Abstract
Purpose
Image‐guided radiotherapy provides images not only for patient positioning but also for online adaptive radiotherapy. Accurate delineation of organs‐at‐risk (OARs) on Head and Neck (H&N) CT and MR images is valuable to both initial treatment planning and adaptive planning, but manual contouring is laborious and inconsistent. A novel method based on the generative adversarial network (GAN) with shape constraint (SC‐GAN) is developed for fully automated H&N OARs segmentation on CT and low‐field MRI.
Methods and material
A deep supervised fully convolutional DenseNet is employed as the segmentation network for voxel‐wise prediction. A convolutional neural network (CNN)‐based discriminator network is then utilized to correct predicted errors and image‐level inconsistency between the prediction and ground truth. An additional shape representation loss between the prediction and ground truth in the latent shape space is integrated into the segmentation and adversarial loss functions to reduce false positivity and constrain the predicted shapes. The proposed segmentation method was first benchmarked on a public H&N CT database including 32 patients, and then on 25 0.35T MR images obtained from an MR‐guided radiotherapy system. The OARs include brainstem, optical chiasm, larynx (MR only), mandible, pharynx (MR only), parotid glands (both left and right), optical nerves (both left and right), and submandibular glands (both left and right, CT only). The performance of the proposed SC‐GAN was compared with GAN alone and GAN with the shape constraint (SC) but without the DenseNet (SC‐GAN‐ResNet) to quantify the contributions of shape constraint and DenseNet in the deep neural network segmentation.
Results
The proposed SC‐GAN slightly but consistently improve the segmentation accuracy on the benchmark H&N CT images compared with our previous deep segmentation network, which outperformed other published methods on the same or similar CT H&N dataset. On the low‐field MR dataset, the following average Dice's indices were obtained using improved SC‐GAN: 0.916 (brainstem), 0.589 (optical chiasm), 0.816 (mandible), 0.703 (optical nerves), 0.799 (larynx), 0.706 (pharynx), and 0.845 (parotid glands). The average surface distances ranged from 0.68 mm (brainstem) to 1.70 mm (larynx). The 95% surface distance ranged from 1.48 mm (left optical nerve) to 3.92 mm (larynx). Compared with CT, using 95% surface distance evaluation, the automated segmentation accuracy is higher on MR for the brainstem, optical chiasm, optical nerves and parotids, and lower for the mandible. The SC‐GAN performance is superior to SC‐GAN‐ResNet, which is more accurate than GAN alone on both the CT and MR datasets. The segmentation time for one patient is 14 seconds using a single GPU.
Conclusion
The performance of our previous shape constrained fully CNNs for H&N segmentation is further improved by incorporating GAN and DenseNet. With the novel segmentation method, we showed that the low‐field MR images acquired on a MR‐guided radiation radiotherapy system can support accurate and fully automated segmentation of both bony and soft tissue OARs for adaptive radiotherapy.
Keywords: fully convolutional DenseNet, generative adversarial network, head and neck images, shape representation loss
1. Introduction
Head and Neck (H&N) cancer is the fifth most common cancer diagnosed worldwide and the eighth most common cause of cancerous death.1 H&N cancer is typically treated with chemoradiotherapy. For the radiotherapy component, intensity‐modulated radiation therapy (IMRT) is preferred due to its superior critical organ sparing and target dose homogeneity.2 Accurate delineation of organs at risks (OARs) is a prerequisite for high‐quality IMRT and is usually performed manually by oncologists and dosimetrists. The process not only is tedious but also suffers from substantial intra‐ and interobserver variabilities.3 The time‐consuming process is further challenged by the need to adapt H&N treatment plans due to the interfractional changes commonly occur to these patients.4 The anatomical changes can be observed in daily cone beam CT (CBCT) images for patient positioning, but better quality fan beam simulation CTs are commonly acquired to more accurately assess the anatomical changes, delineate the OARs, and perform dose calculation in off‐line adaptive radiotherapy planning. Recent advances in MR‐guided radiotherapy (MRgRT) provide H&N images with superior soft tissue contrast on the daily basis,5 further paving the path to online adaptive radiotherapy, where automated segmentation of the OARs is more urgently needed.
A straightforward strategy to segment OARs is by performing deformable registration between the established H&N atlases and the target image. The OARs annotations in the atlas can then be propagated to the target image. Due to its potential to perform segmentation without user interaction, atlas‐based segmentation methods have attracted considerable attention. Han et al.6 incorporated object shape information from atlases and employed a hierarchical atlas registration for H&N CT images segmentation. Bondiau et al.7 evaluated the performance of atlas‐based segmentation on the brainstem in a clinical radiotherapy context. However, atlas‐based methods can be susceptible to anatomical variations, where additional post‐processing steps are required for refinement. Alternatively, active shape or appearance model8, 9‐based segmentation methods can guide the surface formation by restricting the segmentation results to anatomically plausible shapes described by the pretrained statistical model10 and are commonly employed as post‐processing techniques for atlas‐based methods. For example, Fritscher et al.10 combined statistical appearance models, geodesic active contours, and multiple atlases to segment OARs in H&N CT images. Nevertheless, without supervision, most of the model‐based methods are sensitive to initialization and insufficiently robust to intersubject shape variations of OARs.11
Recently, methods based on deep learning, particularly the convolutional neural networks (CNNs), have demonstrated the potential for medical image segmentation, target detection, registration, and other tasks.12, 13, 14, 15, 16, 17, 18, 19, 20 Specifically for H&N OARs CT segmentation, Ibragimov et al.21modeled the task as multisegmentation subtasks and trained 13 CNNs. The trained networks were then sequentially applied to 2D patches of the test image in a sliding window fashion to locate the expected H&N OARs, which were then refined using the Markov random field algorithm. The patch‐based segmentation method suffers from computational redundancy is inefficient and unable to learn global features.22 The post‐processing step requires additional parameter tuning, preventing the process from being fully automated. Despite its theoretical appeal, deep learning segmentation in the 3D domain is still a largely unsolved problem. 3D CNN requires substantially more training samples than their 2D counterparts, thus easily suffer from overfitting, vanishing‐gradient problem, and high computational cost.23 To overcome these challenges, we developed a novel automated H&N OARs segmentation method that combines the fully convolutional residual network (FC‐ResNet) with a shape representation model (SRM). The SRM network trained to capture the 3D OARs shape features were used to constrain the FC‐ResNet. We showed that superior H&N segmentation performance to state‐of‐the‐art methods could be achieved with the dual neural nets with a relatively small training dataset.24
In addition to CT, automated H&N segmentation was also performed on MR images, which have a increasing importance in MR‐guided radiation therapy that supports online adaptive planning and delivery. Urban et al.25 proposed a random forest classifier that incorporates atlas and image features from multiparametric MR images for H&N OARs segmentation. The study was limited to three soft tissue organs. Kieselmann et al.26 took an atlas approach to segment the parotids, spinal cord, and mandible on T1‐weighted MR. Other than the limited organs in these studies, they were based on high‐field diagnostic multiparametric MR images while MR‐guided radiation therapy more often relies on lower magnetic fields and is more limited in variety of sequences. To our best knowledge, there has not been an automated segmentation study on the low‐field H&N MR images from MR‐guided radiotherapy systems.
In this study, we aim to further improve the segmentation performance from previous work and then test the improved deep segmentation network on low‐field MR images from MRgRT.
2. Materials and method
In this study, the shape constrained (SC) generative adversarial network (GAN), which we refer to as SC‐GAN, is used to further improve the segmentation accuracy of the previously introduced SRM method. The overall architecture of our proposed SC‐GAN is illustrated in Fig. 1. SC‐GAN consists of three tightly integrated modules. A 3D fully convolutional DenseNet is designed as the network for segmenting H&N OARs. The Dice's index is used to provide deep supervision on the DenseNet to alleviate the severe class imbalance due to the small OARs size and improve its optimization performance. Then, a pretrained 3D convolutional autoencoder‐based shape representation model is employed as a regularizer in the training stage to strengthen the shape consistency of predictions of the segmentation network with its ground truth in the latent shape space. Inspired by the GAN,27 a CNN‐based discriminative network is employed to supervise and enforce the segmentation network to produce more accurate predictions. With the coupling structure, the segmentation network benefits from the shape constraint from SRM and adversarial learning with the discriminative network. Moreover, the proposed SC structure prevents GAN from overfitting and make it more stable to train.
Figure 1.
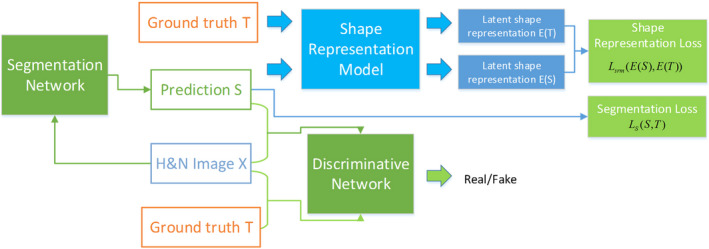
The overall structure of the proposed SC‐GAN network. [Color figure can be viewed at wileyonlinelibrary.com]
2.A. 3D Fully Convolutional DenseNet
U‐Net28 architecture and its extensions29, 30 are widely utilized in medical image analysis. The U‐net architecture comprises an analysis path that learns deep features of the input and a synthesis path performs segmentation based on these learned features. Skip connections allow high‐resolution features from the analysis path to propagate to the corresponding layers in the synthesis path of the network, which improves the performance significantly.16 Deeper networks learn more representative features and result in better performance. However, more layers can significantly impede the propagation of gradients, which is known as vanishing gradient.31 Deep residual network32 partly addresses this problem by introducing skip connections bypassing each residual blocks, but then the residual architecture can easily result in a large number of redundant features to impede the information flow as these connections are incorporated into networks by summation.23
Dense blocks33 are used in the analysis path of our segmentation network by hierarchically extracting the abstract representations of the input to facilitate gradients propagate to preceding layers and improve the network performance. Within each dense block, layers are directly connected with all their preceding layers by concatenation, which is shown in Fig. 2(a). In dense block, the size of the output channel for each convolution layer is commonly referred to as the growth rate k. In other words, the number of feature maps grows k per layer. In our model, there are five dense blocks, each consisting of four dense units with a growth rate of 12, where the dense unit is regarded as one layer. Within each dense block, one convolutional layer with a stride of 2 is used to reduce the resolution of the feature volumes. To further limit the size of the parameter space, bottleneck layers (convolution layer with the 1 × 1×1 kernel) are employed to half the number of feature volumes.
Figure 2.
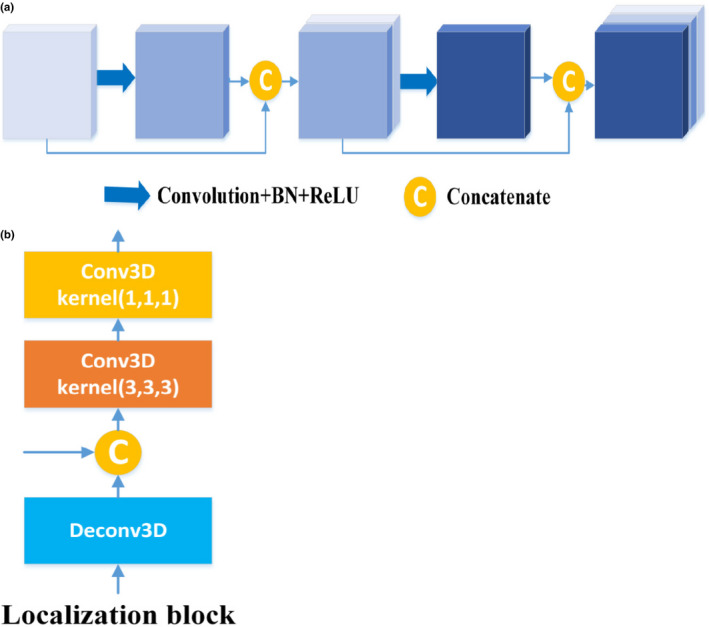
The structure of dense block and localization block. (a) Dense block. (b) Localization block. [Color figure can be viewed at wileyonlinelibrary.com]
In the synthesis path of the segmentation network, feature volumes are concatenated with the feature volumes from the analysis path and then passed to the next localization block. As illustrated in Fig. 2 (b), each localization block consists of a 3 × 3 × 3 convolutional layer, which is followed by a 1 × 1 × 1 convolutional layer with half number of feature volumes. Within each localization block, deconvolutional layers (stride 2) are employed for resolution restoration. In the model, each convolutional layer is followed by batch normalization (BN)34 and a rectified linear unit.35
Multiscale features fusion and the deep supervision technique are utilized in our proposed segmentation network to integrate more fine details for accurate segmentation of small organs (e.g., optical chiasm and optical nerves) and speed up network convergence. Specifically, deep supervision22 is employed in the synthesis path by integrating feature volumes of different scales at different levels of the network. The feature volumes are combined via element‐wise summation after upscaling to the same image resolution using deconvolutional layers to form the final predictions of the network. Deep supervision serves as a strong regularization to boost gradient back‐propagation by guiding the training of the lower layers in the network. The overall architecture of the proposed segmentation network is illustrated in Fig. 3.
Figure 3.
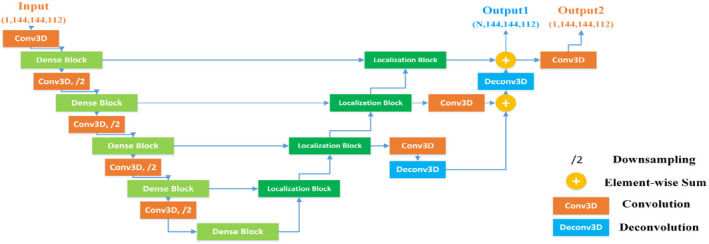
The architecture of the segmentation network. N represents the number of organs to be segmented. For concise illustration, batch normalization and rectified linear unit are omitted from this figure. [Color figure can be viewed at wileyonlinelibrary.com]
It is worth noting that the output channels for H&N images correspond to different OARs. The architecture of the segmentation network is adapted to better integrate into the discriminative network and SRM. To satisfy the input channel size of SRM, combat potential difficulties in capturing the shape characteristics of each organ separately, especially small organs, and avoid being dominated by the predictions when they are fed into the discriminative network, an additional 1 × 1 × 1 convolutional layer with the filter number of 1 is employed on the multichannel output of the segmentation network. Consequently, the segmentation network has two outputs with different channel sizes. One single‐channel output is designated for the discriminative network and SRM and the other multichannel output with each channel correspondences to each H&N OAR for segmentation network. At the testing stage, the multichannel output is employed as the segmentation result for the H&N multiorgan image segmentation task.
2.B. Shape representation model
Learning and incorporating the shape characteristics of the OARs are of great importance when solving the image‐wise prediction problems.36 As shown in our previously study,24 a SRM increases the robustness and stability of the segmentation network without depending on an extensive patient dataset. Here, we constructed a similar model and employed it as prior information in the training stage of SC‐GAN.
Shape representation model predicts label maps from the segmentation network and then encourages them to follow the shape characteristics of the ground truth. Considering that the stacked convolutional autoencoder can learn a latent representation from the original input in the encoder, the encoder is employed as SRM to encode the segmentation and the ground truth. The architecture of SRM is illustrated in Fig. 4.
Figure 4.
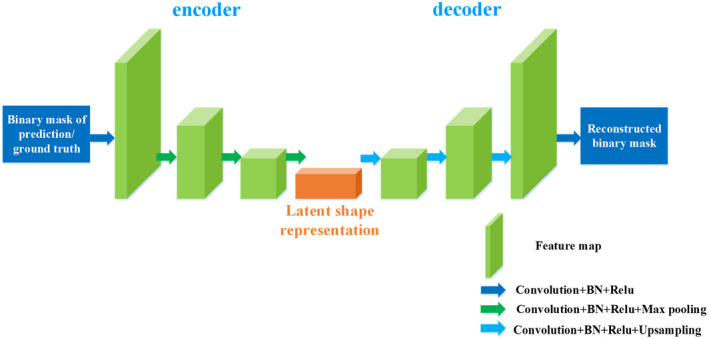
The architecture of shape representation model. [Color figure can be viewed at wileyonlinelibrary.com]
To better capture the latent shape characteristics of the target organs, SRM is trained on the binary shape masks of the H&N OARs. In the training stage, the encoder block aims to project it onto the latent shape representation space and then obtain the higher order shape representation of the H&N OARs, while decoder works on accurate reconstruction. Thus, the SRM training objective function can be built as follows:
| (1) |
where θ SRM denotes all trainable parameters of SRM, E(·) and D(·) represent encoder and decoder, respectively. L Dice is formulated as a Dice coefficient loss function. At the testing stage, the shape representation loss between the predicted segmentation S and the ground truth T is incorporated into the objective function of the segmentation network to constrain its training and minimize the shape deviation between S and T.
Different from the loss terms utilized in the previously study,24 the reconstruction loss between D(E(S)) and the ground truth T is removed from the objective function of the segmentation network for the following reasons. First, the segmentation network optimization can focus on the voxel‐level consistency (segmentation loss), latent shape space consistency (shape representation loss), and higher order spatial consistency (adversarial loss). Second, it was determined that the reconstruction loss between D(E(S)) and the ground truth T contributes little to improve the performance of the segmentation network due to the accumulated error from the encoder, the decoder, and the segmentation network.
2.C. Discriminative network
On par with the segmentation network, a CNN‐based network is employed as our discriminative network as shown in Fig. 5. The discriminative network plays an adversarial role to prompt the global‐level field of view of the segmentation network and strengthen the higher order spatial consistency of its predictions.37 Our discriminator comprises four convolutional layers with BNs, four max pooling layers, and one fully connection layer. During training, the discriminative network receives the image pair of original H&N image and prediction from the segmentation network or the ground truth with the same number of channels, outputs a single scalar indicating whether the label map is from the segmentation network or the ground truth. With the supervision from the discriminator, the segmentation network is further prompted to produce correct predictions.
Figure 5.
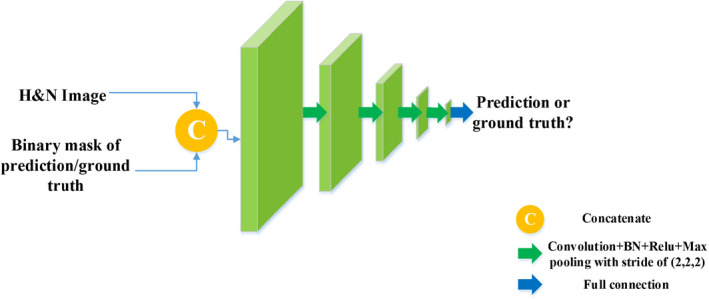
The architecture of the discriminative network. [Color figure can be viewed at wileyonlinelibrary.com]
2.D. SC‐GAN
As illustrated in Fig. 1, our proposed SC‐GAN consists of three tightly integrated networks, shape representation model, segmentation network, and discriminative network. Shape representation model is pretrained with the ground truth of the training dataset, then employed as a regularizer to constrain the predictions of the segmentation network during training. The training process of our proposed SC‐GAN follows the optimization procedure of the original GAN.27 The objective of the original GAN is to minimize the probability of the predictions by the generative network G to be recognized while maximizing the probability of the discriminative network D making a wrong decision.38 The objective function is defined as:
| (2) |
where denotes the true data samples, D(z) represents the probability that z came from the true data rather than generated data. denotes the prior input noise of generative network, while G(v) represents the generated data.
For multiclass segmentation task, the objective function is formulated as follows:
| (3) |
Here, λ adv is set to keep the balance of adversarial learning. θ s and θ D represent the trainable parameters of the segmentation network S and discriminative network D, respectively. X and T denote the original H&N images and ground truths, respectively. As for our proposed SC‐GAN, considering the integration of the shape representation loss between the segmentations S(X) and T, and optimization of the single‐channel output and multichannel output from the network S, the objective function of the proposed SC‐GAN thus is constructed as:
| (4) |
where E(·) represents encoder block in SRM. E (S(X)) and E (T) represent the latent shape representation of the predictions and the ground truths. S(X)1 and S (X) N denote the single‐channel output and multi‐channel output, respectively. Similarly, T 1 and T N denote the single‐channel ground truths (i.e., binary masks of the OARs) and multichannel ground truths (i.e., categorized ground truths), respectively. λ srm is the weight of the shape representation loss term used in the training stage. To alleviate the severe class imbalance, the segmentation loss Ls is formulated as a multiclass Dice coefficient loss, which encourages the segmentation network to make right voxel‐wise class label predictions. Shape representation loss L SRM is formulated as a cross entropy loss function. The third term L D is a binary cross‐entropy and defines as follows:
| (5) |
where z and z ′ denotes the label and the output of the discriminative networks.
Similar to the primary GAN, the optimization procedure for our SC‐GAN can be simultaneously decomposed into the segmentation network optimization and the discriminative network optimization, respectively.27
During training, the segmentation network attempts to make right voxel‐wise class label predictions that deceive the discriminative network D as the ground truth. Therefore, the segmentation network can be optimized by:
| (6) |
The discriminative can be optimized by:
| (7) |
3. Experiments
3.A. Experimental datasets and preprocessing
The performance of the modified segmentation network SC‐GAN was first benchmarked on the public database used in the previous study. It is then applied to low‐field 0.35T H&N MRI images acquired on an MRgRT system (Meridian, ViewRay, Oakwood Village, Ohio).
3.A.1. H&N CT Dataset
We tested our proposed method on the Public Domain Database for Computational Anatomy (PDDCA) version 1.4.1. The original CT data were derived from the Radiation Therapy Oncology Group (RTOG) 0522 study, a multi‐institutional clinical trial led by Kian Ang.39 This dataset contains 48 patients CT volumes with anisotropic pixel spacing ranging from 0.76 to 1.76 mm and an interslice thickness ranging from 1.25 to 3.0 mm. Thirty‐two of the 48 patients in the database with the complete manual labeling of nine structures, including the brainstem, optical chiasm, mandible, parotid glands (both left and right), optical nerves (both left and right), and submandibular glands (both left and right), were used in this study. The test on a public database not only helped us to develop the segmentation networks but also placed the proposed method in a frame of reference that can be cross‐compared with competing segmentation methods.
3.A.2. H&N MRI Dataset
The H&N MRI dataset containing 25 H&N MRI volumes with the manual labeling of the brainstem, optical chiasm, larynx, mandible, pharynx, parotid glands (both left and right), and optical nerves (both left and right) was used to test the efficacy of automated segmentation using the proposed method. The patient data were collected from UCLA hospital under an Institutional Review Board (IRB) approved protocol. The image sequence is TrueFISP. The voxel size and image resolution are 1.5 × 1.5 × 1.5 mm3 and 334 × 300 × 288, respectively.
3.A.3. Preprocessing
As preprocessing, we first normalized the CT and MRI dataset, respectively, so each volume has zero mean and unit variance. To further homogenize the data, reduce memory consumption, and increase computational speed, all CT H&N images were resampled to isotropic resolution of 1.5 mm3 × 1.5 mm3 × 1.5 mm3 and then cropped to keep only the part of the patient with OARs relevant to the study, resulting in a uniform matrix size of 144 × 144 × 112, which was used for both training and testing. MRI images were cropped to meet the uniform matrix size of 144 × 144 × 112 without any changes on the resolution because they have the same resolution already.
The splits of training/testing for H&N CT and MRI images are 22/10 and 15/10, respectively. Three subjects were each held out from the CT and MR training set for independent validation and fine tune of the hyperparameters.
3.B. Comparison algorithms
To illustrate the contributions of each module, we trained two other networks for comparison in this study. The details of the comparison networks are summarized in Table 1. It is worth noting that the architecture of FC‐ResNet is same as the FC‐DenseNet with all of the dense blocks are replaced by the residual blocks. The two networks were implemented on Tensorflow 1.8.0 library on Python 2.7 using the same configurations and datasets as SC‐GAN‐DenseNet.
Table 1.
Summary of the trained networks
| Network | Segmentation network | Discriminative network | Objective function for segmentation network |
|---|---|---|---|
| SC‐DenseNet | FC‐DenseNet | / | L s + λ srm L SRM |
| GAN | FC‐ResNet | CNN | L s + λ adv L D |
| SC‐GAN‐ResNet | FC‐ResNet | CNN | L S + λ srm L SRM + λ adv L D |
| SC‐GAN‐DenseNet (proposed) | FC‐DenseNet | CNN | L s + λ srm L SRM + λ adv L D |
3.C. Implementation details
We implemented the proposed method using Tensorflow. The training of the SRM and SC‐GAN took approximately 2 and 20 h, respectively, using an NVIDIA GeForce GTX 1080 GPU with 8GB memory with the mini‐batch size of 1 for SC‐GAN and 4 for SRM. In the testing phase, the total processing time of one 3D H&N scan of size 144 × 144 × 112 was 14 s, which was a significant improvement compared with the conventional segmentation methods. Adam optimizers were utilized to optimize the three networks, respectively. The learning rates for the segmentation network and the discriminative network were initialized as lr s = 1e − 3 and lr D = 1e − 4. Regarding SRM, the initial learning rate was set tolr srm = 5e − 4. To ensure that the discriminative network sufficiently influences the segmentation network, the initial learning rate employed in the discriminative network lr D was smaller than lr S to slow down the convergence of the discriminative network. lr s and lr D were divided by a factor of 5 every 10 epochs when the validation loss stopped improving to prevent overfitting. Moreover, an early stopping strategy was also utilized if there was no improvement in the validation loss after 50 epochs. The same decay strategy and early stopping strategy were also performed on lr srm and the training of SRM. Additionally, the weights of shape representation loss term λ srm and adversarial loss term λ adv in the objective function were both set as 0.1 based on the performance on the validation set.
3.D. Learning curves
There are 72,912 trainable parameters in SRM. However, SRM is pretrained and does not need to be retrained during the training of SC‐GAN. GAN consists of a generator and a discriminator. The numbers of trainable parameters in the generator (i.e., FC‐DenseNet) and the discriminator are 1,999,118 and 1,177,153, respectively. To address the scarcity of the data available in this study, data augmentation, dropout layers, and early stopping strategies were adopted to prevent overfitting. During training, each volume was rotated by one of the three angles (90°, 180°, 270°) and randomly scaled between 0.8 and 1.2 on the fly for data augmentation. With data augmentation, we increase the number of the original training date by fivefold. A dropout layer with the dropout rate of 0.3 was utilized in each convolutional layer in the segmentation network and the discriminator to further reduce overfitting. Additionally, learning rate decay and early stopping strategies were utilized when the validation loss stopped decreasing. The learning curves of SRM, segmentation network, and discriminator are shown in Fig. 6. The test loss of the generator and SRM consistently decreases as the training loss goes down, demonstrating that no serious overfitting is observed with such small datasets.
Figure 6.
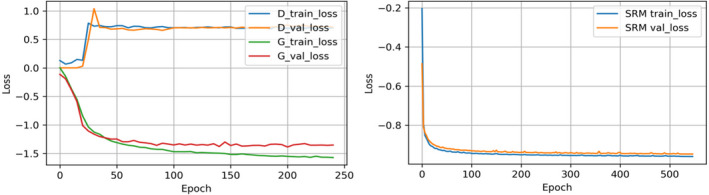
Learning curves of the discriminator, the generator (i.e., segmentation network), and the shape representation model. [Color figure can be viewed at wileyonlinelibrary.com]
3.E. Evaluation metrics
For each test image, we used five evaluation metrics to quantitatively evaluate the performance of the proposed framework against manual segmentation. The segmentation evaluation metrics are defined as below:
Dice Similarity Coefficient (DSC)40:
Positive Predictive Value (PPV):
Sensitivity (SEN):
Average Surface Distance (ASD):
95%Maximum Surface Distance (95%SD): 95%SD is based on the calculation of 95th percentile of the distances between the boundary points of A and B, which is expected to eliminate the impact of a small subset of incorrect segmentations on the evaluation of the overall segmentation quality.
In these metrics, A and B refer to the manual and automatic segmentation, respectively. d(z, A) denotes the minimum distance of voxel z on the automatically segmented organ surface B to the voxels on the ground truth surface A, d(u, B) denotes the minimum distance of voxel u on the ground truth surface A to the voxels on the automatically segmented organ surface B.
3.F. H&N CT segmentation results
3.F.1. Qualitative and quantitative evaluation
The qualitative and quantitative evaluation results on PDDCA dataset are listed in Fig. 7 and Tables 2 and 3, respectively. It can be observed that SC‐GAN‐DenseNet has achieved accurate segmentation on all nine CT H&N OARs. Moreover, quantitative evaluation results in Tables 2 and 3 illustrate that SC‐GAN‐DenseNet consistently outperforms GAN, SC‐DenseNet, and SC‐GAN‐ResNet on all H&N OARs showing the benefits of incorporating SRM, dense connectivity as well as adversarial learning. As demonstrated in the second column in Fig. 7 GAN without being constrained by SRM can lead to false‐positive islands and undersegmented OARs, which are rectified by incorporating shape representation loss between the prediction of the segmentation network and ground truth as shown in the third column in Fig. 7. Furthermore, leveraging the latent shape representation learned by SRM, the predictions of the segmentation network is robust to interpatient shape variations.
Figure 7.
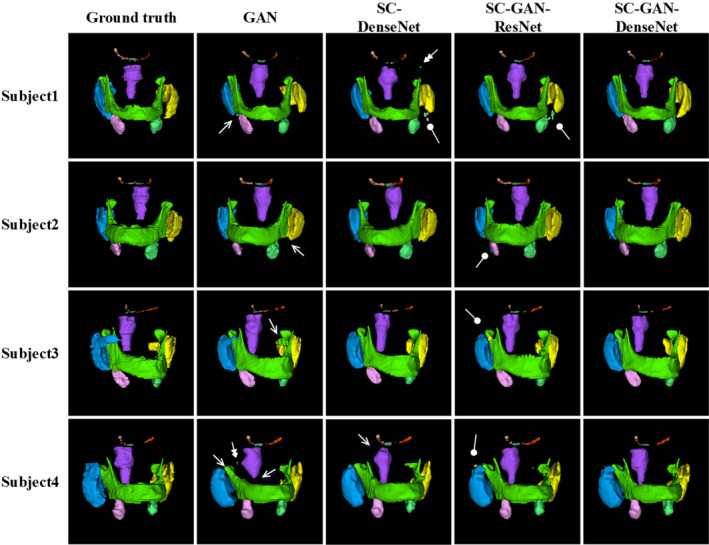
Examples of the H&N CT segmentation results by generative adversarial network (GAN), shape constraint (SC)‐DenseNet, SC‐GAN‐ResNet, and SC‐GAN‐DenseNet. The first column shows the ground truth, the second, third, fourth, and fifth columns present the segmentation results by GAN, SC‐DenseNet, SC‐GAN‐ResNet, and SC‐GAN‐DenseNet, respectively. Brainstem (purple), optical chiasm (dark green), mandible (green), left and right optical nerves (orange and light orange), left and right parotid glands (blue and yellow), left and right submandibular glands (pink and light green). The single, double, and blunt arrows denote false‐positive islands, undersegmentations, and mis‐segmentations, respectively. [Color figure can be viewed at wileyonlinelibrary.com]
Table 2.
Quantitative evaluation results on PDDCA dataset (DSC, PPV, and SEN). The best performer of each metric and each organ is bolded
| Organ | DSC (%) | PPV (%) | SEN(%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | |
| BS | 83.75 ± 2.0 | 85.09 ± 2.2 | 85.34 ± 4.1 | 86.72 ± 2.9 | 80.43 ± 4.6 | 83.78 ± 5.0 | 83.92 ± 4.4 | 87.92 ± 4.2 | 87.95 ± 5.6 | 86.67 ± 1.9 | 87.21 ± 4.8 | 85.78 ± 4.2 |
| OC | 55.85 ± 12 | 54.47 ± 13 | 57.54 ± 10 | 59.16 ± 9.7 | 54.42 ± 10.4 | 56.01 ± 11 | 54.87 ± 10 | 54.72 ± 10.3 | 58.85 ± 12 | 55.25 ± 10 | 62.79 ± 12 | 68.29 ± 7.5 |
| MA | 92.03 ± 1.9 | 92.85 ± 1.4 | 93.48 ± 0.9 | 93.91 ± 1.3 | 95.01 ± 2.4 | 95.92 ± 1.7 | 94.61 ± 2.5 | 96.82 ± 1.7 | 89.37 ± 3.9 | 90.01 ± 3.4 | 92.50 ± 2.5 | 91.25 ± 2.7 |
| LO | 63.90 ± 7.9 | 64.09 ± 7.7 | 64.59 ± 6.3 | 66.38 ± 4.8 | 60.13 ± 5.3 | 59.91 ± 5.5 | 63.48 ± 6.9 | 69.61 ± 7.8 | 69.83 ± 10 | 69.84 ± 12 | 67.38 ± 7.8 | 65.15 ± 4.1 |
| RO | 63.58 ± 6.2 | 63.80 ± 5.3 | 69.49 ± 6.8 | 69.91 ± 4.3 | 54.12 ± 6.5 | 55.70 ± 5.5 | 58.97 ± 8.3 | 64.24 ± 7.5 | 77.19 ± 7.9 | 75.27 ± 8.8 | 85.29 ± 7.7 | 78.40 ± 8.3 |
| LP | 83.56 ± 1.9 | 84.77 ± 4.0 | 84.94 ± 1.9 | 85.49 ± 1.7 | 86.95 ± 5.4 | 85.02 ± 4.7 | 86.48 ± 5.0 | 87.24 ± 3.3 | 80.89 ± 4.4 | 84.85 ± 5.9 | 83.91 ± 4.4 | 84.02 ± 3.5 |
| RP | 84.11 ± 3.8 | 84.62 ± 4.5 | 85.36 ± 3.2 | 85.77 ± 2.4 | 84.11 ± 5.1 | 83.29 ± 5.2 | 84.76 ± 4.9 | 84.14 ± 4.4 | 84.40 ± 5.2 | 86.63 ± 7.8 | 86.26 ± 4.5 | 87.84 ± 4.8 |
| LS | 76.84 ± 8.3 | 78.8 ± 7.5 | 79.99 ± 5.6 | 80.65 ± 5.0 | 80.43 ± 10 | 84.38 ± 6.1 | 83.14 ± 6.3 | 84.71 ± 9.3 | 75.02 ± 8.1 | 75.55 ± 7.8 | 78.17 ± 6.7 | 78.29 ± 8.0 |
| RS | 76.24 ± 7.4 | 78.73 ± 8.0 | 79.16 ± 8.2 | 81.86 ± 4.9 | 81.13 ± 10 | 79.74 ± 7.3 | 81.11 ± 6.0 | 80.62 ± 3.9 | 72.56 ± 7.8 | 79.15 ± 8.9 | 78.43 ± 8.2 | 79.09 ± 6.9 |
BS, Brainstem; OC, Optic Chiasm; MA, Mandible; LO, Left Optic nerve; RO, Right Optic nerve; LP, Left Parotid; RP, Right Parotid; LS, Left Submandibular gland; RS, Left Submandibular gland.
Table 3.
Quantitative evaluation results on PDDCA dataset (ASD and 95%SD)
| Organ | ASD (mm) | 95%SD (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | |
| BS | 1.50 ± 0.24 | 1.51 ± 0.37 | 1.46 ± 0.43 | 1.41 ± 0.31 | 3.85 ± 0.61 | 3.98 ± 1.08 | 3.81 ± 0.90 | 3.62 ± 0.75 |
| OC | 1.89 ± 1.72 | 1.49 ± 0.92 | 1.41 ± 0.79 | 1.27 ± 0.34 | 4.50 ± 1.99 | 4.18 ± 1.99 | 3.78 ± 0.86 | 3.77 ± 1.18 |
| MA | 0.80 ± 0.79 | 0.68 ± 0.55 | 0.54 ± 0.12 | 0.55 ± 0.14 | 2.17 ± 1.08 | 1.88 ± 0.59 | 2.19 ± 0.72 | 2.09 ± 0.63 |
| LO | 0.91 ± 0.44 | 1.35 ± 0.97 | 1.10 ± 1.32 | 0.73 ± 0.22 | 2.30 ± 1.07 | 2.75 ± 1.78 | 2.22 ± 1.35 | 2.03 ± 0.47 |
| RO | 1.90 ± 1.41 | 1.00 ± 0.15 | 1.12 ± 0.53 | 0.78 ± 0.16 | 2.80 ± 1.21 | 2.93 ± 1.47 | 2.36 ± 0.16 | 2.09 ± 0.41 |
| LP | 2.12 ± 1.81 | 2.21 ± 1.42 | 1.50 ± 0.92 | 1.39 ± 0.38 | 4.66 ± 2.83 | 4.58 ± 3.34 | 4.45 ± 2.63 | 4.62 ± 2.70 |
| RP | 2.41 ± 2.81 | 2.00 ± 0.27 | 2.09 ± 1.57 | 1.40 ± 0.37 | 4.47 ± 2.84 | 4.29 ± 3.18 | 4.23 ± 2.17 | 3.80 ± 1.10 |
| LS | 2.02 ± 1.51 | 1.62 ± 0.34 | 1.38 ± 0.99 | 1.44 ± 1.02 | 4.58 ± 0.38 | 4.20 ± 0.97 | 4.60 ± 0.44 | 4.50 ± 2.24 |
| RS | 3.52 ± 2.66 | 2.23 ± 1.13 | 1.91 ± 1.27 | 1.56 ± 0.55 | 4.63 ± 2.74 | 4.74 ± 2.84 | 4.05 ± 1.42 | 3.95 ± 2.72 |
BS, Brainstem; OC, Optic Chiasm; MA, Mandible; LO, Left Optic nerve; RO, Right Optic nerve; LP, Left Parotid; RP, Right Parotid; LS, Left Submandibular gland; RS, Left Submandibular gland. Bold fonts denote the best performers.
There are remaining mis‐segmented volumes using SC‐GAN‐ResNet due to the intrinsic limitations of ResNet. By overcoming the limitation using dense connectivity, the segmentation performance is further improved using SC‐GAN‐DenseNet as shown in the fourth column of Fig. 7.
3.F.2. Comparison with state‐of‐the‐art methods
In Tables 4 and 5, we compare SC‐GAN‐DenseNet with five state‐of‐the‐art H&N segmentation methods based on a hierarchical atlas,6 an active appearance model,41 a patch‐based CNN,21 a hierarchical vertex regression method,11 and our previous study using SRM and FC‐ResNet.24 It is worth noting that the segmentation performance reported in Ref.11, 24, 41 was evaluated on the same PDDCA dataset with our proposed SC‐GAN‐DenseNet, which enables a direct comparison. Moreover, the active appearance model‐based segmentation method41 proposed by Mannion et al. was the winner of the MICCAI 2015 Head and Neck Auto Segmentation Grand Challenge. The experimental results in Tables 4 and 5 demonstrate that the proposed segmentation method outperforms the conventional atlas‐based, model‐based, and CNN‐based method on the Dice's index and segmentation speed. Moreover, the paired Student's t‐test indicate that the improvement between SC‐GAN and SRM‐FC‐ResNet24 (P = <0.05) on 7 OARs are statistically significant, as indicated by asterisks in Table 4. The comparison results with the SRM constrained FC‐ResNet indicate that adversarial training and dense connectivity further benefit the segmentation networks performance the in H&N multiorgan segmentation task. The improvement in DSC using our method in comparison to the model‐based method,41 and the hierarchical vertex regression method11 is not significant, but our segmentation is two orders of magnitude faster.
Table 4.
Comparison of segmentation accuracy between the state‐of‐the‐art methods and our method (DSC %)
| Organ/Method | Han6 | Mannion41 | Ibragimov21 | Wang11 | Tong24 | SC‐GAN‐DenseNet (proposed) |
|---|---|---|---|---|---|---|
| Brainstem | 82 | 87 ± 4 | Unavailable | 90.3 ± 3.8 | 86.97 ± 2.95 | 86.72 ± 2.92 |
| Optical chiasm | Unavailable | 35 ± 16 | 37.4 ± 13.4 | Unavailable | 58.35 ± 10.28a | 59.16 ± 9.76 |
| Mandible | 89 | 93 ± 1 | 89.5 ± 3.6 | 94.4 ± 1.3 | 93.67 ± 1.21 | 93.91 ± 1.32 |
| Left optical nerve | Unavailable | 63 ± 5 | 63.9 ± 6.9 | Unavailable | 65.31 ± 5.75a | 66.38 ± 4.83 |
| Right optical nerve | Unavailable | 63 ± 5 | 64.5 ± 7.5 | Unavailable | 68.89 ± 4.74a | 69.91 ± 4.38 |
| Left parotid | 82 | 84 ± 7 | 76.6 ± 6.1 | 82.3 ± 5.2 | 83.49 ± 2.29a | 85.49 ± 1.78 |
| Right parotid | 82 | 84 ± 7 | 77.9 ± 5.4 | 82.9 ± 6.1 | 83.18 ± 1.45a | 85.77 ± 2.44 |
| Left submandibular | 69 | 78 ± 8 | 69.7 ± 13.3 | Unavailable | 75.48 ± 6.49a | 80.65 ± 5.08 |
| Right submandibular | 69 | 78 ± 8 | 73.0 ± 9.2 | Unavailable | 81.31 ± 6.45a | 81.86 ± 4.96 |
The statistical significance (P < 0.05). Bold fonts denote the best performers.
Table 5.
Comparison of runtime between the state‐of‐the‐art methods and our method
3.G. H&N MRI segmentation results
Figure 8 and Tables 6 and 7 present the H&N MRI qualitative and quantitative evaluation results, respectively. Segmentation accuracies consistent with that on the PDDCA CT dataset are observed with moderate differences between organs related to different CT‐MR contrast. For instance, superior brainstem segmentation accuracy is achieved on MR due to its better contrast in MR while mandible segmentation accuracy is lower on MR due to the low bone signal. Although the Dice's index of the chiasm, optical nerves and parotids SC‐GAN‐ResNet segmentations on the MR were not better than on the CT, the 95%SD is still superior using the MR. The comparison among GAN, SC‐DenseNet, SC‐GAN‐ResNet, and SC‐GAN‐DenseNet on the MR images is consistent with the CT segmentation with SC‐GAN‐DenseNet being the best in both Dice's index and surface agreement, followed by SC‐GAN‐ResNet, SC‐DenseNet, and GAN.
Figure 8.
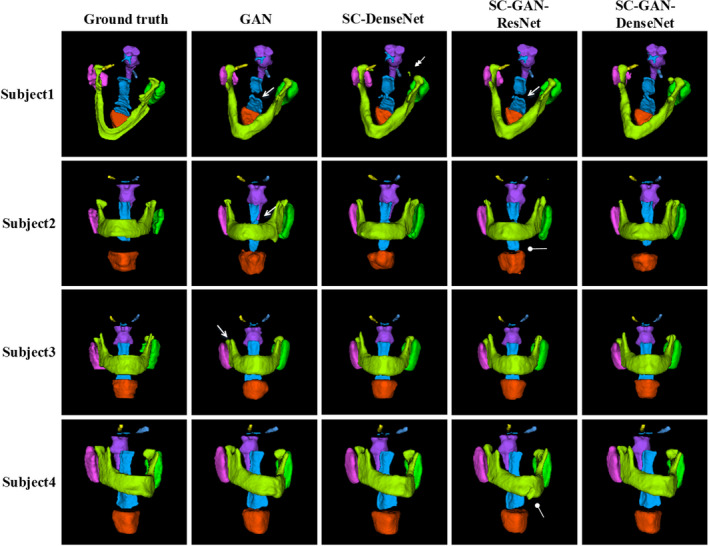
Examples of the H&N MRI segmentation results by generative adversarial network (GAN), shape constraint (SC)‐DenseNet, SC‐GAN‐ResNet, and SC‐GAN‐DenseNet. The first column shows the ground truth, the second, third, fourth, and fifth columns present the segmentation results by GAN, SC‐DenseNet, SC‐GAN‐ResNet, and SC‐GAN‐DenseNet, respectively. Brainstem (purple), optical chiasm (blue), larynx (orange), mandible (grass green), left and right optical nerves (yellow and light blue), left and right parotid glands (pink and green), pharynx (blue). The single, double, and blunt arrows denote false‐positive islands, undersegmentations, and mis‐segmentations, respectively. [Color figure can be viewed at wileyonlinelibrary.com]
Table 6.
Quantitative evaluation results on H&N MRI dataset (DSC, PPV, and SEN)
| Organ | DSC (%) | PPV (%) | SEN (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | |
| BS | 90.49 ± 1.8 | 90.54 ± 2.63 | 90.86 ± 1.2 | 91.57 ± 2.85 | 90.20 ± 3.9 | 87.07 ± 6.8 | 88.01 ± 3.8 | 88.72 ± 5.6 | 90.80 ± 2.6 | 95.00 ± 3.2 | 94.16 ± 2.9 | 94.91 ± 2.5 |
| OC | 53.16 ± 12. | 56.45 ± 10.4 | 57.59 ± 10. | 58.92 ± 7.22 | 59.05 ± 12 | 65.10 ± 8.4 | 63.06 ± 12 | 62.67 ± 8.1 | 50.93 ± 13 | 53.0 ± 5.0 | 54.19 ± 12 | 60.77 ± 5.2 |
| LA | 74.09 ± 6.3 | 75.19 ± 4.5 | 76.51 ± 6.7 | 79.86 ± 5.31 | 70.91 ± 8.3 | 69.04 ± 8.4 | 74.17 ± 6.0 | 77.30 ± 5.6 | 85.22 ± 6.1 | 87.01 ± 8.0 | 85.31 ± 7.6 | 88.40 ± 5.9 |
| MA | 80.59 ± 6.9 | 80.34 ± 7.9 | 80.97 ± 7.1 | 81.64 ± 4.44 | 78.48 ± 6.8 | 78.98 ± 3.6 | 78.20 ± 8.8 | 78.50 ± 4.3 | 84.71 ± 5.5 | 86.5 ± 4.6 | 85.98 ± 5.8 | 86.65 ± 6.0 |
| LO | 63.97 ± 4.9 | 68.05 ± 6.5 | 66.27 ± 4.1 | 71.65 ± 4.46 | 76.16 ± 6.9 | 68.31 ± 7.1 | 75.10 ± 7.6 | 76.20 ± 6.5 | 58.64 ± 9.2 | 70.18 ± 6.1 | 60.65 ± 4.7 | 67.95 ± 3.2 |
| RO | 65.12 ± 7.0 | 64.55 ± 3.2 | 68.55 ± 6.5 | 69.31 ± 6.58 | 68.83 ± 8.8 | 70.06 ± 4.2 | 75.11 ± 2.9 | 70.77 ± 5.1 | 66.23 ± 5.7 | 63.48 ± 10 | 64.26 ± 9.6 | 68.42 ± 6.6 |
| LP | 83.62 ± 6.4 | 81.64 ± 5.6 | 84.43 ± 5.1 | 86.48 ± 5.01 | 79.27 ± 5.0 | 74.63 ± 5.2 | 78.42 ± 6.1 | 82.63 ± 6.7 | 90.03 ± 5.5 | 91.92 ± 6.1 | 92.74 ± 4.4 | 90.57 ± 5.8 |
| RP | 81.54 ± 6.1 | 80.74 ± 2.9 | 81.94 ± 5.3 | 82.48 ± 5.34 | 76.93 ± 7.7 | 71.59 ± 7.9 | 77.61 ± 8.0 | 79.87 ± 6.9 | 90.59 ± 4.8 | 97.13 ± 4.4 | 90.21 ± 4.5 | 89.79 ± 5.2 |
| PH | 68.31 ± 8.3 | 68.35 ± 8.6 | 69.52 ± 7.6 | 70.60 ± 7.62 | 66.26 ± 9.3 | 63.63 ± 9.0 | 63.52 ± 8.1 | 68.83 ± 7.5 | 72.28 ± 8.4 | 74.33 ± 3.5 | 78.09 ± 6.9 | 73.48 ± 3.0 |
BS, Brainstem; OC, Optic Chiasm; LA, Larynx; MA, Mandible; LO, Left Optic nerve; RO, Right Optic nerve; LP, Left Parotid; RP, Right Parotid; PH, Pharynx. Bold fonts denote the best performers.
Table 7.
Quantitative evaluation results on H&N MRI dataset (ASD and 95%SD)
| Organ | ASD (mm) | 95%SD (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | GAN | SC‐DenseNet | SC‐GAN‐ResNet | SC‐GAN‐DenseNet | |
| BS | 0.74 ± 0.15 | 0.76 ± 0.22 | 0.73 ± 0.13 | 0.68 ± 0.29 | 1.68 ± 0.83 | 2.09 ± 0.72 | 1.28 ± 0.47 | 2.08 ± 1.28 |
| OC | 0.81 ± 0.36 | 0.85 ± 1.02 | 0.81 ± 1.09 | 0.78 ± 0.44 | 2.88 ± 1.72 | 2.80 ± 1.97 | 2.24 ± 1.90 | 2.05 ± 1.37 |
| LA | 1.97 ± 1.14 | 1.81 ± 1.46 | 1.70 ± 1.60 | 1.76 ± 1.37 | 4.89 ± 2.68 | 4.27 ± 1.35 | 3.20 ± 2.68 | 3.92 ± 2.56 |
| MA | 1.15 ± 0.50 | 1.05 ± 0.72 | 1.16 ± 0.45 | 1.13 ± 0.48 | 2.87 ± 1.15 | 2.65 ± 1.73 | 3.02 ± 1.29 | 2.72 ± 1.31 |
| LO | 1.50 ± 0.31 | 0.98 ± 0.62 | 1.09 ± 1.34 | 0.44 ± 0.26 | 2.44 ± 0.55 | 1.54 ± 1.23 | 1.95 ± 2.34 | 1.48 ± 0.44 |
| RO | 1.50 ± 2.26 | 0.64 ± 0.19 | 0.51 ± 0.20 | 0.86 ± 0.53 | 2.77 ± 0.77 | 1.68 ± 0.44 | 1.40 ± 0.44 | 1.56 ± 0.43 |
| LP | 4.05 ± 1.72 | 3.17 ± 0.91 | 2.25 ± 2.39 | 1.00 ± 0.39 | 6.31 ± 3.68 | 6.47 ± 3.38 | 4.94 ± 2.71 | 2.58 ± 1.39 |
| RP | 4.23 ± 1.95 | 3.47 ± 1.53 | 1.49 ± 1.01 | 1.42 ± 1.18 | 6.48 ± 3.91 | 4.82 ± 2.76 | 3.89 ± 2.33 | 3.56 ± 2.10 |
| PH | 1.35 ± 0.68 | 1.64 ± 1.21 | 1.27 ± 1.12 | 0.97 ± 0.45 | 5.60 ± 2.12 | 4.23 ± 2.15 | 4.33 ± 2.28 | 2.84 ± 1.31 |
BS, Brainstem; OC, Optic Chiasm; LA, Larynx; MA, Mandible; LO, Left Optic nerve; RO, Right Optic nerve; LP, Left Parotid; RP, Right Parotid; PH, Pharynx). Bold fonts denote the best performers.
4. Discussion
In this work, we improved our previous shape representation model constrained segmentation network24 and developed a novel SC generative adversarial network (SC‐GAN‐DenseNet) for H&N OARs segmentation. SC‐GAN‐DenseNet combines the advantages of the powerful (i.e., SRM) for 3D shape regularization, fully convolutional DenseNet for accurate segmentation, and adversarial training for fast and accurate multiorgan segmentation correcting segmentation errors. Direct comparison among GAN, SC‐GAN‐ResNet, and SC‐GAN‐DenseNet was conducted on both H&N CT and MRI dataset and demonstrates that SC‐GAN‐DenseNet can consistently achieve more accurate segmentation on a total of 11 H&N OARs with varying sizes, morphological complexities, and image contrast.
Compared with other state‐of‐the‐art H&N segmentation methods on public H&N CT PDDCA dataset, SC‐GAN‐DenseNet outperforms SC‐GAN and atlas‐based,6 CNN‐based21 methods by a significant margin on both segmentation performance and speed. The proposed SC‐GAN‐DenseNet segmentation network also outperforms our previous segmentation network using SRM‐FC‐ResNet.24 Although the segmentation performance using our method is not statistically different than the model‐based41 and hierarchical vertex regression methods,11 our method is two orders of magnitude faster. We consider the time difference critical in adaptive radiotherapy applications.
The remarkable performance of SC‐GAN is due to the following technical aspects that are worth discussing.
4.A. Segmentation network
Comparison of the segmentation results of SC‐GAN‐ResNet and SC‐GAN‐DenseNet in Figs. 7 and 8 and Tables 2, 3, 6, and 7 reveals the contribution of fully convolutional DenseNet. The segmentation accuracy of SC‐GAN‐DenseNet consistently outperforms SC‐GAN‐ResNet on all OARs, demonstrating the superiority of dense connectivity to medical image segmentation tasks. Dense connectivity not only can boost information flow and gradients propagation through the network but reduce network parameters by allowing the network to reuse features efficiently. The network thus becomes easier to train and less prone to overfitting even with a small training dataset.
4.B. Adversarial training
An additional loss function from the discriminative network that distinguishes between ground truth and prediction segmentation is incorporated into the objective function for the segmentation network update. The comparison between the segmentation results of shape representation model constrained FC‐ResNet24 and SC‐GAN‐DenseNet in Table 6 indicates that adversarial training can further improve the performance of the segmentation network. With adversarial training, the network can capture the inconsistencies that voxel‐wise loss function cannot.
4.C. Shape representation model
The remaining mis‐segmented volumes using SC‐GAN‐ResNet are mainly due to the limitations of the segmentation network. SRM is pretrained on the binary masks of the ground truth in the dataset. Thus, with the strong shape constraint from SRM, the segmentation network can approximately distinguish the area being processed is target organs or background. However, it is difficult to make correct class label prediction in the ambiguous areas. Furthermore, latent anatomy characteristics learned by SRM can have a direct influence on the segmentation network performance. When the testing patient has substantially different anatomies, an incorrect SRM may reduce the segmentation accuracy. A robust categorized shape representation model, which potentially needs modifications on the network architecture and more patient data to train, is a key point for the future study.
As the central focus of this study, we evaluated the efficacy of the improved segmentation network on the low‐field MR H&N data. Although in principle, MR provides superior soft tissue contrast that should aid the automated segmentation, the low‐field single‐parametric MR images suffer from low signal‐to‐noise ratio and substantial susceptibility artifacts due to metals and air cavities in this region. It is encouraging to demonstrate that good segmentation accuracy can be achieved for a wide range of H&N OARs including the bony structure. While the Dice's index of the automated mandible contour is lower on MR than on CT, the fully automated segmentation is usable without additional manual edition as indicated by the low average and 95% surface distances. It is worthnoting that for low‐field MR segmentation, SC‐GAN‐DenseNet is the only method achieves 95%SD under 4 mm for all organs. This low surface distance is important to reduce the uncertainties in dose received by these organs. At the same time, the benefit of superior soft tissue contrast is demonstrated in the brainstem, optical nerves, chiasm, and parotids in terms of better surface agreement. This result is particularly important in the era of MR‐guided radiotherapy where 3D images with better soft‐tissue contrast than CBCT are available at the time of treatment.
5. Conclusion
We present a modified shape representation model constrained DenseNet by adversarial training (i.e., SC‐GAN‐DenseNet) for H&N multiple organs segmentation. By combining the strengths of SRM, DenseNet, and adversarial training, the novel SC‐GAN‐DenseNet is shown superior to other state‐of‐the‐art methods in accuracy and computational efficiency using small training datasets. For the low‐field MR guidance images, SC‐GAN‐DenseNet was able to robustly perform fully automated segmentation on a variety of organs‐at‐risk, despite the low signal‐to‐noise afforded by this modality.
Acknowledgments
Nuo Tong is supported in part by the CSC Chinese Government Scholarship. This work was also supported by NIH Grants R01CA230278 and R01CA188300.
References
- 1. Rorke MAO, Ellison MV, Murray LJ, Moran M, James J, Anderson LA. Human papillomavirus related head and neck cancer survival: a systematic review and meta‐analysis. Oral Oncol. 2012;48:1191–1201. [DOI] [PubMed] [Google Scholar]
- 2. Gutiontov SI, Shin EJ, Lok B, Lee NY, Cabanillas R. Intensity‐modulated radiotherapy for head and neck surgeons. Head Neck. 2016;38(Suppl 1):E2368–E2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nelms BE, Tomé WA, Robinson G, Heeler JW. Variations in the contouring of organs at risks: test case from a patient with oropharyngeal cancer. Int J Radiat Oncol Biol Phys. 2012;82:368–378. [DOI] [PubMed] [Google Scholar]
- 4. Castelli J, Simon A, Lafond C, et al. Adaptive radiotherapy for head and neck cancer. Acta Oncol. (Madr). 2018;57:1284–1292. [DOI] [PubMed] [Google Scholar]
- 5. Pollard JM, Wen Z, Sadagopan R, Wang J. The future of image‐guided radiotherapy will be MR guided. Br J Radiol. 2017;90:20160667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han X, Hoogeman MS, Levendag PC, et al. Atlas‐based auto‐segmentation of head and neck CT images. Medical Image Computing and Computer‐Assisted Intervention. 2008:434–441. [DOI] [PubMed]
- 7. Bondiau PY, Malandain G, Chanalet S, et al. Atlas‐based automatic segmentation of MR images: validation study on the brainstem in radiotherapy context. Int J Radiat Oncol Biol Phys. 2005;61:289–298. [DOI] [PubMed] [Google Scholar]
- 8. Cootes T. Active appearance models. IEEE Pattern Anal Mach Intell. 2001;23:681–685. [Google Scholar]
- 9. Cootes TF, Taylor CJ, Cooper DH, Graham J. Active shape models‐their training and application. Comput Vis Image Underst. 1995;61:38–59. [Google Scholar]
- 10. Fritscher KD, Peroni M, Zaffino P, Spadea MF, Schubert R, Sharp G. Automatic segmentation of head and neck CT images for radiotherapy treatment planning using multiple atlases, statistical appearance models, and geodesic active contours. Med Phys. 2014;41:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Z, Wei L, Wang L, Gao Y, Chen W, Shen D. Hierarchical vertex regression‐based segmentation of head and neck CT images for radiotherapy planning. IEEE Trans Image Process. 2018;27:923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arindra A, Setio A, Ciompi F, et al. Pulmonary nodule detection in CT images: false positive reduction using multi‐view convolutional networks. IEEE Trans Med Imaging. 2016;35:1160–1169. [DOI] [PubMed] [Google Scholar]
- 13. Kawahara J, BenTaieb A, Hamarneh G. Deep features to classify skin lesions. Int Symp Biomed Imaging. 2016;1397–1400.
- 14. Dou Q, Chen H, Yu L, et al. Automatic Detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging. 2016;35:1182–1195. [DOI] [PubMed] [Google Scholar]
- 15. Li X, Chen H, Qi X, Dou Q, Member S. H‐DenseUNet : hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging. 2018;37:2663–2674. [DOI] [PubMed] [Google Scholar]
- 16. Chen L, Bentley P, Mori K, et al. DRINet for medical image segmentation. IEEE Trans Med Imaging. 2018;37:2453–2462. [DOI] [PubMed] [Google Scholar]
- 17. Moeskops P, Viergever MA, Mendrik AM, De Vries LS, Benders MJNL, Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016;35:1252–1261. [DOI] [PubMed] [Google Scholar]
- 18. Mateus D, Simonovsky M, Navab N, Komodakis N. A deep metric for multimodal registration. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2016. 2016;9902:10–18. [Google Scholar]
- 19. Oktay O, Bai W, Lee M, et al. Multi‐input cardiac image super‐resolution using convolutional neural networks. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2016. 2016:246–254.
- 20. Shah A, Conjeti S, Navab N, Katouzian A. Deeply learnt hashing forests for content based image retrieval in prostate MR images. Med. Imaging 2016 Image Process. 2016;9784:978414. [Google Scholar]
- 21. Ibragimov B, Xing L. Segmentation of organs‐at‐risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kayalibay B, Jensen G, van dSP. CNN‐based segmentation of medical imaging data. arXiv Prepr. arXiv1701.03056 2017.
- 23. Zhang R, Zhao L, Lou W, et al. Automatic segmentation of acute ischemic stroke from DWI using 3D fully convolutional DenseNets. IEEE Trans Med Imaging. 2018;0062:1. [DOI] [PubMed] [Google Scholar]
- 24. Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multi‐organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys. 2018;45:4558–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Urban S, Tanacs A. Atlas‐based global and local RF segmentation of head and neck organs on multimodal MRI images. Proceedings of the 10th International Symposium on Image and Signal Processing and Analysis. 2017;(Ispa):99–103.
- 26. Kieselmann JP, Kamerling CP, Burgos N, et al. Geometric and dosimetric evaluations of atlas‐ based segmentation methods of MR images in the head and neck region. Phys Med Biol 2018;63:145007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodfellow IJ, Pouget‐abadie J, Mirza M, Xu B, Warde‐farley D. Generative adversarial nets. Advances in Neural Information Processing Systems. 2014;2672–2680.
- 28. Navab N, Hornegger J, Wells WM, Frangi AF. U‐Net: convolutional networks for biomedical image segmentation. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2015. 2015;234–241.
- 29. Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U‐Net : learning dense volumetric segmentation from sparse annotation. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2016. 2016;424–432.
- 30. Milletari F. V‐Net : fully convolutional neural networks for volumetric medical image segmentation. 3D Vision (3DV), 2016 Fourth International Conference on IEEE. 2016;567–573.
- 31. Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. International Conference Artificial Intelligence and Statistics. 2010;249–256.
- 32. He K, Sun J. Deep residual learning for image recognition. IEEE Conference on Computer Vision and Pattern Recognition 2016;770–778.
- 33. Huang G, Van Der ML, Weinberger KQ. Densely connected convolutional networks. IEEE Conference on Computer Vision and Pattern Recognition 2017;4700–4708.
- 34. Szegedy C, Com SG. Batch normalization : accelerating deep network training by reducing internal covariate shift. ICML. 2015;448–456.
- 35. Hinton GE. Rectified linear units improve restricted Boltzmann machines. ICML. 2010;807–814.
- 36. Ravishankar H, Venkataramani RB, Thiruvenkadam S, Sudhakar P. Learning and incorporating shape models for semantic segmentation. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2017. Cham, Switzerland: Springer. 2017;203–211. [Google Scholar]
- 37. Han Z, Wei B, Mercado A, Leung S, Li S. Spine‐GAN : semantic segmentation of multiple spinal structures. Med Image Anal. 2018;50:23–35. [DOI] [PubMed] [Google Scholar]
- 38. Mahasseni B, Lam M, Todorovic S. Unsupervised video summarization with adversarial LSTM networks. IEEE Conference on Computer Vision and Pattern Recognition. 2017;202–211.
- 39. Ang KK, Zhang Q, Rosenthal DI, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. 2014;32:2940–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dice LR. Measures of the amount of ecologic association between species. Ecology. 2009;26:297–302. [Google Scholar]
- 41. Mannion‐haworth R, Bowes M, Ashman A, Guillard G, Brett A, Vincent G. Fully automatic segmentation of head and neck organs using active appearance models. MIDAS J. 2016; http://www.midasjournal.org/browse/publication/967.


