Abstract
Purpose of Review:
Experimental and analytical advances have enabled systematic, high-resolution studies of humoral immune responses, and are beginning to define mechanisms of immunity to HIV.
Recent findings:
High-throughput, information-rich experimental and analytical methods, whether genomic, proteomic, or transcriptomic have firmly established their value across a diversity of fields. Consideration of these tools as trawlers in “fishing expeditions” has faded as “data-driven discovery” has come to be valued as an irreplaceable means to develop fundamental understanding of biological systems. Collectively, studies of HIV-1 infection and vaccination including functional, biophysical, and biochemical humoral profiling approaches have provided insights into the phenotypic characteristics of individual and pools of antibodies. Relating these measures to clinical status, protection/efficacy outcomes, and cellular profiling data using machine learning has offered the possibility of identifying unanticipated mechanisms of action and gaining insights into fundamental immunological processes that might otherwise be difficult to decipher.
Summary:
Recent evidence establishes that systematic data collection and application of machine learning approaches can identify humoral immune correlates that are generalizable across distinct HIV-1 immunogens and vaccine regimens and translatable between model organisms and the clinic. These outcomes provide a strong rationale supporting the utility and further expansion of these approaches both in support of vaccine development as well as more broadly in defining mechanisms of immunity.
Keywords: antibody, systems serology, effector function, machine learning
Introduction
The challenges that traditional vaccine development approaches have encountered in the face of a global epidemic point toward the value of novel experimental and analytical approaches in support of HIV vaccine research. Specifically, “systems serology”, a method devised to offer an unbiased and comprehensive assessment of the humoral immune response, is one such alternative for evaluating and guiding the development of vaccines. This approach aims to not only experimentally capture, but to analytically leverage, the incredible diversity of antibody activities and characteristics observed between subjects and within individuals that may relate to protection (1–3). Paired with machine learning tools, novel high-resolution experimental data offers an opportunity to reduce dependence on anticipated mechanisms and correlates of protection, and instead to broaden investigation to support development of a more fundamental understanding of the coordinated aspects of humoral immune responses and their importance. This toolkit offers the prospect of improving differentiation between candidate vaccines, understanding coordinated aspects of humoral immunity, and perhaps most critically for identifying aspects of the antibody response that serve as hallmarks, if not mechanisms, of protective immunity.
An interest in antibodies
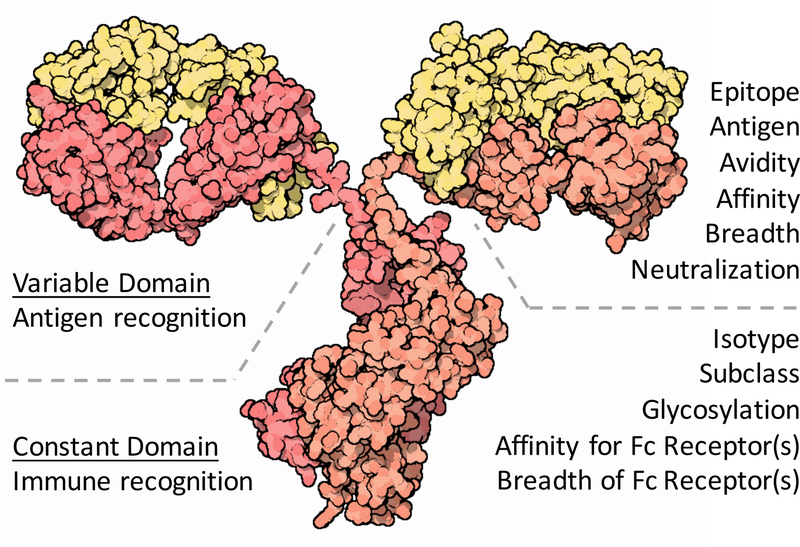
Systems serology aims to effectively characterize and learn from the vast biodiversity in antibody profiles that result from antigenic exposure(s). To illustrate the merits of such an approach, consider that more unique antibody types likely exist in a single germinal center in a single subject than might reasonably be tested in years of passive infusion experiments. Thus, millions of individual antibodies, which may each differ in Fv and Fc character (Figure 1), can lead to a remarkable combinatorial diversity that may reveal critical insights into immune mechanisms of infection control. The diversity of solutions explored and the numerous interactions between components makes their parsing challenging, but results in a rich landscape which can be mined for valuable insights into antibody mediated anti-viral activities. Development of improved experimental and analytical frameworks in the characterization of serum constituents offers the prospect of developing both a more nuanced and a more complete understanding of mechanisms of immunity.
Figure 1. Features of the antibody response.
The Ig variable domain (Fv), responsible for pathogen recognition, varies in terms of its antigen and epitope specificity, and the avidity, affinity, and breadth of recognition, as well as whether that recognition results in neutralization of infectivity. The constant, or crystallizable domain (Fc), responsible for innate immune recognition, varies in terms of its isotype, subclass, and glycosylation profile, which impact the antibody’s affinity for Fc receptors and the class(es) of Fc receptors able to interact. Image from doi:10.2210/rcsb_pdb/mom_2001_9.
The focus on antibody responses stems from the importance of this aspect of adaptive immunity in accomplishing vaccine-mediated protection from infection (4). However, that many vaccines induce robust antibody responses but lack efficacy indicates that while the presence of pathogen-specific antibodies may be necessary, it is not sufficient, and there are specific qualitative attributes of some but not all antibodies that are associated with immunity. The proven ability of neutralizing antibodies to protect from infection establishes this qualitative attribute to be of particular importance (5). However, an accumulation of evidence from clinical and animal model studies suggests that other antibody attributes and activities can also meaningfully relate and mechanistically contribute to resistance to infection (6–8). In brief, despite distinct viral vectors and immunogens, diverse candidate vaccines have consistently identified a role for pathogen-specific binding antibodies, sometimes associated with individual anti-viral effector functions, as correlates of immunity. Passive transfer experiments have likewise supported a role for antibody activities other than neutralization in some (9–12) but not all contexts (13, 14). Seeking to better understand the mechanism(s) of action of “binding” antibodies, their specific Fv and Fc characteristics, and their ability to be elicited and result in complete protection has been a major motivation in the development of expansive antibody profiling tools (summarized in (1, 2)).
The value in holistic assessment of antibody activity does not reside in the setting of HIV alone; it has been observed that for most approved vaccines, humoral response magnitude or even neutralization potency alone do not equate to protective immunity (4). Instead, across a variety of pathogens, evidence supports a critical role for numerous and diverse functional attributes of the antibody response, collectively referred to as antibody effector functions (6, 15–23). The immune system explores diverse antibody variants that recognize diverse epitopes on diverse antigens, and couples this Fv domain diversity with Fc domain isotype, subclass, and glycosylation profile variation, which results in differences in interactions with innate immune receptors, such as Fc receptors (Figure 1). These receptors, some high affinity, some low affinity, some activating, some inhibitory, are variably expressed on innate immune effector cells poised to respond to the signal they receive from the antibodies bound to a pathogen or infected cell (15, 16). In this way, antibodies act at the intersection of the adaptive and innate arms of the immune system, providing an adaptive means to engage innate immune effector cells to clear pathogens and infected cells. Given that the activity of polyclonal antibody pools derives from the cumulative traits of the antibodies present, some which may collaborate and some which may compete with each other (24, 25), it is difficult to imagine an analytical approach more suited to deconstructing this mixture into components that can be related to relevant clinical, genetic, or functional characteristics than machine learning (ML). Just as polyclonal serum antibody neutralization profiles can be deconvoluted into components similar to known broadly neutralizing monoclonals (26, 27), qualitative attributes of the antibody Fc such as isotype, subclass, and glycosylation profile, drive differential effector function potency in ways that algorithms can reliably reconstruct. Statistically-principled analytical procedures are beginning to provide insights into antibody response profiles and relationships between features of the humoral response, functions of the humoral response, and outcomes of interest (Table 1); the remarkable combinatorial diversity of millions of individual antibodies of differing Fv and Fc character is beginning to reveal mechanisms of infection control.
Table 1:
Examples of studies in which ML has been employed to derive insights into humoral immunity.
| Species | Observations | Reference |
|---|---|---|
| Correlates of Immunity | ||
| Human | Relationships between humoral response features and established correlates of reduced risk. | (24, 64) |
| NHP | Identification of correlates of vaccine efficacy. | (39, 40, 44, 65–71) |
| Group Differences | ||
| Human | Differences between subjects receiving different vaccines. | (24, 64) |
| Differences in humoral response among controllers and progressors. | (50, 72) | |
| Differences in humoral responses among subjects who do versus do not develop broadly neutralizing antibodies. | (42, 73, 74) | |
| Differences in humoral response observed among subjects differing by genotype. | (43) | |
| NHP | Differences in humoral response among subjects immunized with different vaccines. | (39, 40, 70) |
| Differences in humoral response among subjects immunized with different adjuvants. | (44, 48, 65) | |
| immune responses in male and female macaques in response to vaccination | (75) | |
| Differences in humoral response among subjects immunized by different routes. | (39) | |
| Antibody Activities | ||
| Human | Biophysical features associated with diverse effector functions among vaccine recipients. | (24, 45, 49, 76) |
| Biophysical features associated with diverse effector functions among HIV infected subjects. | (43, 44, 46, 50, 77) | |
| NHP | Biophysical antibody features associated with diverse effector functions among vaccinated subjects. | (39, 75) |
Insights from clinical studies
Vaccine efficacy trials provide key opportunities to develop much-needed insights into the relationships between humoral response characteristics and infection outcomes. Diverse vaccine concepts have been evaluated, ranging from immunization with adjuvanted protein per VAX003 and VAX004, adenovirus-vectored HIV gene delivery as in HVTN 505, and combination prime-boost regimens, such as RV144. To date, only RV144 demonstrated efficacy, showing a modest but statistically significant reduction in the risk of infection among vaccine recipients as compared to controls (28). However, even for vaccines that are not efficacious, or for immunogenicity studies in which efficacy is not a primary endpoint, there may be opportunities to deepen our understanding of the response differences between infected and uninfected vaccine recipients. Case control studies conducted in these contexts have defined virological, genetic, and immunological associations with risk of infection (29–34).
“Inclusive excellence” in correlates analysis
Based on the identification of additional correlates in secondary and subsequent analyses (2, 35–38), the case of the RV144 trial makes clear that if additional or alternative immunological response outputs had been considered, additional and/or alternative correlates may have been identified. Because these correlates serve as key beachheads downstream in vaccine research, our understanding of protective immunity benefits from inclusive data collection that captures the diversity of features that may impact the anti-viral profile of the humoral immune response. In theory, because it is possible that multiple mechanisms exist whereby protection from infection can be achieved, correlates have the potential to be local or global, serving as either narrow or broad indicators of efficacy across diverse regimens, immunogens, geographic, and genetic settings. Given the significant human and capital resources dedicated to preclinical and clinical vaccine evaluation, systematic and inclusive analysis of responses in vaccine studies can help to maximize the return on this investment for any given study, as well as enable robust meta-analyses across studies.
Promisingly, such broad evaluation of correlates, with hundreds or thousands of measured features of the response, has begun to demonstrate generalizability across different candidate vaccines (Table 1). For example, evaluation of an SIV vaccine comprised of an adenovirus prime and protein boost identified IgG antibodies specific to the HIV envelope protein that interacted with FcgRIIa, associated with monocyte-mediated phagocytosis activity, and IgA antibodies specific to envelope, which were associated with neutrophil-mediated phagocytosis (39). These correlates, and the specific model learned from this adenovirus vaccine study generalized to a second NHP immunization and protection study, an RV144-like regimen that was found to protect against SHIV challenge (40). Further, two of these four correlates, monocyte-mediated phagocytosis and envelop-specific antibody binding to FcgRIIa, have now been observed to correlate with reduced risk of infection in the HVTN505 human vaccine study (41). In summary, broad experimental evaluation of humoral response combined with data mining supported identification of correlates of protection that transcend vaccine, regimen, challenge virus, and species.
Tradeoffs between scientific “bang” and statistical “buck”
Traditional correlates analysis approaches seek to accommodate competing objectives: they attempt to balance decreasing statistical power with the increased likelihood of identifying correlates of protection as more response variables are included for analysis. As an increasing number of potential correlates are evaluated, it becomes more likely that a biologically meaningful correlate will be considered, but also more likely that a response will happen to correlate with outcome by chance. While high confidence in the identified correlates requires statistical rigor, it is important to ensure that this standard does not unintentionally reduce the insights afforded by any given study. Though a necessary constraint to traditional correlates study design, predetermination of features to be used in correlates analysis also raises the risk of confirmation bias, which can be considered here to reflect the tendency to prioritize assessment of response characteristics that are “expected” to relate to protection. Fortunately, alternative approaches have been developed to complement traditional correlates analysis, approaches that can provide equal analytical rigor in the context of richer datasets.
Defining Signatures of Immunity using Machine Learning
Robust, statistically principled analytical tools have emerged over the past two decades to define “rules” for relationships between variables by learning from patterns within comprehensive datasets (Figure 2). These analytical approaches, collectively referred to as machine learning (ML), have developed at the intersection of artificial intelligence and statistics. ML approaches are becoming as integral to the biological sciences as other fields, where, for example, they annotate the content of images, improve recommendations to consumers, and assist in digital assistant natural language processing. ML approaches offer a means to systematically identify “signatures” or patterns observed within data sets that appear reproducibly. Relevant to systems serology, these tools can be used to identify relationships across populations of subjects (42), among variable antibody pools (43), or differentially protected individuals (44). For example, they can define relationships between antibody activities (45), biophysical properties and degree of protection (39), or define types of antibodies that are either often or infrequently co-induced (43) (Table 1). Because generalizable patterns imply biological relevance, such “pattern finding” analyses offer the possibility of defining new mechanistic connections.
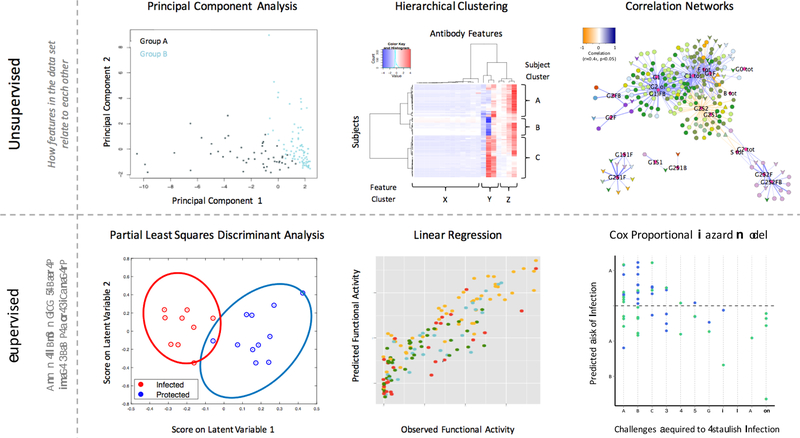
Figure 2. Examples of humoral immune response analysis supported by machine learning.
Unsupervised methods (top) including principal component analysis (left), hierarchical clustering (center), and correlation networks (right) depict the main aspects of variation in, magnitudes of, and underlying relationships between humoral response features and subjects. Supervised methods (bottom) model how data can be used to predict parameters of interest, such as group (left), antibody activity (center), or challenge outcome (right).
Unsupervised ML
Unsupervised analysis methods are often described as a means to let data self-organize. These approaches include clustering and principal component analyses, which provide an overall view of similarities and differences across a dataset (Figure 2). These “agenda-free” analysis methods can thereby further elucidate classes of responses and classes of responders. Understanding relationships between measured features of the immune response is also valuable in and of itself, as correlation is expected to result from co-regulation. For example, correlations have been observed between functionally potent IgG1 and IgG3 responses, as well as between the more immunologically inert IgG2 and IgG4 subclasses (46). Linking this growing understanding of correlated aspects of the antibody response to transcriptional programs may extend these insights to define regulatory pathways associated with the generation of responses with these characteristics (44, 47, 48).
Supervised ML
In contrast, supervised approaches have an explicit goal: to use response data to learn predictive models for a given characteristic of interest. These methods seek to define how well a given dataset can inform accurate classification or regression predictions, and which measurements are responsible for these predictions (Figure 2). They leverage known examples to make extrapolations to unknown cases, seeking to reveal meaningful biological relationships by learning historical associations and trends. Further, these approaches rely on the presumption that in the biological setting, past performance will be an indicator of future results. Use of the humoral response profile to differentiate between clinical classes such as HIV controllers and progressors, between relatively susceptible or resistant subjects in challenge experiments, or between cases and controls are common applications (Table 1). These ML approaches can be used to model infection risk, such as predicting the number of challenges likely to result in infection, as well as to learn relationships between features, such as models of antibody functions developed from biophysical antibody features.
Semi-supervised ML
Though not yet explored in this context, semi-supervised approaches, which were developed to address situations in which class labels are definitively known only for a subset of the samples, may have significant value in human correlates studies. For a number of pathogens, human challenge experiments are unethical, and observing protection from infection is therefore dependent on real-world infectious agent exposures that may or may not occur. Accordingly, when response features between infected (cases) and uninfected (controls) subjects are compared, it is expected that the group of uninfected subjects will in fact be comprised of a mixture of individuals who were exposed and protected, some who were not exposed but would have been protected, and some “dilutive” subjects who were not exposed but would have been infected if exposed (the positive but unlabeled). For a marginally effective vaccine regimen such as RV144, a minority of participants were protected and the majority of samples among the control group are expected to dilute analysis. Semi-supervised approaches offer the ability to use response data to infer the identity of these dilutive “positive unlabeled” subjects such that they could then be excluded from correlates analysis. It thereby offers the potential for higher confidence in the validity of identified correlates, and the prospect of identifying additional correlates. It will be interesting to evaluate and understand how this approach might be practically deployed on vaccine case control studies and to define the impact it might have on correlates analysis.
Using data to drive discovery
While there are legitimate reasons to disfavor overly complex analytical approaches, there are robust analytical means to identify whether a dataset is “too wide” (too many features, too few subjects), or a method “too powerful” (good at memorization), which can provide confidence that observations arise from biologically meaningful relationships rather than from chance occurrences. Complementary analytical tools capable of evaluating the quality of ML models from a statistical perspective have evolved alongside these learning algorithms and provide a means to establish confidence or impart skepticism (Figure 3).
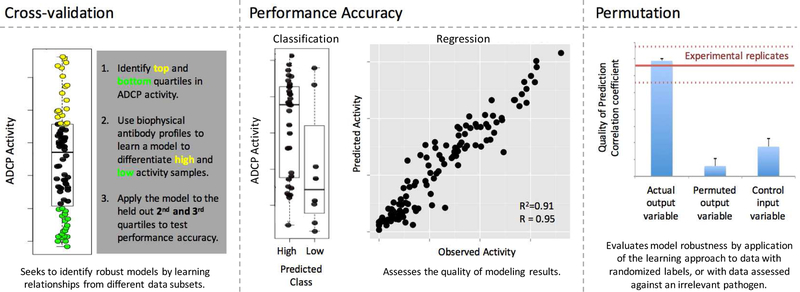
Figure 3. Evaluating model accuracy and robustness.
Left. In repeated cross-validation, data is split into different training and test sets. Models learned from training data are applied to the held-out test data. The example shown learns rules about ADCP activity from the extremes and applies these rules to the samples with intermediate activity. Center. Performance accuracy can be defined by characterizing the degree of agreement between model and observation for classification and regression models. Right. Permutation testing, in which the performance of randomized data is compared to actual data, offers a means to gauge model robustness. In this case, actual data can be used to model antibody activity with similar fidelity as the experimental data can be experimentally replicated, and significantly better than when models are learned from permuted data, or from control data, such as that relating to a different pathogen.
Rigor and reproducibility
First, repeated cross validation can be employed to determine the sensitivity of either a model’s performance or the features the model relies on to changes in the samples used for training. Using this approach, a subset of the data is used to train the model and the remainder is left out in order to test the generalizability of the model. In this way, cross-validation assesses the ability of the data to make predictions about unseen examples. It is important to note that because preclinical vaccine studies are often comprised of relatively small numbers of subjects, the simple act of omitting one, as would be done in “leave one out”, or omitting N subjects, as would be done in N-fold cross-validation, can cause a feature with a significant relationship to a group or an outcome in the complete dataset to be dropped. Because such immune features are not sufficiently strong predictive indicators, they are unlikely to contribute to cross-validated models. Thus, in this way, cross-validated models can be more robust and rigorous than traditional statistical tests carried out on complete datasets.
Permutation tests, in which data labels are randomly shuffled prior to model building, can serve as a negative control for the computational analysis by setting a baseline benchmark for performance. Permutation randomizes the input data such that existing relationships are lost; understanding the quality of models learned from randomly scrambled data serves as a means to critically evaluate the potential biological significance of the model built on the actual data. If similarly well-performing models can be learned from scrambled data, then the validity of models learned from the actual data, or the appropriateness of the modeling approach, should be questioned. While it is expected in principle that randomized data will yield models with random performance, in practice, the likelihood of observing random performance is impacted by study size and data composition as well as the modeling approach, making permutation testing an important aspect of robustness testing.
Of course, further experiments serve as the most rigorous test of model validity. Confirmation in a validation cohort by repeating the original experiment with a new set of subjects is a gold standard that is not always feasible. Additionally, while good predictive performance in an independent validation cohort is consistent with expectations for mechanistically relevant relationships, because it can be observed independent of a causal relationship, this type of validation does not directly address mechanism. Other, more targeted types of follow-up experiments might be better suited to demonstrate mechanistic validity. For example, the biological validity of contributing features has been confirmed in depletion experiments in a number of studies (24, 39, 49).
The last area in which the rigor and reproducibility of ML analyses could perhaps be improved is data and code availability. Beyond the additional quality control and exploratory analysis that widespread data and code sharing would enable, easy access to shared resources will facilitate methodological consistency and increase analysis efficiency by reducing the collective coding resources spent addressing similar problems.
Challenges and Rewards in Application of ML to humoral response characterization
It has been our experience that diverse ML approaches can achieve consistent performance and rely upon consistent features, that models can perform similarly well as experimental replicates and much better than with randomized or control features, and that they can recapitulate known aspects of antibody immunobiology as well as define new ones (49, 50). However, models learned from humoral responses alone are expected not to perfectly capture outcomes, particularly for correlates studies, as anatomical, host genetic, viral genetic, and stochastic factors are also at play. The impact of these factors might be most easily appreciated from the non-uniformity of challenge outcomes among control (ie: sham-immunized) subjects. Nonetheless, both model quality, the degree of agreement between models and observations, and model utility, or biological validity, are important endpoints (Figure 4).
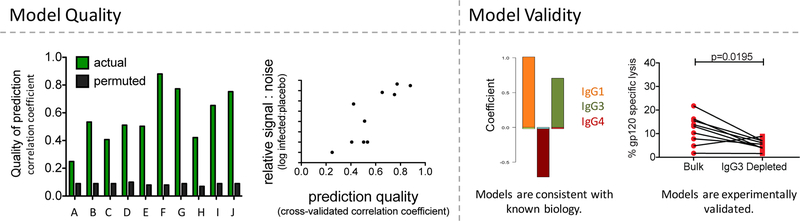
Figure 4. Evaluating model quality and validity.
Left. Prediction quality, as defined by correlation coefficient between cross-validated models learned from actual (green) and permuted (black) data for a set of ten antibody effector functions (A-J). While performance varies considerably (0.2 < r < 0.9) for models learned from real data, permuted data consistently yields essentially random performance (r ~0.1). Relationship between prediction quality and assay signal-to-noise, which suggests that models may be limited by performance characteristics of cell-based assays such as signal to noise (here) and assay reproducibility (50). Right. The biological validity of the models can be supported by prior knowledge, such as the reliance of NK activation predictions antibody types known to have these activities (IgG1 and IgG3) (49), and experimentally confirmed, such as by observing decreased activity when IgG3s are depleted (24).
Choosing a method
While a wide range of ML methods are readily available for implementation, choosing the most applicable method requires answering a few fundamental questions about the modeling task and data. Firstly, choice of the best modeling method can depend on whether protection is treated as a discrete or continuous variable. When protection is treated as a discrete variable, such as infection status, classification approaches can be used to distinguish between different levels of protection. If protection is treated as a continuous variable (e.g., time to infection, viral load, etc.), regression approaches can be used to directly model the measurements of protection. Since regression approaches are capable of identifying small-scale differences in protection, they are best suited for cases where the measurement has fine-grained variations. On the other hand, if the protection variable is multimodal, it may be easier to discretize the protection variable around the modes and perform classification (2, 44). In studies involving multiple vaccine groups, classification approaches can also be used to distinguish between vaccine groups and identify vaccine-specific signatures. Furthermore, in studies with a sufficient number of samples per immunization group, modeling protection within each group can help identify vaccine-specific correlates of protection (39).
Considering response features
Though high-resolution antibody data offers an excellent opportunity for fine-grained exploration into mechanisms of protection, it also poses challenges in building useful ML models. The number of antibody properties or “features” that are profiled is often much higher than the number of subjects in vaccine studies, which results in a data matrix/tensor that is too wide, or “rank deficient”. Naïve modeling with such rank deficient matrices is likely to produce models that memorize (overfit) the data and use redundant features. Therefore, it is necessary to reduce the dimensionality of the matrix by employing techniques for feature filtering and feature selection (51). Feature filtering discards features that are too noisy or that do not meet certain quality criteria, which can be defined by domain experts and can be specific to the experimental approach used for generating the data. Feature filtering is typically performed on the data before applying an ML method, and can therefore be task-agnostic. Feature selection, in contrast, minimizes the redundancy among features employed by the model by discarding features that do not contribute significantly to protection. The threshold for determining the significance of contribution of features can be set by assessing its impact on the prediction performance. Hence, feature selection is also used as a technique to identify the most significant features contributing to protection. When data from multiple sources/experiments are used for modeling protection, it is important for the models to account for variations in such data. Performing data normalization and standardization before integrating data from different sources can help reduce the impact of such variation on modeling. Since the ability of the models to identify reliable correlates of protection is directly linked to the quality of the input data, it is essential to include a combination of data normalization, feature filtering, and feature selection techniques when training ML models.
Modeling and Evaluation
A common challenge in using ML methods for modeling protection is determining the complexity of the model. Though complex methods tend to achieve higher prediction accuracy than simpler models, this improvement can come at the cost of interpretability. As more complex functions are used to better model the relationship between protection and data, the mode and extent of contribution by the features to protection can become unclear. In cases where interpretability and accuracy are equally important, a simple but accurate model with a straightforward methodology is preferable to a complex model with slightly better accuracy. Complex models also tend to employ more parameters than simpler models, and hence are more prone to overfitting the training data. Contrary to expectation, increasing the complexity of a model can sometimes reduce prediction performance, especially when data is limited. Choosing the complexity of the model is therefore a balancing-act between performance and interpretability.
The accuracy and credibility of an ML method are critical for establishing the mechanistic value of the resulting biological insights. Hence, it is important to evaluate and compare the predictive performance of the models using robust evaluation metrics. For example, metrics like balanced accuracy or receiver operating characteristic (ROC) curves provide a realistic estimate of classification performance even when class sample sizes in the data are skewed. While cross-validation is typically used to evaluate the predictive performance in vaccine studies with limited samples, it is also useful to provide an estimate of the variation in prediction performance. Beyond reporting the model performance, it is also essential to identify which features played a critical role in the model’s performance. While feature selection techniques are often used to identify the most important features, they may not fully represent the multiple pathways through which protection is achieved. For example, a feature selection technique may minimize redundancy in the model by selecting one feature from among a set that are equally predictive of protection. Therefore, identifying such co-correlates via post-prediction analysis rather than reporting a single feature is key. This practice can reduce interpretive over-reliance on specific features that reflect or serve as a proxy for a broader group.
New Frontiers
Experimental resolution and throughput continue to improve generating more nuanced information as well as a growing subject diversity and overall number of functional and phenotypic profiles of humoral responses. For example, single cell transcriptomics and B cell sequencing have matured from methods appropriate for individual cases to more generalizable approaches (52, reviewed in 53). Coupling next-generation sequencing to mass-spectrometry of pathogen-specific antibody CDRs has provided a means to relate the antibody transcriptome to the proteome (54), and novel microfluidics platform-based analysis of secreted antibodies from single cells promises further high-resolution insights into the phenotypic and functional aspects of subjects’ humoral responses (55). Phage and microarray-based peptide libraries based on the virome (56, 57) have enabled significant insights into histories and impacts of viral exposures. Even the structural resolution at which humoral responses can be defined has undergone dramatic improvements, as cryo-electron microscopy can now be practically deployed on polyclonal sera (58).
Concerning analytics, new algorithms are constantly being developed, though with a trend toward increasing complexity. As an illustrative example, neural nets and deep learning approaches are beginning to offer unparalleled insights into single cell transcriptional data (59, 60). These and other new approaches have the potential to result in similar enhancements to model performance in antibody characterization, though the considerations of accuracy versus interpretability outlined here apply. While there are reasons to suppose that such models may be more robust than singular correlates, they are less likely to lend themselves to “just so stories” about protection.
There are already clues about what the future of integrated data and a more holistic approach may have to offer. Studies evaluating the impact of the microbiome have driven intense interest in broader and unanticipated connections in immunology. Another such area in which this holistic approach seems to have taken root is the resurgent interest in understanding trained immunity (61–63). Studies of this concept, also referred to as “heterologous” immunity, reflect a growing appreciation that vaccination toward specific pathogens can alter aspects of the immune system relevant to other vaccines/pathogens.
Conclusion
Tools that enable comprehensive dissection of humoral immune responses have the potential to experimentally capture the vast biodiversity that exists within these responses, and use this data to analytically elucidate mechanisms of immunity. Using such “systems serology” approaches, immunologists have begun to define protective humoral response signatures in vaccinated and naturally infected subjects in HIV and other diseases. Recent evidence shows that systematic data collection and application of machine learning approaches can identify correlates that are generalizable across very distinct immunogens and vaccine regimens. Further, these machine learning-defined correlates have been shown to be translatable between model organisms and the clinic, providing a strong case for their utility and further advancement in support of vaccine development. We anticipate that systematic antibody profiling, particularly in combination with other -omics data types, will expand our understanding of the diversity of possible antibody responses, connect these responses to cellular mechanisms and pathways, and identify the key aspects of different immunization strategies that result in protection.
Key points.
Comprehensive and unbiased analysis of immune responses can support data-driven discovery.
Machine learning tools offer a complementary approach to traditional correlates analysis.
Statistically principled approaches to big data are beginning to define humoral mechanisms of vaccine-mediated immunity.
Acknowledgements:
We would like to thank Drs. Chris Bailey-Kellogg, Hao D. Cheng, Ickwon Choi, Jishnu Das, and Karen Dowell for generating representative visualizations.
Financial support and sponsorship:
M.E.A. acknowledges support from the Bill and Melinda Gates Foundation OPP1146996; NIH NIAID and NIGMS 1P01AI120756, P20GM104416, 1R01AI129801, and 1R01AI131975.
Footnotes
Conflicts of Interest:
The authors of this manuscript have declared the following potential competing interests: M.E.A is a recipient of funding from the Bill and Melinda Gates Foundation, The National Institute of Allergy and Infectious Diseases of the National Institutes of Health, and The National Institute of General Medical Science of the National Institutes of Health.
References and Recommended Reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Arnold KB, Chung AW. Prospects from systems serology research. Immunology 2018;153(3):279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung AW, Alter G. Systems serology: profiling vaccine induced humoral immunity against HIV. Retrovirology 2017;14(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackerman ME, Barouch DH, Alter G. Systems serology for evaluation of HIV vaccine trials. Immunol Rev 2017;275(1):262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol 2010;17(7):1055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascola JR. Passive transfer studies to elucidate the role of antibody-mediated protection against HIV-1. Vaccine 2002;20(15):1922–5. [DOI] [PubMed] [Google Scholar]
- 6.Crowley AR, Ackerman ME. Mind the Gap: How interspecies variability in IgG and its receptors may complicate comparisons of human and non-human primate effector function. Frontiers in Immunology 2019;in press.*This review lists many studies showing relationships between antibody effector function and anti-viral outcomes in animal models and humans, and describes differences in antibody and antibody receptor biology between rhesus macaques and humans.
- 7.Corey L, Gilbert PB, Tomaras GD, et al. Immune correlates of vaccine protection against HIV-1 acquisition. Sci Transl Med 2015;7(310):310rv7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomaras GD, Plotkin SA. Complex immune correlates of protection in HIV-1 vaccine efficacy trials. Immunol Rev 2017;275(1):245–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bournazos S, Klein F, Pietzsch J, et al. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 2014;158(6):1243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halper-Stromberg A, Lu CL, Klein F, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 2014;158(5):989–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hessell AJ, Hangartner L, Hunter M, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 2007;449(7158):101–4. [DOI] [PubMed] [Google Scholar]
- 12.Horwitz JA, Bar-On Y, Lu CL, et al. Non-neutralizing Antibodies Alter the Course of HIV-1 Infection In Vivo. Cell 2017;170(4):637–48 e10.*This study demonstrated the ability of a non-neutralizing antibody to exert evolutionary pressure on the HIV-1 virus in vivo.
- 13.Parsons MS, Lee WS, Kristensen AB, et al. Fc-dependent functions are redundant to efficacy of anti-HIV antibody PGT121 in macaques. J Clin Invest 2019;129(1):182–91.*This study showed that effector function may not contribute to protection afforded by highly broad and potent neutralizing antibodies. This is also an important observation in light of data from mouse models of HIV infection that have suggested a role of Fc-mechanisms in the antiviral activity of PGT121.
- 14.Moldt B, Shibata-Koyama M, Rakasz EG, et al. A nonfucosylated variant of the anti-HIV-1 monoclonal antibody b12 has enhanced FcgammaRIIIa-mediated antiviral activity in vitro but does not improve protection against mucosal SHIV challenge in macaques. J Virol 2012;86(11):6189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nimmerjahn F, Gordan S, Lux A. FcgammaR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol 2015;36(6):325–36. [DOI] [PubMed] [Google Scholar]
- 16.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 2008;8(1):34–47. [DOI] [PubMed] [Google Scholar]
- 17.Bouharoun-Tayoun H, Oeuvray C, Lunel F, et al. Mechanisms underlying the monocyte-mediated antibody-dependent killing of Plasmodium falciparum asexual blood stages. J Exp Med 1995;182(2):409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiLillo DJ, Tan GS, Palese P, et al. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 2014;20(2):143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn BM, Yu WH, Karim MM, et al. A Role for Fc Function in Therapeutic Monoclonal Antibody-Mediated Protection against Ebola Virus. Cell Host Microbe 2018;24(2):221–33 e5.*This study uses ML to model the importance of antibody effector function in the setting of ebola infection.
- 20.Jegerlehner A, Schmitz N, Storni T, et al. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol 2004;172(9):5598–605. [DOI] [PubMed] [Google Scholar]
- 21.Kohl S, Loo LS. Protection of neonatal mice against herpes simplex virus infection: probable in vivo antibody-dependent cellular cytotoxicity. J Immunol 1982;129(1):370–6. [PubMed] [Google Scholar]
- 22.Shi J, McIntosh RS, Pleass RJ. Antibody- and Fc-receptor-based therapeutics for malaria. Clin Sci (Lond) 2006;110(1):11–9. [DOI] [PubMed] [Google Scholar]
- 23.Warfield KL, Swenson DL, Olinger GG, et al. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. J Infect Dis 2007;196 Suppl 2:S430–7. [DOI] [PubMed] [Google Scholar]
- 24.Chung AW, Ghebremichael M, Robinson H, et al. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci Transl Med 2014;6(228):228ra38. [DOI] [PubMed] [Google Scholar]
- 25.Tomaras GD, Ferrari G, Shen X, et al. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proceedings of the National Academy of Sciences of the United States of America 2013;110(22):9019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doria-Rose NA, Altae-Tran HR, Roark RS, et al. Mapping Polyclonal HIV-1 Antibody Responses via Next-Generation Neutralization Fingerprinting. PLoS Pathog 2017;13(1):e1006148.*This study reports a widely used ML-based approach to define the epitope-specificity of neutralizing antibodies present in polyclonal serum samples.
- 27.Georgiev IS, Doria-Rose NA, Zhou T, et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 2013;340(6133):751–6. [DOI] [PubMed] [Google Scholar]
- 28.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 2009;361(23):2209–20. [DOI] [PubMed] [Google Scholar]
- 29.Edlefsen PT, Rolland M, Hertz T, et al. Comprehensive sieve analysis of breakthrough HIV-1 sequences in the RV144 vaccine efficacy trial. PLoS computational biology 2015;11(2):e1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fong Y, Shen X, Ashley VC, et al. Modification of the Association Between T-Cell Immune Responses and Human Immunodeficiency Virus Type 1 Infection Risk by Vaccine-Induced Antibody Responses in the HVTN 505 Trial. J Infect Dis 2018;217(8):1280–8.*This study is an example of case control analysis in the setting of vaccine that did not meet overall efficacy endpoint criteria.
- 31.Janes HE, Cohen KW, Frahm N, et al. Higher T-Cell Responses Induced by DNA/rAd5 HIV-1 Preventive Vaccine Are Associated With Lower HIV-1 Infection Risk in an Efficacy Trial. J Infect Dis 2017;215(9):1376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li SS, Gilbert PB, Tomaras GD, et al. FCGR2C polymorphisms associate with HIV-1 vaccine protection in RV144 trial. J Clin Invest 2014;124(9):3879–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prentice HA, Tomaras GD, Geraghty DE, et al. HLA class II genes modulate vaccine-induced antibody responses to affect HIV-1 acquisition. Sci Transl Med 2015;7(296):296ra112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolland M, Edlefsen PT, Larsen BB, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 2012;490(7420):417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez LG, Martinez DR, deCamp AC, et al. V1V2-specific complement activating serum IgG as a correlate of reduced HIV-1 infection risk in RV144. PLoS One 2017;12(7):e0180720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates NL, Liao HX, Fong Y, et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med 2014;6(228):228ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zolla-Pazner S, deCamp A, Gilbert PB, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS One 2014;9(2):e87572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haynes BF, Gilbert PB, McElrath MJ, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 2012;366(14):1275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackerman ME, Das J, Pittala S, et al. Route of immunization defines multiple mechanisms of vaccine-mediated protection against SIV. Nat Med 2018.**This study shows generalization of correlates of protection identified for an adenovirus-vectored vaccine evaluated in NHP to an RV144-like regimen tested in NHP. A subset of correlates have been further supported in humans.
- 40.Bradley T, Pollara J, Santra S, et al. Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge. Nat Commun 2017;8:15711.**This study identifies correlates of protection and distinguish between immunization groups, but also serves as an independent validation cohort for the model of protection learned from an adenovirus-protein vaccine regimen.
- 41.Neidich SD, Fong Y, Li SS, et al. Antibody Fc-Effector Functions Decrease HIV-1 Acquisition Risk in Vaccinated Humans. in revision **This study validates the correlates of protection observed in two earlier NHP studies in the clinic.
- 42.Richardson SI, Chung AW, Natarajan H, et al. HIV-specific Fc effector function early in infection predicts the development of broadly neutralizing antibodies. PLoS Pathog 2018;14(4):e1006987.*This study defines differences early in the immune response that differentiate subjects who will go on to develop broadly neutralizing antibodies from those who do not.
- 43.Lai JI, Licht AF, Dugast AS, et al. Divergent antibody subclass and specificity profiles but not protective HLA-B alleles are associated with variable antibody effector function among HIV-1 controllers. Journal of virology 2014;88(5):2799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaccari M, Gordon SN, Fourati S, et al. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 2016;22(7):762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Ferrari G, Alter G, et al. Diversity of Antiviral IgG Effector Activities Observed in HIV-Infected and Vaccinated Subjects. J Immunol 2016;197(12):4603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ackerman ME, Mikhailova A, Brown EP, et al. Polyfunctional HIV-Specific Antibody Responses Are Associated with Spontaneous HIV Control. PLoS Pathog 2016;12(1):e1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fourati S, Ribeiro SP, Blasco Tavares Pereira Lopes F, et al. Integrated systems approach defines the antiviral pathways conferring protection by the RV144 HIV vaccine. Nat Commun 2019;10(1):863.*This study relates transcriptional profiles to humoral responses and vaccine efficacy outcomes.
- 48.Francica JR, Zak DE, Linde C, et al. Innate transcriptional effects by adjuvants on the magnitude, quality, and durability of HIV envelope responses in NHPs. Blood Adv 2017;1(25):2329–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi I, Chung AW, Suscovich TJ, et al. Machine Learning Methods Enable Predictive Modeling of Antibody Feature:Function Relationships in RV144 Vaccinees. PLoS computational biology 2015;11(4):e1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alter G, Dowell KG, Brown EP, et al. High-resolution definition of humoral immune response correlates of effective immunity against HIV. Mol Syst Biol 2018;14(3):e7881.**This study uses humoral response profiling to differentiate HIV infected subjects who progress versus those who control virus, and defines the features of functionally potent antibodies. The features identified match biological intuition and some have been experimentally validated.
- 51.Guyon I, Andr, #233, et al. An introduction to variable and feature selection. J Mach Learn Res 2003;3:1157–82. [Google Scholar]
- 52.Briney B, Inderbitzin A, Joyce C, et al. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 2019;566(7744):393–7.**This paper defines the extent of similarity and differences among immunoglublin sequences across diverse subjects. It represents a technical tour de force and sheds light on the potential value and generalizability of observations related to antibody sequencing data.
- 53.Kwong PD, Chuang GY, DeKosky BJ, et al. Antibodyomics: bioinformatics technologies for understanding B-cell immunity to HIV-1. Immunol Rev 2017;275(1):108–28.*This paper is an instructive review of antibody sequencing approaches and their value to immunobiology and vaccine development and evaluation.
- 54.Lee J, Paparoditis P, Horton AP, et al. Persistent Antibody Clonotypes Dominate the Serum Response to Influenza over Multiple Years and Repeated Vaccinations. Cell Host Microbe 2019;25(3):367–76 e5.*This study matches influenza-specific antibody sequences to corresponding protein products in serum and tracks clonal families over time in a subject. Studies such as this have led to important insights in the diversity of serum antibody-ome and also shed light on potential differences in antibody sequence repertoires between different B cell types.
- 55.Eyer K, Doineau RCL, Castrillon CE, et al. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat Biotechnol 2017;35(10):977–82.**This paper reports a microfluidic approach for encapsulation and evaluation of antibodies secreted from single B cells. This study reports secretion rates and affinities, but the approach may be adaptable to other assessments of interest.
- 56.Sanchez-Lockhart M, Reyes DS, Gonzalez JC, et al. Qualitative Profiling of the Humoral Immune Response Elicited by rVSV-DeltaG-EBOV-GP Using a Systems Serology Assay, Domain Programmable Arrays. Cell Rep 2018;24(4):1050–9 e5. [DOI] [PubMed] [Google Scholar]
- 57.Xu GJ, Kula T, Xu Q, et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015;348(6239):aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bianchi M, Turner HL, Nogal B, et al. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 2018;49(2):288–300 e8.*This paper describes how polyclonal serum samples can be evaluated for their epitope specificity using electron microscopy. Determination of epitopes recognized by mixtures of antibodies has been a challenging goal in the past, and this advance may be enabling.
- 59.Lopez R, Regier J, Cole MB, et al. Deep generative modeling for single-cell transcriptomics. Nat Methods 2018;15(12):1053–8.**This paper describes an impactful advance in analysis of single cell RNA sequencing data enabled by Bayesian deep learning.
- 60.Way GP, Greene CS. Bayesian deep learning for single-cell analysis. Nat Methods 2018;15(12):1009–10. [DOI] [PubMed] [Google Scholar]
- 61.Messina NL, Zimmermann P, Curtis N. The impact of vaccines on heterologous adaptive immunity. Clin Microbiol Infect 2019.*This is one of a set of reviews related to heterologous or trained immunity.
- 62.Sanchez-Ramon S, Conejero L, Netea MG, et al. Trained Immunity-Based Vaccines: A New Paradigm for the Development of Broad-Spectrum Anti-infectious Formulations. Front Immunol 2018;9:2936.*This is one of a set of reviews related to heterologous or trained immunity.
- 63.Uthayakumar D, Paris S, Chapat L, et al. Non-specific Effects of Vaccines Illustrated Through the BCG Example: From Observations to Demonstrations. Front Immunol 2018;9:2869.*This is one of a set of reviews related to heterologous or trained immunity.
- 64.Chung AW, Kumar MP, Arnold KB, et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell 2015;163(4):988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fouts TR, Bagley K, Prado IJ, et al. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proceedings of the National Academy of Sciences of the United States of America 2015. [DOI] [PMC free article] [PubMed]
- 66.Jones AT, Shen X, Walter KL, et al. HIV-1 vaccination by needle-free oral injection induces strong mucosal immunity and protects against SHIV challenge. Nat Commun 2019;10(1):798.*This study explores route and mode of immunization in NHP. ML approaches identify T cell responses and antibody-dependent cellular viral inhibition as correlates of protection.
- 67.Malherbe DC, Mendy J, Vang L, et al. Combination Adenovirus and Protein Vaccines Prevent Infection or Reduce Viral Burden after Heterologous Clade C Simian-Human Immunodeficiency Virus Mucosal Challenge. J Virol 2018;92(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sui Y, Lewis GK, Wang Y, et al. Mucosal vaccine efficacy against intrarectal SHIV is independent of anti-Env antibody response. J Clin Invest 2019;129(3):1314–28.**This study links the notion of trained immunity to the setting of HIV infection by demonstrating that vaccine-associated alterations in the gut microbiome can relate to protection outcomes, as opposed to directly result from adaptive immunity.
- 69.Barouch DH, Alter G, Broge T, et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 2015;349(6245):320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parks C G-106 Mucosal vaccination with a replication-competent VSV-HIV chimera delivering Env trimers protects rhesus macaques from rectal SHIV infection. JAIDS Journal of Acquired Immune Deficiency Syndromes 2017;74:58. [Google Scholar]
- 71.Singh S, Ramirez-Salazar EG, Doueiri R, et al. Control of Heterologous Simian Immunodeficiency Virus SIVsmE660 Infection by DNA and Protein Coimmunization Regimens Combined with Different Toll-Like-Receptor-4-Based Adjuvants in Macaques. J Virol 2018;92(15).*This study finds an association between mucosal antibody responses and protection from infection, serving as a good reminder of the importance of immunity at the front line, and the value of mucosal Ig profiling as a complement to serum Ig evaluation.
- 72.Sadanand S, Das J, Chung AW, et al. Temporal variation in HIV-specific IgG subclass antibodies during acute infection differentiates spontaneous controllers from chronic progressors. AIDS 2018;32(4):443–50.*This study identifies differences in the timing of induction of IgG2 and IgG3 responses targeting envelope as discriminators between progressors and controllers.
- 73.Lofano G, Gorman MJ, Yousif AS, et al. Antigen-specific antibody Fc glycosylation enhances humoral immunity via the recruitment of complement. Sci Immunol 2018;3(26).**This study utilizes observations from ML applied to a cohort of subjects that do and do not raise broadly neutralizing antibodies and goes on to experimentally associate the phenotypic differences with a mechanism whereby humoral immunity can be enhanced. It is an exciting example of discovery of new biology from patterns identified using ML.
- 74.Cheng HD, Grimm SK, Gilman MS, et al. Fine epitope signature of antibody neutralization breadth at the HIV-1 envelope CD4-binding site. JCI Insight 2018;3(5).*This study defines amino-acid level signatures of the epitopes of CD4bs-specific neutralizing antibodies in a panel of monoclonal antibodies as well as in a set of polyclonal serum samples.
- 75.Miller-Novak LK, Das J, Musich TA, et al. Analysis of Complement-Mediated Lysis of Simian Immunodeficiency Virus (SIV) and SIV-Infected Cells Reveals Sex Differences in Vaccine-Induced Immune Responses in Rhesus Macaques. J Virol 2018;92(19).*This NHP immunization study follow up focuses on differences in the immune response associated with sex, and in doing so, points to this variable as potentially important modifier of vaccine responses and/or efficacy.
- 76.Barouch DH, Tomaka FL, Wegmann F, et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase ½a clinical trial (APPROACH) and in rhesus monkeys (NHP 13–19). Lancet 2018;392(10143):232–43.*This study reports on the immunogenicity of a novel HIV vaccine in humans as well as its corresponding profile and protection outcomes in an NHP model.
- 77.Dugast AS, Stamatatos L, Tonelli A, et al. Independent evolution of Fc- and Fab-mediated HIV-1-specific antiviral antibody activity following acute infection. Eur J Immunol 2014;44(10):2925–37. [DOI] [PMC free article] [PubMed] [Google Scholar]