S A (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lescoute A & Westhof E Topology of three-way junctions in folded RNAs. RNA 12, 83–93 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reblova K, Sponer J & Lankas F Structure and mechanical properties of the ribosomal L1 stalk three-way junction. Nucleic Acids Res 40, 6290–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trabuco LG et al. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J Mol Biol 402, 741–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fei J, Kosuri P, MacDougall DD & Gonzalez RL Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell 30, 348–59 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Cornish PV et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A 106, 2571–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valle M et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10, 899–906 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Schmeing TM et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–94 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korostelev A, Trakhanov S, Laurberg M & Noller HF Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–77 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Lill R, Robertson JM & Wintermeyer W Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry 25, 3245–55. (1986). [DOI] [PubMed] [Google Scholar]
- 30.Yusupov M et al. Crystal Structure of the Ribosome at 5.5 Å Resolution. Science 292, 883–896 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Selmer M et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–42 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Liu Q & Fredrick K Contribution of intersubunit bridges to the energy barrier of ribosomal translocation. Nucleic Acids Res 41, 565–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komoda T et al. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J Biol Chem 281, 32303–9 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Wasserman MR, Alejo JL, Altman RB & Blanchard SC Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol 23, 333–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spirin AS [On the mechanism of ribosome function. The hypothesis of locking-unlocking of subparticles]. Dokl Akad Nauk SSSR 179, 1467–70 (1968). [PubMed] [Google Scholar]
- 36.Savelsbergh A et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell 11, 1517–23 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Gao YG et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khade PK & Joseph S Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol 18, 1300–2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu G et al. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon-anticodon duplex. Nat Struct Mol Biol 21, 817–24 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Samaha RR, Green R & Noller HF A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome [published erratum appears in Nature 1995 Nov 23;378(6555):419]. Nature 377, 309–14 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Kim DF & Green R Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell 4, 859–64 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Julian P et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A 105, 16924–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agirrezabala X et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 32, 190–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Altman RB & Blanchard SC Insights into the molecular determinants of EF-G catalyzed translocation. RNA 17, 2189–200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woese C Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature 226, 817–820 (1970). [DOI] [PubMed] [Google Scholar]
- 46.Spirin AS The ribosome as a conveying thermal ratchet machine. J Biol Chem 284, 21103–19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frank J & Gonzalez RL Jr. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem 79, 381–412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spirin AS Ribosomal translocation: facts and models. Prog Nucleic Acid Res Mol Biol 32, 75–114 (1985). [DOI] [PubMed] [Google Scholar]
- 49.Dorner S, Brunelle JL, Sharma D & Green R The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol 13, 234–41 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunkle JA et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brilot AF, Korostelev AA, Ermolenko DN & Grigorieff N Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A 110, 20994–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J, Gagnon MG, Bulkley D & Steitz TA Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 160, 219–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salsi E, Farah E, Netter Z, Dann J & Ermolenko DN Movement of elongation factor G between compact and extended conformations. J Mol Biol 427, 454–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belardinelli R et al. Choreography of molecular movements during ribosome progression along mRNA. Nat Struct Mol Biol 23, 342–8 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Tourigny DS, Fernandez IS, Kelley AC & Ramakrishnan V Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jenner LB, Demeshkina N, Yusupova G & Yusupov M Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17, 555–60 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Guo Z & Noller HF Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A 109, 20391–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ermolenko DN & Noller HF mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol 18, 457–62 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]