Milk undergoes sustained contact with the built environment during processing into finished dairy products. This contact has the potential to influence the introduction, viability, and growth of microorganisms within the milk. Currently, the population dynamics of bacteria in milk undergoing processing are not well understood. Therefore, we measured for total and viable bacterial composition and cell numbers in milk over time and at different processing points in a cheese manufacturing facility in California. Our results provide new perspectives on the dramatic variations in microbial populations in milk during processing even over short amounts of time. Although some of the changes in the milk microbiota were predictable (e.g., reduced viable cell numbers after pasteurization), other findings could not be easily foreseen based on knowledge of bacteria contained in raw milk or when the equipment was last cleaned. This information is important for predicting and controlling microbial spoilage contaminants in dairy products.
KEYWORDS: bovine milk, built environment, cheese, dairy, fermentation, microbiota, processing
ABSTRACT
We set out to identify the viable and total bacterial content in milk as it passes through a large-scale, dairy product manufacturing plant for pasteurization, concentration, separation, blending, and storage prior to cheese manufacture. A total of 142 milk samples were collected from up to 10 pieces of equipment for a period spanning 21 h on two collection dates in the spring and late summer of 2014. Bacterial composition in the milk was determined by 16S rRNA marker gene, high-throughput DNA sequencing. Milk samples from the late summer were paired such that half were treated with propidium monoazide (PMA) to enrich for viable cells prior to quantification by PCR and identification by DNA sequence analysis. Streptococcus had the highest median relative abundance across all sampling sites within the facility on both sampling dates. The proportions of Anoxybacillus, Thermus, Lactococcus, Lactobacillus, Micrococcaceae, and Pseudomonas were also elevated in some samples. Viable cells detected by PMA treatment showed that Turicibacter was enriched after high-temperature short-time pasteurization, whereas proportions of Staphylococcus were significantly reduced. Using clean-in-place (CIP) times as a reference point, Bacillus, Pseudomonas, and Anoxybacillus were found in high relative proportions in several recently cleaned silos (<19 h since CIP). At later times (>19 h after CIP), 10 of 11 silos containing elevated viable cell numbers were enriched in Acinetobacter and/or Lactococcus. These results show the tremendous point-to-point and sample-dependent variations in bacterial composition in milk during processing.
IMPORTANCE Milk undergoes sustained contact with the built environment during processing into finished dairy products. This contact has the potential to influence the introduction, viability, and growth of microorganisms within the milk. Currently, the population dynamics of bacteria in milk undergoing processing are not well understood. Therefore, we measured for total and viable bacterial composition and cell numbers in milk over time and at different processing points in a cheese manufacturing facility in California. Our results provide new perspectives on the dramatic variations in microbial populations in milk during processing even over short amounts of time. Although some of the changes in the milk microbiota were predictable (e.g., reduced viable cell numbers after pasteurization), other findings could not be easily foreseen based on knowledge of bacteria contained in raw milk or when the equipment was last cleaned. This information is important for predicting and controlling microbial spoilage contaminants in dairy products.
INTRODUCTION
Modern agricultural methods and food processing facilities are under increasing pressure to provide enough food for the world’s population, which is expected to reach as many as 9.9 billion people by 2050 (1). Although processing methods have undergone significant improvements such that food can be prepared in higher quantities and with greater levels of safety and quality, foodborne illness is still a significant public health threat, and approximately one-third of all food is lost to waste and spoilage defects (2). The microbial contents of foods prepared in small volumes can be informative (3–9); however, there remains the need to accurately monitor and control spoilage microbes in facilities designed for large-scale processing (10–13). This is evident in dairy processing, which results in numerous food products that are highly vulnerable to quality defects and spoilage.
The culturable microbes in raw (unpasteurized) bovine milk have been intensively studied in efforts to understand and control the quality and safety of fluid milk and dairy products (14, 15). However, culture-based methods are limited in their capacity to identify and quantify different bacterial taxa, in part because not all microorganisms are easily cultivable. With the emergence of techniques targeting nucleic acids, the microbial composition in raw milk was found to be more complex than was previously understood (13). On the other hand, an important drawback of using molecular methods is that they do not typically discriminate between viable and dead cells. This issue is particularly relevant to foods like milk that are pasteurized and where the majority of microorganisms are either injured or dead. To selectively detect living cells, DNA cross-linking agents such as propidium monoazide (PMA) have been applied (16–19).
We previously reported that milk produced and transported in tanker trucks in central California contains a highly diverse assemblage of bacteria that is subject to modification over relatively short periods of time in storage silos at dairy processing facilities (20). Here, we characterized the bacterial diversity in milk as it passes through different processing stages at a large-scale dairy facility prior to the final pasteurization step used in cheese manufacture. These stages encompass the pasteurization, concentration, separation, blending, and storage steps that occur prior to the initiation of cheese fermentations (Fig. 1). Changes in total and viable (PMA-treated) bacterial composition were measured over time in individual pieces of equipment on different sampling dates.
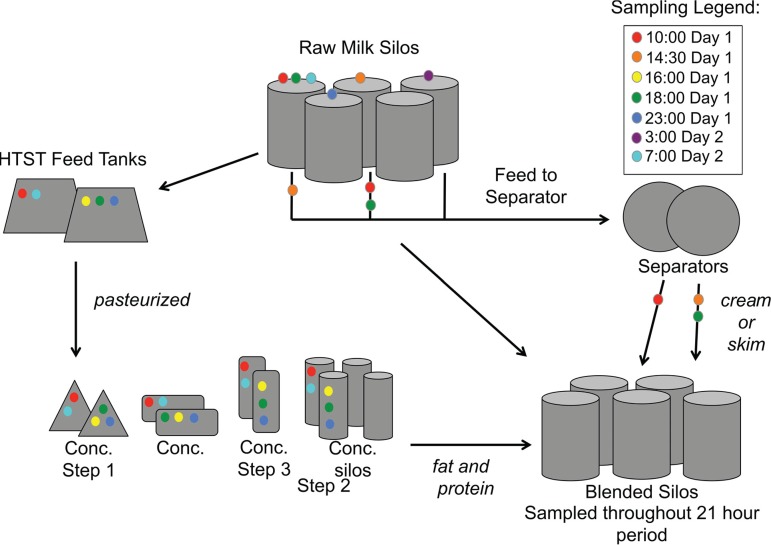
FIG 1.
Diagram of milk sample collection. Milk was collected from actively operating equipment over a 21-h period at the indicated times. Concentration step 3 was not sampled during the spring dates. Arrows indicate the direction that milk was transported through the processing facility. Conc., concentration.
RESULTS
Predominant bacteria in milk during processing.
Members of the Streptococcus genus were present in the highest median relative abundance in milk collected in both spring (April 2014) and late summer (September 2014) dates (Fig. 2). This genus was found in milk at each of the processing steps. It also comprised more than 10% of the milk microbiota for the majority of the samples from both spring (see Fig. S1 in the supplemental material) and late summer (Fig. 3). The representative sequence from the most abundant Streptococcus OTU shared 100% nucleotide identity with Streptococcus thermophilus and Streptococcus salivarius. A conclusive species-level assignment was not possible because of the high level of 16S rRNA gene nucleotide conservation (>99.9% percent) between these two species.
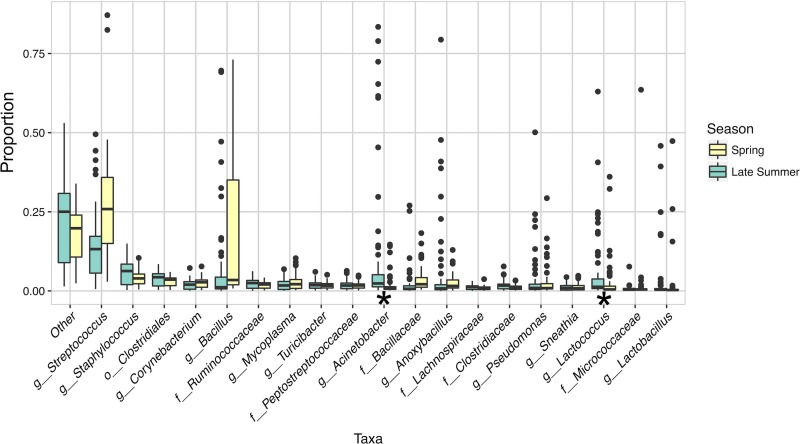
FIG 2.
Bacteria present in milk during processing. Box plots of taxa detected at >0.01 average relative abundance during milk storage, pasteurization, concentration, and separation processing steps are shown for two sampling dates in 2014 (“Spring” indicates sampling performed on 1 and 2 April 2014 and “Late Summer” indicates 29 and 30 September 2014). These samples were not PMA treated prior to analysis. Milk microbiota from concentration step 3 were not included here because they were not tested in the spring. Proportions were estimated from an OTU table rarefied at 9,000 sequences per sample. The “Other” category represents taxa that were present at <0.01 mean relative abundance in the data set. LefSe analysis was performed with season as class and equipment type as subclass. Boxes define the interquartile range of taxonomic abundance and the individual points are outliers. Significant differences in taxa between seasons are indicated by an asterisk. Taxonomic levels are abbreviated in the x axis according to the following: “o” = order, “f” = family, and “g” = genus.
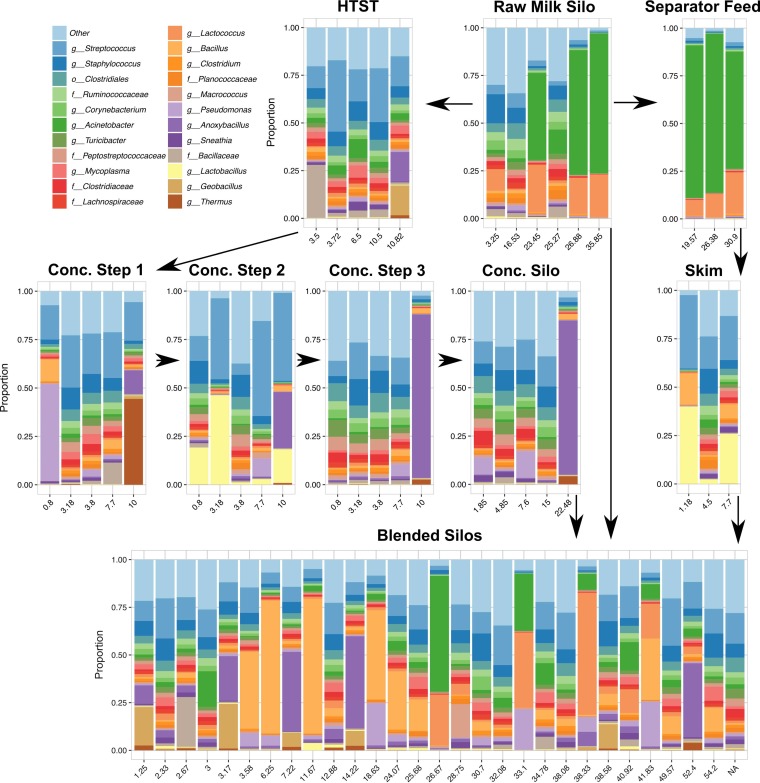
FIG 3.
Dominant bacterial taxa in milk during the late summer sampling date. Bacterial taxa present at >0.01 average relative abundance within the data set after rarefaction to 9,000 sequences per sample are shown. Taxa present at <0.01 average relative abundance were grouped into the category “Other”. Each bar graph is labeled with the piece of equipment that samples were collected from. The number of hours that each piece of equipment had been running prior to sample collection is shown on the x axis. Arrows indicate the direction that the milk moves through the facility.
Bacteria present in <1% mean relative abundance, designated the “other” category, also comprised a large fraction of the total microbiota (Fig. 2), a result that is indicative of the diverse microbial composition of milk. Among the taxa that were present in at least a 1% mean relative abundance, certain bacteria were more ubiquitously present, while others were enriched in particular pieces of equipment (see Fig. S1 [spring] in the supplemental material and Fig. 3 [late summer]). Comparisons between the different collection dates also showed that Acinetobacter (linear discriminant analysis [LDA] effect size = 2.84, P = 5.13 × 10−7) and Lactococcus (LDA effect size = 2.57, P = 6.40 × 10−7) were significantly more abundant in milk from the late summer compared to the spring (Fig. 2).
Conditionally rare taxa are those organisms that are typically rare but occasionally become highly abundant and contribute a greater amount to microbial community dynamics than is apparent from their overall low proportional abundance (21). Although our data set was not perfectly appropriate for the calculation of binomial distributions of taxa over time (21), several genera followed this trend. Anoxybacillus was present at a very low median relative abundance within the facility (an average of 1.6% in spring and 0.8% in late summer [Fig. 2]) but represented more than 40% of total bacteria detected in four milk samples from the late summer. These samples were collected downstream of pasteurization after equipment had been running for more than 7 h and included milk from the following: concentration step 3 after that piece of equipment had been running for 10 h (10:00 in the morning), a concentration silo after operating for 22.5 h (also at 10:00 in the morning), a blended silo sampled after 7.2 h (20:03 in the evening), and a blended silo after 14.2 h (21:23 in the evening) of operation (Fig. 3 and see Fig. S2 in the supplemental material). Equipment upstream of concentration step 3, including the high-temperature short-time (HTST) feed tank, and concentration step 1 also harbored a >10% relative abundance of Anoxybacillus after 10 h of operation (at 10:00 in the morning).
Other conditionally rare taxa included Thermus, Lactococcus, Lactobacillus, Micrococcaceae, and Pseudomonas (Fig. 2, Fig. 3, and Fig. S1). In the late summer, Thermus was enriched in milk from concentration step 1, a processing point that occurs immediately after HTST pasteurization. Lactobacillus was enriched at concentration step 2 on both collection dates and in skim milk in the separator in late summer. Lactococcus levels, on the other hand, were highest in raw milk and blended silos on both dates. A high level of Micrococcaceae was observed in a raw milk silo in the spring, whereas the highest relative abundances of Pseudomonas were observed in the concentration steps (a concentration silo in spring and concentration step 1 in late summer) (Fig. 3 and Fig. S1).
The predominance of Pseudomonas so soon after pasteurization merited further investigation. The majority (82.4%) of pseudomonads identified in concentration step 1 in the summer were defined by a single operational taxonomic unit (OTU). The representative sequence from this OTU shared 99% DN sequence identity with Pseudomonas thermotolerans and Pseudomonas jinjuensis (Fig. S3). Interestingly, Pseudomonas OTUs from the blended silos shared 100% sequence identity with other Pseudomonas species (Fig. S3).
Viable bacterial cell number and diversity changes postpasteurization according to PMA treatment.
To optimize the PMA protocol for detection of viable bacteria recovered from milk, living and heat-killed L. casei BL23 cells were suspended in ultra-high-temperature (UHT)-treated milk, washed with phosphate-buffered saline (PBS), and subjected to different PMA concentrations and incubation times. Exposure for 5 min to a concentration of 25 μM PMA was selected for use because this protocol was sufficient to render the dead cells to be minimally detectable (2.23 × 107 total cell reduction, representing approximately 99.9% of the untreated control cells) while retaining the maximum number of living cells (Fig. S4).
All milk samples collected in the late summer were measured for both viable (PMA-treated) and total (no-PMA treatment) bacterial cell numbers by quantitative PCR (qPCR). These results showed that milk treated with PMA contained between 29 cells/ml and 4.3 × 105 cells/ml (average = 1.6 × 104 cells/ml; median = 1.2 × 103 cells/ml) (Fig. S5A). The total bacterial cell numbers were similar, with a range of 52 cells/ml to 4.7 × 105 cells/ml (average = 1.4 × 104 cells/ml; median = 1.9 × 103). PMA-enriched and total bacterial cell counts were highly positively correlated (Pearson r = 0.9814) (Fig. S5B).
We next compared viable (PMA-treated) and total (no-PMA treatment) cell numbers and beta-diversities for milk collected pre- and postpasteurization. For the raw, unpasteurized milk, the estimates of viable and total bacterial cell numbers were equivalent (Fig. 4A). This result was consistent with the lack of any detectable differences in bacterial diversity, according to the weighted UniFrac distance metric, between the viable and total cell fractions of each milk sample (Fig. 4B).
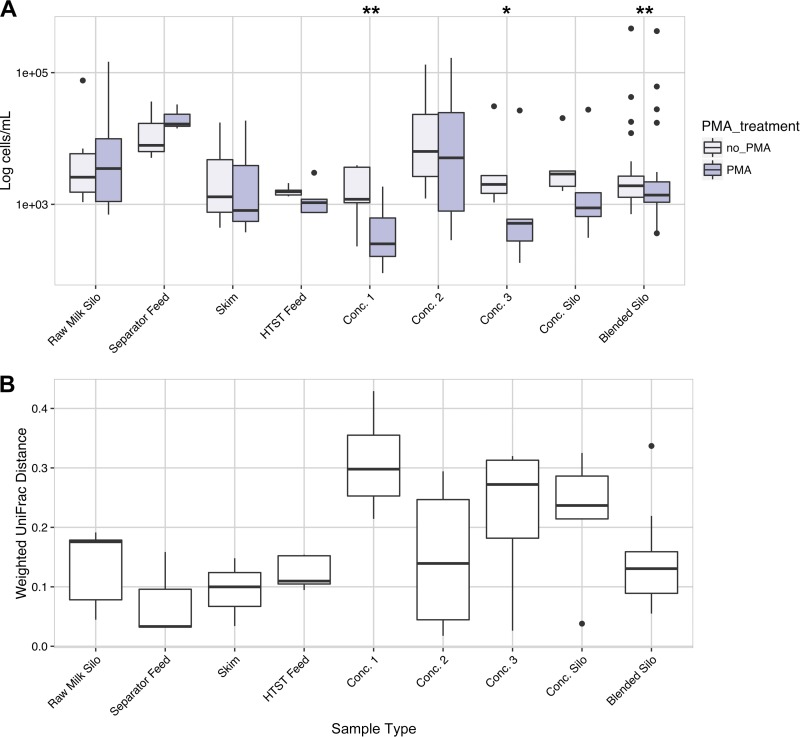
FIG 4.
PMA treatment results in significant effects on milk collected postpasteurization. (A) Weighted UniFrac distance between the PMA treated and untreated milk samples. (B) Bacterial cell counts (log10 transformed) estimated by qPCR for PMA-treated (PMA) or untreated (no PMA) milk collected during the late summer. Significant differences between cell count estimates for PMA-treated and untreated samples were determined by the paired t test, assuming equal variance, and are indicated by asterisks. *, P < 0.05; **, P < 0.01.
After pasteurization, the viable and total bacterial populations were distinct (Fig. 4). For milk collected from most pieces of equipment after pasteurization (concentration step 1 to the concentration silos), the qPCR estimates of the viable bacterial cell numbers were lower than found for total cell counts, thereby indicating high proportions of dead cells (Fig. 4A). Higher median UniFrac distances were also found between the bacteria in pasteurized milk from concentration step 1, concentration step 3, and concentration silos (Fig. 4B). Concentration step 2 differed from the others because it contained higher cell numbers, even after the exclusion of dead cells by PMA (Fig. 4). The bacterial diversities at concentration step 2 were also more similar between the viable and total cell fractions (Fig. 4B). Lastly, although the blended silos had significantly lower bacterial cell counts after PMA treatment, the viable cell community composition was similar to the total (Fig. 4).
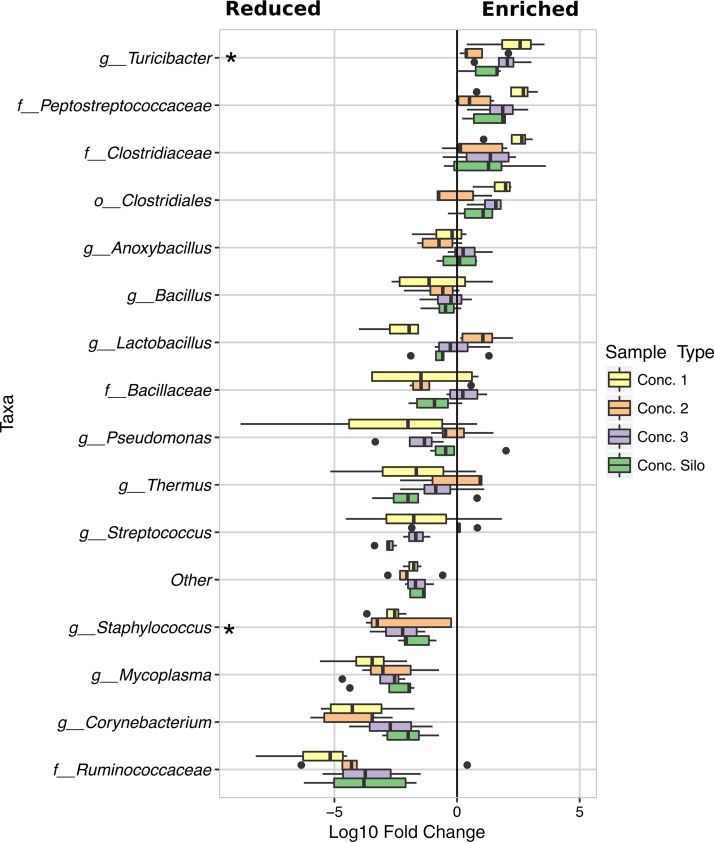
Pasteurization enriches for certain bacteria in the Firmicutes phylum.
Turicibacter, a genus in the Firmicutes phylum, was significantly enriched (LDA effect size = 2.57, P = 0.01406) among the viable cells found in pasteurized milk (concentration steps 1, 2, and 3 and concentration silo) (Fig. 5). This result was confirmed by Turicibacter genus-specific qPCR (Fig. S6). Although Turicibacter was the only taxon that reached statistical significance for enrichment after PMA treatment, Firmicutes in the Peptostreptococcaceae and Clostridiaceae families, as well as other members of the Clostridia class, also showed a trend toward enrichment. Notably, proportions of endospore-forming bacteria such as Anoxybacillus, Bacillus, and Thermus exhibited very little change with PMA exposure, indicating that they were viable at the time of sampling. Certain Bacilli that do not produce endospores also retained viability, including Streptococcus and Lactobacillus, two genera that also dominated the bacterial communities in samples from concentration step 2 (Fig. 3 and 5).
FIG 5.
Microbial taxa impacted by pasteurization as detected by PMA treatment. The fold change in the proportions of bacterial taxa between PMA-treated and untreated milk collected from equipment after pasteurization. Prior to the fold change calculations for each taxa (PMA-treated aliquot/untreated aliquot), sequence counts of 0 were replaced with 1. Unchanged OTU counts from each sample were analyzed using LefSe with PMA treatment set as class and the collection event (one PMA-treated aliquot and one untreated aliquot from each sample were compared) set as subclass. Pairwise comparisons were made only with subclasses of the same name. A significant difference (LDA effect size > 2, P < 0.005) between the PMA-treated and untreated aliquots for all four pieces of equipment is indicated by an asterisk.
Conversely, the proportions of other Bacilli and Clostridia in pasteurized milk were lower in the viable (PMA-treated) cell population than in the total (no-PMA) cell population. Staphylococcus (class Bacilli) cell proportions were among those taxa that were significantly reduced (LDA effect size = 2.31, P = 0.00014) (Fig. 5). The relative abundances of bacteria in the Ruminococcaceae family (class Clostridia) were also lower, although not significantly. In addition, the proportions of Corynebacterium (class Actinobacteria) were less, as were those for other bacteria in that class, leading to a significant reduction at the class (LDA effect size = 2.27, P = 0.00036) and order (Actinomycetales) levels (LDA effect size = 2.26, P = 0.00026).
Time since CIP results in variable outcomes in bacterial cell numbers and diversity.
Milk processing equipment is regularly cleaned. Clean-in-place (CIP) times for different pieces of equipment (raw milk silo, separator feed, concentration silo, and blended silo) were collected for the late summer sampling dates, thereby enabling comparisons between CIP times and bacterial composition. To inform our understanding of how viable bacterial communities might be impacted by CIP times, only PMA-treated milk samples were used for this analysis. Similar conclusions were reached when all (viable and dead) bacterial cells were examined (Fig. S7).
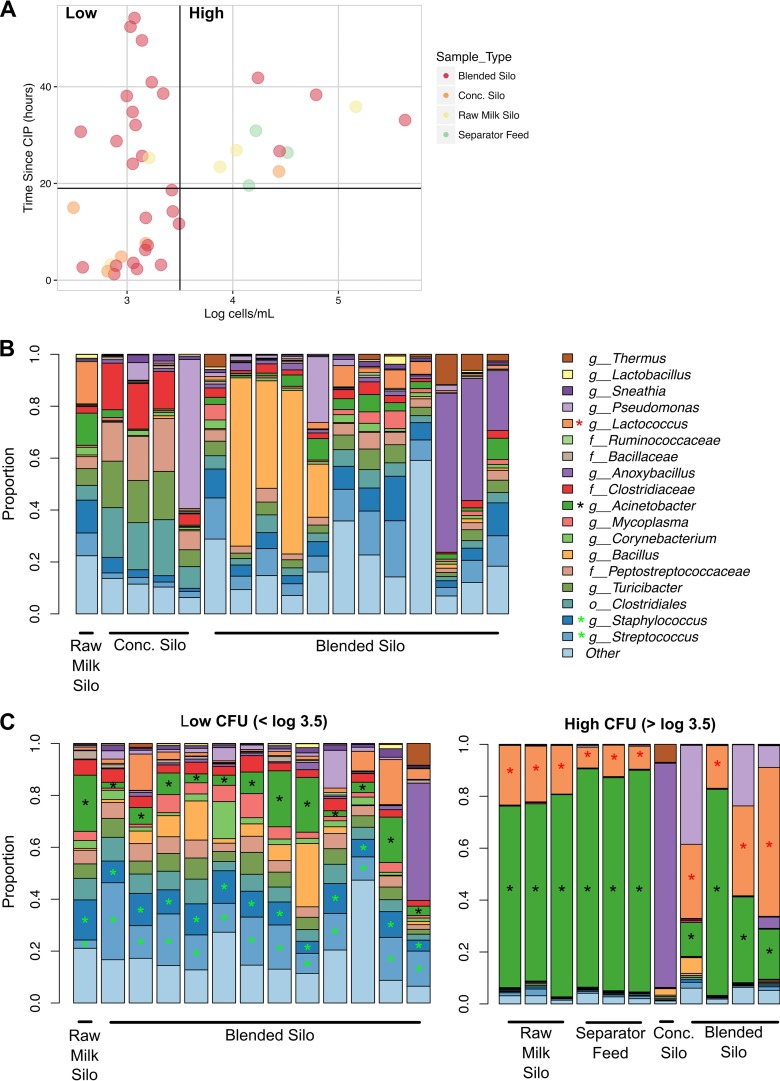
Comparisons of viable cell numbers to CIP times showed that silos and separator feed equipment that had been cleaned within 19 h of sampling had ≤3,200 bacterial cells/ml (Fig. 6A). Dominant (viable) taxa from milk in processing equipment <19 h postcleaning included Bacillus, Pseudomonas, and Anoxybacillus (Fig. 6B).
FIG 6.
Comparison of CIP and bacterial cell numbers and diversity. PMA-treated milk collected from silos on the late summer collection dates are shown. (A) Estimated viable bacterial cell counts compared to the number of hours since clean in place (CIP) for each milk sample. A vertical line at 3,200 cells/ml and a horizontal line at 19 h since CIP delineate the cutoff for low and high bacterial loads in silos, respectively. (B) Microbial community structure of individual milk samples collected from silos with <19 h since CIP. (C) Microbial community structure of individual milk samples collected from silos with >19 h since CIP and low (left panel) or high (right panel) bacterial loads. A black asterisk indicates significant (LDA effect size > 2, P < 0.05) enrichment in both low- and high-cell-number groups relative to recently cleaned silos (<19 h CIP). Green and red asterisks indicate enrichment in milk containing low and high numbers of cells relative to recently cleaned silos, respectively.
After 19 h since CIP, the milk samples could be divided into two groups: (i) milk in which the bacterial load did not increase above 3,200 cells/ml even 54 h after CIP and (ii) milk in which cell numbers increased over time after CIP (Fig. 6A). The bacteria in milk at processing steps that maintained low cell numbers, even with long durations of time since CIP (Fig. 6A and C), were diverse and contained numerous taxa. This milk was also significantly enriched with Acinetobacter, Streptococcus, and Staphylococcus relative to milk from recently cleaned equipment (<19 h prior) (Fig. 6B). In comparison, milk that harbored increased quantities of bacteria was dominated by only a few taxa (>70% of the total community). Both Acinetobacter and Lactococcus were highly enriched compared to both recently cleaned equipment and equipment that maintained low cell numbers after CIP (Fig. 6, Fig. S8A). Further analysis showed that the most abundant Lactococcus OTU (median 92% of the total Lactococcus OTUs) was closely related to Lactococcus lactis and Lactococcus taiwanensis. The estimated cell amounts of Bacillus and Anoxybacillus, two potential spoilage agents, were similar between silos that had low and high total bacterial cell counts (Fig. S8).
DISCUSSION
Initial dairy processing steps frequently involve milk pasteurization, separation, and concentration, followed by blending to reach specific ratios of fat and protein (22). In this study, we identified the consistently present, as well as the more variable, bacterial taxa found in bovine milk during processing intended for cheese manufacture. We also showed how the bacterial content in milk at each of those processing steps changes over short periods of time within and between individual pieces of equipment. Identification and quantification of the viable bacteria confirmed the impacts of pasteurization at the level of individual processing steps and for specific taxa. This variation in bacterial diversity has important consequences for predicting how the microorganisms in milk affect product quality.
Streptococcus was the most abundant bacterial genus in the milk on both sampling dates, as was previously observed for raw milk in tanker trucks delivered to the same facility (20). Because this genus was consistently found in milk from all pieces of equipment, including among the viable cells measured after pasteurization, our findings support the work of others showing the persistence of this genus in milk during the manufacture of cheese and other dairy products (13, 23).
Also consistent with prior findings (20) was the large fraction of bacteria that were rare (<1% relative abundance) (“other” in Fig. 1). Other bacteria were conditionally rare, such that they were only sporadically dominant. Those taxa included the dairy-relevant genera Thermus, Anoxybacillus, Lactococcus, Lactobacillus, Micrococcaceae, and Pseudomonas. The high proportions of these bacteria could be due to their predominance in some shipments of raw milk, their enrichment or introduction in particular pieces of equipment, or a combination of these factors. This “conditionally rare” phenomenon was observed previously in other environments, including air, large bodies of water, human skin and gastrointestinal tract, and brewery wastewater treatment facilities (21).
In the milk examined here, Thermus was enriched in milk at concentration step 1 at 10 h of operation. Concentration step 1 is the step that immediately follows pasteurization. Irrespective of the entry point of this organism into the milk, this finding is consistent with the thermotolerant properties of the Thermus species. Thermus is important in dairy processing because it was shown to cause pink discoloration in cheese (24, 25). Although the enrichment of Thermus could be a cause for concern, our data indicate that this genus is likely controlled by current cleaning protocols or is only an occasional contaminant, because it was not present throughout the sample collection period. Instead, Thermus proportions constituted only 0.03% of the total bacterial population in milk from concentration step 1 a couple of hours later (16:00 in the afternoon), after a CIP on the same day.
Anoxybacillus was also a conditionally rare member of the milk microbiota. This genus was enriched in milk sampled at the same time and on the same day from concentration steps 2 and 3 and the concentration silo, along with a couple of other time points in blended silos. Anoxybacillus has been frequently found as a contaminant of dairy processing facilities (26) and particularly in milk powders (27, 28). High endospore counts caused by this genus can lead to rejection of milk powder due to presumed poor hygiene (27). Thermophiles such as Anoxybacillus in general can cause product spoilage (29–31). Like Thermus, Anoxybacillus seems to be relatively well controlled as the relative abundance of this genus was at <0.8% by 16:00 in all concentration steps, after CIP occurred. However, because the proportions of Anoxybacillus and Thermus were not significantly reduced by PMA treatment, even immediately after pasteurization, the results suggest that these taxa, when present, were likely viable.
Pseudomonas was sporadically enriched in the viable and total cell fractions of milk collected from the concentration steps following pasteurization, as well as from blended silos. The presence of Pseudomonas postpasteurization is a concern in dairy processing plants because of the proteases, lipases, and lecithinases produced by these bacteria, which are responsible for decreased shelf life and spoilage of fluid milk (32) and cheese (33) even after cell death has occurred. Pseudomonas is generally understood to be sensitive to standard milk pasteurization protocols (34, 35). Therefore, the (re)introduction of Pseudomonas might have also occurred, considering that pasteurized milk is vulnerable to microbial contamination. Such a possibility is consistent with prior studies (13, 35–37), including reports wherein Pseudomonas was found in water used throughout the plant (35, 37). Although the most abundant Pseudomonas OTUs in pasteurized milk were highly similar to the thermotolerant species P. thermotolerans (38), confirmation requires further investigation.
Because dead or inactive bacteria can be detected by standard 16S rRNA gene sequencing methods, we also applied PMA to enrich for living, or viable, cells in the milk. PMA-based detection relies on the presence of an intact cell membrane to prevent binding to genomic DNA. PMA treatment was especially appropriate for detection of live bacterial cells after pasteurization given that this compound was shown to be particularly effective for differentiating living from dead cells when cell death was caused by heat stress (39). A limitation of this approach is the tendency for increased or decreased diffusion of PMA through the bacterial cell membrane, depending on the general physiological state and extracellular properties of the organism (e.g., exopolysaccharide, peptidoglycan, etc.) (18, 40). Moreover, in the presence of high numbers of living cells, PMA can result in overestimates in viable cell numbers. However, the high correlation between estimated bacterial cell counts before and after PMA treatment and the lack of enrichment of only a few taxonomic groups suggest that the selected protocol, at worst, reduced the living fraction proportionally relative to the total community. At best, and consistent with the enrichment of thermodurics in pasteurized milk after PMA treatment, the PMA method allowed a good approximation of bacterial taxa that were viable both pre- and postpasteurization.
The largest changes in both total cell counts and overall microbial community structure with PMA treatment occurred in samples downstream of pasteurization. This was expected because pasteurization is likely to kill the majority of bacteria present. Several, primarily endospore-forming, members of the Firmicutes phylum were enriched among the viable cells following HTST treatment. Bacteria in the Turicibacter genus, an endospore-forming member of the class Erysipelotrichia (41), were significantly higher in all subsequent treatment steps measured. Although Turicibacter was previously detected in pasteurized milk (13, 42), the impact of this organism on dairy product quality is not known. On the other hand, the proportions of other Firmicutes, including members of the Staphylococcus genus, were significantly reduced in milk after pasteurization. Other Firmicutes and Actinobacteria were only modestly (nonsignificantly and sometimes negatively) affected.
Milk in concentration step 2 was an outlier in our comparisons, and the microbiota in that piece of equipment did not exhibit a substantial change in microbial community structure or a significant decrease in estimated bacterial cell counts following PMA exposure. Instead, these milk samples contained a slight increase in cell numbers and a different community structure, enriched in Streptococcus and Lactobacillus compared to the surrounding equipment. Although the reason for this is not clear, it is possible that these two genera were concentrated in concentration step 2 but were not retained or were significantly diluted at subsequent steps.
Because CIP times were available for equipment sampled on the late summer collection dates, we were able to make direct comparisons between the time since cleaning and bacterial populations present in the milk. The milk generally contained low bacterial cell numbers (<3.5 log10 cells/ml), independent of CIP. However, cell numbers were increased in several samples retrieved more than 19 h after CIP. Acinetobacter and/or Lactococcus comprised the majority of viable bacteria in most of those samples. Because both of these genera were also enriched in raw milk silos tested on other dates (20), it is possible that a high abundance of Acinetobacter and Lactococcus is a facility-specific characteristic. Because the predominance of these organisms could not be directly linked to time since CIP, these bacteria might reside in biofilms which cannot be completely removed by cleaning.
Our findings suggest that equipment-specific trends in microbial composition should be identified and closely monitored when the time after CIP exceeds 19 h. However, it should be noted that elevated bacterial numbers might not necessarily be associated with spoilage. Instead, the presence of Lactococcus might contribute positively to cheese development given that the DNA sequences in this data set share a high level of nucleotide identity with L. lactis. Moreover, bacteria more commonly recognized as spoilage agents (e.g., Bacillus or Anoxybacillus) were proportionally more abundant, with estimated cell numbers unchanged, in milk containing low total quantities of bacteria.
In conclusion, we found that bacterial populations are highly dynamic over spatial and temporal scales in milk undergoing common processing steps. Although some of the changes in the milk microbiota are predictable (e.g., reduced viable cell numbers after pasteurization), other differences would not be easily estimated based on knowledge of the bacteria contained in raw milk or CIPs. These other differences may be due to facility- or equipment-specific bacterial communities that should be identified within individual manufacturing facilities to allow location-specific monitoring for spoilage-associated organisms. Because 16S rRNA marker gene surveys are limited by their capacity to identify some bacteria to the species level (e.g., Streptococcus, Pseudomonas, and Lactococcus), this limitation should be taken into account. Although we were able to rule out some species among these genera, other primer sets or surveys targeting protein-encoding genes might provide additional resolution. However, even with these limitations, the findings here provide targets within the facility (e.g., blended silos) where more strain level/species level monitoring could potentially be used to predict product spoilage. Further studies, including culture-based studies, are needed within this and other dairy processing facilities to determine the cause of this microbial variation and ultimately establish the relative impact of different milk-associated microbiomes on product quality.
MATERIALS AND METHODS
Sample collection and processing.
A total of 71 milk samples were collected on 1 and 2 April 2014 (referred to as the spring season dates), and another 71 samples were collected on 29 and 30 September 2014 (referred to as the late summer dates). Milk was collected directly from 8 (spring) to 10 (late summer) types of equipment (Fig. 1). The equipment is used for pasteurization, concentration, separation, blending, and storage of milk prior to the final pasteurization step before curd production. Milk from actively operating equipment was collected once every 1.5 to 4.5 h for a total of three to six collection times for each piece of equipment. Blended silos were the exception and were randomly sampled throughout 21-h periods to obtain a total of 30 and 29 samples, respectively, on the spring and late summer collection dates. Concentration step 3 was sampled during the late summer only. The amount of time that the equipment had been operating prior to sampling varied from 0.8 to 54.2 h. To the best of our ability, we attempted to collect the same batches of milk as it flowed through the facility; however, because of the scale of the operation, it is likely that more than one batch was examined on each of the collection dates.
Milk was collected from each piece of equipment as previously described (20) into 200- to 400-ml food-grade or clinical-grade sterile bags and stored at 4°C until the end of the day. Samples were transported overnight on ice in hard sided insulated containers and were received within 12 to 32 h after collection. Upon receipt, each milk fraction for the late summer date was mixed by shaking and inversion and divided into two 30-ml aliquots for measurement with or without PMA treatment. One 30-ml aliquot was obtained for samples collected for the spring season. Each of the aliquots was centrifuged at 13,000 × g at 4°C for 5 min. Cream samples were mixed with an equal quantity of PBS (pH 7.2; 137 mM NaCl, 2.68 mM KCl, 10.1 mM Na2HPO4, 1.76 mM KH2PO4) by vortexing prior to centrifugation. For all samples, the fat layer and supernatant were removed. Cell pellets were then suspended in PBS and centrifuged again prior to storage at –80°C.
Propidium monoazide methods.
The selective binding of propidium monoazide (PMA) to DNA of dead rather than to living bacterial cell populations can be affected by the type of cells being assayed and the matrix in which the cells are contained. Therefore, we optimized a propidium monoazide protocol using Lactobacillus casei in UHT milk. L. casei BL23 (43) (provided by Vicente Mondero, IATA-CSIC, Spain) was grown to exponential phase (108 cells/ml) in Lactobacilli MRS broth (Becton Dickinson, Franklin Lakes, NJ). A fraction of the cells was heated to 80°C for 5 min for inactivation. Each of the resulting cultures was washed with PBS and inoculated into UHT-treated milk at a 1:100 ratio for a final concentration of 106 cells/ml. Serial dilutions of the suspensions were immediately plated on MRS agar for enumeration of viable L. casei. The cells were then washed in PBS again and suspended in PBS to mimic methods for collecting bacteria from milk as described above. Each washed culture was then divided into 500-μl aliquots into which either PMA at a final concentration of 25 or 50 μM PMA or an equal volume of water (untreated control) was added. The suspensions were incubated in duplicate in the dark with shaking at 200 rpm on a gyratory shaker (model G2; New Brunswick Scientific Co., Inc., Edison, NJ) for 5, 15, or 30 min as previously described (19). Subsequently, the cells were exposed to a 500-W halogen light bulb held 20 cm away for 5 min. During that time, the cells were kept on ice and rotated at 1-min intervals. Excess PMA was removed by centrifugation at 13,000 × g for 2 min, followed by washing in PBS before the cells were collected again by centrifugation at 13,000 × g for 4 min.
For detection of the living fraction of bacteria in milk collected at the commercial processor, cell pellets recovered from the late summer milk samples were exposed to PMA prior to freezing at –80°C. For PMA treatment, the cells were suspended in 500 μl of PBS, and 6.25 μl of a freshly prepared 2 mM PMA stock solution was added to achieve a final concentration of 25 μM PMA. These suspensions were treated as described above prior to DNA extraction.
DNA extraction.
DNA was extracted from cells collected from milk using the PowerFood microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) as previously described (20).
Estimation of total bacterial cell numbers and Turicibacter by quantitative real-time PCR.
Bacterial cell numbers were estimated using quantitative PCR (qPCR) targeting bacterial 16S rRNA genes with primers UniF (GTGSTGCAYGGYYGTCGTCA) and UniR (ACGTCRTCCMCNCCTTCCTC) (44, 45) as previously described (20). Reactions were prepared with a 400 nM primer mixture and SsoFast Evagreen Supermix with Low ROX (Bio-Rad Laboratories, Inc.). Thermocycling was performed in a 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA). Paired t tests assuming equal variance were performed in R (www.r-project.org) on log10-transformed values of cells/ml estimated using qPCR for milk collected from within the dairy processing facility before and after PMA treatment.
Turicibacter cell numbers were estimated by qPCR using the primers TuriciF (CAGACGGGGACAACGATTGGA) and TuriciR (TACGCATCGTCGCCTTGGTA) targeting a conserved region in Turicibacter 16S rRNA genes (46). The standard curve for Turicibacter quantification was from a PCR product that was gel purified (Wizard SV gel and PCR cleanup system; Promega, Madison, WI). The PCR product was obtained using the primers TuriciF and TuriciR and milk collected at concentration step 1 as the template. The PCR amplicon DNA was quantified with a Qubit 3.0 fluorometer using a Qubit double-stranded DNA (dsDNA) HS assay kit (Life Technologies, Eugene, OR) and sequenced to confirm that only Turicibacter DNA was amplified (46). 16S rRNA gene copies determined by qPCR were divided by 3 to account for the three 16S rRNA genes in the Turicibacter sanguinis genome, and this value was used to estimate Turicibacter cell numbers.
For the development of the PMA assay, DNA was extracted from L. casei BL23 grown to stationary phase at 37°C in MRS broth. The concentration of extracted DNA was determined using a Quant-iT PicoGreen dsDNA assay kit (Molecular Probes) according to the manufacturer’s instructions. Dilutions of the genomic DNA were used to construct a qPCR standard curve employed in the development of the PMA treatment method. Estimates of the total number of L. casei genome copies/ml were based on the known genome size of 3.1Mbp. The second standard curve, used for estimation of total cell number in milk samples, was prepared from exponential-phase L. casei BL23 collected from the manufacturing facility. For that standard curve, concurrent with DNA extraction, viable cell numbers of L. casei were estimated by plating serial dilutions of the culture onto MRS agar for colony enumeration.
16S rRNA gene sequencing and analysis.
DNA sequencing was performed as previously described (20). Briefly, primers F515 and R806 were used to amplify the V4 region of 16S rRNA genes (47) with Ex Taq DNA polymerase (TaKaRa, Otsu, Japan). Pooled PCR amplicons were sequenced by 250-bp paired-end sequencing on an Illumina MiSeq at the University of California, Davis, CA (http://dnatech.genomecenter.ucdavis.edu/). An 8-bp barcode present on the 5′ end of primer F515 was used to demultiplex the DNA sequence reads during analysis.
FASTQ files were analyzed with QIIME version 1.9.1 (48) as previously described (20). OTU counts generated using QIIME were filtered to remove OTUs occurring at <0.005% relative abundance (49). OTU counts were adjusted by rarefaction at a depth of 9,000 sequences per sample for further analysis. Fewer than 9,000 sequence reads were obtained for each cream sample, and therefore bacterial community analysis was not performed on these samples. However, they were included in cell/ml estimations. To determine statistically significant differences in taxonomic abundance between experimental groups, rarefied OTU counts were summed by taxonomic level. Taxa present at less than 1% relative abundance in all samples were combined into a category termed “Other.” Differential taxonomic abundances between experimental groups were then analyzed using LefSe (50).
Because the spring and late summer samples were sequenced in different MiSeq runs, PCR amplicons from five milk samples were included in each run to examine for batch effects. The gPCA.batchdetect function in the gPCA package in R (51) showed no significant effect of sequencing run (1,000 permutations, P = 0.719) between spring and late summer. Therefore, no batch correction was performed. Moreover, storage at 4°C overnight prior to DNA extraction did not result significant changes to bacterial composition. This was confirmed by a lack of effect size greater than 2 according to LefSe for 28 raw milk samples processed immediately after collection compared to 54 samples, collected on the same date, that were first placed at 4°C overnight prior to extraction.
Data availability.
The DNA sequences for this study are publicly available through the Qiita database (https://qiita.ucsd.edu) under study ID 10485 and in the European Nucleotide Archive under accession number ERP015209.
Supplementary Material
ACKNOWLEDGMENTS
The California Dairy Research Foundation grant “Bacterial Signatures of Milk Safety and Quality” funded this study. The funding agency did not participate in study design, data collection, or interpretation of the data. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). The USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00270-19.
REFERENCES
- 1.Keneda T, Bietsch K. 2016. 2016 world population data sheet. Population Reference Bureau, Washington, DC: http://www.prb.org/Publications/Datasheets/2016/2016-world-population-data-sheet.aspx. [Google Scholar]
- 2.Alexander P, Brown C, Arneth A, Finnigan J, Moran D, Rounsevell M. 2017. Losses, inefficiencies, and waste in the global food system. Agric Systems 153:190–200. doi: 10.1016/j.agsy.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bokulich NA, Mills DA. 2013. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl Environ Microbiol 79:5214–5223. doi: 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dugat-Bony E, Garnier L, Denonfoux J, Ferreira S, Sarthou AS, Bonnarme P, Irlinger F. 2016. Highlighting the microbial diversity of 12 French cheese varieties. Int J Food Microbiol 238:265–273. doi: 10.1016/j.ijfoodmicro.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 5.Duthoit F, Godon JJ, Montel MC. 2003. Bacterial community dynamics during production of registered designation of origin Salers cheese as evaluated by 16S rRNA gene single-strand conformation polymorphism analysis. Appl Environ Microbiol 69:3840–3848. doi: 10.1128/AEM.69.7.3840-3848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randazzo CL, Torriani S, Akkermans ADL, de Vos WM, Vaughan EE. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl Environ Microbiol 68:1882–1892. doi: 10.1128/AEM.68.4.1882-1892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacerda ICA, Gomes FCO, Borelli BM, Faria CLL, Franco GR, Mourao MM, Morais PB, Rosa CA. 2011. Identification of the bacterial community responsible for traditional fermentation during sour cassava starch, cachaca and minas cheese production using culture-independent 16s rRNA gene sequence analysis. Braz J Microbiol 42:650–657. doi: 10.1590/S1517-83822011000200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang WX, Qiao ZW, Shigematsu T, Tang YQ, Hu C, Morimura S, Kida K. 2005. Analysis of the bacterial community in Zaopei during production of Chinese Luzhou-flavor liquor. J I Brewing 111:215–222. doi: 10.1002/j.2050-0416.2005.tb00669.x. [DOI] [Google Scholar]
- 9.Stellato G, De Filippis F, La Storia A, Ercolini D. 2015. Coexistence of lactic acid bacteria and potential spoilage microbiota in a dairy processing environment. Appl Environ Microbiol 81:7893–7904. doi: 10.1128/AEM.02294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo R, Xue T, Weeks M, Turner MS, Bansal N. 2016. Inhibition of bacterial growth in sweet cheese whey by carbon dioxide as determined by culture-independent community profiling. Int J Food Microbiol 217:20–28. doi: 10.1016/j.ijfoodmicro.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Feng XM, Dong HH, Yang P, Yang RJ, Lu J, Lv J, Sheng J. 2016. Culture-dependent and -independent methods to investigate the predominant microorganisms associated with wet processed coffee. Curr Microbiol 73:190–195. doi: 10.1007/s00284-016-1047-3. [DOI] [PubMed] [Google Scholar]
- 12.Lee SM, Lee S, Singh D, Oh JY, Jeon EJ, Ryu HS, Lee DW, Kim BS, Lee CH. 2017. Comparative evaluation of microbial diversity and metabolite profiles in doenjang, a fermented soybean paste, during the two different industrial manufacturing processes. Food Chem 221:1578–1586. doi: 10.1016/j.foodchem.2016.10.135. [DOI] [PubMed] [Google Scholar]
- 13.Quigley L, McCarthy R, O’Sullivan O, Beresford TP, Fitzgerald GF, Ross RP, Stanton C, Cotter PD. 2013. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J Dairy Sci 96:4928–4937. doi: 10.3168/jds.2013-6688. [DOI] [PubMed] [Google Scholar]
- 14.Machado SG, Bagliniere F, Marchand S, Van Coillie E, Vanetti MC, De Block J, Heyndrickx M. 2017. The biodiversity of the microbiota producing heat-resistant enzymes responsible for spoilage in processed bovine milk and dairy products. Front Microbiol 8:302. doi: 10.3389/fmicb.2017.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnelly CW. 2014. Cheese and microbes. ASM Press, Washington, DC. [Google Scholar]
- 16.Soejima T, Iida K, Qin T, Taniai H, Seki M, Yoshida S. 2008. Method to detect only live bacteria during PCR amplification. J Clin Microbiol 46:2305–2313. doi: 10.1128/JCM.02171-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erkus O, de Jager VCL, Geene RTCM, van Alen-Boerrigter I, Hazelwood L, van Hijum SAFT, Kleerebezem M, Smid EJ. 2016. Use of propidium monoazide for selective profiling of viable microbial cells during Gouda cheese ripening. Int J Food Microbiol 228:1–9. doi: 10.1016/j.ijfoodmicro.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Nocker A, Cheung CY, Camper AK. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live versus dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 67:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Moyne AL, Harris LJ, Marco ML. 2013. Assessments of total and viable Escherichia coli O157:H7 on field and laboratory grown lettuce. PLoS One 8:e70643. doi: 10.1371/journal.pone.0070643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kable ME, Srisengfa Y, Laird M, Zaragoza J, McLeod J, Heidenreich J, Marco ML. 2016. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. mBio 7:e00836-16. doi: 10.1128/mBio.00836-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N, Gilbert JA. 2014. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5:e01371. doi: 10.1128/mBio.01371-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tetra Pak Processing Systems. 2015. Dairy processing handbook. Tetra Pak Processing Systems AB, Lund, Sweden: https://dairyprocessinghandbook.com/. [Google Scholar]
- 23.Delgado S, Rachid CT, Fernandez E, Rychlik T, Alegria A, Peixoto RS, Mayo B. 2013. Diversity of thermophilic bacteria in raw, pasteurized and selectively-cultured milk, as assessed by culturing, PCR-DGGE and pyrosequencing. Food Microbiol 36:103–111. doi: 10.1016/j.fm.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 24.Quigley L, O’Sullivan DJ, Daly D, O’Sullivan O, Burdikova Z, Vana R, Beresford TP, Ross RP, Fitzgerald GF, McSweeney PL, Giblin L, Sheehan JJ, Cotter PD. 2016. Thermus and the pink discoloration defect in cheese. mSystems 1:e00023-16. doi: 10.1128/mSystems.00023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daly DFM, McSweeney PLH, Sheehan JJ. 2012. Pink discolouration defect in commercial cheese: a review. Dairy Sci Technol 92:439–453. doi: 10.1007/s13594-012-0079-0. [DOI] [Google Scholar]
- 26.Caspers MP, Boekhorst J, Abee T, Siezen RJ, Kort R. 2013. Complete genome sequence of Anoxybacillus flavithermus TNO-09.006, a thermophilic sporeformer associated with a dairy-processing environment. Genome Announc 1:e00010-13. doi: 10.1128/genomeA.00010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess SA, Lindsay D, Flint SH. 2010. Thermophilic bacilli and their importance in dairy processing. Int J Food Microbiol 144:215–225. doi: 10.1016/j.ijfoodmicro.2010.09.027. [DOI] [PubMed] [Google Scholar]
- 28.Postollec F, Mathot AG, Bernard M, Divanac’h ML, Pavan S, Sohier D. 2012. Tracking spore-forming bacteria in food: from natural biodiversity to selection by processes. Int J Food Microbiol 158:1–8. doi: 10.1016/j.ijfoodmicro.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Trmcic A, Martin NH, Boor KJ, Wiedmann M. 2015. A standard bacterial isolate set for research on contemporary dairy spoilage. J Dairy Sci 98:5806–5817. doi: 10.3168/jds.2015-9490. [DOI] [PubMed] [Google Scholar]
- 30.Sadiq FA, Li Y, Liu T, Flint S, Zhang G, Yuan L, Pei Z, He G. 2016. The heat resistance and spoilage potential of aerobic mesophilic and thermophilic spore forming bacteria isolated from Chinese milk powders. Int J Food Microbiol 238:193–201. doi: 10.1016/j.ijfoodmicro.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Lucking G, Stoeckel M, Atamer Z, Hinrichs J, Ehling-Schulz M. 2013. Characterization of aerobic spore-forming bacteria associated with industrial dairy processing environments and product spoilage. Int J Food Microbiol 166:270–279. doi: 10.1016/j.ijfoodmicro.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Meng L, Zhang Y, Liu H, Zhao S, Wang J, Zheng N. 2017. Characterization of Pseudomonas spp. and associated proteolytic properties in raw milk stored at low temperatures. Front Microbiol 8:2158. doi: 10.3389/fmicb.2017.02158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales P, Fernández-García E, Nuñez M. 2005. Volatile compounds produced in cheese by Pseudomonas strains of dairy origin belonging to six different species. J Agric Food Chem 53:6835–6843. doi: 10.1021/jf050717b. [DOI] [PubMed] [Google Scholar]
- 34.Wang CY, Wu HD, Lee LN, Chang HT, Hsu YL, Yu CJ, Yang PC, Hsueh PR. 2006. Pasteurization is effective against multidrug-resistant bacteria. Am J Infect Control 34:320–322. doi: 10.1016/j.ajic.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Chiesa F, Lomonaco S, Nucera D, Garoglio D, Dalmasso A, Civera T. 2014. Distribution of Pseudomonas species in a dairy plant affected by occasional blue discoloration. Ital J Food Safety 3:1722. doi: 10.4081/ijfs.2014.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dogan B, Boor KJ. 2003. Genetic diversity and spoilage potentials among Pseudomonas spp. isolated from fluid milk products and dairy processing plants. Appl Environ Microbiol 69:130–138. doi: 10.1128/AEM.69.1.130-138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown NM, Arbon J, Redpath C. 2000. Contamination of milk-bank samples with Pseudomonas aeruginosa during pasteurization by penetration of organisms through the screw lid during cooling. J Hosp Infect 46:321–322. doi: 10.1053/jhin.2000.0811. [DOI] [PubMed] [Google Scholar]
- 38.Manaia CM, Moore ER. 2002. Pseudomonas thermotolerans sp. nov., a thermotolerant species of the genus Pseudomonas sensu stricto. Int J Syst Evol Microbiol 52:2203–2209. doi: 10.1099/00207713-52-6-2203. [DOI] [PubMed] [Google Scholar]
- 39.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. 2007. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol 73:5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emerson JB, Adams RI, Roman CMB, Brooks B, Coil DA, Dahlhausen K, Ganz HH, Hartmann EM, Hsu T, Justice NB, Paulino-Lima IG, Luongo JC, Lymperopoulou DS, Gomez-Silvan C, Rothschild-Mancinelli B, Balk M, Huttenhower C, Nocker A, Vaishampayan P, Rothschild LJ. 2017. Schrodinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5:86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auchtung TA, Holder ME, Gesell JR, Ajami NJ, Duarte RT, Itoh K, Caspi RR, Petrosino JF, Horai R, Zarate-Blades CR. 2016. Complete genome sequence of Turicibacter sp. strain H121, isolated from the feces of a contaminated germ-free mouse. Genome Announc 4:e00114-16. doi: 10.1128/genomeA.00114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh AM, Crispie F, Kilcawley K, O’Sullivan O, O’Sullivan MG, Claesson MJ, Cotter PD. 2016. Microbial succession and flavor production in the fermented dairy beverage kefir. mSystems 1:e00052-16. doi: 10.1128/mSystems.00052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maze A, Boel G, Zuniga M, Bourand A, Loux V, Yebra MJ, Monedero V, Correia K, Jacques N, Beaufils S, Poncet S, Joyet P, Milohanic E, Casaregola S, Auffray Y, Perez-Martinez G, Gibrat JF, Zagorec M, Francke C, Hartke A, Deutscher J. 2010. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J Bacteriol 192:2647–2648. doi: 10.1128/JB.00076-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, Arai H, Tanimoto I, Nishimura F, Takashiba S. 2003. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene, and total bacteria. FEMS Immunol Med Microbiol 39:81–86. doi: 10.1016/S0928-8244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- 45.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM, Steiner JM, Cook AK, Toresson L. 2012. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 7:e51907. doi: 10.1371/journal.pone.0051907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reese S. 2013. gPCA: batch effect detection via guided principal components analysis, vR package version 1.0. https://CRAN.R-project.org/package=gPCA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA sequences for this study are publicly available through the Qiita database (https://qiita.ucsd.edu) under study ID 10485 and in the European Nucleotide Archive under accession number ERP015209.