Abstract
Proteomic and imaging markers have been widely studied as potential biomarkers for diagnosis, monitoring and prognosis of Alzheimer’s disease. In this study, we used Alzheimer Disease Neuroimaging Initiative dataset and performed parallel independent component analysis on cross sectional and longitudinal proteomic and imaging data in order to identify the best proteomic model for diagnosis, monitoring and prediction of Alzheimer disease (AD).
We used plasma proteins measurement and imaging data from AD and healthy controls (HC) at the baseline and 1 year follow-up. Group comparisons at baseline and changes over 1 year were calculated for proteomic and imaging data. The results were fed into parallel independent component analysis in order to identify proteins that were associated with structural brain changes cross sectionally and longitudinally. Regression model was used to find the best model that can discriminate AD from HC, monitor AD and to predict MCI converters from non-converters.
We showed that five proteins are associated with structural brain changes in the brain. These proteins could discriminate AD from HC with 57% specificity and 89% sensitivity. Four proteins whose change over 1 year were associated with brain structural changes could discriminate AD from HC with sensitivity of 93%, and specificity of 92%. This model predicted MCI conversion to AD in 2 years with 94% accuracy. This model has the highest accuracy in prediction of MCI conversion to AD within the ADNI-1 dataset. This study shows that combination of selected plasma protein levels and MR imaging is a useful method in identifying potential biomarker.
Keywords: Alzheimer’s Disease, Tensor Based Morphometry, Proteomics, Magnetic Resonance Imaging, Biomarkers, Parallel ICA
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disorder. Emerging diagnostic techniques using multiple imaging modalities have contributed tremendously to our knowledge of the disease in recent years (Perrin et al., 2009), but AD’s pathophysiology still remains largely elusive. Emerging disease-modifying strategies for AD necessitate accurate biomarkers for early diagnosis, monitoring and prognosis more than ever before. Magnetic resonance imaging (MRI) measures have been shown to predict conversion of mild cognitive impairment (MCI) to AD (deToledo-Morrell et al., 2004; Jack et al., 1999; Risacher et al., 2010; Whitwell et al., 2008) and cognitive decline in elderly (Mungas et al., 2002; Rusinek et al., 2003). Similarly proteomic indices have been shown to have diagnostic value in AD (Britschgi and Wyss-Coray, 2009; Hye et al., 2006; Ray et al., 2007) and predict MCI conversion to AD (Ray et al., 2007). There are potential advantages of proteomics over imaging as biomarkers in AD. For example, a recent study on familial AD estimated that changes in Aβ amyloid and tau protein levels may occur 5 years before brain atrophy is detectable in MRI (Bateman, 2012). A group of investigators recently used multivariate approach for analysing proteomic profile in Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset and found that apoE, B-type natriuretic peptide, C-reactive protein, and pancreatic polypeptide are significantly different between MCI and AD group (Hu et al., 2012). They also showed that CSF Aβ42 levels and t-tau/Aβ42 ratios correlated with the number of apoE4 alleles, plasma levels of B-type natriuretic peptide and pancreatic polypeptide (Hu et al., 2012). However it is not clear if any of these proteins are associated with changes in the brain, as they did not use any imaging data in their analysis.
Combining imaging markers with proteomics (i.e. Imaging Proteomics) provides an opportunity to identify proteins that are specifically associated with changes in the brain (Mattay et al., 2008; Meyer-Lindenberg and Weinberger, 2006). In addition, this approach may provide further mechanistic insights into pathophysiological mechanisms of AD, which in turn could serve as new targets for therapeutic strategies (Britschgi and Wyss-Coray, 2009).
Data mining in a large dataset with many different variables is prone to the problem of over fitting, and is all the more challenging due to the within-modality correlations with mass univariate techniques (Pearlson, 2009). Simply reducing the number of features considered does not necessarily address the problem, as the relationship between variables may also provide crucial information; for example, the discovery of Metabolic Syndrome X (Eckel et al., 2005) was based on a set of closely correlated physiological variables.
Parallel independent component analysis (PICA) greatly diminishes this methodological problem (Liu et al., 2008). PICA is an unsupervised multivariate algorithm to extract independent within-modality patterns with strongest between-modality connections when more than one modality is available. The intermodal associations between resulting components are then further identified and quantified. These components can be later compared on a component-wise basis between experimental groups. PICA has been applied widely in neurosciences research such as assessing relationships between electroencephalography (EEG), structural and functional MRI (fMRI), with single-nucleotide polymorphism (SNP) array (Jagannathan et al., 2010; Liu et al., 2009a; Liu et al., 2009b), EEG with fMRI (Wu et al., 2010) and positron emission tomography (PET) with structural MRI (Tosun et al., 2011). Here, we used PICA to explore the relationship between structural MRI data and proteomics in ADNI dataset. The goal was to determine proteins that were associated with brain structural changes and therefore could have potential value as biomarkers for diagnosis and monitoring AD as well as predicting MCI conversion to AD.
Methods
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative database (adni.loni.ucla.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org. Here, we used ADNI-1 and ADNI Plasma QC Multiplex Data.
Subjects
The eligibility criteria for the inclusion of participants are described at: http://adni.loni.ucla.edu/wp-content/uploads/2010/09/ADNI_GeneralProceduresManual.pdf. Subjects are divided into the following groups:
Healthy Controls (HC): Mini-Mental State Examination (MMSE) scores between 24-30, Clinical Dementia Rating (CDR) of 0, non-depressed and non-MCI.
MCI subjects: MMSE scores between 24-30, with memory complaint, objective memory loss measured by education adjusted scores on Wechsler Memory Scale Logical Memory II, a CDR of 0.5, no significant levels of impairment in other cognitive domains, and preserved activities of daily living.
Mild AD subjects: MMSE scores between 20-26, CDR of 0.5-1.0, meeting NINCDS/ADRDA criteria for probable AD (McKhann et al., 1984).
In this study, we chose AD and HC subjects from ADNI database who had baseline and 12-month-follow-up plasma proteins measurement and had pre-processed quality checked structural MRI data. For the MCI patients, only those who had baseline plasma proteins levels and had been followed up for at least two years were selected (n=300), of whom 110 subjects converted to AD during the follow up period.
Targeted Multiplex Proteomics
Procedure of plasma protein data collection and measurement is explained in detail elsewhere (http://adni.loni.ucla.edu/wp-content/uploads/2010/11/BC_Plasma_Proteomics_Data_Primer.pdf). In brief, plasma proteins were measured in a subset of EDTA plasma samples (obtained in the morning following an overnight fast) at baseline and 1 year follow up, using a 190 analyte multiplex immunoassay panel. The panel, referred to as the human discovery map, was developed on the Luminex xMAP platform by Rules-Based Medicine (RBM) to contain proteins previously reported in the literature to be altered as a result of cancer, cardiovascular disease, metabolic disorders, and inflammation. In addition, RBM partnered with Satoris (Inc., California, USA) to include plasma proteins previously reported to change in patients with Alzheimer’s disease (Ray et al., 2007). Assays have been qualified based on the least detectable dose, precision, cross-reactivity, dilutional linearity, and spike recovery. Results of analyses on 148 analytes, which passed quality control, were used in this study.
MRI Acquisition and Preprocessing
High-resolution T1-weighted MRI scans were acquired on 1.5 Tesla MRI scanners from Siemens, General Electric Healthcare, and Philips Medical Systems with the standard ADNI MRI protocol (Jack et al., 2008). Each subject was scanned with a sagittal 3D MP-RAGE sequence, with acquisition parameters: inversion time (TI)/repetition time (TR): 1000/2400 ms; flip angle: 8°; 24 cm field of view; 192×192×166 acquisition matrix, and a voxel size of 1.25× 1.25×1.2 mm3. In plane, zero-filled reconstruction yielded a 256×256 matrix for a reconstructed voxel size of 0.94×0.94×1.20 mm3, later reconstructed to 1 mm isotropic voxels. The scan quality was evaluated by the ADNI MRI quality control centre at the Mayo Clinic following standardized criteria. Images were calibrated with phantom-based geometric corrections to ensure consistency among scans acquired at different sites. Image corrections were applied using a processing pipeline at the Mayo Clinic, consisting of: (1) correction of geometric distortion due to gradient non-linearity (Jovicich et al., 2006), i.e., “gradwarp”, (2) “B1 correction” for adjustment of image intensity inhomogeneity due to B1 non-uniformity (Jack et al., 2008), (3) “N3” bias field correction for reducing residual intensity inhomogeneity (Sled et al., 1998), and (4) geometrical scaling for removing scanner and potential session specific calibration errors using a phantom scan acquired for each subject (Gunter et al., 2009). All original image files as well as images with all of these corrections are available to the general scientific community at http://www.loni.ucla.edu/ADNI/Data/.
Voxel-Based Morphometry
For the cross-sectional (baseline) study, data pre-processing and voxel-based morphometry (VBM) were performed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm) and MATLAB v7.11 (Math-Works, Natick, MA, USA). First, tissue classification was carried out on MR images to separate gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) using the standard unified segmentation model in SPM8 (Ashburner and Friston, 2005). Second, GM population templates were generated from the entire image dataset using the diffeomorphic anatomical registration using exponentiated Lie algebra (DARTEL) technique (Ashburner, 2007). Third, after an initial affine registration of the GM DARTEL templates to the tissue probability maps in Montreal Neurological Institute (MNI) space (http://www.mni.mcgill.ca/), non-linear warping of GM images was performed to the DARTEL GM template in MNI space. Fourth, images were then modulated to ensure that relative volumes of GM were preserved following the spatial normalization procedure. Finally, images were smoothed with an 8 mm full width at half maximum Gaussian kernel. To identify group differences in the modulated GM a two-sample t-test was used at each voxel. To find differences that were spatially extended, we used an inference method on the t-test images called Threshold Free Cluster Enhancement (TFCE) (Smith and Nichols, 2009). TFCE is an extension of cluster-wise inference that does not depend on a cluster-forming threshold. The risk of false positives over the brain was controlled with the family wise error rate (5% level) using a permutation method with 5000 permutations (Nichols and Holmes, 2002), (FSL version 4.1.9, http://www.fmrib.ox.ac.uk/fsl/). A binary mask was created from voxels showing significant between group differences. Pre-processed GM images and the resulting masks were used as inputs for parallel ICA.
Tensor-Based Morphometry
Tensor based morphometry (TBM) was used to assess changes over time i.e. for longitudinal analysis mainly using SPM8 software package. Pre-processing stages for TBM have been described in detail elsewhere (Kipps et al., 2005). Briefly, the follow up T1-weighted MRI image scan was co-registered with the baseline image. A high-dimensional deformation field was used to warp the corrected late image to match the baseline image for each subject (Ashburner and Friston, 2000). The amount of volume change was quantified by taking the determinant of the gradient of deformation at the single-voxel level (Jacobian determinants). The following formula was applied to the segmented gray matter image obtained from the first scan and the Jacobian determinant map: (Jacobian value−1) × GM. The resulting product image represented a measure of the gray matter specific volume change between the baseline and follow up scans. The normalization parameters were estimated by matching the customized gray matter template that was created earlier with the segmented gray matter image from the first scan and were then applied to the product image (Ashburner and Friston, 1999). Normalized images were smoothed using an 8 mm isotropic Gaussian kernel. Finally, the smoothed images were multiplied by an inclusive gray matter mask. The value of each voxel in pre-processed image shows volume loss or expansion during 1-year follow up. Statistical analysis was performed using permutation testing as described above. Binary mask was created from voxels showing significant group difference (AD<HC) in the pre-processed images (p < 0.05 FWE-corrected) during one year. The resulting binary mask and pre-processed images from one year longitudinal TBM were used as inputs for monitoring disease progression (see below).
Parallel Independent Component Analysis (PICA)
We used PICA to explore the relationship between plasma proteins and regional brain atrophy simultaneously. The details of PICA has been described elsewhere (Liu et al., 2009b). Briefly ICA estimates sets of variables (components) that are maximally independent; specifically, it minimizes the mutual information between pairs of components. PICA, for two modalities, simultaneously optimizes the independence of components in each modality and the similarity of subject weights between the modalities. Thus two sets of components are produced; one for each modality, but a subset of the components will be defined by subject weights that are similar between the two modalities (Liu et al., 2009b). The number of components in each analysis was estimated by Akaike Information Criterion and Minimum Distance Length (Calhoun et al., 2001). All of the components were normalized to Z score to remove scaling and thresholded at z > 1.7 for visualization purposes. The Pearson correlation coefficients were computed for every possible pair of subject weights between the two modalities, and significant relationships determined using false discovery rate (PFDR < 0.05).
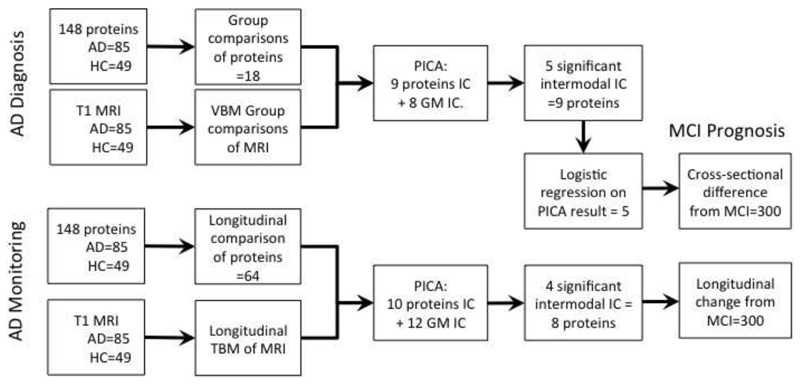
Following steps of analyses (see Figure 1) were carried out:
-
1-
To identify proteomic makers that may have potential value as biomarkers for diagnosis of AD, we first used a two-sample t-test to find significant differences (increase or decrease) in plasma protein concentration between AD and HC groups as well as in their GM density separately. Then PICA was run on those proteins and brain regions that showed significant differences (increase or decrease) between groups (after correction for multiple comparisons, FWE for images, and FDR for proteomics. The result provides proteins that are closely associated with modulated GM at the baseline.
-
2-
To identify proteins that may have potential value as biomarkers for monitoring AD progression, we first calculated TBM maps of AD and HC subjects over one year showing longitudinal structural brain changes. We then calculated plasma concentration difference (increase or decrease) of the proteins for each individual over one year. A two-sample t-test on the longitudinal protein concentration changes and TBM results to identify between groups difference (corrected with FWE, p<0.05). Finally PICA was run on the TBM and protein changes (increase or decrease) over 1 year in AD subjects only. This analysis identified proteins whose changes over one year correlated with the structural changes of the brain in AD.
-
3-
To identify proteins that may have potential value as biomarkers to determine the prognosis of MCI, we first carried out logistic regression (using stepwise forward Wald method) on the resulted proteomic factors of the cross-sectional analysis and corrected that for age and sex, to develop the best model for discriminating AD from HC participants. The rational for this model was that proteins that strongly correlated with brain structural changes were likely to be useful potential biomarkers for diagnosis of AD. In addition, protein changes might precede brain structural changes or/and cognitive changes. Also, we aimed to produce a totally data-driven model rather than using a priori. A similar approach was applied to the longitudinal data. We used this model to identify MCI subjects who converted to AD. Receiver operating characteristic (ROC) curves was graphed and areas under the curves (AUC) along with its 95% confidence intervals (CI) were computed. Note that MCI subjects were not used for developing the “predictive” model to obtain unbiased estimates of MCI conversion accuracy.
Fig.1.
Schematic representation of steps of data analysis.
Results
Data from total of 49 HC, 300 MCI (of which 110 converted to AD within 2 years), and 85 AD patients were analysed. Participants’ demographic and cognitive data are summarized in Table 1. Schematic presentation of steps of analysis and the results is shown in Figure 1. Plasma level of 18 proteins that showed cross sectional group difference at the baseline and 64 proteins that showed significant longitudinal difference between groups in a year are shown in Table S1, S2, S3, S4 (supplementary data).
Table 1. Demographic and cognitive characterestics of subjects.
| Groups | HC | MCI | AD | F/χ2 (p-values) |
| Number | 49 | 300 | 85 | |
| Age | 75.1 (5.7) | 74.7 (7.2) | 75.2 (7.8) | 0.171(0.84) |
| Percent Female | 44.9% | 35.3% | 44.7% | 3.73(0.15) |
| Education (yrs) | 15.5 (2.9) | 15.8 (2.9) | 15.1 (3.1) | 1.41 (0.24) |
| MMSE | 29.0 (1.2) | 27.1 (1.7) | 23.5 (1.9) | 196 (<0.001) |
| ADAS-Cog | 9.7 (4.3) | 18.2 (6.3) | 28.7(7.6) | 151(<0.001) |
HC=Healthy Control; MCI=Mild Cognitive Impairment; AD=Alzheimer’s disease; MMSE=Mini–mental state examination; ADAS-Cog=Alzheimer's Disease Assessment Scale-cognitive subscale.
Cross-Sectional Analysis at Baseline
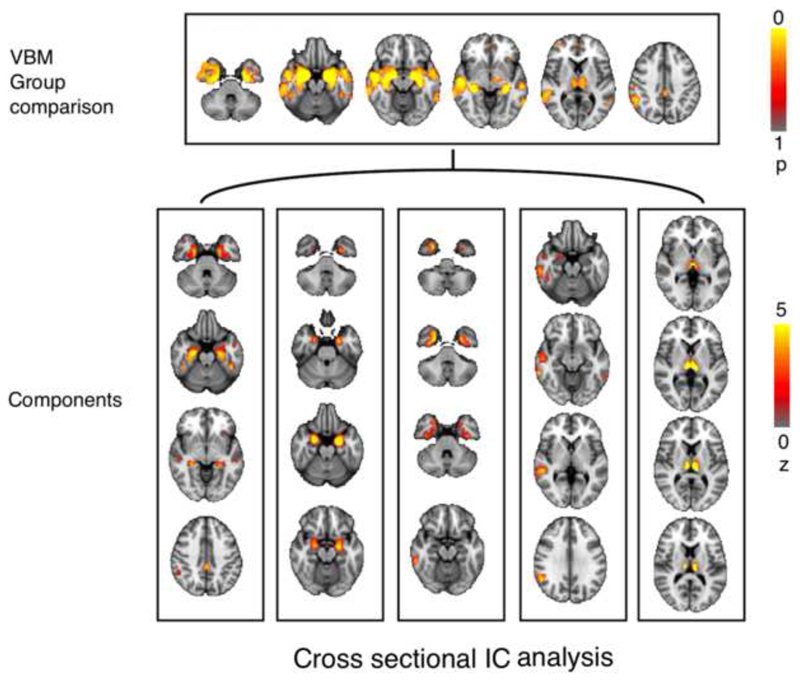
Eighteen proteins exhibited significant differences (p < 0.05, FDR-corrected) between AD and HC subjects (Table S1). The cross-sectional VBM analysis found significantly reduced (p<0.05, FWE-corrected) gray matter in AD subjects relative to HC in several brain regions, including hippocampus, amygdala, thalamic nuclei, posterior cingulate-precuneus junctions, inferior, middle and superior temporal gyri, parahippocampal/fusiform cortices and right angular gyrus (Figure 2). PICA showed that 8 imaging components (components that were resulted from running ICA on imaging data) associated with 9 protein components (Table S2). Of the 8×9=72 possible intermodal pairwise correlations, 5 components had statistically significant relationship (PFDR < 0.05) (Figure 2). Modulated GM in hippocampus, amygdala and several other temporal lobe structures along with posterior cingulate cortex and thalamus was associated with decreased interleukin-16 (IL-16), and apolipoproteinE (apoE) as well as increased thyroxin-binding globulin (TBG), and alpha-2-macroglobulin levels. Modulated GM in right inferior and middle temporal gyri was associated with increased brain natriuretic peptide (BNP), pancreatic polypeptide (PPP), and peptide YY (PYY) concentrations. Thalamic GM was significantly correlated with decreased IgM, as well as increased Eotaxin-3, and PYY levels. Among the 5 significant intermodal components, a total of 9 proteins had significant Z-scores (PFDR < 0.05). The proteins and brain regions in these components are summarized in Table 2.
Fig 2.
Results of VBM group comparison (top) using t-test corrected for multiple comparison using TFC thresholded at p>0.05. Bottom shows the result of parallel independent component analysis at the baseline thresholded at z>1.7.
Table 2. Plasma proteins and structural MRI components.
| Study | Plasma Proteins | Z | Prot. IC | sMRI IC | Brain Regions (|z|>2.3) | r (p) |
|---|---|---|---|---|---|---|
| Cross-sectional | Interleukin-16 | 2.8 | A | A | Hippocampus, Posterior Cingulate, Inferior and Middle Temporal Gyrus, Thalamic nuclei | 0.41 (<0.0001) |
| Thyroxine-Binding Globulin | 1.7 | B | Amygdala, Anterior Hippocampus, Temporal Fusiform Cortex | 0.30 (0.002) |
||
| Alpha-2-Macroglobulin | 1.7 | |||||
| Apolipoprotein E | 1.7 | C | Fusiform Cortices, parahippocampal gyri, Right Inferior Temporal gyrus | 0.22 (0.02) |
||
| Brain Natriuretic Peptide | 2.6 | B | D | Right Inferior and middle Temporal Gyrus | -0.22 (0.02) |
|
| Pancreatic Polypeptide | 2.1 | |||||
| Peptide YY | -2.0 | |||||
| IgM | 2.0 | C | E | Thalamic Nuclei | -0.22 (0.02) |
|
| Eotaxin-3 | 1.7 | |||||
| Peptide YY | -1.6 | |||||
| Longitudinal | Bone Morphogenetic Protein 6 | 4.0 | A | A | Right fusiform cortex | -0.30 (0.04) |
| Matrix Metalloproteinase-9 | 3.1 | |||||
| Neuronal Cell Adhesion Molecule | 3.1 | |||||
| Fibroblast Growth Factor 4 | 2.5 | |||||
| Matrix Metalloproteinase-10 | -2.4 | |||||
| Superoxide Dismutase 1-Soluble | 1.7 | |||||
| von Willebrand Factor | 6.9 | B | B | Cingulate Gyrus | 0.30 (0.04) |
|
| C | Left fusiform cortex | -0.27 (0.04) |
||||
| E-selectin | 7.8 | C | D | Amygdala | 0.26 (0.04) |
Longitudinal Analysis
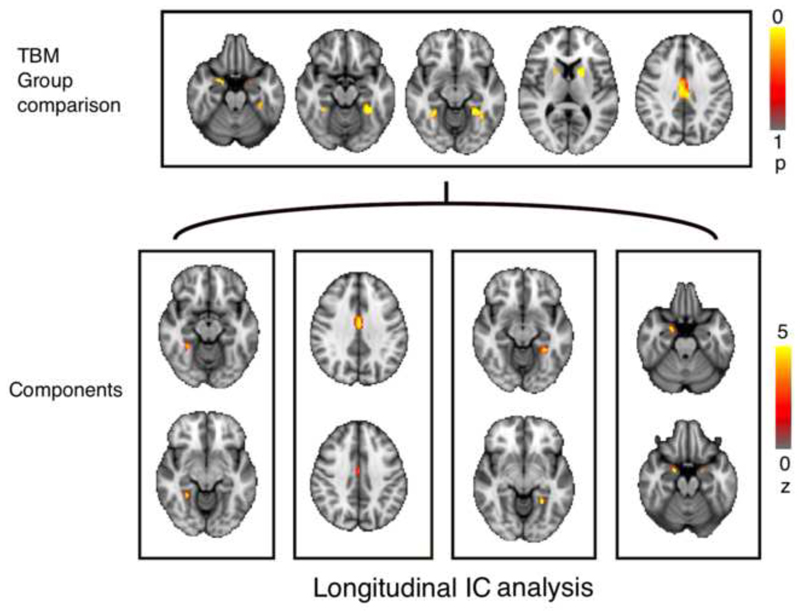
sixty-four proteins showed significant difference in their changes over 1 year in AD when compared with controls (PFDR < 0.05) (Table S1). Similarly, caudate, putamen, amygdala, fusiform cortex, posterior cingulate, lingual, and parahippocampal gyri, and right anterior cingulate and right middle temporal gyri showed significantly (PFWE< 0.05) more gray matter loss in AD than healthy subjects over 1 year follow up (Figure 3). PICA on AD subjects (n=85) identified 10 proteomic components associated with 12 structural imaging components (Table S1, S3 and S5). Of the 10×12=120 possible intermodal pairwise correlations, 4 components were significant (PFDR < 0.05) (Figure 3). In AD, right fusiform cortex atrophy rate was associated with decreased bone morphogenetic protein 6 (BMP6), and fibroblast growth factor 4 (FGF4), while these factors increased in healthy controls. Right fusiform atrophy was also associated with less reduction in AD (compared to normal reduction) of neuronal cell adhesion molecule (NCAM), and matrix metalloproteinase-10 (MMP-10). In addition, increased matrix metalloproteinase-9 (MMP-9), and superoxide dismutase 1- soluble (SOD-1) levels were associated with right fusiform atrophy. Cingulate gyrus and left fusiform cortex atrophy rate were associated with more reduction (compared with normal reduction) of von willebrand factor (vWF) level. Atrophy rate of amygdala was associated with reduced E-selectin level (Table 2). Among the 4 significant intermodal components, a total of 8 proteins had significant Z-scores significant (PFDR < 0.05).
Fig 3.
Results of TBM group comparison (top) using using t-test corrected for multiple comparison using TFC thresholded at p>0.05. Bottom shows the result of parallel independent component analysis over 1 year thresholded at z>1.7.
Diagnostic and Prognostic Utility of the Potential Biomarkers
The 9 proteins, which significantly correlated with regional brain atrophy at the baseline, were entered into the stepwise logistic regression and 5 proteins were found to be capable of meaningfully classifying AD from control (Table 3). This model discriminated AD from normal subjects with specificity of 62% and sensitivity of 93% (AUC=0.917, CI=[0.86,0.96], SD=0.02, p<0.0001). Cross validation using discriminant analysis with leave one out (DALOO) showed sensitivity of 89.4%, and specificity of 57%. When this model was applied on MCI patients (n=342) at the baseline, 64.5% (40 out of 62) of converters to AD were correctly identified.
Table 3. Comparing the results of different logistic regressions.
|
1st Logistic regression |
2nd Logistic regression |
3rd Logistic regression |
|
|---|---|---|---|
| Diagnostic | A2Macro | BNP | BNP |
| Angiotensinogen | IL16 | IL16 | |
| Apo AII | PYY | PYY | |
| Apo B | TBG | TBG | |
| Apo E | APO E | APO E | |
| Calcitonin | PLGF | ||
| CK-MB | SGOT | ||
| Factor VII | |||
| FASLG Receptor | |||
| FSH | |||
| HGF | |||
| IL16 | |||
| Interferon gamma | |||
| Induced Protein 10 | |||
| MCP2 | |||
| MMP9 | |||
| PLGF | |||
| PYY | |||
| Resistin | |||
| SGOT | |||
| Vitronectin | |||
| Sens., Spec. (DALOO) |
100%, 100% (89.2%, 79%) |
94.1%, 81.6% (91.8%, 81.6%) |
92.9, 62.4% (89.4%, 57%) |
| Monitoring | BMP6 | BMP6 | BMP6 |
| B2M | Eselectin | Eselectin | |
| CLU | MMP10 | MMP10 | |
| MMP10 | NrCAM | NrCAM | |
| NrCAM | Leptin | ||
| PLGF | PLGF | ||
| TFF3 | TFF3 | ||
| Thrombospondin1 | VCAM | ||
| Sens., Spec. (DALOO) |
100%, 100% (94.1%, 75.5%) |
100%, 100% (91.8%, 93.9%) |
92.9%, 69.4% (92.9%, 91.8%) |
1st Logistic regression on 148 proteins.
2nd Logistic regression on proteins that showed significant difference between groups (18 in cross sectional and 64 in longitudinal analysis). (no MRI data used)
3rd Logistic regression on proteins identified from running PICA on the result of 2nd logistic regression and between group comparisons of VBM (9 in cross sectional and 8 in longitudinal).
In addition, the four proteins whose changes over time correlated with structural brain changes were used as a model to discriminate AD from control. This model had 92.9% sensitivity and 69.4% specificity. Cross validation of this model using DALOO showed sensitivity of 92.9%, and specificity of 91.8%. We, then, used this model to predict MCI converters and found that the model correctly identified 93.5% (58 out of 62) of the converters. When ApoE (a well known risk factor for AD) was removed from the model, the sensitivity and specificity of the model for discriminating AD from control dropped to %85 and %50 respectively and the predictive power of MCI convertors to AD fell to 56%. Adding age and sex to the model did not affect the result significantly.
Discussion
Protein changes at baseline
There are differences in GM in AD patients relative to controls in hippocampus, amygdala, thalamic nuclei, posterior cingulate-precuneus junctions, inferior, middle and superior temporal gyri, parahippocampal/fusiform cortices and right angular gyrus. This is consistent with previous studies (Echavarri et al., 2011; Karas et al., 2003; Shiino et al., 2006; Singh et al., 2006). We also found that plasma levels of 18 proteins were significantly changed in AD subjects when compared with control group. Among these proteins the levels of IL-16, TBG, α2-Macroglobulin, apoE, BNP, PPP, PYY, IgM, and Eotaxin-3 were associated with regional brain atrophy at baseline. Current biological evidence supports relevance of these proteins to pathophysiology of Alzheimer disease. For example, many studies showed the role of apoE in AD particularly through its catalytic action on amyloid Aβ (Potter and Wisniewski, 2012). In our study, apoE plasma level was associated with atrophy of hippocampus, posterior cingulate, inferior and middle temporal gyri, thalamic nuclei, amygdala, temporal fusiform cortex, and parahippocampal gyri. This is consistent with previous studies showing positive correlation between apoE level and GM density (Gupta et al., 2011), and apoE level and cognitive function (Yasuno et al., 2012a). A more recent study in a non-demented elderly population showed that persons with the APOEε4 allele have a smaller hippocampal volume (den Heijer et al., 2012). Another study showed that APOEε4 carriers exhibited greater atrophy of medial temporal lobe structures (Wolk and Dickerson, 2010). A task related fMRI study on AD patients, in which subjects were asked to detect novel items from previously learned items, showed that subjects with APOEε4 demonstrated less signal change in hippocampus (Johnson et al., 2006). Plasma apoE levels were also found to correlate with Aβ burden (Gupta et al., 2011) and cognitive performance in the aging population (Yasuno et al., 2011; Yasuno et al., 2012b).
Previous studies showed α2-Macroglobulin (A2M) increase in plasma (Hye et al., 2006) and its association with neural metabolism in hippocampus (Thambisetty et al., 2008) of patients with AD. In our study, association of A2M with atrophy of medial temporal lobe structures may be a representation of amyloid deposition and subsequent cell loss in these areas that often occur in AD. There are also independent studies showing A2M elevation in temporal cortex of AD patients (Wood et al., 1993). A2M is induced by inflammatory cytokines released by neural cells (Strauss et al., 1992) and is a marker for brain blood barrier breakdown (Cucullo et al., 2003). In addition, we found that IL-16 level was associated with atrophy of hippocampus, posterior cingulate, inferior and middle temporal gyri, and thalamus. Together these findings support an inflammatory process in early AD, which may be a useful target for future therapies. Interestingly, plasma IL-16 was reported to increase in mild and moderate AD but not in severe AD cases (Motta et al., 2007), suggesting that these proteins may only be useful markers at the early stages of the disease.
IgM, another inflammatory marker, was increase in AD group. Increased IgM in patients with AD has been previously reported (Kell et al., 1996; Soares et al., 2012) however there is no evidence to support the specificity of IgM for a pathologic process in thalamus in AD. IgM increase may be an epiphenomenon related to an inflamatory or infectious process in AD subjects (Miklossy, 2008).
We found that BNP level was associated with right inferior and middle temporal gyri atrophy. We found no evidence in the literature to support a specific link between BNP and these structures. However, a previous report showed independent association of raised N-terminal pro-hormone of BNP (NT-proBNP) level with poor cognitive performance, even in subjects with no history of prior stroke or coronary heart disease (Daniels et al., 2011). BNP is co-secreted from strained cardiac myocytes and has been associated with congestive heart failure. A study on a cohort of dementia showed that high plasma BNP levels was associated with vascular dementia, whereas they failed to demonstrate a significant increase in the AD subjects (Kondziella et al., 2009). Using a multiplex platform, it was shown that elevated NT-proBNP in CSF could discriminate mildly demented subjects from cognitively normal controls (Craig-Schapiro et al., 2011). Our findings indicate that BNP may not be a specific marker for AD (Soares et al., 2012) but it might indicate a vascular component for AD development (Zlokovic, 2011).
Thyroxine-Binding Globulin is produced by the liver and binds thyroxine in blood. TBG level change may be associated with thyroid dysfunction that is common in the elderly. Low and high tertile of serum thyrotropin concentration has been shown to increase risk of Alzheimer disease in women (Tan et al., 2008).
Our study showed that pancreatic peptides and Eotaxin-3 significantly change in AD patients. This is consistent with previous reports (Craig-Schapiro et al., 2011; Koide et al., 1995; Soares et al., 2012; Westin et al., 2012). However, we found no evidence to support specific contribution of pancreatic peptides and Eotaxin-3 in thalamic pathology in AD (as showed in our study). Reduction of pancreatic peptides binding sites have been reported in the temporal cortex and hippocampus of AD subjects (Martel et al., 1990), though there are reports against pancreatic peptide changes in AD (Wikkelso et al., 1991). Recently, it was shown that increased CSF concentration of Eotaxin-3 was associated with conversion of MCI to AD (Westin et al., 2012). Eotaxin-3, along with other CSF biomarkers, could enhance power of tau/Aβ42 ratio to discriminate between subjects with CDR=0 and CDR>0 (Craig-Schapiro et al., 2011). Others showed correlation of CSF Aβ42 levels and t-tau/Aβ42 ratios with the number of apoE4 alleles, plasma levels of B-type natriuretic peptide and pancreatic polypeptide have bee demonstrated (Hu et al., 2012). Our study partly confirmed the result of the latter study however there are difference in the methodology. For example, we only identified proteins that are significantly associated with brain structural changes therefore reducing the chance of epiphenomena in our result.
Taking together, current available evidence is not sufficient to suggest that these peptides would be useful potential biomarkers for detection of AD.
Protein Interval Changes
We found that plasma level of 64 proteins changed within 1 year in AD more than that seen in controls. However, only changes of eight proteins were associated with regional brain atrophy including the right and left fusiform cortices, cingulate gyrus and amygdala. These proteins were BMP6, MMP-9, NCAM, FGF4, MMP-10, SOD-1, vWF, and E-selectin. Previous reports showed that BMP6, MMP-9, FGF4, NCAM and SOD-1 were linked to regulation of adult neurogenesis (Boutin et al., 2009; Crews et al., 2010; Faiz et al., 2006; Ingraham et al., 2011; Kosaka et al., 2006), a neural replenishment mechanism that is believed to be impaired in AD (Lazarov et al., 2010). Moreover, it has been shown that MMP-9 and SOD-1 have anti-Aβ amyloid properties (Murakami et al., 2011; Yan et al., 2006). Several previous studies found no significant difference in plasma MMP-9 level between healthy controls and AD patients at baseline (Lim et al., 2011; Lorenzl et al., 2003; Martin-Aragon et al., 2009), whereas one study reported significantly decreased MMP-9 in AD (Horstmann et al., 2010) and another showed that MMP-9/biliverdin reductase ratio could predict MCI to AD progression (Mueller et al., 2010). It has been demonstrated that CSF concentrations of MMP-10 in association with that of Eotaxin-3, could enhance differentiation of demented patients from non-demented healthy controls (Craig-Schapiro et al., 2011).
We showed that Von Willebrand factor (vWF) changes over time showed negative correlation with left fusiform atrophy and positive correlation with cingulate gyri atrophy. vWF is a marker of endothelial dysfunction (Vischer, 2006) which, in a recent meta-analysis, showed to be associated with all types of dementias particularly of vascular type (Quinn et al., 2011) supporting the notion implicating disturbance of endothelial activation in AD (Grammas, 2011; Zlokovic, 2011).
Potential proteomic biomarkers for diagnosis of AD
We showed that combination of plasma proteins levels of IL-16, TBG, BNP, PYY, and ApoE can discriminate AD patients from healthy individuals with sensitivity of 89% and specificity of 57%. However, changes of BMP6, Eselectin, MMP10 and NrCAM over a year can provide a more accurate model for AD diagnosis with 93% sensitivity and 92% specificity. This is superior to any other diagnostic test thus far, for example, when compared with CSF Aβ42 (Sensitivity 80%, specificity 82%), CSF Tau protein (Sensitivity 80%, specificity 83%), MRI (Sensitivity 83%, specificity 89%), FDG-PET (Sensitivity 90%, specificity 89%), and SPECT (Sensitivity 80%, specificity 85%).
Potential Proteomic biomarkers for predicting MCI conversion to AD
We found that combination of plasma proteins levels of IL-16, TBG, BNP, PYY, and ApoE can predict MCI converters with 62.5% accuracy. This is not better than currently available measures such as brain MRI, which has 73% sensitivity and 81% specificity. However changes in BMP6, Eselectin, MMP10 and NrCAM can predict 93.5% of MCI converters, which is superior to any marker currently available for this purpose.
These findings show a real potential for use of these proteins for early diagnosis and monitoring AD as well as predict MCI conversion to AD. Clearly blood tests are more convenient and probably cheaper than imaging for diagnosis and disease monitoring.
Conclusion
This report is the most comprehensive linkage study between proteomics and structural brain atrophy in Alzheimer’s disease. We showed that combination of selected plasma protein levels and MRI may be a useful method in identifying potential biomarker for diagnosis and monitoring of AD, as well as for predicting MCI conversion to AD. Further study is required to evaluate suitability of these proteins as biomarkers in differentiating AD from other types of dementia.
Supplementary Material
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
References
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Human Brain Mapping. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bateman R. The dominantly inherited Alzheimer's network trials: an opportunity to prevent Alzheimer's disease. Program and Abstracts of the Alzheimer's Association International Conference; Vancouver, British Columbia Canada. 2012. F3-04. [Google Scholar]
- Boutin C, Schmitz B, Cremer H, Diestel S. NCAM expression induces neurogenesis in vivo. Eur J Neurosci. 2009;30:1209–1218. doi: 10.1111/j.1460-9568.2009.06928.x. [DOI] [PubMed] [Google Scholar]
- Britschgi M, Wyss-Coray T. Blood protein signature for the early diagnosis of Alzheimer disease. Arch Neurol. 2009;66:161–165. doi: 10.1001/archneurol.2008.530. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig-Schapiro R, Kuhn M, Xiong C, Pickering EH, Liu J, Misko TP, Perrin RJ, Bales KR, Soares H, Fagan AM, Holtzman DM. Multiplexed immunoassay panel identifies novel CSF biomarkers for Alzheimer's disease diagnosis and prognosis. PLoS ONE. 2011;6:e18850. doi: 10.1371/journal.pone.0018850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews L, Adame A, Patrick C, Delaney A, Pham E, Rockenstein E, Hansen L, Masliah E. Increased BMP6 levels in the brains of Alzheimer's disease patients and APP transgenic mice are accompanied by impaired neurogenesis. J Neurosci. 2010;30:12252–12262. doi: 10.1523/JNEUROSCI.1305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucullo L, Marchi N, Marroni M, Fazio V, Namura S, Janigro D. Blood-brain barrier damage induces release of alpha2-macroglobulin. Mol Cell Proteomics. 2003;2:234–241. doi: 10.1074/mcp.M200077-MCP200. [DOI] [PubMed] [Google Scholar]
- Daniels LB, Laughlin GA, Kritz-Silverstein D, Clopton P, Chen WC, Maisel AS, Barrett-Connor E. Elevated natriuretic peptide levels and cognitive function in community-dwelling older adults. Am J Med. 2011;124:670 e671–678. doi: 10.1016/j.amjmed.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Heijer T, van der Lijn F, Ikram A, Koudstaal PJ, van der Lugt A, Krestin GP, Vrooman HA, Hofman A, Niessen WJ, Breteler MM. Vascular risk factors, apolipoprotein E, and hippocampal decline on magnetic resonance imaging over a 10-year follow-up. Alzheimers Dement. 2012 doi: 10.1016/j.jalz.2011.07.005. [DOI] [PubMed] [Google Scholar]
- deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, Wuu J, Turner DA. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging. 2004;25:1197–1203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Echavarri C, Aalten P, Uylings HB, Jacobs HI, Visser PJ, Gronenschild EH, Verhey FR, Burgmans S. Atrophy in the parahippocampal gyrus as an early biomarker of Alzheimer's disease. Brain Struct Funct. 2011;215:265–271. doi: 10.1007/s00429-010-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- Faiz M, Acarin L, Peluffo H, Villapol S, Castellano B, Gonzalez B. Antioxidant Cu/Zn SOD: expression in postnatal brain progenitor cells. Neurosci Lett. 2006;401:71–76. doi: 10.1016/j.neulet.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Grammas P. Neurovascular dysfunction, inflammation and endothelial activation: implications for the pathogenesis of Alzheimer's disease. J Neuroinflammation. 2011;8:26. doi: 10.1186/1742-2094-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter JL, Bernstein MA, Borowski BJ, Ward CP, Britson PJ, Felmlee JP, Schuff N, Weiner M, Jack CR. Measurement of MRI scanner performance with the ADNI phantom. Med Phys. 2009;36:2193–2205. doi: 10.1118/1.3116776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta VB, Laws SM, Villemagne VL, Ames D, Bush AI, Ellis KA, Lui JK, Masters C, Rowe CC, Szoeke C, Taddei K, et al. Plasma apolipoprotein E and Alzheimer disease risk: the AIBL study of aging. Neurology. 2011;76:1091–1098. doi: 10.1212/WNL.0b013e318211c352. [DOI] [PubMed] [Google Scholar]
- Horstmann S, Budig L, Gardner H, Koziol J, Deuschle M, Schilling C, Wagner S. Matrix metalloproteinases in peripheral blood and cerebrospinal fluid in patients with Alzheimer's disease. Int Psychogeriatr. 2010;22:966–972. doi: 10.1017/S1041610210000827. [DOI] [PubMed] [Google Scholar]
- Hu WT, Holtzman DM, Fagan AM, Shaw LM, Perrin R, Arnold SE, Grossman M, Xiong C, Craig-Schapiro R, Clark CM, Pickering E, et al. Plasma multianalyte profiling in mild cognitive impairment and Alzheimer disease. Neurology. 2012 doi: 10.1212/WNL.0b013e318266fa70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hye A, Lynham S, Thambisetty M, Causevic M, Campbell J, Byers HL, Hooper C, Rijsdijk F, Tabrizi SJ, Banner S, Shaw CE, et al. Proteome-based plasma biomarkers for Alzheimer's disease. Brain. 2006;129:3042–3050. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- Ingraham CA, Park GC, Makarenkova HP, Crossin KL. Matrix metalloproteinase (MMP)-9 induced by Wnt signaling increases the proliferation and migration of embryonic neural stem cells at low O2 levels. J Biol Chem. 2011;286:17649–17657. doi: 10.1074/jbc.M111.229427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan K, Calhoun VD, Gelernter J, Stevens MC, Liu J, Bolognani F, Windemuth A, Ruano G, Assaf M, Pearlson GD. Genetic associations of brain structural networks in schizophrenia: a preliminary study. Biol Psychiatry. 2010;68:657–666. doi: 10.1016/j.biopsych.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Schmitz TW, Trivedi MA, Ries ML, Torgerson BM, Carlsson CM, Asthana S, Hermann BP, Sager MA. The influence of Alzheimer disease family history and apolipoprotein E epsilon4 on mesial temporal lobe activation. J Neurosci. 2006;26:6069–6076. doi: 10.1523/JNEUROSCI.0959-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Karas GB, Burton EJ, Rombouts SA, van Schijndel RA, O'Brien JT, Scheltens P, McKeith IG, Williams D, Ballard C, Barkhof F. A comprehensive study of gray matter loss in patients with Alzheimer's disease using optimized voxel-based morphometry. Neuroimage. 2003;18:895–907. doi: 10.1016/s1053-8119(03)00041-7. [DOI] [PubMed] [Google Scholar]
- Kell SH, Allman RM, Harrell LE, Liu T, Solvason N. Association between Alzheimer's disease and bound autochthonous IgM on T cells. J Am Geriatr Soc. 1996;44:1362–1365. doi: 10.1111/j.1532-5415.1996.tb01408.x. [DOI] [PubMed] [Google Scholar]
- Kipps CM, Duggins AJ, Mahant N, Gomes L, Ashburner J, McCusker EA. Progression of structural neuropathology in preclinical Huntington's disease: a tensor based morphometry study. J Neurol Neurosurg Psychiatry. 2005;76:650–655. doi: 10.1136/jnnp.2004.047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide S, Onishi H, Hashimoto H, Kai T, Yamagami S. Plasma neuropeptide Y is reduced in patients with Alzheimer's disease. Neurosci Lett. 1995;198:149–151. doi: 10.1016/0304-3940(95)11973-z. [DOI] [PubMed] [Google Scholar]
- Kondziella D, Gothlin M, Fu M, Zetterberg H, Wallin A. B-type natriuretic peptide plasma levels are elevated in subcortical vascular dementia. Neuroreport. 2009;20:825–827. doi: 10.1097/WNR.0b013e328326f82f. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Kodama M, Sasaki H, Yamamoto Y, Takeshita F, Takahama Y, Sakamoto H, Kato T, Terada M, Ochiya T. FGF-4 regulates neural progenitor cell proliferation and neuronal differentiation. FASEB J. 2006;20:1484–1485. doi: 10.1096/fj.05-5293fje. [DOI] [PubMed] [Google Scholar]
- Lazarov O, Mattson MP, Peterson DA, Pimplikar SW, van Praag H. When neurogenesis encounters aging and disease. Trends Neurosci. 2010;33:569–579. doi: 10.1016/j.tins.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim NK, Villemagne VL, Soon CP, Laughton KM, Rowe CC, McLean CA, Masters CL, Evin G, Li QX. Investigation of matrix metalloproteinases, MMP-2 and MMP-9, in plasma reveals a decrease of MMP-2 in Alzheimer's disease. J Alzheimers Dis. 2011;26:779–786. doi: 10.3233/JAD-2011-101974. [DOI] [PubMed] [Google Scholar]
- Liu J, Demirci O, Calhoun VD. A Parallel Independent Component Analysis Approach to Investigate Genomic Influence on Brain Function. IEEE Signal Process Lett. 2008;15:413–416. doi: 10.1109/LSP.2008.922513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kiehl KA, Pearlson G, Perrone-Bizzozero NI, Eichele T, Calhoun VD. Genetic determinants of target and novelty-related event-related potentials in the auditory oddball response. Neuroimage. 2009a;46:809–816. doi: 10.1016/j.neuroimage.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp. 2009b;30:241–255. doi: 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzl S, Albers DS, Relkin N, Ngyuen T, Hilgenberg SL, Chirichigno J, Cudkowicz ME, Beal MF. Increased plasma levels of matrix metalloproteinase-9 in patients with Alzheimer's disease. Neurochem Int. 2003;43:191–196. doi: 10.1016/s0197-0186(03)00004-4. [DOI] [PubMed] [Google Scholar]
- Martel JC, Alagar R, Robitaille Y, Quirion R. Neuropeptide Y receptor binding sites in human brain. Possible alteration in Alzheimer's disease. Brain Res. 1990;519:228–235. doi: 10.1016/0006-8993(90)90082-m. [DOI] [PubMed] [Google Scholar]
- Martin-Aragon S, Bermejo-Bescos P, Benedi J, Felici E, Gil P, Ribera JM, Villar AM. Metalloproteinase's activity and oxidative stress in mild cognitive impairment and Alzheimer's disease. Neurochem Res. 2009;34:373–378. doi: 10.1007/s11064-008-9789-3. [DOI] [PubMed] [Google Scholar]
- Mattay VS, Goldberg TE, Sambataro F, Weinberger DR. Neurobiology of cognitive aging: insights from imaging genetics. Biol Psychol. 2008;79:9–22. doi: 10.1016/j.biopsycho.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Miklossy J. Chronic inflammation and amyloidogenesis in Alzheimer's disease -- role of Spirochetes. J Alzheimers Dis. 2008;13:381–391. doi: 10.3233/jad-2008-13404. [DOI] [PubMed] [Google Scholar]
- Motta M, Imbesi R, Di Rosa M, Stivala F, Malaguarnera L. Altered plasma cytokine levels in Alzheimer's disease: correlation with the disease progression. Immunol Lett. 2007;114:46–51. doi: 10.1016/j.imlet.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Mueller C, Zhou W, VanMeter A, Heiby M, Magaki S, Ross MM, Espina V, Schrag M, Dickson C, Liotta LA. The heme degradation pathway is a promising serum biomarker source for the early detection of Alzheimer's disease. Journal of Alzheimer's Disease. 2010;19:1081–1091. doi: 10.3233/JAD-2010-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungas D, Reed BR, Jagust WJ, DeCarli C, Mack WJ, Kramer JH, Weiner MW, Schuff N, Chui HC. Volumetric MRI predicts rate of cognitive decline related to AD and cerebrovascular disease. Neurology. 2002;59:867–873. doi: 10.1212/wnl.59.6.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Murata N, Noda Y, Tahara S, Kaneko T, Kinoshita N, Hatsuta H, Murayama S, Barnham KJ, Irie K, Shirasawa T. SOD1 (copper/zinc superoxide dismutase) deficiency drives amyloid beta protein oligomerization and memory loss in mouse model of Alzheimer disease. J Biol Chem. 2011;286:44557–44568. doi: 10.1074/jbc.M111.279208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human Brain Mapping. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlson G. Multisite collaborations and large databases in psychiatric neuroimaging: advantages, problems, and challenges. Schizophr Bull. 2009;35:1–2. doi: 10.1093/schbul/sbn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer's disease. Nature. 2009;461:916–922. doi: 10.1038/nature08538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H, Wisniewski T. Apolipoprotein E: Essential Catalyst of the Alzheimer Amyloid Cascade. Int J Alzheimers Dis. 2012;2012 doi: 10.1155/2012/489428. 489428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn T, Gallacher J, Deary I, Lowe G, Fenton C, Stott D. Association between circulating hemostatic measures and dementia or cognitive impairment: systematic review and meta-analyzes. Journal of Thrombosis and Haemostasis. 2011;9:1475–1482. doi: 10.1111/j.1538-7836.2011.04403.x. [DOI] [PubMed] [Google Scholar]
- Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, Friedman LF, Galasko DR, Jutel M, Karydas A, Kaye JA. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- Risacher SL, Shen L, West JD, Kim S, McDonald BC, Beckett LA, Harvey DJ, Jack CR, Jr, Weiner MW, Saykin AJ. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol Aging. 2010;31:1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinek H, De Santi S, Frid D, Tsui WH, Tarshish CY, Convit A, de Leon MJ. Regional brain atrophy rate predicts future cognitive decline: 6-year longitudinal MR imaging study of normal aging. Radiology. 2003;229:691–696. doi: 10.1148/radiol.2293021299. [DOI] [PubMed] [Google Scholar]
- Shiino A, Watanabe T, Maeda K, Kotani E, Akiguchi I, Matsuda M. Four subgroups of Alzheimer's disease based on patterns of atrophy using VBM and a unique pattern for early onset disease. Neuroimage. 2006;33:17–26. doi: 10.1016/j.neuroimage.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Singh V, Chertkow H, Lerch JP, Evans AC, Dorr AE, Kabani NJ. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer's disease. Brain. 2006;129:2885–2893. doi: 10.1093/brain/awl256. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- Soares HD, Potter WZ, Pickering E, Kuhn M, Immermann FW, Shera DM, Ferm M, Dean RA, Simon AJ, Swenson F, Siuciak JA, et al. Plasma Biomarkers Associated With the Apolipoprotein E Genotype and Alzheimer Disease. Arch Neurol. 2012:1–8. doi: 10.1001/archneurol.2012.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss S, Bauer J, Ganter U, Jonas U, Berger M, Volk B. Detection of interleukin-6 and alpha 2-macroglobulin immunoreactivity in cortex and hippocampus of Alzheimer's disease patients. Lab Invest. 1992;66:223–230. [PubMed] [Google Scholar]
- Tan ZS, Beiser A, Vasan RS, Au R, Auerbach S, Kiel DP, Wolf PA, Seshadri S. Thyroid function and the risk of Alzheimer disease: the Framingham Study. Arch Intern Med. 2008;168:1514–1520. doi: 10.1001/archinte.168.14.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambisetty M, Hye A, Foy C, Daly E, Glover A, Cooper A, Simmons A, Murphy D, Lovestone S. Proteome-based identification of plasma proteins associated with hippocampal metabolism in early Alzheimer's disease. J Neurol. 2008;255:1712–1720. doi: 10.1007/s00415-008-0006-8. [DOI] [PubMed] [Google Scholar]
- Tosun D, Schuff N, Mathis CA, Jagust W, Weiner MW. Spatial patterns of brain amyloid-beta burden and atrophy rate associations in mild cognitive impairment. Brain. 2011;134:1077–1088. doi: 10.1093/brain/awr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vischer UM. von Willebrand factor, endothelial dysfunction, and cardiovascular disease. J Thromb Haemost. 2006;4:1186–1193. doi: 10.1111/j.1538-7836.2006.01949.x. [DOI] [PubMed] [Google Scholar]
- Westin K, Buchhave P, Nielsen H, Minthon L, Janciauskiene S, Hansson O. CCL2 Is Associated with a Faster Rate of Cognitive Decline during Early Stages of Alzheimer's Disease. PLoS ONE. 2012;7:e30525. doi: 10.1371/journal.pone.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Shiung MM, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr MRI patterns of atrophy associated with progression to AD in amnestic mild cognitive impairment. Neurology. 2008;70:512–520. doi: 10.1212/01.wnl.0000280575.77437.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikkelso C, Ekman R, Westergren I, Johansson B. Neuropeptides in cerebrospinal fluid in normal-pressure hydrocephalus and dementia. Eur Neurol. 1991;31:88–93. doi: 10.1159/000116653. [DOI] [PubMed] [Google Scholar]
- Wolk DA, Dickerson BC. Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer's disease. Proc Natl Acad Sci U S A. 2010;107:10256–10261. doi: 10.1073/pnas.1001412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JA, Wood PL, Ryan R, Graff-Radford NR, Pilapil C, Robitaille Y, Quirion R. Cytokine indices in Alzheimer's temporal cortex: no changes in mature IL-1 beta or IL-1RA but increases in the associated acute phase proteins IL-6, alpha 2-macroglobulin and C-reactive protein. Brain Res. 1993;629:245–252. doi: 10.1016/0006-8993(93)91327-o. [DOI] [PubMed] [Google Scholar]
- Wu L, Eichele T, Calhoun VD. Reactivity of hemodynamic responses and functional connectivity to different states of alpha synchrony: a concurrent EEG-fMRI study. Neuroimage. 2010;52:1252–1260. doi: 10.1016/j.neuroimage.2010.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Hu X, Song H, Yin K, Bateman RJ, Cirrito JR, Xiao Q, Hsu FF, Turk JW, Xu J, Hsu CY, et al. Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. J Biol Chem. 2006;281:24566–24574. doi: 10.1074/jbc.M602440200. [DOI] [PubMed] [Google Scholar]
- Yasuno F, Tanimukai S, Sasaki M, Hidaka S, Ikejima C, Yamashita F, Kodama C, Mizukami K, Michikawa M, Asada T. Association Between Cognitive Function and Plasma Lipids of the Elderly After Controlling for Apolipoprotein E Genotype. Am J Geriatr Psychiatry. 2011 doi: 10.1097/JGP.0b013e318211819b. [DOI] [PubMed] [Google Scholar]
- Yasuno F, Tanimukai S, Sasaki M, Hidaka S, Ikejima C, Yamashita F, Kodama C, Mizukami K, Michikawa M, Asada T. Association between cognitive function and plasma lipids of the elderly after controlling for apolipoprotein E genotype. Am J Geriatr Psychiatry. 2012a;20:574–583. doi: 10.1097/JGP.0b013e318211819b. [DOI] [PubMed] [Google Scholar]
- Yasuno F, Tanimukai S, Sasaki M, Ikejima C, Yamashita F, Kodama C, Hidaka S, Mizukami K, Asada T. Effect of plasma lipids, hypertension and APOE genotype on cognitive decline. Neurobiol Aging. 2012b doi: 10.1016/j.neurobiolaging.2011.12.028. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nat Rev Neurosci. 2011;12:723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.