The concentration of Hg in the biosphere has increased dramatically over the last century as a result of industrial activities. The microbial conversion of inorganic Hg to MeHg is a global public health concern due to bioaccumulation and biomagnification of MeHg in food webs. Exposure to neurotoxic MeHg through the consumption of fish represents a significant risk to human health and can result in neuropathies and developmental disorders. Anaerobic microbial communities in sediments and periphyton biofilms have been identified as sources of MeHg in aquatic systems, but the associated biomolecular mechanisms are not fully understood. In the present study, we investigate the biochemical mechanisms and kinetics of MeHg formation by HgcAB in sulfate-reducing bacteria. These findings advance our understanding of microbial MeHg production and may help inform strategies to limit the formation of MeHg in the environment.
KEYWORDS: HgcAB, anaerobic bacteria, environmental microbiology, enzyme kinetics, mercury methylation, methylmercury
ABSTRACT
Methylmercury (MeHg) is a potent bioaccumulative neurotoxin that is produced by certain anaerobic bacteria and archaea. Mercury (Hg) methylation has been linked to the gene pair hgcAB, which encodes a membrane-associated corrinoid protein and a ferredoxin. Although microbial Hg methylation has been characterized in vivo, the cellular biochemistry and the specific roles of the gene products HgcA and HgcB in Hg methylation are not well understood. Here, we report the kinetics of Hg methylation in cell lysates of Desulfovibrio desulfuricans ND132 at nanomolar Hg concentrations. The enzymatic Hg methylation mediated by HgcAB is highly oxygen sensitive, irreversible, and follows Michaelis-Menten kinetics, with an apparent Km of 3.2 nM and Vmax of 19.7 fmol · min−1 · mg−1 total protein for the substrate Hg(II). Although the abundance of HgcAB in the cell lysates is extremely low, Hg(II) was quantitatively converted to MeHg at subnanomolar substrate concentrations. Interestingly, increasing thiol/Hg(II) ratios did not impact Hg methylation rates, which suggests that HgcAB-mediated Hg methylation effectively competes with cellular thiols for Hg(II), consistent with the low apparent Km. Supplementation of 5-methyltetrahydrofolate or pyruvate did not enhance MeHg production, while both ATP and a nonhydrolyzable ATP analog decreased Hg methylation rates in cell lysates under the experimental conditions. These studies provide insights into the biomolecular processes associated with Hg methylation in anaerobic bacteria.
IMPORTANCE The concentration of Hg in the biosphere has increased dramatically over the last century as a result of industrial activities. The microbial conversion of inorganic Hg to MeHg is a global public health concern due to bioaccumulation and biomagnification of MeHg in food webs. Exposure to neurotoxic MeHg through the consumption of fish represents a significant risk to human health and can result in neuropathies and developmental disorders. Anaerobic microbial communities in sediments and periphyton biofilms have been identified as sources of MeHg in aquatic systems, but the associated biomolecular mechanisms are not fully understood. In the present study, we investigate the biochemical mechanisms and kinetics of MeHg formation by HgcAB in sulfate-reducing bacteria. These findings advance our understanding of microbial MeHg production and may help inform strategies to limit the formation of MeHg in the environment.
INTRODUCTION
Mercury (Hg) occurs naturally in the environment and is released in part from natural sources, such as geothermal activity and volcanism. However, large quantities of Hg are released as a result of anthropogenic activities, such as mining operations, coal combustion, and other industrial processes (1). In a complex global cycle, Hg is transformed biotically and abiotically among several major forms, including elemental Hg(0), mercuric Hg(II) and methylmercury (MeHg). MeHg is the most prevalent organomercurial and is a potent neurotoxin (2); thus, human exposure to MeHg is a public health concern. MeHg bioaccumulates in the food web, and humans are exposed to this neurotoxin through their diet, particularly by consuming Hg-contaminated fish. Certain anaerobic bacteria and archaea are capable of converting Hg to MeHg. Here, we refer to these microorganisms as Hg methylators. The physiological role of microbial MeHg production is unclear, as Hg methylation apparently does not impart resistance to Hg toxicity (3). Interestingly, several Hg methylators have been shown to simultaneously methylate Hg and demethylate MeHg (4). Hg methylation has been studied in environmental samples and in pure cultures of Hg methylators (5–8). However, Hg methylation rates vary significantly with different strains, as well as in the presence of organic matter and/or thiolates (9, 10).
Early investigations of Hg methylation in cell extracts of a methanogen suggested that a methyl group could be transferred from methylcobalamin to Hg by both enzymatic and nonenzymatic processes (11–13). Nonenzymatic Hg methylation by methylcobalamin (Equation 1) is most favorable at pH 4.5 (12, 13).
| (1) |
Following the identification and isolation of the sulfate-reducing bacterium Desulfovibrio desulfuricans LS as an environmentally relevant Hg methylator (14), a series of studies investigated associations between metabolic pathways, corrinoids, and Hg methylation (15–18). Based on incorporation of 14C from radiolabeled precursors into MeHg, the reductive acetyl-coenzyme A (CoA) pathway was implicated in Hg methylation (15, 18). High levels of 14C incorporation into MeHg were observed in cultures supplemented with l-[3-14C]serine and [14C]formate (18). In addition, studies with [5-14CH3]tetrahydrofolate indicated that methyltetrahydrofolate (CH3-H4folate) may serve as the methyl donor to form MeHg (17). However, enzyme activities associated with the reductive acetyl-CoA pathway were several orders of magnitude lower in D. desulfuricans LS than in acetogens (18). 57Co labeling experiments, corrinoid extractions, and mass spectrometry identified cobalamin as the major corrinoid in D. desulfuricans LS (16). Additional studies implicated a 40-kDa corrinoid protein in enzymatic Hg methylation, but the specific protein involved was not identified or further characterized (17). As a result of these findings, it was proposed that Hg methylation is a two-step process involving (i) transfer of a methyl group from CH3-H4folate to a corrinoid protein, followed by (ii) transfer of the methyl group from the methylcorrinoid to Hg(II) to form MeHg (17). Later studies suggested that Hg methylation may be independent of the acetyl-CoA pathway, as incomplete-oxidizing sulfate reducing bacteria that do not use the acetyl-CoA pathway for metabolism are still able to methylate Hg (19).
The genetic basis of Hg methylation was not well understood until recently (20). The hgcAB gene pair was shown to be required for Hg methylation in the sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 and in the iron-reducing bacterium Geobacter sulfurreducens PCA (20). Deletion of either gene resulted in a complete loss of Hg methylation activity. To date, all strains of bacteria and archaea with hgcAB genes that have been assayed for Hg methylation are capable of producing MeHg (3, 6).
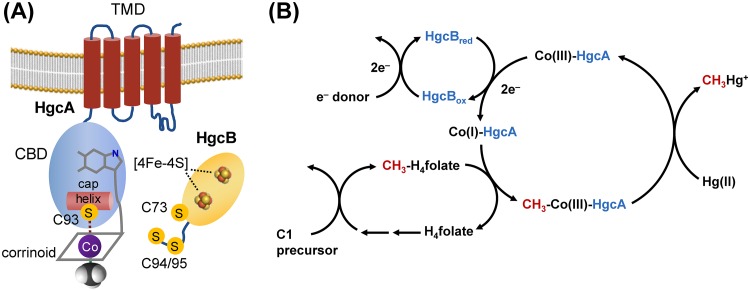
Based on sequence analysis and homology modeling it was predicted that hgcA encodes a protein consisting of a corrinoid-binding domain (CBD) facing the cytosol and a transmembrane domain (TMD) anchored in the cytoplasmic membrane (Fig. 1A) (20). The CBD of HgcA is homologous to the CBD of the corrinoid iron-sulfur protein (CFeSP) from the reductive acetyl-CoA (Wood-Ljungdahl) pathway of carbon fixation (20), but the TMD has no detectable sequence similarity to that of any known protein (20). The CBD of HgcA contains a strictly conserved Cys residue (C93) that is critical for Hg methylation activity in vivo (21). The gene hgcB almost always appears immediately downstream of hgcA and encodes a ferredoxin with two [4Fe-4S] cluster-binding motifs (20). HgcB has a unique architecture among ferredoxins, consisting of two [4Fe-4S] cluster-binding motifs (CX2CX2CX3CP), an additional strictly conserved Cys residue (C73), and a pair of conserved cysteines at the C terminus (C94 and C95) (Fig. 1A) (20, 21). It is not known whether HgcA and HgcB form a multiprotein complex. However, we refer here to these proteins collectively as HgcAB to emphasize that both are required for Hg methylation.
FIG 1.
Structural features and proposed mechanism of HgcAB-mediated Hg methylation. (A) Cartoon representation of sequence-based structural models of HgcA and HgcB. Key components and features are labeled as follows: TMD, transmembrane domain; CBD, corrinoid-binding domain; [4Fe-4S], iron-sulfur cluster; C, cysteine). (B) Proposed roles of HgcA and HgcB in Hg methylation (C1 precursor, one-carbon compound; Co(I)/(III), cobalt center of a corrinoid cofactor and its oxidation state; H4folate, tetrahydrofolate; CH3-H4folate, 5-methyltetrahydrofolate; Hg(II), mercuric mercury; CH3Hg+, methylmercury.
A general mechanism for Hg methylation by HgcAB is shown (Fig. 1B). The reaction cycle begins with HgcB providing low-potential electrons to the oxidized corrinoid cofactor of HgcA to generate the supernucleophilic Co(I) state. The Co(I)-corrinoid then accepts a methyl group from a methyl donor, such as CH3-H4folate, to form a CH3-Co(III)-corrinoid. The methylcorrinoid then transfers its methyl group to a Hg(II) substrate, either directly or through a ligand exchange reaction (22, 23), to form CH3Hg+. After methyl transfer, the corrinoid is in an oxidized state. Thus, every turnover cycle requires low-potential electrons, donated by HgcB, to reduce the corrinoid to the Co(I) state. How the Hg(II) substrate enters cells and interacts with HgcAB is currently unknown, but uptake and transport of Hg(II) into the cytoplasm are essential and may limit methylation rates observed in vivo (24, 25).
To evaluate the kinetics of Hg methylation mediated by HgcA and HgcB independent of transport processes, we performed Hg methylation assays in cell lysates of D. desulfuricans ND132. ND132 is a sulfate-reducing bacterium that is often used as a model organism for studying Hg methylation due to robust Hg methylation by ND132 and its similarity to D. desulfuricans LS (3, 26). The present study using ND132 builds on the foundation of earlier studies conducted with the strain D. desulfuricans LS, which was not sequenced and is considered lost (15–18). Owing to advances in MeHg analysis, we perform experiments at much lower and more environmentally relevant levels of Hg (0.5 to 60 nM compared to the 0.5 to 8 mM) previously used for studies with D. desulfuricans LS (17). We follow Hg methylation rates in ND132 cell lysates over time and study the effects of pH, temperature, and total protein concentration. We determine initial rates of Hg methylation in cell lysates as a function of Hg(II) concentration and calculate apparent kinetic parameters (Km and Vmax) for the ND132 wild type (WT) and compare results to those from an hgcAB deletion mutant (ΔhgcAB). Furthermore, we evaluate the contributions of enzymatic Hg methylation and nonenzymatic Hg methylation, as well as whether the Hg methylation reaction is reversible or a separate MeHg demethylation pathway exists in ND132. We also conduct a set of targeted experiments to gain insights into aspects of the proposed mechanism of Hg methylation by HgcA and HgcB. Specifically, we investigate the sensitivity of Hg methylation to oxygen exposure, evaluate a potential role of ATP in Hg methylation and determine the effect of supplementation of the cellular metabolites CH3-H4folate and pyruvate, which are proposed methyl and electron donors, respectively.
RESULTS
Time dependence of Hg methylation and MeHg demethylation.
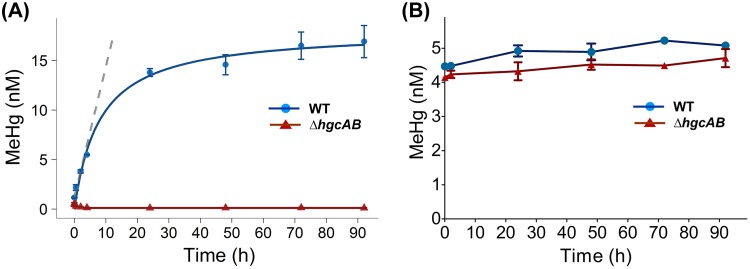
The time course for the conversion of Hg(II) to MeHg in cell lysates of ND132 WT and ΔhgcAB was determined over a period of up to 92 h. The levels of MeHg produced in WT cell lysates increased at a rate of 1.31 ± 0.14 nM/h within the first 2 h, decreasing over time and plateauing between 24 and 92 h (Fig. 2A). At the 92-h time point, 56.4% of the added Hg(II) was converted to MeHg. No significant MeHg production was observed in ΔhgcAB cell lysates over the course of the experiment. MeHg levels in ΔhgcAB cell lysates were <0.5% of MeHg in WT cell lysates at 92 h, confirming that Hg methylation in ND132 cell lysates is strictly an HgcAB-dependent process. In addition, the lack of significant MeHg production in ΔhgcAB cell lysates indicates that nonenzymatic methylation mediated by methylcorrinoids, either free (Equation 1) or associated with other proteins, did not contribute significantly to MeHg formation in the cell lysates. Initial rates of Hg methylation measured over the first 2 h after Hg(II) addition were used for subsequent experiments.
FIG 2.
Time dependence of Hg methylation and MeHg demethylation in ND132. (A) Hg methylation in cell lysates of ND132 WT (blue) and ΔhgcAB (red) (1.5 mg/ml total protein concentration) under strictly anaerobic conditions at 32°C in the presence of 30 nM Hg(II). The blue line shows a nonlinear fit of the concentration data and the gray dashed line shows a linear fit (R2 = 0.98) for time points between 0 and 2 h to determine initial rates. (B) MeHg concentrations in ND132 cell lysates of WT (blue) and ΔhgcAB (red) as a function of time under a similar experimental setup as that in panel A but with 5 nM MeHg as the substrate. Error bars represent standard deviation between duplicate sets of samples (n = 2).
To determine whether a fraction of Hg was reduced to Hg(0) over the course of the experiment and to close the Hg mass balance, we also measured the total Hg (THg) for every time point. No significant changes in THg levels were observed, eliminating the possibility that Hg was lost through reduction to Hg(0) over the course of the experiment (Fig. S1).
To determine whether MeHg is demethylated in ND132 cell lysates, we incubated WT cell lysates with 5 nM MeHg and measured MeHg levels over time up to 92 h. Considering that demethylation may be independent of HgcAB, we compared the demethylation of MeHg in cell lysates of both ND132 WT and the ΔhgcAB mutant. No demethylation was observed for either strain (Fig. 2B).
pH, temperature, and total protein concentration dependence of Hg methylation.
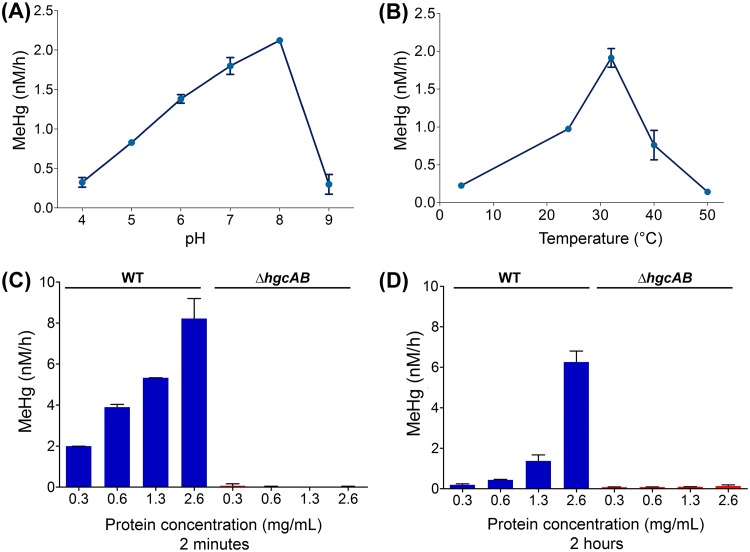
To provide further evidence that Hg methylation in ND132 cell lysates is an enzymatic process, we determined Hg methylation rates as a function of pH, temperature, and total protein concentration. The rate of MeHg production in WT cell lysates increased by a factor of 6.5 between pH 4.0 (0.32 ± 0.06 nM/h) and pH 8.0 (2.12 ± 0.03 nM/h) (Fig. 3A). Hg methylation rates in WT cell lysates varied by an order of magnitude between 4°C (0.22 ± 0.00 nM/h) and 32°C (1.91 ± 0.12 nM/h), and between 32°C and 50°C (0.14 ± 0.01 nM/h), with a fairly narrow optimum centered around 32°C (Fig. 3B).
FIG 3.
Dependence of Hg methylation rates on pH, temperature, and total protein concentration. Hg methylation rates in cell lysates of ND132 WT measured as a function of (A) pH (4, 5, 6, 7, 8, and 9), (B) temperature (4, 24 [room temperature], 32, 40, and 50°C), and (C and D) total protein concentration. Samples were incubated with 30 nM Hg(II) and harvested for MeHg analysis at 2 h, except in panel C, where samples were harvested at 2 min after Hg(II) addition. pH 7, 32°C, and strictly anaerobic conditions correspond to standard experimental conditions. Error bars represent standard deviation between duplicate sets of samples (n = 2).
To investigate the effect of total protein concentration on the rate of Hg methylation in cell lysates, we measured MeHg production rates at 2 min (Fig. 3C) and 2 h (Fig. 3D) over a range of up to ∼8 times the initial total protein concentration (0.3 mg/ml to 2.6 mg/ml). The highest total protein concentration achievable is limited by the volume of buffer required to resuspend cell pellets prior to cell lysis. As expected, the rate of MeHg formation increased with increasing concentrations of total protein. Although the initial rates determined within 2 min of the addition of Hg(II) were higher and increased linearly in response to total protein concentration, methylation rates determined at 2 h increased exponentially with increasing total protein concentrations.
Enzyme kinetics of HgcAB-mediated Hg methylation.
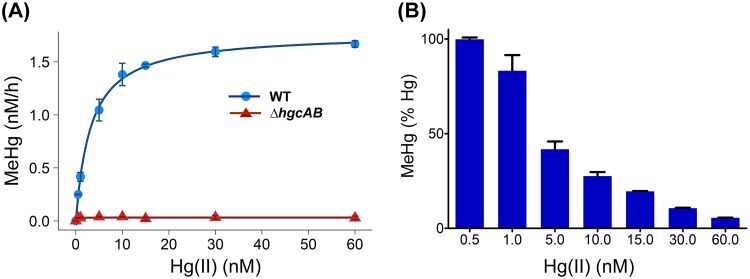
To examine the effects of increasing substrate concentrations on the rate of MeHg production, we incubated ND132 cell lysates (1.5 mg/ml total protein concentration) with Hg(II) (added as HgCl2) as a substrate at concentrations ranging from 0.5 nM to 60 nM. ΔhgcAB cell lysates were used as a negative control. Initial MeHg formation rates followed Michaelis-Menten kinetics with a maximum rate Vmax of 19.7 (± 0.35) fmol · mg total protein−1 · min−1 and a Km of 3.2 nM (± 0.26) for Hg(II) (Fig. 4A). CH3-H4folate is the presumed methyl donor for MeHg. As described in the following section, CH3-H4folate levels were not rate limiting under the current experimental conditions. Furthermore, the addition of dithiothreitol (DTT) at concentrations above 2 mM did not affect the observed Hg methylation rates (Fig. S2). At low concentrations of Hg(II), 0.5 nM and 1 nM, nearly 100% of the added Hg(II) was converted to MeHg (Fig. 4B).
FIG 4.
Dependence of Hg methylation rates on Hg substrate concentration. (A) Initial rates of Hg methylation in ND132 cell lysates (1.5 mg/ml total protein) in response to increasing concentrations of added Hg(II) from 0.5 nM to 60 nM at 32°C for 2 h under strictly anaerobic conditions for WT (blue circles) and ΔhgcAB strains (red triangles). Hg methylation rates in WT cell lysates were fitted to the Michaelis-Menten equation with a Vmax of 1.77 ± 0.03 nM/h and Km of 3.2 ± 0.26 nM (blue line). (B) Percentage of Hg(II) converted to MeHg within 2 h as a function of initial Hg(II) concentration. Error bars represent standard deviation between duplicate set of samples (n = 2).
Reverse transcription-PCR (RT-PCR) data from ND132 (20) and proteomics studies in ND132 (27) and G. sulfurreducens PCA (28) indicated that the abundance of HgcA and HgcB in Hg methylators is extremely low. Owing to the low abundance and resultant difficulties in determining accurate concentrations of HgcA and HgcB in the cell lysates, we used the obtained kinetic parameters to estimate turnover numbers (kcat), catalytic efficiencies (kcat/Km), and free energies of activation based on transition state theory (29, 30) for a range of enzyme concentrations expressed as a fraction of total cellular protein concentration (Table 1, Fig. S3). It should be noted that these values are calculated solely for the purpose of gaining further insights into the functioning of HgcAB based on estimated kinetic parameters. Additionally, since the value for Vmax reported here is normalized to the total protein in the lysates, the Vmax of the pure enzyme is expected to be higher. Nevertheless, the value of Km, which is independent of the enzyme concentration, suggests that HgcA and HgcB readily bind Hg(II) at very low concentrations.
TABLE 1.
Turnover numbers, catalytic efficiencies, and activation free energies of enzymatic Hg methylation for a range of postulated enzyme concentrations
| [E]a (% of total protein) | kcat (s−1) | kcat/Km (M−1 · s−1) | ΔG‡b (kcal · mol−1) |
|---|---|---|---|
| 0.1 | 1.2 × 10−5 | 4 × 103 | 24.1 |
| 0.01 | 1.2 × 10−4 | 4 × 104 | 22.8 |
| 0.001 | 1.2 × 10−3 | 4 × 105 | 21.4 |
| 0.0001 | 1.2 × 10−2 | 4 × 106 | 20.1 |
a[E], concentration of HgcA, expressed as a fraction of total cell protein.
bΔG‡, activation free energy.
Effect of oxygen and cellular metabolites on Hg methylation.
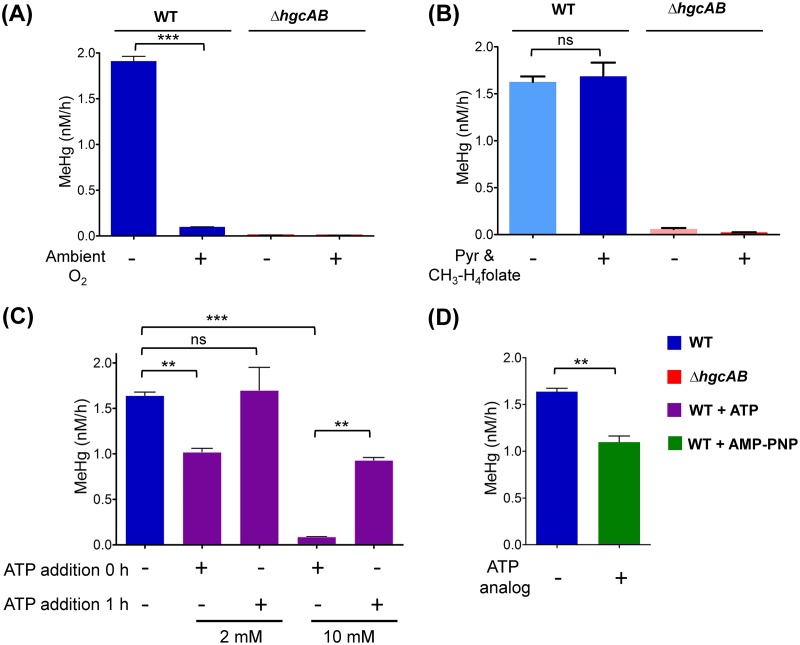
Based on the current mechanistic hypothesis, each MeHg formation turnover cycle requires the transfer of low-potential electrons to reduce the corrinoid of HgcA to the Co(I) state to accept a methyl group from a methyl donor (Fig. 1B). The presence of oxygen would raise the redox potential in solution and rapidly oxidize any Co(I) due to the low midpoint potential of the Co(I)/Co(II) couple (31, 32). Therefore, corrinoid-dependent methyl transfer reactions are highly redox sensitive. Exposure to oxygen would interrupt the transfer of methyl groups to the corrinoid and inhibit MeHg formation. To test the effect of oxygen on Hg methylation, MeHg production in ND132 cell lysates was measured under both aerobic (exposed to ambient oxygen levels) and anaerobic conditions (standard experimental conditions in a glove box under N2 and <0.6 ppm O2). MeHg formation was inhibited by 95% (P < 0.001) in the presence of ambient oxygen (Fig. 5A). As expected, no effect of ambient oxygen on MeHg formation was observed in the ΔhgcAB cell lysates. Thus, the observed oxygen sensitivity of HgcAB-mediated Hg methylation in cell lysates of WT ND132 is consistent with the proposed requirement for low-potential electrons (i.e., <−450 mV) for the corrinoid in HgcA to achieve the Co(I) state (31, 32). Additionally, the presence of oxygen may also affect the oxidation state of the two [4Fe-4S] clusters in HgcB.
FIG 5.
Effect of ambient oxygen and key cellular metabolites on Hg methylation. (A) Hg methylation rates in WT ND132 cell lysates and ΔhgcAB under aerobic (exposed to ambient oxygen) and anaerobic conditions (N2 environment, <0.6 ppm O2). (B) Hg methylation rates in WT ND132 cell lysates and ΔhgcAB under unamended conditions compared to the Hg methylation rates for samples amended with 16.8 μM CH3-H4folate and 10 mM pyruvate (Pyr). (C) Effect of 2 mM ATP and 10 mM ATP on the concentration of MeHg produced at 2 h in the presence of 30 nM Hg(II) compared to that in the unamended sample. ATP was supplemented before the addition of Hg(II) (ATP addition at 0 h) or 1 h after the addition of Hg(II) (ATP addition at 1 h). (D) Effect of 5 mM nonhydrolyzable ATP analog AMP-PNP on MeHg production compared to the unamended sample. Data were analyzed with a two-sample t test. Not significant (ns), P > 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Error bars represent the standard deviation between duplicates (n = 2).
CH3-H4folate was previously proposed as a potential methyl donor required for the formation of MeHg. Furthermore, no Hg methylation was detected in cell lysates of D. desulfuricans LS without the addition of 10 mM pyruvate, in which pyruvate was proposed to play a role in the generation of reductant such as reduced ferredoxin (15, 17, 18). To determine whether the exogenous addition of 16.8 μM CH3-H4folate and 10 mM pyruvate, as used in previous studies, leads to increased Hg methylation, we compared the Hg methylation rates under our standard experimental conditions (i.e., unamended) to that of samples amended with pyruvate and CH3-H4folate. No significant changes in the rate of MeHg formation were observed in response to the combined addition of 16.8 μM CH3-H4folate and 10 mM pyruvate (P > 0.05) (Fig. 5B). Because the Hg(II) concentration used for the current experiments (30 nM) is much lower than the levels used for assays with D. desulfuricans LS (0.7 mM), we propose that the availability of pyruvate and CH3-H4folate were not limiting factors during MeHg formation under our experimental conditions. In addition, involvement of a physiological methyl donor other than CH3-H4folate cannot be ruled out.
Previous studies have proposed a role for ATP driving the uptake or methylation of Hg (24). To determine whether Hg methylation in cell lysates is ATP-dependent, we measured the effect of exogenously added ATP on MeHg formation independent of uptake processes. ATP was added to cell lysates at a concentration of 2 mM, which is close to the average physiological ATP concentration determined for Escherichia coli cells (33), and in excess, 10 mM. ATP was added at 0 h, i.e., before adding Hg. In a separate set of samples, ATP was supplemented 1 h after Hg was added (ATP at 1 h). Samples were harvested 2 h after the addition of Hg(II) in both cases. In selecting the timing of ATP supplementation, we assumed that a substantial amount of endogenous ATP present in the lysate would decrease after 1 h, and that the addition of exogenous ATP at that time point would increase MeHg production if Hg methylation is ATP dependent. The 1 h time point is within the initial, linear rate period of the Hg methylation progress curve (Fig. 2). Interestingly, MeHg production decreased significantly upon addition of exogenous ATP at concentrations of 2 mM and 10 mM (P < 0.05), by 38% and 95%, respectively, compared to that of unamended samples (Fig. 5C). Addition of 2 mM ATP at 1 h did not significantly increase MeHg production. However, addition of 10 mM ATP after 1 h significantly decreased the total amount of MeHg formed after 2 h. The amount of MeHg produced after supplementation of 10 mM ATP was about 56% of that formed under unamended conditions.
To rule out the possibility that the decrease in MeHg production in the presence of ATP may have arisen from the loss of Hg(II) through ATP-dependent reduction to Hg(0), we determined the concentration of THg in ND132 cell lysates after 2 h. No significant loss in THg was observed in samples with or without added ATP (Fig. S4), indicating that the addition of ATP did not result in a loss of Hg(II) through reduction to Hg(0).
Genomic analysis of Hg methylators indicated the presence of a gene encoding a reductive activator of corrinoid proteins (RACo) in several Hg methylators (Table S1, Fig. S5). RACo harbors an ATP-binding site and has been shown to form a complex with CFeSP (34). Methylation of CFeSP occurs only in the Co(I) state, which can be oxidized sporadically to the inactive Co(II) state every 100–2,000 turnovers (35). RACo serves as the activator of inactive Co(II)-CFeSP to active Co(I)-CFeSP by coupling the hydrolysis of ATP to the reduction of Co(II) to Co(I) (36). In Moorella thermoacetica and Carboxydothermus hydrogenoformans, the gene encoding RACo is located in close proximity to the genes encoding enzymes of the reductive acetyl-CoA (Wood-Ljungdahl) pathway (35). Although D. desulfuricans ND132 lacks CFeSP and does not have a complete Wood-Ljungdahl pathway (20), we hypothesize that a RACo homolog, commonly present among many methylators, may serve a similar ATP-dependent corrinoid activation role for HgcA.
We also evaluated the effect of the addition of the nonhydrolyzable ATP analog adenylyl-imidodiphosphate (AMP-PNP) on MeHg formation in ND132 cell lysates. AMP-PNP was added at a concentration of 5 mM, an amount that is expected to compete with endogenous ATP for ATP hydrolysis sites of most ATPases (37). The addition of AMP-PNP resulted in a decrease of MeHg production by 33% (P < 0.05) compared to that in the unamended control (Fig. 5D). This effect on MeHg production is similar to the decrease observed after addition of ATP. However, the mechanism by which both ATP and AMP-PNP decrease MeHg production is unclear.
DISCUSSION
Why bacteria and archaea methylate mercury remains a mystery, as methylation apparently does not impart these organisms with mercury resistance (3). Microbial Hg methylation has been the subject of investigation since the late 1960s (11, 38). The discovery of hgcAB offered a first glimpse at the unique bioinorganic chemistry involved in the methylation of Hg. In this study, we investigated the cellular Hg methylation mediated by HgcAB. We measured the contributions of enzymatic and nonenzymatic processes to Hg methylation in cell lysates of the anaerobic model Hg methylator D. desulfuricans ND132 and investigated the effect of various biochemical parameters and metabolites on Hg methylation. The mechanistic hypothesis of Hg methylation (Fig. 1B) was developed based on the sequences of HgcA and HgcB, as well as early investigations of Hg methylation in D. desulfuricans LS (15–18).
As expected, methylation activity was detected in lysates of the WT, but not in lysates of the ΔhgcAB strain, confirming that HgcA and HgcB are essential for converting inorganic Hg(II) to MeHg (Fig. 2A). It should be noted that ND132 and other strains of sulfate-reducing bacteria have been reported to simultaneously methylate Hg(II) and demethylate MeHg (3, 4). In our experiments with ND132 cell lysates, however, we did not observe demethylation of added MeHg over a period of several days (Fig. 2B). Therefore, we conclude that Hg methylation by HgcAB is irreversible and that there is no other mechanism independent of HgcAB for MeHg demethylation in experiments with ND132 cell lysates that may impact the Hg methylation rate parameters.
The rate of MeHg production in WT DN132 cell lysates was 6.5 times higher at pH 8.0 than at pH 4.0 (Fig. 3A). This result is in contrast to the pH-rate profile for the nonenzymatic methylation of Hg(II) by methylcobalamin (Equation 1) in which the highest Hg methylation rate was observed at pH 4.5, and decreased significantly at pH 6.0 and higher (13). In addition to pH, the Hg methylation rates in WT cell lysates were also dependent on temperature. The highest rate of MeHg production in WT DN132 cell lysates was centered around 32°C with a fairly narrow optimum, decreasing by an order of magnitude at temperatures of 22°C and below or 40°C and above (Fig. 3B). The temperature dependence is consistent with the optimal growth temperature of 32°C for ND132 cultures and a lack of any substantial growth at 45°C and above (3). The observed pH and temperature dependencies suggest that MeHg is predominantly formed by an enzymatic process dependent on HgcAB under the experimental conditions.
At the lowest lysate concentration (0.3 mg/ml protein), the initial rate of MeHg formation determined over a period of 2 min after Hg(II) addition was higher than the rate determined over a 2-h period. However, the rate increased linearly with the concentration of the lysate in the 2-min experiments, while it increased exponentially with lysate concentration in the 2-h experiments (Fig. 3C and D). The linear increase in the 2-min experiments indicates that the rates of MeHg formation were limited only by the overall enzyme concentration. However, the exponential increase of Hg methylation rate at higher lysate concentrations in the 2-h experiments suggests that the rate is also dependent on the concentration of a secondary substrate present in the lysates, which may have been depleted over time. While this secondary substrate is currently unknown, our findings are consistent with previous studies with D. desulfuricans LS in which the rate of MeHg formation also increased exponentially with increasing cell lysate concentrations, suggesting that Hg methylation involves two or more components, such as a methyl donor, electron donor, and/or Hg(II) substrate (17). Nevertheless, Hg(II) substrate concentration dependence under the conditions tested in our experiments followed Michaelis-Menten kinetics (Fig. 4A). Although the estimated turnover numbers (kcat, ∼1 × 10−5 to 1 × 10−2 s−1) are very low compared to those of an average enzyme (Table 1), the low Km of 3.2 nM yields catalytic efficiencies (kcat/Km, ∼4 × 103 to 4 × 106 M−1 · s−1) that fall within reasonable ranges (39).
The chemical speciation of Hg(II) is a critical factor that may impact apparent methylation rates due to the high affinity of Hg(II) for biological thiolates (SR−) with stability constants (log β) of up to 45 for Hg(II)-bis-thiolate [Hg(SR)2] complexes (40). Intracellular thiolates comprise thiol functional groups on proteins (i.e., cysteine) and small molecular thiols. The concentration of thiols in cells can range from 0.1 to 10 mM (40), which is approximately 3 orders of magnitude higher than the Hg(II) concentrations used for methylation assays (0.5 nM to 60 nM). Therefore, Hg(II)-thiolate complexes are expected to be the dominant species in cells, as thiolates tend to outcompete any other ligands under these conditions. We evaluated the effect of thiolate levels by comparing methylation rates in the presence of 2 to 10 mM DTT. Our data show that high thiolate/Hg(II) ratios did not impact Hg methylation rates significantly (Fig. S2). Consequently, the concentration of DTT was maintained at 2 mM in all cell lysate experiments to keep Hg(II) speciation identical between experiments. It should be noted that although the thermodynamic stability of Hg(SR)2 species is extremely high, ligand exchange of such complexes with other free thiolates is rapid (41). Therefore, the low Km and quantitative conversion of Hg(II) to MeHg at high thiolate/Hg(II) ratios (Fig. 4B) are remarkable and suggest the existence of an active site that may harbor thiolate ligands to bind Hg(II) with high affinity, possibly involving a ligand exchange mechanism, similar to mechanisms previously described for MerA and MerB (42, 43). A series of conserved cysteines identified in sequences of HgcB (C94, C95, and C73) may facilitate such ligand exchange processes. Indeed, site-directed mutagenesis experiments in ND132 showed that a C73A mutation eliminates Hg methylation in vivo, but only one of the two C-terminal cysteines (C94 or C95) is essential for activity (21).
Based on the hypothesized roles for HgcA and HgcB (Fig. 1B), in addition to a Hg(II) substrate, Hg methylation requires both a source of electrons to regenerate the Co(I) state of the corrinoid cofactor and a methyl donor. Although the source of electrons is unknown, HgcB likely assumes a key role in shuttling electrons from an unidentified electron donor to the corrinoid cofactor of HgcA. If the intracellular molar ratio of HgcA to HgcB remains constant, the generation of electrons may become rate limiting as it depends on active metabolic processes, which are expected to decrease in the cell lysates over time. Furthermore, the immediate methyl donor to HgcA remains unknown, as supplementation of CH3-H4folate did not result in an increased rate of Hg methylation over a period of 2 h (Fig. 5B).
If MeHg formation is dependent on energy derived from ATP hydrolysis, such as the ATP-dependent corrinoid activation by a reductive activator (RACo), adding ATP or a nonhydrolyzable ATP would be expected to affect MeHg production. Our experiments showed that supplementation of ATP leads to a substantial decrease in Hg methylation activity in cell lysates (Fig. 5C). A similar decrease in Hg methylation was observed with the competitive nonhydrolyzable analog AMP-PNP (Fig. 5D). Thus, we could not confirm the hypothesis that reductive activation by RACo or a similar ATP-dependent activator is essential for Hg methylation. Alternatively, ATP may affect Hg speciation or impact other, yet to be identified, cellular components related to Hg methylation.
Several key observations in this study are consistent with results obtained from studies with D. desulfuricans LS. The majority (>95%) of cellular Co in D. desulfuricans LS was reported to be associated with macromolecules (16). In ND132 cell lysates, no nonenzymatic Hg methylation was observed and, in the ΔhgcAB mutant, Hg methylation was negligible, indicating that HgcAB is indeed responsible for all Hg methylation. A 40-kDa protein, being the only major Co-containing protein in D. desulfuricans LS (17), agrees with the molecular mass of ND132 HgcA (36.7 kDa without its corrinoid cofactor) or with a HgcAB complex (1:1 molar ratio; ∼49 kDa), although it is not known whether HgcA and HgcB form a stable complex.
However, there are several differences between the experimental conditions used in the present study and those performed previously with D. desulfuricans LS. While the typical concentration of Hg(II) in our experiments was 30 nM, which is similar to levels used for methylation experiments with whole cells (25), experiments with D. desulfuricans LS were conducted at Hg(II) levels in the mM range (0.5 to 8 mM) (17). Hg toxicity to ND132 cells was apparent through decreased cell density at Hg concentrations above ∼5 μM (3). Detection limits for the analysis of MeHg by inductively coupled plasma mass spectrometry (ICP-MS) following the current standard Environmental Protection Agency (EPA) methods are in the low pM range (44). The vastly improved detection limits for MeHg enable our studies at significantly lower Hg levels, which are more physiologically relevant and dramatically reduce potential impacts of Hg toxicity to cellular proteins. These advances also allowed us to evaluate Hg methylation kinetics in lysates without supplementing essential metabolites. For example, no Hg methylation was observed with D. desulfuricans LS cell extracts unless 10 mM pyruvate and 16.8 μM CH3-H4folate were added (18). Under our experimental conditions, we observed no effect on Hg methylation activity in ND132 cell lysates of adding an exogenous methyl donor (CH3-H4folate) or electron source (pyruvate). These differences in the experimental design are reflected in differences between the Vmax and Km values previously determined for D. desulfuricans LS (Vmax = 0.728 nmol min−1 · mg protein−1 and Km = 0.872 mM) and those described here (Vmax = 19.7 fmol · min−1 · mg total protein−1 and Km = 3.2 nM). Differences in the inhibition of methylation activity by ambient oxygen (45% for D. desulfuricans LS [17], 95% in this study) suggest that nonenzymatic methylation may have been more prominent in studies with D. desulfuricans LS and may have contributed to overall MeHg production. Here, we showed that essentially all of the MeHg formed in ND132 is a product of enzymatic activity mediated by HgcAB.
Inhibition of Hg methylation by oxygen is consistent with the proposed HgcAB-mediated turnover cycle (Fig. 1B). Exposure of cell lysates to ambient oxygen could inactivate either FeS-containing HgcB or corrinoid-containing HgcA or oxidize the active Co(I)-corrinoid to Co(II) or Co(III). As a consequence, in the presence of oxygen, HgcA would not be able to accept a methyl group, thus interrupting the turnover cycle. Nevertheless, further studies to identify the chemistry of the specific corrinoid cofactor and direct evidence for its participation in Hg methylation are needed.
We speculate that the fraction of HgcA is equivalent to approximately 0.0004% of the total protein concentration in cell lysates (see supplemental material), which would correspond to approximately 1 in 220,000 proteins in the cell lysate. Such low abundance would explain difficulties in identifying HgcA protein fragments and low transcript levels in previously published studies (20, 27, 28). The calculated kinetic parameters resulting from this HgcA abundance estimate are kcat = 3 × 10−3 s−1, and kcat/Km = 9 × 105 M−1 · s−1. Consequently, we speculate that the specific activity of HgcA is close to 4 nmol · min−1 · mg−1 HgcA. Comparing the current estimated rate constant for the HgcAB-mediated enzymatic Hg methylation to previously published nonenzymatic rate constants for methylcobalamin, the enzymatic reaction is approximately 2,500 times faster at pH 7.0 than the nonenzymatic reaction, which is fastest at pH 4.5 (12, 13). Although the cofactor-replete expression and purification of HgcA and HgcB presents significant challenges, studies with purified HgcA and HgcB will be essential to validate the postulated kinetic parameters.
In conclusion, we demonstrate that cell lysates containing HgcA and HgcB exhibit Hg methylation activity even at subnanomolar Hg(II) concentrations, which is remarkable considering Hg(II) is known to form thermodynamically stable complexes with the abundant pool of cellular thiols present in cells. Future studies aimed at characterizing the molecular structures and functions of HgcA and HgcB are essential to delineate the interplay between methyl donor, electron donor, and the Hg(II) substrate in cellular Hg methylation. Taking into consideration that deletion of hgcAB does not significantly alter the growth of Hg methylators, understanding the cellular biochemistry of Hg methylation could help inform strategies to limit MeHg formation in Hg-methylating bacteria and archaea and thus curtail the formation of this potent neurotoxin in the environment.
MATERIALS AND METHODS
Strains and culture conditions.
Desulfovibrio desulfuricans ND132 cells (wild-type [WT] or ΔhgcAB mutant) were grown in modified minimal organic yeast medium containing 40 mM fumarate, 40 mM pyruvate, and 1.2 mM thioglycolate and 1 mM cysteine HCl as reducing agents for 3 days (optical density at 600 nm [OD600] = 0.3) at 32°C under strictly anaerobic conditions (<0.6 ppm O2) in 2-liter medium bottles capped with butyl rubber stoppers (21). For the preparation of ND132 cell lysates, all of the following steps were carried out under strictly anaerobic conditions inside a LabMaster Pro glove box (MBraun, Stratham, NH) under an N2 atmosphere maintained below 0.6 ppm O2. The cultures were harvested at 4°C by centrifugation for 20 min at 7,500 × g in centrifuge bottles with sealing closures. Pellets were washed 2 times with deoxygenated phosphate-buffered saline (PBS; pH 7.0). Cell pellets were resuspended in cold resuspension solution (Pierce EDTA-free protease inhibitor [Thermo Fisher Scientific, Waltham, MA] Benzonase nuclease [MilliporeSigma, St. Louis, MO], and 2 mM dithiothreitol [DTT] in PBS [pH 7.0]) and disrupted on ice by four cycles of sonication (50 sec, 70% power) with a Sonic Dismembrator 250 (Thermo Fisher Scientific, Waltham, MA) under minimal light to prevent photolytic degradation of methylcorrinoids. Unlysed cells were removed by centrifugation at 30,000 × g for 1 h at 4°C. Removal of unlysed cells was confirmed by microscopic examination with an Axioscope 2 (Carl Zeiss Microscopy LLC, Thornwood, NY). Total protein concentration of cell lysates was determined by the Bradford method with bovine serum albumin (MilliporeSigma, St. Louis, MO) as the standard, and the lysates were stored in aliquots at −80°C in amber glass vials capped with Teflon-lined silicone stoppers and sealed. Cell lysates were used within 2 weeks of preparation.
Methylation assays.
Methylation assays were performed under strictly anaerobic conditions in the dark at 32°C inside an anaerobic glove box at a total protein concentration of 1.5 mg/ml for 2 h (standard conditions), unless otherwise specified. To avoid any potential variability that may result from differences in the thiolate concentrations in the cell lysates, the concentration of DTT was kept constant at 2 mM for all experiments. Cell lysates of ND132 WT and ΔhgcAB were equilibrated under the desired experimental conditions for 2 min before adding aliquots of a freshly prepared 1.5 μM (300 ppb) HgCl2 stock solution to a final concentration of 30 nM (6 ppb), unless otherwise specified. The reactions were stopped at the desired time points by adding 0.5% (vol/vol) trace-metal grade H2SO4 (Thermo Fisher Scientific, Waltham, MA), and the samples were moved immediately to a −20°C freezer and stored until MeHg analysis. All experiments were performed in duplicate. The reaction buffer, PBS (pH 7.0; without cell lysates) and ΔhgcAB cell lysates were used as controls. Samples were processed similarly for total Hg (THg) analysis. Experiment-specific variations used, while keeping all other parameters constant, were as follows. For the time dependence experiments, aliquots were removed from the reaction mixture before the addition of Hg(II) (0 h), and following the addition of Hg(II) at 2 min, 30 min, 2, 4, 24, 48, 72, and 92 h. For the pH dependence experiments, the cell lysates were diluted in PBS buffer and solutions were adjusted to pH values of 4.0, 5.0, 6.0, 7.0, 8.0, and 9.0, respectively. For the temperature dependence experiments, the samples were preequilibrated before the addition of Hg(II) for 5 min at the desired temperatures, 4°C, room temperature (24°C), 32, 40, and 50°Crespectively, and, following the addition of Hg(II), the samples were incubated for 2 h. For the protein concentration dependence experiments, the cell lysates were diluted to final protein concentrations of 0.3, 0.6, 1.3, and 2.6 mg/ml, respectively. For the substrate concentration dependence experiments, cell lysates were spiked with Hg(II) to final concentrations of 0.5, 1, 5, 10, 15, 50 and 60 nM with stock solutions of 50 nM, 0.5 and 1.5 μM. The effects of pyruvate and CH3-H4folate were tested by adding a freshly prepared sodium pyruvate stock solution (500 mM) to a final pyruvate concentration of 10 mM and a freshly prepared 10 mg/ml stock solution of CH3-H4folate (MilliporeSigma, St. Louis, MO) to a final concentration of 16.8 μM. The samples were equilibrated with pyruvate and CH3-H4folate for 2 min before the addition of Hg(II). To test the oxygen sensitivity, sets of WT and ΔhgcAB cell lysates were incubated at standard conditions inside the glove box (<0.6 ppm O2), and other sets were preequilibrated outside the glove box (at ambient O2) for 2 min before the addition of Hg(II). The incubation was continued for 2 h under the same conditions after the addition of Hg(II). All other experimental parameters and procedures were as described above.
The effect of ATP was tested by adding a freshly prepared stock solution of ATP disodium salt (MilliporeSigma, St. Louis, MO) to a set of vials before the addition of Hg(II) (ATP at 0 h) and 1 h after the addition of Hg(II) (ATP at 1 h) to achieve final ATP concentrations of 2 mM and 10 mM. The reaction was stopped at 2 h, as described previously. To test the effect of a nonhydrolyzable ATP analog, a freshly prepared stock solution of adenosine 5′-(β,γ-imido)triphosphate lithium salt hydrate (MilliporeSigma, St. Louis, MO) was added to cell lysates to a final concentration of 5 mM and incubated for 5 min before initiating the reaction by addition of Hg(II).
Demethylation assays.
Demethylation potentials were measured in the cell lysates of WT and ΔhgcAB ND132; the samples were processed under standard conditions described above, except that 5 nM MeHg was used as the substrate instead of Hg(II). A 250 nM MeHg working stock solution was freshly prepared from a 1 ppm methylmercury standard solution (Brooks Rand Instruments, Seattle, WA).
Total mercury analysis.
All samples were treated with 5% (vol/vol) BrCl overnight, followed by 5-min incubation with 30% (wt/vol) NH2OH HCl before the total mercury (THg) analysis. THg was analyzed by reduction with 0.8% (wt/vol) stannous chloride (SnCl2) on an RA-915+ mercury analyzer (Ohio Lumex, USA). An aliquot of Hg-containing sample was added to an excess of 0.8% (wt/vol) SnCl2 and purged with ultrahigh-purity N2. The emerging Hg(0) was quantified by a cold vapor atomic absorption spectroscopy (CV-AAS) Zeeman effect Hg analyzer (Lumex RA-915+; Ohio Lumex Company, Inc. Twinsburg, OH), which was calibrated with a set of Hg standards (Brooks Rand Instruments, Seattle, WA).
Methylmercury analysis.
All samples were processed using a nonaqueous extraction method (45) followed by a modified version of EPA method 1630, described previously (44). Briefly, Me200Hg was added to all the samples as an internal standard, methylmercury (MeHg) was then extracted with an acidic KBr solution, followed by extraction of MeHg with CH2Cl2. After phase separation, CH2Cl2 was volatilized completely to release MeHg into a pure aqueous phase. MeHg samples were further processed by distillation, ethylation, and trapping on a Tenax column via N2-purging. Following thermal desorption and separation by gas chromatography, MeHg was detected by ICP-MS. The recovery of spiked MeHg standards was 100% ± 10%, and the detection limit was about 3 × 10−5 nM MeHg.
Statistical analysis.
GraphPad Prism (GraphPad Software, La Jolla, CA) and the R software environment for statistical computing and graphics (https://www.r-project.org/) were used to analyze and plot the data.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xiangping Yin and Linduo Zhao at Oak Ridge National Laboratory (ORNL) for technical assistance in mercury and methylmercury analyses.
This research was sponsored by the Office of Biological and Environmental Research, Office of Science, U.S. Department of Energy (DOE), as part of the Critical Interfaces Science Focus Area at Oak Ridge National Laboratory, which is managed by UT-Battelle, LLC, for the DOE under contract DE-AC05-00OR22725. The manuscript was authored by UT-Battelle, LLC, under contract no. DE-AC05-00OR22725 with the U.S. Department of Energy.
The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (https://www.energy.gov/downloads/doe-public-access-plan).
We declare that we have no conflicts of interest regarding the contents of this article.
S.S.D., J.D.W., S.W.R., J.M.P., and A.J. designed the research. S.S.D. and A.J. performed the experiments and analyses. S.S.D., K.W.R., J.M.P., J.D.W., S.W.R., and A.J. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00438-19.
REFERENCES
- 1.United Nations Environment Programme. 2013. Global Mercury Assessment 2013: sources, emissions, releases, and environmental transport. UNEP Chemicals Branch, Geneva, Switzerland. [Google Scholar]
- 2.Castoldi AF, Coccini T, Manzo L. 2003. Neurotoxic and molecular effects of methylmercury in humans. Rev Environ Health 18:19–31. [DOI] [PubMed] [Google Scholar]
- 3.Gilmour CC, Elias DA, Kucken AM, Brown SD, Palumbo AV, Schadt CW, Wall JD. 2011. Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl Environ Microbiol 77:3938–3951. doi: 10.1128/AEM.02993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridou R, Monperrus M, Gonzalez PR, Guyoneaud R, Amouroux D. 2011. Simultaneous determination of mercury methylation and demethylation capacities of various sulfate-reducing bacteria using species-specific isotopic tracers. Environ Toxicol Chem 30:337–344. doi: 10.1002/etc.395. [DOI] [PubMed] [Google Scholar]
- 5.Ranchou-Peyruse M, Monperrus M, Bridou R, Duran R, Amouroux D, Salvado JC, Guyoneaud R. 2009. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiol J 26:1–8. doi: 10.1080/01490450802599227. [DOI] [Google Scholar]
- 6.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somenahally AC, Johs A, Hurt RA Jr, Bailey KL, Elias DA. 2013. Mercury methylation by novel microorganisms from new environments. Environ Sci Technol 47:11810–11820. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 7.Graham AM, Bullock AL, Maizel AC, Elias DA, Gilmour CC. 2012. Detailed assessment of the kinetics of Hg-cell association, Hg methylation, and methylmercury degradation in several Desulfovibrio species. Appl Environ Microbiol 78:7337–7346. doi: 10.1128/AEM.01792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyes A, Mason RP, Kim EH, Sunderland E. 2006. Mercury methylation in estuaries: insights from using measuring rates using stable mercury isotopes. Mar Chem 102:134–147. doi: 10.1016/j.marchem.2005.09.018. [DOI] [Google Scholar]
- 9.Zhao L, Chen H, Lu X, Lin H, Christensen GA, Pierce EM, Gu B. 2017. Contrasting effects of dissolved organic matter on mercury methylation by Geobacter sulfurreducens PCA and Desulfovibrio desulfuricans ND132. Environ Sci Technol 51:10468–10475. doi: 10.1021/acs.est.7b02518. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer JK, Morel FMM. 2009. High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat Geosci 2:123–126. doi: 10.1038/ngeo412. [DOI] [Google Scholar]
- 11.Wood JM, Kennedy FS, Rosen CG. 1968. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium. Nature 220:173. doi: 10.1038/220173a0. [DOI] [PubMed] [Google Scholar]
- 12.Bertilsson L, Neujahr HY. 1971. Methylation of mercury compounds by methylcobalamin. Biochemistry 10:2805–2808. [DOI] [PubMed] [Google Scholar]
- 13.DeSimone RE, Penley MW, Charbonneau L, Smith SG, Wood JM, Hill HA, Pratt JM, Ridsdale S, Williams RJ. 1973. The kinetics and mechanism of cobalamin-dependent methyl and ethyl transfer to mercuric ion. Biochim Biophys Acta 304:851–863. doi: 10.1016/0304-4165(73)90232-8. [DOI] [PubMed] [Google Scholar]
- 14.Compeau GC, Bartha R. 1985. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol 50:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berman M, Chase T, Bartha R. 1990. Carbon flow in mercury biomethylation by Desulfovibrio desulfuricans. Appl Environ Microbiol 56:298–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi SC, Bartha R. 1993. Cobalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS. Appl Environ Microbiol 59:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi SC, Chase T Jr, Bartha R. 1994. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Appl Environ Microbiol 60:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi SC, Chase T Jr, Bartha R. 1994. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Appl Environ Microbiol 60:4072–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekstrom EB, Morel FM, Benoit JM. 2003. Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria. Appl Environ Microbiol 69:5414–5422. doi: 10.1128/AEM.69.9.5414-5422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parks JM, Johs A, Podar M, Bridou R, Hurt RA Jr, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L. 2013. The genetic basis for bacterial mercury methylation. Science 339:1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 21.Smith SD, Bridou R, Johs A, Parks JM, Elias DA, Hurt RA Jr, Brown SD, Podar M, Wall JD. 2015. Site-directed mutagenesis of HgcA and HgcB reveals amino acid residues important for mercury methylation. Appl Environ Microbiol 81:3205–3217. doi: 10.1128/AEM.00217-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Riccardi D, Beste A, Smith JC, Parks JM. 2014. Mercury methylation by HgcA: theory supports carbanion transfer to Hg(II). Inorg Chem 53:772–777. doi: 10.1021/ic401992y. [DOI] [PubMed] [Google Scholar]
- 23.Demissie TB, Garabato BD, Ruud K, Kozlowski PM. 2016. Mercury methylation by cobalt corrinoids: relativistic effects dictate the reaction mechanism. Angew Chem Int Ed Engl 55:11503–11506. doi: 10.1002/anie.201606001. [DOI] [PubMed] [Google Scholar]
- 24.Schaefer JK, Rocks SS, Zheng W, Liang L, Gu B, Morel FMM. 2011. Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria. Proc Natl Acad Sci U S A 108:8714–8719. doi: 10.1073/pnas.1105781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu YR, Lu X, Zhao LD, An J, He JZ, Pierce EM, Johs A, Gu BH. 2016. Effects of cellular sorption on mercury bioavailability and methylmercury production by Desulfovibrio desulfuricans ND132. Environ Sci Technol 50:13335–13341. doi: 10.1021/acs.est.6b04041. [DOI] [PubMed] [Google Scholar]
- 26.Gilmour CC, Bullock AL, McBurney A, Podar M, Elias DA. 2018. Robust mercury methylation across diverse methanogenic archaea. mBio 9:e02403-17. doi: 10.1128/mBio.02403-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian C, Chen HM, Johs A, Lu X, An J, Pierce EM, Parks JM, Elias DA, Hettich RL, Gu BH. 2018. Quantitative proteomic analysis of biological processes and responses of the bacterium Desulfovibrio desulfuricans ND132 upon deletion of its mercury methylation genes. Proteomics 18:e1700479. doi: 10.1002/pmic.201700479. [DOI] [PubMed] [Google Scholar]
- 28.Qian C, Johs A, Chen H, Mann BF, Lu X, Abraham PE, Hettich RL, Gu B. 2016. Global proteome response to deletion of genes related to mercury methylation and dissimilatory metal reduction reveals changes in respiratory metabolism in Geobacter sulfurreducens PCA. J Proteome Res 15:3540–3549. doi: 10.1021/acs.jproteome.6b00263. [DOI] [PubMed] [Google Scholar]
- 29.Lienhard GE. 1973. Enzymatic catalysis and transition-state theory. Science 180:149–154. doi: 10.1126/science.180.4082.149. [DOI] [PubMed] [Google Scholar]
- 30.Eyring H. 1935. The activated complex in chemical reactions. J Chem Phys 3:107–115. doi: 10.1063/1.1749604. [DOI] [Google Scholar]
- 31.Schumacher W, Holliger C, Zehnder AJ, Hagen WR. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett 409:421–425. doi: 10.1016/S0014-5793(97)00520-6. [DOI] [PubMed] [Google Scholar]
- 32.Lexa D, Saveant JM. 1983. The electrochemistry of vitamin B12. Acc Chem Res 16:235–243. doi: 10.1021/ar00091a001. [DOI] [Google Scholar]
- 33.Yaginuma H, Kawai S, Tabata KV, Tomiyama K, Kakizuka A, Komatsuzaki T, Noji H, Imamura H. 2014. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci Rep 4:6522. doi: 10.1038/srep06522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meister W, Hennig SE, Jeoung JH, Lendzian F, Dobbek H, Hildebrandt P. 2012. Complex formation with the activator RACo affects the corrinoid structure of CoFeSP. Biochemistry 51:7040–7042. doi: 10.1021/bi300795n. [DOI] [PubMed] [Google Scholar]
- 35.Ragsdale SW, Pierce E. 2008. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta 1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hennig SE, Jeoung JH, Goetzl S, Dobbek H. 2012. Redox-dependent complex formation by an ATP-dependent activator of the corrinoid/iron-sulfur protein. Proc Natl Acad Sci U S A 109:5235–5240. doi: 10.1073/pnas.1117126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brady ST. 1985. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature 317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 38.Jensen S, JernelÖV A. 1969. Biological methylation of mercury in aquatic organisms. Nature 223:753. doi: 10.1038/223753a0. [DOI] [PubMed] [Google Scholar]
- 39.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. 2011. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 40.Cardiano P, Falcone G, Foti C, Sammartano S. 2011. Sequestration of Hg2+ by some biologically important thiols. J Chem Eng Data 56:4741–4750. doi: 10.1021/je200735r. [DOI] [Google Scholar]
- 41.Cheesman BV, Arnold AP, Rabenstein DL. 1988. Nuclear magnetic resonance studies of the solution chemistry of metal complexes. 25. Hg(thiol)3 complexes and Hg(II)-thiol ligand exchange kinetics. J Am Chem Soc 110:6359–6364. doi: 10.1021/ja00227a014. [DOI] [Google Scholar]
- 42.Benison GC, Di Lello P, Shokes JE, Cosper NJ, Scott RA, Legault P, Omichinski JG. 2004. A stable mercury-containing complex of the organomercurial lyase MerB: catalysis, product release, and direct transfer to MerA. Biochemistry 43:8333–8345. doi: 10.1021/bi049662h. [DOI] [PubMed] [Google Scholar]
- 43.Lian P, Guo HB, Riccardi D, Dong AP, Parks JM, Xu Q, Pai EF, Miller SM, Wei DQ, Smith JC, Guo H. 2014. X-ray structure of a Hg2+ complex of mercuric reductase (MerA) and quantum mechanical/molecular mechanical study of Hg2+ transfer between the C-terminal and buried catalytic site cysteine pairs. Biochemistry 53:7211–7222. doi: 10.1021/bi500608u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.U.S. Environmental Protection Agency. 1998. Method 1630: methyl mercury in water by distillation, aqueous ethylation, purge and trap, and CVAFS. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 45.Bloom NS, Colman JA, Barber L. 1997. Artifact formation of methyl mercury during aqueous distillation and alternative techniques for the extraction of methyl mercury from environmental samples. Fresen J Anal Chem 358:371–377. doi: 10.1007/s002160050432. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.