Endo- and exo-type xylanases depolymerize xylan and play crucial roles in the assimilation of xylan in bacteria and fungi. Exoxylanases release xylose from the reducing or nonreducing ends of xylooligosaccharides; this is generated by the activity of endoxylanases. β-Xylosidase, which hydrolyzes xylose residues on the nonreducing end of a substrate, is well studied. However, the function of reducing-end xylose-releasing exoxylanases (Rex), especially in fungal cellulolytic systems, remains unclear. This study revealed the mode of xylan hydrolysis by Rex from the cellulolytic fungus Talaromyces cellulolyticus (Xyn30A), which belongs to the glycoside hydrolase family 30-7 (GH30-7). A conserved residue related to Rex activity is found in the substrate-binding site of Xyn30A. These findings will enhance our understanding of the function of GH30-7 Rex in the cooperative hydrolysis of xylan by fungal enzymes.
KEYWORDS: Talaromyces cellulolyticus, exoxylanase, glycoside hydrolase family 30, lignocellulose, xylan, xylooligosaccharide
ABSTRACT
In this study, we characterized the mode of action of reducing-end xylose-releasing exoxylanase (Rex), which belongs to the glycoside hydrolase family 30-7 (GH30-7). GH30-7 Rex, isolated from the cellulolytic fungus Talaromyces cellulolyticus (Xyn30A), exists as a dimer. The purified Xyn30A released xylose from linear xylooligosaccharides (XOSs) 3 to 6 xylose units in length with similar kinetic constants. Hydrolysis of branched, borohydride-reduced, and p-nitrophenyl XOSs clarified that Xyn30A possesses a Rex activity. 1H nuclear magnetic resonance (1H NMR) analysis of xylotriose hydrolysate indicated that Xyn30A degraded XOSs via a retaining mechanism and without recognizing an anomeric structure at the reducing end. Hydrolysis of xylan by Xyn30A revealed that the enzyme continuously liberated both xylose and two types of acidic XOSs: 22-(4-O-methyl-α-d-glucuronyl)-xylotriose (MeGlcA2Xyl3) and 22-(MeGlcA)-xylobiose (MeGlcA2Xyl2). These acidic products were also detected during hydrolysis using a mixture of MeGlcA2Xyln (n = 2 to 14) as the substrate. This indicates that Xyn30A can release MeGlcA2Xyln (n = 2 and 3) in an exo manner. Comparison of subsites in Xyn30A and GH30-7 glucuronoxylanase using homology modeling suggested that the binding of the reducing-end residue at subsite +2 was partially prevented by a Gln residue conserved in GH30-7 Rex; additionally, the Arg residue at subsite −2b, which is conserved in glucuronoxylanase, was not found in Xyn30A. Our results lead us to propose that GH30-7 Rex plays a complementary role in hydrolysis of xylan by fungal cellulolytic systems.
IMPORTANCE Endo- and exo-type xylanases depolymerize xylan and play crucial roles in the assimilation of xylan in bacteria and fungi. Exoxylanases release xylose from the reducing or nonreducing ends of xylooligosaccharides; this is generated by the activity of endoxylanases. β-Xylosidase, which hydrolyzes xylose residues on the nonreducing end of a substrate, is well studied. However, the function of reducing-end xylose-releasing exoxylanases (Rex), especially in fungal cellulolytic systems, remains unclear. This study revealed the mode of xylan hydrolysis by Rex from the cellulolytic fungus Talaromyces cellulolyticus (Xyn30A), which belongs to the glycoside hydrolase family 30-7 (GH30-7). A conserved residue related to Rex activity is found in the substrate-binding site of Xyn30A. These findings will enhance our understanding of the function of GH30-7 Rex in the cooperative hydrolysis of xylan by fungal enzymes.
INTRODUCTION
Plant cell wall polysaccharides, such as cellulose and hemicellulose, are promising sources of biofuels and chemicals. Enzymatic hydrolysis is an important process used in conversion of these polysaccharides. Xylan, a major component of hemicellulose, is composed mainly of β-1,4-d-xylopyranose polymers decorated with side chain residues such as α-1,2- and/or α-1,3-linked l-arabinofuranose and α-1,2-linked 4-O-methyl-d-glucuronic acid (MeGlcA). Due to the heterogeneous nature of xylan, its degradation requires combined and synergistic activity of different enzymes, including endoxylanases (EC 3.2.1.8), β-xylosidases (EC 3.2.1.37), α-l-arabinofuranosidases (EC 3.2.1.55), α-glucuronidases (EC 3.2.1.131), ferulic acid esterases (EC 3.1.1.73), and acetylxylan esterases (EC3.1.1.72) (1, 2).
Endoxylanase and β-xylosidase are the core enzymes used to degrade xylan into monomers. In the CAZy database (http://www.cazy.org), endoxylanases are classified mostly into glycoside hydrolase family 10 (GH10) and GH11. Endoxylanases randomly cleave the β-1,4 linkage in the xylan main chain in an endo manner. This activity, which generates xylose, xylobiose (Xyl2), and xylooligosaccharides (XOSs), including branched XOSs, depends on the type of side chain residues present in xylan (3–6). Xyl2 and XOSs are further cleaved into xylose by β-xylosidase. β-Xylosidase releases xylose from the nonreducing ends of XOSs; thus, β-xylosidase does not hydrolyze the xylose residue in the reducing end of branched XOS (7, 8). Conversely, certain bacterial GH8 xylanases are characterized as reducing-end xylose-releasing exo-oligoxylanases (Rex) (EC 3.2.1.156) (9–11). GH8 Rex liberates xylose from the reducing ends of XOSs via an inverting catalytic mechanism (9). This can potentially increase the yield of xylose in hydrolysis of pretreated lignocellulose because Rex readily hydrolyzes branched XOSs that are resistant to enzymatic hydrolysis by endoxylanases and β-xylosidases (11).
Although GH8 xylanases are not found in fungal hemicellulases, XYN IV, isolated from the cellulolytic fungus Trichoderma reesei, is the only enzyme exhibiting Rex activity (12). XYN IV releases xylose from glucuronoxylan, arabinoxylan, and XOS. Endoxylanase activity for rhodymenan (an algal unsubstituted β-1,3-β-1,4-xylan) is also observed in XYN IV (12). XYN IV belongs to subfamily 7 of GH30 (GH30-7); however, the catalytic and structural properties of GH30-7 Rex are very limited because most GH30 xylanases (both bacterial GH30-8 and fungal GH30-7 enzymes) are characterized as glucuronoxylanases. Glucuronoxylanases are types of endoxylanases that strictly recognize the α-1,2-linked MeGlcA substitution on glucuronoxylans (13–20). Recently, we determined the crystal structure of GH30-7 glucuronoxylanase (Xyn30B isolated from Talaromyces cellulolyticus) and found Arg-46, which is required for the recognition of MeGlcA substitution (20). In contrast, this Arg residue is not conserved in XYN IV, which has enzyme activities that differ from those of glucuronoxylanase (20).
T. cellulolyticus is a promising cellulolytic fungus for biomass hydrolysis and production of cellulase (21, 22). GH3 β-xylosidase (Bxy3A), GH10 endoxylanase (Xyl10A), and six GH11 endoxylanases (Xyl11A to Xyl11C and Xyl11E to Xyl11G), which are related to the hydrolysis of the xylan main chain, have been identified and characterized in T. cellulolyticus (23–26). Furthermore, two GH30-7 xylanases, Xyn30A and Xyn30B, have been detected in a culture containing birchwood glucuronoxylan (20). Xyn30B has been characterized as a bifunctional enzyme possessing both glucuronoxylanase and exoxylobiohydrolase activities (20). Xyn30A may be a putative exoxylanase, as indicated by the high similarity of its sequence to that of XYN IV (77%); however, Xyn30A has not been fully characterized. Here, we report on the catalytic properties of Xyn30A. The mode of action of Xyn30A for xylan hydrolysis was investigated using various XOSs and hydrolysates from endoxylanase as substrates. Furthermore, the Rex activity of Xyn30A is discussed based on the comparison of subsite conformation between Xyn30A and Xyn30B by the homology-modeling method.
RESULTS AND DISCUSSION
Expression and purification of Xyn30A.
A recombinant Xyn30A was overexpressed using a T. cellulolyticus homologous expression system (27) and purified to electrophoretic homogeneity (Fig. 1). Xyn30A was identified and verified by analyzing masses of peptide fragments obtained by treating purified Xyn30A with trypsin (see Fig. S1 in the supplemental material). The molecular weight of Xyn30A, obtained using SDS-PAGE, was slightly greater than the 49,356 calculated using the amino acid sequence excluding the deduced N-terminal sequence (Fig. 1). The average molecular weight of Xyn30A was determined as 56,352 by time of flight mass spectrometry (TOF-MS). These results suggest that Xyn30A is glycosylated at several sites; five Asn residues (Asn-119, Asn-163, Asn-299, Asn-393, and Asn-433) were predicted to be the putative N-glycosylation sites (Fig. 2). Gel filtration chromatography showed that the molecular weight was approximately 117,000, suggesting that Xyn30A forms a dimer (see Fig. S2 in the supplemental material). With the exception of Xyn30A and XYN IV isolated from T. reesei, there are currently no reports on GH30 xylanases forming a dimer structure (28).
FIG 1.
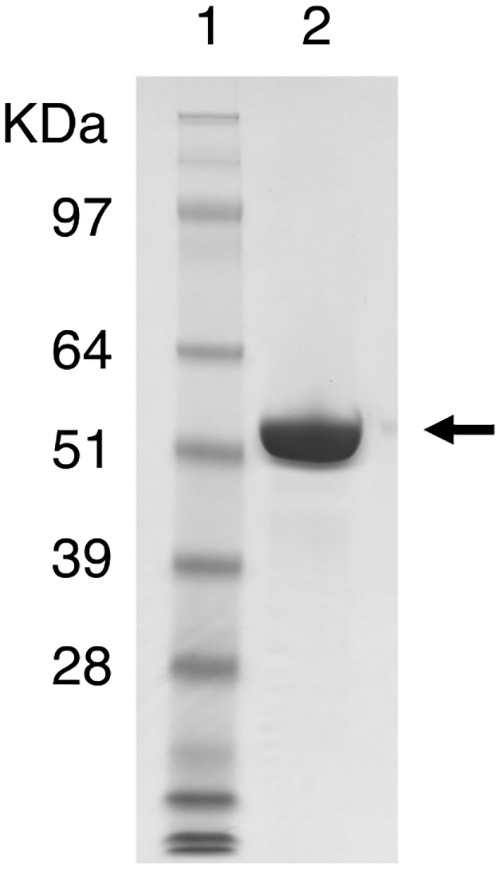
SDS-PAGE analysis of purified Xyn30A protein. Lanes: 1, molecular mass standards; 2, purified Xyn30A (10 μg protein). An arrow indicates the position of Xyn30A.
FIG 2.
Multiple-sequence alignments of GH30-7 and GH30-8 xylanases. Amino acid sequences of T. cellulolyticus Xyn30A (NCBI protein accession no. GAM43270), T. cellulolyticus Xyn30B (GAM36763), T. reesei XYN IV (AAP64786), T. reesei XYN VI (EGR45006), Erwinia chrysanthemi EcXynA (AAB53151), and Bacillus subtilis BsXynC (CAA97612) were aligned using the Clustal Omega server (46). The features are shown as follows: a putative N-terminal signal peptide of Xyn30A (highlighted in black), conserved Arg residues in GH30-7 (highlighted in red), putative N-glycosylation sites of Xyn30A (arrows), catalytic Glu residues (highlighted in gray), Phe-201 and Gln-272 of Xyn30A and corresponding residues in GH30-7 (red characters), conserved Arg residues in GH30-8 (red box), and a β2-α2 loop in Xyn30B (blue characters). The signal sequence was predicted using the SignalP 4.1 server in sensitive mode for D-cutoff values (http://www.cbs.dtu.dk/services/SignalP/). N-Glycosylation sites were predicted using the NetNglyc server (http://www.cbs.dtu.dk/services/NetNGlyc/).
Hydrolysis of xylan and XOS by Xyn30A.
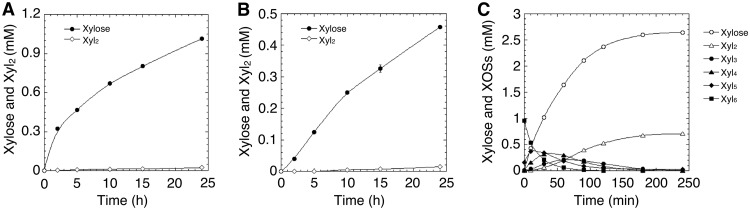
Purified Xyn30A exhibited a slight xylanase activity on birchwood glucuronoxylan (0.224 U/mg), beechwood glucuronoxylan (0.162 U/mg), and wheat arabinoxylan (0.279 U/mg). The activities were 2 to 4 orders of magnitude lower than the known activities of GH10 and GH11 endoxylanases (Xyl10A and Xyl11C, respectively) and than that of the GH30 glucuronoxylanase (Xyn30B) from T. cellulolyticus (20, 24, 29). Unlike these xylanases, Xyn30A produced xylose as the major product throughout xylan hydrolysis (Fig. 3A and B). The amount of Xyl2 was slightly increased during the reaction, but linear XOSs (Xyln, where n = 3 to 6) were hardly detectable by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). These results indicate that Xyn30A possesses exoxylanase activity, which releases xylose.
FIG 3.
Hydrolysis of xylan and Xyl6 by Xyn30A. Hydrolysis was carried out in a reaction mixture containing 10 μg ml−1 purified Xyn30A and 10 mg ml−1 birchwood glucuronoxylan (A), 10 mg ml−1 wheat arabinoxylan (B), or 1 mM Xyl6 in 50 mM sodium acetate (pH 4.0) (C). The reaction mixtures containing xylan and Xyl6 were incubated at 40 and 45°C, respectively. Concentrations of xylose and Xyl2 were determined by HPAEC-PAD.
Linear XOSs (Xyln, where n = 3 to 6) were appropriate substrates for detecting the exoxylanase activity of Xyn30A (Table 1). Xylotriose (Xyl3) was hydrolyzed to xylose and Xyl2 with a specific activity of 28.1 U/mg. No β-xylosidase activity for Xyl2 or 4-nitrophenyl-β-xylopyranoside (pNP-Xyl) was detected in Xyn30A. The optimum pH and temperature for hydrolysis of Xyl3 were estimated to be around pH 3.5 and 55°C, respectively (see Fig. S3 in the supplemental material). Xyn30A retained more than 90% activity after incubation for 30 min at 45°C at a pH range of 3.5 to 7.0 and was stable for 24 h at temperatures under 50°C at pH 4.0.
TABLE 1.
Substrate specificity of Xyn30A toward XOSsa
| Substrate | Relative activity (%) |
|---|---|
| Xyl3 | 100 |
| Xyl4 | 79 |
| Xyl5 | 81 |
| Xyl6 | 83 |
| MeGlcA3Xyl4 | 91 |
| 2-Ara3Xyl3 | 80 |
| 3-Ara3Xyl4 | 98 |
| 2,3-Di-Ara3Xyl3 | 100 |
| BR-Xyl3 | 0.13 |
| BR-MeGlcA3Xyl3 | 0.057 |
| pNP-Xyl2 | 44 |
Assays were carried out at 45°C in 50 mM sodium acetate buffer (pH 4.0) with a 2 mM concentration of each substrate. The specific activity for Xyl3 (28.1 U/mg) was taken as 100%.
Time-dependent changes in xylose and XOSs during hydrolysis of xylohexaose (Xyl6) show that Xyn30A released xylose in an exo manner. Xylopentaose (Xyl5) accumulated first; it was then hydrolyzed to xylotetraose (Xyl4) and Xyl3 and to xylose and Xyl2 at the end of the reaction (Fig. 3C). The molar ratio of xylose to Xyl2 in the reaction mixture was 4:1, indicating that Xyl2 was delivered from the two terminal residues remaining after digestion in an exo manner. This is consistent with the processive mode of hydrolysis used by the characterized GH8 Rex (10, 11).
Substrate specificity of Xyn30A.
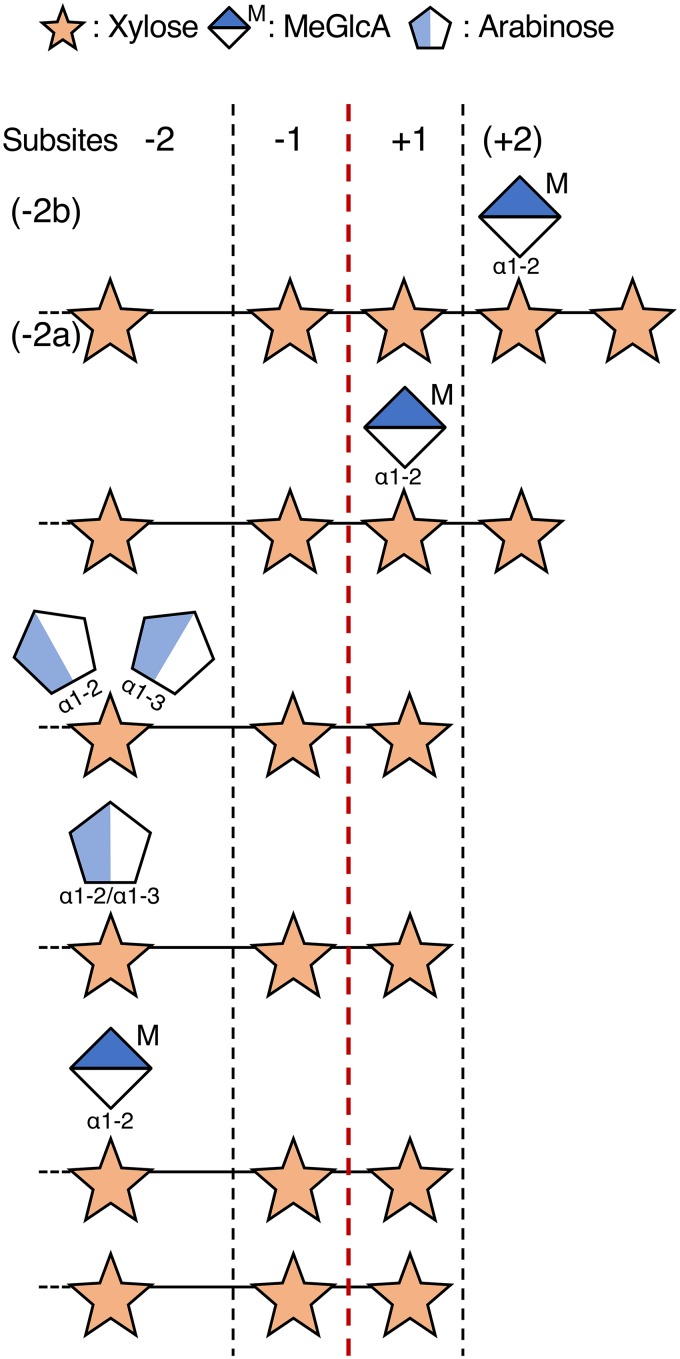
To clarify the directional selectivity of digestion in an exo manner, we examined the substrate specificity of Xyn30A using various types of XOSs. Our results indicate that Xyn30A released xylose from both MeGlcA- and α-l-arabinofuranosyl (Ara)-XOSs with 80 to 100% of relative activity for Xyl3 (Table 1). The relative activities for borohydride-reduced (BR)-XOSs, which are xylotriitol (BR-Xyl3) and 23-(4-O-methyl-α-d-glucuronyl)-xylotriitol (BR-MeGlcA3Xyl3), were 0.13 and 0.057% of that for Xyl3, respectively. Using BR substrates, Xyn30A released xylitol but not xylose (see Fig. S4 in the supplemental material). These results show that Xyn30A acts at the reducing end of XOSs and has no specificity for substrate side chains at subsite −2 (Fig. 4).
FIG 4.
Cleavage patterns of linear and branched XOSs in hypothetical subsites of Xyn30A. Points of cleavage are indicated by the red dashed line.
The low activity of Xyn30A on BR substrates indicates that substrate recognition for the reducing end of a substrate at subsite +1 is important for catalytic activity of Xyn30A. Xyn30A degraded pNP-β-d-xylobioside (pNP-Xyl2) into pNP and Xyl2 with higher relative activity than that shown by Xyn30A for BR substrates (Table 1). This result also confirms that Xyn30A hydrolyzes the glycosyl linkage at the reducing end. In contrast, Xyn30A showed no activity for pNP-β-lactoside and pNP-β-cellobioside, although these substrates are detectable during hydrolysis by Xyl10A (29); this suggests that subsites −1 and −2 of Xyn30A strictly recognize the xylan main chain. Subsite −2 of GH30 glucuronoxylanase is further divided into subsites −2a and −2b, which recognize the main chain and the MeGlcA side chain, respectively (20, 30, 31). Xyn30A broadly recognizes the various types of Ara3- and MeGlcA3-XOSs, indicating that the structure of subsite −2b is different in Xyn30A and GH30 glucuronoxylanase; these structural differences are described below (see “3D structure model of Xyn30A”). Based on substrate recognition for the reducing end of XOSs (Table 1) and hydrolysis profiles of xylan and XOSs (Fig. 3), we concluded that T. cellulolyticus Xyn30A possesses Rex activity.
Rex activity is well known in bacterial GH8 exoxylanases (9, 32). However, unlike GH30-7 Rex, GH8 Rex exhibits poor activity on polymeric xylan and is designated an exo-oligoxylanase (9). Analyses of kinetic properties of GH8 Rex indicate that Xyl3 is the most suitable substrate for this enzyme (9, 11, 33). The Km values of GH8 Rex (BH2105) isolated from Bacillus halodurans increase with increased degree of polymerization in XOS; this results in decreased kcat/Km values. On the other hand, no significant difference was observed in kinetic constants for linear XOSs of Xyn30A; the kcat/Km of Xyl6 was 1.7 times greater rather than that of Xyl3 (Table 2). These results suggest that Xyn30A is suitable for the hydrolysis of longer XOSs.
TABLE 2.
Kinetic constants of Xyn30A
| XOS | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| Xyl3 | 0.38 | 27.5 | 71.5 |
| Xyl4 | 0.29 | 21.0 | 72.1 |
| Xyl5 | 0.21 | 20.8 | 98.5 |
| Xyl6 | 0.18 | 21.1 | 118 |
Stereochemistry of Xyl3 hydrolysate.
GH8 Rex can utilize only the β-anomer of a substrate, forming β-xylose and α-XOS in an inverting catalytic mechanism via a single-displacement reaction (9, 34). To reveal the α/β-anomer selectivity of Xyn30A, we used 1H nuclear magnetic resonance (1H NMR) to analyze the anomeric composition of the hydrolysate obtained from Xyl3 using Xyn30A.
The 1H NMR spectra were complicated due to peaks from xylose, Xyl2, and Xyl3 overlapping in the spectra (see Fig. S5 to S7 and Table S1 in the supplemental material); thus, for estimations for each α/β-anomeric ratio, see “1H NMR analysis of xylotriose hydrolysate” in the supplemental material. The released α- and β-xylose products were present at the ratio of 3.2:6.8, with 50% of Xyl3 consumption in a 5-min reaction. This ratio was similar to the equilibrate anomeric ratio (α/β ratio = 3.5:6.5) of Xyl3. These results suggest that the Rex activity of Xyn30A is not dependent on recognizing the anomeric structure of a substrate but does show a slight preference for the β-anomer. On the other hand, 92% of newly formed Xyl2 was detected as β-anomer at 50% Xyl3 consumption (see Table S2 and Fig. S8 in the supplemental material). The proportion of β-anomer slowly decreased by 83% in a 33-min reaction where 96% of the substrate was consumed; this was likely due to slow anomerization to the α-anomer. These results confirm that Xyn30A uses a retaining catalytic mechanism similar to that used by known GH30 xylanases (12, 35). Analysis of the anomeric composition of the Xyl3 hydrolysate revealed that Xyn30A and GH8 differ in substrate recognition and catalytic mechanisms.
Molecular structure of acidic XOS products generated by Xyn30A.
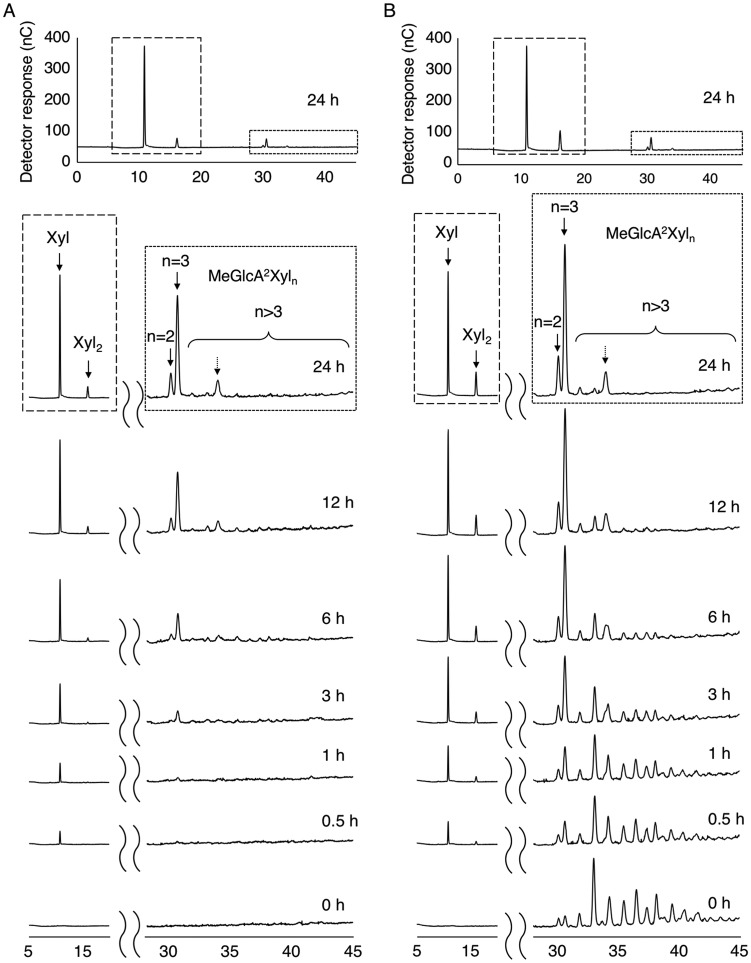
During hydrolysis of beechwood glucuronoxylan by Xyn30A, acidic XOSs were accumulated (Fig. 5A). The two products, presented as n = 2 and 3 in Fig. 5A, showed the same elution profiles as those of commercial 22-(4-O-methyl-α-d-glucuronyl)-xylobiose (MeGlcA2Xyl2) and 22-(MeGlcA)-xylotriose (MeGlcA2Xyl3) standards, respectively (data not shown). Structures of acidic XOSs in the reaction mixture were further defined using mass spectrometry.
FIG 5.
HPAEC-PAD profiles of Xyn30A products. (A) Time course analysis of the hydrolysis of beechwood glucuronoxylan by Xyn30A. (B) Time course analysis of the hydrolysis of MeGlcA2Xyln (n = 2 to 14) mixture by Xyn30A. Hydrolysis was performed at 45°C using a mixture (pH 4.0) consisting of 200 μg ml−1 Xyn30A and 10 mg ml−1 beechwood xylan (A) or 9 mg ml−1 MeGlcA2Xyln mixture (B).
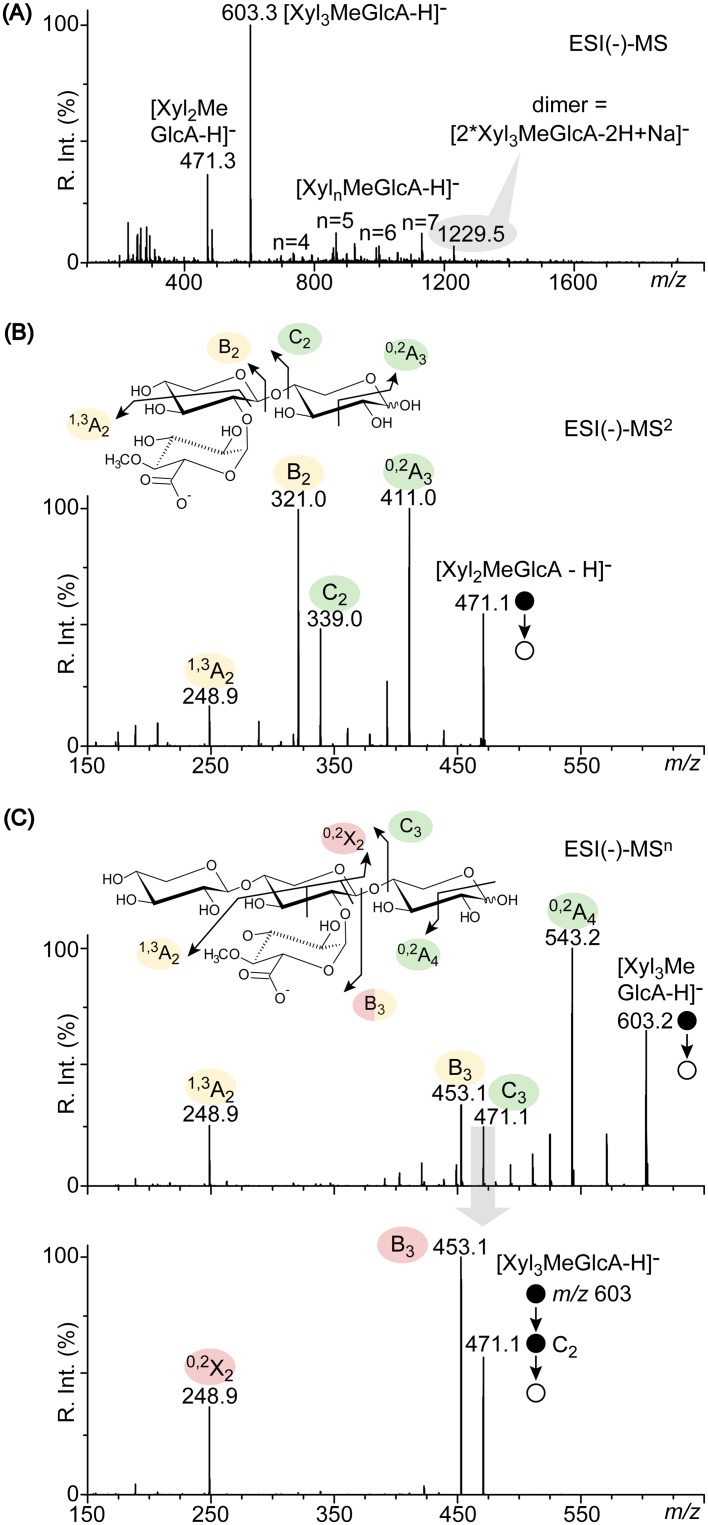
The negative-ion-mode electrospray ionization [ESI(−)] mass spectrum of hydrolysate from Xyn30A is shown in Fig. 6A. The negative-ion mode favors the detection of acidic XOSs carrying a MeGlcA moiety upon the deprotonation of the -COOH group (6). The shortest detected XOS was Xyl2MeGlcA at m/z 471, and the major XOS was Xyl3MeGlcA at m/z 603 (consisting of two and three xylose units, respectively). These results agree with those obtained using HPAEC-PAD analysis (Fig. 5A). The major XOS also formed a dimer at m/z 1,229 (Fig. 6A); this likely occurred via two deprotonated Xyl3MeGlcA adducted by one sodium cation, which formed a singly charged noncovalent dimer [see Fig. S9 in the supplemental material for its ESI(−)-MS2 spectrum]. Longer XOSs of four to seven xylose units were barely detected in the background. Conversely, an ion corresponding to the peak detected in HPAEC-PAD analysis (Fig. 5A, dotted arrow) was not detected in the ESI(−)-MS spectrum, suggesting that this product was an unknown XOS possessing no acidic substituent.
FIG 6.
Structural analysis of hydrolysis products obtained from beechwood glucuronoxylan. (A) ESI(−)-MS spectra of acidic XOSs produced by Xyn30A. (B) ESI(−)-MS2 spectra of the shortest Xyl2MeGlcA. (C) ESI(−)-MSn spectra of the major Xyl3MeGlcA. Structures of XOSs are depicted in insets using pairs of diagnostic product ions proposed previously (6).
Multistage mass spectrometry in negative-ion mode is used to analyze the structures of acidic XOSs. This technique relies on one-way and stepwise depolymerization of the xylose chain by glycosidic bond cleavage at the reducing ends of ions triggered by MSn selection/activation (6). Pairs of diagnostic product ions instantly reveal the shape of each activated species and the original architecture of the acidic XOS; this provides insights into the mode of action of the enzyme. The ESI(−)-MS2 spectrum of [Xyl2MeGlcA-H]− displayed the pair C2/0,2A3 formed by glycosidic bond cleavage/cross-ring cleavage at the reducing end (Fig. 6B, green circles) and pair B2/1,3A2 formed by dehydration of C2/internal cross-ring cleavage (yellow circles); these pairs are archetypal of a glucuronic acid located one unit away from the reducing end of the activated species. At two units long, the shortest XOS from Xyn30A carries a MeGlcA moiety at its nonreducing end (MeGlcA2Xyl2 [Fig. 6B, inset]).
The same two pairs of diagnostic product ions are detected in the ESI(−)-MS2 spectra of deprotonated Xyl3MeGlcA (C3/0,2A4 [Fig. 6C, green circles] and B3/1,3A2 [yellow circles]), showing that MeGlcA is located one unit away from its reducing end. This was validated by the ESI(−)-MS3 analysis of the C3 spectrum in Xyl3MeGlcA, which displays only one pair of B3/0,2X2 formed by dehydration of C3/internal cross-ring cleavage that retains the -COO− group (red circles); this indicates MeGlcA at the reducing end of C3 with no isomeric forms present (6). The major three-unit-long XOS from Xyn30A thereby carried the acidic group on the middle xylose unit (MeGlcA2Xyl3 [Fig. 6C, inset]). These results show that the major acidic XOSs produced by Xyn30A were MeGlcA2Xyl3 and MeGlcA2Xyl2. MeGlcA2Xyl3 was not further hydrolyzed upon prolonged incubation by Xyn30A (data not shown).
Evaluation of Xyn30A activity using hydrolysate of glucuronoxylan treated with Xyn30B.
Rex activity generally releases xylose at the reducing end; therefore, Rex activity likely did not generate the MeGlcA2Xyl3 and MeGlcA2Xyl2 detected during hydrolysis of beechwood glucuronoxylan. This indicates that Xyn30A may act in both exo and endo manners. To determine how MeGlcA2Xyl3 and MeGlcA2Xyl2 are produced by Xyn30A, we examined the hydrolytic activity of Xyn30A using a hydrolysate of beechwood glucuronoxylan pretreated with Xyn30B; this hydrolysate contained a mixture of acidic XOSs consisting mainly of MeGlcA2Xyln (n = 2 to 14) (Fig. 5B, 0 h) (20).
We initially expected that the release of xylose from MeGlcA2Xyln would be prevented by the presence of a branched reducing end; however, the levels of MeGlcA2Xyl3 and MeGlcA2Xyl2 continually increased immediately after the reaction was initiated (Fig. 5B). These results indicate that Xyn30A can hydrolyze the reducing end of acidic XOS at MeGlcA2Xyl3 or MeGlcA2Xyl2 (Fig. 4). This reaction proceeded slower than that using linear XOSs (Fig. 3C). XOS intermediates (Xyln, n = 3 to 6) during the reaction were hardly detectable. If Xyn30A degrades MeGlcA2Xyln in an endo manner, a temporal increase in the levels of short-branched XOSs should be observed. However, the levels of acidic XOSs longer than MeGlcA2Xyl4 were uniformly decreased during the reaction as indicated by their peak area on a chromatogram (see Fig. S10 in the supplemental material). Thus, Xyn30A is proposed to release MeGlcA2Xyl3 or MeGlcA2Xyl2 on the reducing end in an exo manner. In addition, Xyn30A easily generates XOSs which possess the branched terminal end corresponding to MeGlcA2Xyl2 (Table 1; Fig. 4). These results explain why hydrolysis of beechwood glucuronoxylan by Xyn30A is accompanied by accumulation of MeGlcA2Xyl3 and MeGlcA2Xyl2 (Fig. 5A). Tenkanen et al. have reported that XYN IV generates Ara2Xyl3 from arabinoxylan as a major Ara-XOS product (12). Hydrolysis of arabinoxylan by Xyn30A is likely similar to that of glucuronoxylan.
Xylan hydrolysis via combined activity of Xyn30A and endoxylanase.
Xyn30A and various xylanolytic enzymes, including Xyn30B, GH10 and GH11 endoxylanases, and GH3 β-xylosidase, have been detected in a culture of T. cellulolyticus containing birchwood glucuronoxylan (20). How Xyn30A contributes to xylan hydrolysis was examined using beechwood glucuronoxylan pretreated with Xyl10A or Xyl11C purified from recombinant T. cellulolyticus strains. The major acidic XOSs in the hydrolysates produced by Xyl10A and Xyn11C were identified as MeGlcA3Xyl3 and MeGlcA3Xyl4, respectively, using ESI(−)-MSn (6). When these hydrolysates were further digested by Xyn30A, all the produced XOSs were one unit shorter after the second digestion by Xyn30A (Fig. 7). The structures of major XOSs were evaluated by multistage MSn tracking of the pairs of diagnostic product ions (see Fig. S11 in the supplemental material). The results indicated that the second digestion using Xyn30A shortened the acidic XOS generated during the first digestion by Xyl10A or Xyl11C (Fig. 7, insets, black and red dashed lines).
FIG 7.
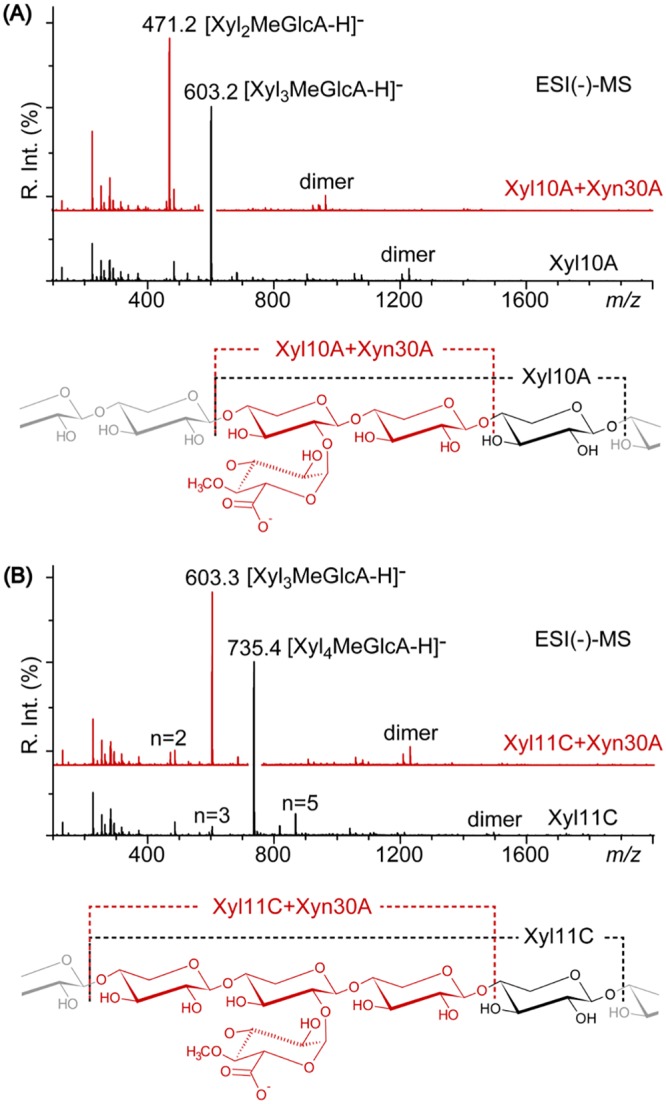
Structural analysis of products obtained from hydrolysis of beechwood glucuronoxylan by endoxylanase and Xyn30A. (A) ESI(−)-MS spectra of acidic XOS generated by Xyl10A (black line, bottom) and by a second digestion with Xyn30A (red line, top); (B) ESI(−)-MS spectra of acidic XOSs generated by Xyl11C (black line, bottom) and by a second digestion with Xyn30A (red line, top). The structures of XOSs are depicted in the insets.
Hydrolyzing beechwood glucuronoxylan and wheat arabinoxylan using both Xyn30A and an endoxylanase significantly increased the yield of xylose without decreasing that of Xyl2 (Table 3). This occurred because Xyn30A hydrolyzed the reducing ends of branched and linear XOSs. It should be noted that the reducing ends in branched XOSs cannot be degraded by β-xylosidase, which releases xylose from the nonreducing ends. Thus, using Xyn30A with endoxylanase and β-xylosidase may increase the xylose yield during hydrolysis of xylan.
TABLE 3.
Effect of supplementation with Xyn30A on xylan hydrolysis by endoxylanase
| Enzyme(s) | Mixture ratioa | Yield (mM) from: |
|||
|---|---|---|---|---|---|
| Beechwood glucuronoxylan |
Wheat arabinoxylan |
||||
| Xylose | Xyl2 | Xylose | Xyl2 | ||
| Xyl10A | 1 | 5.91 | 11.0 | 2.44 | 3.42 |
| Xyl10A-Xyn30A | 0.8/0.2 | 9.08 | 10.8 | 5.91 | 3.22 |
| Xyl11C | 1 | 3.19 | 7.43 | 1.14 | 1.04 |
| Xyl11C-Xyn30A | 0.8/0.2 | 9.10 | 7.37 | 5.91 | 1.07 |
Enzymes were added to the reaction mixture at a total protein loading of 5 mg enzyme per g xylan.
3D structure model of Xyn30A.
The xyn30A gene encodes the Xyn30A protein, consisting of 475 amino acid residues. The amino acid sequence of Xyn30A shows 77% similarity to that of XYN IV from T. reesei (12) (Fig. 2). Xyn30A shares amino acid sequence identity with fungal GH30-7 glucuronoxylanases, such as Xyn30B (38%) from T. cellulolyticus and XYN VI (34%) form T. reesei (18), but is less similar to the bacterial GH30-8 enzymes, such as XynA from Erwinia chrysanthemi (EcXynA) (24%) and XynC from Bacillus subtilis (BsXynC) (24%) (13, 36). Two conserved catalytic residues known in GH30 xylanase, a general acid/base residue and a nucleophilic residue, were found to correspond with Glu-198 and Glu-290, respectively, in Xyn30A (Fig. 2, gray highlights). Xyn30A and XYN IV lack the Arg residue conserved in GH30-7 glucuronoxylanases (Fig. 2, red highlights) and GH30-8 glucuronoxylanases (Fig. 2, red box). This observation is consistent with the fact that both Xyn30A and XYN IV are not MeGlcA appendage-dependent xylanases.
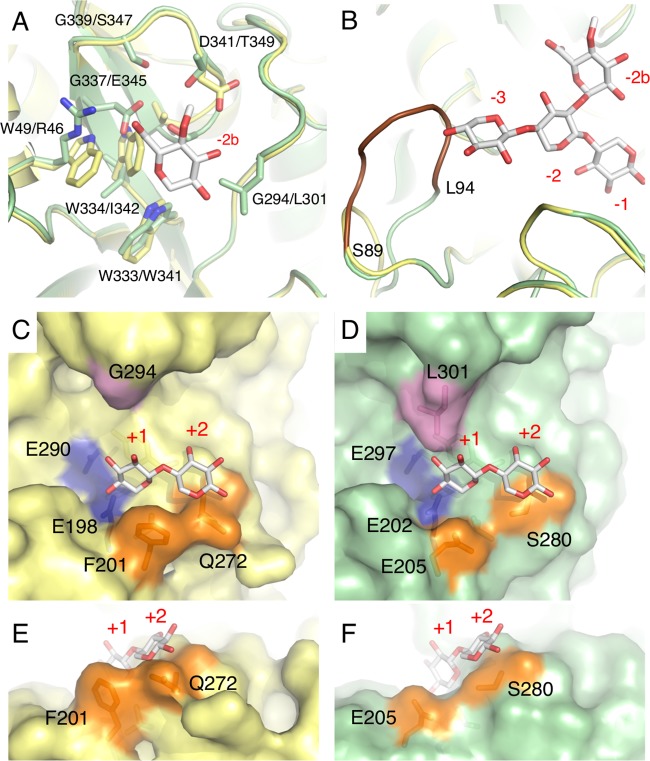
To examine the structural and catalytic differences between GH30 Rex and glucuronoxylanase, a three-dimensional (3D) structural model of Xyn30A was constructed by homology modeling using a structure of Xyn30B as a template. The residues forming subsite −2b in Xyn30A differ significantly from those of Xyn30B; the only exception is Trp-341, which corresponds to Trp-333 in Xyn30A (Fig. 8A). Arg-46, which is a critical residue for glucuronoxylanase activity of Xyn30B, is replaced with Trp-49. Other residues of Xyn30B, such as Leu-301, Glu-345, and Ser-347, are also replaced with glycine in the corresponding positions. These replacements may lead to reduced appendage specificity at the −2b subsite, providing Xyn30A with broad substrate specificity for branched XOSs (Table 1).
FIG 8.
3D structural analysis of Xyn30A. The Xyn30A homology model is compared with the crystal structure of Xyn30B (PDB no. 6IUJ) (20). The superimposed model structures of Xyn30A and Xyn30B, with MeGlcA2Xyl3 at negative subsites (A and B) and Xyl2 at positive subsites (C to F), are based on the crystal structure of EcXynA complexed with MeGlcA2Xyl3 (PDB no. 2Y24) and Clostridium thermocellum Xyn30A (CtXyn30A) complexed with xylobiose (PDB no. 5A6M), respectively. Atoms are depicted as follows: C in Xyn30A (yellow) and C in Xyn30B (green), C in ligands (white), O (red), and N (blue). (A) MeGlcA and amino acid residues in subsite −2b of Xyn30A and Xyn30B are shown in order of Xyn30A/Xyn30B. (B) The β2-α2 loops of Xyn30A and Xyn30B containing MeGlcA2Xyl3 are depicted at subsites −3 to −1. The extended loop of Xyn30B (amino acid residues are shown as blue characters in Fig. 2) is depicted in brown. (C and D) Structures of positive subsites in Xyn30A (C) and Xyn30B (D) with xylobiose at the +1 and +2 subsites. Two catalytic Glu residues, Phe-201 and Gln-272, and Gly-294 of Xyn30A are shown in blue, orange, and purple, respectively; the equivalent residues of Xyn30B are shown using the same colors. (E and F) Different view of positive subsites in Xyn30A (E) and Xyn30B (F).
The β2-α2 loop of Xyn30B forms a partial roof structure at subsite −3 and interrupts introduction of the nonreducing end of linear XOSs at subsite −3 (Fig. 2, blue characters, and Fig. 8B, brown loop) (20). This structural feature of Xyn30B is responsible for its exoxylobiohydrolase activity, which releases Xyl2 from the nonreducing end of linear XOSs. We confirm that the β2-α2 loop of Xyn30A is shorter than that of Xyn30B because Xyn30A lacks the residues corresponding to Ser-89 to Leu-94 of Xyn30B (Fig. 2 and 8B). This observation supports the fact that Xyn30A has similar kinetic constants for XOSs that are longer than Xyl3 (Table 2).
The structure of subsites +1 and +2 in Xyn30A contains an open space that is similar to that of Xyn30B, although subsite +2 in GH8 Rex is blocked by a loop structure (37). A comparison of Xyn30A and Xyn30B shows that Glu-205, Ser-280, and Leu-301 in Xyn30B are replaced with Phe-201, Gln-272, and Gly-294 in Xyn30A, respectively (Fig. 8C and D). Phe-201 and Gln-272 may interact with a xylose residue at subsite +1, and a bulky side chain of Gln-272 may interrupt the binding of xylose residue at subsite +2 (Fig. 8E and F). It should be noted that Gln-272 is conserved only in Xyn30A and XYN IV (Fig. 2). This observation suggests that Gln-272 is important for Rex activity and is a promising target for further characterization. However, whether interaction with a xylose residue at subsite +2 is interrupted only by Gln-272 remains unclear. A dimer structure, observed in Xyn30A and XYN IV, raises the possibility that the amino acid residue contributing to Rex activity is contained in another subunit. Further studies using crystal structures are needed to answer this question. The release of MeGlcA2Xyl2 and MeGlcA2Xyl3 by Xyn30A may be explained by the open space around the C-2 position of xylose in subsite +1 and +2, respectively (Fig. 4 and 8C). However, in this study, how these branched terminal residues are incorporated at subsites +1 and +2 remains unknown.
To our knowledge, a GH30 xylanase possessing Rex activity has been found only in XYN IV from T. reesei (12); therefore, this study reports the second example of a fungal GH30-7 Rex. It should be noted that both T. reesei and T. cellulolyticus are representative cellulase producers (22, 38). The GH30-7 Rex may also be distributed in other fungal cellulolytic systems; however, it is difficult to specifically detect Rex activity in a crude enzyme using a conventional xylanase assay (such as assays used to determine reducing sugars). Homology modeling of Xyn30A suggests that GH30-7 Rex possesses a conserved Gln-272 at subsite +2 but does not possess the Arg residue conserved in GH30-7 glucuronoxylanase (which is Arg-46 in Xyn30B). Based on these observations, we assessed the distribution of putative GH30-7 Rex proteins. Genes encoding such proteins were found to be distributed in 24 Ascomycota fungi, including the genera Talaromyces, Trichoderma, Byssochlamys, Fonsecaea, Cladophialophora, Phialophora, Exophiala, Rachicladosporium, Thielavia, Hortaea, Stagonospora, Parastagonospora, Pyrenophora, Stemphylium, Alternaria, Exserohilum, Bipolaris, Aureobasidium, Baudoinia, Scytalidium, Pyrenophora, Cercospora, Paraphaeosphaeria, and Coniochaeta. These results suggest that Rex-type proteins may constitute a new subgroup of GH30-7 xylanases.
In conclusion, we characterized the mode of action of fungal GH30-7 Rex based on molecular structures of xylan and XOS hydrolysates. Our results indicate that the Rex activity of T. cellulolyticus Xyn30A releases xylose and branched XOSs from the reducing end of glucuronoxylan in an exo manner. The results obtained using homology modeling provide insights into the specific structure of substrate-binding sites in GH30-7 Rex and predict the presence of putative GH30-7 Rex distributed in Ascomycota fungi. Treatment with GH30-7 Rex can contribute to cooperative hydrolysis of xylan using fungal cellulolytic systems.
MATERIALS AND METHODS
Chemicals.
Birchwood xylan was obtained from Sigma-Aldrich (St. Louis, MO, USA). Beechwood xylan and wheat arabinoxylan (low viscosity) were purchased from Megazyme (Wicklow, Ireland). XOSs, such as linear Xyln (n = 3 to 6), MeGlcA2Xyl2, MeGlcA2Xyl3, MeGlcA3Xyl4, 23-α-l-arabinofuranosyl-xylotriose (2-Ara3Xyl3), 33-α-l-arabinofuranosyl-xylotetraose (3-Ara3Xyl4), 23,33-di-α-l-arabinofuranosyl-xylotriose (2,3-di-Ara3Xyl3), BR-Xyl3, and BR-MeGlcA3Xyl3, were obtained from Megazyme. pNP-Xyl2 was obtained from Megazyme, and pNP-Xyl, pNP-β-lactoside, and pNP-β-cellobioside were obtained from Sigma-Aldrich. Xylose and Xyl2 were obtained from Fujifilm Wako Pure Chemical Corporation (Osaka, Japan).
Strains and culture conditions.
T. cellulolyticus CF-2612 (FERM BP-10848) was maintained on potato dextrose agar (Difco, Detroit, MI, USA) plates (39). The T. cellulolyticus YP-4 uracil autotroph was maintained on potato dextrose agar plates containing uracil and uridine at final concentrations of 1 g liter−1 each (27). The prototrophic transformants of T. cellulolyticus YP-4 expressing recombinant protein Xyn30A were maintained on MM agar (1% [wt/vol] glucose, 10 mM NH4Cl, 10 mM potassium phosphate [pH 6.5], 7 mM KCl, 2 mM MgSO4, and 1.5% [wt/vol] agar) plates (40). The recombinant enzyme was produced using a soluble starch medium containing 2% (wt/vol) soluble starch (Fujifilm Wako) and 0.2% (wt/vol) urea as described previously (27).
Plasmid construction and fungal transformation.
The plasmid pANC202 (27), which contains the pyrF gene and the glucoamylase (glaA) promoter and terminator regions, was used to construct the plasmid expressing recombinant Xyn30A. Escherichia coli DH5α (TaKaRa Bio, Kyoto, Japan) was used for DNA procedures. The xyn30A gene (1,580 bp, including three introns) was amplified from the CF-2612 genome using the forward primer 5′-ATTGTTAACAATATGCGGCACCCAATCCCTATTC, with the HpaI site (underlined), and the reverse primer 5′-AATCCTGCAGGCTACTCCAACCAAAACACCTGCACC, with the SbfI site (underlined); primer design was based on the genomic sequence of T. cellulolyticus registered in DDBJ/EMBL/GenBank (DF933843.1) (25). The expression plasmid pANC214 was constructed by ligating the xyn30A fragment, which had been digested with HpaI/SbfI, into the EcoRV/SbfI site of pANC202. The presence and location of the ligated gene fragment were verified by DNA sequencing.
The plasmid pANC214 was transformed into protoplasts of T. cellulolyticus YP-4 by nonhomologous integration into the host chromosomal DNA (40). The recombinant Xyn30A-producing strain was selected based on the amount of recombinant protein in culture supernatant that was visualized by SDS-PAGE using NuPAGE 4 to 12% Bis-Tris gels (Invitrogen, Carlsbad, CA, USA); the recombinant Xyn30A-producing strain was subsequently designated Y214.
Purification of Xyn30A.
Purification of Xyn30A from the culture supernatant of the Y214 strain was performed using an ÄKTA purifier chromatography system (GE Healthcare, Buckinghamshire, United Kingdom) at room temperature. The culture supernatant was filtered through a 0.22-μm polyethersulfone membrane and desalted using a HiPrep 26/10 desalting column (GE Healthcare) that had been equilibrated with 20 mM 2-(N-morpholino)ethanesulfonic acid (pH 6.5). The desalted sample was applied to a Resource Q anion-exchange column (6 ml; GE Healthcare) that had been equilibrated with the same buffer, and protein peaks were eluted with a linear gradient of 25 to 150 mM NaCl (25 column volumes) at a flow rate of 4 ml min−1. Fractions containing Xyn30A were confirmed by SDS-PAGE and pooled. The samples, adjusted to a final concentration of 0.85 M (NH4)2SO4, were subjected to Source 15PHE (10 ml; GE Healthcare) hydrophobic-interaction chromatography. For this, we used a 0.8 to 0.3 M (NH4)2SO4 gradient (using 25 column volumes) in 20 mM sodium acetate buffer (pH 5.5) at a flow rate of 2.5 ml min−1. The fractions containing the target protein were pooled, and the buffer in the pooled sample was replaced with 20 mM sodium acetate buffer (pH 4.0) by ultrafiltration using Vivaspin 20-5K (Sartorius, Göttingen, German). The samples were applied to a Resource S cation-exchange column (6 ml; GE Healthcare) that had been equilibrated with 20 mM sodium acetate buffer (pH 4.0); protein peaks were eluted with a linear gradient of 75 to 275 mM NaCl (using 30 column volumes) at a flow rate of 4 ml min−1. Xyn30A fractions were pooled and concentrated by ultrafiltration using Vivaspin 20-5K. The purified enzyme was preserved in a 20 mM sodium acetate buffer (pH 4.5) containing 0.01% NaN3 at 4°C. The protein concentration was determined with a bicinchoninic acid protein assay kit (Thermo Scientific, Rockford, IL, USA) using bovine serum albumin (Thermo Scientific) as the protein standard.
Enzyme activity assays.
All enzyme activity assays were performed in triplicate. Xylanase activity was measured by assaying the reducing sugars released after reacting xylanase with 10 mg ml−1 soluble xylan. The enzymatic reaction was carried out in 50 mM sodium acetate buffer (pH 4.0) at 45°C for 30 min. The concentration of reducing sugars was determined using 3,5-dinitrosalicylic acid (DNS) (41). Enzyme activities against XOSs and BR-XOSs were measured in a reaction mixture containing purified Xyn30A and 2 mM substrate in 50 mM sodium acetate (pH 4.0); the reactions were carried out at 45°C for 10 and 30 min, respectively. The reaction was stopped via incubation at 99°C for 5 min. The released product (xylose or xylitol) was assessed using HPAEC-PAD. Enzymatic activity against pNP-based chromogenic glycosides was assayed in 50 mM sodium acetate (pH 4.0) using 2 mM substrate. The reaction mixture was incubated at 45°C for 10 min, and the reaction was stopped by adding Na2CO3 at a final concentration of 0.67 M. The concentration of released 4-nitrophenol was determined by measuring the absorbance at 420 nm. One unit of enzyme activity for each substrate was defined as the amount of enzyme that catalyzed the release of 1 μmol of product per minute.
Xyn30A was characterized with respect to pH and temperature under standard assay conditions using 2 mM Xyl3. The optimal pH values and pH stabilities were examined using McIlvaine buffer (pH 2.0 to 7.0) (42). To examine stability, samples of Xyn30A were preincubated in McIlvaine buffer at pH values ranging from 2.0 to 8.0 for 30 min at 45°C, and residual activity was subsequently measured. The optimal reaction temperature was examined at 40 to 70°C. To evaluate thermal stability, samples of Xyn30A were preincubated in 50 mM sodium acetate (pH 4.0) at 30 to 65°C for 30 min or at 4 to 65°C for 24 h, and residual activity was then measured.
Kinetic parameters were evaluated using 0.05 to 2 mM (each) Xyl3, Xyl4, Xyl5, and Xyl6. The reaction was performed at 45°C in 50 mM sodium acetate, pH 4.0. The enzyme concentration in the reaction mixtures was 2.0 nM. The molar concentration of Xyn30A was calculated by using the theoretical molecular weight of 49,356, which was calculated from the amino acid sequence excluding a deduced N-terminal sequence. Kinetic constants were determined by using the nonlinear least-squares data fitting method in Microsoft Excel 2016 (Microsoft, Redmond, WA, USA) (43).
HPAEC-PAD analysis of XOSs, xylose, and xylitol was performed as described previously using a Dionex ICS-3000 ion chromatography system equipped with a CarboPac PA1 (Dionex, Sunnyvale, CA, USA) (20).
MS.
The molecular mass of purified Xyn30A was evaluated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) with a Spiral TOF JMS-S3000 (JEOL, Tokyo, Japan). The purified sample was applied to the MALDI target plate after dilution into a mixture containing 0.5% (wt/vol) sinapinic acid, 0.1% trifluoroacetic acid, and 25% acetonitrile. Monovalent and bivalent ions from conalbumin (molecular weight, 75,000), included in the gel filtration calibration kit HMW (GE Healthcare), were used for external mass calibration. Instrument control, data acquisition, and data analysis were conducted using MSTornado (JEOL).
Acidic XOSs were molecularly and structurally analyzed by electrospray ionization multistage mass spectrometry in negative-ion mode using ESI(−)-MSn (n = 1 to 4) and an amaZon SL-STT2 ion trap (Bruker, Bremen, Germany). Methanolic solutions of XOSs were introduced into the ionization source using infusion mode and a syringe pump at a flow rate of 10 μl min−1. The apparatus was operated in enhanced-resolution mode (mass range, 100 to 2,000 m/z; scanning rate, 8,100 m/z per second). The width of the selection window was set at 1 Da in MS2 to obtain clean isotopic selection, and at 2 Da in MS3 and MS4 to obtain sufficient signal intensity. The amplification of the excitation was set according to the experiment to reach a survival yield at ∼15 to 30%. Instrument control, data acquisition, and data processing were performed using Compass 1.3 SR2 provided by Bruker. mMass 5.5.0.0 (44) and Illustrator CC (Adobe Inc., San Jose, CA, USA) were used for data processing and artwork.
1H NMR analysis.
The stereochemistry of glycoside bond cleavage was characterized with 1H NMR spectroscopy using Xyl3 as the substrate. All chemicals and enzymes were prepared using D2O. The reaction mixture, consisting of 1.34 mg ml−1 Xyn30A, 22 mM Xyl3, 28.5 mM sodium acetate buffer (pH 4.0), and 1.47 mM sodium citrate, was mixed in an NMR tube. 1H NMR spectra were recorded at various intervals on a JNM-ECS400 (JEOL, Tokyo, Japan) at 23°C. Prior to starting the reaction, the 1H NMR spectrum (0 min) was measured without adding the enzyme solution. The reaction was performed in a relatively concentrated state, compared to the standard assay condition, to ensure that the hydrolysis of the substrate proceeded more rapidly than anomerization of the products. Sodium citrate in the mixture was used as internal standard for comparison of the peak areas.
Hydrolysis of acidic XOSs by Xyn30A.
Samples of acidic XOSs from beechwood glucuronoxylan were prepared using GH10 xylanase from T. cellulolyticus (Xyl10A), GH11 xylanase from T. cellulolyticus (Xyl11C), or GH30-7 glucuronoxylanase from T. cellulolyticus (Xyn30B). The purified recombinant enzymes were prepared as described previously (20, 23, 24).
Hydrolysis of beechwood xylan using Xyl10A or Xyl11C was carried out in a 100-μl mixture containing 10 mg ml−1 substrate and 100 μg ml−1 enzyme in 50 mM sodium acetate buffer (pH 4.0); the reaction was conducted at 45°C for 24 h and stopped via incubation at 99°C for 5 min. Then, 90 μl of mixture was hydrolyzed with 90 μg ml−1 Xyn30A at 45°C for 24 h in a final volume of 100 μl. The acidic XOSs in the hydrolysate were analyzed by ESI(−)-MSn.
Hydrolysis of beechwood xylan using Xyn30B was carried out in a 100-μl mixture containing 10 mg ml−1 substrate and 625 ng ml−1 enzyme in 50 mM sodium acetate buffer (pH 4.0); the reaction was conducted at 40°C for 12 h. Then, 90 μl of mixture was hydrolyzed with 200 μg ml−1 Xyn30A at 45°C in a final volume of 100 μl. The reaction was stopped at appropriate time intervals (0 to 24 h), and the hydrolysate was analyzed by HPAEC-PAD.
Synergistic hydrolysis of xylan.
A reaction mixture containing xylanase and either 10 mg ml−1 beechwood xylan or 10 mg ml−1 wheat arabinoxylan was incubated in 50 mM sodium acetate (pH 5.0) at 45°C for 24 h. A xylanase mixture was added to the reaction mixture at 5 mg/g-xylan of protein using the following combinations (ratio): Xyl10A alone, Xyl10A-Xyn30A (0.8:0.2), Xyl11C alone, and Xyl11C-Xyn30A (0.8:0.2). The concentrations of xylose and Xyl2 in the mixtures were determined by HPAEC-PAD.
Homology modeling.
The homology model structure of Xyn30A was based on the crystal structure of Xyn30B from T. cellulolyticus (20) (PDB no. 6IUJ). The homology model was constructed using Modeller software (45). Figures depicting molecular structures were generated with open-source PyMol molecular graphic system, version 1.8 (Schrödinger, New York, NY, USA).
Supplementary Material
ACKNOWLEDGMENTS
We thank Maya Ishii, Benchaporn Inoue, and Miho Yoshimi for their technical assistance.
This work was supported by a Basic Research Funding grant from the National Institute of Advanced Industrial Science and Technology.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00552-19.
REFERENCES
- 1.Biely P, Singh S, Puchart V. 2016. Towards enzymatic breakdown of complex plant xylan structures: state of the art. Biotechnol Adv 34:1260–1274. doi: 10.1016/j.biotechadv.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Dodd D, Cann I. 2009. Enzymatic deconstruction of xylan for biofuel production. Glob Change Biol Bioenergy 1:2–17. doi: 10.1111/j.1757-1707.2009.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juturu V, Wu JC. 2012. Microbial xylanases: engineering, production and industrial applications. Biotechnol Adv 30:1219–1227. doi: 10.1016/j.biotechadv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Biely P, Vršanská M, Tenkanen M, Kluepfel D. 1997. Endo-β-1,4-xylanase families: differences in catalytic properties. J Biotechnol 57:151–166. doi: 10.1016/S0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- 5.Kolenová K, Vršanská M, Biely P. 2006. Mode of action of endo-β-1,4-xylanases of families 10 and 11 on acidic xylooligosaccharides. J Biotechnol 121:338–345. doi: 10.1016/j.jbiotec.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Fouquet T, Sato H, Nakamichi Y, Matsushika A, Inoue H. 2019. Electrospray multistage mass spectrometry in the negative ion mode for the unambiguous molecular and structural characterization of acidic hydrolysates from 4-O-methylglucuronoxylan generated by endoxylanases. J Mass Spectrom 54:213–221. doi: 10.1002/jms.4321. [DOI] [PubMed] [Google Scholar]
- 7.Jordan DB, Wagschal K. 2010. Properties and applications of microbial β-d-xylosidases featuring the catalytically efficient enzyme from Selenomonas ruminantium. Appl Microbiol Biotechnol 86:1647–1658. doi: 10.1007/s00253-010-2538-y. [DOI] [PubMed] [Google Scholar]
- 8.Tenkanen M, Luonteri E, Teleman A. 1996. Effect of side groups on the action of β-xylosidase from Trichoderma reesei against substituted xylo-oligosaccharides. FEBS Lett 399:303–306. doi: 10.1016/S0014-5793(96)01313-0. [DOI] [PubMed] [Google Scholar]
- 9.Honda Y, Kitaoka M. 2004. A family 8 glycoside hydrolase from Bacillus halodurans C-125 (BH2105) is a reducing end xylose-releasing exo-oligoxylanase. J Biol Chem 279:55097–55103. doi: 10.1074/jbc.M409832200. [DOI] [PubMed] [Google Scholar]
- 10.Hong P-Y, Iakiviak M, Dodd D, Zhang M, Mackie RI, Cann I. 2014. Two new xylanases with different substrate specificities from the human gut bacterium Bacteroides intestinalis DSM 17393. Appl Environ Microbiol 80:2084–2093. doi: 10.1128/AEM.03176-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valenzuela SV, Lopez S, Biely P, Sanz-Aparicio J, Pastor F. 2016. The glycoside hydrolase family 8 reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 is active on branched xylooligosaccharides. Appl Environ Microbiol 82:5116–5124. doi: 10.1128/AEM.01329-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenkanen M, Vršanská M, Siika-Aho M, Wong DW, Puchart V, Penttilä M, Saloheimo M, Biely P. 2013. Xylanase XYN IV from Trichoderma reesei showing exo- and endo-xylanase activity. FEBS J 280:285–301. doi: 10.1111/febs.12069. [DOI] [PubMed] [Google Scholar]
- 13.St John FJ, Rice JD, Preston JF. 2006. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J Bacteriol 188:8617–8626. doi: 10.1128/JB.01283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vršanská M, Kolenová K, Puchart V, Biely P. 2007. Mode of action of glycoside hydrolase family 5 glucuronoxylan xylanohydrolase from Erwinia chrysanthemi. FEBS J 274:1666–1677. doi: 10.1111/j.1742-4658.2007.05710.x. [DOI] [PubMed] [Google Scholar]
- 15.Valenzuela SV, Diaz P, Pastor FI. 2012. Modular glucuronoxylan-specific xylanase with a family CBM35 carbohydrate-binding module. Appl Environ Microbiol 78:3923–3931. doi: 10.1128/AEM.07932-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padilha IQM, Valenzuela SV, Grisi T, Diaz P, de Araújo DAM, Pastor FI. 2014. A glucuronoxylan-specific xylanase from a new Paenibacillus favisporus strain isolated from tropical soil of Brazil. Int Microbiol 17:175–184. doi: 10.2436/20.1501.01.220. [DOI] [PubMed] [Google Scholar]
- 17.St John FJ, Crooks C, Dietrich D, Hurlbert J. 2016. Xylanase 30 A from Clostridium thermocellum functions as a glucuronoxylan xylanohydrolase. J Mol Catal B Enzym 133:S445–S451. doi: 10.1016/j.molcatb.2017.03.008. [DOI] [Google Scholar]
- 18.Biely P, Puchart V, Stringer MA, Mørkeberg Krogh K. 2014. Trichoderma reesei XYN VI—a novel appendage-dependent eukaryotic glucuronoxylan hydrolase. FEBS J 281:3894–3903. doi: 10.1111/febs.12925. [DOI] [PubMed] [Google Scholar]
- 19.Maehara T, Yagi H, Sato T, Ohnishi-Kameyama M, Fujimoto Z, Kamino K, Kitamura Y, St John FJ, Yaoi K, Kaneko S. 2018. GH30 glucuronoxylan-specific xylanase from Streptomyces turgidiscabies C56. Appl Environ Microbiol 84:e018501-17. doi: 10.1128/AEM.01850-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamichi Y, Fouquet T, Ito S, Watanabe M, Matsushika A, Inoue H. 2019. Structural and functional characterization of a bifunctional GH30-7 xylanase B from the filamentous fungus Talaromyces cellulolyticus. J Biol Chem 294:4065–4078. doi: 10.1074/jbc.RA118.007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue H, Decker SR, Taylor LE II, Yano S, Sawayama S. 2014. Identification and characterization of core cellulolytic enzymes from Talaromyces cellulolyticus (formerly Acremonium cellulolyticus) critical for hydrolysis of lignocellulosic biomass. Biotechnol Biofuels 7:151. doi: 10.1186/s13068-014-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii T, Inoue H, Yano S, Sawayama S. 2018. Strain improvement for industrial production of lignocellulolytic enzyme by Talaromyces cellulolyticus, p 135–154. In Fang X, Qu Y (ed), Fungal cellulolytic enzymes. Springer Singapore, Singapore. [Google Scholar]
- 23.Kishishita S, Yoshimi M, Fujii T, Taylor LE II, Decker SR, Ishikawa K, Inoue H. 2014. Cellulose-inducible xylanase Xyl10A from Acremonium cellulolyticus: purification, cloning and homologous expression. Protein Expr Purif 94:40–45. doi: 10.1016/j.pep.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe M, Inoue H, Inoue B, Yoshimi M, Fujii T, Ishikawa K. 2014. Xylanase (GH11) from Acremonium cellulolyticus: homologous expression and characterization. AMB Express 4:27. doi: 10.1186/s13568-014-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii T, Koike H, Sawayama S, Yano S, Inoue H. 2015. Draft genome sequence of Talaromyces cellulolyticus strain Y-94, a source of lignocellulosic biomass-degrading enzymes. Genome Announc 3:e00014-15. doi: 10.1128/genomeA.00014-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanna M, Yano S, Inoue H, Fujii T, Sawayama S. 2011. Enhancement of β-xylosidase productivity in cellulase producing fungus Acremonium cellulolyticus. AMB Express 1:15. doi: 10.1186/2191-0855-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue H, Fujii T, Yoshimi M, Taylor LE II, Decker SR, Kishishita S, Nakabayashi M, Ishikawa K. 2013. Construction of a starch-inducible homologous expression system to produce cellulolytic enzymes from Acremonium cellulolyticus. J Ind Microbiol Biotechnol 40:823–830. doi: 10.1007/s10295-013-1286-2. [DOI] [PubMed] [Google Scholar]
- 28.Parkkinen T, Hakulinen N, Tenkanen M, Siika-Aho M, Rouvinen J. 2004. Crystallization and preliminary X-ray analysis of a novel Trichoderma reesei xylanase IV belonging to glycoside hydrolase family 5. Acta Crystallogr D Biol Crystallogr 60:542–544. doi: 10.1107/S0907444903029123. [DOI] [PubMed] [Google Scholar]
- 29.Inoue H, Kishishita S, Kumagai A, Kataoka M, Fujii T, Ishikawa K. 2015. Contribution of a family 1 carbohydrate-binding module in thermostable glycoside hydrolase 10 xylanase from Talaromyces cellulolyticus toward synergistic enzymatic hydrolysis of lignocellulose. Biotechnol Biofuels 8:77. doi: 10.1186/s13068-015-0259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St John FJ, Hurlbert JC, Rice JD, Preston JF, Pozharski E. 2011. Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase. J Mol Biol 407:92–109. doi: 10.1016/j.jmb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Urbániková L, Vršanská M, Mørkeberg Krogh KBR, Hoff T, Biely P. 2011. Structural basis for substrate recognition by Erwinia chrysanthemi GH30 glucuronoxylanase. FEBS J 278:2105–2116. doi: 10.1111/j.1742-4658.2011.08127.x. [DOI] [PubMed] [Google Scholar]
- 32.Juturu V, Wu JC. 2014. Microbial exo-xylanases: a mini review. Appl Biochem Biotechnol 174:81–92. doi: 10.1007/s12010-014-1042-8. [DOI] [PubMed] [Google Scholar]
- 33.Lagaert S, Van Campenhout S, Pollet A, Bourgois TM, Delcour JA, Courtin CM, Volckaert G. 2007. Recombinant expression and characterization of a reducing-end xylose-releasing exo-oligoxylanase from Bifidobacterium adolescentis. Appl Environ Microbiol 73:5374–5377. doi: 10.1128/AEM.00722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollet A, Delcour JA, Courtin CM. 2010. Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit Rev Biotechnol 30:176–191. doi: 10.3109/07388551003645599. [DOI] [PubMed] [Google Scholar]
- 35.Linares-Pasten JA, Aronsson A, Karlsson EN. 2018. Structural considerations on the use of endo-xylanases for the production of prebiotic xylooligosaccharides from biomass. Curr Protein Pept Sci 19:48–67. doi: 10.2174/1389203717666160923155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurlbert JC, Preston JF. 2001. Functional characterization of a novel xylanase from a corn strain of Erwinia chrysanthemi. J Bacteriol 183:2093–2100. doi: 10.1128/JB.183.6.2093-2100.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fushinobu S, Hidaka M, Honda Y, Wakagi T, Shoun H, Kitaoka M. 2005. Structural basis for the specificity of the reducing end xylose-releasing exo-oligoxylanase from Bacillus halodurans C-125. J Biol Chem 280:17180–17186. doi: 10.1074/jbc.M413693200. [DOI] [PubMed] [Google Scholar]
- 38.Peterson R, Nevalainen H. 2012. Trichoderma reesei RUT-C30—thirty years of strain improvement. Microbiology 158:58–68. doi: 10.1099/mic.0.054031-0. [DOI] [PubMed] [Google Scholar]
- 39.Fang X, Yano S, Inoue H, Sawayama S. 2009. Strain improvement of Acremonium cellulolyticus for cellulase production by mutation. J Biosci Bioeng 107:256–261. doi: 10.1016/j.jbiosc.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 40.Fujii T, Iwata K, Murakami K, Yano S, Sawayama S. 2012. Isolation of uracil auxotrophs of the fungus Acremonium cellulolyticus and the development of a transformation system with the pyrF gene. Biosci Biotechnol Biochem 76:245–249. doi: 10.1271/bbb.110498. [DOI] [PubMed] [Google Scholar]
- 41.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 42.MacIlvaine TC. 1921. A buffer solution for colorimetric comparison. J Biol Chem 49:183–186. [Google Scholar]
- 43.Kemmer G, Keller S. 2010. Nonlinear least-squares data fitting in Excel spreadsheets. Nat Protoc 5:267–281. doi: 10.1038/nprot.2009.182. [DOI] [PubMed] [Google Scholar]
- 44.Strohalm M, Kavan D, Novák P, Volný M, Havlíček V. 2010. MMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal Chem 82:4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- 45.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 15:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.