Mycoplasma bovis is the main causal species of bovine mycoplasmal disease and leads to significant economic losses because of its severe symptoms, strong infectivity, and refractoriness. As for mastitis, culling cows with intramammary infections is a general countermeasure to prevent spreading. The conventional antimicrobial susceptibility test for mycoplasma is time-consuming and troublesome, but no quick and easy method for grasping the antimicrobial susceptibility of the causal strain exists at present. Treatment without antimicrobial susceptibility information may be one reason why M. bovis infection is refractory. Detecting a mutation involved in decreased susceptibility to antimicrobial agents of the causal strain makes it possible to easily select suitable antimicrobials for treatment, and this technique will help improve the cure rate and prevent the overuse of ineffective antimicrobial agents. In this study, we developed a technique to quickly and easily assess antimicrobial susceptibility based on the genetic characteristics of M. bovis strains in Japan.
KEYWORDS: Mycoplasma bovis, antimicrobial resistance, hybridization probe, multilocus sequence typing
ABSTRACT
Mycoplasma bovis isolates belonging to the sequence type 5 (ST5) group, the dominant group in Japan since 1999, were low susceptible to 16-membered macrolides and tetracyclines and were confirmed to have a guanine-to-adenine transition mutation at position 748 in the 23S rRNA gene (rrl) and adenine-to-thymine transversion mutations at positions 965 and 967 in the 16S rRNA gene (rrs) (Escherichia coli numbering). Moreover, isolates of ST93 and ST155, members of the ST5 group, were low susceptible to lincosamides and azithromycin and showed an adenine-to-guanine transition mutation at position 2059 of rrl. Isolates of ST93 were additionally low susceptible to spectinomycin and showed a cytosine-to-adenine transversion mutation at position 1192 of rrs. Strains of the ST5 group seem to spread to Japan and Europe from North America with imported cows, while strains of ST93 and ST155 originated in Japan. Melting curve analysis using hybridization probes revealed the existence of point mutations involved in decreased susceptibility to macrolides, lincosamides, and spectinomycin, as demonstrated by changes in the melting curve shape and/or decreases in the melting peak temperature, so the susceptibility to these antimicrobials can be assessed on the same day. For decreased susceptibility to fluoroquinolones to exist, nonsynonymous mutations in the DNA gyrase gene (gyrA) and topoisomerase IV gene (parC) had to coexist. The combination of amino acid substitutions of serine at position 83 in gyrA and serine at position 80 in parC resulted in particularly low susceptibility to fluoroquinolones.
IMPORTANCE Mycoplasma bovis is the main causal species of bovine mycoplasmal disease and leads to significant economic losses because of its severe symptoms, strong infectivity, and refractoriness. As for mastitis, culling cows with intramammary infections is a general countermeasure to prevent spreading. The conventional antimicrobial susceptibility test for mycoplasma is time-consuming and troublesome, but no quick and easy method for grasping the antimicrobial susceptibility of the causal strain exists at present. Treatment without antimicrobial susceptibility information may be one reason why M. bovis infection is refractory. Detecting a mutation involved in decreased susceptibility to antimicrobial agents of the causal strain makes it possible to easily select suitable antimicrobials for treatment, and this technique will help improve the cure rate and prevent the overuse of ineffective antimicrobial agents. In this study, we developed a technique to quickly and easily assess antimicrobial susceptibility based on the genetic characteristics of M. bovis strains in Japan.
INTRODUCTION
Bovine disease caused by Mycoplasma bovis, e.g., calf pneumonia, arthritis, genital disease, mastitis, and otitis, is a problem all over the world and occasionally results in massive outbreaks (1, 2). In Japan, mastitis due to M. bovis is recognized as the main mycoplasmal disease on dairy farms and in some cases has led to significant economic losses because of severe symptoms, strong infectivity, and refractoriness (2). No effective vaccine is available for the control of M. bovis infection, so antimicrobial treatment is the major therapeutic tool, and bacterial examination of bulk milk is the typical preventive method (2).
Although the antimicrobials that are potentially effective against bovine mycoplasmal infection are mostly protein synthesis inhibitors (e.g., aminoglycosides, macrolides, lincosamides, tetracyclines, phenicols, and pleuromutilins) and nucleic acid synthesis inhibitors (e.g., fluoroquinolones), mycoplasmal strains with low susceptibility to some of these antimicrobials have recently been emerging, and the spread of these strains may make antimicrobial therapies more difficult (3–5). Proper prescription of antimicrobial agents will lead not only to improvement of the cure rate but also to prevention of the overuse of ineffective antimicrobials; however, the traditional antimicrobial susceptibility test for mycoplasmas is time-consuming and labor-intensive, so it is not helpful at emergency clinics (6, 7). Hence, it is desirable to develop a method for quick and easy assessment of the antimicrobial susceptibility of the causal strain. In M. bovis or Mycoplasma californicum, specific mutations located in the 16S rRNA gene (rrs), 23S rRNA gene (rrl), DNA gyrase gene (gyrA), and topoisomerase IV gene (parC) were found in strains with low susceptibility to various antimicrobial agents (7–9). Reconfirming the universality of the roles of these mutations and exploiting them may lead to the development of a method that can compensate for the drawbacks of conventional methods (7). Melting curve analysis using hybridization probes is a useful method for detecting target single nucleotide polymorphisms (SNPs); high sensitivity and rapidity are virtues of this method (7, 10).
Multilocus sequence typing (MLST) is a genotyping method that has excellent universal comparability and is useful for characterizing the genetic background and for phylogenetic and epidemiological study (11). In addition, we can assess the bacteriological characteristics or epidemic of each sequence type (ST) by annotating the various characteristics of each strain (possession of pathogenic genes, origin, isolating date, isolated host, geographical area, etc.). By combining genotyping by MLST with information regarding antimicrobial susceptibility, we may assess the reasons for the emergence, invasion, or spread of various strains with low susceptibility.
In this study, we investigated the change in genotypes by MLST of M. bovis strains isolated in Japan over a period of 26 years and clarified the effect of their changes on the susceptibility to various antimicrobial agents. We investigated a variety of mutation points that are suggested to be involved in decreased susceptibility to antimicrobial agents in Japan and reconfirmed their roles in antimicrobial susceptibility. Subsequently, we attempted to establish a probe-based genetic method for the rapid detection of these mutations (9).
RESULTS
Evolutionary relationships and population structure of M. bovis field isolates in Japan.
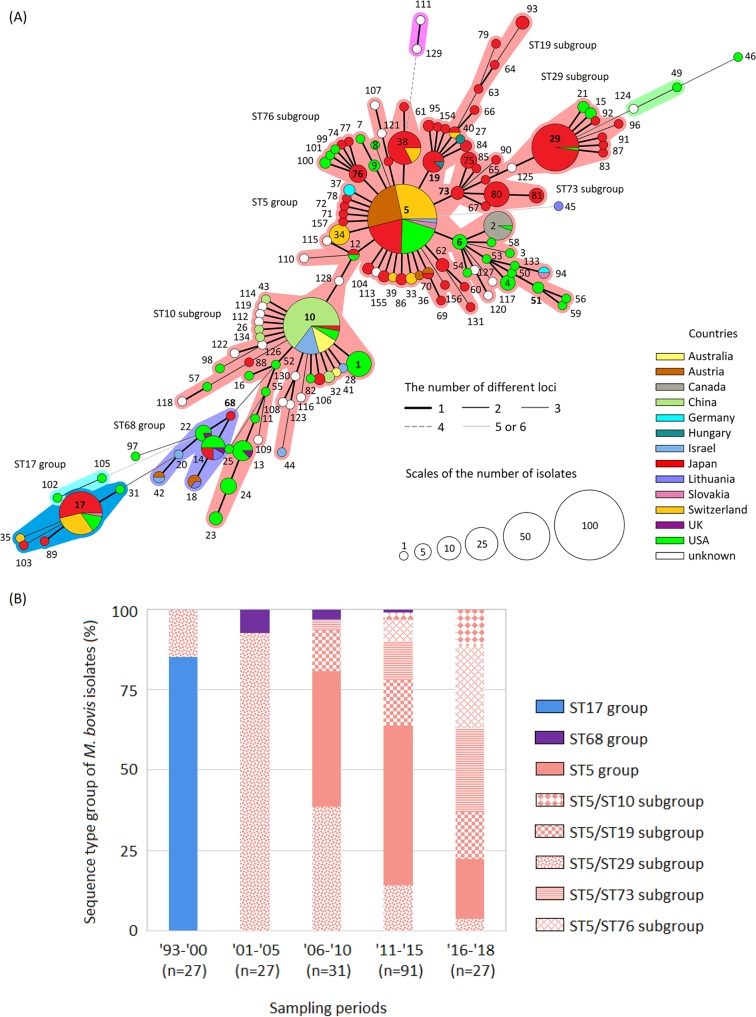
The 203 isolates analyzed gave a total of 52 different STs that were divided into three ST groups (i.e., the ST5 group, ST17 group, and ST68 group) using Based Upon Related Sequence Types (BURST) and minimum spanning tree (MST) analyses (Fig. 1A). There were five subcentral STs (i.e., ST10, ST19, ST29, ST73, and ST76) in the ST5 group, which was the largest ST group (Fig. 1A). Of 52 STs, 42 were specific to Japanese isolates (Fig. 1A). As for the foreign distribution of each ST group or ST subgroup that was confirmed in Japan, the ST5 group, including the ST19, ST29, ST73, and ST76 subgroups, was mainly confirmed in strains from the United States and Europe. However, most of the strains from Australia, China, and Israel belonged to the ST10 subgroup. Strains belonging to the ST68 group were isolated from the United States, Israel, and Europe. Most strains belonging to the ST17 group were isolated from the United States and Europe before 1998 (Fig. 1A). As for the chronological change of each ST group or ST subgroup in Japan, most of the isolates showed STs belonging to the ST17 group until 1998, but the dominant ST group shifted completely to the ST5 group after 1999 (Fig. 1B; see also Table S1 in the supplemental material). Regarding the appearance of subgroups in the ST5 group, the ST29 subgroup was dominant until 2005, but the appearance of isolates other than the ST29 subgroup has been increasing since then (Fig. 1B and Table S1). Isolates belonging to the ST10 subgroup first emerged in 2014, and their appearance has been increasing (Fig. 1B and Table S1). Isolates belonging to the ST68 group were sporadically confirmed from 2002 to 2011 (Fig. 1B). Strains of ST5, ST14, ST17, and ST29 were already isolated in the United States before their first confirmation in Japan, and strains of ST10 were also isolated in the United States, China, Israel, and Australia before their first confirmation in Japan.
FIG 1.
(A) Minimum spanning tree representing the evolutionary relationship between M. bovis sequence types (STs) by multilocus sequencetyping. Numbers indicate STs, and the central ST and subcentral STs of each ST group are indicated in bold. A group of STs in which two or fewer loci differ from each other is considered an ST group and is surrounded by a specific color. The size of the circles represents the population size, and the circular chart in each circle indicates the proportion of countries of origin in each ST. The thickness and dotting of the lines indicate the number of different loci between the STs; a thicker line denotes a closer distance than a thin line, and a thin line denotes a closer distance than a dotted line. Among the whole-genome-analyzed strains, strain PG45T was identified as a member of ST17, and all strains from China were members of the ST10 subgroup (Hubei-1, 08M, CQ-W70, HB0801, NM2012, and Ningxia-1). (B) Chronological change of genotypes by MLST of M. bovis isolates in Japan. Numbers on the x axis represent years, with the last two digits shown for each year.
Relationship between genotypes by MLST and susceptibility to antimicrobial agents.
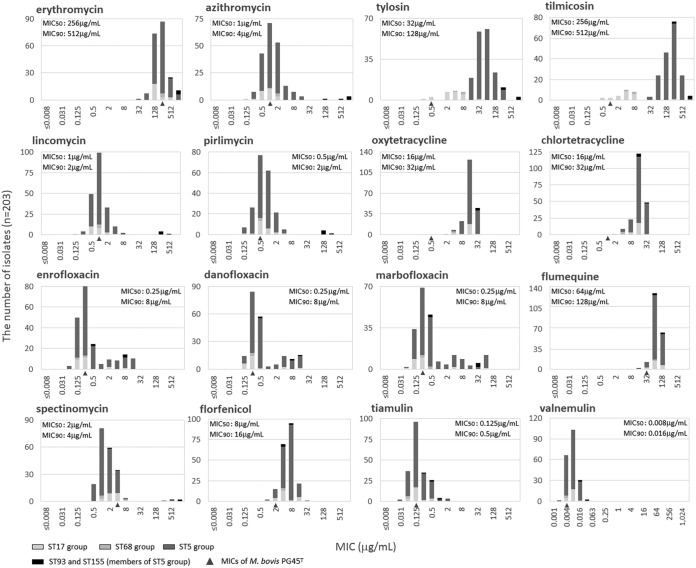
The MIC distributions of the 16 tested antimicrobial agents against M. bovis field isolates, the MIC50 and MIC90 values, and the MICs for strain PG45T (MICPG45T) (a quality control reference isolate) are shown in Table 1 and Fig. 2. The antimicrobial to which the isolates exhibited the highest susceptibility was valnemulin, with MIC50 and MIC90 values of 0.008 μg/ml and 0.016 μg/ml, respectively (12). The antimicrobial to which the isolates exhibited the highest susceptibility after valnemulin was tiamulin, with MIC50 and MIC90 values of 0.125 μg/ml and 0.5 μg/ml, respectively. The MICs of florfenicol, tiamulin, and valnemulin exhibited normal distributions, and the MIC for strain PG45T also fit into each MIC range, so all of the M. bovis isolates were considered to maintain their susceptibility to these antimicrobial agents. On the other hand, M. bovis is considered to have natural resistance to erythromycin (a 14-membered macrolide) and flumequine, and the MICs for strain PG45T (MICs, 256 and 32 μg/ml) and the MIC50 (MICs, 256 and 64 μg/ml) and MIC90 (MICs, 512 and 128 μg/ml) of the field isolates were extremely high. M. bovis isolates that show low susceptibility to tetracyclines seem to have spread since the 1990s in Japan. The MICs of oxytetracycline and chlortetracycline for strain PG45T were 0.5 and 1 μg/ml, respectively; however, their MIC ranges for field isolates were higher values (MICs, 2 to 32 μg/ml), and the MIC50 and MIC90 values were 16 and 32 μg/ml, respectively. Moreover, the MICs of 16-membered macrolides (e.g., tylosin and tilmicosin) and fluoroquinolones other than flumequine exhibited clear bimodal distributions. Isolates highly susceptible to tylosin and tilmicosin with MIC values of ≤8 μg/ml are members of ST14 and the ST17 group, and isolates with low susceptibility such that their MICs for tylosin and tilmicosin are ≥16 μg/ml are members of ST68 and the ST5 group (Tables S1 and S2). As for fluoroquinolones other than flumequine, there was no clear correlation between MIC and genotypes by MLST, and both the high MIC groups and low MIC groups consisted of various STs (Tables S1 and S2). Most of the isolates exhibited high susceptibility to azithromycin (a 15-membered macrolide), lincosamides, and spectinomycin; however, isolates with low susceptibility were sporadically confirmed. All three isolates of ST93 were less susceptible to azithromycin (MIC, 1,024 μg/ml), lincosamides (MICs, 128 to 512 μg/ml), spectinomycin (MICs, 512 to 1,024 μg/ml), and fluoroquinolones (MICs, 8 to 32 μg/ml) than most of the remaining isolates except for isolates of ST155 (Tables S1 and S2). Both isolates of ST155 were less susceptible to azithromycin (MICs, 128 to 512 μg/ml) and lincosamides (MICs, 128 to 256 μg/ml) than all of the remaining isolates except for isolates of ST93 (Tables S1 and S2). Other than these isolates, two isolates of ST103 and ST29 were less susceptible to spectinomycin (MICs, 256 to 512 μg/ml) than all of the remaining isolates except for isolates of ST93 (Tables S1 and S2).
TABLE 1.
Antimicrobial susceptibility profiles of M. bovis field isolates from 203 cases of bovine infection in Japan from 1993 to 2018
| Antimicrobial agent | MIC data (μg/ml) |
|||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | MICPG45T | |
| Erythromycin | 32–1,024 | 256 | 512 | 256 |
| Azithromycin | 0.125–1,024 | 1 | 4 | 1 |
| Tylosin | 0.25–1,024 | 32 | 128 | 0.5 |
| Tilmicosin | 0.5–1,024 | 256 | 512 | 1 |
| Lincomycin | 0.125–512 | 1 | 2 | 1 |
| Pirlimycin | 0.125–256 | 0.5 | 2 | 0.5 |
| Oxytetracycline | 2–32 | 16 | 32 | 0.5 |
| Chlortetracycline | 2–32 | 16 | 32 | 1 |
| Enrofloxacin | 0.063–16 | 0.25 | 8 | 0.25 |
| Danofloxacin | 0.125–16 | 0.25 | 8 | 0.25 |
| Marbofloxacin | 0.063–64 | 0.25 | 8 | 0.25 |
| Flumequine | 16–128 | 64 | 128 | 32 |
| Spectinomycin | 0.5–1,024 | 2 | 4 | 4 |
| Florfenicol | 1–32 | 8 | 16 | 2 |
| Tiamulin | 0.031–2 | 0.125 | 0.5 | 0.125 |
| Valnemulin | 0.002–0.031 | 0.008 | 0.016 | 0.004 |
FIG 2.
Relationship between antimicrobial susceptibility distribution and genotypes by MLST of M. bovis field isolates in Japan. MIC values of strain PG45T are indicated with a triangle under the bar graph. M. bovis showed natural resistance to erythromycin and flumequine. Isolates of ST93 (a member of the ST5 group) showed low susceptibility to spectinomycin, macrolides, lincosamides, tetracyclines, and fluoroquinolones. Moreover, isolates of ST155 (a member of the ST5 group) showed low susceptibility to macrolides, lincosamides, and tetracyclines. Although ST14 (a member of the ST68 group) and isolates in the ST17 group (oldcomer) showed high susceptibility to 16-membered macrolides, ST68 and isolates in the ST5 group (newcomer) showed low susceptibility to these agents. Susceptibility to tetracyclines was on a declining trend overall. Isolates that showed low susceptibility to fluoroquinolones emerged sporadically, and their ST groups were varied. On the other hand, susceptibility to pleuromutilins was high, and no isolate with low susceptibility was found.
Relationship between mutations in target genes and susceptibility to antimicrobial agents.
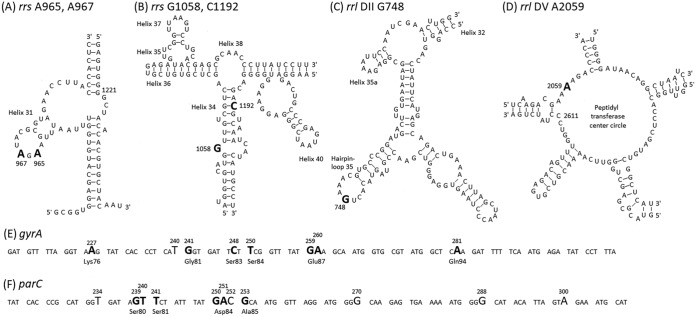
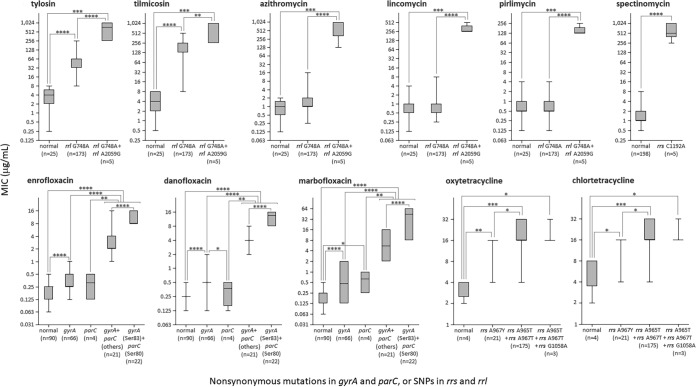
The SNPs in target genes (i.e., rrs, rrl, gyrA, and parC) that were confirmed in M. bovis field isolates in Japan are shown in Fig. 3. The adenines at positions 965 and 967 (A965 and A967, respectively) in helix 31 of both rrs genes were thymines (A965T and A967T) in all isolates belonging to the ST5 group and isolates of ST68 and ST103 (Table S1). Moreover, both rrs A967T were confirmed in all three isolates of ST14, and both rrs genes had cytosine (A967C) in 18 isolates belonging to the ST17 group (Table S1). The guanine at position 1058 (G1058) in helix 34 of either or both rrs genes was adenine (G1058A) in two isolates of ST5 and one isolate of ST67 (Table S1). It has been suggested that SNPs at these positions are involved in decreased susceptibility to tetracyclines (oxytetracycline MICs, from 2 to 4 μg/ml [normal isolates] to 4 to 32 μg/ml [isolates with A965T and/or A967Y]; chlortetracycline MICs, from 2 to 8 μg/ml [normal isolates] to 4 to 32 μg/ml [isolates with A965T and/or A967Y]) (8). Overall, isolates in which the presence of either combination of SNPs was confirmed showed a significantly low susceptibility to oxytetracycline and chlortetracycline; however, some isolates possessing SNPs showed the same MIC values as normal isolates (Fig. 4). Moreover, there were no clear results as to whether the presence of rrs G1058A would affect the decreased susceptibility to tetracyclines (Fig. 4).
FIG 3.
Single nucleotide polymorphisms (SNPs) in target genes that were confirmed in M. bovis field isolates in Japan. (A and B) The two-dimensional structure of helix 31 and helix 34 of 16S rRNA (rrs). (C and D) Hairpin-loop 35 in domain II and the peptidyl transferase center circle in domain V of 23S rRNA (rrl) of M. bovis PG45T (GenBank accession no. CP002188). Each SNP is indicated in large font and bold. (E and F) The DNA sequences of the quinolone resistance-determining region of the DNA gyrase gene (gyrA) (E) and topoisomerase IV gene (parC) (F) of M. bovis PG45T. Of the SNPs indicated in large font, the SNPs which cause nonsynonymous mutations are in bold, and the original amino acids are shown under the triplet. The nucleotides and amino acids are numbered on the basis of the E. coli sequence (NCBI RefSeq accession no. NC_002655).
FIG 4.
Relationship between the existence of nonsynonymous mutations or SNPs in target genes and MICs to each antimicrobial agent. The box-and-whisker plot indicates the maximum, upper quartile, median, lower quartile, and minimum MIC value. As for fluoroquinolones, gyrA (Ser83) + parC (Ser80) refers to isolates in which the coexistence of amino acid substitutions at Ser83 in gyrA and Ser80 in parC was confirmed, and gyrA + parC (others) refers to isolates in which combinations of other amino acid substitutions were confirmed. Significant differences were evaluated by the Mann-Whitney U test. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
The cytosine at position 1192 (C1192) in helix 34 of either or both rrs genes was adenine (C1192A) in one isolate of ST29, one isolate of ST103, and all three isolates of ST93 (Table S1). These five isolates which possessed rrs C1192A showed significantly low susceptibility to spectinomycin (P < 0.0001) (spectinomycin MICs, from 0.5 to 8 μg/ml [normal isolates] to 256 to 1,024 μg/ml [isolates with C1192A]) (Fig. 4) (9).
The guanine at position 748 (G748) in hairpin-loop 35 of either or both rrl genes was adenine (G748A) in all isolates belonging to the ST5 group and one isolate each of ST17 and ST68 (Table S1). The adenine at position 2059 (A2059) in the peptidyl transferase center circle of either or both rrl genes was guanine (A2059G) in all five isolates of ST93 and ST155 (Table S1). It is suggested that the SNP at rrl G748 is involved in decreased susceptibility to 16-membered macrolides, and the SNP at rrl A2059 is involved in decreased susceptibility to macrolides and lincosamides (7, 9). Significantly low susceptibility to tylosin and tilmicosin was observed in the 173 isolates in which rrl G748A was confirmed (P < 0.0001) (tylosin MICs, from 0.25 to 8 μg/ml [normal isolates] to 8 to 256 μg/ml [isolates with G748A]; tilmicosin MICs, from 0.5 to 8 μg/ml [normal isolates] to 8 to 512 μg/ml [isolates with G748A]), and an additional decrease in susceptibility was observed in five isolates in which the coexistence of rrl A2059G was confirmed (P < 0.0001 or <0.05) (tylosin and tilmicosin MICs, 256 to 1,024 μg/ml) (Fig. 4). On the other hand, isolates in which rrl G748A was confirmed did not show a significant change in susceptibility to azithromycin or lincosamides (azithromycin MICs, from 0.125 to 2 μg/ml [normal isolates] to 0.25 to 16 μg/ml [isolates with G748A]; lincomycin MICs, from 0.125 to 4 μg/ml [normal isolates] to 0.25 to 8 μg/ml [isolates with G748A]; pirlimycin MICs, from 0.125 to 4 μg/ml [normal isolates] to 0.125 to 4 μg/ml [isolates with G748A]), but five isolates in which rrl A2059G coexists showed significantly low susceptibility to azithromycin and lincosamides (P < 0.0001) (azithromycin MICs, 128 to 1,024 μg/ml; lincomycin MICs, 256 to 512 μg/ml; pirlimycin MICs, 128 to 256 μg/ml) (Fig. 4).
These results suggested that a nonsynonymous mutation in the quinolone resistance-determining region (QRDR) of gyrA or parC is often confirmed in isolates with low susceptibility to fluoroquinolones (9). In gyrA of 109 isolates, nonsynonymous mutations (A227T, G241A, C248W, T250C, G259M, A260T, and A281T) resulting in amino acid substitutions (Lys76Met, Gly81Ser, Ser83Phe, Ser83Tyr, Ser84Pro, Glu87Lys, Glu87Gln, Glu87Val, and Gln94Leu) were observed (Table S1). Of these nonsynonymous mutations, SNP C248W, which causes an amino acid substitution of serine at position 83 (Ser83), was observed in 90 isolates and was the most frequent SNP in gyrA (Table S1). Moreover, SNP T240C, a silent mutation located between these nonsynonymous mutations, was observed in two isolates (Table S1). In parC of 47 isolates, nonsynonymous mutations (G239T, T240A, T241C, G250H, A251K, and G253C) resulting in amino acid substitutions (Ser80Ile, Ser80Arg, Ser81Pro, Asp84Tyr, Asp84His, Asp84Asn, Asp84Val, Asp84Gly, and Ala85Pro) were observed (Table S1). Of these nonsynonymous mutations, SNPs G239T and T240A, which cause an amino acid substitution of serine at position 80 (Ser80), were observed in 24 isolates and 4 isolates, respectively. G239T was the most frequent nonsynonymous mutation in parC (Table S1). Moreover, SNP C252T, a silent mutation located between these nonsynonymous mutations, was observed in 159 isolates (Table S1). In 66 isolates in which a nonsynonymous mutation was confirmed in only the QRDR of gyrA, significantly low susceptibility to fluoroquinolones was observed (P < 0.0001), but isolates showing MICs overlapping the MICs of normal strains were also observed (enrofloxacin MICs, 0.063 to 0.5 μg/ml [normal isolates] to 0.125 to 1 μg/ml [isolates with a nonsynonymous mutation in the QRDR of gyrA]; danofloxacin MICs, 0.125 to 0.5 μg/ml [normal isolates] to 0.125 to 2 μg/ml [isolates with a nonsynonymous mutation in the QRDR of gyrA]; marbofloxacin MICs, 0.063 to 0.5 μg/ml [normal isolates] to 0.125 to 2 μg/ml [isolates with a nonsynonymous mutation in the QRDR of gyrA]) (Fig. 4). Moreover, four isolates in which nonsynonymous mutations were confirmed in only the QRDR of parC did not show significantly low susceptibility to enrofloxacin or danofloxacin (enrofloxacin MICs, 0.125 to 0.5 μg/ml; danofloxacin MICs, 0.125 to 0.5 μg/ml; marbofloxacin MICs, 0.25 to 1 μg/ml) (Fig. 4). On the other hand, 43 isolates in which nonsynonymous mutations were confirmed in the QRDRs of both gyrA and parC showed obvious and significantly low susceptibility to fluoroquinolones (P < 0.0001) (enrofloxacin MICs, 1 to 16 μg/ml; danofloxacin MICs, 2 to 16 μg/ml; marbofloxacin MICs, 2 to 64 μg/ml) (Fig. 4). In particular, the coexistence of amino acid substitutions at Ser83 in gyrA and Ser80 in parC resulted in significantly lower susceptibility to fluoroquinolones than did other combinations of amino acid substitutions (P < 0.0001) (enrofloxacin MICs, 8 to 16 μg/ml; danofloxacin MICs, 8 to 16 μg/ml; marbofloxacin MICs, 8 to 64 μg/ml) (Fig. 4).
Rapid detection of mutations associated with decreased susceptibility to antimicrobials by melting curve analysis using hybridization probes.
The hybridization probe is a useful tool for detecting SNPs, but it simultaneously detects meaningless SNPs, such as silent mutations. For five SNPs without silent mutations around the target SNPs, i.e., rrs A965T and/or A967Y, rrs C1192A, rrl G748A, and rrl A2059G, a detection method using hybridization probes was examined. On the other hand, detection by DNA sequencing was investigated as a way to detect target SNPs in the QRDRs of gyrA and parC with silent mutations around the target SNPs. The primers designed in this study sufficiently amplified three DNA regions of M. bovis field isolates and strain PG45T containing five target SNPs involved in low susceptibility to tetracyclines, spectinomycin, macrolides, and lincosamides (Table 2). The primers for the amplification of the QRDRs of gyrA and parC also sufficiently amplified these DNA regions of M. bovis field isolates and strain PG45T. On the other hand, these DNA regions of other non-M. bovis organisms (eight mycoplasma-type strains, two acholeplasma-type strains, and nine bacterial strains that cause bovine mastitis) were not amplified by PCR using these primers. Moreover, the mixing of these other microbials did not affect the melting curve analysis using hybridization probes or DNA sequencing of the QRDR. In this study, the detection limits of the PCR and DNA extraction method were suggested to be 126 CFU/ml for the milk samples with somatic cell counts (SCCs) of 20 × 103 and 716 × 103 cell/ml, respectively, but decreased to 1,260 CFU/ml for the milk samples with SCCs of 1,600 × 103 and 3,000 × 103 cell/ml, respectively. Storage at −20°C or 4°C for 7 days resulted in a further reduction in the detection limit of each sample to 1/10.
TABLE 2.
Primers and probes used in melting curve genotyping analysis performed by hybridization probe or DNA sequencing for detecting mutations involved in reduced susceptibility to tetracyclines, spectinomycin, macrolides, lincosamides, and fluoroquinolones
| Target mutation(s) | Primer or probe | Sequence (5′ to 3′) | Annealing temp (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| rrs A965 and/or A967 | Forward primer | GCA TAG GAA ATG ATG CTA CC | 61 | 808 | This study |
| Reverse primer | TGC TCC ATG TCA CCA CTT C | ||||
| Hybridization probe | CGG TGG AGC ATG TGG TTT AA-FITC | ||||
| LC Red640-TTG AAG ATA CGC GTA GAA CC-phosphate | |||||
| rrs C1192 | Forward primer | Same as rrs A965 and/or A967 | 61 | 808 | This study |
| Reverse primer | Same as rrs A965 and/or A967 | ||||
| Hybridization probe | CCG AGT AAT CGG GAG GAA GG-FITC | ||||
| LC Red640-GGG GAC GAC GTC AAA TCA TC-phosphate | |||||
| rrl DII G748 | Forward primer | AGC TTT TGG GAA GAA GCG | 65 | 836 | This study |
| Reverse primer | TTA CAT TGT CGG CGC AAG G | ||||
| Hybridization probe | CCC TAA GTG GAG GGT CGA AC-FITC | ||||
| LC Red640-GTA GTA CAC TGA AAC GTG CC-phosphate | |||||
| rrl DV A2059 | Forward primer | GGA CTT TTG TCT GAA TTT GCC | 61 | 609 | This study |
| Reverse primer | GTT GAC TCC ACT GCT ACT G | ||||
| Hybridization probe | GAA AAC GCT GGG TTC CCG CA-FITC | ||||
| LC Red640-CAA GAC GAA AAG ACC CCA TG-phosphate | |||||
| QRDR in gyrA | gyrA-F primer | GACGAATCATCTAGCGAG | 59 | 531 | 21 |
| gyrA-R primer | GCCTTCTAGCATCAAAGTAGC | ||||
| QRDR in parC | parC-F primer | GAGCAACAGTTAAACGATTTG | 59 | 488 | 21 |
| parC-R primer | GGCATAACAACTGGCTCTT |
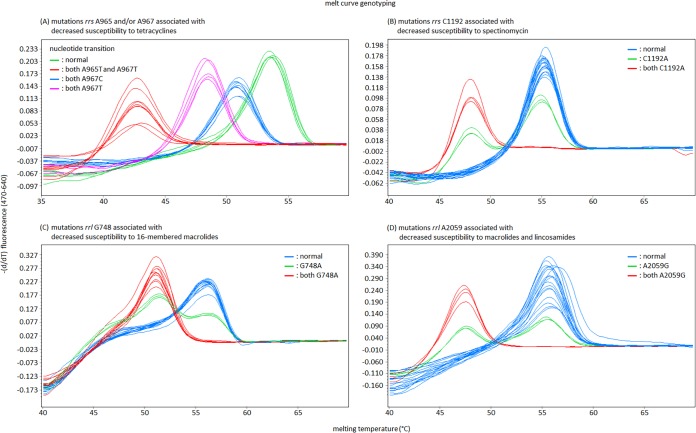
The numbers of rrs-rrl operons vary among microbial species, but there are usually two operons in the genome of M. bovis as with many bovine mycoplasma (GenBank accession no. CP002188) (13). In the melting curve analysis using hybridization probes, a single melting peak at a lower-than-normal temperature is observed for the case in which the mutations occur together at target SNPs in both rrs-rrl genes. In this case, the temperature of the melting peaks detecting each SNP decreased from 53.53 ± 0.12°C to 50.86 ± 0.09°C, 48.45 ± 0.1°C, and 43.05 ± 0.32°C (SNP at rrs A965 and/or A967), from 55.46 ± 0.40°C to 48.39 ± 0.08°C (SNP at rrs C1192), from 56.04 ± 0.13°C to 50.99 ± 0.17°C (SNP at rrl G748), and from 55.79 ± 0.62°C to 47.58 ± 0.06°C (SNP at rrl A2059) (Fig. 5). However, the melting curve changed to a bimodal curve if the mutation occurred at the target SNP in either rrs-rrl gene (Fig. 5). These changes in the melting curve were automatically detected by the analytical software of the real-time PCR machine (Fig. 5).
FIG 5.
The results of melt-curve genotyping using hybridization probes for detecting mutations associated with decreased susceptibility to tetracyclines, spectinomycin, lincosamides and/or macrolides in M. bovis.
DISCUSSION
Mutations associated with decreased susceptibility to many antimicrobial families were previously investigated in field isolates or laboratory-derived mutants of various mycoplasmal species (5, 7, 9, 14–16). Mutations rrs A965T and A967T, rrs C1192A, rrl G748A, and rrl A2059G were confirmed in Japanese M. bovis isolates that showed low susceptibility to tetracyclines, spectinomycin, macrolides, and lincosamides, and these mutations were also confirmed in Hungarian isolates which showed low susceptibility to these antimicrobial agents (5, 9). Moreover, mutations rrs A965T and A967T, rrl G748A, and rrl A2059G were confirmed in the genome data from Chinese isolates that belong to the ST10 subgroup (i.e., Hubei-1, 08M, CQ-W70, HB0801, NM2012, and Ningxia-1). Of these mutations, rrl G748A was almost entirely absent in isolates belonging to the ST17 group that show high susceptibility to 16-membered macrolides; however, members of the ST17 group have only rarely been isolated. On the other hand, the mutation rrl G748A was confirmed in all isolates belonging to the ST5 group, the members of which show low susceptibility to 16-membered macrolides. Mutations involved in decreased susceptibility to tetracyclines, rrs A965T and A967T (8), are also genetic characteristics of the ST5 group. According to the MLST database, the ST5 group is currently confirmed everywhere in the world. The ST5 group became the dominant group worldwide in the 2000s. Tetracyclines and macrolides had been used as first-line antimicrobial agents for the treatment of bovine mycoplasmal disease, so the low susceptibility to these antimicrobial agents caused by mutations rrs A965T and A967T and rrl G748A may be one reason that the ST5 group became the dominant group in the world. It is speculated that strains belonging to the ST5 group with these mutations originate from North America. Strains of ST29 and ST5, the main STs of the ST5 group in Japan, were first confirmed in the United States in 1994 and 2000, respectively, prior to being confirmed in Japan in 1996 and 2006, respectively; the United States and Canada were the main countries from which live cows were imported at that time (Fig. S1) (17). Rosales et al. suggested that the extensive livestock trade led to international expansion of specific M. bovis strains (11). Hence, the genetic background research and antimicrobial susceptibility data for M. bovis suggested that tetracyclines and 16-membered macrolides may already be inappropriate to use as first-line antimicrobial agents for treating M. bovis infection (5, 9, 18). On the other hand, Mycoplasma bovigenitalium or Mycoplasma californicum showed adequate susceptibility to tetracyclines and 16-membered macrolides (4, 7). The mutation rrl A2059G caused decreased susceptibility to macrolides and lincosamides and additionally caused significantly lower susceptibility to 16-membered macrolides than did rrl G748A (5, 9, 19). It has been observed in many mycoplasma species that SNPs located at rrl A2058 and rrl A2062 also cause similar susceptibility changes (5, 7, 9, 14–16). In Mycoplasma pneumoniae, mutations at these positions have harmful effects on growth (20). This effect is presumed to be one reason why M. bovis of ST93 and ST155 with the mutation rrl A2059G did not spread widely. The mutation rrs C1192A caused decreased susceptibility to spectinomycin and has only been confirmed in five isolates of ST29, ST93, and ST103 in Japan. M. bovis cases showing low susceptibility to spectinomycin are rare in Japan, but they are widespread in Europe, North America, and Israel (2, 3, 9, 18). The use of spectinomycin against M. bovis was widespread in these countries, and this seems to have led to the current situation, whereas spectinomycin is generally used to treat poultry diseases and not bovine diseases in Japan. M. bovis cases showing low susceptibility to fluoroquinolones were sporadically observed in Japan. Most of the STs in these cases were previously confirmed to be susceptible prior to the isolation of an isolate with low susceptibility, or they were specific to Japan. Hence, these strains with low susceptibility to fluoroquinolones were speculated to emerge with the use of fluoroquinolone agents for the treatment of cattle in Japan. In M. bovis, essential mutations leading to decreased susceptibility to fluoroquinolones were located in the QRDR of gyrA or parC, and mutations in gyrB or parE appear to be irrelevant to changes in susceptibility (21). Furthermore, the coexistence of nonsynonymous mutations in both gyrA and parC led to more greatly decreased susceptibility than mutations in either gyrA or parC alone. In particular, amino acid substitutions at Ser83 in gyrA and Ser80 in parC form a combination that most effectively leads to decreased susceptibility, and mutations at these positions seem to be observed frequently in field isolates (9). The coexistence of amino acid substitutions at Ser83 in gyrA and Ser80 in parC was also confirmed in the genome data from a Chinese strain, Ninxia-1. These SNPs could be easily reproduced through fewer than 10 passages in a medium supplied with the respective antimicrobial agent (9).
Florfenicol and pleuromutilins are speculated to be useful antimicrobial agents for the treatment of M. bovis infection, not only in Japan but also in many other countries (9, 18). Although it is suggested that SNPs associated with decreased susceptibility to florfenicol and pleuromutilins were located at several positions in rrl genes, i.e., rrl C2035, rrl G2448, rrl C2500, and rrl G2506 (9), these SNPs have not been observed in Japanese field isolates. In Japan, florfenicol is applied to bovine pneumonia and porcine diseases, but pleuromutilins are commonly used only for porcine diseases. The formation and cleavage of base pairs between positions rrl A2057 and rrl C2611 are crucial factors in the change in susceptibilities to erythromycin and lincosamides (7). Many bovine mycoplasmas which do not form base pairs between these positions show natural resistance to erythromycin and susceptibility to lincosamides. Conversely, mycoplasma species forming a base pair between these positions are sensitive to erythromycin and show natural resistance to lincosamides, i.e., M. pneumoniae, Mycoplasma gallisepticum, Ureaplasma parvum, and Ureaplasma urealyticum (22–24). Flumequine also appears to be ineffective for the treatment of M. bovis infection, but its resistance mechanism is unknown. Unlike M. bovis and M. californicum, Mycoplasma bovirhinis, an indigenous mycoplasma in the bovine nasal cavity, has adequate susceptibility to flumequine, and so it may be useful for the isolation of M. bovis or M. californicum from bovine nasal samples containing huge quantities of M. bovirhinis (7).
Hybridization probes are typically used in SNP analysis, and a real-time PCR assay can accurately differentiate mutations of target DNA regions by measuring the melting temperature of the probe-amplicon hybrid, even if there are few nucleotide differences or only one such difference (10). The method presented in this study successfully detected target mutations, and all mutations were shown as shifts in the melting-peak temperature or changes in the shape of the melting curve. If possible, DNA sequencing of the mutated domain is recommended for more accurate judgment (10). The total time required for this method was only 3 h, and the cost of the reagent was $2.40 per sample. On the other hand, mutations involved in fluoroquinolone resistance were detected by direct DNA sequencing of the QRDR of gyrA and parC (21). These SNPs are scattered in the QRDR of gyrA and parC, and some of them are silent mutations (9). In such cases, even if DNA sequencing of the target genes is more time-consuming than SNP analysis using hybridization probes, it may be easier than the traditional method. In experimental intramammary infections caused by M. bovis, M. bovis persisted in milk for approximately 2 weeks, and its concentration in udder tissue remained very high (103 to ∼108 CFU/ml) (25). If a fresh milk sample was used, M. bovis was usually detectable by this method (detection limit, 126 to 1,260 CFU/ml). Storage of the milk sample led to a decrease in the sensitivity of this method, and the survivability of the mycoplasma was also greatly reduced by storage (26). When it is impossible to test immediately, culture samples that can be expected to contain a large number of M. bovis bacteria should be prepared at the same time (26).
In the present study, we established a rapid and easy method for possibly predicting the reduced susceptibility of tetracyclines, spectinomycin, macrolides, and lincosamides against M. bovis. Guidelines for the effective and safe use of spectinomycin, lincosamides, and pleuromutilins, which are currently impossible to use in bovine mastitis treatment in Japan, are still to be developed. The route and dosage need to be examined, as does the severity of mastitis that can be successfully treated with antimicrobial therapy. In particular, the treatment of infected but asymptomatic cows, which are common in outbreaks of bovine mycoplasmal mastitis, is an important issue that must be addressed in order to avoid culling and the consequent economic losses (27).
Macrolides and tetracyclines are recognized as first-line antimicrobial agents for the treatment of bovine mycoplasmal disease in Japan, but M. bovis strains of the ST5 group, the members of which show low susceptibility to these antimicrobial agents, have been spreading in Japan since 1999. In addition, M. bovis strains which show low susceptibility to lincosamides, spectinomycin, or fluoroquinolones sporadically emerged. Specific SNPs were confirmed in isolates with low susceptibility to tetracyclines, macrolides, lincosamides, and spectinomycin, and a melting curve analysis method using a hybridization probe was established for the quick and easy detection of these SNPs. This technique will help in the selection of useful antimicrobials for the treatment of bovine mastitis caused by M. bovis.
MATERIALS AND METHODS
Mycoplasmal isolates, antimicrobial agents, and susceptibility testing.
A total of 203 M. bovis isolates were collected from bovine samples obtained between 1993 and 2018 in Japan. The methods used to isolate and identify these isolates have been described previously (27). The susceptibilities of the M. bovis isolates to 16 antimicrobials approved for therapeutic applications in veterinary use in Japan were examined. These consisted of an aminoglycoside (spectinomycin), macrolides (erythromycin, azithromycin, tylosin, and tilmicosin), lincosamides (lincomycin and pirlimycin), tetracyclines (oxytetracycline and chlortetracycline), a phenicol (florfenicol), fluoroquinolones (enrofloxacin, danofloxacin, marbofloxacin, and flumequine), and pleuromutilins (tiamulin and valnemulin). For quality control of the test and for later evaluation, M. bovis PG45T was also added to this study, and the MIC values of the antimicrobial agents against each isolate were determined by the agar microdilution method according to the method recommended by Hannan (6).
MLST, phylogenetic analysis, and analysis of mutations involved in decreased susceptibility to antimicrobial agents.
M. bovis genomic DNAs were prepared from logarithmic-phase broth cultures by using an InstaGene matrix (Bio-Rad Laboratories, Hercules, CA), according to the manufacturer’s instructions. All PCR amplifications were conducted using PrimeSTAR GXL DNA polymerase (TaKaRa Bio, Inc., Otsu, Japan), and the products were purified using a LaboPass PCR purification kit (Cosmo Genetech, Seoul, South Korea) and sequenced on a 3130 Genetic Analyzer (Applied Biosystems) using a BigDye Terminator (version 3.1) ready reaction cycle sequencing kit (Applied Biosystems). MLST was performed with the oligonucleotide primers described previously (28), and these primers have also been posted on the MLST website (https://pubmlst.org/mbovis/). The allele number and ST were determined using the MLST website, and the central and subcentral STs of each ST group were determined by using BURST in the MLST website (https://pubmlst.org/bigsdb?db=pubmlst_mbovis_isolates&page=query). The possible evolutionary relationship between the isolates and M. bovis population structure was determined using the BioNumerics software version 7.5 (Applied Maths, Sint-Martens-Latem, Belgium) and was evaluated by a minimum spanning tree (MST) analysis. The MST was created based on a database of the isolates used in this study and isolates registered on the MLST website.
Oligonucleotide primers for the PCR amplification and sequencing of target genes (i.e., the 16S and 23S rRNA genes [rrs and rrl]) were designed from the M. bovis PG45T genome (GenBank accession no. CP002188) (13). Oligonucleotide primers for the QRDR of gyrA and parC were used as reported in a previous study (21). The sequences of the oligonucleotide primers are shown in Table S3. Sequence editing, consensus, and alignment were performed using GENETYX version 13 (Tokyo, Japan). The numbering of the nucleotide positions and the amino acid positions throughout the article is based on the rrs, rrl, gyrA, and parC genes of Escherichia coli (21, 29, 30), unless otherwise indicated.
Rapid detection of mutations associated with changed susceptibility to antimicrobials by melting curve analysis using hybridization probes.
Extraction of DNA from milk samples was described previously (7). For the detection of target mutations, a 50-μl amplicon was prepared using PrimeSTAR GXL DNA polymerase with 1.5 μl of DNA template from the milk sample, or at least 10 ng of genomic DNA. The PCR primers for amplification of the target regions were designed by comparative analysis of the rrs and rrl sequences between M. bovis (GenBank accession numbers CP002188, CP002058, CP002513, CP005933, CP011348, CP019639, and CP023663) and various Mollicutes bacteria (GenBank accession numbers AB182581, AMWK01000000, AORH00000000, AP013353, AP014631, AP014657, AP017902, AP018135, AUAL01000000, BX293980, CP000896, CP002107, CP002108, CP007154, CP007229, CP007521, CP009770, CP011096, FP236530, FR668087, FUXF00000000, JFDP00000000, JNJU00000000, NC_000908, NC_000912, NC_011374, NR_076192, and X68421). The primer sequences, annealing temperatures, and amplicon sizes are shown in Table 2. The PCR amplification conditions consisted of 35 cycles of 98°C for 10 s, the annealing temperature for 15 s, and 68°C for 60 s/kb of amplicon size. The melting curve analysis assay for the detection of target mutations was carried out in a 20-μl solution of the 2-fold-diluted PCR product containing 0.2 μM each hybridization probe. A total of four hybridization probe sets against target mutations were also designed from the M. bovis PG45T genome (GenBank accession number CP002188) (13). These consisted of a donor probe whose 3′ end was labeled with fluorescein isothiocyanate (FITC) and an acceptor probe whose 5′ end was labeled with LC Red640. The hybridization probe sequences are also shown in Table 2. The protocol for the melting curve analysis using a LightCycler 480 system II (Roche Diagnostics GmbH, Mannheim, Germany) consisted of a single cycle of 95°C for 60 s (20°C/s), 35°C for 60 s (20°C/s), and 70°C for 0 s (0.05°C/s). The analytical results were calculated using the analytical software included with the real-time PCR machine (operated in automatic mode), i.e., LightCycler3 480 SW 1.5.1 and Exor4 for XDMS_R (Roche Diagnostics GmbH). The species specificity of this method was confirmed on genomic DNA samples of nine bovine mycoplasmal strains (M. alkalescens PG51T, M. arginini G230T, M. bovigenitalium PG11T, M. bovirhinis PG43T, M. bovoculi M165/89T, M. canadense 275 CT, M. californicum ST-6T, M. dispar 462/2T, and PG45T), two acholeplasmal strains (Acholeplasma axanthum S743T and Acholeplasma laidlawii PG8T), and nine strains belonging to bacterial species that can cause bovine mastitis (E. coli ATCC 12810, Enterococcus faecalis ATCC 19433, Enterococcus faecium NCTC 7171, Staphylococcus aureus ATCC 12600, Staphylococcus epidermidis ATCC 146, Streptococcus agalactiae NCTC 11360, Streptococcus dysgalactiae NCDO 2023, Streptococcus uberis ATCC 19436, and Streptococcus parauberis DSM 6631). The SCCs in the milk samples were checked using a DeLaval cell counter (Cardiff, UK). Bovine milk samples with various SCCs (20 × 103, 716 × 103, 1,600 × 103, and 3,000 × 103 cell/ml) and serial dilutions of strain PG45T (1.26 × 100 to 1.26 × 107 CFU/ml) were prepared to confirm the sensitivity of this method, and the effects of two milk-sample storage conditions were also evaluated (−20°C or 4°C for 7 days).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a 2017–2021 FY research project of the Ministry of Agriculture, Forestry and Fisheries of Japan: Improving Animal Disease Prevention Technologies to Combat Antimicrobial Resistance.
We thank the staff of the Livestock Hygiene Service Center, the Animal Research Center Hokkaido Research Organization, the Federation of Hokkaido Agricultural Mutual Aid Associations, Kyodoken Institute, and staff or retired staff of the National Institute of Animal Health (M. Eguchi, I. Uchida, K. Nishimori, and K. Tanaka) for their help with the collection of M. bovis isolates and bovine samples from mycoplasmal diseases. Moreover, pirlimycin was kindly supplied by Zoetis Japan and Eiken Chemical Co., Ltd.
We declare that there are no functional or other relationships that might lead to a conflict of interest.
All authors have seen and approved the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00575-19.
REFERENCES
- 1.Hewicker-Trautwein M, Feldmann M, Kehler W, Schmidt R, Thiede S, Seeliger F, Wohlsein P, Ball HJ, Buchenau I, Spergser J, Rosengarten R. 2002. Outbreak of pneumonia and arthritis in beef calves associated with Mycoplasma bovis and Mycoplasma californicum. Vet Rec 151:699–703. [PubMed] [Google Scholar]
- 2.Maunsell FP, Woolums AR, Francoz D, Rosenbusch RF, Step DL, Wilson DJ, Janzen ED. 2011. Mycoplasma bovis infections in cattle. J Vet Intern Med 25:772–783. doi: 10.1111/j.1939-1676.2011.0750.x. [DOI] [PubMed] [Google Scholar]
- 3.Gerchman I, Levisohn S, Mikula I, Lysnyansky I. 2009. In vitro antimicrobial susceptibility of Mycoplasma bovis isolated in Israel from local and imported cattle. Vet Microbiol 137:268–275. doi: 10.1016/j.vetmic.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Kawai K, Higuchi H, Iwano H, Iwakuma A, Onda K, Sato R, Hayashi T, Nagahata H, Oshida T. 2014. Antimicrobial susceptibilities of Mycoplasma isolated from bovine mastitis in Japan. Anim Sci J 85:96–99. doi: 10.1111/asj.12144. [DOI] [PubMed] [Google Scholar]
- 5.Lerner U, Amram E, Ayling RD, Mikula I, Gerchman I, Harrus S, Teff D, Yogev D, Lysnyansky I. 2014. Acquired resistance to the 16-membered macrolides tylosin and tilmicosin by Mycoplasma bovis. Vet Microbiol 168:365–371. doi: 10.1016/j.vetmic.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 6.Hannan PC. 2000. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. International Research Programme on Comparative Mycoplasmology. Vet Res 31:373–395. doi: 10.1051/vetres:2000100. [DOI] [PubMed] [Google Scholar]
- 7.Hata E, Nagai K, Murakami K. 2019. Mutations associated with change of susceptibility to lincosamides and/or macrolides in field and laboratory-derived Mycoplasma californicum strains in Japan, and development of a rapid detection method for these mutations. Vet Microbiol 229:81–89. doi: 10.1016/j.vetmic.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 8.Amram E, Mikula I, Schnee C, Ayling RD, Nicholas RA, Rosales RS, Harrus S, Lysnyansky I. 2015. 16S rRNA gene mutations associated with decreased susceptibility to tetracycline in Mycoplasma bovis. Antimicrob Agents Chemother 59:796–802. doi: 10.1128/AAC.03876-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulyok KM, Kreizinger Z, Wehmann E, Lysnyansky I, Bányai K, Marton S, Jerzsele Á, Rónai Z, Turcsányi I, Makrai L, Jánosi S, Nagy SÁ, Gyuranecz M. 2017. Mutations associated with decreased susceptibility to seven antimicrobial families in field and laboratory-derived Mycoplasma bovis strains. Antimicrob Agents Chemother 61:e01983-16. doi: 10.1128/AAC.01983-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyon E. 2001. Mutation detection using fluorescent hybridization probes and melting curve analysis. Expert Rev Mol Diagn 1:92–101. doi: 10.1586/14737159.1.1.92. [DOI] [PubMed] [Google Scholar]
- 11.Rosales RS, Churchward CP, Schnee C, Sachse K, Lysnyansky I, Catania S, Iob L, Ayling RD, Nicholas RA. 2015. Global multilocus sequence typing analysis of Mycoplasma bovis isolates reveals two main population clusters. J Clin Microbiol 53:789–794. doi: 10.1128/JCM.01910-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stipkovits L, Ripley PH, Tenk M, Glávits R, Molnár T, Fodor L. 2005. The efficacy of valnemulin (Econor) in the control of disease caused by experimental infection of calves with Mycoplasma bovis. Res Vet Sci 78:207–215. doi: 10.1016/j.rvsc.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Wise KS, Calcutt MJ, Foecking MF, Röske K, Madupu R, Methé BA. 2011. Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523). Infect Immun 79:982–983. doi: 10.1128/IAI.00726-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi H, Nakajima H, Shimizu Y, Eguchi M, Hata E, Yamamoto K. 2005. Macrolides and lincomycin susceptibility of Mycoplasma hyorhinis and variable mutation of domain II and V in 23S ribosomal RNA. J Vet Med Sci 67:795–800. doi: 10.1292/jvms.67.795. [DOI] [PubMed] [Google Scholar]
- 15.Lysnyansky I, Gerchman I, Flaminio B, Catania S. 2015. Decreased susceptibility to macrolide-lincosamide in Mycoplasma synoviae is associated with mutations in 23S ribosomal RNA. Microb Drug Resist 21:581–589. doi: 10.1089/mdr.2014.0290. [DOI] [PubMed] [Google Scholar]
- 16.Pereyre S, Gonzalez P, de Barbeyrac B, Darnige A, Renaudin H, Charron A, Raherison S, Bébéar C, Bébéar CM. 2002. Mutations in 23S rRNA account for intrinsic resistance to macrolides in Mycoplasma hominis and Mycoplasma fermentans and for acquired resistance to macrolides in M. hominis. Antimicrob Agents Chemother 46:3142–3150. doi: 10.1128/AAC.46.10.3142-3150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Animal Quarantine Service, Ministry of Agriculture, Forestry and Fisheries. 2017. Annual report of Animal Quarantine Service 1980–2017. Animal Quarantine Service, Narita, Japan: (In Japanese.) [Google Scholar]
- 18.Gautier-Bouchardon AV, Ferré S, Le Grand D, Paoli A, Gay E, Poumarat F. 2014. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS One 9:e87672. doi: 10.1371/journal.pone.0087672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong LC, Gao D, Jia BY, Wang Z, Gao YH, Pei ZH, Liu SM, Xin JQ, Ma HX. 2016. Antimicrobial susceptibility and molecular characterization of macrolide resistance of Mycoplasma bovis isolates from multiple provinces in China. J Vet Med Sci 78:293–296. doi: 10.1292/jvms.15-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narita M. 2013. Diagnosis, treatment and prevention of infectious diseases. Topics: I. Countermeasures against epidemic infectious diseases: 4. Macrolide-resistant Mycoplasma pneumoniae. Nihon Naika Gakkai Zasshi 102:2823–2830. (In Japanese.) doi: 10.2169/naika.102.2823. [DOI] [PubMed] [Google Scholar]
- 21.Lysnyansky I, Mikula I, Gerchman I, Levisohn S. 2009. Rapid detection of a point mutation in the parC gene associated with decreased susceptibility to fluoroquinolones in Mycoplasma bovis. Antimicrob Agents Chemother 53:4911–4914. doi: 10.1128/AAC.00703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bébéar C, Pereyre S, Peuchant O. 2011. Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiol 6:423–431. doi: 10.2217/fmb.11.18. [DOI] [PubMed] [Google Scholar]
- 23.Li BB, Shen JZ, Cao XY, Wang Y, Dai L, Huang SY, Wu CM. 2010. Mutations in 23S rRNA gene associated with decreased susceptibility to tiamulin and valnemulin in Mycoplasma gallisepticum. FEMS Microbiol Lett 308:144–149. doi: 10.1111/j.1574-6968.2010.02003.x. [DOI] [PubMed] [Google Scholar]
- 24.Pereyre S, Métifiot M, Cazanave C, Renaudin H, Charron A, Bébéar C, Bébéar CM. 2007. Characterisation of in vitro-selected mutants of Ureaplasma parvum resistant to macrolides and related antibiotics. Int J Antimicrob Agents 29:207–211. doi: 10.1016/j.ijantimicag.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Bocklisch H, Kreusel S, Bryś A, Pfützner H. 1991. Experimental infection of the udder of ewes due to Mycoplasma bovis. Zentralbl Veterinarmed B 38:385–390. [DOI] [PubMed] [Google Scholar]
- 26.Boonyayatra S, Fox LK, Besse TE, Sawant A, Gay JM. 2010. Effects of storage methods on the recovery of Mycoplasma species from milk samples. Vet Microbiol 144:210–213. doi: 10.1016/j.vetmic.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Hata E, Suzuki K, Hanyu H, Itoh M, Higuchi H, Kobayashi H. 2014. Molecular epidemiology of cases of Mycoplasma californicum infection in Japan. Appl Environ Microbiol 80:7717–7724. doi: 10.1128/AEM.02488-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Register KB, Thole L, Rosenbush RF, Minion FC. 2015. Multilocus sequence typing of Mycoplasma bovis reveals host-specific genotypes in cattle versus bison. Vet Microbiol 175:92–98. doi: 10.1016/j.vetmic.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Petrov AS, Bernier CR, Hershkovits E, Xue Y, Waterbury CC, Hsiao C, Stepanov VG, Gaucher EA, Grover MA, Harvey SC, Hud NV, Wartell RM, Fox GE, Williams LD. 2013. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res 41:7522–7535. doi: 10.1093/nar/gkt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.