Abstract
Aging alters gene expression of growth and remodeling factors in human skeletal muscle both at rest and in response to acute resistance exercise. Physiol Genomics 32: 393–400, 2008. First published December 11, 2007; doi:10.1152/physiolgenomics.00191.2007.—The purpose of this investigation was to compare expression of genes that function in inflammation and stress, cell structure and signaling, or remodeling and growth in skeletal muscle of young (32 ± 7 yr, n =15) and elderly (72 ±5 yr, n = 16) healthy subjects before and after a bout of resistance leg exercises. A real-time RT-PCR method was used to screen 100 transcripts in v. lateralis biopsies obtained before and 72 h postexercise. The screen identified 15 candidates for differential expression due to aging and/or exercise that were measured quantitatively. The median levels of four mRNAs (insulin-like growth factor-1 and its binding protein IGFBP5, ciliary neurotrophic factor, and the metallopeptidase MMP2) were significantly affected by aging and were greater (1.6- to 2.3-fold, P ≤0.05) in the young than elderly muscle at both time points. The median levels of three mRNAs were significantly (P ≤0.05) affected by exercise in the young. The metallopeptidase inhibitor TIMP1 and [H9251]-cardiac actin mRNAs increased 2-fold and 6.5-fold, respectively, and GDF8 (myostatin) mRNA decreased by 50%. However, elderly muscle did not display any significant changes in gene expression postexercise. Thus, aging muscle shows decreased levels at rest and an impaired response to exercise for a number of mRNAs for factors potentially involved in muscle growth and remodeling. Future studies must determine the functional importance of these gene expression changes to protein synthesis, satellite cell activity, and other processes that are directly involved in the mechanisms of muscle hypertrophy.
Keywords: growth factors, metallopeptidase, ciliary neurotrophic factor, alpha cardiac actin, myostatin
sarcopenia, the gradual loss of skeletal muscle mass during aging, is thought to be caused by the combined effects of age-associated decreases in physical activity, nutritional intake, and hormone and neural stimulation of the muscle (16). The overt causes of sarcopenia likely have a negative influence on the two primarily discussed mechanisms by which muscle mass changes: muscle protein turnover (synthesis and breakdown) and function of muscle stem cells (satellite cells) (3, 54). In elderly subjects compared with young, protein synthesis has been reported as decreased and protein breakdown increased in resting muscle, and the synthetic response to resistance exercise was blunted (4, 53, 58). Similarly, satellite cell content in resting muscle and their ability to proliferate after exercise is decreased in elderly muscle (17, 47, 60). However, there are reports conflicting with both of these sets of conclusions (49,62). The differences may be due to study design or reflective of the complex number of additional cell types and factors that affect muscle metabolism and satellite cell function.
Immune cells are postulated to contribute to repair after muscle damage, including that which occurs with resistance exercise, by mediating a progression from an acute inflamma-tory response that may influence protein turnover to a local environment that promotes myogenesis through the provision of growth factors (57). Furthermore, chronic inflammation is associated with aging and is implicated in sarcopenia (50). For this reason, we chose to examine macrophage function in muscle at rest and 72 h postresistance exercise in groups of healthy young and elderly subjects (44). The selection of an appropriate postexercise time point had been investigated previously (15). At rest, elderly muscle compared with young at least tended to possess significantly fewer macrophages even though whole muscle of the elderly contained significantly higher mRNA levels for the cytokines IL-1β and IL-10. Post-exercise, functional markers indicated that macrophages were activated and cytokine expression increased, but these responses were exclusive to the young group, which is consistent with another report (29). These results suggest that decreased macrophage numbers and increased cytokine levels even in healthy elderly may be detrimental to resting muscle and diminish the functional response to exercise. Thus, in addition to protein turnover and satellite cell function, macrophage activities represent a third general means by which aging affects human muscle for which gene expression profiling may identify the factors affected or involved.
Gene expression profiling has primarily used microarrays to survey thousands of genes in human skeletal muscle and has been used to compare young and elderly muscle at rest, and in response to acute resistance exercise and exercise training (23, 34, 40, 48, 67). However, due to the quantitative limitations of microarrays, the results were typically “molecular signatures” comprising changes in many genes from a wide variety of cellular pathways that should be further investigated in prospective studies. For this reason, we developed a custom real-time reverse transcriptase polymerase chain reaction (RT RT-PCR) screening method as a targeted approach to gene expression profiling. The primary aim of this investigation was to use this method to determine if specific genes are differentially expressed 72 h after resistance exercise and/or affected by aging. The secondary aim was to determine if age affected gene expression in resting muscle. RT RT-PCR was used to screen 100 transcripts representing genes that we hypothesize are important to muscle due to their function in inflammation and stress, cell structure and signaling, or remodeling and growth. A large number of proteases were included due to their potential role in remodeling and their widespread ability to activate other factors (56). The RT RT-PCR screen identified 15 candidate genes that were quantitatively analyzed for altered expression in muscle at rest or in response to resistance exercise.
METHODS
Subjects.
Young adult and elderly males (Table 1) were recruited from the community by newspaper advertisements and word of mouth. All subjects gave written informed consent approved by the Institutional Review Board of the University of Arkansas for Medical Sciences (UAMS). These subjects and their data have been previously compared though three subjects from the previous study were not included here due to limited sample quantities (44). All subjects were healthy, sedentary, nonsmokers. Subjects were not taking anti-inflammatory medications or analgesics chronically and were required to refrain from such or participating in any unusual physical activity for at least 3 days prior to and throughout the protocol. Dietary control was not included in the study but is acknowledged as a potential source of variability in the measured responses to exercise.
Table 1.
Physical characteristics of young and elderly male subjects
| Young (n =15) | Elderly (n =16) | P | |
|---|---|---|---|
| Age, yr | 32±7 | 72±5 | <0.001 |
| Height, cm | 177.5±7.4 | 178.9±8.7 | 0.63 |
| Weight, kg | 83.5±14.6 | 84.5±10.6 | 0.83 |
| BMI, kg/m2 | 26.4±3.5 | 26.4±1.9 | 0.93 |
| Curl 1-RM, Nm | 249.6±44.1 | 197.6±34.3 | <0.001 |
| Extension 1-RM, Nm | 283.1±40.8 | 224.5±34.8 | <0.001 |
| Press 1-RM, Nm | 1,585.0±268.8 | 1,579.7±195.0 | 0.95 |
Values are means ± SD. BMI, body mass index; 1-RM, one-repetition maxiumum.
Resistance exercise protocol.
Following 10 min of light cycling for warm-up, each subject’s one-repetition maximum (1-RM) was obtained for bilateral leg press, curl, and extension (Keiser, Fresno, CA) (Table 1). Subjects rested for 5 min and then completed three sets of eight repetitions followed by a fourth set to voluntary failure for each of the three exercises with resistance of 80% 1-RM. Subjects were given 2 min of rest between sets and 5 min between exercises. Subjects were allowed to walk and perform light stretching during the rest periods. Verbal encouragement was consistently given to all subjects during the resistance exercise. The purpose of the chosen exercise protocol was to mimic a standard resistance exercise session that is commonly performed chronically to increase muscle mass and strength.
Muscle biopsy.
Muscle biopsy samples were taken from the vastus lateralis muscle just prior to the exercise session and again 72 h postexercise. The first biopsy was taken from the subject’s nondominant leg, and the second biopsy was taken from the dominant leg. Tissue was obtained after local anesthetic (lidocaine HCl 1%, 3 ml) with the use of a 5 mm Bergstrom needle with suction (21). The muscle was cleansed of excess blood, connective tissue, and fat and frozen in liquid N2. Samples were stored at −80°C until analysis.
RNA purification.
Frozen muscles were powdered using a stainless-steel pulverizer and liquid N2. Total RNA was isolated using the Totally RNA kit according to the supplied protocol (Ambion, Austin, TX). The muscle was disrupted in a scaled volume of lysis buffer using a high-speed electric homogenizer. Purified RNA was resus-pended in 50 μl of nuclease-free water and then treated with DNase using DNA-free reagents (Ambion). RNA concentration and integrity were determined using an Agilent 2100 Bioanalyzer and the RNA 6000 Nano Labchip kit (Agilent Technologies, Palo Alto, CA).
RT RT-PCR screening method.
Aliquots of cDNA were pooled to represent four groups: young and elderly skeletal muscle pre- and 72 h postresistance exercise. The four pools contained samples from 10 subjects each and were used in an initial screen for gene expression differences in 100 transcripts (available in a supplemental file) by RT RT-PCR. The 10 young and elderly subject samples chosen to make up the cDNA pools were those having the most cDNA remaining from a prior study. All aspects of RT RT-PCR were performed using the reverse transcription and SYBR-based PCR reagents and the amplification and detection systems of Applied Biosystems along with their recommended standard conditions (Applied Biosystems, Foster City, CA) (ABI). Specific modifications to the manufacturer’s standard conditions were as follows. Screening reactions contained 150 nM of each primer set and 10 ng of RNA equivalents of cDNA. Control reactions, measuring glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and 18s ribosomal RNA (rRNA), required slightly different conditions of 250 nM or 300 nM of primers and 4 ng or 1 ng of RNA equivalents of cDNA, respectively. Gene expression between pools (assayed singly) was normalized to 18s, but this varied <10% between pools and was chosen over GAPDH, which varied up to twofold between pools. For the screening, relative quantitation for each transcript was calculated as a fold-change relative to results for the young preexercise pool by the 2−ΔΔCt method of analysis (38).
Quantitative RT RT-PCR.
The custom RT RT-PCR screening method identified potentially differentially expressed transcripts (selection criteria described in RESULTS) to be quantified in the individual samples of the entire study population using standard curves (assayed in triplicate) as described previously (15). SYBR green-based assay parameters for each specific primer set were optimized to ≥90% efficiency and ≤12% interassay variation with the exceptions being reactions for connective tissue growth factor (18% variation) and ciliary neurotrophic factor (CNTF, 76% efficiency and 20% variation). Individual subject samples were then assayed singly due to limited sample and the number of transcripts of interest. Gene expression results were calculated using standard curves (log of the ng RNA equivalents of cDNA vs. cycle number) generated from serial dilutions of a pooled muscle cDNA remaining from a previous study. Data were normalized to 18s rRNA and presented as relative units or gene expression relative to the standard curves. Melting curves and controls lacking template were used to verify the single product specificity of all RT RT-PCR reactions.
Statistical analysis.
The physical characteristics of subjects were compared using a two-sample t-test. Differences in transcript levels were analyzed using a nonparametric approach since the variables were typically not normally distributed. The Mann-Whitney test was used to identify differences between the young and elderly groups at pre- and post-exercise. The Wilcoxon signed-ranks test was used within groups to identify differences in pre- vs. post-exercise measurements. An adjustment for multiple comparisons was made using a Benjamini-Hochberg false discovery rate-controlling procedure for the proportion of erroneous significances (5%) based on 15 mRNAs being quantified for each group (68). All P values before and after adjustment are available in a supplemental file.1 Significance was taken at P ≤ 0.05.
RESULTS
Physical characteristics.
The young and elderly subject groups were not different for anthropometric measurements; however, the young achieved significantly greater 1-RMs for two of three leg exercises (Table 1). There was no difference in average height, weight, or body mass index between the young (32 ±7 yr) and elderly (72 ±5 yr). While the 1-RMs for leg press were similar (P = 0.95), the young subjects had significantly higher ~20%) 1-RM for leg curls (P <0.001) and extensions (P <0.001).
Gene expression screening by RT RT-PCR.
The abundance of transcripts representing 100 genes (all data presented in the supplemental file) were quantified by RT RT-PCR using pools of cDNA made from young (n = 10) and elderly (n = 10) skeletal muscle at rest and 72 h after a standard bout of resistance exercise. First, due to the quantity of RNA necessary to accurately quantify rarely expressed transcripts, transcripts having a threshold cycle for RT RT-PCR comparison ≥30 were excluded from the priority analyses. This was a practical decision based on our experience that has been validated by others (64). Another transcript was eliminated based on melting curve data that created a concern for reaction specificity. This eliminated 41 of the 100 transcripts. Transcript results were then analyzed by the 2−ΔΔCt method (38) to calculate fold-differences relative to the young muscle pre-exercise pool, which was assigned a value of 1.0. This custom RT RT-PCR screening method identified 15 transcripts that displayed relative between-pool differences of ≤0.5 or ≥1.5, suggesting that they were differentially expressed in skeletal muscle during aging and/or the 72 h response to resistance exercise (Table 2). The potential differences identified by the screening method between young and elderly muscle before, after, or in response to acute resistance exercise were then examined by quantitative RT RT-PCR to assay individual samples for the entire study population.
Table 2.
Relative expression of transcripts (listed alphabetically) identified from whole skeletal muscle potentially affected by aging and/or exercise using a RT RT-PCR screening method
| Young | Elderly | ||||
|---|---|---|---|---|---|
| Transcript, Accession No. | Ct | Pre | Post | Pre | Post |
| α-Cardiac actin (ACTC1), NM_005159 | 23.94 | 1.0 | 8.0 | 1.4 | 3.1 |
| Atrogin-1 or F-box protein 32 (MAFbx), NM 058229 | 28.9 | 1.0 | 0.8 | 0.4 | 0.5 |
| Ciliary neurotrophic factor (CNTF) NM 000614 | 28.18 | 1.0 | 1.2 | 0.5 | 0.7 |
| Connective tissue growth factor (CTGF), NM_001901 | 27.04 | 1.0 | 0.6 | 0.6 | 0.5 |
| Early growth response protein-1 (EGR1), NM 001964 | 26.99 | 1.0 | 1.8 | 0.6 | 0.6 |
| Fibroblast growth factor-6 (FGF6), NM_020996 | 26.02 | 1.0 | 1.3 | 0.5 | 0.9 |
| Fibronectin (FN1), X02761 | 25.57 | 1.0 | 0.6 | 0.1 | 0.3 |
| Growth and differentiation factor 8 or myostatin (GDF8), NM_005259 | 25.54 | 1.0 | 0.5 | 0.5 | 0.4 |
| Insulin-like growth factor binding protein-5 (IGFBP5), NM_000599 | 21.66 | 1.0 | 0.8 | 0.5 | 0.6 |
| Insulin-like growth factor-1 (IGF1), NM_000618 ‘ | 27.71 | 1.0 | 0.6 | 0.3 | 0.4 |
| Matrix metallopeptidase-2 (MMP2), NM_004530 | 25.06 | 1.0 | 1.3 | 0.5 | 0.8 |
| Osteosarcoma viral oncogene homolog (c-fos), AY212879 | 29.33 | 1.0 | 0.9 | 0.3 | 0.4 |
| TIMP metallopeptidase inhibitor-1 (TIMP1), X03124 | 27.83 | 1.0 | 2.4 | 0.9 | 1.4 |
| Vascular endothelial growth factor-A (VEGFA), M32977 | 25.38 | 1.0 | 1.2 | 0.5 | 0.9 |
| Vascular endothelial growth factor-D (VEGFD), NM_004469 | 28.26 | 1.0 | 0.9 | 0.5 | 0.6 |
Transcript name, abbreviation, and sequence are available using the provided accession number at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=gene. Ct is threshold real-time (RT) RT-PCR cycle for detection of transcript in young muscle pre-exercise pool. Transcript levels are presented as the fold-difference relative to the young muscle pre-exercise pool with differences ≤0.5 or ≥1.5 in boldface. Pre, pre-exercise; Post, post-exercise.
Age-dependent differences in gene expression.
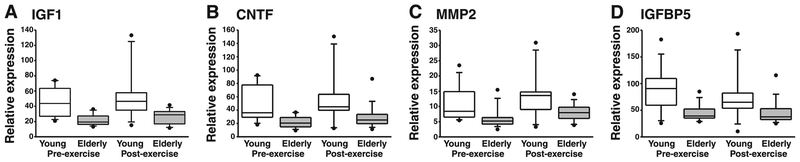
The influence of aging on expression of the 15 transcripts shown in Table 2 was quantified for young and elderly skeletal muscle samples obtained at rest or 72 h after resistance exercise (Table 3). The abundance of 4 [insulin-like growth factor (IGF)1, CNTF, matrix metallopeptidase (MMP)2, and IGF binding protein (IGFBP)5] of the 15 transcripts was consistently greater (P <0.05) in young than elderly muscle at both time points and ranged from medians that were 1.6- to 2.3-fold different (Table 3 and Fig. 1). Also as predicted by the screening method, six other transcripts were higher (P ≤ 0.05) in young than elderly muscle before exercise but they did not display the consistency of being affected by age in the post-exercise sample [c-fos, FN1, growth and differentiation factor (GDF)8, MAFbx, VEGFA, and VEGFD] (Table 3).
Table 3.
Median mRNA levels (quantitative relative expression) of skeletal muscle transcripts affected by aging and/or exercise as measured by quantitative RT RT-PCR
| Young, n = 15 | Elderly, n = 16 | ||||
|---|---|---|---|---|---|
| Transcript, Accession No. | Pre Post | Pre Post | Sig | ||
| α-Cardiac actin (ACTC1), NM_005159 | 0.52 | 3.38 | 0.47 | 1.35 | C |
| Atrogin-1 or F-box protein 32 (MAFbx), NM 058229 | 20.85 | 14.66 | 12.22 | 11.03 | A |
| Ciliary neurotrophic factor (CNTF) NM 000614 | 36.00 | 44.95 | 20.52 | 25.15 | A, B |
| Connective tissue growth factor (CTGF), NM_001901 | 4.46 | 3.66 | 3.27 | 3.19 | |
| Early growth response protein-1 (EGR1), NM 001964 | 2.90 | 2.58 | 2.07 | 2.50 | |
| Fibroblast growth factor-6 (FGF6), NM_020996 | 6.52 | 7.95 | 4.11 | 6.99 | |
| Fibronectin (FN1), X02761 | 5.95 | 10.22 | 3.60 | 6.43 | A |
| Growth and differentiation factor 8 or myostatin (GDF8), NM_005259 | 49.33 | 23.61 | 25.79 | 14.80 | A, C |
| Insulin-like growth factor binding protein-5 (IGFBP5), NM_000599 | 90.66 | 64.95 | 39.17 | 37.63 | A, B |
| Insulin-like growth factor-1 (IGF1), NM_000618 ‘ | 43.67 | 46.50 | 19.29 | 28.77 | A, B |
| Matrix metallopeptidase-2 (MMP2), NM_004530 | 8.41 | 13.58 | 5.21 | 7.95 | A, B |
| Osteosarcoma viral oncogene homolog (c-fos), AY212879 | 0.46 | 0.70 | 0.24 | 0.16 | A |
| TIMP metallopeptidase inhibitor-1 (TIMP1), X03124 | 12.54 | 24.74 | 12.85 | 15.72 | C |
| Vascular endothelial growth factor-A (VEGFA), M32977 | 8.70 | 8.54 | 4.92 | 5.79 | A |
| Vascular endothelial growth factor-D (VEGFD), NM_004469 | 18.07 | 15.35 | 7.25 | 7.38 | A |
Significance (Sig) was accepted at P ≤ 0.05. Significance is noted by A, B, or C for the following comparisons: A = young vs. elderly pre-exercise, B = young vs. elderly post-exercise, C = pre- vs. post-exercise in young, and there were no significant differences found for pre- vs. post-exercise in elderly
Fig. 1.
The effects of aging on the skeletal muscle transcripts for IGF1 (A), ciliary neurotrophic factor (CNTF, B), matrix metallopeptidase (MMP2, C), and IGF binding protein-5 (IGFBP5, D). Data are presented as box plots denoting the medians (line), the 25th and 75th percentiles (boxes), and the 10th and 90th percentiles (whiskers), along with outliers (dots). Between-group significance, with young being greater than elderly, was seen in both pre (white boxes, P ≤0.05)- and post-resistance (gray boxes, P ≤ 0.05) exercise for each transcript. Expression data were determined by quantitative real-time RT-PCR and normalized to 18s rRNA levels.
Exercise-induced changes in gene expression.
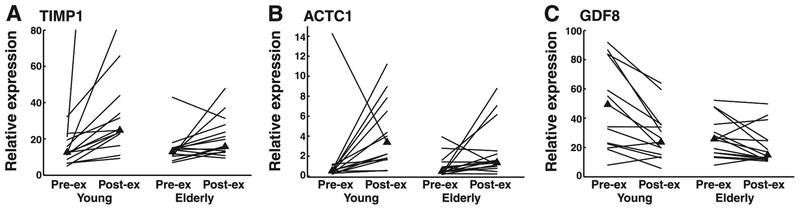
The data for the 15 transcripts of interest were also analyzed for exercise induced change by comparing pre- and post-exercise expression within the young and elderly groups. Three mRNAs were significantly (P ≤0.05) affected by exercise in young muscle. Median tissue inhibitor of metalloproteinase (TIMP)1 and ACTC1 expression increased 2-fold and 6.5-fold respectively, whereas growth and differentiation factor-8 (GDF8) median expression post-exercise decreased to 50% of pre-exercise levels (Table 3 and Fig. 2). However, the response to exercise for these three mRNAs was not significant in elderly muscle. The screening results also predicted that EGR1 would be induced by exercise in the young though this result was not confirmed.
Fig. 2.
The effects of exercise on the median (▲) and individual subject expression (line) of transcripts for tissue inhibitor of metalloproteinase (TIMP1, A), α-cardiac actin isoform (ACTC1, B), and growth and differentiation factor-8 (GDF8, C). Post-exercise (Post-ex) expression was significantly different than pre-exercise (Pre-ex) for the young (P ≤ 0.05) but not the elderly for all 3 genes. Expression data were determined by quantitative real-time RT-PCR and normalized to 18s rRNA levels.
Immunohistochemistry was performed to determine if the exercise-induced changes in mRNA levels were paralleled by changes in protein. Analysis of multiple cross-sections of skeletal muscle from all groups did not show differences in staining using GDF8 antibodies, and differences in TIMP1 were only detected for the young subject that displayed the greatest mRNA increase after exercise (data not shown). ACTC1 was excluded from this analysis due to the lack of a suitable antibody.
DISCUSSION
The main finding of this investigation is that a targeted approach to gene expression profiling using RT RT-PCR identified a number of genes potentially involved in remodeling and growth of skeletal muscle that are differentially expressed between young adults and healthy but weaker elderly subjects. Significant differences and consistent results indicated that aging and/or the 72 h response to resistance exercise-affected levels of mRNAs for the growth factors CNTF, IGF1, and its binding protein IGFBP5, along with the negative regulator of muscle growth GDF8 (myostatin), and for genes that may be associated with muscle remodeling including the metallopeptidase MMP2 and its inhibitor TIMP1, and the developmental actin isoform, ACTC1. Significant differences were also seen for the transcription factor c-fos, the growth factors VEGFD and VEGFA and FGF6, and two other genes associated with remodeling, fibronectin and MAFbx (atrogin), but the effects of aging were less consistent and these genes will be excluded from the detailed discussion.
IGF axis.
IGF1 may be the most extensively discussed growth factor with respect to skeletal muscle because it is thought to regulate muscle mass in response to hormonal and mechanical stimulation through its IGF-1Ea and MGF isoforms respectively (1, 24). The data presented here indicate that median mRNA levels for IGF1 and IGFBP5 in resting young muscle are more than twice that of elderly muscle. Our IGF1 assay did not discriminate between isoforms and the results agree with a study that used a similar assay (66). As IGF-1Ea levels have been shown to be 100-fold greater in resting muscle than MGF, we assume that our results are predominantly due to the IGF-1Ea isoform. However, those studies that specifically measured IGF-1Ea and MGF for healthy groups of young and elderly did not record significant differences (30, 36).
Although different at baseline between the young and elderly, IGF1 and IGFBP5 were unchanged after acute exercise. It has been suggested that young and elderly subjects need to be stratified based on their level of hypertrophic response to exercise training for the positive relationship between IGF-1Ea mRNA and training-induced hypertrophy, as well as the predictive value of the response to acute exercise, to be apparent(43). However, the mechanistic role of IGF1 is still unclear since the association between human muscle IGF1 and parameters such as myosin heavy chain isoform distribution, myofibrillar protein synthesis, or myofiber size at rest or after acute exercise have been modest at best (30, 36, 66). Even less is known about the possible function of IGFBP5 under these circumstances, but in general, IGFBP5 can augment, inhibit, or act independently of IGF1 to control cell proliferation, differentiation, and survival (5). To our knowledge, IGFBP-5 has only been examined once in human skeletal muscle and was not different between able-bodied subjects and spinal cord injury patients (6). In rodents, muscle IGFBP5 has been shown to decrease with age and to be regulated by hypertrophic stimuli though results vary with the experimental paradigm (6,55). Other members of the IGF axis assayed in our study included IGF2 and IGFBP3 though they did not meet the screening criteria for differential expression.
CNTF.
Our results identified the CNTF transcript as being 1.8-fold higher in young than aged muscle at rest and after a bout of resistance exercise. CNTF is a cytokine of the IL-6 family that could influence skeletal muscle through multiple functions (61). CNTF was first identified in muscle lysates from rats due to its ability to promote neuron survival (18). CNTF production by the sciatic nerve in aged rats was decreased and strongly correlated with decreased muscle twitch and tetanic tension, which could be improved by exogenous CNTF (27). Administration of CNTF also prevented the effects of atrophy models on specific muscles (22, 31). However, other studies suggest that CNTF can affect muscle excitation and reverse insulin resistance, and when used in muscle cultures CNTF regulates stem cell differentiation and protein synthesis, suggesting numerous mechanisms through which the age-associated decrease in CNTF identified here could contribute to sarcopenia (11, 45, 63, 65). Given these findings together with genetic studies showing that variations of CNTF and its receptor influence muscle strength and function (2, 13, 14, 51) and that CNTF is being used in clinical trials for neurodegenerative disease (20, 41), it is surprising that CNTF expression in human skeletal muscle has not been extensively quantified. Unfortunately, side-effects of weight loss and anorexia seen in the trial for treatment of neurodegeneration led to CNTF being tested in obesity. Thus, exogenous CNTF not being a likely candidate for treatment of sarcopenia emphasizes the importance of investigating resistance exercise training as a natural means to increase CNTF levels locally in muscle of the elderly to the physiological levels observed in younger adults.
MMP and tissue inhibitor.
MMPs are a family of proteolytic enzymes that are regulated by a set of tissue inhibitors and function in tissue remodeling, including that of skeletal muscle, through the cleavage of the extracellular matrix and other factors that regulate cell behavior (7, 10). The panel of genes investigated here contained >15 MMPs and TIMPs. Of significance, MMP2 mRNA levels were lower in elderly muscle than young, but the response to exercise was variable in both groups. By contrast, TIMP1 showed a more consistent re sponse to exercise, increasing in 27 of 31 subjects, although the magnitude was typically greater in the young than elderly. Consistent with these results, TIMP1 mRNA has been shown to decrease in human muscle with disuse (59). Thus, MMP2 and TIMP1 are clearly regulated at the transcriptional level by aging and resistance exercise but vary widely between individuals. As with all the gene products discussed, an expanded time course of their response to exercise will be an important part of future investigations of their regulation and function.
MMP2 and TIMP1 are expressed in human skeletal muscle and have been hypothesized to function in the angiogenic response that occurs shortly after exercise (52). Contrary to our results for resistance exercise, TIMP1 was not induced with cycling exercise, but MMP9 was induced, suggesting that MMPs and TIMPs display specificity to the mode of exercise, a hypothesis that is supported by animal exercise studies (9,37). With respect to aging muscle, decreased resting levels and a dampened response to exercise for MMP2 and TIMP1 may diminish satellite cell function since in culture MMP2 is produced by human satellite cells and has been shown to be necessary for migration of mouse cells (19, 26).
α-Cardiac actin.
The multigene family of actins provide the major structural component of the thin filament of the myofibril(8). The [H9251]-skeletal muscle (ACTA1) and α-cardiac (ACTC1) actin isoforms are both expressed in skeletal muscle and were included in our transcript analysis (28). ACTA1 was the far more abundant transcript (data not shown) but did not meet the screening criteria for differential expression, whereas the ACTC1 mRNA increased after exercise for a median response of 6.5-fold in the young. However, the increase in ACTC1 after exercise in the elderly did not reach significance. These results are consistent with work showing induction of ACTC1 expression during muscle regeneration after trauma or during pathology and appears characteristic of activated satellite cells and regenerating myofibers (42). In fact, muscle regeneration has been hypothesized to recapitulate aspects of fetal skeletal muscle development where ACTC1 is highly expressed but is soon largely replaced by ACTA1 (33). However, our exercise paradigm did not result in overt muscle trauma or obvious satellite cell activation (data not shown), suggesting for the first time that ACTC1 is a sensitive marker of the muscle response to resistance exercise. Importantly, the dampened response of ACTC1 in the elderly may be a marker of diminished responsiveness and potentially for the known decline in the regenerative potential of satellite cells (12).
GDF8.
GDF8 (myostatin) has been extensively studied as a negative regulator of skeletal muscle growth since deletion of the gene resulted in dramatic muscle growth in mice and cattle (25, 39). We found that the GDF8 mRNA significantly decreased 72 h after resistance exercise in the young (52%), but the decrease in the elderly (42%) lost significance after a P value adjustment for multiple comparisons. Another study found GDF8 mRNA levels decreased to a similar degree in young and elderly subjects at 24 h after exercise, but this response was impaired in elderly women, and a study of just women showed that their decrease in GDF8 was the same at 4 h after exercise (35, 46). Our study was limited by only comparing the GDF8 response to exercise between young and elderly males, but combining these data at different time points suggests that resistance exercise depresses GDF8 expression for the longest duration (up to 72 h or longer) in young males, but in elderly males GDF8 returns to resting levels sometime between 24 and 72 h, a response that may be similar to young females though this is not known. Elderly females have the shortest duration, with GDF8 likely returning to resting between 4 and 24 h. Thus, the duration of GDF8 downregulation may be one of the molecular events that controls muscle hypertrophy in response to chronic resistance training. GDF8 downregulation after resistance exercise was correlated with lean muscle mass of the thigh and was associated with regulation of the factors that control satellite cell activation and proliferation (35). This implies that satellite cells may not respond to exercise in the elderly due to the shortened duration of the GDF8 response. However, it has been shown that the duration can be extended in the elderly by training (32) and suggests that the optimal exercise routine for the elderly to maintain GDF8 suppression could involve, at least initially, more frequent exercise sessions.
In summary, a custom method for gene expression profiling using screening and quantitative RT RT-PCR identified a number of mRNAs that are expressed at lower levels in elderly compared with young muscle and do not show the same magnitude of responsiveness to exercise as seen in young muscle. We hypothesize that these gene expression differences in the elderly may diminish the levels of factors needed for skeletal muscle growth and remodeling. IGF1 and GDF8 are currently popular factors of interest in skeletal muscle, whereas relatively little is known about the regulation or function of CNTF, MMP2/TIMP1, and ACTC1 in human skeletal muscle particularly with respect to aging. The elderly subjects of this study were originally characterized as showing elevated expression of the proinflammatory cytokine IL-1β, and though there is evidence in the literature to suggest that inflammation negatively regulates a number of the factors identified, that remains to be determined. Due to the abundance of the gene products discussed here, they are likely primarily derived from the muscle fiber. However, the satellite cell may also be a source, and because of its role in muscle regeneration and hypertrophy, it is almost certainly a target for some of these factors. Their relative importance to other cell types or general processes in muscle such as protein synthesis must also be defined. Moreover, further studies are required to address several challenges that include associating mRNA levels with protein levels or activity, understanding the significance of the acute vs. the training response, understanding whether variabil ity between individuals is a difference in the time course of response, and whether each of these variables is sex specific.
Supplementary Material
ACKNOWLEDGMENTS
We thank Todd Trappe and Chad Carroll, now at Ball St University, and William Evans and Jonathan Harvey of UAMS for assistance with the clinical protocol.
GRANTS
This research was supported in part by funds provided to the UAMS Microarray Facility through Act 1 of The Arkansas Tobacco Settlement Proceeds Act of 2000; by National Center for Research Resources grants through the BRIN/INBRE Program (P20 RR-16460) and the General Clinical Research Center at the University of Arkansas for Medical Sciences (M01-RR14288); and by National Institute on Aging Grant AG012411 to C. A. Peterson.
Footnotes
Article published online before print. See web site for date of publication (http://physiolgenomics.physiology.org).
The online version of this article contains supplemental material.
REFERENCES
- 1.Adamo ML, Farrar RP. Resistance training, and IGF involvement in the maintenance of muscle mass during the aging process. Ageing Res Rev 5: 310–331, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Arking DE, Fallin DM, Fried LP, Li T, Beamer BA, Xue QL, Chakravarti A, Walston J. Variation in the ciliary neurotrophic factor gene and muscle strength in older Caucasian women. J Am Geriatr Soc 54: 823–826, 2006. [DOI] [PubMed] [Google Scholar]
- 3.Baar K, Nader G, Bodine S. Resistance exercise, muscle loading/ unloading and the control of muscle mass. Essays Biochem 42: 61–74, 2006. [DOI] [PubMed] [Google Scholar]
- 4.Balagopal P, Rooyackers OE, Adey DB, Ades PA, Nair KS. Effects of aging on in vivo synthesis of skeletal muscle myosin heavy-chain and sarcoplasmic protein in humans. Am J Physiol Endocrinol Metab 273: E790–E800, 1997. [DOI] [PubMed] [Google Scholar]
- 5.Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. Biochem J 395: 1–19, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bickel CS, Slade JM, Haddad F, Adams GR, Dudley GA. Acute molecular responses of skeletal muscle to resistance exercise in able-bodied and spinal cord-injured subjects. J Appl Physiol 94: 2255–2262, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta 1477: 267–283, 2000. [DOI] [PubMed] [Google Scholar]
- 8.Buckingham M, Alonso S, Barton P, Cohen A, Daubas P, Garner I, Robert B, Weydert A. Actin and myosin multigene families: their expression during the formation and maturation of striated muscle. Am J Med Genet 25: 623–634, 1986. [DOI] [PubMed] [Google Scholar]
- 9.Carmeli E, Moas M, Lennon S, Powers SK. High intensity exercise increases expression of matrix metalloproteinases in fast skeletal muscle fiigh. Exp Physiol 90: 613–619, 2005. [DOI] [PubMed] [Google Scholar]
- 10.Carmeli E, Moas M, Reznick AZ, Coleman R. Matrix metalloproteinases and skeletal muscle: a brief review. Muscle Nerve 29: 191–197, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Mao Z, Liu S, Liu H, Wang X, Wu H, Wu Y, Zhao T, Fan W, Li Y, Yew DT, Kindler PM, Li L, He Q, Qian L, Wang X, Fan M. Dedifferentiation of adult human myoblasts induced by ciliary neurotrophic factor in vitro. Mol Biol Cell 16: 3140–3151, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conboy IM, Rando TA. Aging, stem cells and tissue regeneration: lessons from muscle. Cell Cycle 4: 407–410, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Conwit RA, Ling S, Roth S, Stashuk D, Hurley B, Ferrell R, Metter EJ. The relationship between ciliary neurotrophic factor (CNTF) genotype and motor unit physiology: preliminary studies. BMC Physiol 5: 15, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Mars G, Windelinckx A, Beunen G, Delecluse C, Lefevre J, Thomis MA. Polymorphisms in the CNTF and CNTF receptor genes are associated with muscle strength in men and women. J Appl Physiol 102: 1824–1831, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Dennis RA, Trappe TA, Simpson P, Carroll C, Huang BE, Nagarajan R, Bearden E, Gurley C, Duff GW, Evans WJ, Kornman K, Peterson CA. Interleukin-1 polymorphisms are associated with the inflammatory response in human muscle to acute resistance exercise. J Physiol 560: 617–626, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty TJ. Invited review: Aging and sarcopenia. J Appl Physiol 95: 1717–1727, 2003. [DOI] [PubMed] [Google Scholar]
- 17.Dreyer HC, Blanco CE, Sattler FR, Schroeder ET, Wiswell RA. Satellite cell numbers in young and older men 24 hours after eccentric exercise. Muscle Nerve 33: 242–253, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Ebendal T Comparative screening for ciliary neurotrophic activity in organs of the rat and chicken. J Neurosci Res 17: 19–24, 1987. [DOI] [PubMed] [Google Scholar]
- 19.El Fahime E, Torrente Y, Caron NJ, Bresolin MD, Tremblay JP. In vivo migration of transplanted myoblasts requires matrix metalloproteinase activity. Exp Cell Res 258: 279–287, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Ettinger MP, Littlejohn TW, Schwartz SL, Weiss SR, McIlwain HH, Heymsfield SB, Bray GA, Roberts WG, Heyman ER, Stambler N, Heshka S, Vicary C, Guler HP. Recombinant variant of ciliary neurotrophic factor for weight loss in obese adults: a randomized, dose-ranging study. JAMA 289: 1826–1832, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Evans WJ, Phinney SD, Young VR. Suction applied to a muscle biopsy maximizes sample size. Med Sci Sports Exerc 14: 101–102, 1982. [PubMed] [Google Scholar]
- 22.Fraysse B, Guillet C, Huchet-Cadiou C, Camerino DC, Gascan H, Leoty C. Ciliary neurotrophic factor prevents unweighting-induced functional changes in rat soleus muscle. J Appl Physiol 88: 1623–1630, 2000. [DOI] [PubMed] [Google Scholar]
- 23.Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, Kandarian SC. Identification of a molecular signature of sarcopenia. Physiol Genomics 21: 253–263, 2005. [DOI] [PubMed] [Google Scholar]
- 24.Goldspink G Mechanical signals, IGF-I gene splicing, and muscle adaptation. Physiology (Bethesda) 20: 232–238, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17: 71–74, 1997. [DOI] [PubMed] [Google Scholar]
- 26.Guerin CW, Holland PC. Synthesis and secretion of matrix-degrading metalloproteases by human skeletal muscle satellite cells. Dev Dyn 202: 91–99, 1995. [DOI] [PubMed] [Google Scholar]
- 27.Guillet C, Auguste P, Mayo W, Kreher P, Gascan H. Ciliary neurotrophic factor is a regulator of muscular strength in aging. J Neurosci 19: 1257–1262, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunning P, Ponte P, Blau H, Kedes L. alpha-skeletal and alpha-cardiac actin genes are coexpressed in adult human skeletal muscle and heart. Mol Cell Biol 3: 1985–1995, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamada K, Vannier E, Sacheck JM, Witsell AL, Roubenoff R. Senescence of human skeletal muscle impairs the local inflammatory cytokine response to acute eccentric exercise. FASEB J 19: 264–266, 2005. [DOI] [PubMed] [Google Scholar]
- 30.Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J Physiol 547: 247–254, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helgren ME, Squinto SP, Davis HL, Parry DJ, Boulton TG, Heck CS, Zhu Y, Yancopoulos GD, Lindsay RM, DiStefano PS. Trophic effect of ciliary neurotrophic factor on denervated skeletal muscle. Cell 76: 493– 504, 1994. [DOI] [PubMed] [Google Scholar]
- 32.Hulmi JJ, Ahtiainen JP, Kaasalainen T, Pollanen E, Hakkinen K, Alen M, Selanne H, Kovanen V, Mero AA. Postexercise myostatin and activin IIb mRNA levels: effects of strength training. Med Sci Sports Exerc 39: 289–297, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Ilkovski B, Clement S, Sewry C, North KN, Cooper ST. Defining alpha-skeletal and alpha-cardiac actin expression in human heart and skeletal muscle explains the absence of cardiac involvement in ACTA1 nemaline myopathy. Neuromuscul Disord 15: 829–835, 2005. [DOI] [PubMed] [Google Scholar]
- 34.Jozsi AC, Dupont-Versteegden EE, Taylor-Jones JM, Evans WJ, Trappe TA, Campbell WW, Peterson CA. Molecular characteristics of aged muscle reflect an altered ability to respond to exercise. Int J Sport Nutr Exerc Metab 11: S7–S13, 2001. [DOI] [PubMed] [Google Scholar]
- 35.Kim JS, Cross JM, Bamman MM. Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. Am J Physiol Endocrinol Metab 288: E1110–E1119, 2005. [DOI] [PubMed] [Google Scholar]
- 36.Kim JS, Kosek DJ, Petrella JK, Cross JM, Bamman MM. Resting and load-induced levels of myogenic gene transcripts differ between older adults with demonstrable sarcopenia and young men and women. J Appl Physiol 99: 2149–2158, 2005. [DOI] [PubMed] [Google Scholar]
- 37.Koskinen SO, Wang W, Ahtikoski AM, Kjaer M, Han XY, Komulainen J, Kovanen V, Takala TE. Acute exercise induced changes in rat skeletal muscle mRNAs and proteins regulating type IV collagen content. Am J Physiol Regul Integr Comp Physiol 280: R1292–R1300, 2001. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-delta delta C(T)] method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 39.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387: 83–90, 1997. [DOI] [PubMed] [Google Scholar]
- 40.Melov S, Tarnopolsky MA, Beckman K, Felkey K, Hubbard A. Resistance exercise reverses aging in human skeletal muscle. PLoS ONE 2: e465, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller RG, Bryan WW, Dietz MA, Munsat TL, Petajan JH, Smith SA, Goodpasture JC. Toxicity and tolerability of recombinant human ciliary neurotrophic factor in patients with amyotrophic lateral sclerosis. Neurology 47: 1329–1331, 1996. [DOI] [PubMed] [Google Scholar]
- 42.Moll R, Holzhausen HJ, Mennel HD, Kuhn C, Baumann R, Taege C, Franke WW. The cardiac isoform of alpha-actin in regenerating and atrophic skeletal muscle, myopathies and rhabdomyomatous tumors: an immunohistochemical study using monoclonal antibodies. Virchows Arch 449: 175–191, 2006. [DOI] [PubMed] [Google Scholar]
- 43.Petrella JK, Kim JS, Cross JM, Kosek DJ, Bamman MM. Efficacy of myonuclear addition may explain differential myofiber growth among resistance-trained young and older men and women. Am J Physiol Endocrinol Metab 291: E937–E946, 2006. [DOI] [PubMed] [Google Scholar]
- 44.Przybyla B, Gurley C, Harvey JF, Bearden E, Kortebein P, Evans WJ, Sullivan DH, Peterson CA, Dennis RA. Aging alters macrophage properties in human skeletal muscle both at rest and in response to acute resistance exercise. Exp Gerontol 41: 320–327, 2006. [DOI] [PubMed] [Google Scholar]
- 45.Ramirez BU, Retamal L, Vergara C. Ciliary neurotrophic factor (CNTF) affects the excitable and contractile properties of innervated skeletal muscles. Biol Res 36: 303–312, 2003. [DOI] [PubMed] [Google Scholar]
- 46.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Myogenic gene expression at rest and after a bout of resistance exercise in young (18–30 yr) and old (80–89 yr) women. J Appl Physiol 101: 53–59, 2006. [DOI] [PubMed] [Google Scholar]
- 47.Renault V, Thornell LE, Eriksson PO, Butler-Browne G, Mouly V. Regenerative potential of human skeletal muscle during aging. Aging Cell 1: 132–139, 2002. [DOI] [PubMed] [Google Scholar]
- 48.Roth SM, Ferrell RE, Peters DG, Metter EJ, Hurley BF, Rogers MA. Influence of age, sex, and strength training on human muscle gene expression determined by microarray. Physiol Genomics 10: 181–190, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth SM, Martel GF, Ivey FM, Lemmer JT, Metter EJ, Hurley BF, Rogers MA. Skeletal muscle satellite cell populations in healthy young and older men and women. Anat Rec 260: 351–358, 2000. [DOI] [PubMed] [Google Scholar]
- 50.Roth SM, Metter EJ, Ling S, Ferrucci L. Inflammatory factors in age-related muscle wasting. Curr Opin Rheumatol 18: 625–630, 2006. [DOI] [PubMed] [Google Scholar]
- 51.Roth SM, Schrager MA, Ferrell RE, Riechman SE, Metter EJ, Lynch NA, Lindle RS, Hurley BF. CNTF genotype is associated with muscular strength and quality in humans across the adult age span. J Appl Physiol 90: 1205–1210, 2001. [DOI] [PubMed] [Google Scholar]
- 52.Rullman E, Rundqvist H, Wagsater D, Fischer H, Eriksson P, Sundberg CJ, Jansson E, Gustafsson T. A single bout of exercise activates matrix metalloproteinase in human skeletal muscle. J Appl Physiol 102: 2346–2351, 2007. [DOI] [PubMed] [Google Scholar]
- 53.Sheffield-Moore M, Paddon-Jones D, Sanford AP, Rosenblatt JI, Matlock AG, Cree MG, Wolfe RR. Mixed muscle and hepatic derived plasma protein metabolism is differentially regulated in older and younger men following resistance exercise. Am J Physiol Endocrinol Metab 288: E922–E929, 2005. [DOI] [PubMed] [Google Scholar]
- 54.Solomon AM, Bouloux PM. Modifying muscle mass-the endocrine perspective. J Endocrinol 191: 349–360, 2006. [DOI] [PubMed] [Google Scholar]
- 55.Spangenburg EE, Abraha T, Childs TE, Pattison JS, Booth FW. Skeletal muscle IGF-binding protein-3 and −5 expressions are age, muscle, and load dependent. Am J Physiol Endocrinol Metab 284: E340–E350, 2003. [DOI] [PubMed] [Google Scholar]
- 56.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol 17: 463–516, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tidball JG. Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol 288: R345–R353, 2005. [DOI] [PubMed] [Google Scholar]
- 58.Trappe T, Williams R, Carrithers J, Raue U, Esmarck B, Kjaer M, Hickner R. Influence of age and resistance exercise on human skeletal muscle proteolysis: a microdialysis approach. J Physiol 554: 803–813, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urso ML, Scrimgeour AG, Chen YW, Thompson PD, Clarkson PM. Analysis of human skeletal muscle after 48 h immobilization reveals alterations in mRNA and protein for extracellular matrix components. J Appl Physiol 101: 1136–1148, 2006. [DOI] [PubMed] [Google Scholar]
- 60.Verdijk LB, Koopman R, Schaart G, Meijer K, Savelberg HH, van Loon LJ. Satellite cell content is specifically reduced in type II skeletal muscle fibers in the elderly. Am J Physiol Endocrinol Metab 292: E151– E157, 2007. [DOI] [PubMed] [Google Scholar]
- 61.Vergara C, Ramirez B. CNTF, a pleiotropic cytokine: emphasis on its myotrophic role. Brain Res Brain Res Rev 47: 161–173, 2004. [DOI] [PubMed] [Google Scholar]
- 62.Volpi E, Sheffield-Moore M, Rasmussen BB, Wolfe RR. Basal muscle amino acid kinetics and protein synthesis in healthy young and older men. JAMA 286: 1206–1212, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang MC, Forsberg NE. Effects of ciliary neurotrophic factor (CNTF) on protein turnover in cultured muscle cells. Cytokine 12: 41– 48, 2000. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, Chan F, Gonzalez C, Zhang L, Samaha RR. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics 7: 59, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watt MJ, Dzamko N, Thomas WG, Rose-John S, Ernst M, Carling D, Kemp BE, Febbraio MA, Steinberg GR. CNTF reverses obesity-induced insulin resistance by activating skeletal muscle AMPK. Nat Med 12: 541–548, 2006. [DOI] [PubMed] [Google Scholar]
- 66.Welle S, Bhatt K, Shah B, Thornton C. Insulin-like growth factor-1 and myostatin mRNA expression in muscle: comparison between 62–77 and 21–31 yr old men. Exp Gerontol 37: 833–839, 2002. [DOI] [PubMed] [Google Scholar]
- 67.Welle S, Brooks AI, Delehanty JM, Needler N, Thornton CA. Gene expression profile of aging in human muscle. Physiol Genomics 14: 149–159, 2003. [DOI] [PubMed] [Google Scholar]
- 68.Westfall PH, Institute SAS. Multiple Comparisons and Multiple Tests: Using the SAS System. Cary, NC: SAS Institute, 1999, p. xiv, 397. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.