The quality and safety of vegetable fermentations are dependent on the activities of LAB naturally present in the phyllosphere. Despite their critical role in determining the success of fermentation, the processes that determine the abundance and diversity of LAB in vegetables used for fermentation are poorly characterized. Our work demonstrates that the limited ability of LAB to grow in the cabbage phyllosphere environment may constrain their abundance on cabbage leaves. These results suggest that commercial fermentation of Napa cabbage proceeds despite low and variable abundances of LAB across different growing regions. Propagule limitation may also explain ecological distributions of other rare members of phyllosphere microbes.
KEYWORDS: 16S, cabbage, community assembly, lactic acid bacteria, microbiome, phyllosphere
ABSTRACT
Patterns of phyllosphere diversity have become increasingly clear with high-throughput sequencing surveys, but the processes that control phyllosphere diversity are still emerging. Through a combination of lab and field experiments using Napa cabbage and lactic acid bacteria (LAB), we examined how dispersal and establishment processes shape the ecological distributions of phyllosphere bacteria. We first determined the abundance and diversity of LAB on Napa cabbage grown at three sites using both culture-based approaches and 16S rRNA gene amplicon sequencing. Across all sites, LAB made up less than 0.9% of the total bacterial community abundance. To assess whether LAB were low in abundance in the Napa cabbage phyllosphere due to a limited abundance in local species pools (source limitation), we quantified LAB in leaf and soil samples across 51 vegetable farms and gardens throughout the northeastern United States. Across all sites, LAB comprised less than 3.2% of the soil bacterial communities and less than 1.6% of phyllosphere bacterial communities. To assess whether LAB are unable to grow in the phyllosphere even if they dispersed at high rates (establishment limitation), we used a gnotobiotic Napa cabbage system in the lab with experimental communities mimicking various dispersal rates of LAB. Even at high dispersal rates, LAB became rare or completely undetectable in experimental communities, suggesting that they are also establishment limited. Collectively, our data demonstrate that the low abundance of LAB in phyllosphere communities may be explained by establishment limitation.
IMPORTANCE The quality and safety of vegetable fermentations are dependent on the activities of LAB naturally present in the phyllosphere. Despite their critical role in determining the success of fermentation, the processes that determine the abundance and diversity of LAB in vegetables used for fermentation are poorly characterized. Our work demonstrates that the limited ability of LAB to grow in the cabbage phyllosphere environment may constrain their abundance on cabbage leaves. These results suggest that commercial fermentation of Napa cabbage proceeds despite low and variable abundances of LAB across different growing regions. Propagule limitation may also explain ecological distributions of other rare members of phyllosphere microbes.
INTRODUCTION
The phyllosphere, the aboveground portion of plants that can be colonized by microbes, plays important roles in the productivity of agricultural systems, as well as the safety and quality of food (1–3). Various bacteria, fungi, and other microbes colonize the phyllosphere, where they are antagonists, commensals, or mutualists of their hosts (4–7). Many recent studies have described patterns of phyllosphere microbial diversity using high-throughput sequencing (4, 8–16). An extensive body of experimental work has characterized the dynamics of phyllosphere species and populations, including many plant pathogens and biocontrol agents (17, 18). A mechanistic understanding of the specific ecological processes that explain patterns of phyllosphere community composition is still emerging.
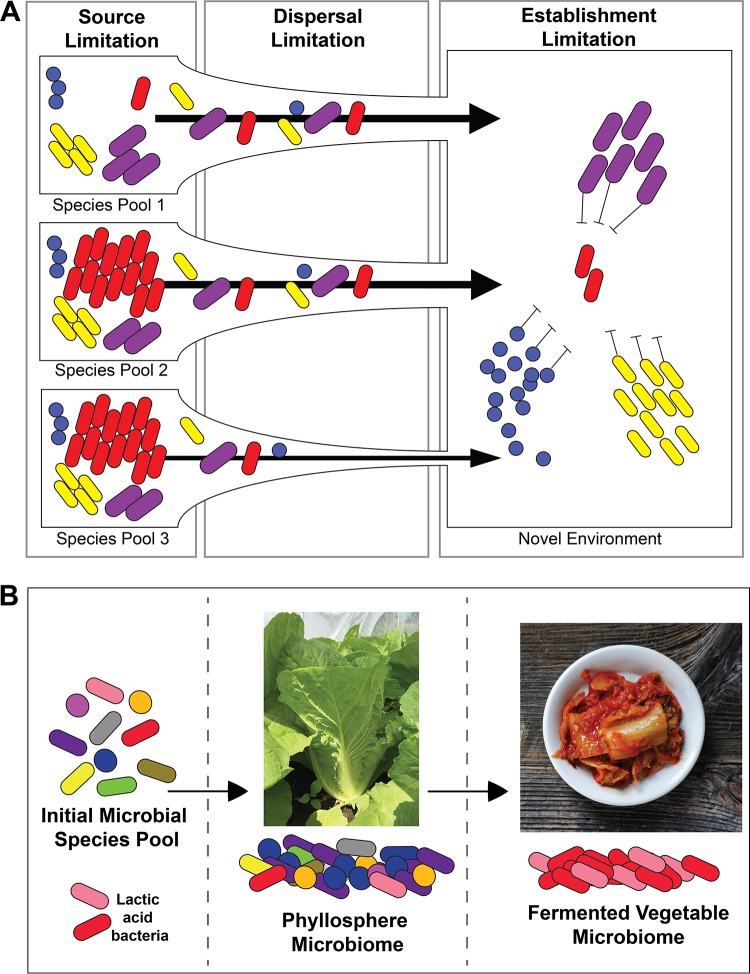
Dispersal may be one ecological process that shapes phyllosphere microbiome diversity and function. When a microbe is present in a local species pool, its abundance in the phyllosphere may be constrained by how many cells successfully reach and colonize available plant leaf habitat. In plant and animal ecology, species are considered propagule limited when propagules (seeds, spores, larvae, etc.) fail to reach all suitable habitat patches at saturating densities (19–22). While the term propagule is not widely used in microbiology, a microbial propagule is a cell or other biological unit (e.g., spore, group of cells in a biofilm) that can generate a new microbial population when disseminated (23). Propagule limitation can be divided into three different components: (i) source limitation, where not enough propagules are locally produced to colonize all potential habitats; (ii) dispersal limitation, where enough propagules are produced, but they cannot reach all available habitats; and (iii) establishment limitation, where local population sizes are constrained by availability of viable niche spaces and not by propagule abundance (22) (Fig. 1A). Several studies in other microbiomes, including the built environment (24), tree roots (25), and aquatic communities (26), have provided observational evidence for propagule limitation. Many studies in the phyllosphere have tracked the dispersal and movement of microbial species and populations (17), but we are unaware of studies that have experimentally tested the significance of propagule limitation as a driver of phyllosphere microbiome community composition.
FIG 1.
Conceptual overview of propagule limitation and the farm-to-ferment assembly of fermented vegetable microbiomes. (A) The three types of propagule limitation are source, dispersal, and establishment. Source limitation is where not enough propagules are present in local species pools to disperse to the phyllosphere. Dispersal limitation is where enough propagules are produced in local species pools, but they cannot reach all available habitats due to lack of dispersal opportunities. Establishment limitation is where local population sizes are constrained by the availability of habitats that can be colonized, for example, by competition with other phyllosphere bacteria. (B) Local species pools consist of bacteria that could disperse to cabbage leaves to form the phyllosphere microbiome. When chopped up and fermented, the phyllosphere microbiome is reassembled to become the fermented vegetable microbiome.
Lactic acid bacteria (LAB) can be found in the phyllosphere and are widely known for their beneficial impacts in food systems (27). In addition to fermenting vegetables to make products such as sauerkraut, kimchi, and natural pickles (28–30), plant-associated LAB have the potential to impact human health through increasing the nutritional quality of raw food materials (31). Despite their widespread use in food fermentation, surprisingly little is known about the ecology of LAB in the phyllosphere (32, 33). Most studies have focused on the dynamics of LAB during fermentation (28, 34, 35), and the processes that determine LAB distributions in the phyllosphere are largely unknown.
Lactic acid bacteria are essential for successful vegetable fermentations, and variation in the initial abundance and composition of LAB may impact the outcome of cabbage fermentation (36–39). In both large- and small-scale production, vegetable fermentations are not usually inoculated with defined starter cultures and instead rely on LAB present in the phyllosphere (40). Microbes from local species pools in the farms where cabbages are grown colonize cabbage leaves during phyllosphere microbiome assembly. During the fermentation process, the phyllosphere microbiome undergoes a second stage of community assembly, leading to a new fermented vegetable microbiome (Fig. 1B). Some studies have detected LAB in soil and water (41, 42), but the sources of LAB cells in agricultural systems and their dispersal dynamics are poorly characterized.
Here, we introduce cabbage leaves as a model system for determining the ecological processes shaping phyllosphere bacterial community composition. We used Napa cabbage (Brassica rapa subsp. pekinensis, also known as Chinese cabbage), which is grown around the world to be consumed fresh, as well as for fermentation into kimchi and other fermented vegetable products (43–45). It is a fast-growing species that can be easily grown in plant tissue culture and seeds can be sterilized to create gnotobiotic plants. Using field- and lab-grown plants, we first determined that LAB are low in abundance in the Napa cabbage phyllosphere (NCP). We then used field sampling and an experimental approach with gnotobiotic cabbages to determine that establishment limitation explains the low abundance of LAB in the NCP. Our work demonstrates the utility of the Napa cabbage system as a model to link patterns with processes in phyllosphere microbiomes and illustrates the importance of dispersal processes in explaining species distributions in microbial communities.
RESULTS
LAB are in low abundance in the Napa cabbage phyllosphere.
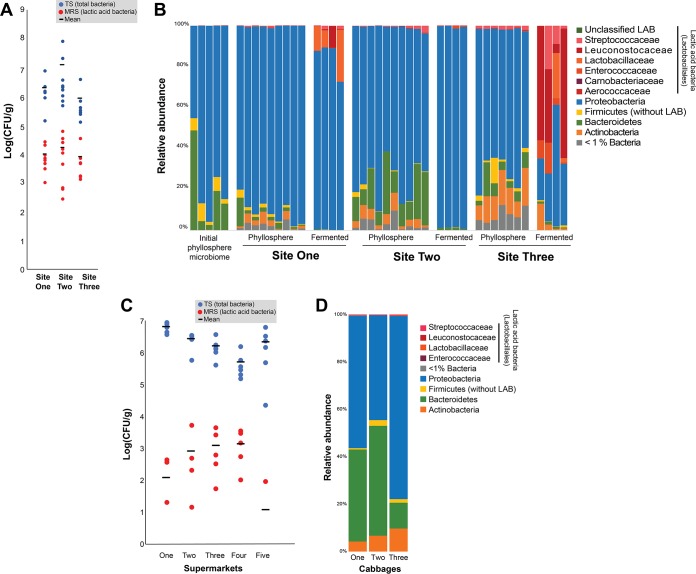
To estimate the abundance and diversity of LAB in the NCP, we grew Napa cabbages at three different field sites in the Boston, MA, area (see Fig. S2 in the supplemental material). After 2 months in the field (August through October), cabbages were returned to the lab where we determined phyllosphere bacterial community composition using 16S rRNA gene amplicon sequencing. We also plated cabbage leaf homogenates on de Man, Rogosa, and Sharpe (MRS) agar, which is selective for many LAB taxa (46), and tryptic soy (TS) agar, which enables the growth of many phyllosphere community members (47). We paired these culture-based approaches with amplicon sequencing of the 16S rRNA gene (V4 variable region) to determine the relative abundance (RA) of LAB as all reads assigned to the order Lactobacillales compared to the total number of bacterial reads in each sample. Plating demonstrated that LAB were low in abundance in the NCP, with average total cultural abundances of 0.76% at site 1, 0.51% at site 2, and 1.09% at site 3 (Fig. 2A). Amplicon sequencing also demonstrated the low abundance of LAB with RAs of 0.30% at site 1, 0.89% at site 2, and 1.68% at site 3 (Fig. 2B).
FIG 2.
Microbial diversity and LAB abundance in the Napa cabbage phyllosphere. (A) Abundance of total culturable bacteria (TS agar in blue) and lactic acid bacteria (MRS agar in red) in the Napa cabbage phyllosphere at three sites in the Boston, MA, area. Site 1, n = 9; site 2, n = 10; site 3, n = 7. (B) Relative abundance of bacterial phyla and Lactobacillales families identified in the Napa cabbage phyllosphere using amplicon sequencing of the 16S rRNA gene. Each column represents an individual cabbage showing the initial phyllosphere microbiome of the lab-grown cabbage (n = 5), site 1 phyllosphere (n = 9) and fermented (n = 4), site 2 phyllosphere (n = 10) and fermented (n = 4), and site 3 (n = 7) and fermented (n = 4). Members of the Lactobacillales are shown at the family level in order to indicate the prevalence of major LAB groups. (C) Abundance of total culturable bacteria (TS agar in blue) and lactic acid bacteria (MRS agar in red) in the Napa cabbage phyllosphere from cabbages purchased at five supermarkets. For each supermarket, n = 6. (D) Relative abundance of phyla identified in the Napa cabbage phyllosphere purchased from one of the five supermarkets. Each column represents an individual cabbage. As with panel B, members of the Lactobacillales are shown at the family level in order to indicate the prevalence of major LAB groups.
Amplicon sequencing enabled us to determine not just the relative abundance of LAB but also the diversity of LAB genera present in the NCP. Members of the Lactobacillaceae, Leuconostocaceae, and Streptococcaceae were detected in low numbers on the cabbages from all three sites, including the genera Lactobacillus, Fructobacillus, Leuconostoc, Lactococcus, and Streptococcus (see Table S1 in the supplemental material). These genera contain species that are important for vegetable fermentation, including Leuconostoc mesenteroides, a common heterofermentative LAB detected in the early stages of many fermented vegetables (48), and Lactobacillus plantarum, a homofermentative LAB that is typically found in the later stages of fermentation (48). Although the NCP reads assigned to LAB were low, all of the main LAB families essential for fermenting vegetables were detected at all three sites.
Sequence-based data on cabbage phyllosphere bacterial diversity are very limited (43, 49), and our amplicon sequence data help fill this gap. Across the three sites, Proteobacteria dominated the amplicon sequence data sets, with Bacteroidetes, Firmicutes (non-LAB), and Actinobacteria making up smaller fractions of the NCP at each site (Fig. 2B, Table S2). This phylum-level composition is similar to the phyllosphere microbiomes of other leafy vegetables (47, 50–52) and a range of other plant species where Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes dominate (3, 50, 53–55). The most abundant bacterial genera detected across the three sites were Sphingomonas, Pseudomonas, and Methylobacterium, which are also abundant taxa in many phyllosphere microbiomes (4, 56). While the same broad taxonomic groups of bacteria were found across the three sites, there were significant differences in phyllosphere community composition across all three sites (permutational multivariate analysis of variance [PERMANOVA]; F = 34.02, P < 0.001), suggesting that differences in local species pools can drive variation in NCP assembly across sites.
To determine how LAB abundances in the NCP change from phyllosphere to fermentation, we measured fermentation potential of NCP material from each of the three sites. Leaves were chopped into small pieces, combined with salt (2% [wt/wt]), and fermented at 24°C for 14 days. At site 1 and site 3, the RAs of LAB in the community increased as the cabbage was fermented (Fig. 2B), and we observed characteristic declines in pH in our small-scale fermentations (see Fig. S1 in the supplemental material). The increase in LAB was particularly striking at site 3, where the LAB went from ∼1% RA in phyllosphere samples to an RA of >50% in three of the four fermented samples (Fig. 2B). The fermentation of site 2 phyllosphere samples diverged from site 1 and site 3 in that site 2 communities were dominated by Proteobacteria and had very small amounts of LAB at the end of fermentation. As with the phyllosphere communities, the composition of fermented bacterial communities was significantly different across sites (PERMANOVA; F = 21.43, P < 0.01), suggesting that initial phyllosphere composition can translate into differences in final fermentation community composition.
To confirm that LAB are consistently low in abundance in the NCP and that our findings were not an artifact of our experimental manipulations, field sites, or cabbage variety, we used the same culture-based and amplicon sequencing approaches to measure LAB in Napa cabbages purchased from five supermarkets in Boston, MA. Using culture-based methods, LAB had a low abundance in all of the sampled cabbages with a total cultural abundance between 0.004 and 0.78% (Fig. 2C). Using amplicon sequencing on cabbages from one of the supermarkets, the RA of LAB in the NCP was found to be 0.16%, supporting the culture-based results (Fig. 2D; see Table S3 in the supplemental material). Together, these results indicate that LAB are not abundant in the NCP both in experimental cabbages grown in our field sites and in commercially available cabbages. Building on this observation, we next sought to determine ecological explanations for the rarity of LAB.
LAB are not abundant in local species pools across the northeastern United States.
One explanation for why LAB have a low abundance in the Napa cabbage microbiome is that they are rare in local species pools in farms and gardens, or source limited (Fig. 1A). A limited abundance of LAB in species pools would provide few opportunities for dispersal of LAB cells to the NCP. While there are many potential species pools that could be sources of LAB, such as water (57), insects (58, 59), soil (16), and leaves (60), we selected and tested the two species pools that we predicted could be the main reservoir for the phyllosphere microbial community: soil and leaves.
To determine whether LAB are source limited, we sampled soil and leaves from 51 sites throughout the northeastern United States (Fig. S2) and used selective plating and amplicon sequencing to determine LAB abundance in the species pools. Sites 1, 2, and 3 from above were included in this species pool survey. Sampling locations were small farms or community gardens where cruciferous vegetables, including cabbage, were being grown or had been grown in the recent past. At each site, soil and leaves were collected from five randomly selected locations and were then pooled and homogenized to give a site-specific sample. Soil samples were taken from the top 5 cm of soil after dead plant litter was removed. Leaf samples consisted of a mix of vegetation, including crop and weed species (Amaranthus retroflexus, Portulaca oleracea, Cerastium arvense, Chenopodium album, and Ambrosia artemisiifolia). We did not directly sample cabbage leaves because each site had a range of cabbages of different species, varieties, and ages and because some sites were not growing cabbage while we were sampling. Weed species growing within each site provided a common potential species pool that could be consistently sampled. We used the same paired culture-based plating and 16S rRNA gene amplicon sequencing as described above to determine the RA of LAB in the NCP to estimate the RA of LAB in soil and leaf species pools.
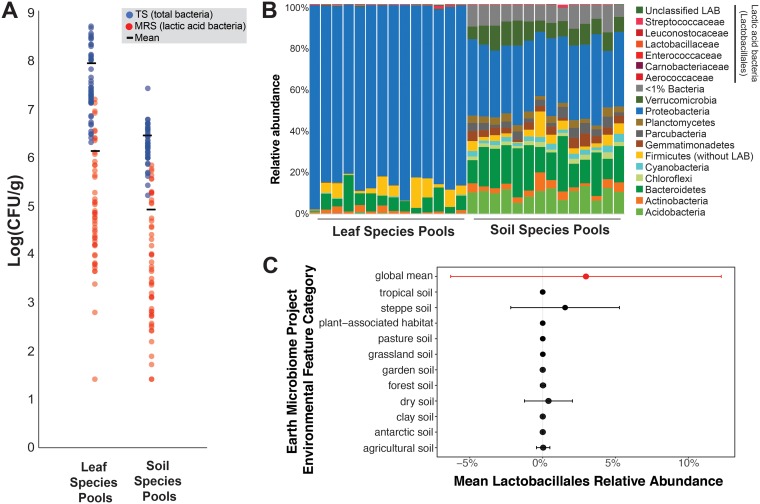
Across all 51 sites, LAB had a low abundance in both the leaf and soil species pools. With culture-based methods, the mean RAs of LAB were 1.55% in the leaves and 3.20% in the soil (Fig. 3A). Using 16S rRNA gene amplicon sequencing across a subset (n = 14) of the soil and leaf samples from across the geographic range sampled, we found a much lower RA of LAB, where leaves had an RA of 0.32% and soil had an RA of 0.14% (Fig. 3B; Table S4). The lower RA of LAB detected using amplicon sequencing is likely due to an inability to culture many leaf and soil bacteria on TS agar and incomplete selectivity of the MRS agar leading to undercounting of non-LAB community members relative to LAB. This survey across the northeastern United States indicated that LAB are rare members in the sampled species pools of soil and leaves. These species pools could be the main reservoir for the Napa cabbage microbiome and the scarcity of LAB in these species pools may contribute to an overall low abundance of LAB in environments where Napa cabbages grow.
FIG 3.
LAB are rare in species pools. (A) Abundance of total culturable bacteria (TS agar in blue) and lactic acid bacteria (MRS agar in red) in leaf and soil species pools from 51 farms through the northeastern United States. Each point represents CFU data collected from a pool of five leaf or soil samples from each site. (B) Relative abundance of bacteria found in leaf samples and soil samples at a subset of farms from panel A, as determined with amplicon sequencing of the 16S rRNA gene (V4 region). As with Fig. 2, members of the Lactobacillales are shown at the family level in order to indicate the prevalence of major LAB groups. Each column represents sequence data collected from a pool of five leaf or soil samples from each site. Data from 14 sites are presented. (C) Relative abundance of Lactobacillales in amplicon sequence data collected as part of the Earth Microbiome Project. Points indicate means, and error bars indicate one standard deviation.
To determine whether LAB that colonize NCP can be found in the soil and leaf species pools, we compared LAB detected in the NCP phyllosphere data collected from sites 1 to 3 with our survey of soil and leaf samples across the 51 sites. All of the LAB taxa identified in the NCP were also detected in the soil and leaf species pools (Table S1). However, there were LAB identified in these species pools which were not detected in the NCP. For instance, Vagococcus and Enterococcus (Enterococcaceae) were present in one of the soil samples, but not present in any of the sampled NCPs. Facklamia and Aerococcus (Aerococcaceae), as well as Alloiococcus, Desemzia, Marinilactibacillus, Dolosigranulum, Granulicatella, and Atopoistipes (Carnobacteriaceae), were present in a few of the sampled species pools (both soil and leaves) but were rarely detected in the NCP. These data indicate that the sampled species pools could be reservoirs of LAB that can eventually colonize cabbage leaves.
LAB are in low abundance across a global sample of soil and leaf species pools.
To further examine the distribution and abundance of LAB in the environment, we determined the relative abundance of taxa from the order Lactobacillales in a recently published global survey of bacterial communities (61). At a global scale, LAB were in low abundance with a mean RA of ∼2.0% (standard deviations, ±9.2%). In the majority of soils and plant-associated systems, LAB were particularly rare with average RAs of <0.001% except in steppe (mean RA = 3.7%) and dry soils (mean RA = 1.6%) (Fig. 3C). Outside of soil- and plant-associated systems, LAB were present in low relative abundance across multiple ecosystems (mean RA < 0.01%; Fig. S2). Several hot spots for LAB abundance were observed in insect-associated habitats (mean RA = 22.5%), human-associated habitats (mean RA = 15.1%), bird nests (mean RA = 11.1%), coral reefs (mean RA = 10.9%), animal-associated habitats (mean RA = 9.3%), and city environments (mean RA = 7.8%). These global data suggest that the abundance of LAB is constrained across many plant and soil environments and that the rarity of LAB is not unique to the Napa cabbage phyllosphere environment.
Gnotobiotic and field-grown Napa cabbages demonstrate that LAB are establishment limited.
In addition to being source limited, establishment limitation could also constrain the abundance of LAB in the NCP (Fig. 1A). Despite being in low abundance in the field environment, it is possible that LAB could be capable of rapid growth once they arrive in the NCP if they can easily become established. However, if LAB cannot colonize the leaf due to poor growth, weak competitive abilities, or an inability to tolerate the phyllosphere environment, they will remain at a low abundance in the cabbage phyllosphere.
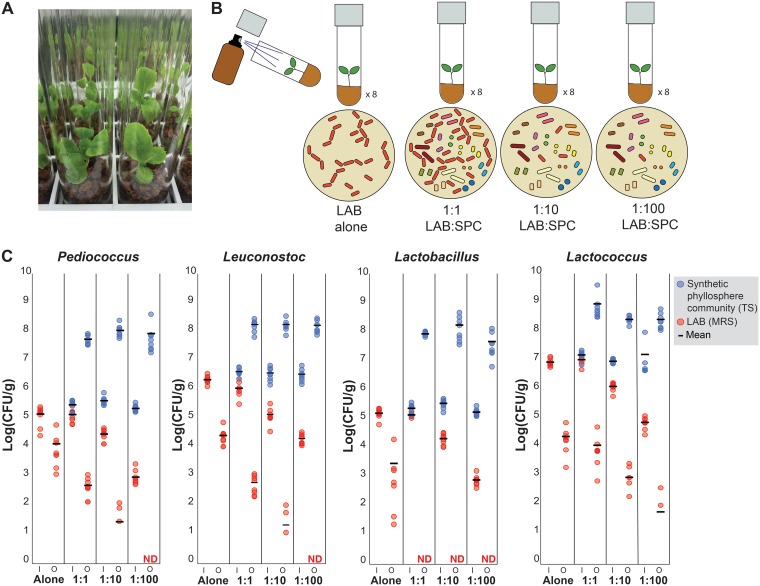
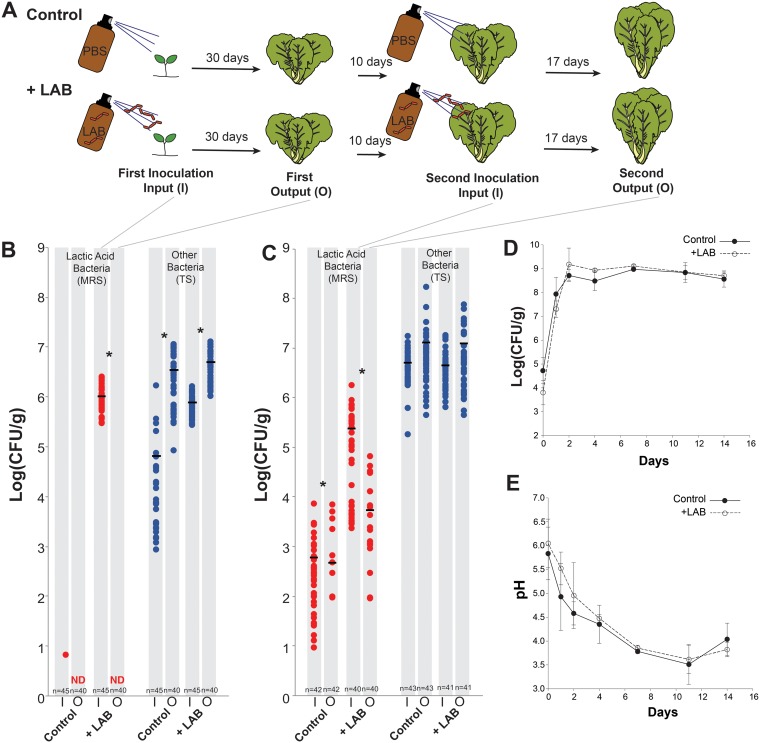
To test whether LAB are establishment limited, we developed gnotobiotic Napa cabbages. These cabbages were prepared by surface-sterilizing Napa cabbage seeds and planting them into sterilized glass tubes containing Murashige and Skoog (MS) basal salt broth and calcined clay (Fig. 4A). After the cabbages had grown for a week, we inoculated them with pure cultures of LAB, along with various levels of LAB mixed with a synthetic phyllosphere community (SPC) consisting of 14 bacterial taxa representing the common phyla of bacteria detected in the NCP (Table 1). Four different lactic acid bacteria were tested: Leuconostoc mesenteroides strain BN10, Lactobacillus plantarum strain MKR2, Lactococcus lactis strain D119, and Pediococcus pentosaceus strain B6N. These bacteria represent some of the most common genera and species of LAB that are consistently found in fermented vegetable products (28, 62, 63). The ability of LAB to establish in the NCP was measured by comparing their abundance at the start of the experiment (input) to their abundance after 10 days of growth in the NCP (output). Three different ratios of LAB relative to the SPC were inoculated onto the Napa cabbages to determine how initial LAB abundance impacts establishment: a 1:1, a 1:10, and a 1:100 initial starting ratio of LAB to SPC (Fig. 4B). The 1:100 dilution results in LAB abundances that approximate levels detected in our field experiments described above. The treatment with high levels of LAB relative to the SPC (1:1) was designed to mimic high rates of LAB dispersal to the NCP (Fig. 4B).
FIG 4.
Gnotobiotic cabbages demonstrate that LAB are establishment limited in the Napa cabbage phyllosphere. (A) Photograph of gnotobiotic Napa cabbages used in experiments. (B) Overview of experimental design showing four treatments (LAB grown alone, LAB grown 1:1 with synthetic phyllosphere community [SPC], LAB grown 1:10 with SPC, and LAB grown 1:100 with SPC). Treatments were applied using sterile brown amber spray bottles. Four lactic acid bacteria were used in these experiments: Pediococcus pentosaceus strain B6N, Leuconostoc mesenteroides strain BN10, Lactobacillus plantarum strain MKR2, and Lactococcus lactis strain D119. (C) Abundance of the SPC (TS agar in blue) and LAB (MRS agar in red) in the input (I) inoculum and output (O) after 10 days of growth in the NCP is shown. Input and output were significantly different from one another in all tested LAB species (t test, P < 0.5, corrected for repeat sampling using a Hochberg correction; n = 8; ND, not detected).
TABLE 1.
Bacterial isolates used to construct the synthetic phyllosphere community in gnotobiotic cabbages
| Strain | Phylum | Family | Genus/species | NCBI accession no. of 16S rRNA gene sequence | Isolation source |
|---|---|---|---|---|---|
| BSC5 | Alphaproteobacteria | Rhizobiaceae | Unknown Rhizobiaceae species | MK308543 | NCPa |
| BSC6 | Gammaproteobacteria | Enterobacteriaceae | Pantoea sp. | MK308544 | NCP |
| BSC12 | Gammaproteobacteria | Enterobacteriaceae | Pantoea sp. | MK308545 | NCP |
| BSC13 | Actinobacteria | Microbacteriaceae | Microbacterium sp. | MK308546 | NCP |
| BSC14 | Bacteroidetes | Sphingobacteriaceae | Pedobacter sp. | MK308547 | NCP |
| BSC44 | Alphaproteobacteria | Caulobacteraceae | Brevundimonas sp. | MK308548 | NCP |
| BSC45 | Gammaproteobacteria | Pseudomonadaceae | Pseudomonas sp. | MK308549 | NCP |
| BSC46 | Actinobacteria | Micrococcaceae | Unknown Micrococcaceae species | MK308550 | NCP |
| BSC51 | Gammaproteobacteria | Xanthomonadaceae | Xanthomonas sp. | MK308551 | NCP |
| BSC57 | Betaproteobacteria | Alcaligenaceae | Achromobacter sp. | MK308552 | NCP |
| BSC58 | Alphaproteobacteria | Sphingomonadaceae | Sphingomonas sp. | MK308553 | NCP |
| GR2 | Actinobacteria | Microbacteriaceae | Curtobacterium sp. | MK308554 | NCP |
| GR11 | Actinobacteria | Microbacteriaceae | Unknown Microbacteriaceae species | MK308555 | NCP |
| GR29 | Bacteroidetes | Sphingobacteriaceae | Sphingobacterium sp. | MK308556 | NCP |
| BN10 | Firmicutes | Leuconostocaceae | Leuconostoc mesenteroides | MK329278 | NCP |
| MKR2 | Firmicutes | Lactobacillaceae | Lactobacillus brevis | MK329281 | Fermented vegetable product |
| B6N | Firmicutes | Lactobacillaceae | Pediococcus pentosaceus | MK329280 | NCP |
| D119 | Firmicutes | Streptococcaceae | Lactococcus lactis | MK329277 | Farm soil |
NCP, Napa cabbage phyllosphere.
All four of the LAB tested decreased in abundance when sprayed onto Napa cabbages as pure cultures, suggesting that LAB cannot grow alone on Napa cabbage leaves and instead die over time (Fig. 4C). Decreases in viable LAB cells ranged from ∼3 log in L. lactis to ∼1 log in P. pentosaceus (Fig. 4C). In the presence of the SPC, the decline of LAB was even greater, with all LAB decreasing when inoculated at a 1:1 ratio with the SPC. For example, in the presence of the SPC, there was an ∼2-log-fold decrease in viable P. pentosaceus cells. Dilution of the LAB (the 1:10 and 1:100 LAB:SPC treatments) resulted in a greater decrease in LAB abundances (Fig. 4C). The most dramatic decrease was seen in Lactobacillus plantarum, which when coinoculated with the SPC was not detected at any of the three LAB dilutions. In two of the tested LAB, L. mesenteroides and P. pentosaceus, no bacteria were detected after being applied at the lowest (1:100) dilution. In contrast to the decrease in abundance seen in all of the LAB sprayed onto the NCP, the SPC increased in abundance in all treatments, demonstrating that the experimental conditions were conducive to bacterial growth. The SPC had an average abundance of 1.6 × 108 CFU/g across all community treatments, which is comparable to levels detected in field-grown cabbages (Fig. 2A).
To confirm that LAB are also establishment limited in a field setting with larger cabbages and background levels of bacterial dispersal, 3-week-old Napa cabbage seedlings were inoculated with a mixed community of LAB (Leuconostoc mesenteroides strain BN10, Lactobacillus plantarum strain MKR2, and Pediococcus pentosaceus strain B6N; here referred to as “+LAB”) or phosphate-buffered saline (PBS; here referred to as “Control”) and then planted into three treatment and three control beds in an experimental garden at Tufts University in Medford, MA, in mid-May (Fig. 5A). We selected these three LAB because they are naturally resistant to vancomycin, as are many members of the Lactobacillales (64, 65), and can therefore be easily tracked after being applied to plants in field settings. After a month of growth, a leaf was removed from each cabbage, and the RA of the LAB was estimated using culture-based approaches. Ten days later, the cabbages were sprayed again with the LAB mix or PBS, and the abundance of LAB was determined after 17 days.
FIG 5.
LAB are establishment limited in the phyllosphere of field-grown Napa cabbages. (A) Overview of the experimental design. Napa cabbages were grown in the lab and were sprayed with either PBS as a control or an equal mix of three LAB (Pediococcus pentosaceus strain B6N, Leuconostoc mesenteroides strain BN10, and Lactobacillus plantarum strain MKR2) for the +LAB treatment. Input data were collected right after inoculation, and output data were collected after 30 days of growth in the field. After 10 days of growth, cabbages were inoculated again with the same treatments. Input data were collected at the second inoculation, and output data were collected after 17 more days of growth. (B) Abundance of LAB (MRS agar in red) and other phyllosphere bacteria (TS agar in blue) in the input (I) inoculum and output (O) leaf harvests for the first period of the experiment. An asterisk indicates a significant difference between input (I) and output (O) bacterial densities (t test, P < 0.5, corrected for repeat sampling using a Hochberg correction). The number of replicates is indicated at the bottom of the graph. ND, not detected. (C) Same as panel B, but for the second set of inputs and outputs. (D) Abundance of LAB during fermentation of cabbages from the Control and +LAB treatments in panels B and C. Points indicate mean values (n = 3). Error bars represent one standard deviation. (E) pH during fermentation of cabbages from the Control and +LAB treatments in panels B and C. Points indicate mean values (n = 3). Error bars represent one standard deviation. Control and +LAB treatments were not significantly different from one another in panel D or E (see the text for repeated-measures ANOVA statistics).
As with our lab-grown gnotobiotic cabbages, the LAB did not grow well in the phyllosphere of field-grown plants. After the initial inoculation, +LAB cabbages had an average LAB density of 1.0 × 106 CFU/g, and only one out of 45 Control cabbages had detectable LAB (6 CFU/g; Fig. 5B). After 1 month, no LAB were detected in both the Control and the +LAB cabbages, demonstrating the same rapid die-off of LAB in the phyllosphere observed in laboratory conditions (Fig. 5B). At the time of the second inoculation, Control cabbages had background densities of LAB similar to what were observed in our field-planted cabbages described above. This background LAB growth is likely due to colonization from local species pools or movement of LAB between treatment and control beds. The second inoculation raised the LAB density to an average density of 2.5 × 105 log CFU/g in the +LAB treatment, but LAB levels decreased to an average of 5.5 × 103 log CFU/g about 2 weeks later (Fig. 5C).
To determine whether supplementing field cabbages with LAB changed fermentation outcomes, we harvested Control and +LAB cabbages and fermented them in small (118-ml) sterile glass jars. Both Control and +LAB cabbages had similar fermentation dynamics, with no significant differences in total LAB abundance (Fig. 5D) or acidification (Fig. 5E) over the 2 weeks of fermentation (LAB abundance repeated-measures ANOVA, F1,4 = 0 .01, P = 0.95; acidification repeated-measures ANOVA, F1,4 = 0.18, P = 0.69). These results demonstrate that low levels of LAB are sufficient for a successful fermentation to proceed and that supplementation of LAB in the field is unlikely to impact fermentation outcomes.
DISCUSSION
Lactic acid bacteria are critical for successful vegetable fermentation and fermented vegetable producers rely on naturally occurring LAB during the production of these foods. While a number of common LAB species are detected in fermented products, there are few studies which investigate the ecology of these LAB in either the vegetable phyllosphere or the field environment (66, 67). We used culture-based and sequencing methods to show that LAB are in low abundance in the phyllosphere of experimental and commercially available Napa cabbages. Using lab and field experiments, we investigated whether two components of propagule limitation, source and establishment limitation, could explain the low abundance of LAB in the NCP. Source limitation was tested by conducting a survey of two different species pools across 51 sites throughout the northeastern United States. This survey found that LAB were rare in both soil and leaf species pools, and this result held true at a global scale. By inoculating LAB into the NCP, it was also shown that they are unable to establish or grow, providing evidence that establishment limitation could also restrict their abundance. Surprisingly, instead of finding evidence for growth of LAB in the phyllosphere, we found that they die over time, both alone and when growing with other phyllosphere bacteria. Our work demonstrates that while LAB are consistently found on Napa cabbage plants, they are always at low abundance and have limited capabilities to grow in the phyllosphere.
The culture-based and amplicon sequencing results indicate that LAB are consistently in low abundance in the Napa cabbage phyllosphere. Our study only used varieties of Napa cabbage (Brassica rapa subsp. pekinensis), and there are many other Brassica species and cultivars that are fermented including green, red, and savoy cabbage (all cultivars of Brassica oleracea). The different phylloplane structure and chemistry of these species and cultivars could influence the ability of LAB to grow and establish, and LAB may not be establishment limited for all cabbage varieties or all plant species. However, previous studies do support our observation of limited colonization by LAB in other phyllosphere systems. Metagenomic studies that were not specifically designed to study the ecology of LAB also found that Lactobacillales are generally in low abundance in the phyllosphere (51, 52, 68). Culture-based studies from the 1960s noted that LAB are in low abundance on the surfaces of cucumbers, beets, carrots, and other vegetables (67). A recent survey of a fermentation production facility found that LAB are in low levels in the green cabbage phyllosphere (49). Collectively, these studies demonstrate a consistently low abundance of LAB across other species and varieties of cabbage, as well as other vegetables.
Our survey of soil and leaf species pools from 51 sites demonstrates that LAB are rare in agroecosystems of the northeastern United States. We focused on two species pools, soil and leaves, which we predicted would be the main inoculation source for the phyllosphere microbiome. Soil was thought to be a potential species pool as previous research has shown the phyllosphere is typically a subset of the bacteria detected in the rhizosphere (16). Leaves were also considered a potential species pool for the phyllosphere microbiome since neighboring plants growing in proximity to Napa cabbages could facilitate dispersal of microbes as plant leaves brush against one another or as other vectors (farm tools, humans, insects, etc.) move bacteria from one plant to another. LAB were in low abundance in both of these tested species pools, which demonstrates the potential for source limitation of LAB in agroecosystems.
We acknowledge that there are other potential reservoirs of LAB that might colonize cabbage leaves that were not sampled in this study. For example, water (57), air (60), and herbivores that feed on cabbage leaves (59, 69) could all contain LAB. Insects could be an especially important species pool to consider as certain insects, including Scaptomyza spp. and Mamestra brassicae, have been reported to vector bacteria between plants as they feed on leaves (58, 70). Honeybee species have been shown to have LAB in their intestinal tracts (59), and although bees do not usually feed on leaves, they could have other interactions with plant leaves that might transfer bacteria into the phyllosphere. Our analysis of the Earth Microbiome Project data revealed that members of the Lactobacillales were highest in abundance in insect-associated systems, as well as in association with birds, and were low in abundance in water, air, and soil. This suggests that herbivory or excretion from birds may contribute LAB to phyllosphere microbiomes. Future experiments that experimentally manipulate insect herbivory in the Napa cabbage phyllosphere could help assess the contribution of this microbial species pool to dispersal dynamics of LAB.
LAB were not able to grow well in the phyllosphere, and the levels decreased despite inoculating the cabbages with large amounts of LAB. Initial LAB inoculum lost viability over time and fell to levels comparable to those detected in the Napa cabbages planted out at the three field sites. These data suggest that there may be a low carrying capacity for LAB on the leaf or that there is a constant influx of LAB which do not survive once they arrive in the phyllosphere. It is not surprising that LAB do not colonize the phyllosphere in high levels since they lack many of the microbial traits that are found in phyllosphere-adapted bacteria. For example, many phyllosphere bacteria are pigmented and use these pigments to protect against high levels of UV light on leaf surfaces (71–73). LAB generally lack photoprotective pigments (74) and are likely poorly equipped to deal with damage caused by UV.
Studies using next-generation sequencing have shown a common pattern in microbial community composition where a few common species make up the majority of the community, along with a “long tail” of rare species (75, 76). These rare species might play a disproportionate role in ecosystem functioning or could function only when the environment changes (77). Conditional rarity describes bacterial taxa that remain in low abundance until environmental conditions enable them to rapidly increase in abundance (78) and may be aptly applied to LAB in a farm-to-ferment framework (Fig. 1B). While the agroecological function of LAB was not tested in this study, it is possible that they remain rare in the environment but shift to an increased abundance after plant decomposition at the end of the growing season when plants are harvested. LAB are also important in creating silage where plants are fermented in anaerobic conditions on a farm (79, 80). Release of plant sugars and a shift to an anaerobic environment are two of the main environmental changes which are also present in fermented vegetable production. LAB are not good at persisting in the Napa cabbage phyllosphere, but the anaerobic, salty, and sugar-rich conditions of the fermentation environment allow these bacteria to become dominant members of the fermented food microbiome.
This research focused exclusively on the bacteria that make up the NCP microbiome. Most plants are colonized by fungi, protists, and other microbes that may contribute to the ecological distributions of LAB in the phyllosphere. For example, many fungi can live in the phyllosphere of cabbages as pathogens (81, 82), and the phyllosphere yeast Rhodotorula is known to cause defects in fermented vegetable products (83, 84). Future work should determine the contributions of these other microbes in the phyllosphere and fermentation microbiome assembly process.
We developed gnotobiotic Napa cabbages to dissect microbial community dynamics in the phyllosphere. Our model system integrating germ-free plants with a culture collection spanning the diversity of cabbage phyllosphere bacteria adds to the growing number of gnotobiotic systems being used to study how microbial communities assemble in the phyllosphere and rhizosphere (13, 16, 85). We acknowledge that young cabbages in test tubes with artificial soil medium do not fully recapitulate the dynamics of full-sized field-grown cabbages, but our results in the lab were supported by similar field experiments. The Napa cabbage phyllosphere model provides future opportunities to explore how other ecological processes, including microbe-microbe interactions, explain patterns of phyllosphere diversity and impact the quality of fermented vegetables.
MATERIALS AND METHODS
Determining the abundance of LAB in the Napa cabbage phyllosphere.
Kaboko F1 hybrid organic Napa cabbage seeds (High Mowing Seeds) were surface sterilized by soaking in 10% bleach for 40 min and then rinsed eight times with sterile deionized water. Seeds were sown in plant pots (5.5 × 5.5 × 5 cm) and watered as necessary with autoclaved (121°C for 20 min) deionized water. Pots were filled with Sunshine mix 1 (Sun Gro Horticulture) that was autoclaved (121°C for 20 min) to reduce its microbial load. Pots were then placed in sterile Sun bags (pore size 0.02 μm; Sigma-Aldrich) to minimize contamination with microbes from the lab environment. Pots were kept under light racks (full-spectrum T5 fluorescent bulbs) with a 16-h light cycle at 24°C.
After cabbages had grown at least three true leaves (about 4 weeks of growth), they were planted out at three sites in mid-August. Site 1 was an urban community garden with a mix of vegetables and flowers in Boston, MA; site 2 was a raised garden bed used regularly for plant research projects at Tufts University (Medford, MA); and site 3 was a small, rural farm used for vegetable production about 25 miles outside Boston (Lincoln, MA) (Fig. S1). To help reduce variation in soil conditions between sites, peat pots (Jiffy pots 10.16 cm wide by 10.16 cm deep) containing Sunshine mix soil were first placed into the site in a grid formation (∼25-cm spacing between pots), and the cabbages were planted into the peat pots. All cabbages were watered as needed throughout the growing season. Each plant was fertilized with 50 ml of fertilizer (3:4:4 N:P:K, 5% calcium, 1% magnesium, 2% sulfur) prepared in a concentration of 15 g/liter and filter sterilized before application.
After 2 months (mid-October), the cabbages were cut at the base of the head and placed in a Ziploc bag for transporting back to the lab. In the lab, leaf fragments were taken from each cabbage for amplicon sequencing and for plating. For amplicon sequencing, leaf fragments (∼4 cm2) were taken from three randomly selected leaves from across the plant (both inner and outer leaves) and frozen at –80°C for processing at a later date. In addition, three leaf fragments per plant were taken for culture-dependent analyses. The leaf fragments were homogenized with 500 μl of phosphate-buffered saline (PBS) in a 1.5-ml microcentrifuge tube using a sterile micropestle. The cabbage homogenate was then plated on tryptic soy (TS) agar plates containing cycloheximide (100 mg/liter) to estimate total cultural phyllosphere bacteria and de Man, Rogosa, and Sharpe (MRS) agar plates containing cycloheximide (100 mg/liter) to estimate total LAB abundance. After a week of aerobic growth at 24°C, colonies were counted from TS and MRS plates to determine CFU per gram of cabbage.
After samples of Napa cabbage were taken for plating and sequencing, the remaining cabbage was shredded using a Cuisinart DLC-2ABC Mini-Prep Plus food processor for ∼2 min, and 2% (wt/wt) salt was added. Cabbage homogenate was then compacted into sterile 5-ml screw-top vials (Axygen Scientific) and maintained at 24°C for 14 days. On days 1, 2, 4, 7, 10, and 14, four replicate ferments were harvested. Ferments were homogenized in a Whirl-Pak bag. A 100-μl sample of the liquid ferment was then frozen at –20°C for DNA extraction and amplicon sequencing (see below). We acknowledge that these small-scale fermentations do not completely reflect the conditions in home or industrial fermentations, where the production environment or other ingredients may introduce LAB (43, 86). However, these assays do provide an estimate of the fermentation potential of vegetable materials.
To determine the LAB abundance and diversity in commercial Napa cabbages, we purchased cabbages from five different supermarkets in the Boston, MA, area. For each supermarket sampled, three Napa cabbages were purchased on the same day and returned to the lab, where 20 g of cabbage from each of four outer leaves was placed into 118-ml Whirl-Pak bags. In addition, 20 g of cabbage was taken from four inner leaves (leaves which were not exposed to environment) and also placed into 118-ml Whirl-Pak bags. Napa cabbage leaves were then homogenized in 50 ml of 1× PBS for 1 min per bag. The cabbage homogenate was then plated on TS agar plates containing 21.6 mg/liter of natamycin to estimate total cultural phyllosphere bacteria and MRS agar plates also containing 21.6 mg/liter of natamycin to estimate total LAB abundance.
Amplicon sequencing of the 16S rRNA gene.
DNA was extracted from leaf fragments or ferment liquid using the MoBio Powersoil kit (Qiagen) according to the manufacturer’s instructions. To generate amplicon libraries, a portion of the 16S rRNA gene was amplified using the primers 515F (GTGYCAGCMGCCGCGGTAA) and 806R (GGACTACNVGGGTWTCTAAT) (87). Golay barcodes (12 bp) were incorporated in reverse primer constructs to allow for multiplexing. Promega PCR master mix was used for duplicate PCRs in 25-μl reaction mixtures with 0.2 μM concentrations of each primer and 1 μl of DNA template using the following thermocycler conditions: an initial denaturation at 95°C for 5 min; 35 cycles of 45 s at 95°C, 60 s at 50°C, and 90 s at 72°C; and a final elongation at 72°C for 10 min. PCR amplicons were pooled, cleaned and normalized using SequalPrep normalization plates (Life Technologies). Equimolar concentrations were pooled for sequencing on an Illumina MiSeq in the CU Boulder BioFrontiers Sequencing Center using the v2 300-cycle kit (Illumina, Inc.).
QIIME (88) and UPARSE (89) were used to process amplicon sequencing data, as described by Andrei et al. (90), with some modifications. Sequences were demultiplexed using the Golay barcodes via QIIME v1.9.1 (89). The following options were used to obtain raw forward and reverse fastq read files: split_libraries_fastq.py -q 0 –max_bad_run_length 250 –min_per_read_length_fraction 0.0001 –sequence_max_n 250 –store_demultiplexed_fastq. Paired ends were merged in usearch v8 (91). Data from two independent runs on an Illumina MiSeq were combined. Reads were quality filtered, and operational taxonomic unit (OTU) tables were constructed using the UPARSE pipeline (89). OTUs were clustered at 97% sequence similarity with de novo chimera detection enabled. The following parameters were used to modify the UPARSE pipeline: the –minh option of -uchime_ref was set to 1.5 for reference-based chimera removal to reduce the false-positive detection of chimeras; the OTU table was generated by mapping quality-filtered reads back to the OTU seeds by setting the following –usearch_global parameters: -maxaccepts 0 -maxrejects 0, in order to avoid overinflation of specific OTU counts and ensure that individual reads are correctly mapped to their respective OTUs. Consensus taxonomy was assigned using QIIME v1.9.1 (88) on a custom database sourced from SILVA v128 (92). Because LAB are rare in the Napa cabbage phyllosphere, we did not rarefy the amplicon sequencing data as this could lead to the loss of rare LAB reads.
Identifying LAB in local species pools.
To determine whether soil or neighboring plants are major reservoirs of LAB, we quantified the abundance of LAB at 51 farms or community gardens throughout the northeastern United States using culture-based and amplicon sequencing approaches. Farmers were recruited using local extension agencies and regional farm networks. From each farm, five randomly selected sites were sampled, and 20 g of leaf material and 20 g of soil from the top 5 cm of soil were collected. Samples were kept at 4°C for 1 or 2 days before being processed at the lab. Slurries of either leaf or soil were made by placing the collected material into a Whirl-Pak bag together with 50 ml of PBS and then macerating the sample for 5 min. Equal volumes (10 ml) of each of the five soil and five leaf samples from each site were pooled to create a pooled leaf mix and a pooled soil mix.
A culture-based approach was used to determine the abundance of LAB in all leaf and soil samples. Soil and leaf homogenates were plated onto both MRS and TS plates containing 21.6 mg/liter natamycin. Colonies were counted after aerobic incubation at 24°C for 1 week, and the ratios of CFU growing on MRS plates to TS plates were used to provide an estimate of the RA of LAB in the phyllosphere community. LAB colonies were distinguished from Enterobacteriaceae colonies which can grow on MRS plates as LAB are small white, creamy, and/or translucent colonies, whereas Enterobacteriaceae colonies are large, viscous, and cream colored. Amplicon sequencing was performed on a subsample (14 of 51 sites) of leaf and soil slurries to confirm the plating data. DNA was extracted using a MoBio Powersoil DNA extraction kit, and amplicon libraries were prepared and sequenced as previously described.
Gnotobiotic Napa cabbages.
Calcined clay (Turface) was washed repeatedly with tap water to remove fine particles and dust. The clay was then autoclaved (121°C for 20 min) to reduce bacterial load. Cabbage growth chambers were prepared by adding 10 g of dry calcined clay and 10 ml of Murashige and Skoog (MS) basal salt broth (4.4 g/liter of basal salt to water) to glass test tubes (15 cm by 2.5 cm). Tubes were covered with 22-mm Magenta two-way test tube caps (Sigma-Aldrich) and autoclaved at 121°C for 60 min. Kakabo Napa cabbage seeds were sterilized by placing 100 seeds in a 1.5-ml microcentrifuge tube, vortex mixing the seeds for 5 min in 1 ml of 70% ethanol, and then vortex mixing them again for 5 min in 1 ml of 50% bleach. Ethanol and bleach were rinsed away using four washes in sterile deionized water with vortex mixing for 5 min. The seeds were left soaking in the sterile deionized water for at least 1 h to help soften any remaining seed coat. Using sterilized tweezers, one sterile seed was added to each prepared tube, and the lid was firmly replaced. Tubes were placed under light racks (full-spectrum T5 fluorescent bulbs) with a 16-h light cycle at 24°C. The relative humidity inside the test tubes was 55%.
Creating a synthetic phyllosphere community for Napa cabbage.
A synthetic phyllosphere community (SPC) was created by sampling Napa cabbage leaf samples collected from the cabbage field experiment previously described (sites 1, 2, and 3). We isolated a range of phyllosphere bacteria by plating leaf homogenates onto the following media: yeast extract peptone dextrose, tryptic soy, Pseudomonas isolation, M17, and de Man, Rogosa, and Sharpe. Single colonies of different morphotypes were streaked out and identified using 16S rRNA gene sequencing with the primers 27f (AGAGTTTGATCCTGGCTCAG) and 1492r (GGTTACCTTGTTACGACTT) (93). Identities and NCBI accession numbers of strains are given in Table 1. Strains were made into experimental glycerol stocks by growing each strain as an overnight liquid culture (TS broth) and pelleting the overnight culture at 3,000 × g for 5 min at 4°C. The supernatant was discarded, and the pellet was then washed with 1× PBS before being resuspended in 15% glycerol in 1× PBS.
Inoculating gnotobiotic cabbages to test establishment limitation.
Inocula of all 14 members of the SPC and four LAB (Leuconostoc mesenteroides strain BN10, Lactobacillus plantarum strain MKR2, Lactococcus lactis strain D119, and Pediococcus pentosaceus strain B6N) were prepared from frozen glycerol stocks. These glycerol stocks were prepared by growing each strain on solid media (TS agar for SPC strains and MRS agar for LAB) and then scraping cells of each of the strains into sterile 15% glycerol before storage at –80°C. Multiple tubes were prepared for each strain, and one tube was thawed and plated before experiment setup to determine viable cell densities (CFU/μl).
All of the SPC members were diluted in 1× PBS to 10,000 CFU/μl, and 8 ml of each stock was combined to create a solution containing equal concentrations of each of the 14 members. Each of the four LAB was also diluted in PBS to 10,000 CFU/μl. Multiple treatments with different mixing ratios of LAB to SPC were prepared: 1:1, 1:10, and 1:100. These ratios were prepared by mixing equal volumes of the full-strength LAB and SPC inocula (1:1) or by mixing 10-fold dilutions of the LAB inoculum with the full-strength SPC inoculum for the 1:10 and 1:100 concentrations. Inocula of each of the LAB alone were also prepared for a total of 16 treatments: each LAB alone and each LAB in a mix with the SPC in ratios of 1:1, 1:10, and 1:100. A negative-control bottle containing just 1× PBS was also prepared to test for contamination of sterile plants, bottles, or reagents.
All 16 treatments were sprayed onto plants using amber Boston round glass bottles (59.15 ml) with an atomizer. Bottles and atomizers were sterilized by soaking them in a 30% bleach solution for at least 30 min and then rinsing them with sterile deionized water. Three pumps of inoculum (∼600 μl) were applied to each cabbage using the atomizer with 16 replicate cabbages per treatment. After spraying, half the cabbages (n = 8) were harvested to enumerate the input bacteria inoculated. To harvest, cabbages were removed from the tubes with sterilized tweezers, and roots were cut off using sterilized dissection scissors. The entire mass of the cabbage was weighed in a 1.5-ml microcentrifuge tube, and 400 μl of 1× PBS was added to each tube. Leaves were then homogenized with a sterile micropestle, and the cabbage slurry was then diluted and plated onto TS agar and MRS agar plates to quantify total phyllosphere bacteria and LAB abundance, respectively. Cabbages were left to grow at 24°C under light racks, as described earlier, for 10 days. All remaining cabbages in each treatment were harvested in the same manner as the inputs to get output counts on TS agar and MRS agar.
Testing establishment limitation in the field.
To test whether LAB are establishment limited in a field setting, seeds of the Napa cabbage variety “Bilko F1 OG” were surface sterilized as described above and planted in pots containing Sunshine mix soil. We switched seed varieties in this experiment (from the Kakabo variety used above) due to a limited availability of Kakabo seeds. Pilot experiments comparing the growth of LAB and SPC isolates found no differences between the Bilko F1 OG and Kakabo varieties. For our field experiments, we did not use the sterile tissue culture approach described above because preliminary experiments suggested cabbages grown in ambient lab conditions had no detectable LAB. We did not attempt to control the composition of other phyllosphere bacteria in this experiment. After the cabbages had grown under light racks (same conditions described above) for 3 weeks, they were inoculated with a mixed community of LAB, including Leuconostoc mesenteroides BN10, Lactobacillus plantarum MKR2, and Pediococcus pentosaceus B6N (+LAB), or sprayed with PBS as a control (Control). The inoculum was added as described above for gnotobiotic lab plants, where three pumps of inoculum (∼600 μl) were sprayed using an atomizer attached to a sterile glass bottle. This is the “first inoculation” in Fig. 5. One leaf was removed from each replicate cabbage postinoculation (n = 45 per treatment) to determine the density of LAB inoculated per gram of cabbage (on MRS agar) and the density of background phyllosphere bacteria (on TS agar). The cabbage leaf was weighed in a 1.5-ml microcentrifuge tube, and then 400 μl of PBS was added. The cabbage leaf was homogenized with a sterile micropestle, and the cabbage slurry was then diluted and plated on MRS agar plates with 21.6 mg/liter of natamycin to inhibit fungi and 100 mg/liter of vancomycin to reduce the growth of Enterobacteriaceae. The cabbages were then planted into raised beds (183 × 122 × 38 cm) filled with a mix of field soil and composted cow manure in an experimental garden at Tufts University in mid-August. Three beds were assigned as controls, and three were assigned as treatments. After a month of growth (mid-September), a leaf was removed from each replicate cabbage (n = 40), placed into a 29.57-ml Whirl-Pak filter bag (Nasco, Modesto, CA), and taken back to the lab for processing. This is the “first output” in Fig. 5. Approximately 0.4 g of the cabbage leaf was placed into a 1.5-ml microcentrifuge tube with 400 μl of PBS and homogenized with a sterile micropestle. The homogenized cabbage slurry was then diluted and plated on MRS plates with 21.6 mg/liter of natamycin to inhibit fungi and 100 mg/liter of vancomycin to reduce the growth of Enterobacteriaceae (94). Ten days after the first harvest, the cabbages were sprayed again with LAB inoculum or PBS (“second inoculation” in Fig. 5), and the abundance of LAB was tracked after 17 days (early October, “second output” in Fig. 5) via plating, as was just described.
Fermentation of field-grown cabbages.
Field-grown cabbages from the Control and +LAB treatments were fermented in 4-oz. (∼118 ml) sterile glass canning jars with metal screw-top lids. Cabbages from each Control or +LAB plot were pooled (Control bed 1, 13 cabbages; Control bed 2, 15 cabbages; Control bed 3, 15 cabbages; +LAB bed 1, 15 cabbages; +LAB bed 2, 12 cabbages; +LAB bed 3, 14 cabbages) and finely sliced using a knife sterilized with 70% ethanol. This chopped cabbage was then mixed for 5 min with salt to obtain a 2% (wt/wt) salt concentration. Equal amounts of the cabbage mix were distributed into the sterile jars and tightly compacted, and the lids were then sealed. Jars were incubated in the dark at 24°C. LAB abundance and pH were measured at 0, 1, 2, 4, 7, and 14 days. At each time point, the jar of sauerkraut was emptied into a sterile 118.29-ml Whirl-Pak bag and fully mixed to homogenize the sample. LAB abundance was determined by plating cabbage juice from the ferment onto MRS agar. The pH was determined by inserting a pH probe into the Whirl-Pak bag with ∼20 ml of liquid.
Analysis of the global distribution of LAB.
To further understand the ecology of LAB at a global scale, we investigated their relative abundance in the recently published Earth Microbiome Project (61). A rarefied OTU table (10,000 sequences per sample) that was generated from closed reference OTU clustering was retrieved from the EMP website (ftp://ftp.microbio.me/emp/release1). The table was filtered to identify members of the order Lactobacillales in QIIME and analyzed in R.
Statistical analysis.
Statistical analyses were performed using R v3.5.1. All CFU data were log transformed prior to analysis. PERMANOVAs were conducted using the vegan package. Repeated-measures analysis was carried out using the lme4 package and the post hoc test ANOVA was carried out with the car package. Student t tests were corrected for repeat sampling using a Hochberg correction.
Data availability.
Partial 16S rRNA gene sequences of bacterial isolates have been deposited in NCBI (see Table 1 for accession numbers). Amplicon sequence data have been deposited in the NCBI database as BioProject PRJNA510140 with Sequence Read Archive accession numbers SAMN10612113 to SAMN10612199.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to the numerous farmers and community gardeners who provided access to their land for various aspects of this work. We are especially grateful to Erik Jacobs for providing farm access and support for pilot experiments. All amplicon library preparation and sequencing were performed by Jonah Ventures. Megan Biango-Daniels, Casey Cosetta, and Freddy Lee provided helpful feedback on previous versions of the manuscript. Elizabeth Landis provided support with plant care. Kinsey Drake, Claire Forgan, and Emily Van Doren provided research support in the lab.
This study was supported by USDA NIFA grant 2017-67013-26520.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00269-19.
REFERENCES
- 1.Brandl MT. 2006. Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol 44:367–392. doi: 10.1146/annurev.phyto.44.070505.143359. [DOI] [PubMed] [Google Scholar]
- 2.Heaton JC, Jones K. 2008. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: a review. J Appl Microbiol 104:613–626. doi: 10.1111/j.1365-2672.2007.03587.x. [DOI] [PubMed] [Google Scholar]
- 3.Rastogi G, Coaker GL, Leveau J. 2013. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol Lett 348:1–10. doi: 10.1111/1574-6968.12225. [DOI] [PubMed] [Google Scholar]
- 4.Vorholt JA. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 5.Müller DB, Vogel C, Bai Y, Vorholt JA. 2016. The plant microbiota: systems-level insights and perspectives. Annu Rev Genet 50:211–234. doi: 10.1146/annurev-genet-120215-034952. [DOI] [PubMed] [Google Scholar]
- 6.Schoelz JE, Stewart LR. 2018. The role of viruses in the phytobiome. Annu Rev Virol 5:93–111. doi: 10.1146/annurev-virology-092917-043421. [DOI] [PubMed] [Google Scholar]
- 7.Leach JE, Triplett LR, Argueso CT, Trivedi P. 2017. Communication in the phytobiome. Cell 169:587–596. doi: 10.1016/j.cell.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 8.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim S-T, Weigel D, Kemen EM. 2016. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peñuelas J, Terradas J. 2014. The foliar microbiome. Trends Plant Sci 19:278–280. doi: 10.1016/j.tplants.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA. 2010. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J 4:719–728. doi: 10.1038/ismej.2010.9. [DOI] [PubMed] [Google Scholar]
- 11.Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. 2014. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet 10:e1004283. doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. 2014. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 5:e00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Glavina del Rio T, Jones CD, Tringe SG, Dangl JL. 2015. Plant microbiome: salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 14.Bodenhausen N, Horton MW, Bergelson J. 2013. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beattie GA. 2015. Microbiomes: curating communities from plants. Nature 528:340–341. doi: 10.1038/nature16319. [DOI] [PubMed] [Google Scholar]
- 16.Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, Hüttel B, McHardy AC, Vorholt JA, Schulze-Lefert P. 2015. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 17.Kinkel LL. 1997. Microbial population dynamics on leaves. Annu Rev Phytopathol 35:327–347. doi: 10.1146/annurev.phyto.35.1.327. [DOI] [PubMed] [Google Scholar]
- 18.Andrews JH, Harris RF. 2000. The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol 38:145–180. doi: 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- 19.Primack RB, Miao SL. 1992. Dispersal can limit local plant distribution. Conserv Biol 6:513–519. doi: 10.1046/j.1523-1739.1992.06040513.x. [DOI] [Google Scholar]
- 20.Cain ML, Milligan BG, Strand AE. 2000. Long-distance seed dispersal in plant populations. Am J Bot 87:1217–1227. doi: 10.2307/2656714. [DOI] [PubMed] [Google Scholar]
- 21.Turnbull LA, Crawley MJ, Rees M. 2000. Are plant populations seed-limited? A review of seed sowing experiments. Oikos 88:225–238. doi: 10.1034/j.1600-0706.2000.880201.x. [DOI] [Google Scholar]
- 22.Clark CJ, Poulsen JR, Levey DJ, Osenberg CW. 2007. Are plant populations seed limited? A critique and meta-analysis of seed addition experiments. Am Nat 170:128–142. doi: 10.1086/518565. [DOI] [PubMed] [Google Scholar]
- 23.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreås L, Reysenbach A-L, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 24.Adams RI, Miletto M, Taylor JW, Bruns TD. 2013. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J 7:1262–1273. doi: 10.1038/ismej.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peay KG, Garbelotto M, Bruns TD. 2010. Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91:3631–3640. doi: 10.1890/09-2237.1. [DOI] [PubMed] [Google Scholar]
- 26.Bell T. 2010. Experimental tests of the bacterial distance–decay relationship. ISME J 4:1357–1365. doi: 10.1038/ismej.2010.77. [DOI] [PubMed] [Google Scholar]
- 27.Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol 15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 28.Rhee SJ, Lee J-E, Lee C-H. 2011. Importance of lactic acid bacteria in Asian fermented foods. Microb Cell Fact 10(Suppl 1):S5. doi: 10.1186/1475-2859-10-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyung KH, Medina Pradas E, Kim SG, Lee YJ, Kim KH, Choi JJ, Cho JH, Chung CH, Barrangou R, Breidt F. 2015. Microbial ecology of watery kimchi. J Food Sci 80:M1031–M1038. doi: 10.1111/1750-3841.12848. [DOI] [PubMed] [Google Scholar]
- 30.Plengvidhya V, Breidt F Jr, Lu Z, Fleming HP. 2007. DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl Environ Microbiol 73:7697–7702. doi: 10.1128/AEM.01342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montaño A, Casado FJ, de Castro A, Sánchez AH, Rejano L. 2004. Vitamin content and amino acid composition of pickled garlic processed with and without fermentation. J Agric Food Chem 52:7324–7330. doi: 10.1021/jf040210l. [DOI] [PubMed] [Google Scholar]
- 32.Minervini F, Celano G, Lattanzi A, Tedone L, De Mastro G, Gobbetti M, De Angelis M. 2015. Lactic acid bacteria in durum wheat flour are endophytic components of the plant during its entire life cycle. Appl Environ Microbiol 81:6736–6748. doi: 10.1128/AEM.01852-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golomb BL, Marco ML. 2015. Lactococcus lactis metabolism and gene expression during growth on plant tissues. J Bacteriol 197:371–381. doi: 10.1128/JB.02193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuyts S, Van Beeck W, Oerlemans EFM, Wittouck S, Claes IJJ, De Boeck I, Weckx S, Lievens B, De Vuyst L, Lebeer S. 2018. Carrot juice fermentations as man-made microbial ecosystems dominated by lactic acid bacteria. Appl Environ Microbiol 84:e00134-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Cagno R, Coda R, De Angelis M, Gobbetti M. 2013. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol 33:1–10. doi: 10.1016/j.fm.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Gangopadhyay H, Mukherjee S. 1971. Effect of different salt concentrations on the microflora and physico-chemical changes in sauerkraut fermentation. J Food Sci Technol Mysore 8:127–131. [Google Scholar]
- 37.Trail AC, Fleming HP, Young CT, Mcfeeters RF. 1996. Chemical and sensory characterization of commercial sauerkraut. J Food Quality 19:15–30. doi: 10.1111/j.1745-4557.1996.tb00402.x. [DOI] [Google Scholar]
- 38.Mcfeeters RF. 2004. Fermentation microorganisms and flavor changes in fermented foods. J Food Sci 69:FMS35–FMS37. doi: 10.1111/j.1365-2621.2004.tb17876.x. [DOI] [Google Scholar]
- 39.Harris LJ. 1998. The microbiology of vegetable fermentations, p 45–72. In Wood BJB. (ed), Microbiology of fermented foods. Springer, New York, NY. [Google Scholar]
- 40.Yoon SS, Barrangou-Poueys R, Breidt F Jr, Klaenhammer TR, Fleming HP. 2002. Isolation and characterization of bacteriophages from fermenting sauerkraut. Appl Environ Microbiol 68:973–976. doi: 10.1128/AEM.68.2.973-976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y-S, Yanagida F, Shinohara T. 2005. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett Appl Microbiol 40:195–200. doi: 10.1111/j.1472-765X.2005.01653.x. [DOI] [PubMed] [Google Scholar]
- 42.Lauzon HL, Ringø E. 2011. Prevalence and application of lactic acid bacteria in aquatic environments, p 601–639. In Lahtinen S, Salminen S, Ouwehand A, von Wright A (ed), Lactic acid bacteria: microbiological and functional aspects. CRC Press, Boca Raton, FL. [Google Scholar]
- 43.Lee SH, Jung JY, Jeon CO. 2015. Source tracking and succession of kimchi lactic acid bacteria during fermentation. J Food Sci 80:1871–1877. [DOI] [PubMed] [Google Scholar]
- 44.Jeong SH, Lee HJ, Jung JY, Lee SH, Seo H-Y, Park W-S, Jeon CO. 2013. Effects of red pepper powder on microbial communities and metabolites during kimchi fermentation. Int J Food Microbiol 160:252–259. doi: 10.1016/j.ijfoodmicro.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Patra JK, Das G, Paramithiotis S, Shin H-S. 2016. Kimchi and other widely consumed traditional fermented foods of Korea: a review. Front Microbiol 7:1493. doi: 10.3389/fmicb.2016.01493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J Appl Bacteriol 23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 47.Williams TR, Moyne A-L, Harris LJ, Marco ML. 2013. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS One 8:e68642. doi: 10.1371/journal.pone.0068642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zabat MA, Sano WH, Wurster JI, Cabral DJ, Belenky P. 2018. Microbial community analysis of sauerkraut fermentation reveals a stable and rapidly established community. Foods 7:77. doi: 10.3390/foods7050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Einson JE, Rani A, You X, Rodriguez AA, Randell CL, Barnaba T, Mammel MK, Kotewicz ML, Elkins CA, Sela DA. 2018. A vegetable fermentation facility hosts distinct microbiomes reflecting the production environment. Appl Environ Microbiol 84:e01680-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams TR, Marco ML. 2014. Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors. mBio 5:e01564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leff JW, Fierer N. 2013. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One 8:e59310. doi: 10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau J. 2012. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J 6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 54.Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA. 2012. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6:1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. 2013. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS One 8:e78613. doi: 10.1371/journal.pone.0078613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE. 2016. The phyllosphere: microbial jungle at the plant-climate interface. Annu Rev Ecol Evol Syst 47:1–24. doi: 10.1146/annurev-ecolsys-121415-032238. [DOI] [Google Scholar]
- 57.Yanagida F, Chen Y-S, Yasaki M. 2007. Isolation and characterization of lactic acid bacteria from lakes. J Basic Microbiol 47:184–190. doi: 10.1002/jobm.200610237. [DOI] [PubMed] [Google Scholar]
- 58.Lilley AK, Hails RS, Cory JS, Bailey MJ. 2006. The dispersal and establishment of pseudomonad populations in the phyllosphere of sugar beet by phytophagous caterpillars. FEMS Microbiol Ecol 24:151–157. doi: 10.1016/S0168-6496(97)00054-8. [DOI] [Google Scholar]
- 59.Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC. 2012. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 7:e33188. doi: 10.1371/journal.pone.0033188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindemann J, Upper CD. 1985. Aerial dispersal of epiphytic bacteria over bean plants. Appl Environ Microbiol 50:1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, Earth Microbiome Project Consortium. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong T, Guan Q, Song S, Hao M, Xie M. 2012. Dynamic changes of lactic acid bacteria flora during Chinese sauerkraut fermentation. Food Control 26:178–181. doi: 10.1016/j.foodcont.2012.01.027. [DOI] [Google Scholar]
- 63.Tamminen M, Joutsjoki T, Sjöblom M, Joutsen M, Palva A, Ryhänen E-L, Joutsjoki V. 2004. Screening of lactic acid bacteria from fermented vegetables by carbohydrate profiling and PCR-ELISA. Lett Appl Microbiol 39:439–444. doi: 10.1111/j.1472-765X.2004.01607.x. [DOI] [PubMed] [Google Scholar]
- 64.Campedelli I, Mathur H, Salvetti E, Clarke S, Rea MC, Torriani S, Ross RP, Hill C, O’Toole PW. 2018. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl Environ Microbiol 85:e01738-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swenson JM, Facklam RR, Thornsberry C. 1990. Antimicrobial susceptibility of vancomycin-resistant Leuconostoc, Pediococcus, and Lactobacillus species. Antimicrob Agents Chemother 34:543–549. doi: 10.1128/AAC.34.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamont JR, Wilkins O, Bywater-Ekegärd M, Smith DL. 2017. From yogurt to yield: potential applications of lactic acid bacteria in plant production. Soil Biol Biochem 111:1–9. doi: 10.1016/j.soilbio.2017.03.015. [DOI] [Google Scholar]
- 67.Daeschel MA, Andersson RE, Fleming HP. 1987. Microbial ecology of fermenting plant materials. FEMS Microbiol Lett 46:357–367. doi: 10.1111/j.1574-6968.1987.tb02472.x. [DOI] [Google Scholar]
- 68.Dees MW, Lysøe E, Nordskog B, Brurberg MB. 2015. Bacterial communities associated with surfaces of leafy greens: shift in composition and decrease in richness over time. Appl Environ Microbiol 81:1530–1539. doi: 10.1128/AEM.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welte CU, de Graaf RM, van den Bosch TJM, Op den Camp HJM, van Dam NM, Jetten M. 2016. Plasmids from the gut microbiome of cabbage root fly larvae encode SaxA that catalyses the conversion of the plant toxin 2-phenylethyl isothiocyanate. Environ Microbiol 18:1379–1390. doi: 10.1111/1462-2920.12997. [DOI] [PubMed] [Google Scholar]
- 70.Groen SC, Humphrey PT, Chevasco D, Ausubel FM, Pierce NE, Whiteman NK. 2016. Pseudomonas syringae enhances herbivory by suppressing the reactive oxygen burst in Arabidopsis. J Insect Physiol 84:90–102. doi: 10.1016/j.jinsphys.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bringel F, Couée I. 2015. Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front Microbiol 6:486. doi: 10.3389/fmicb.2015.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacobs JL, Carroll TL, Sundin GW. 2005. The role of pigmentation, ultraviolet radiation tolerance, and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microb Ecol 49:104–113. doi: 10.1007/s00248-003-1061-4. [DOI] [PubMed] [Google Scholar]
- 74.Carr FJ, Chill D, Maida N. 2002. The lactic acid bacteria: a literature survey. Crit Rev Microbiol 28:281–370. doi: 10.1080/1040-840291046759. [DOI] [PubMed] [Google Scholar]
- 75.Lynch MDJ, Neufeld JD. 2015. Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 76.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, Küsel K, Rillig MC, Rivett DW, Salles JF, van der Heijden MGA, Youssef NH, Zhang X, Wei Z, Hol W. 2017. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J 11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N, Gilbert JA. 2014. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5:e01371-14. doi: 10.1128/mBio.01371-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ni K, Wang Y, Cai Y, Pang H. 2015. Natural lactic acid bacteria population and silage fermentation of whole-crop wheat. Asian-Australas J Anim Sci 28:1123–1132. doi: 10.5713/ajas.14.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Driehuis F, Oude Elferink S, Van Wikselaar PG. 2001. Fermentation characteristics and aerobic stability of grass silage inoculated with Lactobacillus buchneri, with or without homofermentative lactic acid bacteria. Grass Forage Sci 56:330–343. doi: 10.1046/j.1365-2494.2001.00282.x. [DOI] [Google Scholar]
- 81.El-Mohamedy R, El-Mougy N. 2009. Occurrence of Pythium rot of Chinese cabbage in Egypt and its biocontrol measures. J Plant Protection Res 49:19. [Google Scholar]
- 82.O’Hara NB, Rest JS, Franks SJ. 2016. Increased susceptibility to fungal disease accompanies adaptation to drought in Brassica rapa. Evolution 70:241–248. doi: 10.1111/evo.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fred EB, Peterson WH. 1922. The production of pink sauerkraut by yeasts. J Bacteriol 7:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barth M, Hankinson TR, Zhuang H, Breidt F. 2009. Microbiological spoilage of fruits and vegetables, p 135–183. In Sperber WH, Doyle MP (ed), Compendium of the microbiological spoilage of foods and beverages. Springer, New York, NY. [Google Scholar]
- 85.Niu B, Paulson JN, Zheng X, Kolter R. 2017. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci U S A 114:E2450–E2459. doi: 10.1073/pnas.1616148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zabat MA, Sano WH, Cabral DJ, Wurster JI, Belenky P. 2018. The impact of vegan production on the kimchi microbiome. Food Microbiol 74:171–178. doi: 10.1016/j.fm.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gregory Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 90.Andrei A-Ş, Robeson MS II, Baricz A, Coman C, Muntean V, Ionescu A, Etiope G, Alexe M, Sicora CI, Podar M, Banciu HL. 2015. Contrasting taxonomic stratification of microbial communities in two hypersaline meromictic lakes. ISME J 9:2642–2656. doi: 10.1038/ismej.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 92.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mathur S, Singh R. 2005. Antibiotic resistance in food lactic acid bacteria: a review. Int J Food Microbiol 105:281–295. doi: 10.1016/j.ijfoodmicro.2005.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Partial 16S rRNA gene sequences of bacterial isolates have been deposited in NCBI (see Table 1 for accession numbers). Amplicon sequence data have been deposited in the NCBI database as BioProject PRJNA510140 with Sequence Read Archive accession numbers SAMN10612113 to SAMN10612199.