Methylotrophic metabolism has gained huge attention considering its broad application in ecology, agriculture, industries, and human health. The genus Mycobacterium comprises both pathogenic and nonpathogenic species. Several members of this genus are known to utilize methanol as the sole carbon source for growth. Although various pathways underlying methanol utilization have been established, the regulation of methylotrophic metabolism is not well studied. In the present work, we explore the regulation of methanol metabolism in M. smegmatis and discover a dedicated two-component system (TCS), MnoSR, that is involved in its regulation. We show that the loss of MnoSR renders the bacterium incapable of utilizing methanol and 1,3-propanediol as the sole carbon sources. Additionally, we establish that MnoS acts as the common sensor for the alcohols in M. smegmatis.
KEYWORDS: methylotrophic metabolism, Mycobacterium, alcohol metabolism, histidine kinase, methanol oxidation, two-component system
ABSTRACT
Mycobacterium smegmatis and several other mycobacteria are able to utilize methanol as the sole source of carbon and energy. We recently showed that N,N-dimethyl-p-nitrosoaniline (NDMA)-dependent methanol dehydrogenase (Mno) is essential for the growth of M. smegmatis on methanol. Although Mno from this bacterium shares high homology with other known methanol dehydrogenases, methanol metabolism in M. smegmatis differs significantly from that of other described methylotrophs. In this study, we dissect the regulatory mechanism involved in the methylotrophic metabolism in M. smegmatis. We identify a two-component system (TCS), mnoSR, that is involved in the regulation of mno expression. We show that the MnoSR TCS is comprised of a sensor kinase (MnoS) and a response regulator (MnoR). Our results demonstrate that MnoS undergoes autophosphorylation and is able to transfer its phosphate to MnoR by means of phosphotransferase activity. Furthermore, MnoR shows specific binding to the putative mno promoter region in vitro, thus suggesting its role in the regulation of mno expression. Additionally, we find that the MnoSR system is involved in the regulation of MSMEG_6239, which codes for a putative 1,3-propanediol dehydrogenase. We further show that M. smegmatis lacking mnoSR is unable to utilize methanol and 1,3-propanediol as the sole carbon source, which confirms the role of MnoSR in the regulation of alcohol metabolism. Our data, thus, suggest that the regulation of mno expression in M. smegmatis provides new insight into the regulation of methanol metabolism, which furthers our understanding of methylotrophy in mycobacteria.
IMPORTANCE Methylotrophic metabolism has gained huge attention considering its broad application in ecology, agriculture, industries, and human health. The genus Mycobacterium comprises both pathogenic and nonpathogenic species. Several members of this genus are known to utilize methanol as the sole carbon source for growth. Although various pathways underlying methanol utilization have been established, the regulation of methylotrophic metabolism is not well studied. In the present work, we explore the regulation of methanol metabolism in M. smegmatis and discover a dedicated two-component system (TCS), MnoSR, that is involved in its regulation. We show that the loss of MnoSR renders the bacterium incapable of utilizing methanol and 1,3-propanediol as the sole carbon sources. Additionally, we establish that MnoS acts as the common sensor for the alcohols in M. smegmatis.
INTRODUCTION
Methanol is one of the major C1 compounds found in nature and is a crucial carbon source for methylotrophic bacteria (1–3). Methanol metabolism has remained a topic of interest due to its wide range of industrial and agricultural applications (4). Methylotrophs also play an important role in interaction with plants to execute promising ecological applications (5). Methanol dehydrogenase is required for the conversion of methanol into formaldehyde, which is the primary and critical step in methanol utilization as the sole carbon source (2, 3). Notwithstanding the differences in the biochemical and structural properties of methanol dehydrogenases from different bacteria, several studies on the regulation of methylotrophic metabolism have shown that methanol dehydrogenases are overproduced by bacteria during growth on methanol (6–13). This suggests that the production of methanol dehydrogenase is the underlying cellular response to the presence of methanol in the extracellular environment and is conserved among the majority of methylotrophs. Thus, it is both important and interesting to explore the regulatory mechanism(s) involved in C1 metabolism.
The mode of upregulation of methanol dehydrogenase expression in the presence of methanol differs among the known methylotrophs and involves different mechanisms. Earlier studies have suggested the involvement of a two-component system (TCS) in the regulation of methanol dehydrogenase expression in Gram-negative methanol-utilizing bacteria, such as Methylobacterium extorquens and Paracoccus denitrificans (14–16). In addition to the C1 compounds, the expression of methanol dehydrogenase has also been shown to be regulated by the lanthanides (17, 18). In Mycobacterium sp. strain JC1, methanol dehydrogenase expression has been shown to be positively regulated by the TetR family of transcriptional regulator MdoR; additionally, MdoR has also been reported to be essential for the growth of bacteria on methanol (19). In methylotrophs, methanol dehydrogenase is not the only protein overproduced during growth on methanol. Several reports suggest that in methylotrophic metabolism, genes involved in the serine cycle and RuMP pathway are overexpressed in M. extorquens and Bacillus methanolicus, respectively (13, 20, 21). Whereas QscR, a LuxR family of transcriptional regulators, has been shown to regulate serine cycle genes in M. extorquens, little is known about the regulation of RuMP cycle genes in B. methanolicus (20). Thus, there are multiple means by which bacteria regulate methanol metabolism genes in the presence of methanol.
Growth of Mycobacterium smegmatis and other mycobacteria is supported by a range of carbon sources, and the metabolic pathways for their utilization in the majority of cases have been elucidated (22). We have previously shown that M. smegmatis harbors an N,N-dimethyl-p-nitrosoaniline (NDMA)-dependent methanol dehydrogenase (Mno), which is required for methanol utilization by the bacterium (23). However, the factors governing mno expression in the presence of methanol are not known. In the present study, we identify and characterize the factors that regulate methanol metabolism in M. smegmatis. Among the various regulators found in M. smegmatis that control gene expression, the TetR family of transcription regulators (TFTRs) is the most studied (24), and several of the characterized TFTRs present in M. smegmatis are required for the regulation of oxidoreductases (24). Additionally, TCSs in M. smegmatis are also known to regulate genes involved in essential cellular processes, such as nutrient acquisition, physiological response to hypoxia, and virulence in certain cases (25–27). Here, we identify and characterize a two-component system, MnoSR, which is involved in methylotrophic metabolism regulation in M. smegmatis. We report that the MnoSR TCS is composed of a sensor kinase, MnoS, which phosphorylates its cognate response regulator, MnoR. Together, these two proteins in the presence of methanol in the culture medium regulate mno expression. Our data suggest that the mnoSR two-component system is dedicated for the metabolism of alcohols in M. smegmatis. Our study forms the first report on the identification of a TCS involved in the regulation of an alcohol dehydrogenase in M. smegmatis, which will further enhance our understanding of the regulation of methylotrophic metabolism in bacteria.
RESULTS
Identification of a two-component system involved in the regulation of methanol metabolism in Mycobacterium smegmatis.
We previously showed that M. smegmatis produces an NDMA-dependent methanol dehydrogenase (Mno), encoded by MSMEG_6242 or mno, that is essential for bacterial growth on methanol as the sole carbon source (23). We further showed that mno is induced in the presence of methanol in the culture medium, irrespective of the presence of glucose (as an additional carbon source) (23). Here, we asked what regulates mno expression and carried out a detailed in silico analysis of the M. smegmatis genome available at Mycobrowser (https://mycobrowser.epfl.ch) (28). We identified a gene, MSMEG_6244, that codes for a putative TetR family of transcriptional regulators (TFTRs) and is divergent to mno (Fig. 1). This is similar to a previous report that suggested the involvement of MdoR, a TFTR, in the regulation of methanol dehydrogenase of Mycobacterium strain JC1 (19). However, we could not find any significant sequence similarity between MSMEG_6244 and MdoR from Mycobacterium JC1 (data not shown). Nevertheless, to verify the role of MSMEG_6244 in mno expression regulation, we generated an MSMEG_6244 knockout in M. smegmatis by following the method as described previously (23) and confirmed the same by PCR (see Fig. S1 in the supplemental material) and DNA sequencing. We found that a deletion of MSMEG_6244 hampered neither the growth of M. smegmatis on methanol nor the production of Mno (Fig. 2A and B), suggesting that MSMEG_6244 has no role in methanol metabolism in M. smegmatis.
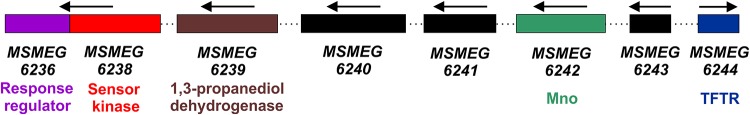
FIG 1.
Arrangement of mnoSR two-component system and the neighboring genes on the M. smegmatis mc2155 genome. The distribution of various genes on the M. smegmatis genome is shown. The direction of gene expression is marked with arrows. Genes considered for the present study are shown in different colors and are labeled; their protein products are also mentioned as sensor kinase (MSMEG_6238), response regulator (MSMEG_6236), Mno (MSMEG_6242), putative 1,3-propanediol dehydrogenase (MSMEG_6239), and a TetR family of transcriptional regulator (MSMEG_6244) divergent to mno.
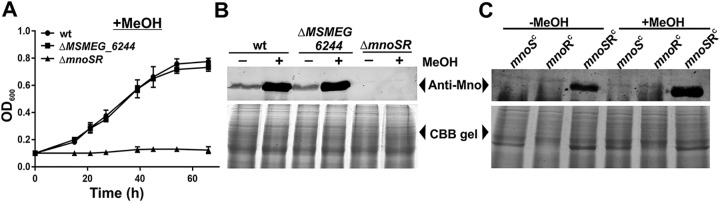
FIG 2.
mnoSR regulates mno expression and is essential for methanol-dependent growth of M. smegmatis. (A) Growth of wild-type (wt), ΔMSMEG_6244, and ΔmnoSR strains of M. smegmatis in the presence of methanol (+MeOH) as the sole carbon source. OD600 of the culture medium was recorded at the given time points and plotted. Time 0 represents the addition of methanol at an OD600 of ∼0.1. The error bars denote the standard deviation in the readings among the three sets of independent experiments. An increase in OD600 is observed only in the case of the wt and ΔMSMEG_6244 strains. (B) Western blotting data to examine Mno production in the presence (+) and absence (–) of methanol (MeOH). Two percent glucose is present in all of the cases. Mno production is observed in the presence of methanol in the wild-type (wt) and ΔMSMEG_6244 strains but not in the mnoSR knockout (ΔmnoSR) strain. (C) Western blot of Mno production in the absence (–MeOH) and presence (+MeOH) of methanol after complementing the mnoSR knockout with mnoS (mnoSC), mnoR (mnoRC), or both mnoS and mnoR (mnoSRC). Methanol-dependent induction of Mno is observed only when complementation is carried out with both mnoS and mnoR. In both panels B and C, the Coomassie blue (CBB)-stained gel is shown to confirm protein loading.
Our in silico analysis of the M. smegmatis genome also revealed the presence of a putative two-component system (TCS) formed by the products of two genes coding for a sensor kinase (MSMEG_6238; hereafter referred to as mnoS) and a response regulator (MSMEG_6236; hereafter referred to as mnoR) in the vicinity of mno (Fig. 1). A DOOR database (29) analysis confirmed that both of the genes form an operon (mnoSR). Furthermore, the stop codon of mnoS overlaps with the start codon of mnoR. TCSs are generally known to be present as operons in mycobacteria and other bacteria (30, 31). Methanol oxidation in several methylotrophs has been shown to be regulated by TCSs (14, 15). Thus, in order to validate whether mno in M. smegmatis is regulated by this TCS, we proceeded with constructing an mnoSR knockout strain of M. smegmatis (ΔmnoSR), having the deletion of both MSMEG_6236 and MSMEG_6238. The knockout was prepared by following the method as described previously (23, 32) and confirmed by PCR (see Fig. S1) and DNA sequencing. Interestingly, our M. smegmatis ΔmnoSR strain is unable to grow when methanol is present as the sole carbon source (Fig. 2A). Additionally, in the ΔmnoSR strain, methanol-dependent Mno production is completely lost as judged by Western blotting (Fig. 2B). These results suggest that Mno production in M. smegmatis is regulated by the MnoSR TCS. To further confirm that both MnoS and MnoR are together required for Mno production and that there is no other cross-reacting TCS, we performed the complementation of M. smegmatis ΔmnoSR by expressing either mnoS (pADatMnoS) or mnoR (pADatMnoR) or both mnoS and mnoR (pADatMnoSR) from an acetamide-inducible promoter (33, 34). Our data show that Mno production could be restored only when the cells were transformed with pADatMnoSR, which coexpressed mnoS and mnoR (Fig. 2C). This, thus, confirms that MnoS and MnoR together form a cognate TCS, which is required for Mno production. The complementation data further rule out the possibility of any cross-reacting TCS component. In these experiments, the expression of MnoS, MnoR, and MnoSR from the pADatMnoS, pADatMnoR, and pADatMnoSR vectors, respectively, in ΔmnoSR cells was confirmed by Western blotting using an anti-His antibody (see Fig. S2 in the supplemental material). Taken together, our data strongly indicate that the MnoSR TCS carries out the positive regulation of Mno and is essential for the growth of M. smegmatis on methanol.
Since the mnoSR knockout and complementation data correctly identified them as the TCS involved in mno regulation, we did not attempt to decipher the role of other genes, such as MSMEG_6240, MSMEG_6241, and MSMEG_6243, present in the vicinity of mno (Fig. 1). While MSMEG_6240 and MSMEG_6241 code for conserved hypothetical protein and putative ATPase, respectively, MSMEG_6243 (located upstream of mno) encodes a hypothetical protein containing the DUF1348 domain. It is likely that the products of these genes are involved in other aspects of mycobacterial physiology, and an examination of them is beyond of the scope of this study.
MnoS and MnoR form the cognate proteins of the two-component system.
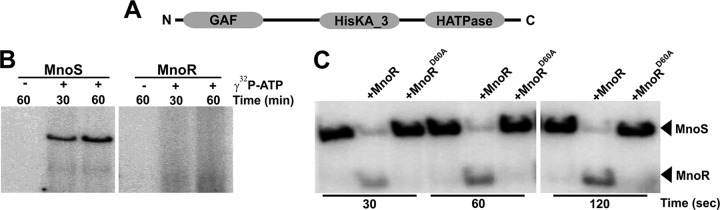
TCSs are involved in the signal transduction process in bacteria. The signaling between the factors of the TCS generally occurs by the autophosphorylation of sensor kinase, which then transfers the phosphate group to its response regulator protein. In most cases, the regulatory protein then binds to DNA and modulates the target gene expression (35). Our in silico analysis, carried out by the Conserved Domain Database (CDD) and Simple Modular Architecture Research Tool (SMART) (36, 37), shows that MnoS contains a GAF domain (named after the cyclic GMP [cGMP]-specific phosphodiesterase, adenylyl cyclase, and FhlA proteins) at its N terminus. This is followed by a histidine kinase domain (HisKA_3) comprising the kinase core and a histidine kinase-like ATPase (HATPase) domain responsible for the ATPase activity of the protein (Fig. 3A). Furthermore, the domain architecture of MnoS was found to be similar to that of other known sensor kinases, viz., Rv2027c and DevS of Mycobacterium tuberculosis (38). In order to confirm whether MnoS is indeed a sensor kinase, we examined the autophosphorylation activity of MnoS in the presence of [γ-32P]ATP. We first cloned, expressed, and purified 6×histidine-tagged MnoS, and purity was checked by SDS-denaturing PAGE (see Fig. S3 in the supplemental material). We next incubated the purified MnoS with [γ-32P]ATP and performed phosphorimaging of the proteins separated by SDS-PAGE. Our data show that MnoS indeed undergoes phosphorylation, thus confirming that MnoS is a sensor kinase that can undergo autophosphorylation (Fig. 3B).
FIG 3.
MnoS shows autophosphorylation and phosphotransferase activity in vitro. (A) Conserved domains present in the MnoS protein from the N to C terminus. GAF domain corresponds to a domain present in cGMP-specific phosphodiesterase, adenylyl cyclase, and FhlA proteins. The histidine kinase domain is represented as HisKA_3, whereas the HATPase domain is responsible for ATPase activity of the protein. (B) Autoradiogram for the autophosphorylation activities of MnoS and MnoR. The phosphorylation activity was carried out for the specified time (in minutes) in the presence or absence of [γ-32P]ATP. The data show that MnoS undergoes autophosphorylation upon incubation with [γ-32P]ATP, whereas MnoR is unable to perform autophosphorylation reaction under similar conditions. (C) Autoradiogram for the phosphotransferase activity of MnoS (presented in all of the lanes) toward either MnoR or MnoRD60A proteins. The position of both MnoS and MnoR proteins in the autoradiogram is marked. The reaction was carried out for the specified time (in seconds). Only the wild-type MnoR undergoes phosphorylation upon incubation with phosphorylated MnoS, which is accompanied by the loss of signal from MnoS.
The phosphotransferase reaction between the sensor kinase and cognate response regulator is the basis of the two-component-based signaling process (39). To confirm whether the MnoR is the response regulator protein that can accept phosphate from the sensor kinase MnoS, we cloned, expressed, and purified MnoR and performed the autophosphorylation and the phosphotransferase reactions in the absence and presence of MnoS. Our data show that MnoR, unlike MnoS, is incapable of demonstrating autophosphorylation activity (Fig. 3B). However, when incubated with the phosphorylated MnoS, MnoR is able to accept the phosphate group from MnoS, resulting in the dephosphorylation of MnoS and consequent phosphorylation of MnoR (Fig. 3C). This experiment confirms the phosphotransferase activity between the two proteins.
Sensor kinase in the majority of cases phosphorylates the conserved aspartate residue on the response regulator protein that eventually undergoes a conformational change and performs required function (35). A previous report suggests that Asp54 in DevR is the key residue that undergoes phosphorylation upon incubation with the cognate sensor kinase DevS, and mutation of Asp54 to valine renders DevR incapable of undergoing phosphorylation (40). A sequence alignment between DevR and MnoR using the EMBOSS Needle pairwise alignment tool (41) revealed the presence of conserved Asp60 in the latter (see Fig. S4 in the supplemental material), which also corresponded to the Asp54 of DevR that has been shown to undergo phosphorylation (40). The phosphorylation site of MnoR was further confirmed to be Asp60 as indicated by the prediction carried out at UniProt (accession number A0R5L8). Thus, in order to examine the specific phosphate acceptor site on the MnoR protein, we generated an MnoR D60A mutant (MnoRD60A) and carried out the phosphotransferase activity. We observed that the MnoRD60A mutant was not phosphorylated by MnoS (Fig. 3C), indicating the significance of the D60 residue in MnoR. Our data, thus, suggest that MnoR is the cognate response regulator of MnoS and that Asp60 is the site for the phosphorylation on MnoR. Thus, together, these proteins form the cognate proteins of the two-component system.
MnoR specifically binds to the mno promoter region.
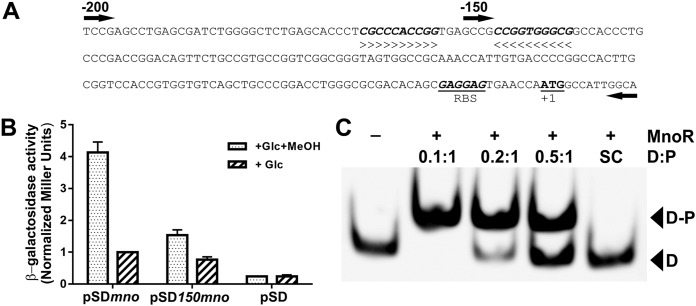
In order to successfully induce mno expression, MnoR must bind to the mno promoter region. We carried out a detailed analysis of 200 bp upstream of the mno translation start site and identified one inverted repeat sequence (Fig. 4A), which is likely the MnoR binding site. We next cloned this DNA segment upstream of a lacZ reporter gene in the promoterless Escherichia coli-Mycobacterium shuttle vector pSD5b (42), thus generating pSDmno, which was used for the transformation of M. smegmatis. Our experiments show the methanol-dependent lacZ expression (measured as β-galactosidase activity) from the 200-bp region, indicating the presence of an inducible promoter upstream of mno (Fig. 4B). We next cloned a 150-bp DNA segment upstream of the mno translation start site in pSD5b in order to truncate the inverted repeat sequence identified above and generated pSD150mno. Interestingly, β-galactosidase activity data suggest that pSD150mno is no longer methanol inducible (Fig. 4B). We conclude that the loss of methanol-dependent induction from pSD150mno is due to the disruption of the inverted repeat sequence.
FIG 4.
MnoR binds specifically to the mno promoter region. (A) DNA sequence upstream of mno start codon ATG (marked as +1). The ∼200-bp region used for the promoter assays and the EMSA is marked as -200, whereas a smaller truncated promoter region of ∼150 bp is marked as -150. The identified inverted repeats are bold and italicized. The putative ribosome binding site for the translation process is marked as RBS. (B) Promoter activity of 200-bp and 150-bp DNA segments upstream of mno translation start site in the presence and absence of methanol (MeOH); 2% glucose (Glc) is present in all of the conditions. Empty vector pSD was used as negative control. β-Galactosidase activity in the form of normalized Miller units is plotted. Higher promoter activity is observed in the case of the 200-bp mno promoter region in the presence of methanol, whereas the 150-bp promoter region shows poor methanol-dependent induction. (C) Electrophoretic mobility shift assay (EMSA) carried out in the absence (–) or presence (+) of MnoR and the ∼200-bp biotinylated mno promoter region. Various mole ratios of DNA/protein (D:P) were used in the experiment. MnoR-bound promoter region complex (D-P) moves more slowly on the nondenaturing acrylamide gel than the free DNA (D). Nonbiotinylated DNA from the same region was used as specific competitor (SC), which confirms that the binding of MnoR to the probe is specific.
We next examined the binding of MnoR to the mno promoter region by carrying out an electrophoretic mobility shift assay (EMSA) with purified MnoR and the biotin-labeled 200-bp DNA segment used in promoter assays. Our data show the successful binding of MnoR to the 200-bp mno promoter region in a concentration-dependent manner (Fig. 4C). Furthermore, this binding is found to be specific since the interaction between DNA and protein could be masked by the use of nonbiotinylated 200 bp mno DNA as a specific competitor (Fig. 4C). Our observations allow us to infer that MnoR binds to the mno promoter region.
MnoSR TCS also regulates MSMEG_6239 expression but is not required for formaldehyde detoxification.
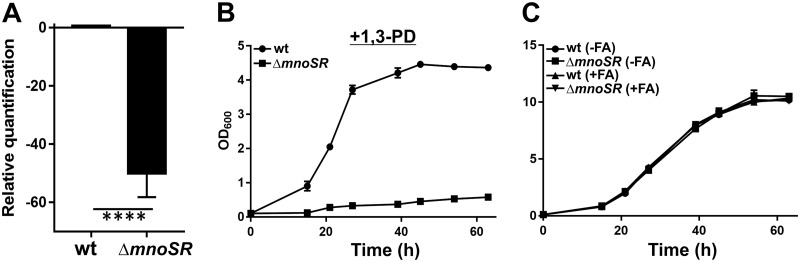
The bioinformatics analysis of the M. smegmatis genome carried out here also revealed MSMEG_6239, which codes for a putative 1,3-propanediol dehydrogenase, in the vicinity of mnoSR. MSMEG_6239 shares 77% sequence similarity with 1,3-propanediol dehydrogenase from Saccharopolyspora erythraea (UniProt accession number A4FCA4). This observation tempted us to monitor the effect of the mnoSR deletion on the expression of MSMEG_6239. We, thus, measured the relative expression of MSMEG_6239 in both wild-type and ΔmnoSR M. smegmatis strains using reverse transcriptase quantitative PCR (RT-qPCR). We observed that the expression of MSMEG_6239 was drastically reduced in the ΔmnoSR strain compared to the level in the wild type (Fig. 5A). Additionally, the growth of the ΔmnoSR strain on 1,3-propanediol as the sole carbon source was found to be hampered compared to that of the wild type, suggesting that the ΔmnoSR strain is unable to utilize 1,3-propanediol (Fig. 5B). Taken together, our data suggest that the MnoSR TCS regulates the expression of both mno and MSMEG_6239, and thus, it is essential for M. smegmatis growth on methanol and 1,3-propanediol.
FIG 5.
mnoSR TCS is essential for MSMEG_6239 expression and 1,3-propanediol utilization but is not required for formaldehyde metabolism. (A) RT-qPCR-based expression data of MSMEG_6239 (which codes for putative 1,3-propanediol dehydrogenase) in both the wild-type (wt) and the ΔmnoSR cells. Drastic downregulation of MSMEG_6239 in the ΔmnoSR strain is observed compared to the wild type. RT-qPCR was performed thrice as independent sets of experiments. The error bars denote the standard deviation among the three experiments. ****, P value = highly significant. (B) Growth of M. smegmatis wild-type (wt) and ΔmnoSR cells in the presence of 1,3-propanediol (+1,3-PD) as the sole carbon source in the culture medium. The growth was monitored by measuring OD600 with time. Plot shows that ΔmnoSR cells are unable to utilize 1,3-propanediol. (C) Growth of wild-type (wt) and ΔmnoSR cells in absence (-FA) and presence (+FA) of 1 mM formaldehyde measured as OD600 with time. Two percent glucose is present in all of the conditions. Growth rates of both the wild-type and the ΔmnoSR cells remain similar at the formaldehyde concentration used in this experiment. In both panels B and C, data presented are an average of three independent experiments with error bars denoting standard deviation.
It is worth mentioning here that the TCS-mediated regulatory mechanism is not just confined to the regulation of methanol oxidation in methylotrophs. A previous report on P. denitrificans suggests that the TCS is known to regulate both methanol and formaldehyde oxidation (14). We, therefore, examined the role of the MnoSR TCS in formaldehyde oxidation by challenging ΔmnoSR and wild-type M. smegmatis strains to sublethal concentrations of formaldehyde (23). Our results show that the loss of mnoSR does not affect the growth of M. smegmatis in the presence of formaldehyde (Fig. 5C). This allows us to conclude that the mnoSR TCS is confined to the regulation of only alcohol metabolism.
MnoSR functions as a bona fide alcohol-sensing two-component system that allows cross-induction of gene expression.
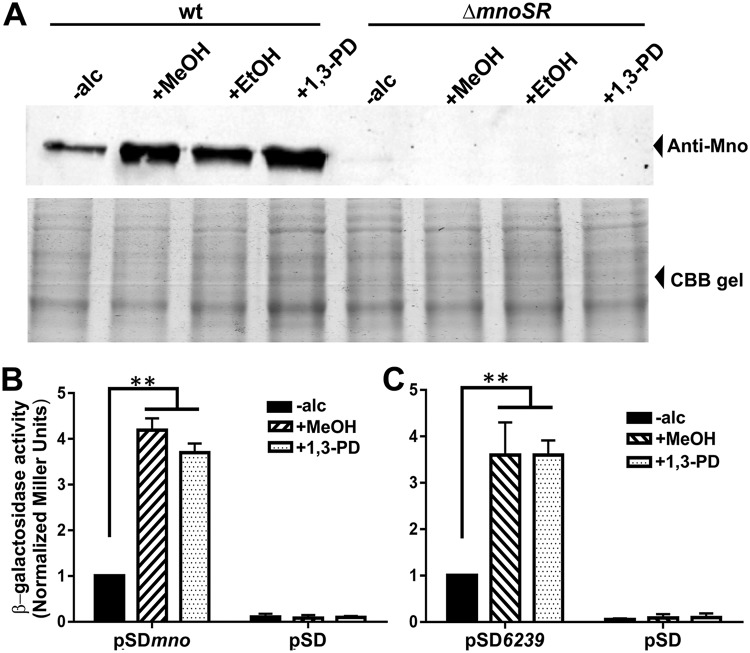
Loss of MnoSR leads to the downregulation of both Mno and putative 1,3-propanediol dehydrogenase production, which strongly indicates that expression of both of the genes is regulated by similar mechanisms. Since, here, one TCS is regulating different alcohol dehydrogenases, conceivably, these alcohol dehydrogenases should express even in the presence of their noncognate alcohols in the culture medium. Therefore, we first assessed the production of Mno in the presence of various alcohols such as methanol, ethanol, and 1,3-propanediol. Interestingly, we observed production of Mno in all of the alcohols (Fig. 6A); moreover, this expression required the presence of MnoSR since in its absence, the alcohol-dependent Mno production was lost in the ΔmnoSR strain (Fig. 6A). These data suggest that MnoSR functions as a dedicated TCS for various alcohols in M. smegmatis. We wish to add here that Mno is able to act upon various different alcohols in the oxidation reaction (23). Thus, it is valid to expect Mno production when an alcohol other than methanol is present in the culture medium, even though Mno is required primarily for methanol utilization in vivo (23).
FIG 6.
MnoSR functions as a bona fide alcohol-sensing two-component system that allows cross-induction of gene expression. (A) Expression of Mno monitored in M. smegmatis wild-type (wt) and ΔmnoSR cells in the absence (-alc) and presence of various alcohols, such as methanol (MeOH), ethanol (EtOH), and 1,3-propanediol (1,3-PD), by Western blotting using anti-Mno antibodies. Mno production occurs in the presence of all of the alcohols examined in this study and is observed only in the wild-type cells and not in the ΔmnoSR cells. CBB gel represents the Coomassie blue-stained SDS-PAGE gel showing the equal amounts of proteins in the samples processed for Western blotting. Promoter assays by measuring the β-galactosidase activity were carried out with an ∼200-bp promoter region of both mno (pSDmno [B]) and MSMEG_6239 (pSD6239 [C]) fused with lacZ in an E. coli-Mycobacterium promoterless shuttle plasmid, pSD5b, in the absence (-alc) and the presence of methanol (+MeOH) or 1,3-propanediol (+1,3-PD). The enzyme activity is presented as normalized Miller units with respect to -alc. The expression from both mno and MSMEG_6239 promoter regions is observed in the presence of either of the alcohols in the culture medium. The promoterless empty plasmid (pSD) acted as a negative control. The assays were performed at least thrice. Error bars depict the standard deviation. **, P = very significant.
We further cloned the ∼200-bp region upstream from the translation start site of MSMEG_6239 in pSD5b (42) ahead of the lacZ reporter gene to generate pSD6239. We next monitored the expression of lacZ from both the mno and MSMEG_6239 promoter regions by carrying out β-galactosidase assays in the absence and presence of methanol and 1,3-propanediol. We observed higher β-galactosidase activities from both promoter regions when the culture medium contained either methanol or 1,3-propanediol, which immediately suggested that both gene promoters respond to all of the alcohols (Fig. 6B and C); very low β-galactosidase activity was observed in the absence of alcohols, which corroborates our Western blotting data. Thus, a significantly higher amount of promoter activity in the presence of either methanol or 1,3-propanediol suggests that MnoSR allows for cross-induction of both mno and MSMEG_6239 genes by the addition of either of the alcohols in the culture medium. This further confirms the involvement of similar regulatory mechanisms for the expression of the two genes.
DISCUSSION
Gene expression regulation in M. smegmatis, a soil-dwelling microbe, is complex and deals with an abundance of regulatory factors, which extends from TFTRs to two-component systems (24, 26, 43). TCSs are of special interest due to their involvement in the survival of Mycobacterium tuberculosis and in establishing a successful infection (26). Various studied mycobacterial TCSs to date justify their presence in both pathogenic and nonpathogenic mycobacterial species and perform a wide range of functions from virulence, hypoxia, stress and survival during infection, nutrient sensing and uptake to development of antibiotic resistance (25–27, 40). Although several Mycobacterium species are able to utilize methanol as the sole source of carbon and energy (44), it is intriguing that they employ different and unique pathways for carbon assimilation during methylotrophic metabolism (23, 44, 45). A previous report suggests that the methanol dehydrogenase in Mycobacterium JC1 is under positive regulation of MdoR, a TFTR (19). Nevertheless, the mechanisms involved in the regulation of methanol metabolism are only poorly understood and have not been explored in detail in the mycobacterial methylotrophs.
Here, we have identified and characterized a two-component system, MnoSR, that is required for the establishment of the methylotrophic metabolism in M. smegmatis. We show that MnoSR not only regulates expression of methanol dehydrogenase gene (mno) but is also required for the expression of MSMEG_6239, which codes for a putative 1,3-propanediol dehydrogenase. Thus, MnoSR is essential for the growth of M. smegmatis on alcohols, such as methanol and 1,3-propanediol, as the sole carbon sources. Additionally, the unaffected growth of the ΔmnoSR strain in the presence of formaldehyde, a key intermediate of methanol metabolism (3), suggests that the regulation of methanol oxidation and formaldehyde detoxification in M. smegmatis differ from these processes in other mycobacterial species and require further exploration.
Although the loss of Mno production in the ΔmnoSR strain in the presence of any of the alcohols suggests that the MnoSR TCS responds to the presence of alcohols in the culture medium, we have previously shown that mno overexpression is observed in the presence of both methanol and formaldehyde (23). Hence, the regulation of Mno in M. smegmatis involving factors other than the MnoSR TCS cannot be ruled out, and it remains to be elucidated if an alcohol is the primary and/or a direct inducer for mno expression. Additionally, it may be hypothesized here that the GAF domain present in the N terminus of MnoS that is similar to DevS is involved in sensing and responding to small secondary messenger molecules, such as cGMP, cAMP, or cyclic-di-GMP (38, 46). Although the GAF domain is present in a number of sensory proteins (39, 46), it is not always necessary that it binds or responds to the cyclic nucleotides (47). Thus, it remains to be seen if global regulation of methylotrophy involves cyclic nucleotides as specific signals. Taken together, we believe that additional factors are involved in the regulation of methylotrophic metabolism as a whole in mycobacteria and that MnoSR forms a dedicated regulatory system for the initial step of methanol metabolism by acting as a sensor for methanol and other alcohols.
Our study presents a comprehensive analysis of the regulation of the utilization of methanol and other alcohols by involving MnoSR as the regulatory proteins. The observation that MnoSR also regulates the gene coding for a putative C3 alcohol dehydrogenase (i.e., 1,3-propanediol) suggests that MnoSR has far-reaching effects on mycobacterial physiology and is likely involved in functions beyond the regulation of methylotrophic metabolism. It is interesting to note that although methanol dehydrogenases from Mycobacterium JC1 and M. smegmatis share high homology, their modes of regulation of expression differ (19, 23). Thus, our current and previous findings together strongly suggest that the regulation of methylotrophic metabolism in M. smegmatis differs from that in other mycobacterial species and appears to be unique among the known methylotrophs (19, 23, 44, 45). We believe that our work on methylotrophy in M. smegmatis will help us to obtain a broader understanding of the gene expression regulation mechanisms functional in mycobacteria.
MATERIALS AND METHODS
Bacterial strain, media, and growth conditions.
Escherichia coli strain XL1-Blue was used for all of the cloning experiments, whereas production of recombinant proteins was performed in the BL21(DE3) strain. Both of the strains were grown in LB broth (Difco) at 37°C with constant shaking at 200 rpm. M. smegmatis strain mc2155 and its derivatives generated here were grown in MB7H9 broth (Difco) containing 2% glucose (as required) or 2% (vol/vol) of the desired alcohols, such as methanol, ethanol, or 1,3-propanediol, as carbon sources, wherever required, along with 0.05% Tween 80. Appropriate antibiotics were added in the media for all of the plasmid-bearing cultures at the concentrations reported elsewhere (23). When required, M. smegmatis cells were induced with 2% acetamide to monitor the expression of recombinant proteins.
Construction of genetic knockouts.
Construction and confirmation of mnoSR and MSMEG_6244 genetic knockouts were carried out by employing the strategy as described previously (23, 32). Upstream and downstream fragments of both of the genes were PCR amplified from M. smegmatis genomic DNA, whereas the Hygr cassette was PCR amplified from the pVV16 vector (obtained through BEI Resources, NIAID, NIH; Naked Plasmid pVV16 for expression in Mycobacterium smegmatis, catalog number NR-13402). Allelic exchange substrate (AES) was then constructed using the primers listed in Table 1. Linear AES DNA fragments were then electroporated in M. smegmatis cells containing pJV53 (kind gift from Graham Hatfull, University of Pittsburgh, USA; Addgene plasmid number 26904). Plasmid curing of pJV53 was performed essentially as described previously (48). Plasmid-cured cells were further used for complementation.
TABLE 1.
List of oligonucleotides used in the present studya
| Oligonucleotide | Sequence (5′–3′) | Purpose |
|---|---|---|
| MnoSFor | ATGGCCGAAGCGGCCCGCACC | Cloning of mnoS in pMS-QS-CHS and pSS4 |
| MnoSRev | GCAGAATGTCGTTGAGACGGTTGAGC | |
| MnoRFor | ATGACCGTCACGACGCGCGAG | Cloning of mnoS in pMS-QS-CHS and pSS4 |
| MnoRRev | GATCAACCCGCGCTTGCTC | |
| MnoSUpFor | CAGGTCGGGGGCCTGCTCGACC | Amplification of mnoS upstream fragment |
| MnoSUpRev | CCACGTACATCACCACAAGCACCCACTCGGCGTCGAGG | |
| MnoSHygFor | CGAGTGGGTGCTTGTGGTGATGTACGTGGCGAACTCC | Amplification of Hygr cassette |
| MnoSHygRev | CGTACACGGCCTGATCCGGGGGGCGTCAGG | |
| MnoSDownFor | CCTGACGCCCCCCGGATCAGGCCGTGTACGCGGCGAGC | Amplification of mnoS downstream fragment |
| MnoSDownRev | GGTGTTCACCGGCGTCGTGCGTTCC | |
| MnoSLongFor | CTGGCGTTCACCAACGCGATCC | PCR confirmation of ΔmnoS |
| MnoSLongRev | CGATCTGACCCCTGACGAACTGTCC | |
| MnoRD60AFor | CGACGTGGTGCTGCTGGCCCTCAAGCTCTCGGCCGGATC | Site directed mutation for generating MnoRD60A |
| 6244Upfor | AGAACTCTAGAGGTGGGCGAGGGTGC | Amplification of MSMEG_6244 upstream fragment |
| 6244UpRev | CGTACATCACCACGCTCGCGTGCCGCGTC | |
| 6244HygFor | GCACGCGAGCGTGGTGATGTACGTGGCGAAC | Amplification of Hygr cassette |
| 6244HygRev | CTAACCGCCTGAGGGATCCGGGGGGCGTC | |
| 6244DownFor | GACGCCCCCCGGATCCCTCAGGCGGTTAG | Amplification of MSMEG_6244 downstream fragment |
| 6244DownRev | GCTGCTGTTCGGGTTTGGGTCGTTC | |
| 6244LongFor | CAGGTCCGGGCAGCTGACACCACGG | PCR confirmation of ΔMSMEG_6244 |
| 6244LongRev | GCTGTTGTCGGTATCGCCACAGCATTACC | |
| rpoBRTFor | TCGATGTCACTGTCCTTCTCGGATC | RT-qPCR for rpoB expression |
| rpoBRTRev | GACCGTCTGGCTCTTGATCTC | |
| mnoRTFor | TCTGCTTGTTGGTGGACTTG | RT-qPCR for mno expression |
| mnoRTRev | GTCGAACCCCAAGGACTACA | |
| 6239RTFor | GAAATCGTGTTCGGCATCGATTCG | RT-qPCR for MSMEG_6239 expression |
| 6239RTRev | CTCCAGACCTGCGGGGTCACG | |
| mnopSDRev | GCATGCCAATGGCCATTGGTTCACTCCTCGCTG | Cloning of 200-bp mno fragment upstream from start codon in pSD5b |
| mno250pSDFor | ACGACCATCTAGAGCCTGAGCGATC | |
| NobiotinpSD | CCACTGCAGTGCATATGGAAGTGATTCC | PCR amplification of probe for EMSA |
| biotinpSD | Biotin-CCACTGCAGTGCATATGGAAGTGATTCC | |
| 6239pSDrev | ACCGCATGCGGCGCGGACTCCACCTGC | Cloning of 200-bp MSMEG_6239 fragment upstream from start codon in pSD5b |
| 6239200pSDFor | GTATGCGACAAGGTGGTCGTCG |
Sequence of each oligonucleotide from 5′ to 3′ is given. The purpose of each oligonucleotide in the present study is also mentioned for easy reference.
Cloning of mnoS and mnoR.
Sequences of mnoS (MSMEG_6238) and mnoR (MSMEG_6236) were obtained from Mycobrowser knowledge base (28). M. smegmatis genomic DNA was used as a template for PCR amplification of the genes using primers given in Table 1. PCR products of the genes mnoS and mnoR were cloned in E. coli expression vector pMS-QS-CHS (49) to generate pADt7MnoS and pADt7MnoR, respectively. Plasmids pMV261 (carrying the hsp60 promoter [50]) and pMVAcet (carrying a mycobacterial acetamide-inducible promoter system [33, 34]) were further modified to yield pSS1 (23) and pSS4, respectively, which helped in blunt-end cloning experiments. MnoS and MnoR were cloned in pSS4 to obtain pADatMnoS and pADatMnoR, respectively. The plasmids were subsequently used to express proteins in mycobacteria. Additionally, coexpression of mnoS and mnoR in the mycobacterial expression vector was achieved by cloning mnoS and mnoR in tandem in pSS4 to generate pADatMnoSR. Colony PCR was performed to screen for positive clones that were further confirmed by DNA sequencing. The MnoRD60A mutant was constructed by performing site-directed mutagenesis as per the described protocol (51) using the primer listed in Table 1 to generate pADt7MnoRD60A.
Real-time PCR for the relative mRNA expression.
To monitor the relative expression of MSMEG_6239 in both the wild type and the ΔmnoSR strain, RNA isolation, cDNA synthesis, and quantitative PCR (qPCR) were performed from the log-phase cells (optical density at 600 nm [OD600] of ∼0.8) essentially as described previously (23). Relative expression level was normalized against the expression of the internal control gene, rpoB. Primers used in the RT-qPCR are listed in Table 1. P values generated from a two-tailed Student’s t test were considered to calculate the level of significance within the experiments performed as three independent sets.
Protein expression and purification.
E. coli BL21(DE3) cells were transformed with pADt7MnoS, pADt7MnoR, and pADt7MnoRD60A for the expression and purification of MnoS, MnoR, and MnoRD60A, respectively. For the expression of recombinant proteins, E. coli BL21(DE3) cells were induced at an OD600 of ∼0.6 by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the protein induction was allowed to take place at 22°C for 12 h. Standard Ni-nitrilotriacetic acid (NTA) column chromatography was performed for the purification of MnoR and MnoRD60A as discussed previously for other proteins (33). MnoS purification was carried out as described elsewhere (52). Both of the eluted proteins were assessed by SDS-PAGE for purity. Purified proteins were dialyzed against buffer containing 40 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1 mM dithiothreitol (DTT), and 40% glycerol, and stored at −20°C until further use. Quantification of proteins was carried out by Bradford assay (Bio-Rad) as per the manufacturer’s instructions.
In vitro kinase and phosphotransferase activities of MnoS.
Purified MnoS and MnoR at a concentration of 10 μM were subjected to autokinase activity by following the method described previously (40). Briefly, the proteins were incubated with 10 μCi of [γ-32P]ATP (Brit, India) in a buffer containing 50 mM Tris-Cl (pH 8.0), 50 mM KCl, 10 mM MgCl2, and 50 μM ATP) at 25°C for 60 min. To assess the phosphotransferase activity, MnoR or MnoRD60A at a concentration of 10 μM was incubated with MnoS in the reaction mixture mentioned above for specified times. In all of the cases, the reaction mixtures were subsequently mixed with SDS loading dye containing 6 M urea, loaded on an SDS-PAGE gel without boiling, and electrophoresed. The gel was further dried and exposed to a phosphor screen, and the autoradiogram was recorded on a Typhoon FLA 9000 phosphorimager (GE Healthcare).
Analysis of expressed proteins by immunoblotting.
To monitor the expression of various proteins, M. smegmatis cells harboring required plasmids were induced at log phase with 2% acetamide, wherever required, and harvested after 5 h. Assessment of the protein expression by immunoblotting was carried out essentially as described before (23). Immunoblotting of Mno was carried out using anti-Mno antibody raised in rabbit (Bioneeds India Pvt. Ltd.). For proteins carrying the hexa-histidine tag, anti-His antibody raised in mouse (Sigma-Aldrich) was used. Blots were further probed with either anti-mouse IgG DyLight 680-conjugated secondary antibody or anti-rabbit IgG DyLight 800-conjugated secondary antibody, as required. Blots were developed on an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA). The protein amounts in all of the samples were quantified by Bradford assay, and equal amounts were loaded on the gel for the analysis of protein expression.
Generation of promoter-reporter constructs and β-galactosidase assay.
To assess mno and MSMEG_6239 promoter activity in the presence and absence of methanol and 1,3-propanediol, putative promoter regions of mno and MSMEG_6239 were cloned in a promoterless E. coli-Mycobacterium shuttle plasmid, pSD5b (42). Approximately 150- and 200-bp regions upstream from the translation start site of mno and a 200-bp region upstream from the translation start site of MSMEG_6239 were PCR amplified using the primers mentioned in Table 1 and cloned in the pSD5b vector between the SphI and XbaI sites to generate pSDmno, pSD150mno, and pSD6239, respectively. Positive clones were verified by DNA sequencing. For the β-galactosidase assay, M. smegmatis cells carrying the required plasmid were grown in the presence or absence of the desired alcohol; log-phase cells were used to carry out the assay by following the method as described before (53, 54).
Interaction of MnoR and mno promoter by electrophoretic mobility shift assay.
EMSA was performed using the biotin-labeled DNA fragment containing the putative mno promoter region. The DNA fragment was PCR amplified from pSDmno and using the primers listed in Table 1; the reverse primers NobiotinpSD and biotinpSD were used to obtain nonbiotinylated or biotinylated DNA fragments, respectively. Purified MnoR and the biotinylated DNA probe were incubated in the binding buffer (40 mM Tris-acetate [pH 8.3], 10 mM magnesium acetate, 1 mM EDTA, 150 mM NaCl, 5% glycerol, and 5 mM 2-mercaptoethanol) for 30 min at 25°C. The DNA-MnoR complex was separated on a 7% nondenaturing acrylamide gel at 10 V/cm for 70 min at 4°C and was further detected by using the LightShift chemiluminescent EMSA kit (Thermo Fisher Scientific) following the manufacturer’s instructions. For the specific competition experiments, nonbiotinylated DNA was added to the reaction mixture before PAGE separation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nimisha Mishra, Gayatri Nimbalkar, and Dilsha C. for help with the experiments. We also thank Surya P. Seniya for the pSS4 plasmid. The pVV16 plasmid was obtained from BEI resources, NIAID, NIH. The pJV53 plasmid was a kind gift from Graham Hatfull, University of Pittsburgh.
A.A.D. thanks the Council of Scientific and Industrial Research (CSIR), Government of India, for the senior research fellowship. This work is supported by intramural funds from IISER Bhopal to V.J.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00535-19.
REFERENCES
- 1.Fall R, Benson AA. 1996. Leaf methanol—the simplest natural product from plants. Trends Plant Sci 1:296–301. doi: 10.1016/1360-1385(96)88175-1. [DOI] [Google Scholar]
- 2.Anthony C. 1982. The biochemistry of methylotrophs. Academic Press, London, England. [Google Scholar]
- 3.Anthony C. 1991. Assimilation of carbon by methylotrophs. Biotechnology 18:79–109. [DOI] [PubMed] [Google Scholar]
- 4.Schrader J, Schilling M, Holtmann D, Sell D, Filho MV, Marx A, Vorholt JA. 2009. Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol 27:107–115. doi: 10.1016/j.tibtech.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Iguchi H, Yurimoto H, Sakai Y. 2015. Interactions of methylotrophs with plants and other heterotrophic bacteria. Microorganisms 3:137–151. doi: 10.3390/microorganisms3020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bystrykh LV, Vonck J, van Bruggen EF, van Beeumen J, Samyn B, Govorukhina NI, Arfman N, Duine JA, Dijkhuizen L. 1993. Electron microscopic analysis and structural characterization of novel NADP(H)-containing methanol: N,N'-dimethyl-4-nitrosoaniline oxidoreductases from the Gram-positive methylotrophic bacteria Amycolatopsis methanolica and Mycobacterium gastri MB19. J Bacteriol 175:1814–1822. doi: 10.1128/jb.175.6.1814-1822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bystrykh LV, Govorukhina NI, van Ophem PW, Hektor HJ, Dijkhuizen L, Duine JA. 1993. Formaldehyde dismutase activities in Gram-positive bacteria oxidizing methanol. J Gen Microbiol 139:1979–1985. doi: 10.1099/00221287-139-9-1979. [DOI] [Google Scholar]
- 8.Van Ophem PW, Van Beeumen J, Duine JA. 1993. Nicotinoprotein [NAD(P)-containing] alcohol/aldehyde oxidoreductases. Purification and characterization of a novel type from Amycolatopsis methanolica. Eur J Biochem 212:819–826. doi: 10.1111/j.1432-1033.1993.tb17723.x. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh R, Quayle JR. 1981. Purification and properties of the methanol dehydrogenase from Methylophilus methylotrophus. Biochem J 199:245–250. doi: 10.1042/bj1990245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt S, Christen P, Kiefer P, Vorholt JA. 2010. Functional investigation of methanol dehydrogenase-like protein XoxF in Methylobacterium extorquens AM1. Microbiology 156:2575–2586. doi: 10.1099/mic.0.038570-0. [DOI] [PubMed] [Google Scholar]
- 11.Skovran E, Palmer AD, Rountree AM, Good NM, Lidstrom ME. 2011. XoxF is required for expression of methanol dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol 193:6032–6038. doi: 10.1128/JB.05367-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park H, Lee H, Ro YT, Kim YM. 2010. Identification and functional characterization of a gene for the methanol: N,N'-dimethyl-4-nitrosoaniline oxidoreductase from Mycobacterium sp. strain JC1 (DSM 3803). Microbiology 156:463–471. doi: 10.1099/mic.0.034124-0. [DOI] [PubMed] [Google Scholar]
- 13.Muller JE, Meyer F, Litsanov B, Kiefer P, Vorholt JA. 2015. Core pathways operating during methylotrophy of Bacillus methanolicus MGA3 and induction of a bacillithiol-dependent detoxification pathway upon formaldehyde stress. Mol Microbiol 98:1089–1100. doi: 10.1111/mmi.13200. [DOI] [PubMed] [Google Scholar]
- 14.Harms N, Reijnders WN, Koning S, van Spanning RJ. 2001. Two-component system that regulates methanol and formaldehyde oxidation in Paracoccus denitrificans. J Bacteriol 183:664–670. doi: 10.1128/JB.183.2.664-670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Springer AL, Morris CJ, Lidstrom ME. 1997. Molecular analysis of mxbD and mxbM, a putative sensor-regulator pair required for oxidation of methanol in Methylobacterium extorquens AM1. Microbiology 143:1737–1744. doi: 10.1099/00221287-143-5-1737. [DOI] [PubMed] [Google Scholar]
- 16.Springer AL, Auman AJ, Lidstrom ME. 1998. Sequence and characterization of mxaB, a response regulator involved in regulation of methanol oxidation, and of mxaW, a methanol-regulated gene in Methylobacterium extorquens AM1. FEMS Microbiol Lett 160:119–124. doi: 10.1111/j.1574-6968.1998.tb12900.x. [DOI] [PubMed] [Google Scholar]
- 17.Masuda S, Suzuki Y, Fujitani Y, Mitsui R, Nakagawa T, Shintani M, Tani A. 2018. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum strain 22A. mSphere 3:e00462-17. doi: 10.1128/mSphere.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochsner AM, Hemmerle L, Vonderach T, Nussli R, Bortfeld-Miller M, Hattendorf B, Vorholt JA. 17 January 2019. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol Microbiol doi: 10.1111/mmi.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H, Ro YT, Kim YM. 2011. MdoR is a novel positive transcriptional regulator for the oxidation of methanol in Mycobacterium sp. strain JC1. J Bacteriol 193:6288–6294. doi: 10.1128/JB.05649-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalyuzhnaya MG, Lidstrom ME. 2005. QscR-mediated transcriptional activation of serine cycle genes in Methylobacterium extorquens AM1. J Bacteriol 187:7511–7517. doi: 10.1128/JB.187.21.7511-7517.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakobsen OM, Benichou A, Flickinger MC, Valla S, Ellingsen TE, Brautaset T. 2006. Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J Bacteriol 188:3063–3072. doi: 10.1128/JB.188.8.3063-3072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baloni P, Padiadpu J, Singh A, Gupta KR, Chandra N. 2014. Identifying feasible metabolic routes in Mycobacterium smegmatis and possible alterations under diverse nutrient conditions. BMC Microbiol 14:276. doi: 10.1186/s12866-014-0276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubey AA, Wani SR, Jain V. 2018. Methylotrophy in mycobacteria: dissection of the methanol metabolism pathway in Mycobacterium smegmatis. J Bacteriol 200:e00288-18. doi: 10.1128/JB.00288-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balhana RJ, Singla A, Sikder MH, Withers M, Kendall SL. 2015. Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions. BMC Genomics 16:479. doi: 10.1186/s12864-015-1696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James JN, Hasan ZU, Ioerger TR, Brown AC, Personne Y, Carroll P, Ikeh M, Tilston-Lunel NL, Palavecino C, Sacchettini JC, Parish T. 2012. Deletion of SenX3-RegX3, a key two-component regulatory system of Mycobacterium smegmatis, results in growth defects under phosphate-limiting conditions. Microbiology 158:2724–2731. doi: 10.1099/mic.0.060319-0. [DOI] [PubMed] [Google Scholar]
- 26.Parish T. 2014. Two-component regulatory systems of Mycobacteria. Microbiol Spectr 2:MGM2-0010-2013. doi: 10.1128/microbiolspec.MGM2-0010-2013. [DOI] [PubMed] [Google Scholar]
- 27.Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR Jr.. 2007. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J Bacteriol 189:5495–5503. doi: 10.1128/JB.00190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res 37:D459–D463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams RHN, Whitworth DE. 2010. The genetic organisation of prokaryotic two-component system signalling pathways. BMC Genomics 11:720. doi: 10.1186/1471-2164-11-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dasgupta N, Kapur V, Singh KK, Das TK, Sachdeva S, Jyothisri K, Tyagi JS. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber Lung Dis 80:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 32.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 33.Pohane AA, Joshi H, Jain V. 2014. Molecular dissection of phage endolysin: an interdomain interaction confers host specificity in Lysin A of Mycobacterium phage D29. J Biol Chem 289:12085–12095. doi: 10.1074/jbc.M113.529594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 35.Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev 22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saini DK, Malhotra V, Tyagi JS. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett 565:75–80. doi: 10.1016/j.febslet.2004.02.092. [DOI] [PubMed] [Google Scholar]
- 39.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS. 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150:865–875. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- 41.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain S, Kaushal D, DasGupta SK, Tyagi AK. 1997. Construction of shuttle vectors for genetic manipulation and molecular analysis of mycobacteria. Gene 190:37–44. doi: 10.1016/S0378-1119(96)00746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton-Foot M, Gey van Pittius NC. 2013. The complex architecture of mycobacterial promoters. Tuberculosis (Edinb) 93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 44.Park SW, Hwang EH, Park H, Kim JA, Heo J, Lee KH, Song T, Kim E, Ro YT, Kim SW, Kim YM. 2003. Growth of mycobacteria on carbon monoxide and methanol. J Bacteriol 185:142–147. doi: 10.1128/JB.185.1.142-147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Z, Orita I, Yin F, Yurimoto H, Kato N, Sakai Y, Izui K, Li K, Chen L. 2010. Overexpression of an HPS/PHI fusion enzyme from Mycobacterium gastri in chloroplasts of geranium enhances its ability to assimilate and phytoremediate formaldehyde. Biotechnol Lett 32:1541–1548. doi: 10.1007/s10529-010-0324-7. [DOI] [PubMed] [Google Scholar]
- 46.Aravind L, Ponting CP. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22:458–459. doi: 10.1016/S0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 47.Lee JM, Cho HY, Cho HJ, Ko IJ, Park SW, Baik HS, Oh JH, Eom CY, Kim YM, Kang BS, Oh JI. 2008. O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis. J Bacteriol 190:6795–6804. doi: 10.1128/JB.00401-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao XJ, Yan MY, Zhu H, Guo XP, Sun YC. 2016. Efficient and simple generation of multiple unmarked gene deletions in Mycobacterium smegmatis. Sci Rep 6:22922. doi: 10.1038/srep22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh MI, Jain V. 2013. Tagging the expressed protein with 6 histidines: rapid cloning of an amplicon with three options. PLoS One 8:e63922. doi: 10.1371/journal.pone.0063922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 51.Erijman A, Dantes A, Bernheim R, Shifman JM, Peleg Y. 2011. Transfer-PCR (TPCR): a highway for DNA cloning and protein engineering. J Struct Biol 175:171–177. doi: 10.1016/j.jsb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Saini DK, Pant N, Das TK, Tyagi JS. 2002. Cloning, overexpression, purification, and matrix-assisted refolding of DevS (Rv 3132c) histidine protein kinase of Mycobacterium tuberculosis. Protein Expr Purif 25:203–208. doi: 10.1006/prep.2002.1628. [DOI] [PubMed] [Google Scholar]
- 53.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 54.Jain V, Sujatha S, Ojha AK, Chatterji D. 2005. Identification and characterization of rel promoter element of Mycobacterium tuberculosis. Gene 351:149–157. doi: 10.1016/j.gene.2005.03.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.