Abstract
The ‘RNA world’, in which RNA molecules stored information and acquired enzymatic properties, has been proposed to have preceded organism life. RNA is now recognized for its central role in biology, with accumulating evidence implicating coding and noncoding (nc)RNAs in myriad mechanisms regulating cellular physiology and disequilibrium in transcriptomes resulting in pathological conditions. Nascently synthesized RNAs are subjected to stringent regulation by sophisticated RNA surveillance pathways. In this review, we integrate these pathways from a developmental viewpoint, proposing RNA surveillance as the convergence of mechanisms that ensure the exact titration of RNA molecules in a spatiotemporally controlled manner, leading to development without the onset of pathological conditions, including cancer.
Shaping the Developmental Transcriptome
The RNAs that ensue from the transcription of coding and ncRNAs (see Glossary) are essential for life and contribute toward diverse known and unknown biologies. Cellular development, with the accompanying rapid synthesis of a plethora of RNAs involved in cell division and differentiation, presents a challenge for quality control, necessitating comprehensive RNA surveillance. Proper orchestration of development depends upon rapid changes in transcriptomic programs between developmentally evolving cells, with the transcriptome being constantly supervised by various pathways. Failure can lead to developmental diseases and malignancies (a list of defects in RNA-processing events that lead to pathological conditions is provided in Table 1). While mRNA processing and mRNA surveillance have been intensively studied for many decades [1], we understand less regarding the surveillance of the noncoding transcriptome. However, a significant portion of the mammalian genome can be transcribed as ncRNAs [2] (Box 1), and it is likely that these noncoding transcripts and/or ncRNA transcription contributes to cellular differentiation during development.
Table 1.
Overview of RNA Surveillance-Linked Pathologies
| Pathology/syndrome | Mechanism | Note on study | Refs |
|---|---|---|---|
| Acute myeloid leukemia | METTL14 and METTL3-dependent RNA methylation | Human patients and mouse models | Vu et al., 2017 Nature medicine; Weng et al., 2018, Cell stem cell |
| Aicardi–Goutières syndrome | Mutations in ADAR1, upregulating interferon-regulated genes and inducing inflammation | Human patients | Rice et al., 2012, Nature Genetics |
| TREX1 mutations activate cGAS/STING through LINE-1 expression, leading to inflammation | Human primary cells | Thomas et al., 2017, Cell Stem Cell | |
| MDA5 mutations lead to its constitutive activation by cellular dsRNA (mainly endogenous retroelement Alu:Alu hybrids) | Biochemistry | Ahmad et al., 2018, Cell | |
| RNase H2 gene mutations activate cGAS/STING pathway and the interferon pathway in an untimely manner | RNaseh2b mutant knock-in mouse model | Mackenzie et al., 2016, EMBO | |
| Amyotrophic lateral sclerosis and frontotemporal lobar degeneration | Defect in siRNA silencing and TE expression | Expression of human TDP-43 protein in Drosophila | Krug et al., 2017, Plos genetics |
| Asymmetric DNA mutagenesis | DNA asymmetric mutations commonly observed in cancers; one class is transcription-coupled | Human patients | Haradhvala et al., 2016, Cell [136] |
| MTR4 and Senataxin helicases in cooperation with RNA exosome prevent R-loop formation and DNA asymmetric mutations | KO cell lines, mouse primary B cells (conditional RNA exosome KO) | Lim et al., 2017, Cell | |
| Cancer | Primary piRNA pathway reactivation coincident with oncogenic transformation of somatic cells | Drosophila model | Fagegaltier et al., 2016, Genes and development [137] |
| Alterations of RNA modifications (m6A and others) | Review | Dai et al., 2018, Cell death and disease | |
| Increase mRNA stability by ARE and GU-rich elements | Reviews | Khabar, 2017 [138], WIREs RNA; Vlasova-St-Louis and Bohjanen, 2017, Cytokines and grow factor review | |
| Cerebellar and corpus callosum hypoplasia | EXOSC8 (RNA exosome) mutations, altering mRNA metabolism | Human patients and zebrafish model | Boczonadi et al., 2014, Nat com |
| Fragile X syndrome | Loss of FMRP protein implicated in human fragile X syndrome. This study showed how FMRP cooperates with ADAR2 to regulate editing of neuronal circuit formation genes | Zebrafish model | Shamay-Ramot et al., 2015, Plos genetics |
| Human Mendelian diseases | Mutations in RNA-binding protein genes | Review | Castello et al., 2013, Trends in genetics |
| Inflammatory myofibroblastic tumors | UPF1 mutations and upregulation of NIK, inducing NF-kB activation and inflammation | Human patients | Lu et al., 2016, JCI |
| Lung adenocarcinoma | ADAR-mediated editing increases FAK kinase mRNA stability and expression, correlating with cancer invasiveness | Human patients | Amin et al., 2017, Science signaling |
| Mild myopathy | APOBEC2 RNA-editing enzyme preferentially expressed in muscles; deficiency leads to decrease in body mass and mild myopathy | APOBEC2-KO mouse model | Sato et al, 2010, JBC |
| Multiple myeloma | Dis3 mutations associated with chromosomal translocations at immunoglobulin heavy chain locus | Human patients | Lionetti et al., 2015, Oncotarget |
| Myotonic dystrophy | RNAs containing microsatellite expansions sequester MBLN2 proteins, perturbing splicing and polyadenylation in brain | Human primary tissues, ‘RNA toxicity hypothesis’ | Goodwin et al., 2015, Cell Rep |
| MBLN3 expression during embryogenesis and muscle regeneration with binding to 3′UTR of cell growth and proliferation genes. Defects in muscle regeneration and function in MBLN3-KO mice | MBLN3-KO mouse model | Poulos et al., 2013, Hum Mol genetics | |
| Myotonic dystrophy type 1 | Alternative splicing and abnormal polyadenylation in muscles due to trinucleotide expansions in RNAs, altering activities of RNA-processing factors, including MBNL proteins | Human samples and KO mouse models | Thomas et al., 2017, Genes and development |
| Pancreatic adenosquamous carcinoma | Somatic mutations of UPF1 (NMD factor) upregulate NMD substrate mRNAs | Human patients | Liu et al., 2014, Nat medicine |
| Perlman syndrome and Wilms tumor susceptibility | DIS3L2 germline mutations, mitotic abnormalities, dysregulated expression of mitotic control proteins | Human patients | Astuti et al., 2012, Nat genetics |
| Pontocerebellar hypoplasia 1 | EXOSC3 (RNA exosome) mutations found in patients. Knockdown experiments in zebrafish perturb embryonic development and brain formation | Human patients and zebrafish model | Wan et al., 2011, Nature Genetics |
| Pontocerebellar hypoplasia 1 like | RBM7 (NEXT complex) mutations, altering gene expression. Knockdown of rbm7 in zebrafish induced defects in motor neurons and cerebellum | Human patients and zebrafish model | Giunta et al., 2016, Human mol genetics |
| R-loop stabilization and genomic instability | EXOSC3 deficiency increases R-loop formation at DNA translocation hotspots | Mouse primary B cells (conditional Exosc3 KO) | Pefanis et al., 2014, Nature |
| R-loop-associated pathologies | Mutations in genes involved in R-loop removal or formation | Review | Richard and Manley, 2017, J Mol Biol. |
| Retinitis pigmentosa, hearing loss, premature aging, short stature, mild intellectual disability and distinctive gestalt | EXOSC2 (RNA exosome) mutations, probably altering RNA metabolism | Human patients | Di Donato et al., 2016, J med genetics |
| RNA-editing and ADAR1-linked pathologies | Implication of editing in different pathologies, cancers, and neuropathies | Review | Song et al., 2016, Genes |
| Systemic lupus erythematosus | RNase H2 gene mutations associated with accumulation of ribonucleotides in genomic DNA and DNA damage, culminating with upregulation of IFN-stimulated genes | Human patients | Günther et al., 2015, J Clin Invest. |
| TREX1 mutations in patients with SLE, with two mutations affecting TREX1 protein subcellular targeting, and probably deregulating its activity | Human patients | Lee-Kirsh et al., 2007, Nat Genet. |
Box 1. The Different Classes of Coding and Noncoding RNAs.
Coding RNAs are processed from pre-mRNAs to mature mRNAs and translated into proteins to fulfill cellular functions, thereby linking information in DNA to active proteins. By contrast, most RNAs do not produce proteins and are referred to as ncRNAs. While transcription from ‘junk DNA’ was initially considered a byproduct and generally treated as ‘transcriptional noise’, it appears that ncRNAs have crucial roles in cellular physiology.
The major categories of ncRNAs are ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), very small RNA (miRNA, siRNA, and piRNA), and long ncRNA (lncRNA). Recent advances in biology have demonstrated unexpected functions for lncRNAs, which regulate diverse physiological events. LncRNAs include various RNAs, such as enhancer RNA (eRNA), promoter-associated antisense RNA, intergenic RNA, and intragenic RNA. Circularization of RNAs is another important aspect of RNA processing, opening the field of circular RNAs, described as potential miRNAs sponges, with critical implications in physiological and pathological processes. Many transposable elements (TEs) are integrated in higher eukaryote genomes and can eventually be expressed. These elements have to be tightly regulated by RNA surveillance because they can be deleterious by introducing genomic instability. In addition to endogenous RNAs, mammalian cells are also challenged by exogenous RNAs that are either transferred by extracellular vesicle cargos resulting from intercellular communication, or from microorganisms, with the latter being mainly viral RNAs.
Cells have to regulate both the transcription and processing of all these different RNAs, with strong quality control ensured by RNA surveillance. Although ‘pervasive’ transcription of genomes is widely reported, tight regulation of gene expression in space and time, culminating in the control of developmental schemes, makes more evolutionary sense.
The recent development of coding RNA and ncRNA transcriptomics and epitranscriptomics [3] has provided evidence for a higher level of RNA diversity. How RNA diversity meaningfully contributes to organism development is the focus of this review. Here, we propose to extend the definition of RNA surveillance beyond its role in quality control to include its implication in post-transcriptional regulation, describing RNA surveillance as the convergence of mechanisms that ensure the exact titration of any RNA molecule in a spatiotemporally controlled manner, leading to development without the onset of pathological conditions.
RNA Diversity and the RNA Surveillance Machinery
RNA surveillance functions to sieve out short-lived, long-lived, and nonfunctional RNAs as per cellular requirements and could be a central mode of gene regulation (Figure 1), culminating in the control of developmental schemes. RNA surveillance pathways, ensuring strong control of their target RNAs (Box 2), have been extensively reviewed elsewhere [1,4] and are briefly described here.
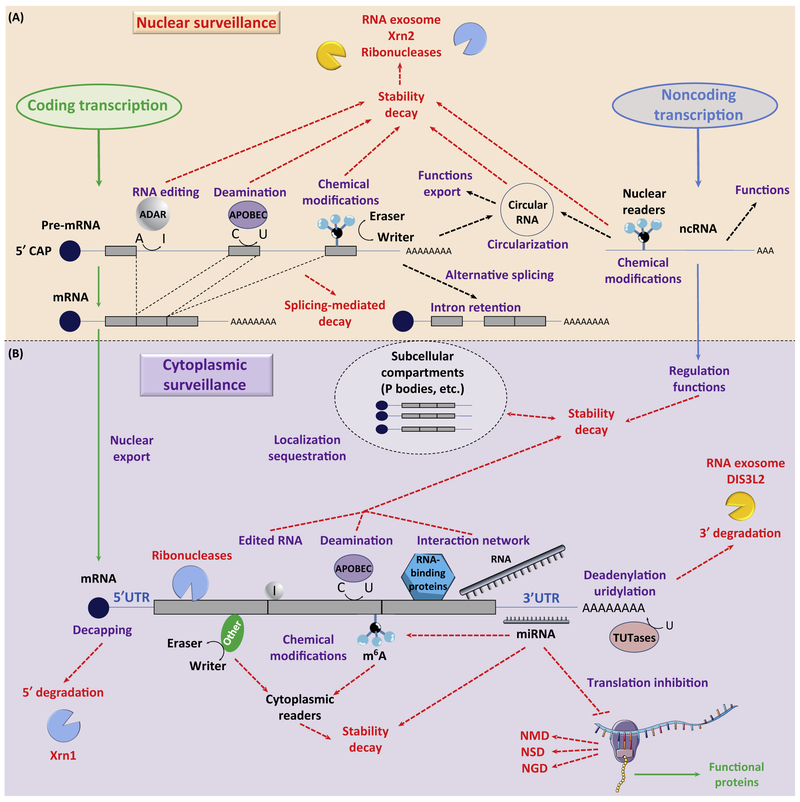
Figure 1. Cellular Context of RNA Processing and Surveillance.
Integrated view of RNA maturation and the different levels of RNA quality control, processing, and surveillance. (A) Nuclear surveillance: maturing mRNAs can undergo various modifications (editing, deamination, or chemical modifications) that affect their stability, decay, splicing, and export. Noncoding (nc)RNAs are also susceptible to chemical modifications and degradation. Circular RNAs can also be produced and potentially exported from the nucleus. (B) mRNA fate is dependent on its modification and interaction with other proteins and/or RNAs. 5′ end decapping leads to RNA degradation by the Xrn1 exonuclease; deadenylation and uridylation trigger 3′ end decay by the RNA exosome or Dis3L2 exonucleases, while other endonucleases can directly cut RNA. This level of cytoplasmic surveillance regulates the concentration of coding RNAs and ncRNAs, which eventually can be delocalized in subcellular compartments. RNAi (e.g., miRNA) fine-tunes gene expression. Nonfunctional transcripts are processed by the nonsense-mediated decay (NMD), nonstop-mediated decay (NSD), or no-go decay (NGD) pathways.
Box 2. The RNA Surveillance Machinery.
RNA surveillance pathways are unusually redundant in terms of their control of their target RNA outcome. Several layers of RNA surveillance work in concert to fine-tune the RNA concentration of any given RNA molecule. This process starts at the chromatin level, where DNA-associated RNAs are unwound and degraded to avoid genomic instability, while nuclear and cytoplasmic surveillance completes this monitoring to control the titration of ncRNA and mRNA levels.
RNA decay is ensured by exoribonucleases that degrade RNAs from their 5′ (XRN family) or 3′ (RNA exosome, Dis3L2) extremities following decapping, polyA shortening, polyuridylation, and so on, induced by specific stimuli or natural turnover. Endoribonucleases [ribonuclease (RNase) A, RNase H, Dis3, Dicer, etc.] can directly cleave the body of RNA molecules and participate in RNA processing. Many cofactors participate in RNA surveillance, including RNA helicases, which unwind RNA molecules, multiprotein complexes, RNA-binding proteins (RBPs), and adaptor proteins, all modulating RNA interactions and degradation. For example, the Ccr4-NOT complex has important roles in both the nucleus and cytoplasm, where it globally contributes to gene expression control, while the transcription and export (TREX) complex regulates mRNA maturation and export, and is essential for cell differentiation and organism development. The THO complex controls both mRNA processing and snoRNA expression in yeast. The Trf4/Air2/Mtr4p polyadenylation (TRAMP) complex interacts with the RNA exosome to monitor the RNA concentration of various substrates, while the SUPV3L1, SKIV2L (SKI) complex is important for cytoplasmic RNA degradation by the RNA exosome complex or the DIS3L2 enzyme. Other important mechanisms participate in mRNA quality control, including splicing-mediated decay, nonsense-mediated decay (NMD), nonstop-mediated decay (NSD), and no-go decay (NGD). RNAi contributes to RNA surveillance using various mechanisms to control mRNA expression and degrade exogenous and TE RNAs. The flow of RNA processing and RNA localization, including its sequestration, are also important parameters contributing to gene expression.
RNA Degradation
mRNAs are protected at their 5′ end by a cap that is removed by a decapping complex, following specific signals or natural turnover. Decapped mRNAs are degraded by the 5′ XRN exonuclease family of proteins, which are highly conserved and have critical functions in RNA surveillance, gene silencing, and nonsense-mediated mRNA decay (NMD) [5]. In yeast, Xnr1 participates in the degradation of antisense ncRNAs, regulating chromatin modifications and gene expression [6]. 5′ RNA processing by decapping and exoribonuclease activity can be initiated co-translationally [7] and is linked to NMD, which participates in the degradation of pervasive noncoding transcripts [8]. At the 3′ end, polyadenylated mRNAs are gradually deadenylated by poly(A) ribonucleases (e.g., PARN) and degraded by the RNA exosome complex or the DIS3L2 enzyme. Dis3L2 has 3′ end exoribonuclease activity, is cytoplasmic, and ensures the degradation of various uridylated RNAs [9]. The RNA exosome has major functions in RNA processing, with emerging functions in the maintenance of genome stability. The RNA exosome has different subunit and cofactor compositions that determine its differential ribonuclease activity [10] and it can degrade RNA unwound from DNA by the RNA helicases Setx and Mtr4 [11,12]. RNAs can be processed by endonucleases, such as RNase H and RNase A, which cleave directly in the body of these molecules. RNase H activity, which cleaves RNA when it is associated with DNA in a DNA/RNA hybrid configuration, is found in all kingdoms of life and is mandatory for DNA replication, DNA repair, and RNA splicing [13]. The Dis3 protein of the RNA exosome complex also has endonuclease activity [14]. Dicer also has endonuclease activity that is implicated in the generation of miRNAs and in RNAi [15], while other endonucleases participate in specific RNA surveillance pathways.
Various Mechanisms Used for RNA Surveillance
RNAs adopt many different secondary structures and interact with other molecules, potentially masking their degradation signals. Therefore, co-factors, in particular helicases, are necessary to unwind RNA for processing [16], while other multiprotein complexes interact with different ribonucleases to finely regulate their activity (e.g., Ccr4-NOT, TREX, THO, TRAMP, SKI, etc.). Other complexes, RNA-binding proteins (RBPs), and adaptor proteins contribute to specific interactions between RNA substrates and RNA decay factors [17]. Transcription generates mRNAs that can include premature termination codons. The NMD pathway detects and degrades these aberrant transcripts, and controls gene expression using UPF proteins along with RNA exo- and endonucleases [18,19]. Other mechanisms monitor transcripts that lack stop codons and remove them by nonstop-mediated decay (NSD) [20], while no-go decay (NGD) degrades transcripts on which ribosomes have stalled. Spliceosome-mediated decay is observed in the context of the expression of non-intronic genes in yeast [21]. Spliceosome-mediated RNA decay is implicated in regulating alternative splicing and intron retention, and occurs in most genes and all tissues due to a central role in cell differentiation [22–24]. RNA silencing is another mechanism that controls gene expression, or degrades exogenous RNAs. ncRNAs are cleaved by the RNase III Dicer [15] to produce miRNAs or small interfering (si)RNAs that are loaded onto the RNA-induced silencing complex (RISC) and the Argonaute (Ago) proteins, which induce inhibition of translation, mRNA cleavage, or degradation of their targets. Germ cells use a piwi-interacting (pi)RNA guide loaded onto PIWI proteins to cleave RNA and promote DNA methylation [25]. miRNAs are themselves subject to degradation by the Tudor-SN endonuclease in human cells [26]. Finally, RNA trafficking and localization participate in the fine-tuning of gene expression. Site-specific mRNA regulation was recently shown in neurons, where the N6-methyladenosine (m6A) eraser FTO is locally translated to regulate m6A RNAs [27].
Influence of RNA Sequence and Modifications on RNA Surveillance
RNA composition modulates their folding, secondary structures, and interactions (Box 3). AU-rich elements (AREs) interact with RBPs that regulate the processing and decay of mRNAs [28]. GU-rich elements are found in genes of signaling components and are upregulated in cancer cells [29]. 3′ untranslated region (UTR) miRNA binding sites are crucial for gene expression regulation and RNAi. mRNA interactions with trans-regulators have key functions in homeostasis, while disruptions lead to neurodevelopmental disorders [30]. RNA composition can be altered by diverse processes, including RNA editing, performed by ADAR proteins that change A to I, with critical implications in gene regulation and diseases [31]. ADAR1- and ADAR2-dependent editing appear to be tissue specific [32], and may be important to confer cell identity and lineage specificity. Some APOBEC proteins can deaminate RNA, resulting in C to U modifications [33], and could be important in the regulation of endogenous or exogenous RNAs by recruiting the RNA exosome to degrade these transcripts.
Box 3. Additional Layers Contributing to RNA Surveillance.
Additional parameters that contribute towards the fine titration of RNA molecules in the cellular milieu must be considered to complete our developmental overview of RNA surveillance. For mRNAs, different intrinsic factors, mainly localized in their 3′ UTR sequences, participate in their stability and surveillance (AU-rich elements, GU-rich elements, miRNA-binding sites, etc.). 3′ capping, 5′ end modifications, and RNA composition itself also strongly orientate RNA molecules to their respective fates. RNAs interact with different molecules, and improvements in biochemistry methods further extend the interactome list every day. A broad range of RNA-binding proteins with important biological functions have been identified, but many processes remain to be discovered. RNA–RNA interactions can be intra- or inter-molecular, with a huge number of possibilities and an interesting dynamic that can have important roles during differentiation and development. Finally, DNA-associated RNAs appear to be transitory, but could have important roles in chromatin remodeling and gene expression regulation.
Coding RNAs and ncRNAs are also subject to editing and chemical modifications. RNA editing by adenosine deaminases acting on RNA (ADAR) proteins directly changes the ribonucleotide sequence from adenosine to inosine (A to I), with emerging functions in physiology and pathology. Some APOBEC proteins can deaminate RNAs, resulting in cytosine to uracil (C to U) alteration and could be implicated in endogenous and exogenous RNA surveillance, participating in the fine-tuning of biological functions. Another expanding field has emerged recently with the study of epitranscriptomes by high-throughput sequencing, and identified more than 100 RNA modifications. The enzymes that write, read, or edit these modifications are now under study, demonstrating the crucial role of these chemical changes in a range of biological functions. Importantly, these RNA modifications are now clearly implicated in pathology, including a large number of cancers. This complex interplay between RNAs, their modifications, and interactions with other molecules, expands the complexity of RNA regulation and surveillance.
RNA modifications regulate their chemical and structural properties [34], with the most abundant m6A implicated in a range of biological processes, including development [35]. Epitranscriptomics [3] has led to the identification of many other modifications, greatly diversifying the RNA species. Specific enzymes catalyze (writers), decipher (readers), or edit (erasers) these RNA alterations, contributing to RNA stability. Uridylation by terminal uridylyl transferases (TUTases) is another modification that affects RNA stability and tags them for degradation [36]. RBPs with critical roles in biological systems have been characterized, suggesting that >10% of proteins have RNA-binding capacity [37]. RBPs regulate pluripotency [38], and mutations are observed in human Mendelian diseases, mainly neurological disorders and cancers [39]. Additionally, complex networks of intra- and intermolecular RNA–RNA interactions impact RNA stability and are dynamically remodeled during human embryonic stem cell (ESC) differentiation [40].
RNA Surveillance during Cellular Differentiation and Embryogenesis
Although RNA surveillance machineries were initially investigated in model organisms, their relevance to development and pathologies is particularly interesting in humans. Indeed, these mechanisms are used to fine-tune gene expression programs during embryogenesis and pluripotency (Figure 2).
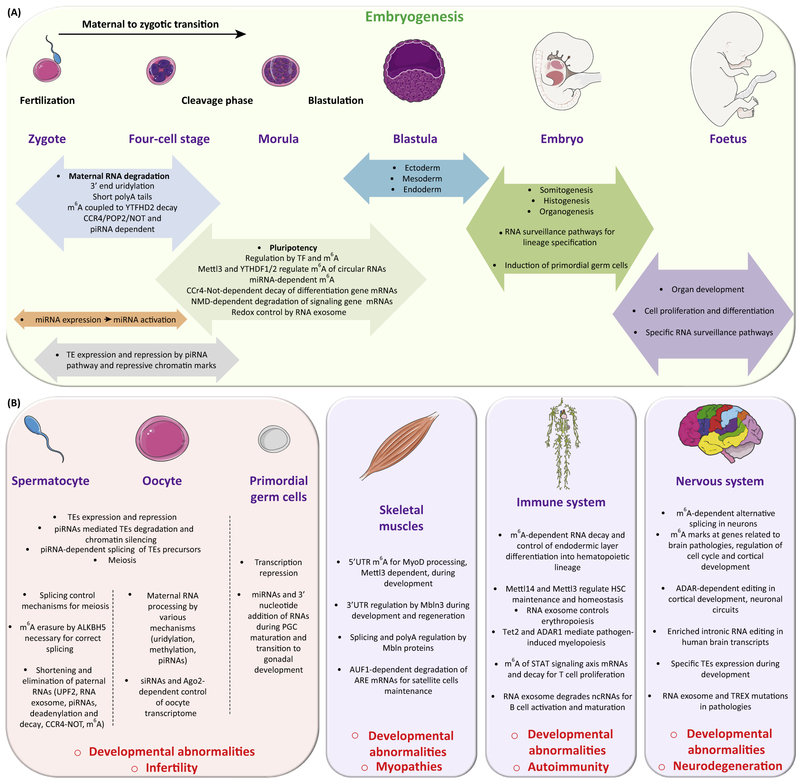
Figure 2. Organism Development and Critical Points of RNA Surveillance.
Overview of RNA-associated events governing organism development. RNA processing and surveillance orchestrate physiological development, from fertilization to tissue and organ maturation. (A) Main steps of embryogenesis and RNA surveillance events. These examples illustrate how RNA surveillance contributes to physiological functions in developing organisms. (B) Different mechanisms of RNA surveillance regulate the development of germ cells, the nervous and immune systems, and skeletal muscles. Defects in these functions lead to developmental abnormalities and associated pathologies. Abbreviations: PGC, primordial germ cell; TE, transposable element; TF, transcription factor. For additional definitions, please see the main text.
Embryogenesis is a paradigm for RNA surveillance since it begins using maternal RNAs but quickly switches to the zygotic transcriptome, a phenomenon called the ‘maternal-to-zygotic transition’ (MZT). The maternal transcriptome is shaped by TUT4 and TUT7 enzymes and uridylation of mRNAs, which facilitates their degradation during mouse oocyte maturation, where these mRNAs have shorter poly(A) tails than those in somatic cells [41]. One-third of maternal RNAs are methylated and, after fertilization, m6A methylation facilitates the decay of these RNAs, which are bound by YTHDF2; this turnover is required for embryonic development in zebrafish [42] and mice [43]. miRNAs are strongly expressed after fertilization, with greater mono- and oligo-adenylation, potentially protecting them from degradation. miRNA function appears to be dynamic, activated at the two-cell stage to regulate developmentally important genes [44]. The CCR4/POP2/NOT deadenylase complex participates in maternal RNA degradation in coordination with the RBP Smaug in Drosophila embryos [45]. Notably, the recruitment of this complex to its RNA target for degradation depends on the piRNA pathway [46]. These events illustrate how RNA-processing pathways can converge to perform a specific task.
At the chromatin level, zygotic DNA is demethylated and, therefore, transposable elements (TEs) may be expressed and must be degraded to avoid genomic instability. Ago2 quickly binds small RNAs to interfere with TE RNAs, followed by the formation of repressive chromatin marks to ensure TE silencing in ESCs [47]. However, this dogma is challenged by evidence demonstrating cellular functions for TEs, such as long interspersed nuclear elements-1 (LINE-1), acting as scaffolding RNA to control gene expression in ESCs and pre-embryos [48]. Many promoters in human and mouse ESCs are transcribed bidirectionally, with coordinated changes during differentiation [49]. DNA sequences, expressing enhancer (e)RNAs, control gene expression and cellular differentiation in the context of chromosome looping and long-range DNA interactions in ESCs, where a superenhancer controls DNA looping at the Nanog locus to maintain pluripotency [50]. Finally, transcription of intragenic enhancers can directly decrease host gene expression by interfering with RNA polymerase II activity and leading to impaired ESC differentiation [51].
In addition to embryogenesis, RNA surveillance can also regulate pluripotency. Zinc finger TF217 (ZFP217) is a key player in maintaining pluripotency via epigenetic and epitranscriptomic regulation, where it interacts with, and sequesters, methyl-transferase-like 3 (Mettl3), decreasing transcript methylation. ZFP217 knockdown in ESCs increased m6A RNA levels, especially of Nanog, Sox2, Klf4, and c-Myc mRNAs, promoting their degradation and inducing cell differentiation [52]. Circular RNAs are themselves methylated; an atlas of these RNAs revealed a specific pattern of methylation in human ESCs, in that they were Mettl3 dependent and interacted with YTHDF1/YTHDF2 readers [53]. A comparison of pluripotent cell methylated transcriptomes revealed gene and cell specificity of m6A, whereby m6A promotes reprogramming to pluripotent cells and, importantly, the sequence specificity of these modifications is regulated by miRNAs in a pairing-dependent manner [54]. Interestingly, miRNA levels can be regulated by the expression of long (l)ncRNAs, as suggested for the ncRNA Cyrano and its complementary sequence, which both modulate the expression of master self-renewal factors and ESC maintenance [55]. Another regulatory network implicating Cyrano, miRNAs, and a circular RNA has been also described in the mammalian brain [56]. These data suggest a complex interplay between the different classes of ncRNA and mRNA in surveillance pathways.
CNOT3, a component of the Ccr4-Not complex, is required for embryonic development; its deficiency increases the poly(A) length, half-life, and steady-state level of differentiation gene mRNAs [57]. In human ESCs, NMD maintains pluripotency by targeting specific genes, including many signaling components, with the TGF-β and BMP axis being important for cell differentiation [58]. RNA exosome controls the redox status of pluripotent stem cells by degrading ARE-containing RNAs, including Gpx2, which is responsible for protection from reactive oxygen species [59], while m6A is crucial for regulating ESCs and pluripotent cells [60], acting in concert with other RNA surveillance mechanisms.
RNA Surveillance during Tissue Development
During development, the three germ layers (ectoderm, mesoderm, and endoderm) give rise to organs after somitogenesis, histogenesis, and organogenesis. Here, we focus on tissues in which RNA surveillance mechanisms have been described (the hematopoietic, nervous, and muscular systems) and address the situation of germ cells, which remodel their chromatin and transcriptional programs (Figure 2).
Immune System
Hematopoiesis generates common myeloid and lymphoid progenitors, which engender blood and immune system cells. A reservoir ensures both stem cell maintenance and differentiation of effector cells. The hematopoietic lineage derives from the endoderm layer, where this transition is controlled by m6A. Mettl3-deficient zebrafish embryos have fewer m6A transcripts, which delays YTHDF2-mediated RNA decay of nocth1a and rhoca [61], genes specifically implicated in this differentiation process. Hematopoietic stem cells (HSCs) express high levels of METTL14, which maintain pluripotency; its deletion induces cell differentiation in HSCs, as well as in acute myeloid leukemia cells, the pathological counterpart of myeloid cells [62]. A similar phenotype is observed with human HSCs, in which depletion of METTL3 impairs stem cell maintenance; by contrast, METTL3 is necessary for the proliferation of acute myeloid leukemia cells, where m6A promotes oncogene translation [63]. tRNAs pseudouridylation chemical modification controls ESCs and their hematopoietic commitment; its deficiency is observed in myelodysplastic syndromes [64]. Myeloid progenitors generate innate immune system cells, thrombocytes, and erythrocytes. The RNA exosome expression level (Exosc8 subunit) is downregulated by the master regulator of erythropoiesis GATA-1, inducing erythrocyte maturation [65]. RNA exosome is also necessary for the expression and signaling of the Kit receptor, preventing erythropoietin-induced differentiation in vivo [66]. Upon infection or development, Tet2 oxidizes 5-methylcytosines of RNAs, modulating the recruitment of the ADAR1 editing enzyme and the expression level of mRNAs, in particular Socs3 and the Jak/Stat axis, inducing an increase in myelopoiesis and mast cell production [67]. This specific level of gene expression regulation further extends the list of mechanisms involved in the control of cellular function.
Lymphoid progenitors produce natural killer cells and T and B lymphocytes, conferring adaptive immunity. Mettl3 has crucial roles in T cell homeostasis and differentiation, where SOCS1, SOCS3, and CISH mRNAs, inhibitors of the STAT signaling axis, are methylated to ensure their decay [68]. Antigen-experienced B cells produce high-affinity antibodies by transcription-coupled DNA alteration processes called class switch recombination (CSR) and somatic hypermutation (SHM) [69,70]. CSR and SHM require a series of steps of ncRNA transcription and coupled RNA processing by the RNA exosome complex in association with Mtr4 and senataxin helicases. In the absence of the RNA exosome complex, CSR and SHM mechanisms are perturbed, leading to weak and/or altered antibody gene diversification [11,71,72]. As a collateral effect of B cell activation, leading to increased transcription and RNA splicing, NMD has a role in degrading potentially deleterious mRNAs in B cells [73]. Finally, the immunoglobulin locus recombination is driven by a 3′ regulatory region (′RR) [74] superenhancer, and transcription of enhancer RNA and 3′RR activation is regulated by the RNA exosome complex [75].
Nervous System
The nervous system derives from the ectoderm, beginning with the formation of the neural tube, giving rise to the brain structures. Methyltransferase components and readers are enriched in the Drosophila nervous system, where they mediate m6A-dependent alternative splicing, which is important for neuronal function and fly behavior in vivo [76]. During cortical development, absence of Mettl14, Mettl3, and m6A prolongs the cell cycle of radial cells in mice. m6A is enriched at genes encoding transcription factors (TFs) and genes implicated in neurogenesis, promoting their degradation. m6A signaling also regulates human cortical neurogenesis in vitro and tags transcripts related to brain pathology risk [77].
The importance of RNA editing in human brain is emerging: different patterns of ADAR-dependent editing have been observed, associated with neuronal maturation and mRNA abundance, potentially influencing miRNA binding during cortical development. These transcripts are related to vesicle or organelle membranes and glutamate signaling, with some perturbations observed in spinal cord injury and glioblastoma [78]. While RNA surveillance usually represses the expression of endogenous retrotransposons, LINE-1 is expressed in the hippocampus during early life, linked to maternal care, and changing the neuronal genome [79]. Another study showed how an RNA splicing event could participate in long-term memory in Drosophila [80]. Finally, a recent study revealed the dynamics of RNA methylation in mouse brain following stress exposure, where an imbalance may be implicated in depressive disorders in humans [81].
Myogenesis
Muscle cells derive from the endoderm layer, where myogenesis requires the key TF MyoD for differentiation and fusion of myoblasts in mature skeletal muscle fibers. In progenitor cells, Mettl3 is required to methylate MyoD mRNAs at the 5′UTR region for processing and skeletal muscle differentiation [82]. During human myogenesis, the NMD factor UPF1 directly promotes the degradation of MyoD protein, linking the mRNA and protein decay mechanisms [83]. After birth, a pool of muscle stem cells, known as satellite cells, allows muscle regeneration in case of injury. The mRNA decay protein AUF1 specifically degrades ARE-containing mRNAs and contributes to the maintenance of these stem cells, while mutations may lead to human myopathies [84].
Germ Cells
Germ cells generate gametes by meiosis, switching from a diploid to haploid state; they are produced by the induction of primordial germ cells (PGCs) from embryonic cells in mammals. PGCs have specific features, including transcription repression and chromatin-state alteration. miRNAs and 3′ nucleotide addition control the RNA molecules necessary for PGC maturation, and the transition to gonadal development in mouse embryos [85]. Germ cells express both lineage-specific TFs and a distinct transcriptional profile, while DNA undergoes global demethylation [86]. This chromatin derepression is potentially perilous because some endogenous TEs can be expressed and must be controlled.
In oocytes, maternal RNAs mainly govern the first steps of zygote maturation and many are then degraded. Maternal RNA degradation is mediated by uridylation and decay [41], and by methylation in combination with their readers. YTHDF2 reader-deficient mice fail to accomplish transcript dosage during oocyte maturation, resulting in female infertility [43]. Mechanistically, YTH domain-containing proteins are important for regulating meiotic genes. In yeast, one YTH domain-containing protein (Mm1), both directly targets meiotic transcripts to the RNA exosome for degradation and tethers them to nuclear foci, enforcing translation inhibition [87]. In mice, YTFHDC2 controls the transition from mitosis to meiosis by binding mitotic transcripts as well as specific piRNA precursors and interacting with granule components [88]. siRNA production also contributes to meiotic control in mouse oocytes, where Ago2 regulates the oocyte transcriptome during meiosis, subsequent chromosome alignment, and, simultaneously, the expression of some transposable elements (TEs) [89].
In male germ cells TDRD6 is highly expressed, interacting with the spliceosome and one methyltransferase; its depletion leads to splicing defects in spermatocytes [90]. The demethylation enzyme ALKBH5 is necessary for male fertility to erase m6A and undertake subsequent accurate splicing, producing some long mRNAs during late meiosis [91]. A global shortening of mRNAs has been previously observed and is necessary during spermatogenesis. This effect is mediated by the NMD component UPF2, which targets and degrades longer 3′UTRs. UPF2 knockout (KO) spermatocytes again accumulate alternatively spliced transcripts, leading to global defects and infertility [92]. This noncanonical role of the NMD pathway could be important in other cells. The RNA exosome complex itself participates in spermatogenesis regulation, during which Exosc10 is dynamically expressed [93].
The CCR4-NOT complex is recruited by the RBP DND1, which binds a motif in the 3′ UTR of the mRNA and destabilizes its targets, regulating apoptosis, inflammation, and pluripotent gene expression in PGCs and spermatogonial stem cells (SSCs) [94]. The RNA methylome atlas revealed the dynamics of m6A modifications, with increased methylation at the pachytene/diplotene spermatocyte and round spermatid stages. Methylation affects the translation efficiency of important genes implicated in SSC proliferation and differentiation, and later for haploid-specific genes [95]. Taken together, these results show the complexity of RNA processing contributing to meiosis.
More globally, RNA surveillance is crucial during normal embryogenesis and development.
RNA Surveillance Deficiencies Leading to Pathologies
RNA surveillance defects lead to associated pathologies during development. Here, we describe some disorders and how RNA processing contributes to maintaining genome integrity (Figure 3 and Table 1).
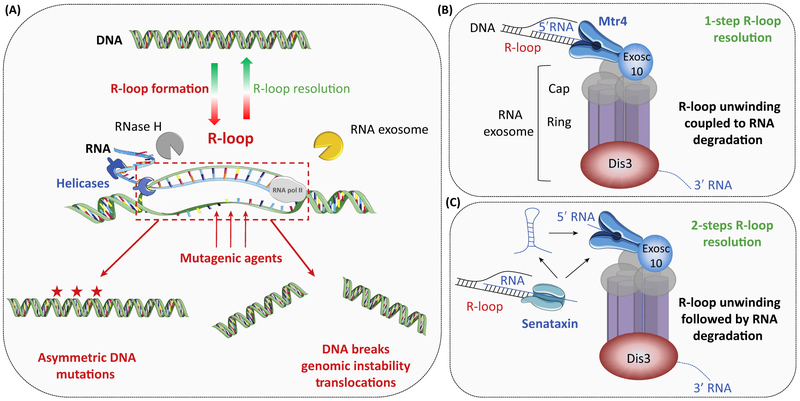
Figure 3. RNA Surveillance of DNA-Associated RNAs.
(A) DNA transcription can create DNA:RNA hybrids, also called R-loops. These structures expose one DNA strand to environmental mutagenic agents (e.g., radiation, chemicals, and enzymes) and must be resolved by RNA surveillance mechanisms (e.g., RNase H or RNA exosome). R-loop persistence results in asymmetric DNA mutations and eventually DNA double-strand breaks and translocations. (B) Cofactors participate in R-loop resolution; the RNA helicase Mtr4 is directly associated with the RNA exosome to unwind DNA-associated RNAs, coupled with RNA decay. (C) Other RNA helicases, such as Senataxin, participate in R-loop unwinding to dissociate DNA-associated RNAs that are subsequently degraded by the RNA exosome.
Immunological Disorders and Autoimmunity
Aicardi–Goutières syndrome (AGS) is a disorder characterized by abnormal inflammation affecting the immune and nervous systems. AGS can be caused by mutations in TREX1, RNase H2 complex, or ADAR1, which upregulate interferon-regulated genes [96]. Mechanistic studies demonstrate that ADAR1 edits endogenous double-strand (ds) RNAs, preventing the activation of the cytosolic dsRNA sensor MDA5 and the interferon pathway [97]. Loss of tolerance is caused by Alu retroelements that form Alu:Alu dsRNA structures and activate MDA5 in the absence of ADAR1-dependent editing [98]. A mouse model of RNase H2 mutation suggested an involvement of the cGAS/STING pathway, with defects in ribonucleotide excision repair, increased DNA damage, and activation of interferon-stimulated genes [99]. RNase H2 mutations are also linked to systemic lupus erythematosus (SLE) [100], and TREX1 deficiencies or mutations have also been implicated in both AGS [101] and SLE [102]. This could be due to retrotransposon expression followed by reverse transcription and accumulation of cytoplasmic nucleic acids in the absence of the nuclease activity of TREX1, or directly linked to the ribonuclease function of this enzyme [103]. Mutations have been identified in the human polynucleotide phosphorylase gene PNPT1 that also converge on interferon pathway activation. This axis is implicated in the processing of mitochondrial dsRNAs, which are usually unwound by SUV3 helicases followed by polynucleotide phosphorylase activity [104]. Several diseases are linked to mutations in RNA exosome genes or its cofactors [105]. Plasma cell disorders, including multiple myeloma, present Dis3 loss-of-function mutations, preferentially associated with chromosomal translocations to the immunoglobulin locus [106], reminiscent of ncRNAs accumulation at translocation hotspots in mice [71].
Neurodegenerative Diseases
Defects in RNA editing are implicated in fragile X syndrome, a frequently inherited form of intellectual disability, caused by the absence of the fragile X mental retardation protein (FMRP). Under normal conditions, ADAR2 interacts with FMRP to regulate editing of genes implicated in neuronal circuit formation, although this editing is increased in Fmrp-deficient zebrafish [107]. RNA editing is also enriched in introns of human brain transcripts, suggesting a conserved role for this function [108]. Some repeat-expansion-associated diseases, such as myotonic dystrophy (neuromuscular), can induce ‘RNA toxicity’ and nervous system pathologies. In brain samples from patients with myotonic dystrophy, RNAs containing microsatellite expansions sequestered the MBLN2 protein and perturbed RNA splicing and polyadenylation, with the same effect observed in Mbln1 KO mice [109].
The RNA exosome is mutated in several human neuropathies: for example, EXOSC3 mutations are implicated in spinal motor neuron disease corresponding to pontocerebellar hypoplasia 1, with a similar phenotype seen in Exosc3-knockdown zebrafish [110]. EXOSC8 mutations induce a progressive disease involving cerebellar and corpus callosum hypoplasia, abnormal myelination, or spinal motor neuron disease. Knockdown experiments showed an increase in ARE mRNAs of myelin proteins related to the pathology [111]. An Exosc2 mutation was identified in a patient with retinitis pigmentosa and mild intellectual disability [112]. RBM7 (part of the NEXT complex) mutation alters RNA metabolism, and knockdown experiments revealed defects in motor neurons and the cerebellum, reminiscent of Exosc3 and Exosc8 [113]. TDP-43 is frequently mutated in human amyotrophic lateral sclerosis and frontotemporal lobar degeneration, inducing defects in siRNA silencing and concomitant TE expression, DNA breaks, and cell death in the Drosophila brain [114].
Myopathies
Myopathies are a group of diseases affecting muscular development and function. Myotonic dystrophy, a class of inherited muscular dystrophies, is caused by triplet expansions (in DMPK or CNBP genes) that create toxic RNAs sequestering MBNL alternative splicing factor proteins. Mbnl3 is expressed during embryogenesis and skeletal muscle regeneration (where relevant exons undergo a prenatal RNA isoform transition) and binds the 3′UTR of cell growth and proliferation genes. Accordingly, Mbnl3-KO mice have age-dependent defects in injury-induced muscle regeneration and muscle function [115]. Alternative splicing and abnormal poly(A) are also detected in the muscles of patients with congenital myotonic dystrophy. Mbnl protein deletions in mouse muscle recapitulated the human symptoms of congenital myopathy with specific defects in RNA splicing [116]. APOBEC2 is preferentially expressed in muscles, with an RNA-editing function. This deaminase is important for regulating muscle development, and APOBEC2-deficient mice have decreased body mass and mild myopathy [117].
Genome Integrity, RNA Surveillance, and Cancer
The emerging field of RNA surveillance provides new perspectives to approach cancer genesis. Transcription of some genomic regions due to sequence context or other reasons can induce transcriptional stalling and associated RNA:DNA hybrid accumulation, also known as R-loops. These structures regulate chromosomal organization, epigenetics, gene expression, DNA replication and repair, and class switch recombination [118]. They are implicated in human diseases, including neuropathies and cancers [119], where R-loops can expose a single-stranded DNA to mutagenic factors. The RNA exosome and its cofactors are necessary for regulating DNA:RNA hybrid levels to suppress asymmetric DNA mutagenesis [11] and associated genomic instability [71,120] (Figure 3). Other RNA-processing pathways participate in R-loop resolution in humans, such as the RNA helicases Aquarius and Senataxin, and topoisomerase I [121]. DNA:RNA hybrid stabilization and associated R-loop formation can lead to dsDNA breaks because of collisions between the stalled transcription and DNA replication machineries [122,123]. These collisions are more detrimental when the transcription–replication machineries collide in a head-on configuration [124], while convergent transcription increases R-loop formation, RNA polymerase stalling, and genomic instability in leukemic cells [71,125–127].
In cancer cells, differential RNA expression can affect any class of RNA, including lncRNAs, miRNAs, and mRNAs themselves, which are normally controlled by RNA surveillance. Somatic mutations in NMD factors have been reported, where UPF1 mutants upregulate NMD substrate transcripts in pancreatic adenosquamous carcinoma [128], and in myofibroblastic tumors. In these cancers, upregulated mRNAs, including NF-kB inducing kinase (NIK), induce NF-kB activation and contribute to inflammation [129]. Germline mutations in the Dis3L2 gene have been found in patients with Perlman syndrome and susceptibility to Wilms’ tumors. Loss of exonuclease activity induces mitotic abnormalities and dysregulated expression of mitotic control proteins [130]. Disequilibrium in m6A modifications has important roles, with oncogenic mutations in methyltransferase, demethylase, or reader genes [131]. The demethylase ALKBH5 is necessary for the proliferation of glioblastoma cells by maintaining the levels of certain mRNAs, in particular FOXM1. Interestingly, the specificity of this gene is conferred by FOXM1 antisense RNA, which works as a guide to target its own mRNAs and induce tumorigenicity [132]. Inversely, the FTO eraser is overexpressed in a subtype of acute myeloid leukemia, where it reduces the expression levels of important genes, contributing to both cell transformation and leukemogenesis [133]. In lung adenocarcinoma, ADAR-mediated editing increases the stability of the FAK kinase mRNA, enhancing its expression, and is correlated with cancer invasiveness [134]. RNA editing also contributes to the epitranscriptome diversity of cancer stem cells [135].
Concluding Remarks
Strong evidence shows that RNA surveillance is a key component of physiological development, regulating early stages of embryogenesis, pluripotency, lineage specification, organogenesis, and the balance of stem cell maintenance and differentiation.
At the cellular level, an intriguing concept is the flow and relationship between these different RNA-processing events (see Outstanding Questions). RNA surveillance starts at the chromatin level with the detection and removal of DNA-associated RNAs and, in addition to its participation in genomic stability, such mechanisms could contribute to gene expression regulation, in concert with epigenomic changes. Nuclear and cytoplasmic RNA surveillances then constantly adjust the RNA concentration using various combinations of mechanisms in different cells. RNA modification incorporation and removal are potentially faster events than transcriptional regulation, providing direct dynamic control of RNA levels. Differential marks and complex RNA surveillance pathways commit coding and ncRNAs to their relevant fates, affecting their kinetics, localization, interactions, and functions. The challenge now is to study these RNA surveillance mechanisms in every type of cell and their impact on homeostasis and differentiation. A more systematic analysis of RNA surveillance pathways in pathologies and cancers should also be considered to explain transcriptomic disorders.
Outstanding Questions.
How do RNA surveillance functions specifically regulate distinct steps of cellular differentiation and development by co- and/or post-transcriptionally acting on mRNAs or ncRNAs? Is this specificity provided by a combination of various RNA surveillance mechanisms? How are RNA surveillance mechanisms regulated during development?
How do RNA modifications specifically tag a subset of RNAs, influencing their processing and decay, but not others? How does a given RNA molecule have different marks depending on cell type?
How do the combinations of RNA modifications (RNA epigenetic code) influence RNA processing? What is the kinetics of RNA modifications?
What are the global functions of RNA-editing events? Does RNA editing also impact cell division and differentiation during organism development?
Is piRNA maturation germ cell specific or does it have a role in other cell types? Are piRNA-like pathways activated in cells to prevent the onset of cancer?
Intercellular RNA exchanges are observed both in physiological and pathological conditions. Does RNA surveillance determine intercellular RNA exchange during embryogenesis and organism development?
While recent data suggest a functional role of some TEs, how does RNA surveillance distinguish functional TEs from undesirable and harmful TEs?
The growing field of RNA surveillance provides great opportunities to better understand all these mechanisms, where RNA-associated events control genome stability, modulate epigenomes, transcriptomes, epitranscriptomes, RNA–RNA interactions, and RNA–protein networks, providing a unique universal system to regulate many biological levels of cellular physiology. The resulting strength and efficiency of these elegant mechanisms has been adopted by evolution to fine-tune global RNA expression during development and beyond.
Highlights.
Emerging functions for RNA surveillance pathways demonstrate their important role in physiological processes, from cellular proliferation to differentiation during development, while defects lead to pathologies.
Complex RNA surveillance pathways are working in concert to regulate RNA levels, with some redundancy and specificity depending on cell types.
RNAs are dynamically edited and remodeled by chemical modifications, strongly influencing their half-life, interactions, localizations, and functions, in particular during dynamic processes such as development.
Dysregulation of RNA surveillance pathways is observed in many cancers, leading to overexpression of different classes of coding RNA and ncRNA.
Acknowledgments
We acknowledge the contribution of Gerson Rothschild, Junghyun Lim, and Orianne Debeaupuis for providing input and critically reading this manuscript. We thank Helen Pickersgill and Katrina Woolcock for editing this manuscript for content and overall input. Research in the Basu lab is supported by grants to B.L. (EMBO fellowship, ALTF 906-2015) and U.B. (NIAID1R01AI099195 and Ro1AI134988), Leukemia and Lymphoma Society, and the Pershing Square Sohn Cancer Research Alliance.
Glossary
- Noncoding (nc)RNA
any class of RNA that does not produce a functional mRNA.
- R-loops
DNA:RNA hybrids. These structures are usually created during transcription and RNA polymerase stalling at specific DNA sequences.
- RNA exosome
an evolutionary conserved multiprotein complex with ribonuclease activity. In humans, it comprises 11 subunits, including two catalytic subunits (Exosc10 and Dis3) and has both 3′ to 5′ exoribonuclease and endoribonuclease activities. The RNA exosome interacts with various complexes and helicases, which fine-tune its activity in the nucleus and cytoplasm.
- RNA processing
a set of processes controlling RNA maturation. These include RNA capping, polyadenylation, splicing, editing, RNA modifications, trafficking, localization, or any additional changes. These different steps are crucial for mRNA and gene expression control, as well as for ncRNAs.
- RNA surveillance
set of processes controlling the RNA level. RNA surveillance is complementary to RNA processing and ensures the exact titration of any RNA molecule in both time and space.
References
- 1.Schoenberg DR and Maquat LE (2012) Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet 13, 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopp F and Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X et al. (2017) Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 [DOI] [PubMed] [Google Scholar]
- 4.Houseley J and Tollervey D (2009) The many pathways of RNA degradation. Cell 136, 763–776 [DOI] [PubMed] [Google Scholar]
- 5.Nagarajan VK et al. (2013) XRN 5′ → 3′ exoribonucleases: structure, mechanisms and functions. Biochim. Biophys. Acta 1829, 590–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.vanDijk EL et al. (2011)XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475, 114–117 [DOI] [PubMed] [Google Scholar]
- 7.Hu W et al. (2009) Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malabat C et al. (2015) Quality control of transcription start site selection by nonsense-mediated-mRNA decay. eLife 4, e06722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ustianenko D et al. (2016) TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J. 35, 2179–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilchert C et al. (2016) The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol 17, 227–239 [DOI] [PubMed] [Google Scholar]
- 11.Lim J et al. (2017) Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell 169, 523–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weick E-M et al. (2018) Helicase-dependent RNA decay illuminated by a cryo-EM structure of a human nuclear RNA exosome-MTR4 complex. Cell 173, 1663–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayerle M et al. (2017)Structural toggle in the RNaseH domain of Prp8 helps balance splicing fidelity and catalytic efficiency. Proc. Natl. Acad. Sci. U. S. A 114, 4739–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebreton A et al. (2008) Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456, 993–996 [DOI] [PubMed] [Google Scholar]
- 15.Bernstein E et al. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 [DOI] [PubMed] [Google Scholar]
- 16.Khemici V and Linder P (2018) RNA helicases in RNA decay. Biochem. Soc. Trans 46, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoms M et al. (2015) The exosome is recruited to RNA substrates through specific adaptor proteins. Cell 162, 1029–1038 [DOI] [PubMed] [Google Scholar]
- 18.Nasif S et al. (2018) Beyond quality control: the role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol 75, 78–87 [DOI] [PubMed] [Google Scholar]
- 19.Popp MW and Maquat LE (2016) Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell 165, 1319–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arribere JA and Fire AZ (2018) Nonsense mRNA suppression via nonstop decay. eLife 7, e33292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volanakis A et al. (2013) Spliceosome-mediated decay (SMD) regulates expression of nonintronic genes in budding yeast. Genes Dev 27, 2025–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Middleton R et al. (2017) IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol 18, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong JJ-L et al. (2013) Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154, 583–595 [DOI] [PubMed] [Google Scholar]
- 24.Vanichkina DP et al. (2018) Challenges in defining the role of intron retention in normal biology and disease. Semin. Cell Dev. Biol 75, 40–49 [DOI] [PubMed] [Google Scholar]
- 25.Svoboda P (2014) Renaissance of mammalian endogenous RNAi. FEBS Lett 588, 2550–2556 [DOI] [PubMed] [Google Scholar]
- 26.Elbarbary RA et al. (2017) Tudor-SN-mediated endonucleolytic decay of human cell microRNAs promotes G1/S phase transition. Science 356, 859–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J et al. (2018) Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Mauriño SM et al. (2017) RNA binding protein regulation and cross-talk in the control of AU-rich mRNA Fate. Front. Mol. Biosci 4, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlasova-St Louis I and Bohjanen PR (2017) Post-transcriptional regulation of cytokine and growth factor signaling in cancer. Cytokine Growth Factor Rev 33, 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wanke KA et al. (2018) Understanding neurodevelopmental disorders: the promise of regulatory variation in the 3′ UTRome. Biol. Psychiatry 83, 548–557 [DOI] [PubMed] [Google Scholar]
- 31.Song C et al. (2016) Functions of the RNA editing enzyme ADAR1 and their relevance to human diseases. Genes 7, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan MH et al. (2017) Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang B et al. (2017) APOBEC: from mutator to editor. J. Genet. Genomics 44, 423–437 [DOI] [PubMed] [Google Scholar]
- 34.Harcourt EM et al. (2017) Chemical and structural effects of base modifications in messenger RNA. Nature 541, 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roundtree IA et al. (2017) Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Almeida C et al. (2017) RNA uridylation: a key posttranscriptional modification shaping the coding and noncoding transcriptome. Wiley Interdiscip. Rev 9, e1440. [DOI] [PubMed] [Google Scholar]
- 37.Turner M and Díaz-Muñoz MD (2018) RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol 19, 120–129 [DOI] [PubMed] [Google Scholar]
- 38.Ye J and Blelloch R (2014) Regulation of pluripotency by RNA binding proteins. Cell Stem Cell 15, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castello A et al. (2013) RNA-binding proteins in Mendelian disease. Trends Genet 29, 318–327 [DOI] [PubMed] [Google Scholar]
- 40.Aw JGA et al. (2016) In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell 62, 603–617 [DOI] [PubMed] [Google Scholar]
- 41.Morgan M et al. (2017) mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 548, 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao BS et al. (2017) m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanova I et al. (2017) The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Q et al. (2016) Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv 2, e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semotok JL et al. (2005) Smaug recruits the CCR4/POP2/ NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol 15, 284–294 [DOI] [PubMed] [Google Scholar]
- 46.Rouget C et al. (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berrens RV et al. (2017) An endosiRNA-based repression mechanism counteracts transposon activation during global DNA demethylation in embryonic stem cells. Cell Stem Cell 21, 694–703.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Percharde M et al. (2018) A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174, 391–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sigova AA et al. (2013) Divergent transcription of long non-coding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci 110, 2876–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blinka S et al. (2016) Super-enhancers at the Nanog locus differentially regulate neighboring pluripotency-associated genes. Cell Rep 17, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cinghu S et al. (2017) Intragenic enhancers attenuate host gene expression. Mol. Cell 68, 104–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguilo F et al. (2015)Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17, 689–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou C et al. (2017) Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep 20, 2262–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen T et al. (2015) m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16, 289–301 [DOI] [PubMed] [Google Scholar]
- 55.Smith KN et al. (2017) Long noncoding RNA moderates microRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Rep 9, 108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleaveland B et al. (2018) A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174, 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng X et al. (2016) CNOT3-dependent mRNA deadenylation safeguards the pluripotent state. Stem Cell Rep 7, 897–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lou C-H et al. (2016) Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Rep 6, 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skamagki M et al. (2017) RNA exosome complex-mediated control of redox status in pluripotent stem cells. Stem Cell Rep 9, 1053–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aguilo F and Walsh MJ (2017) The N6-methyladenosine RNA modification in pluripotency and reprogramming. Curr. Opin. Genet. Dev 46, 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang C et al. (2017) m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 [DOI] [PubMed] [Google Scholar]
- 62.Weng H et al. (2018) METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 22, 191–205.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vu LP et al. (2017) The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med 23, 1369–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guzzi N et al. (2018) Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26 [DOI] [PubMed] [Google Scholar]
- 65.McIver SC et al. (2014) The exosome complex establishes a barricade to erythroid maturation. Blood 124, 2285–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McIver SC et al. (2016) Exosome complex orchestrates developmental signaling to balance proliferation and differentiation during erythropoiesis. eLife 5, e17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Q et al. (2018) Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 554, 123–127 [DOI] [PubMed] [Google Scholar]
- 68.Li H-B et al. (2017) m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shinkura R et al. (2007) Regulation of AID function in vivo. Adv. Exp. Med. Biol 596, 71–81 [DOI] [PubMed] [Google Scholar]
- 70.Hwang JK et al. (2015) Related mechanisms of antibody somatic hypermutation and class switch recombination. Microbiol. Spectr 3, MDNA3–0037-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pefanis E et al. (2014) Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Basu U et al. (2011)The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tinguely A et al. (2012) Cross talk between immunoglobulin heavy-chain transcription and RNA surveillance during B cell development. Mol. Cell. Biol 32, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ju Z et al. (2011) Interaction between the immunoglobulin heavy chain 3′ regulatory region and the IgH transcription unit during B cell differentiation. Mol. Immunol 49, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pefanis E et al. (2015) RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lence T et al. (2016) m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 [DOI] [PubMed] [Google Scholar]
- 77.Yoon K-J et al. (2017)Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171, 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang T et al. (2016) Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci 19, 1093–1099 [DOI] [PubMed] [Google Scholar]
- 79.Bedrosian TA et al. (2018) Early life experience drives structural variation of neural genomes in mice. Science 359, 1395–1399 [DOI] [PubMed] [Google Scholar]
- 80.Gill J et al. (2017) Regulated intron removal integrates motivational state and experience. Cell 169, 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Engel M et al. (2018) The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kudou K et al. (2017) The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biol 7, 170119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feng Q et al. (2017) The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Mol. Cell 67, 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chenette DM et al. (2016) Targeted mRNA decay by RNA binding protein AUF1 regulates adult muscle stem cell fate, promoting skeletal muscle integrity. Cell Rep 16, 1379–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fernández-Pérez D et al. (2018) MicroRNA dynamics at the onset of primordial germ and somatic cell sex differentiation during mouse embryonic gonad development. RNA 24, 287–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strome S and Updike D (2015) Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol 16, 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shichino Y et al. (2018) YTH-RNA-binding protein prevents deleterious expression of meiotic proteins by tethering their mRNAs to nuclear foci. eLife 7, e32155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bailey AS et al. (2017) The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6, e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stein P et al. (2015) Essential role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet 11, e1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akpinar M et al. (2017) TDRD6 mediates early steps of spliceosome maturation in primary spermatocytes. PLoS Genet 13, e1006660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang C et al. (2017) ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. PNAS 115, E325–E333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bao J et al. (2016) UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′ UTR transcripts. PLoS Genet 12, e1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jamin SP et al. (2017) EXOSC10/Rrp6 is post-translationally regulated in male germ cells and controls the onset of spermatogenesis. Sci. Rep 7, 15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamaji M et al. (2017) DND1 maintains germline stem cells via recruitment of the CCR4–NOT complex to target mRNAs. Nature 543, 568–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin Z et al. (2017) Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res 27, 1216–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rice GI et al. (2012) Mutations in ADAR1 cause Aicardi–Goutières syndrome associated with a type I interferon signature. Nat Genet 44, 1243–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liddicoat BJ et al. (2015) RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmad S et al. (2018) Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mackenzie KJ et al. (2016) Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 35, 831–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Günther C et al. (2015) Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest 125, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thomas CA et al. (2017) Modeling of TREX1-dependent auto-immune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell 21,319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee-Kirsch MA et al. (2007) Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet 39, 1065–1067 [DOI] [PubMed] [Google Scholar]
- 103.Yuan F et al. (2015) Human DNA exonuclease TREX1 is also an exoribonuclease that acts on single-stranded RNA. J. Biol. Chem 290, 13344–13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dhir A et al. (2018) Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morton DJ et al. (2018) The RNA exosome and RNA exosome-linked disease. RNA 24, 127–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lionetti M et al. (2015) A compendium of DIS3 mutations and associated transcriptional signatures in plasma cell dyscrasias. Oncotarget 6, 26129–26141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shamay-Ramot A et al. (2015) Fmrp interacts with Adar and regulates RNA editing, synaptic density and locomotor activity in zebrafish. PLoS Genet 11, e1005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huntley MA et al. (2016) Complex regulation of ADAR-mediated RNA-editing across tissues. BMC Genomics 17, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goodwin M et al. (2015) MBNL sequestration by toxic RNAs and RNA misprocessing in the myotonic dystrophy brain. Cell Rep 12, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wan J et al. (2012) Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet 44, 704–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boczonadi V et al. (2014) EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun 5, 4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Di Donato N et al. (2016) Mutations in EXOSC2 are associated with a novel syndrome characterised by retinitis pigmentosa, progressive hearing loss, premature ageing, short stature, mild intellectual disability and distinctive gestalt. J. Med. Genet 53, 419–425 [DOI] [PubMed] [Google Scholar]
- 113.Giunta M et al. (2016) Altered RNA metabolism due to a homozygous RBM7 mutation in a patient with spinal motor neuropathy. Hum. Mol. Genet 25, 2985–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Krug L et al. (2017) Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet 13, e1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Poulos MG et al. (2013) Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice. Hum. Mol. Genet 22, 3547–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thomas JD et al. (2017) Disrupted prenatal RNA processing and myogenesis in congenital myotonic dystrophy. Genes Dev 31, 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sato Y et al. (2010) Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J. Biol. Chem 285, 7111–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chédin F (2016) Nascent connections: R-loops and chromatin patterning. Trends Genet 32, 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Richard P and Manley JL (2017) R loops and links to human disease. J. Mol. Biol 429, 3168–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaillard H and Aguilera A (2016) Transcription as a threat to genome integrity. Annu. Rev. Biochem 85, 291–317 [DOI] [PubMed] [Google Scholar]
- 121.Sollier J et al. (2014) Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.García-Muse T and Aguilera A (2016) Transcription-replication conflicts: how they occur and how they are resolved. Nat. Rev. Mol. Cell Biol 17, 553–563 [DOI] [PubMed] [Google Scholar]
- 123.Hamperl S and Cimprich KA (2016) Conflict resolution in the genome: how transcription and replication make it work. Cell 167, 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hamperl S et al. (2017)Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170, 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meng F-L et al. (2014) Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell 159, 1538–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pannunzio NR and Lieber MR (2016) Dissecting the roles of divergent and convergent transcription in chromosome instability. Cell Rep 14, 1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Heinäniemi M et al. (2016) Transcription-coupled genetic instability marks acute lymphoblastic leukemia structural variation hotspots. eLife 5, e13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu C et al. (2014) The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat. Med 20, 596–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu J et al. (2016) The nonsense-mediated RNA decay pathway is disrupted in inflammatory myofibroblastic tumors. J. Clin. Invest 126, 3058–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Astuti D et al. (2012) Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat. Genet 44, 277–284 [DOI] [PubMed] [Google Scholar]
- 131.Dai D et al. (2018) N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis 9, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang S et al. (2017) m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Z et al. (2017) FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Amin EM et al. (2017) The RNA-editing enzyme ADAR promotes lung adenocarcinoma migration and invasion by stabilizing FAK. Sci. Signal 10, eaah3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jiang Q et al. (2017) RNA editing-dependent epitranscriptome diversity in cancer stem cells. Nat. Rev. Cancer 17, 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Haradhvala NJ et al. (2016) Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 164, 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fagegaltier D et al. (2016) Oncogenic transformation of Drosophila somatic cells induces a functional piRNA pathway. Genes Dev 30, 1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Khabar KS (2017) Hallmarks of cancer and AU-rich elements. Wiley Interdiscip. Rev. RNA 8, e1368. [DOI] [PMC free article] [PubMed] [Google Scholar]