Abstract
DNA and histone proteins define the structure and composition of chromatin. Histone posttranslational modifications (PTMs) are covalent chemical groups capable of modeling chromatin accessibility, mostly due to their ability in recruiting enzymes responsible for DNA readout and remodeling. Mass spectrometry (MS)-based proteomics is the methodology of choice for large-scale identification and quantification of protein PTMs, including histones. High sensitivity proteomics requires online MS coupling with relatively low throughput and poorly robust nano-liquid chromatography (nanoLC) and, for histone proteins, a 2-d sample preparation that includes histone purification, derivatization, and digestion. We present a new protocol that achieves quantitative data on about 200 histone PTMs from tissue or cell lines in 7 h from start to finish. This protocol includes 4 h of histone extraction, 3 h of derivatization and digestion, and only 1 min of MS analysis via direct injection (DI-MS). We demonstrate that this sample preparation can be parallelized for 384 samples by using multichannel pipettes and 96-well plates. We also engineered the sequence of a synthetic “histone-like” peptide to spike into the sample, of which derivatization and digestion benchmarks the quality of the sample preparation. We ensure that DI-MS does not introduce biases in histone peptide ionization as compared to nanoLC-MS/MS by producing and analyzing a library of synthetically modified histone peptides mixed in equal molarity. Finally, we introduce EpiProfileLite for comprehensive analysis of this new data type. Altogether, our workflow is suitable for high-throughput screening of >1000 samples per day using a single mass spectrometer.
Proteomics has become one of the most widely used methodologies in biology and medicine, as identification and quantification of entire cell proteomes has allowed fundamental discoveries in disease etiology and basic science (Beck et al. 2011; Li et al. 2013; Aebersold and Mann 2016; Liu et al. 2016). Nano-liquid chromatography coupled online with tandem mass spectrometry (nanoLC-MS/MS) is nowadays the most sensitive and comprehensive strategy for proteomics analyses. The confidence in identification and quantification of highly complex protein mixtures has become possible due to the high mass accuracy (<5 ppm), high resolution (>60,000), and high sensitivity (<amol) of LC-MS. Moreover, the throughput of such analyses has achieved ranges of 1–3 h per sample (Hebert et al. 2014), including for the analysis of protein posttranslational modifications (PTMs) like phosphorylation (Humphrey et al. 2018).
Histone proteins have been extensively studied in basic science and research associated to disease etiology, as their PTMs play a fundamental role in modeling the chromatin state (Kouzarides 2007). Chromatin remodeling has a direct effect on gene regulation, recruiting enzymes for DNA repair and tuning the chromatin condensation during the cell cycle. Histone PTMs exercise this function both by changing the chemical properties of histone-DNA interactions, e.g., acetylated histones contribute to relax DNA compaction due to reduced positive charges on lysine residues, or by recruiting complexes of protein “readers” that include transcription factors, enzymes, and other structural proteins (Bannister and Kouzarides 2011). Together, the catalysis of a histone modification in a chromatin domain can orchestrate a substantial engagement of cellular factors modulating DNA readout. Aberrant regulation of classes of enzymes defined as histone “writers” (for histone PTM catalysis) or “erasers” (for histone PTM removal) can have effects on the cell phenotype. For instance, mutations in enzymes that are involved in chromatin organization have been found in more than 50% of human cancers (Jones et al. 2016). Abnormal levels of histone PTMs have also been detected in many other diseases, including diabetes (Raciti et al. 2014), PTSD (Dirven et al. 2017), schizophrenia (Ibi and González-Maeso 2015), allergies (Tost 2018), and drug or alcohol addiction (Wimmer et al. 2017). Aging and aging-related diseases such as Alzheimer's are other examples where levels of histone PTMs were found to be different from phenotypes representing optimal fitness (Berson et al. 2018).
Accurate quantification of histone protein levels has also emerging potential for biomarker discovery. For instance, the relative abundance of the histone variant H3.3 is higher as compared to canonical histone H3 in cells that do not proliferate (Maze et al. 2015), indicating a potential role of histone H3.3 as a cellular “clock” to discriminate highly proliferative cells like cancer. Histones with missense mutations have been discovered to be present in cancer tissues; e.g., H3K27M and H3G34V/R mutations occur in selected pediatric brain cancers, and H3K36M or H3G34W/L in pediatric bone tumors (Lohr et al. 2012; Behjati et al. 2013; Bjerke et al. 2013; Qiu et al. 2018). Altogether, the field of “oncohistones” is likely to become a new and important branch of cancer biology.
The analysis of histones and histone PTMs by MS has reached high levels of robustness and reproducibility, with coefficients of variations well below 20% even for low abundance marks (Sidoli et al. 2015b). Since histones are very basic proteins, i.e., highly enriched in lysine and arginine residues, the histone workflow normally includes derivatization of lysine residues to prevent the proteolytic digestion performed by trypsin in generating excessively short peptides (Garcia et al. 2007). The most abundant PTMs on histones are methylations (me1/me2/me3) and acetylation (ac), which are usually analyzed without any requirement for enrichment. However, histones are modified in traces by almost any modification type discovered on any other protein to date (Chen et al. 2007; Peng et al. 2011; Tan et al. 2011, 2014; Zhang et al. 2011; Dai et al. 2014; Huang et al. 2014; Xie et al. 2016). Because of this abundance of different PTMs, it is common to observe isobaric peptides, i.e., peptides carrying the same number and type of modifications but differently localized. By MS, the quantification of such peptides is solved by performing fragment-based MS/MS analysis, mostly now using data-independent acquisition (DIA) methods (Sidoli et al. 2015a,b, 2016b). Our lab has extensively contributed in developing workflows for histone analysis, including derivatization of histones by propionic anhydride (Garcia et al. 2007), MS data acquisition via DIA (Sidoli et al. 2015a,b, 2016b), and software that specifically deals with isobaric peptides (Yuan et al. 2015, 2018). However, this and others’ workflows are still limited in throughput; even at its best pace, the full experimental procedure requires about 2 d of sample preparation and ∼1 h for each LC-MS run. To envision histone analysis via MS as a potential platform for personalized diagnostics or for studies in systems or clinical biology, a much higher throughput is required. In addition, LC separation is a source of carryover and batch effects, as all samples are separated through the same chromatographic column, and one sample contaminating the stationary phase potentially alters the results of all samples injected afterward.
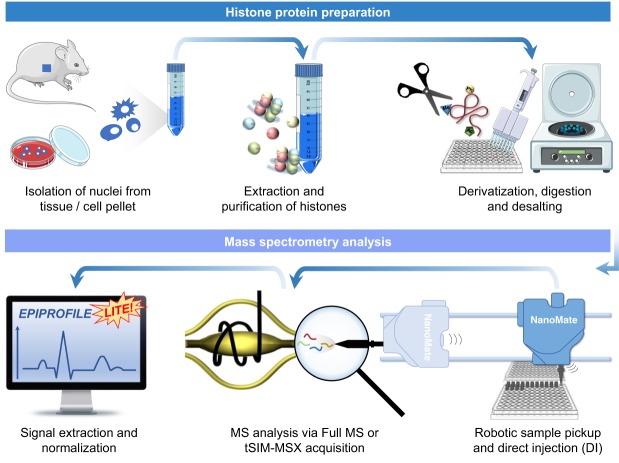
Here, we present a complete optimization of the histone PTM analysis workflow that can be performed in as little as ∼7 h starting from a cell pellet (either tissue or cell culture), and it can be scaled up to ∼400 samples in the same batch. The protocol includes histone extraction from cells in 96-well plates, followed by derivatization using propionic anhydride and digestion using trypsin. After desalting, the analysis of each sample is performed by direct injection using a TriVersa NanoMate (Advion) coupled online with an Orbitrap Fusion Tribrid (Thermo Fisher Scientific) in about 1 min. Raw file processing and peptide quantification are then performed by a newly developed freely available software EpiProfileLite, an adapted version of our EpiProfile software which we routinely use for LC-MS histone experiments (Yuan et al. 2015, 2018). The protocol includes a spike-in of a synthetic peptide mimicking the sequence of endogenous histones, of which complete derivatization and digestion can be quantified for quality control purposes.
Results
In this work, we have tested the performance of a high-throughput workflow for the analysis of histone PTMs via direct injection mass spectrometry (DI-MS) as compared to nanoLC-MS/MS. To validate the method in terms of reliability, robustness, and sensitivity, we performed the following comparisons: (1) We injected by nanoLC-MS/MS and direct injection a library of 61 synthetic peptides to assess that all these peptides are detectable and that there are not any biases in ionization efficiency significantly more pronounced compared to nanoLC-MS/MS. (2) We engineered a synthetic peptide to benchmark the efficiency of our sample preparation, as its byproducts of the derivatization and digestion can still be detected and quantified. We used this peptide to test multiple sample preparation strategies to minimize steps and improve throughput of the full workflow, as with DI-MS, sample preparation becomes the bottleneck of the workflow. (3) We removed the step of peptide N-termini propionylation, as this step is only beneficial to improve peptide hydrophobicity and LC retention, which is no longer needed for DI-MS. (4) We tested multiple types of acquisition methods to verify that data robustness is independent from the acquisition method and it is translatable into mass spectrometers other than the Orbitrap Fusion used in this study. (5) Finally, we proved that our results provide highly comparable results to nanoLC-MS/MS when comparing histone PTMs from brain and liver tissue when using DI-MS.
The optimization of our workflow for histone peptides refers to our state-of-the-art protocol for derivatization of histones which requires propionylation of histones (Sidoli et al. 2016a). To ensure that our workflow is suitable for high-throughput analysis, we optimized a protocol which requires ∼4 h of histone extraction, followed by 3 h of propionylation and digestion, followed by stage tipping (where samples are eluted directly into the buffer for direct injection), followed by about 1 min direct injection (Fig. 1). All of the steps, from the histone extraction to stage-tipping, can be performed with multichannel pipettes in 96-well plates. Stage-tipping was performed using a customized rotor arm holding stage-tips above a collector plate to collect the eluate (Supplemental Fig. 1A). This rotor arm can hold four sets of tips and plates (Supplemental Fig. 1B), allowing for simultaneous stage-tipping of 384 samples. To ensure that the instrument is in optimal condition to analyze samples, we inject digested bovine serum albumin (BSA) and acquire it using a data-dependent acquisition method (DDA) (Supplemental Fig. 1C). Because the analysis does not have chromatographic separation, the instrument will select for MS/MS fragmentation all the signals identified in the full MS (Supplemental Fig. 1D). Subsequently, the analysis of the BSA coverage can be performed using tools like Morpheus (Wenger and Coon 2013); we consider the coverage adequate when >60% (Supplemental Fig. 1E). The instrument should also prove that it can perform multiple injections without issues of signal stability when we re-inject BSA or perform multiple injections of other samples, i.e., for example, quantified peptide H3K9me3 injected 104 times without assistance (Supplemental Fig. 2). Together, the evaluation of the system prior to the experiment is performed by evaluating the sequence coverage of a directly sprayed BSA and repeated injections of the same BSA standard to assess reproducible spray current and stability.
Figure 1.
General workflow of histone analysis using DI-MS.
Detection efficiency assessment by synthetic peptides
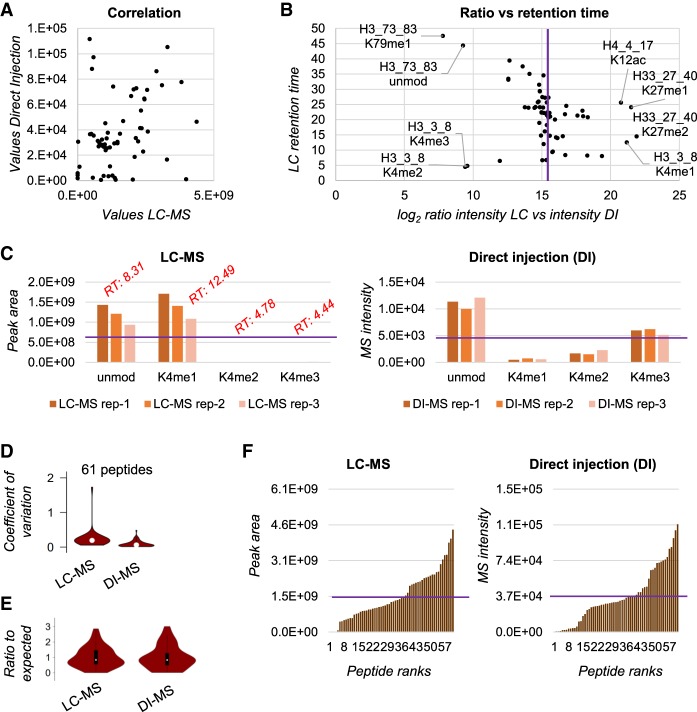
First, we injected a library of 61 isotopically labeled synthetic histone peptides (which we normally use as internal standards), corresponding to all the common modified and unmodified peptides of histone H3 and H4 (Supplemental Table 1). To assess biases in ionization efficiency, we mixed the peptides in all equal molarity (290 fmol/µL) and observed differences in intensity compared to the expected equal intensity for all peptides. The analysis was performed in parallel via nanoLC-MS/MS and via direct injection, showing poor correlation between the two techniques (correlation = 0.31) (Fig. 2A). We did not expect to observe high correlation in this case because high correlation is achieved only when two method are equally capable of quantifying differences in signal intensity of analytes. Since peptides were all mixed in equal molarity, higher or lower signals are only due to biases of various type. Four peptides showed an underestimation by nanoLC-MS/MS: H3K4me2, H3K4me3, H3K79me1, and H3K79unmod (Fig. 2B); the first two are very hydrophilic, while the other two are very hydrophobic. It is reasonable to assume that hydrophilic peptides are not retained properly by nanoLC-MS/MS, and therefore their quantification is underestimated. Regarding the two hydrophobic peptides, they elute in the very last part of the chromatogram where potential interference of nonpeptide contaminants might suppress the actual signal. A side-by-side view of the quantification for three injections of the synthetic peptide TKQTAR (H3 aa 3–8) clearly shows that the very early eluting peptides are underestimated by nanoLC-MS/MS as compared to expected (purple line, Fig. 2C). The three technical replicates performed by direct injection (DI-MS) showed a coefficient of variation significantly smaller than the three technical replicates analyzed by nanoLC-MS/MS (coefficient of variation 8.9% and 23.6%, respectively) (Fig. 2D; Supplemental Fig. 3). This was not surprising, as nano-chromatography is an inevitable source of variability in proteomics analyses. In addition, we speculate that higher reproducibility is also a consequence of the “simper” task required to define signal abundance; when performing LC-MS, the abundance corresponds to the accurate definition of an extracted ion chromatogram, while in DI-MS it is simply the intensity of the signal within a single spectrum. Collectively, our biggest concern was introducing biases by DI-MS significantly larger than nanoLC-MS/MS; our preliminary investigation using a synthetic peptide library verified that the two techniques have at least comparable inaccuracies as compared to expected signal intensity. If anything, DI-MS showed a smaller overall deviation from expected results compared to nanoLC-MS/MS (Fig. 2E,F).
Figure 2.
Detection bias evaluation using a synthetic peptide library. (A) Correlation of the area under the curve extracted for the synthetic peptides by LC-MS (x-axis) and the intensity in the spectrum of the peptides detected by DI-MS (y-axis). The figure is obtained with the average of three technical replicates. (B) Ratio between the area under the curve of the synthetic peptides quantified via LC-MS versus the intensity obtained by DI-MS (x-axis). The y-axis represents the retention time obtained by the LC-MS run. The purple line indicates the expected ratio in case of no bias. (C) Raw abundance of the histone H3 peptide TKQTAR (aa 3–8) obtained by LC-MS (left) and DI-MS (right) for three technical replicates. The purple line represents the theoretical abundance with no bias; the red text indicates the retention time of the LC-MS peaks. (D) Distribution of the coefficient of variation (CV) for three technical replicates for all the 61 synthetic peptides by LC-MS and DI-MS. (E) Ratio between observed abundance and expected abundance for the synthetic peptides. The expected abundance is calculated by averaging all signals as, in theory, they should all provide the same intensity. (F) Distribution of the peptide observed abundance for all 61 peptides by LC-MS (left) and DI-MS (right). Purple line indicates expected intensity.
Quality control by an engineered synthetic ‘histone-like’ peptide sequence
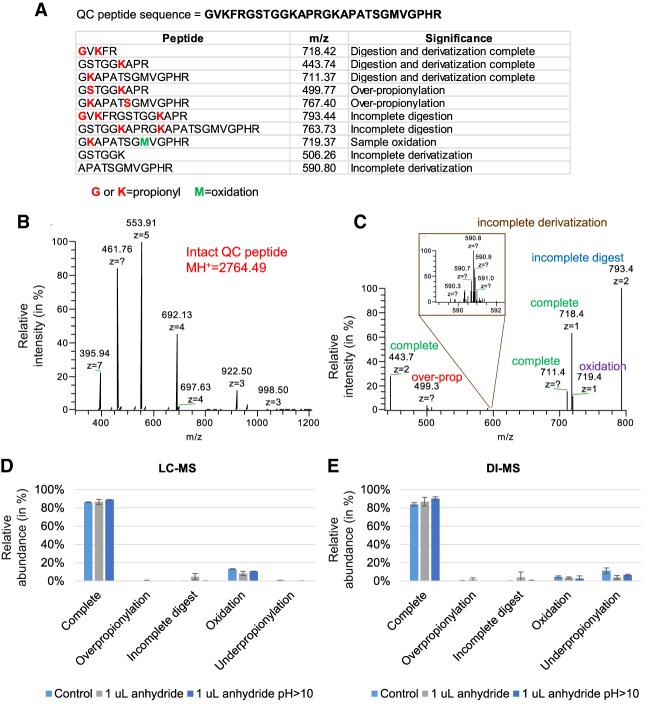
A common gap in histone analysis is a proper quality control (QC) of the sample preparation. To compensate for this issue, we engineered a peptide with a sequence not corresponding to any known histone sequences but with similar physical and chemical properties, i.e., rich in lysine and arginine residues (Fig. 3A). The sequence is not encoded in any database, but it resembles existing histone peptides once digested. This peptide sequence was planned on purpose so that incomplete digestion and/or improper propionylation (under- or overpropionylation) generates peptides that are also detectable within the m/z range of canonical histone peptides. We verified the purity of our peptide by infusing the molecule completely undigested (Fig. 3B), and we verified the sequence by HCD fragmentation and high resolution MS/MS acquisition (Supplemental Fig. 4). We then checked the production of complete and incomplete products by testing a 5-min trypsin digestion (Fig. 3C). Spectra were acquired using a targeted SIM scan which we could multiplex (MSX) in the Orbitrap Fusion acquiring all 10 signals in the same spectrum. Next, we applied the use of the QC peptide by comparing three types of sample preparation, each aiming to minimize the time required to propionylate and digest a purified histone mixture. The sample utilized was mouse embryonic stem cells differentiated for 3 d. As a control, we used the standard protocol for histone derivatization and digestion (Sidoli et al. 2016a) with only 2 h incubation with trypsin and with no N-terminal peptide propionylation. Moreover, we compared two other sample preparations, one including just 1 µL of propionic anhydride for each round of derivatization (to avoid drying before introducing trypsin) and one with an excess of ammonium hydroxide added after propionic anhydride (to assess issues of overpropionylation when pH > 10). All three methods obtained an efficiency of propionylation and digestion estimated to be around 98% when considering the peptide oxidized at the methionine residue as part of the complete products (nanoLC-MS/MS data) (Fig. 3D). Methionine oxidation is considered a negligible issue because all the major peptides quantified in our routine analyses do not contain methionine residues. The comparison with the results obtained with DI-MS analysis (Fig. 3E) had an R2 Pearson correlation >0.98 (Supplemental Table 2). The major observable difference between the two results was a more sensitive detection of the underpropionylated products; this is most likely because chromatographic retention is not a factor during direct injection, and underpropionylated peptides have more hydrophilic properties. In conclusion, we have demonstrated that a sample preparation correctly performed leads to a relative abundance of the completely derivatized and digested spike-in well above 90% (not considering oxidation). We recommend discarding and repeating runs that show poorer yields benchmarked by the “histone-like” spike-in peptide.
Figure 3.
Analysis of the QC peptide. (A) Sequence of the QC peptide (top) and theoretical masses of the products of derivatization and digestion. The indicated m/z was the most intense charge state determined by manual observation of the spectra. (B) Full MS spectrum of the peptide underivatized and undigested. (C) Example of signals detectable after derivatization and digestion. The spectrum was acquired after 5 min digestion (to detect evidence of undigested forms) using a targeted-SIM multiplexed (MSX) scan. (D) Comparison of three sample preparation protocols using the QC peptide detected by LC-MS and (E) DI-MS. Control is defined as 2 h digestion at pH 8 using 5 µL of propionic anhydride; the second protocol used only 1 µL of propionic anhydride, and the third used an excess of ammonium hydroxide to bring the pH > 10. The QC peptide was prepared in a background of endogenous histones purified from mouse stem cells. The error bar represents the standard deviation of three biological replicates.
Acquisition of endogenous histone peptides by variable-size full MS windows
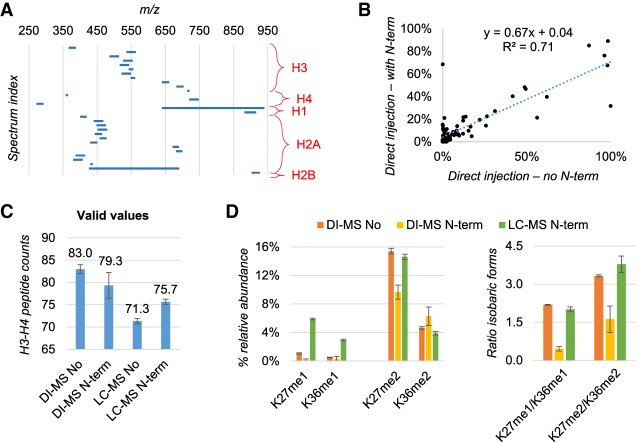
Once we established that the derivatization and digestion protocol were of high efficiency, we created a method to detect peptide groups in small size MS windows (Fig. 4A). Each of these windows included all the unmodified and modified states of peptides sharing the same sequence which, when detected together, can be normalized by the signal of one another to obtain the relative abundance of each modified form. To ensure accurate discrimination between acetylation (42.01 Da) and trimethylation (42.047 Da), we acquired the full MS spectrum using a resolution of 240,000 (Supplemental Fig. 5A). This acquisition could easily detect all the canonical acetylated and methylated states of histone peptides, including low abundance species normally detected only by nanoLC-MS/MS (Supplemental Fig. 5A,B). Because histone peptides have numerous isobaric forms, i.e., peptides sharing the same precursor mass but with different localization of the modification (example in Supplemental Fig. 5C), we programmed the acquisition method to perform targeted MS/MS fragmentation of isobaric peptides which we then used to determine the relative ratio between the isobaric forms using the principle of the Fragment Ion Relative Ratio (Pesavento et al. 2008). This canonical protocol of histone preparation for nanoLC-MS/MS (Sidoli et al. 2016a) contains two rounds of propionylation, including both propionylation of lysine residues prior to digestion and peptide N-terminal propionylation postdigestion. This latter is recommended to increase the hydrophobicity of histone peptides. However, we verified that removing the N-terminal propionylation step improves the detection of the b-ions series of histone peptides, which aid in the discrimination of the isobaric forms at the MS/MS level (Supplemental Fig. 6A–C). First, we verified that we did not introduce significant biases in the estimation of PTM abundances by removing the step of N-terminal propionylation. By comparing the relative abundances estimated by DI-MS of histones extracted from HEK293T cells, we observed a good correlation (R2 = 0.71) by preparing the samples with and without the step of N-terminal derivatization (Fig. 4B). Then, we looked at how many quantifiable peptides we could observe by removing the N-terminal propionylation step; results showed that DI-MS increased the number of peptides with quantification ≠ 0 by removing this step in the sample preparation (Fig. 4C). Understandably, the trend was the opposite for the nanoLC-MS/MS results, as by removing the propionylation, peptides bind less efficiently the C18 column. We also selected two examples of isobaric forms, of which the relative ratio needs to be estimated by using MS/MS fragment ions (Fig. 4D). Data clearly showed that the presence of N-terminal propionylation provides a different ratio estimation between the isobaric forms as compared also to nanoLC-MS/MS (here used as reference).
Figure 4.
Evaluation of detection biases by skipping peptide N-terminal derivatization. (A) Overview of the full MS acquisition windows set up for the DI-MS method. The MS/MS scans of the isobaric forms are not displayed. (B) Relative abundance of endogenous peptides extracted from human HEK293T cells quantified by DI-MS after excluding (x-axis) or including (y-axis) N-terminal peptide derivatization in the protocol. Data are the average of three biological replicates. (C) Number of peptides quantified from histone H3 and H4 with or without N-termini derivatization by DI-Ms and LC-MS. As expected, without N-termini derivatization LC-MS has more issues in binding and resolving histone peptides. (D) Example of isobaric peptides with relative ratio estimated by MS/MS in DI-MS acquisition. The example was selected specifically because these peptides are baseline resolved by LC-MS, which was used as a reference. The ratio was wrongly estimated by DI-MS when using N-terminal derivatization (right).
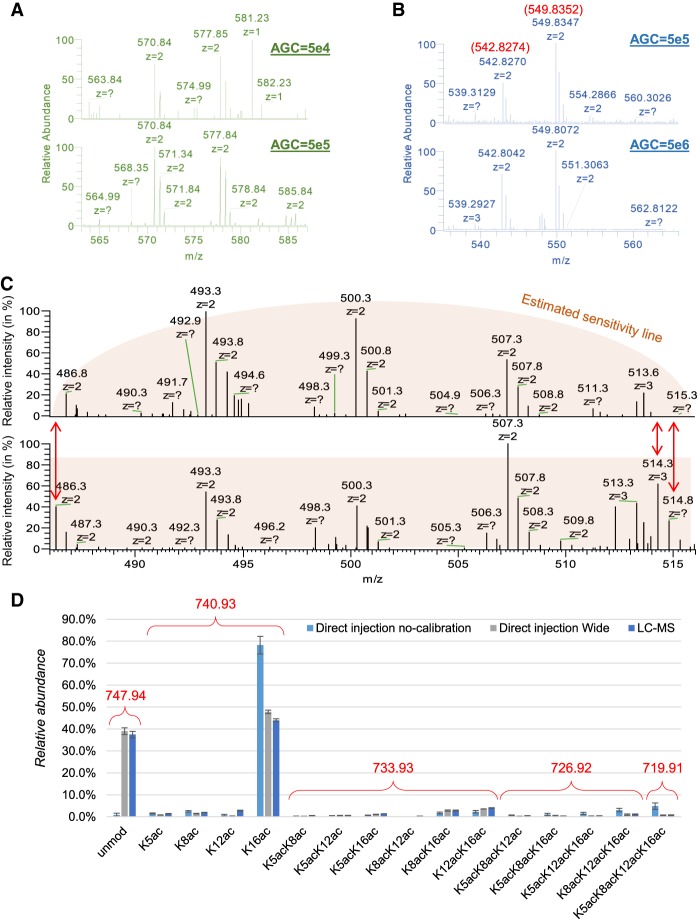
Next, we optimized the acquisition method to ensure the maximum sensitivity and accuracy of the analysis. First, we optimized the automatic gain control (AGC) in the Orbitrap to achieve the best sensitivity while maintaining low potential charge excess into the analyzer, which leads to inaccurate mass detection. We proved that 5 × 105 ions was the ideal target, as 5 × 104 ions led to insufficient sensitivity (Fig. 5A) and 5 × 106 ions created deviations in the mass accuracy (more than 60 ppm) (Fig. 5B). This was of mandatory importance to ensure stringent peak extraction to discriminate acetylation from trimethylation. In addition, we investigated the potential issues caused by nonperfect calibration of the quadrupole for the isolation of the full MS scan. Specifically, the Orbitrap Fusion (Thermo Fisher Scientific) can perform full MS by prefiltering all the ions not desired in the full MS with the quadrupole analyzer. This improves the ion capacity of the full scan, since all the nondesired ions are not accumulated in the ion routing multipole (IRM). However, we verified that noncalibrated wide quadrupole isolation leads to loss of sensitivity at the edges of the isolation window (Fig. 5C). The figure clearly shows that ions at m/z 486.3, 514.3, and 514.8 almost completely disappeared from the spectrum when quadrupole isolation was not properly calibrated. This generates inevitable biases in estimating PTM relative abundances; by quantifying the histone H4 peptide aa 4–17, we noticed that the largest ion (unmodified, m/z 747.94) was underestimated by DI-MS when not calibrated (Fig. 5D). To assess the correct reference, we ran both DI-MS using a larger acquisition window and nanoLC-MS/MS, both showing highly comparable results.
Figure 5.
Optimization of the AGC target and acquisition window for DI-MS. (A) Example displaying how an acquisition using 50,000 ions as AGC is less indicated than 500,000 ions to obtain a clear S/N ratio for the two peptides of histone H3 KQLATKAAR (aa 18–26) unmodified (577.85 m/z) and with one acetyl (570.84 m/z). This sample was derivatized at the peptide N-termini. (B) Example showing that 5,000,000 as AGC target creates issues of mass accuracy as compared to 500,000. The peptides are the same as panel A, but they were not derivatized at the N-termini in this experiment. (C) Example of incorrect (top) and correct (bottom) calibration of the quadrupole wide isolation in an Orbitrap Fusion. The signals at the edges of the acquisition window are lost, creating significant biases in the estimation of peptide abundance. (D) Relative abundance of the peptide of histone H4 GKGGKGLGKGGAKR (aa 4–17) detected using uncalibrated quadrupole wide isolation using DI-MS with narrow acquisition, with an extra 2 m/z acquisition on both ends of the acquisition window and by canonical LC-MS. It is evident that, even without calibration, the acquisition is correctly performed using wide windows and that all isobaric forms of this peptide (which contains four modifiable residues) were resolved.
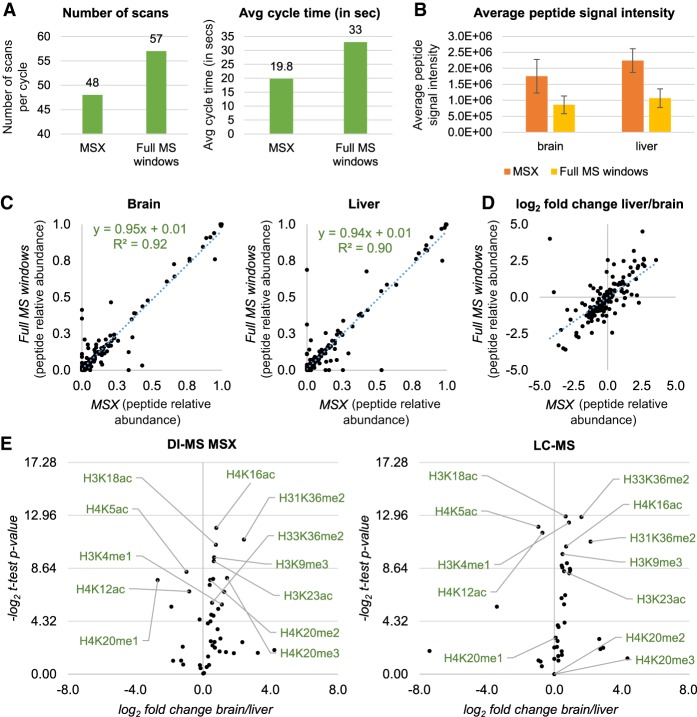
Finally, we tested a fully targeted acquisition method, where each m/z value considered for peptide quantification was not acquired by a full MS scan but by a targeted SIM. This test was performed to assess whether the relative quantification of PTMs is significantly affected by acquiring each signal individually instead of within the same scan. In addition, a targeted method paves the way to the applicability of our setup also for instruments that cannot perform wide quadrupole isolation like the Orbitrap Fusion. To reduce the cycle time, the SIM scans were multiplexed 10 by 10 in MSX scans (full list of scans in Supplemental Table 3), reducing the cycle time further as compared to the method described so far (Fig. 6A). A cartoon representation of the main differences between the acquisition method based on full MS windows and the targeted MSX method is illustrated in Supplemental Figure 7. In addition, we observed a significant increase in sensitivity (calculated using peptide intensities), due to the more focused acquisition range (Fig. 6B). By using both methods, we estimated the relative abundance of 199 histone peptides extracted from mouse brain and liver, achieving excellent correlation values when comparing the two methods (Fig. 6C). As well, when we compared the log2 fold change of each peptide quantified in liver versus the peptide quantified in brain, we obtained very similar values (Fig. 6D). This was reassuring, as performing single MS fills of ions rather than acquiring a range including all the modified peptide forms might have led to differences in quantification accuracy. However, results clearly demonstrate that the two experiments provide very high similarities. Therefore, we adopted the DI-MS method acquired using SIM scans multiplexed (MSX) to estimate which histone PTMs are significantly different in relative abundance between mouse and brain liver. Among others, we identified a higher abundance of selected acetylations, i.e., H4K16ac, H3K23ac, and H3K18ac, and the heterochromatic marker H3K9me3 (Fig. 6E). On the other hand, other acetylations like H4K5ac and H4K12ac were found to be more abundant in liver cells. This also proved that our method can accurately discriminate the relative abundance of isomeric PTMs, i.e., histone modifications of the same type on the same peptide but localized on different amino acid residues. As a final comparison, we injected the same sample in a nanoLC-MS/MS analysis, obtaining high correlation when comparing the relative abundances of peptides acquired with the two acquisition methods (R2 > 0.92 for both tissues) (Supplemental Fig. 8; Supplemental Table 4). We thus focused on the analysis of histones H3 and H4, which have the most modified peptides, obtaining a highly similar volcano plot when deconvoluting the relative abundances of the single histone marks (Fig. 6E; Supplemental Table 5). Minor differences in significantly regulated modifications were observed for the PTMs H4K20me2 and H4K20me3; this can be easily explained by the high hydrophilicity of the peptide carrying these PTMs (aa 20–23, KVLR), which ends up being poorly retained by LC and therefore its significance could not be assessed by nanoLC-MS/MS. Altogether, we implemented a simplified workflow for histone sample preparation combined with a high-throughput DI-MS analysis of histone peptides demonstrating highly comparable results with the current state-of-the-art method but with a throughput almost 100-fold higher.
Figure 6.
Multiplexed SIM acquisition of endogenous histone peptides. (A) Number of scan events per cycle (left) and average time (right) during DI-MS acquisition using tSIM-MSX versus the canonical DI-MS with windows described in Figure 4A. The instrument allows multiplexing up to 10 ions per scan, so the number of scans can be reduced. (B) Average peptide intensity obtained by running a side-by-side acquisition of endogenous histone peptides extracted from mouse brain and liver using DI-MS and the two different acquisition methods. (C) Relative abundance of histone peptides from brain (left) and liver (right) acquired using MSX (x-axis) or full MS windows (y-axis). (D) Log2 fold change of the histone peptides from the two data sets acquired using MSX (x-axis) or full MS windows (y-axis). (E) Volcano plot showing the total relative abundance of single histone modifications from histone H3 and H4 (estimated by summing the relative abundance of all peptides carrying a given modification) in brain versus liver. Data were acquired using DI-MS with MSX (left) and LC-MS (right). Results show the same PTMs as statistically different in abundance between the two tissues, with the exception of H4K20me2 and H4K20me3, due to inaccurate quantification by LC-MS.
Discussion
In this work, we presented the first attempt to analyze hundreds of histone PTMs in a single analysis using <1-min direct injection into a mass spectrometer. The method has been optimized in many aspects, including acquisition parameters and controls for benchmarking the sample preparation. In the end, we demonstrate that this workflow achieves almost identical results to nanoLC-MS/MS, which is currently performed with at least 1-h LC gradients without considering sample loading and column equilibration (Sidoli et al. 2015b).
This approach aims to put a milestone into the high-throughput analysis of cell epigenomes. Histone PTMs play fundamental roles in “flagging” chromatin domains to prevent or promote gene expression, and thus their characterization is fundamental to understand development and disease. Large efforts have been already taken to map histone PTMs in large-scale; one important example is the GCP assay promoted by the LINCS Proteomic Characterization Center for Signaling and Epigenetics. However, to reach goals comparable to the achievements of the genomics community (1000 genomes per day), we need a high throughput at the scale presented in the current work. Our method exploits the fast speed of MS acquisition, highlighting how the real bottleneck in histone analysis is currently chromatographic separation. Running the samples for 1 min allowed the instrument to acquire three cycles of the same spectra, because each cycle lasts about 20 sec (Fig. 6A). With three cycles, the software EpiProfileLite can pick the spectra that have the most complete pattern of ions, discarding, e.g., if in one scan the signal was temporarily unstable. This does not happen routinely, but it is helpful as a precaution. We have tested and do not recommend performing scan averaging, as it unnecessarily slows the duty cycle without gaining extra sensitivity. This rapid analysis allows also for simple spotting of runs that require repetition; since the run has a fixed and small number of spectra, poor quality runs (e.g., in case of signal instability) are easily depicted by the smaller size of the raw file. A low signal leads to fewer signals in the MS and MS/MS spectra, which is reflected in the file size.
Our method still has margins for improvement. For instance, the direct injection of histone peptides could benefit from the addition of supercharging agents, or a wiser or more accurate choice of buffers to maximize efficiency and spray stability, which now we cannot freely test due to the limitation of what is compatible with liquid chromatography. Occasionally, spray stability via direct injection can also be an issue. Thus, investing in this optimization could enhance the robustness of the overall workflow. Nevertheless, we observed a high reproducibility during injections (Supplemental Fig. 1F), indicating that the spray is stable for this type of repetitive injections.
We do not exclude that our work will stimulate more efforts into the analysis or peptide mixtures of reduced complexity without the assistance of chromatographic separation. This is not a novelty, as proteomics can be performed by using MALDI sources, which do not have upfront chromatographic separation. However, it is a fact that the proteomics community largely relies on nanoLC-MS/MS to identify and quantify protein mixtures. Even though there is still a significant gap in sensitivity between direct injection and nanoLC-MS/MS, we are confident that future applications without the use of chromatography will be developed. Chromatography is currently the major cause of technical batch effects, as we also showed by presenting a much lower variation in multiple injections by DI-MS (Fig. 2D).
In conclusion, our goal is to introduce this experimental procedure as the new gold standard in histone analysis, as it has the potential to drastically reduce requirements of instrument time and to potentially eliminate the majority of batch effects caused by chromatographic separation.
Methods
Growth of HEK293T cells
Three biological replicates of human embryonic kidney cells 293 (HEK293) were grown at 37°C in a 5% CO2 atmosphere in Dulbecco's Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) and 100 units/mL penicillin and 100 µg/mL streptomycin.
Mouse stem cell culture and differentiation
Three biological replicates of mouse embryonic stem cells were cultured at 37°C in a 5% CO2 atmosphere on 10-cm plates coated with 0.1% gelatin in Dulbecco's Modified Eagle Medium with 4.5 g/L glucose, L-glutamine, and sodium pyruvate, as well as 1% GlutaMAX, 1% nonessential amino acids, 15% characterized fetal bovine serum, 0.1 mM 2-mercaptoethanol, and supplemented with ESGRO supplement (LIF) to help maintain stem cell pluripotency. After reaching about 70% confluency, the stem cells were differentiated with 10 µM retinoic acid to form embryoid bodies. The embryoid bodies were collected on Day 3 of differentiation for histone extraction.
Mouse brain and liver cell tissue
Brain and liver tissue of three different mice were homogenized in nuclear isolation buffer (NIB) + 0.2% NP-40 Alternative using a dounce homogenizer for 30 strokes, and subsequently histones were extracted from the brain and liver tissue as previously described (Sidoli et al. 2016a).
Histone extraction
Twenty-five microliters of cell pellet were placed into 96-well plates, and nuclei were isolated by using Nuclei Isolation Buffer (NIB) as previously described (Sidoli et al. 2016a). Briefly, the first round of NIB treatment was performed by using NIB + 0.2% NP-40 at a volume buffer:cell pellet of 9:1 to lyse the cell membrane and two subsequent washes without NP-40 to remove detergents. Each step was followed by centrifugation at 600g to collect the pellet. After nuclei were isolated, histone proteins were extracted as described in the protocol of Sidoli et al. (2016a) with minor adjustments. Briefly, histones were acid extracted from nuclei with 0.2 M H2SO4 for 2 h and precipitated with 20% trichloroacetic acid (TCA) for 1 h. The histone pellet was then washed with ice-cold acetone to remove leftover acid.
Histone derivatization and digestion
The derivatization and digestion were performed as previously described (Sidoli et al. 2016a). Briefly, histones were dissolved in 20 µL of 50 mM NH4HCO3, pH 8.0. Derivatization reagent was prepared by mixing the sample with 5 µL of acetonitrile, followed by 5 µL of propionic anhydride and 14 µL of ammonium hydroxide (to balance the pH at 8.0) for 15 min at 37°C. This reaction was performed twice. Histones were then digested with trypsin (enzyme:sample ratio 1:20, 2 h, room temperature) in 50 mM NH4HCO3. Where specified, the derivatization reaction was performed again twice to derivatize peptide N-termini. Where specified (Fig. 3D), only 1 µL of propionic anhydride was used instead of 5, and consequently only 1 µL of ammonium hydroxide instead of 14 µL to adjust the pH. To test overadjustment of pH, only 1 µL of propionic anhydride was used plus 5 µL of ammonium hydroxide instead of 1. Samples were desalted prior to DI-MS or nanoLC-MS/MS analysis by using in-house packed stage-tips. Stage-tips were manufactured by sealing a disk of C18 material at the bottom of a P200 tip, followed by an equivalent of about 0.5 cm of Porous Graphitic Carbon resin (PGC, Hypercarb, Thermo Fisher Scientific) dissolved into acetonitrile. Stage-tips were equilibrated with 50 µL of water + 0.1% TFA, the sample was loaded after adding 1% TFA to the solution, the tips were washed twice with 30 µL of water + 0.1 FA, and elution was performed by using 30 µL of 60% acetonitrile + 0.1% FA. The full procedure was performed by centrifugation to force the solution through the tips with a SpeedVac Plus SC210A (Savant) without using the vacuum pump.
NanoLC-MS
Samples were analyzed by using a nanoLC-MS/MS setup. NanoLC was configured with a 75 µm ID × 17 cm Reprosil-Pur C18-AQ (3 µm; Dr. Maisch GmbH) nano-column packed in-house using an EASY-nanoLC nanoHPLC (Thermo Fisher Scientific). The HPLC gradient was as follows: 2% to 28% solvent B (A = 0.1% formic acid; B = 80% MeCN, 0.1% formic acid) over 45 min, from 28% to 80% solvent B in 5 min, 80% B for 10 min at a flow-rate of 300 nL/min. nanoLC was coupled to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). Data were acquired using a data-independent acquisition method, consisting of a full scan MS spectrum (m/z 300−1100) performed in the Orbitrap at 120,000 resolution (m/Δm for the ion at 200 m/z), followed by 16 MS/MS with windows of 50 m/z using HCD fragmentation and detected in the ion trap. More details on the acquisition method are available in Sidoli et al. (2015b).
Direct injection–MS
For DI-MS, samples were placed in a TriVersa NanoMate (Advion) and acquired either manually or by using a sequence coordinated with MS acquisition by a contact closure. The NanoMate was set up with a spray voltage of 1.7 kV and a gas pressure of 0.5 psi. Samples were acquired in the same Orbitrap Fusion (Thermo Fisher Scientific) as for nanoLC-MS/MS. All scans were acquired in the Orbitrap, at 240,000 resolution for the full MS and at 120,000 resolution for MS/MS. Multiple tests were performed for the AGC target, the MSX acquisition, and the window sizes. The full descriptions of the DI-MS acquisition methods (with windows and with tSIM-MSX) are included as Supplemental Files in the electronic version of the manuscript.
Histone peptide quantification
DIA data obtained from the nanoLC-MS/MS runs were searched using EpiProfile 2.0 (Yuan et al. 2018). The peptide relative ratio was calculated using the total area under the extracted ion chromatograms of all peptides with the same amino acid sequence (including all of its modified forms) as 100%. For isobaric peptides, the relative ratio of two isobaric forms was estimated by averaging the ratio for each fragment ion with different mass between the two species. For DI-MS data, we modified the EpiProfile software and generated a new version named EpiProfileLite (Supplemental Code 1). In DI-MS, histone peptides are collected in MS scans, and isobaric peptides are collected in targeted preset MS/MS scans. EpiProfileLite reads the intensities from MS scans to calculate the percentage of all peptides with the same amino acid sequence. The unique fragment ions in the MS/MS scans are extracted to discriminate isobaric peptide intensities from the MS scans. EpiProfileLite is also available on GitHub at https://github.com/zfyuan/EpiProfileLite, including the user guide. For the analysis of the synthetic peptide library, we used EpiProfile (Yuan et al. 2018) for the LC-MS runs and an in-house R script for the DI-MS runs (Supplemental Code 2). The R script consists of two custom functions based on MSnbase (Gatto and Lilley 2012). Initially, the ms1PeakExtractor function takes an mzML file, extracts all spectra headers, filters them by MS1, and it extracts all candidate spectra from MS1. From the candidate spectra, the ms1PeakMatcher function reads the synthetic peptide sequence table, calculates the theoretical monoisotopic masses by elemental composition masses, extracts all matches from each peptide with m/z, charge, intensity, and delta mass from given mass tolerance in ppm, and finally generates the table format report for the downstream analysis. Data analysis was fully performed by using Microsoft Excel; this way we calculated averages, standard deviations, and two-tail t-tests (statistical confidence assessed when P-value < 0.05).
Data access
All raw mass spectrometry data files from this study have been submitted to the Chorus repository (http://chorusproject.org/) under accession number 1546.
Supplementary Material
Acknowledgments
We thank Dr. William H. Peranteau's lab at the Children's Hospital of Philadelphia for the mouse brain and liver tissue. This work was supported by National Institutes of Health (NIH) grants NIH AI118891, CA196539, and GM110104. Y.K. and B.A.G. are also supported by funding from NIH grant 5T32GM071339-13 and M.L. by grant FAPESP #2017/15835-1. L.M.A. is supported by an NSF-GRFP fellowship, and A.J.A. is supported by NIH R01 grant GM102503.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.247353.118.
References
- Aebersold R, Mann M. 2016. Mass-spectrometric exploration of proteome structure and function. Nature 537: 347–355. 10.1038/nature19949 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. 2011. The quantitative proteome of a human cell line. Mol Syst Biol 7: 549 10.1038/msb.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482. 10.1038/ng.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson A, Nativio R, Berger SL, Bonini NM. 2018. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci 41: 587–598. 10.1016/j.tins.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, Bax DA, Carvalho D, Taylor KR, Vinci M, et al. 2013. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov 3: 512–519. 10.1158/2159-8290.CD-12-0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng JM, Gu W, Zhao YM. 2007. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics 6: 812–819. 10.1074/mcp.M700021-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai LZ, Peng C, Montellier E, Lu ZK, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin FL, et al. 2014. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol 10: 365–370. 10.1038/nchembio.1497 [DOI] [PubMed] [Google Scholar]
- Dirven BCJ, Homberg JR, Kozicz T, Henckens M. 2017. Epigenetic programming of the neuroendocrine stress response by adult life stress. J Mol Endocrinol 59: R11–R31. 10.1530/JME-17-0019 [DOI] [PubMed] [Google Scholar]
- Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. 2007. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc 2: 933–938. 10.1038/nprot.2007.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto L, Lilley KS. 2012. MSnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 28: 288–289. 10.1093/bioinformatics/btr645 [DOI] [PubMed] [Google Scholar]
- Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. 2014. The one hour yeast proteome. Mol Cell Proteomics 13: 339–347. 10.1074/mcp.M113.034769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. 2014. SnapShot: histone modifications. Cell 159: 458–458.e1. 10.1016/j.cell.2014.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SJ, Karayel O, James DE, Mann M. 2018. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat Protoc 13: 1897–1916. 10.1038/s41596-018-0014-9 [DOI] [PubMed] [Google Scholar]
- Ibi D, González-Maeso J. 2015. Epigenetic signaling in schizophrenia. Cell Signal 27: 2131–2136. 10.1016/j.cellsig.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Issa JP, Baylin S. 2016. Targeting the cancer epigenome for therapy. Nat Rev Genet 17: 630–641. 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Li XJ, Hayward C, Fong PY, Dominguez M, Hunsucker SW, Lee LW, McLean M, Law S, Butler H, Schirm M, et al. 2013. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med 5: 207ra142 10.1126/scitranslmed.3007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. 2016. On the dependency of cellular protein levels on mRNA abundance. Cell 165: 535–550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. 2012. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci 109: 3879–3884. 10.1073/pnas.1121343109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Wenderski W, Noh KM, Bagot RC, Tzavaras N, Purushothaman I, Elsasser SJ, Guo Y, Ionete C, Hurd YL, et al. 2015. Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87: 77–94. 10.1016/j.neuron.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Lu ZK, Xie ZY, Cheng ZY, Chen Y, Tan MJ, Luo H, Zhang Y, He W, Yang K, et al. 2011. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics 10: M111.012658 10.1074/mcp.M111.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. 2008. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem 283: 14927–14937. 10.1074/jbc.M709796200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Hu X, Jing Q, Zeng X, Chan KM, Han J. 2018. Mechanism of cancer: oncohistones in action. J Genet Genomics 45: 227–236. 10.1016/j.jgg.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Raciti GA, Nigro C, Longo M, Parrillo L, Miele C, Formisano P, Béguinot F. 2014. Personalized medicine and Type 2 diabetes: lesson from epigenetics. Epigenomics 6: 229–238. 10.2217/epi.14.10 [DOI] [PubMed] [Google Scholar]
- Sidoli S, Lin S, Xiong L, Bhanu NV, Karch KR, Johansen E, Hunter C, Mollah S, Garcia BA. 2015a. Sequential window acquisition of all theoretical mass spectra (SWATH) analysis for characterization and quantification of histone post-translational modifications. Mol Cell Proteomics 14: 2420–2428. 10.1074/mcp.O114.046102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Simithy J, Karch KR, Kulej K, Garcia BA. 2015b. Low resolution data-independent acquisition in an LTQ-Orbitrap allows for simplified and fully untargeted analysis of histone modifications. Anal Chem 87: 11448–11454. 10.1021/acs.analchem.5b03009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Bhanu NV, Karch KR, Wang X, Garcia BA. 2016a. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J Vis Exp (111): e54112 10.3791/54112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Fujiwara R, Garcia BA. 2016b. Multiplexed data independent acquisition (MSX-DIA) applied by high resolution mass spectrometry improves quantification quality for the analysis of histone peptides. Proteomics 16: 2095–2105. 10.1002/pmic.201500527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MJ, Luo H, Lee S, Jin FL, Yang JS, Montellier E, Buchou T, Cheng ZY, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028. 10.1016/j.cell.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MJ, Peng C, Anderson KA, Chhoy P, Xie ZY, Dai LZ, Park J, Chen Y, Huang H, Zhang Y, et al. 2014. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab 19: 605–617. 10.1016/j.cmet.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J. 2018. A translational perspective on epigenetics in allergic diseases. J Allergy Clin Immunol 142: 715–726. 10.1016/j.jaci.2018.07.009 [DOI] [PubMed] [Google Scholar]
- Wenger CD, Coon JJ. 2013. A proteomics search algorithm specifically designed for high-resolution tandem mass spectra. J Proteome Res 12: 1377–1386. 10.1021/pr301024c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer ME, Briand LA, Fant B, Guercio LA, Arreola AC, Schmidt HD, Sidoli S, Han Y, Garcia BA, Pierce RC. 2017. Paternal cocaine taking elicits epigenetic remodeling and memory deficits in male progeny. Mol Psychiatry 22: 1641–1650. 10.1038/mp.2017.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y, et al. 2016. Metabolic regulation of gene expression by histone lysine β-hydroxybutyrylation. Mol Cell 62: 194–206. 10.1016/j.molcel.2016.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZF, Lin S, Molden RC, Cao XJ, Bhanu NV, Wang X, Sidoli S, Liu S, Garcia BA. 2015. EpiProfile quantifies histone peptides with modifications by extracting retention time and intensity in high-resolution mass spectra. Mol Cell Proteomics 14: 1696–1707. 10.1074/mcp.M114.046011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZF, Sidoli S, Marchione DM, Simithy J, Janssen KA, Szurgot MR, Garcia BA. 2018. EpiProfile 2.0: a computational platform for processing epi-proteomics mass spectrometry data. J Proteome Res 17: 2533–2541. 10.1021/acs.jproteome.8b00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZH, Tan MJ, Xie ZY, Dai LZ, Chen Y, Zhao YM. 2011. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol 7: 58–63. 10.1038/nchembio.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.