Abstract
Although plasmids are important for bacterial survival and adaptation, plasmid detection and assembly from genomic, let alone metagenomic, samples remain challenging. The recently developed plasmidSPAdes assembler addressed some of these challenges in the case of isolate genomes but stopped short of detecting plasmids in metagenomic assemblies, an untapped source of yet to be discovered plasmids. We present the metaplasmidSPAdes tool for plasmid assembly in metagenomic data sets that reduced the false positive rate of plasmid detection compared with the state-of-the-art approaches. We assembled plasmids in diverse data sets and have shown that thousands of plasmids remained below the radar in already completed genomic and metagenomic studies. Our analysis revealed the extreme variability of plasmids and has led to the discovery of many novel plasmids (including many plasmids carrying antibiotic-resistance genes) without significant similarities to currently known ones.
Plasmids are extrachromosomal independently replicating DNA molecules that provide their bacterial hosts with additional genetic material important for their survival and adaptation. Before the sequencing era, plasmids were detected based on the various phenotypic changes they provide to their host, such as antibiotic resistance or ability to degrade recalcitrant organic compounds. Sequencing efforts, however, have revealed many cryptic plasmids that do not contribute to the phenotype of the host cell in any obvious way. Although there are about 10,000 plasmids listed in the RefSeq database (Pruitt et al. 2007), many plasmids remain undetected because the task of assembling plasmids from genomic and metagenomic data sets is far from trivial (Antipov et al. 2016; Rozov et al. 2017). We thus conjecture that many classes of plasmids continue to remain unknown the same way as many of the previously unknown classes of viruses that were found in recent studies (Paez-Espino et al. 2016; Roux et al. 2016).
Because plasmids exchange genetic material with the host chromosomes and vary in structure (circular or linear), size (from a thousand to millions of nucleotides), and gene content, it is not clear how to computationally define the concept of a plasmid in such a way that it would be possible to distinguish them from the chromosomes. Also, plasmid assembly is complicated by various repeats that are difficult to resolve using short-read sequencing technologies:
An intra-plasmidic repeat refers to a repeat within a plasmid. Thirty-four percent of plasmids in the RefSeq database contain intra-plasmidic repeats >300 bp, the typical insert size in metagenomic studies.
An inter-plasmidic repeat refers to a repeat shared by multiple plasmids.
A shared repeat refers to a repeat shared between a plasmid and a chromosome. For many isolate samples, shared repeats can be resolved if the plasmid coverage by reads significantly differs from the chromosome coverage (Antipov et al. 2016). It is, however, difficult to resolve such repeats in the case of metagenomic samples with a wide spectrum of chromosome and plasmid coverages across the bacterial community (Rozov et al. 2017) or in the case of isolate samples sequenced during the growth phase (Antipov et al. 2016).
Circular plasmids form uniformly covered cycles within genomic and metagenomic assembly graphs, that is, cycles that have a relatively uniform coverage by reads (with the exception of regions corresponding to intra-plasmidic, inter-plasmidic, and shared repeats). These cycles are difficult to detect because they are “hidden” within a large assembly graph that contains both chromosomal edges (originating from chromosomes) and plasmidic edges (originating from plasmids). Moreover, plasmids with inter-plasmidic repeats form self-overlapping cycles (that traverse edges corresponding to these repeats more than once), thus complicating their detection even further.
plasmidSPAdes (Antipov et al. 2016) and Recycler (Rozov et al. 2017) are plasmid assembly tools that identify plasmids as short uniformly covered cycles in the assembly graph constructed by the SPAdes assembler (Bankevich et al. 2012). Both tools address the complications caused by shared repeats using the difference between the plasmid and chromosome coverages (plasmidSPAdes is limited to isolate genomes, whereas Recycler can work with metagenomes). Although plasmidSPAdes and Recycler revealed a number of novel plasmids, they report many false positives, especially in situations when the chromosome coverage is nonuniform. Arredondo-Alonso et al. (2017) benchmarked these tools on 42 data sets containing short reads sampled from isolate bacterial genomes with 148 plasmids, and estimated that plasmidSPAdes and Recycler have a precision of 0.78 and 0.30, respectively. The low precision and reliance on the uniform coverage makes plasmidSPAdes inapplicable to metagenomic data sets with highly varying coverage across multiple genomes. This is unfortunate because metagenomic data sets represent an untapped source of yet-to-be-discovered plasmids (Jørgensen et al. 2014; Li et al. 2015).
We present the metaplasmidSPAdes algorithm that improves on plasmidSPAdes and Recycler by (1) iteratively extracting subgraphs with gradually increasing coverage from the metagenome assembly graph, (2) finding putative plasmids as uniformly covered cycles in these subgraphs, and (3) verifying the found putative plasmids using a new plasmidVerify tool. We applied plasmidSPAdes+ (plasmidSPAdes complemented by plasmidVerify) and metaplasmidSPAdes to diverse genomic and metagenomic samples and revealed thousands of plasmids that were missed in previous studies, including many plasmids that share no significant similarities with currently known plasmids, as well as plasmids carrying antibiotic-resistance genes (ARGs).
Results
metaplasmidSPAdes workflow
plasmidSPAdes constructs the plasmid graph by removing all edges with coverage similar to the median coverage in the assembly graph. This approach does not work for metagenomes because they have highly nonuniform coverage across various bacterial genomes within a metagenome. metaplasmidSPAdes improves on plasmidSPAdes by resolving dominant plasmids in metagenomes, that is, plasmids with coverage exceeding that of chromosomes and other plasmids, with which they share repeats.
metaplasmidSPAdes uses metaSPAdes (Nurk et al. 2017) for transforming the de Bruijn graph into an assembly graph. It further detects plasmids in the assembly graph by iteratively constructing smaller and smaller subgraphs of the assembly graph and detecting plasmids in these subgraphs. metaplasmidSPAdes removes low-coverage edges (with increasing coverage cutoff at each iteration), uses exSPAnder (Prjibelski et al. 2014) to generate contigs, and detects putative plasmids as cyclic contigs (cyclocontigs) or small connected components in the generated subgraphs.
metaplasmidSPAdes sets a coverage cutoff cov, removes all edges with coverage below cov from the assembly graph, and searches either for a cycle (cyclocontig) supported by the paired-end reads or for a small connected component in the resulting graph. Some of the found cyclocontigs and connected components represent dominant plasmids that were “hidden” in the assembly graph before the removal of low-coverage edges. To reveal more and more hidden plasmids with progressively increasing coverage, metaplasmidSPAdes iteratively increases the coverage cutoff as cov + covadd or as cov*covmult (Fig. 1). Finally, it uses the plasmidVerify tool to check whether contigs and connected components found by metaplasmidSPAdes indeed represent plasmids. The Methods section describes the metaplasmidSPAdes workflow in further detail.
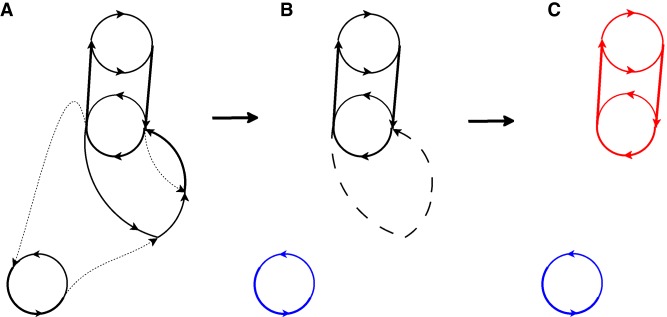
Figure 1.
Iterative plasmid detection in the assembly graph. (A) The assembly graph Graph with three dotted edges representing edges with the lowest coverage. (B) Removal of three edges with the lowest coverage from Graph reveals a plasmid (cyclocontig) shown in blue. The three edges on the graph in A now represent a single dashed edge that has the lowest coverage in Graph. (C) The same graph after the second iteration of metaplasmidSPAdes that removes the dashed edge with the lowest coverage and reveals a plasmid (connected component) shown in red.
Plasmid verification
Each cyclocontig/component reconstructed by metaplasmidSPAdes may contain some chromosomal edges (or even consist entirely of chromosomal edges) arising from phage sequences, transposons, repeats within bacterial chromosomes, etc. We thus developed a plasmidVerify tool that examines the gene content of a cyclocontig and classifies it as plasmidic (chromosomal) using a naive Bayesian classifier. Because plasmids harbor a large variety of genes, plasmidVerify uses a plasmid-specific profile-HMM database to detect remote similarities between cyclocontigs/components detected by metaplasmidSPAdes and known plasmid-specific genes (see Methods section). To construct a set of plasmid-specific HMMs, we formed the PlasmidDatabase data set containing all 9937 plasmids from the RefSeq database (total length 1007 Mb) and the nonPlasmidDatabase data set containing a randomly selected 10% of complete bacterial chromosomes from RefSeq (837 bacterial genomes with total length 3229 Mb).
Analysis of putative novel plasmids found by metaplasmidSPAdes
We annotated some putative novel plasmids found by metaplasmidSPAdes using Prodigal (Hyatt et al. 2010) in metagenomic mode for gene prediction (version 2.6.3), the hmmsearch tool (hmmer.org) with PfamA 30.0 database for gene annotation (version 3.1b2), and the Comprehensive Antibiotic Resistance Database (CARD) (Jia et al. 2017) for predicting ARGs (only “perfect” and “strict” hits).
Benchmarking plasmid verification tools
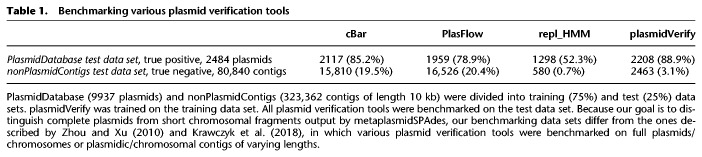
We benchmarked plasmidVerify against three plasmid verification tools (Table 1):
a cBar tool based on 5-mer frequencies (Zhou and Xu 2010),
a PlasFlow tool based on deep neural networks (Krawczyk et al. 2018), and
a repl_HMM approach based on manually curated plasmid replicase HMMs (Jørgensen et al. 2014).
Table 1.
Benchmarking various plasmid verification tools
We did not include PlasmidFinder (Carattoli et al. 2014) in the benchmarking because Arredondo-Alonso et al. (2017) recently showed that it has a very low recall rate (0.36).
To construct a true negative data set for benchmarking, we randomly selected 10% of bacterial genomes from the RefSeq database using the Python random.sample() function. Because most putative plasmids output by metaplasmidSPAdes are shorter than typical bacterial chromosomes, we split all bacterial chromosomes into fragments of length 10 kb and used them as the true negative data set. This procedure resulted in 323,362 sequences (partitioning of PlasmidDatabase into 10-kb-long fragments) that we refer to as nonPlasmidContigs. We selected PlasmidDatabase as the true positive data set for benchmarking.
Table 1 illustrates that plasmidVerify improved on both the true positive and false positive rates compared with the cBar and PlasFlow tools. Although the repl_HMM approach (which uses a small manually curated set of plasmid replicase HMMs) has a lower false positive rate than plasmidVerify, it is not well suited for our goals because it has a low true positive rate and is limited in its ability to detect diverse plasmids; that is, it fails to detect novel plasmid with replicases that significantly differ from the replicases in the curated data set.
To evaluate plasmidVerify's performance on the unseen branches of the microbial tree of life, we performed the following procedure. For each of the four phyla (Firmicutes, Proteobacteria, Cyanobacteria, and Bacteroidetes), we removed all plasmids from the phylum from the training data set, retrained plasmidVerify on the reduced training data set, and tested it on the members of the removed phylum (Supplemental Table S1). The false negative (positive) rates varied from 14.6% to 19.6% (1.3% to 3.6%) across the four analyzed phyla.
We also tested various plasmid verification tools on the set of viral contigs that represent a major source of nonplasmidic circular DNA elements (Supplemental Table S2).
Data sets
We benchmarked metaplasmidSPAdes using one data set with multiple isolate genomes, three mock metagenomic data sets with known bacterial genomes, four metagenomic data sets (with unknown genomes), and one plasmidome data set (all data sets contain paired-end Illumina reads). To infer the set of plasmids in each mock metagenomic data set, we compiled the list of known plasmids from the genomes (including all strains with data present in RefSeq) present in this data set. To check which plasmids from this list are indeed present in the mock sample, we mapped all metagenomic reads to each of these plasmids. We assume that a plasmid is present in the mock data set (reference plasmid) if >95% of its length is covered by metaSPAdes assembly. We used metaSPAdes for this verification because all known metagenomic plasmid detection tools use its assembly graph for plasmid assembly. For information about plasmids in the mock data sets, see Supplemental Table S3. It is worth noting that even though mock metagenomes are usually formed from well-studied genomes, metaplasmidSPAdes was able to reveal some still unknown plasmids even in the mock metagenomes.
Below we provide a brief description of each of the data sets (for detailed information, see Supplemental Table S4, “Information about Benchmarking Data Sets”).
ISOLATES
The ISOLATES data set consists of 21,933 bacterial data sets from the JGI GOLD database (gold.jgi.doe.gov), representing isolate bacterial samples.
HMP
The HMP data set is a mock community of 19 bacterial species, one archaea, and one yeast species studied by The Human Microbiome Project Consortium (The Human Microbiome Project Consortium 2012). Twenty plasmids were originally reported in this data set, but our more stringent approach reduced the number of reference plasmids to 14 (total length ≈854 kb).
MBARC
The Mock Bacteria ARchaea Community (MBARC) data set is a mock microbial community of 23 bacterial and three archaeal species described by Singer et al. (2016). We identified 10 plasmids of total length ≈756 kb in the MBARC data set.
SYNTH
The SYNTH data set is a mock microbial community of 64 diverse bacterial and archaeal species described by Shakya et al. (2013). Shakya et al. (2013) identified 32 plasmids in this data set, but our more stringent approach reduced the number of reference plasmids to 19 (total length ≈1450 kb).
INFANT
The INFANT is a human microbiome data set from an infant's gut described by Bäckhed et al. (2015).
CROHN
The CROHN is a human gut microbiome data set from a patient suffering from Crohn's disease (analyzed by Nurk et al. 2017).
PLASMIDOME
The PLASMIDOME is a plasmid-enriched data set from a microbial community in a biological wastewater treatment reactor described by Shi et al. (2018).
MARINE
The MARINE is a marine sediment metagenome data set collected near the field of active hydrothermal vents in the Atlantic Ocean (Spang et al. 2015).
LAKE
The LAKE is a lake metagenome data set collected at an Indian lake subjected to industrial pollution with fluoroquinolone antibiotics.
Analyzing the ISOLATES data set
We searched for plasmids in the ISOLATES data set with the goal of identifying new plasmids that might have evaded detection in the already completed sequencing projects. We did not benchmark Recycler because Arredondo-Alonso et al. (2017) have already benchmarked plasmidSPAdes and Recycler on diverse isolate data sets.
plasmidSPAdes generated 44,172 plasmidic connected components, including 15,499 cyclocontigs that originated from 7987 out of 21,933 genomes in the ISOLATES data set. To simplify analysis, we limited benchmarking to cyclocontigs and ignored other connected components output by plasmidSPAdes+.
To remove duplicated cyclocontigs from this set, we clustered them based on their k-mer content using Mash (Ondov et al. 2016) and classified plasmids as duplicates if their k-mer compositions differed by <1%. Once duplicates had been removed, 6694 out of the 15,499 identified cyclocontigs were classified as unique. Of these, 2280 cyclocontigs (referred to as plasmidic cyclocontigs) were classified as plasmids by plasmidVerify (Fig. 2). We compared these cyclocontigs against the CARD database of ARGs and detected 356 ARGs in 203 out of 2280 cyclocontigs (see Fig. 2B; for details, see Supplemental Table S5).
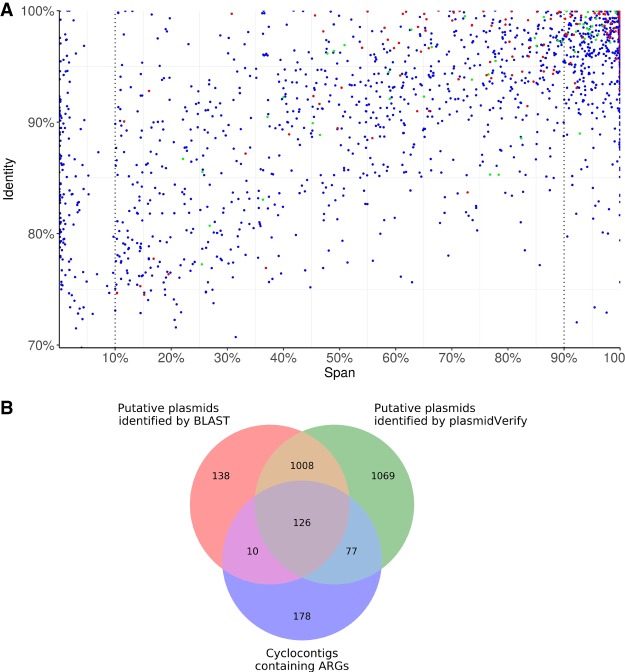
Figure 2.
The scatter plot of the span and identity for all 2280 unique cyclocontigs in the ISOLATES data set reconstructed by plasmidSPAdes+ (A) and the Venn diagram for cyclocontigs identified as plasmids by plasmidVerify, cyclocontigs identified as plasmids by BLAST (span >10%), and cyclocontigs containing ARGs (B). (A) Each dot represents a cyclocontig reported by plasmidSPAdes and verified by plasmidVerify. Red dots represent cyclocontigs containing antibiotic-resistance genes (ARGs). Green dots represent cyclocontigs classified as viral sequences. (B) The Venn diagram illustrates that the HMM-based approach in metaplasmidSPAdes identifies many plasmids with important phenotypes that are missed by a straightforward BLAST-based approach.
To double-check whether a putative cyclocontig originated from a plasmid or a bacterial chromosome, we aligned it against the NCBI nucleotide collection (nr/nt) using the BLAST tool (Altschul et al. 1990) with the e-value threshold 0.001. Cyclocontigs that aligned to the nonplasmidic sequences in the NCBI nucleotide collection (nr/nt) (bacterial chromosomes, viruses, etc.) likely represent false positives, but cyclocontigs that aligned to plasmids (or do not align at all) may represent known or novel plasmids. Thus BLAST alignments can be used as an approximation for the ground truth for additional benchmarking of plasmidVerify, cBar, repl_HMM, and PlasFlow (Supplemental Table S6).
If a cyclocontig aligned to multiple sequences in the NCBI nucleotide collection (nr/nt), we analyzed the one with the maximal BLAST score (alignments to sequences of unknown origin are ignored). BLAST generates either a single alignment that extends over the entire length of the cyclocontig or multiple local alignments. We defined the span of a cyclocontig as the ratio of the total alignment length over the cyclocontig length, and the identity of the cyclocontig as the average percentage of identity across all alignments.
Of 2280 plasmidic cyclocontigs, 1134 and 603 aligned to known plasmids with the span exceeding 10% and 90%, respectively. The remaining 2280 − 1134 = 1146 cyclocontigs can be broken down into the following four categories (for details, see Supplemental Table S5):
255 cyclocontigs ambiguously matched to plasmid/chromosome with a span >10% (putative integrative plasmids);
480 matched bacterial chromosomes (false positive bacterial segments);
31 matched viral sequences (false positive phage segments); and
380 did not match any known plasmids/chromosomes with a span >10% and were classified as novel plasmids.
We analyzed some of the newly identified plasmids in more details (for plasmid maps, see Supplemental Fig. S1):
A 7895-bp-long putative plasmid (from Streptococcus pseudopneumoniae clinical isolate) with span 28% and identity 96% carried an Erm 23S ribosomal RNA methyltransferase, providing resistance to macrolide antibiotics. It also carried a toxin–antitoxin system relB/parE and zeta toxin that may inhibit the cell wall biosynthesis and act as a bacteriocin.
A 53,557-bp-long putative plasmid (from Enterobacter sp. CC120223-11) with span 12% and identity 90% carried an ATP-binding cassette (ABC) antibiotic efflux pump. It contained a toxin–antitoxin system vapB/vapC, genes related to pili and flagella development, and putative members of type IV conjugal transfer systems (Pfam families T4SS_TraI and TraI_2_C), indicating that it is likely self-transferable. It was similar to known plasmids only in the short region containing the parA/parB operon that ensures the accurate partitioning of plasmids after division.
The longest putative novel plasmid in the ISOLATES data set (582 kb) belonged to the halophilic marine gammaproteobacteria Ferrimonas marina, strain DSM 16917. It encoded 685 predicted genes and contained the plasmid replication protein gene repA, as well as an outer membrane phospholipase A1 (OMPLA) essential for bacterial secretion, proteins for flagella formation, and ydaS/ydaT toxin–antitoxin system. It also had some phage signatures such as the phage integrase genes and bacteriophage T4–like capsid assembly protein (Gp20). However, the phage integrase genes do not represent a strong phage marker because they often occur in plasmids.
The shortest putative novel plasmid in the ISOLATES data set (length 1284 bp) encoded a single protein (firmicute plasmid replication protein RepL) and belonged to the fish pathogen Candidatus ichthyocystis 2013Ark19i, a recently described novel intracellular β-proteobacteria (Seth-Smith et al. 2016).
Analyzing the HMP data set
metaplasmidSPAdes reconstructed 21 cyclocontigs in the HMP data set. plasmidVerify classified seven of them as plasmidic, and all of them have corresponding reference plasmids. metaSPAdes and Recycler reconstructed four and six reference plasmids, respectively (Table 2; Supplemental Table S3). metaplasmidSPAdes identified no small uniformly covered connected components in the HMP data set.
Table 2.
Information about reference plasmids reconstructed as cyclocontigs by metaSPAdes, Recycler, and metaplasmidSPAdes (HMP, MBARC, and SYNTH data sets)
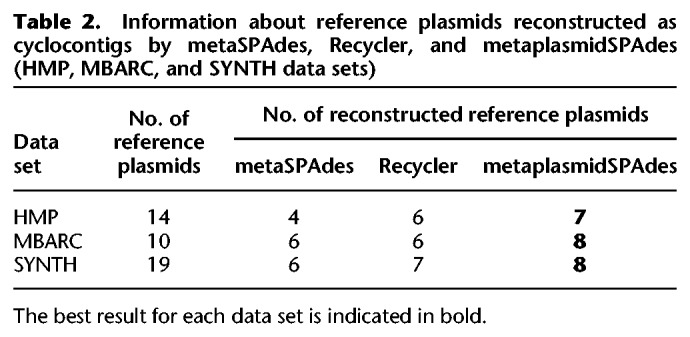
We analyzed why metaplasmidSPAdes missed 14 − 7 = 7 reference plasmids in the HMP data set. Six of them were nondominant plasmids that share repeats with their bacterial hosts or other plasmids (for details, see Supplemental Table S3). The remaining one (dominant plasmid NZ_CP015213.1) was not reconstructed as a single cyclocontig because it had a long intra-repeat. This plasmid was not output as a uniformly covered connected component because it shares >50% of its length with another plasmid (NC_009007.1) and fails the test on the uniformity of coverage as the total length of medial edges (see Methods section) exceeds 80% of the size of this component. For each plasmid that was not assembled in a single cyclocontig by metaplasmidSPAdes, we computed the size and the number of edge count of the largest connected component that contains this plasmid at each iteration of metaplasmidSPAdes (Supplemental Table S7).
Analyzing the MBARC data set
metaplasmidSPAdes reconstructed 32 cyclocontigs, and plasmidVerify classified eight of them as plasmidic. metaplasmidSPAdes assembled eight out of 10 reference plasmids in the MBARC data set into a single cyclocontig (metaSPAdes and Recycler reconstructed six plasmids each). Two remaining plasmids were nondominant plasmids that were missed by metaplasmidSPAdes because their coverage was close to the median coverages of their host chromosomes that share long repeats with these plasmids.
plasmidVerify erroneously classified two out of eight assembled reference plasmids as nonplasmidic: (1) One plasmid from the archaea Natronococcus occultus was misclassified because plasmidVerify is not designed to verify archaeal plasmids, and (2) one short plasmid (of length 2931 bp) did not yield any hits in the Pfam-A database.
Additionally, plasmidVerify classified two cyclocontigs as plasmidic: a 2876-bp-long cyclocontig with a plasmid replication protein that likely represents a novel plasmid (span 19% and identity 76%) and a 53-kb-long cyclocontig that carries a plasmid-specific resolvase gene and aligns to a bacterial chromosome and various plasmids.
Analyzing the SYNTH data set
metaplasmidSPAdes reconstructed 87 cyclocontigs in the SYNTH data set, and plasmidVerify classified 13 of them as plasmidic. metaSPAdes, Recycler, and metaplasmidSPAdes reconstructed six, seven, and eight out of the 19 reference plasmids, respectively. The remaining 11 reference plasmids in the SYNTH data set evaded identification by metaplasmidSPAdes because:
10 of them were nondominant and share long repeats with chromosomes or plasmids with the same or higher coverage (see Supplemental Table S3 for details); and
one dominant plasmid was not output as a cyclocontig because it has inter-plasmidic repeats larger than the library insert size. It was not output as a uniformly covered connected component either because its length (408 kb) exceeds the default threshold for the connected component length (200 kb).
Six out of 13 cyclocontigs that metaplasmidSPAdes classified as plasmidic likely represent still unknown plasmids in the SYNTH community:
Three cyclocontigs have ∼40% span and 80%–93% identity with known plasmids in various Phaeobacter genomes. Two of them (lengths of 22,035 and 5444 bp) were conjugative plasmids carrying mobilization proteins (MobA/MobC), and one of them (of length 11,215 bp) contained a plasmid replicase gene repA, a toxin–antitoxin system parE/parD, and a copper-resistance operon copAB.
One cyclocontig (length of 38,668 bp) did not match any known plasmid/bacterial genomes but carried a plasmid replicase gene.
Two cyclocontigs (lengths of 22,963 and 4103 bp) both had short matches to known plasmids and chromosomes (with span 20% and identity 97%–99%). Because they carry both a replicase gene and conjugal transfer proteins, they likely represent conjugative plasmids.
The remaining two out of 13 cyclocontigs that metaplasmidSPAdes classified as plasmidic aligned to bacterial chromosomes and likely represent false positives (prophages or transposons). plasmidVerify misclassified three reference plasmids (lengths of 16,625, 8368, and 8362 bp) as nonplasmidic because it did not detect any distinctively plasmidic genes within them.
Analyzing the INFANT data set
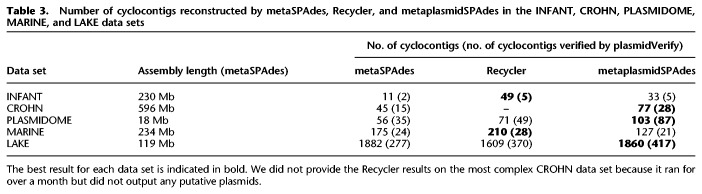
metaplasmidSPAdes reconstructed 33 cyclocontigs in the INFANT data set, and plasmidVerify classified five of them as plasmidic (Table 3):
One of them (length of 4234 bp) matched the pRGFK1358 plasmid with 100% span and 95% identity;
one of them (length of 4608 bp) matched the pRGFK1348 plasmid with 56% span and 95% identity;
two of them (lengths of 3687 and 3338 bp) did not match any known plasmids/chromosomes but harbored the Mob plasmid recombination enzyme and the initiator of plasmid replication Rep3; and
one of them (length of 1553 bp) matched bacterial chromosomes (likely a false positive).
Table 3.
Number of cyclocontigs reconstructed by metaSPAdes, Recycler, and metaplasmidSPAdes in the INFANT, CROHN, PLASMIDOME, MARINE, and LAKE data sets
Analyzing the CROHN data set
metaplasmidSPAdes reconstructed 77 cyclocontigs in the CROHN data set, and plasmidVerify classified 28 of them as plasmidic (Table 3):
Four of them matched known plasmids with 100% span and identity varying from 92%–99%;
14 of them matched known plasmids with spans varying from 21%–79% and identity varying from 78%–97%;
nine of them had a span of <10% and did not have significant matches with any sequences in the nr database; and
one of them aligned to a bacterial chromosome with a span of 38% (likely a false positive).
A 1868-bp-long cyclocontig reconstructed by metaplasmidSPAdes and classified as nonplasmidic by plasmidVerify turned out to be a Streptococcus phage phiJH1301-2, carrying an aminoglycoside-resistance gene (phages were recently shown to carry ARGs) (Balcazar 2014). Although plasmidSPAdes and metaplasmidSPAdes were not designed for viral assembly (there is still no specialized software for viral assembly from genomic and metagenomic data sets), our analysis shows that they are able to detect viruses in genomic and metagenomic data sets.
Analyzing the PLASMIDOME data set
Because the PLASMIDOME data set did not contain information about the reference plasmids, we generated some references for this data set by mapping the assembled PLASMIDOME contigs against the plasmid database (with Mash screen [Ondov et al. 2019], QUAST [Gurevich et al. 2013], and BLAST). This analysis revealed 10 reference plasmids with a total length of ≈100 kb. The fact that the total length of the identified reference plasmids in the PLASMIDOME data set was two orders of magnitude smaller than the total assembly length suggests that most plasmids in the PLASMIDOME data set are not present in the plasmid database.
metaplasmidSPAdes reconstructed 103 cyclocontigs in the PLASMIDOME data set, and plasmidVerify classified 87 of them as plasmidic (Table 3). Seven of these 87 cyclocontigs matched known plasmids with a span >90% (with identity varying from 82%–99%) and 54 have a span exceeding 10% (with identity varying from 75%–99%). Nine out of these 87 cyclocontigs matched a bacterial chromosome or a phage with a span exceeding 10% (likely false positives). The remaining 87 − 7 − 54 − 9 = 17 contigs have spans <10% and were classified as putative novel plasmids.
Analyzing the MARINE data set
metaplasmidSPAdes reconstructed 127 cyclocontigs in the MARINE data set, and plasmidVerify classified 21 of them as plasmidic (Table 3). Three of these cyclocontigs matched known plasmids (one with a 99% span and identity, two with spans of 20% and 60% and identity of 87% and 93%, respectively). Three others matched bacterial chromosomes with spans of 14%, 33%, and 48% and identity of 75%, 100%, and 74%, respectively. The remaining 15 cyclocontigs have spans <10% and were classified as putative novel plasmids.
Analyzing the LAKE data set
metaplasmidSPAdes reconstructed 1860 cyclocontigs in the LAKE data set, and plasmidVerify classified 417 of them as plasmidic (Table 3). Seven of these cyclocontigs matched bacterial chromosomes, 13 matched viral sequences, and nine matched both chromosomes and plasmids and thus likely represent integrative plasmids. Fifty-nine cyclocontigs matched known plasmids with span exceeding 10%, and the remaining 329 cyclocontigs had no significant matches to the NCBI nucleotide collection (nr/nt). The large number of putative plasmids in the LAKE data set (compared with the other data sets we analyzed) may be explained by the fact that the lake was polluted with fluoroquinolones, making plasmids carrying antibiotic resistance and other genes particularly beneficial to the hosts.
Discussion
We showed that plasmidSPAdes+ and metaplasmidSPAdes improve on existing tools for plasmid reconstruction and identify many novel plasmids in diverse genomic and metagenomic data sets. However, even with the improved mechanism of identifying new plasmids, it is still likely that many more plasmids continue to evade detection (false negatives), and some nonplasmidic cyclocontigs end up being reported as plasmids (false positives).
Because some plasmids do not harbor any distinctively plasmidic genes (as defined based on the analysis of known plasmids), the corresponding cyclocontigs are not detected by metaplasmidSPAdes. Users have the option to switch off the plasmidVerify tool and manually analyze all cyclocontigs that fall into this category.
Application of plasmidSPAdes+ and metaplasmidSPAdes to various data sets revealed that many plasmids remain undetected during genomic and metagenomic studies. Moreover, this analysis revealed the enormous variability of plasmids: A large fraction of the found plasmids did not match to any known ones. Even in the already completed sequencing projects (ISOLATES data set), we found 1166 putative plasmidic cyclocontigs with <90% similarity to known ones and without significant hits to viruses or bacterial chromosomes. Ninety-one of these putative plasmids contain ARGs, 246 contain carbohydrate-active enzymes (CAZymes), and 54 contain adhesion-related genes (possibly contributing to horizontal gene transfer). Expansion of the set of known plasmids can help classify them and reflects the evolutionary relationships between plasmids. One can compare plasmid phylogeny with host phylogeny and phenotypic traits and analyze the relationships between resistance type, plasmid replication type, and host type. This information would also be relevant for epidemiological studies. For example, it remains unclear whether resistance dissemination involves a diverse set of plasmids or a single dominant epidemic type. It may correlate with the host range and the type of the ARG (Mathers et al. 2015). metaplasmidSPAdes will help generate a comprehensive data set of plasmids to help address these questions.
Methods
metaplasmidSPAdes workflow
metaplasmidSPAdes uses the default values covadd = 5x and covmult = 1.3. The plasmidVerify module checks whether a cyclocontig or a connected component in the assembly graph originated from a plasmid using a naive Bayesian classifier. To avoid time-consuming read alignments at each iteration, metaplasmidSPAdes aligns paired-end reads against the assembly graph only once and updates the information about the read alignments during the graph modifications. metaplasmidSPAdes pseudocode can be presented as follows:
metaplasmidSPAdes(Reads, covadd, covmult)
Plasmids ← empty set
Graph ← assembly graph of Reads constructed by metaSPAdes
align paired-end reads to Graph and compute coverage of each edge by reads
covmax ← maximum coverage of an edge in Graph
cov ← 0
while cov < covmax
Contigs ← the set of all paths (contigs) in Graph generated by
exSPAnder
for each cyclocontig Cycle in Contigs
add Cycle to the set Plasmids
for each small plasmid-like connected components Component in Graph
if Component contains edges that do not belong to cyclocontigs in Plasmids
add Component to the set Plasmids and remove it from Graph
cov ← max{cov + cadd, cov * cmult}
remove edges with coverage below cov from the assembly graph
iteratively remove dead-end edges from Graph (Antipov et al. 2016)
replace each nonbranching path in Graph with a single edge and recompute its coverage
for each cyclocontig or connected component C in Plasmids
If plasmidVerify(C) = 0
remove С from Plasmids
return PlasmidsplasmidVerify workflow
We predicted genes with Prodigal v2.6.3 (Hyatt et al. 2010) and ran hmmsearch (part of HMMER 3.1b2, http://hmmer.org/) using Pfam-A database v. 30.0 (Finn et al. 2016) on the training data sets from both PlasmidDatabase and nonPlasmidDatabase (7550 plasmids and 242,681 “contigs,” respectively). For each of the two runs and for each HMM, we counted the frequencies of matches (with the bit-score cutoff set to the “noise” level from the Pfam-A database) to PlasmidDatabase and nonPlasmidDatabase, respectively. These frequencies were used to train a naive Bayesian classifier (Friedman et al. 2001). Supplemental Table S8 lists the HMM frequencies in the training data set. Given a cyclocontig, plasmidVerify predicts genes in this contig using Prodigal in the metagenomic mode, runs hmmsearch on the predicted proteins, and classifies the contig as plasmidic or chromosomal by applying the naive Bayesian classifier.
plasmidVerify classified 1%–2% of contigs in the analyzed metagenomic assemblies as plasmidic (Supplemental Table S9). However, because plasmidVerify incorrectly classified a number of chromosomal contigs as plasmidic, plasmidVerify (and other plasmid verification tools) by itself is unable to accurately classify plasmids and thus has to be combined with metaplasmidSPAdes for increased accuracy.
Plasmid-like connected components
We define the size of a connected component in the assembly graph as the total length of its edges. The connected component is called small if its size does not exceed sizemax (default value 200 kb). For each connected component, we compute its median coverage by reads (covmed) as described by Antipov et al. (2016). An edge in a connected component is called medial if its coverage exceeds covmed/α and does not exceed covmed * α (the default value α = 1.3). A connected component is called uniform if the total length of its medial edges exceeds 80% of the size of this component. We classify a small uniform connected component as plasmid-like if its size exceeds 1 kb and if it contains at most two dead-end edges.
Data access
metaplasmidSPAdes results on all mentioned data sets from this study are available at http://data.cab.spbu.ru/index.php/s/tz7mCqDipgbcsbW and as Supplemental File S1. Source code is available at https://github.com/ablab/spades/tree/metaplasmid_3.13.0 and as Supplemental File S2.
Supplementary Material
Acknowledgments
This study was supported by the Russian Science Foundation (grant number 19-16-00049). P.A.P. was supported by NSF/MCB-BSF grant number 1715911. We thank Alexander Shlemov and Yevgeniya Mazur for many helpful comments.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.241299.118.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner P. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32: 3380–3387. 10.1093/bioinformatics/btv688 [DOI] [PubMed] [Google Scholar]
- Arredondo-Alonso S, Willems RJ, van Schaik W, Schürch AC. 2017. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom 3: e000128 10.1099/mgen.0.000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17: 690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Balcazar JL. 2014. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog 10: e1004219 10.1371/journal.ppat.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasmanb H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58: 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acid Res 44: D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. 2001. The elements of statistical learning Springer series in statistics Springer, New York. [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486: 215–221. 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, et al. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45: D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen TS, Xu Z, Hansen MA, Sørensen SJ, Hansen LH. 2014. Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PLoS One 9: e87924 10.1371/journal.pone.0087924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk PS, Lipinski L, Dziembowski A. 2018. PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res 46: e35 10.1093/nar/gkx1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AD, Li LG, Zhang T. 2015. Exploring antibiotic resistance genes and metal resistance genes in plasmid metagenomes from wastewater treatment plants. Front Microbiol 6: 1025 10.3389/fmicb.2015.01025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28: 565–591. 10.1128/CMR.00116-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27: 824–834. 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17: 132 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Starrett GJ, Sappington A, Kostic A, Koren S, Buck CB, Phillippy AM. 2019. Mash Screen: high-throughput sequence containment estimation for genome discovery. bioRxiv 10.1101/557314 [DOI] [PMC free article] [PubMed]
- Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Kyrpides NC. 2016. Uncovering earth's virome. Nature 536: 425–430. 10.1038/nature19094 [DOI] [PubMed] [Google Scholar]
- Prjibelski AD, Vasilinetc I, Bankevich A, Gurevich A, Krivosheeva T, Nurk S, Pham S, Korobeynikov A, Lapidus A, Pevzner PA. 2014. ExSPAnder: a universal repeat resolver for DNA fragment assembly. Bioinformatics 30: i293–i301. 10.1093/bioinformatics/btu266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, Poulos BT, Solonenko N, Lara E, Poulain J, et al. 2016. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537: 689–693. 10.1038/nature19366 [DOI] [PubMed] [Google Scholar]
- Rozov R, Brown Kav A, Bogumil D, Shterzer N, Halperin E, Mizrahi I, Shamir R. 2017. Recycler: an algorithm for detecting plasmids from de novo assembly graphs. Bioinformatics 33: 475–482. 10.1093/bioinformatics/btw651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith HM, Dourala N, Fehr A, Qi W, Katharios P, Ruetten M, Mateos JM, Nufer L, Weilenmann R, Ziegler U, et al. 2016. Emerging pathogens of gilthead seabream: characterisation and genomic analysis of novel intracellular β-proteobacteria. ISME J 10: 1791–1803. 10.1038/ismej.2015.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya M, Quince C, Campbell JH, Yang ZK, Schadt CW, Podar M. 2013. Comparative metagenomic and rRNA microbial diversity characterization using archaeal and bacterial synthetic communities. Environ Microbiol 15: 1882–1899. 10.1111/1462-2920.12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang H, Tian Z, Yang M, Zhang Y. 2018. Characteristics of ARG-carrying plasmidome in the cultivable microbial community from wastewater treatment system under high oxytetracycline concentration. Appl Microbiol Biotechnol 102: 1847–1858. 10.1007/s00253-018-8738-6 [DOI] [PubMed] [Google Scholar]
- Singer E, Andreopoulos B, Bowers RM, Lee J, Deshpande S, Chiniquy J, Ciobanu D, Klenk HP, Zane M, Daum C, et al. 2016. Next generation sequencing data of a defined microbial mock community. Sci Data 3: 160081 10.1038/sdata.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. 10.1038/nature14447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Xu Y. 2010. cBar: a computer program to distinguish plasmid-derived from chromosome-derived sequence fragments in metagenomics data. Bioinformatics 26: 2051–2052. 10.1093/bioinformatics/btq299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.