Abstract
Single nucleotide variation in C13orf31 encoding p.C284R and p.I254V in a protein of unknown function causes systemic juvenile idiopathic arthritis, and associates with risk for leprosy and Crohn’s disease, respectively. Here we set out to uncover the underlying mechanism. C13orf31 formed a complex with fatty acid synthase on peroxisomes and promoted flux through de novo lipogenesis to concomitantly drive high levels of fatty acid oxidation (FAO) and glycolysis, and consequently ATP regeneration. C13orf31-dependent FAO controlled inflammasome activation, mitochondrial and NADPH oxidase-dependent reactive oxygen species production, and bactericidal activity of macrophages. With p.I254V and p.C284R exhibiting decreased and loss of function, respectively, C13orf31 determined organismal resilience to endotoxin shock. We hence discovered a central regulator of macrophage metabolic function and bioenergetic state, which is under evolutionary selection and determines risk for inflammatory and infectious disease.
Immune-related diseases arise from complex environment – gene interaction. While the external triggers remain enigmatic, human genetic studies have revealed the genomic risk landscapes of these conditions. Genes encoding proteins with immune function were predictably identified as plausible candidates at various risk loci. The mechanistic contribution of their polymorphic variants to disease pathophysiology has been experimentally addressed for a few of these genes, though remains largely based on conjecture for most. Furthermore, for a large number of risk loci and –genes it remains entirely unclear how they are mechanistically involved in disease, including some, for which no biological function is known yet at all. We hypothesized that the latter may single out novel mechanisms that are not only important for human health, but for mammalian biology in general.
Two coding polymorphisms in C13orf31 (also known as LACC1), an open reading frame encoding a protein of unknown function, rendered this gene and its product particularly intriguing: firstly, homoallelic carriage of a rare missense mutation at g.43,883,879T>C, leading to p.C284R, has been linked in consanguineous families to systemic juvenile idiopathic arthritis (sJIA), a periodic fever syndrome, and to early-onset Crohn’s disease (EOCD), an inflammatory bowel disease1, 2. Secondly, the haplotype identified by the common rs3764147 (g.43,883,789A>G) single nucleotide polymorphism (SNP), leading to p.I254V, has been associated with increased risk for leprosy, an infection caused by Mycobacterium leprae, and Crohn’s disease (CD)3, 4.
Here we report that the protein encoded by C13orf31 serves as rheostat for the synthesis of endogenous fatty acids and their mitochondrial oxidation, thereby controlling glycolytic activity and overall ATP regeneration. As a consequence, this polymorphic gene determined mitochondrial and NADPH oxidase-dependent reactive oxygen species (ROS) production, bactericidal activity and inflammasome activation in macrophages, prompting us to name this protein FAMIN, fatty acid metabolic – immune nexus.
Results
FAMIN interacts with fatty acid synthase
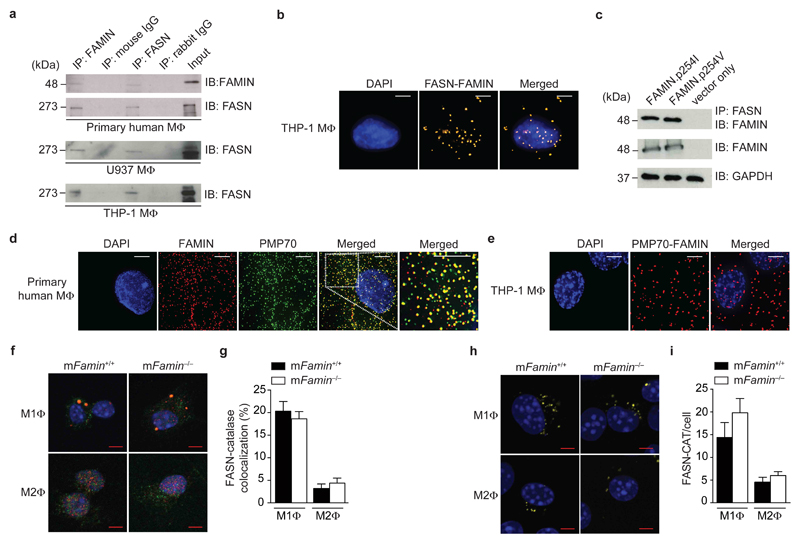
C13orf31 mRNA is most highly expressed in macrophages and predicted to encode a cytoplasmic protein of 430 amino acids (aa) that lacks similarity to other mammalian protein families5, 6. We performed an unbiased proteomic search for interaction partners to begin unravelling FAMIN function. After discounting proteins involved in protein synthesis, folding, degradation and cytoskeletal organization, fatty acid synthase (FASN, EC 2.3.1.85) was the top peptide hit detected with N- and C-terminally Strep-tagged FAMIN expressed as bait in HEK293T cells (Supplementary Fig. 1 and Supplementary Table 1). Endogenous FAMIN reciprocally co-immunoprecipitated with FASN in lysates of primary human monocytes differentiated into macrophages, as well as in lysates from human THP1 and U937 monocytic cell lines differentiated into macrophages (Fig. 1a). Proximity-ligation assays (PLA) further confirmed co-localization of FAMIN and FASN in situ (Fig. 1b). The two common polymorphic variants FAMIN.p254I and FAMIN.p254V exhibited equivalent interaction with FASN in coimmunoprecipitation experiments using HEK293T transfectant lysates (Fig. 1c), further corroborating that FAMIN is in a complex with FASN.
Figure 1. FAMIN interacts with fatty acid synthase and localizes to peroxisomes.
(a) Immunoblots (IB) for FAMIN and FASN after anti-C13orf31, anti-FASN or IgG control immunoprecipitation (IP) of primary human PBMC-derived and PMA-differentiated U937 and THP-1 macrophages (MΦ). (b) Proximity-ligation assay (PLA) of FAMIN and FASN (yellow) in THP-1 macrophages. DAPI, blue; scale bar = 5 μm. (c) IB for FAMIN of lysates and after anti-FASN IP of HEK293 expressing FAMIN.p254I or FAMIN.p254V; GAPDH loading control. (d) Co-localization by immunofluorescence (IF) of FAMIN (red) with PMP70 (green) in primary human macrophages. DAPI nuclear staining, blue; scale bar = 5 μm. Images are enlarged from the demarcated region in Supplementary Fig. 2a; rightmost panel represents further enlargement of the area shown in the white box. (e) PLA of FAMIN and PMP70 (red) in THP-1 macrophages. DAPI, blue; scale bar = 5 μm. (f,g) Co-localization by IF of FASN (red) with catalase (green) in M1 and M2 macrophages. DAPI, blue; scale bar = 5 μm. (h,i) PLA of FASN and catalase (yellow) in M1 and M2 macrophages. DAPI, blue; scale bar = 5 μm. Data are representative of three independent experiments (a–e) or one experiment with three mice and six (f,g) or ten (h,i) cells imaged per sample (f–i; mean ± S.E.M.).
Mammalian FASN is a large cytoplasmic polypeptide multi-enzyme central to de novo lipogenesis (DNL)7. FASN associates with membranes at different subcellular compartments8. We therefore performed confocal microscopy to determine the intracellular localization of FAMIN in macrophages differentiated from primary human monocytes and THP1 cells. FAMIN immunofluorescence (IF) revealed near-perfect co-localization with peroxisome-markers 70-kDa peroxisomal membrane protein (PMP70; encoded by ABCD3) and catalase (Fig. 1d and Supplementary Fig. 2a–c), while little co-localization was present with other organelle markers (Supplementary Fig. 2d,e). Co-localization between FAMIN and PMP70 was confirmed by PLA (Fig. 1e and Supplementary Fig. 2f), corroborating the peroxisomal localization of FAMIN.
Peroxisomes are organelles involved in lipid metabolism, which closely associate with lipid droplets and mitochondria9. Macrophages exhibit metabolic and functional plasticity, epitomized in vitro by culture under non-polarizing (M0), ‘classical’ (M1), and ‘alternative’ (M2) activation conditions10. While aerobic glycolysis is a defining characteristic of ‘inflammatory’ M1 macrophages and their primary mechanism for ATP generation11, ‘regenerative’ M2 macrophages rely on mitochondrial fatty acid oxidation (FAO, also termed β-oxidation) to fuel oxidative phosphorylation (OXPHOS)12. To study FAMIN function, we generated mice with knock-out alleles of 9030625A04Rik (also known as Lacc1), the murine homologue of C13orf31, which we will refer to as ‘mFamin’ (Supplementary Fig. 3)13. Similar to FAMIN, FASN does not contain a peroxisomal localization motif, and has previously been shown to complex with PMP70 on the cytoplasmic side of peroxisomes9, 14. Consistent with this, we found that a proportion of cellular FASN immunoreactivity, 20±3% and 4±2%, co-localized with catalase+ peroxisomes in wild-type M1 and M2 macrophages, respectively, and this was not affected by absence of FAMIN (Fig. 1f,g). PLA confirmed these spatial relationships (Fig. 1h,i). Bone marrow-derived macrophages cultured under M0, M1 and M2 conditions all expressed mFamin mRNA, with highest levels observed when differentiated under M1 conditions (Supplementary Fig. 4a). Fasn mRNA expression was similar in mFamin+/+ and mFamin–/– macrophages, with highest expression observed under M2 conditions (Supplementary Fig. 4b). In addition, FASN protein expression was also equivalent in mFamin+/+ and mFamin–/– M1 and M2 macrophages (Supplementary Fig. 4c). Together, our data indicate that FAMIN and FASN form a complex on peroxisomes, and peroxisomal FASN localization, which is particularly prominent in M1 macrophages, is not impaired in mFamin–/– cells.
FAMIN controls carbon flux through DNL
FASN catalyzes all steps of long-chain saturated fatty acid (LCFA) synthesis (predominantly C16:0 palmitic acid)15. This enzymatic reaction utilizes cytoplasmic acetyl-coenzyme A (CoA), malonyl-CoA (generated from acetyl-CoA by carboxylation), and nicotinamide adenine dinucleotide phosphate (NADPH)15. Cytoplasmic acetyl-CoA is itself generated by ATP citrate lyase (ACLY) following the export of mitochondrial citrate formed during glucose- or amino acid-fuelled Krebs cycle activity16. In consequence, FASN function allows storage of excess energy, derived from glucose and amino acid oxidation, in lipid droplets upon LCFA CoA-activation and –esterification to triacylglycerols (TAGs)17. FASN also synthesizes fatty acyl moieties of membrane lipids and provides the precursors of many other cellular and signaling lipids. In view of the diverse functions of lipid moieties dependent on FASN function, the precise channeling of its products into distinct metabolic pathways is essential15, 17.
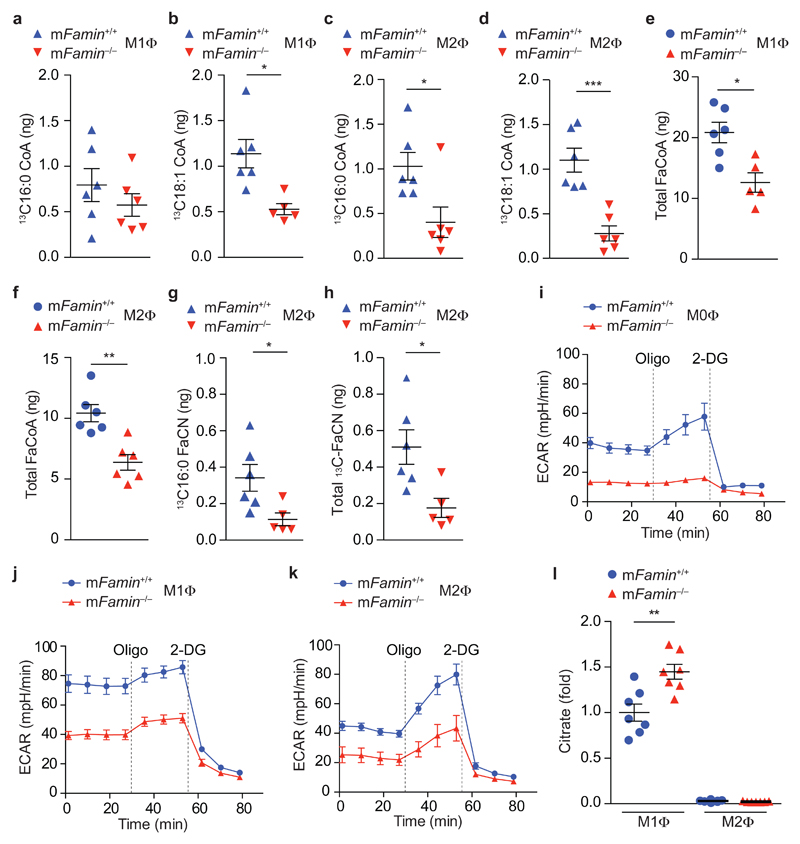
To assess FASN activity in situ, we pulse labeled murine bone marrow-derived macrophages with [13C1,2]-glucose (Supplementary Fig. 5a). Uptake of 13C into total LCFA and C16:0 was below the detection limit (data not shown) in both mFamin+/+ and mFamin–/– M1 and M2 macrophages, consistent with limited turnover and exogenous uptake of the large pools of free fatty acids. However, we did observe 13C incorporation into C16:0 and C18:1 fatty acyl CoA esters, the activated forms of LCFA utilized for lipid synthesis and mitochondrial oxidation, in wild-type M1 and M2 macrophages (Fig. 2a–d). Together, these data imply that newly synthesized fatty acids are immediately CoA-esterified for subsequent use and thus do not determine the abundance of the free fatty acid pool. Furthermore, we discovered that incorporation of 13C into C18:1-CoA in M1 macrophages and C16:0-CoA and C18:1-CoA in M2 macrophages were profoundly reduced in mFamin–/– compared to mFamin+/+ MΦ (Fig. 2a–d). Total LCFA-CoA abundance was also significantly reduced in mFamin–/– compared to mFamin+/+ M1 and M2 macrophages (Fig. 2e,f), while total fatty acid content, including C16:0, was predictably indistinguishable between genotypes (Supplementary table 2). Together, these data indicate that FAMIN determines, via DNL, the fatty acyl CoA availability for processes such as FAO.
Figure 2. FAMIN promotes glycolysis and glucose flux into de novo lipogenesis.
(a–d) Measurement of 13C-labelled C16:0 and C18:1 fatty acyl CoA following a 24 h pulse with 1,2-13C-glucose in mFamin–/– and mFamin+/+ M1 and M2 macrophages (MΦ). (e,f) Measurement of total fatty acyl CoA in M1 and M2 macrophages. (g,h) Measurement of 13C-labelled C16:0 and total fatty acyl carnitine (FaCN) following a 24 h pulse with 1,2-13C-glucose in M2 macrophages. (i–k) Extracellular rate (ECAR) of M0, M1 and M2 macrophages given sequential treatment (dotted vertical lines) with oligomycin (Oligo) and 2-deoxyglucose (2-DG). (l) Citrate levels in M1 and M2 macrophages. Each symbol (a–h, l) represents an individual mouse; small horizontal lines and error bars indicate the mean ± S.E.M. *P < 0.05, **P < 0.01, *** P < 0.001 (Unpaired, two-tailed Student’s t-test; Grubb’s test outlier exclusion). Data are from one experiment with six (a–h) or seven mice (l) or one experiment with three mice representative of three independent experiments (i–k; mean ± S.E.M.).
FAO requires transport of CoA-activated LCFA across mitochondrial membranes as acyl-carnitine esters18. The exchange of CoA for carnitine by cytoplasmic carnitine palmitoyltransferase (CPT1a) is the rate-limiting step of FAO18. Consistent with a reduction in the specific availability of fatty acyl-CoA, incorporation of [13C1,2]glucose-derived 13C into fatty acyl carnitine species (total acyl-Cn and C16:0-Cn) was markedly lower in mFamin–/– compared to mFamin+/+ M2 macrophages (Fig. 2g,h), while incorporation was below the detection limit in M1 macrophages (data not shown). Importantly, while incorporation of glucose-derived 13C into acyl-CoA was substantial after a 24 h pulse (~20-30% of C16:0- and C18:1-CoA in wild-type M2 macrophages), it was low for acyl-Cn species (~4% of total acyl-Cn in wild-type M2 macrophages) at the same time point. Altogether, FAMIN directly controls the flux of glucose-derived carbon into DNL and onwards into acyl-Cn, the latter possibly involving an intermediary step via TAGs12.
FAMIN controls glycolysis via DNL
To determine whether loss of FAMIN consequently influenced glycolytic metabolism in macrophages, we measured the extracellular acidification rate (ECAR), an indicator of glycolysis19, in wild-type and mutant macrophages. We found ECAR was profoundly reduced in mFamin–/– compared to mFamin+/+ M1 macrophages, with a similar proportional decrease from an expectedly lower baseline observed in M0 and M2 macrophages (Fig. 2i–k). The differential in ECAR was further increased upon blockade of the F0 subunit of mitochondrial ATP synthase with oligomycin A (Fig. 2i–k), which stimulates alternative ATP generation via glycolysis20. This observation implies that FAMIN-dependent glycolysis is maximized under conditions where mitochondrial respiration is uncoupled from ATP synthesis. ECAR was abrogated in mFamin–/– and mFamin+/+ macrophages alike upon inhibition of glycolysis with 2-deoxy-D-glucose (Fig. 2i–k). Similarly, ECAR measurements performed in the presence of exogenous pyruvate, hence bypassing glycolysis, were indistinguishable between mFamin–/– and mFamin+/+ macrophages (Supplementary Fig. 5b). Altogether, these data demonstrate that FAMIN determines the baseline and maximum glycolytic capacity of macrophages. Notably, intracellular concentrations of citrate, which itself inhibits glycolysis at the level of phosphofructokinase21, were substantially higher in mFamin–/– compared to mFamin+/+ macrophages differentiated under M1 conditions (Fig. 2l). Krebs cycle intermediates other than citrate remained indistinguishable between genotypes in M1 and M2 macrophages (Supplementary Fig. 5c), indicating absence of direct effects of FAMIN on Krebs cycle activity. Altogether, this indicates that the impairment in glycolysis may be a consequence of defective DNL.
Mitochondrial FAO is promoted by FAMIN
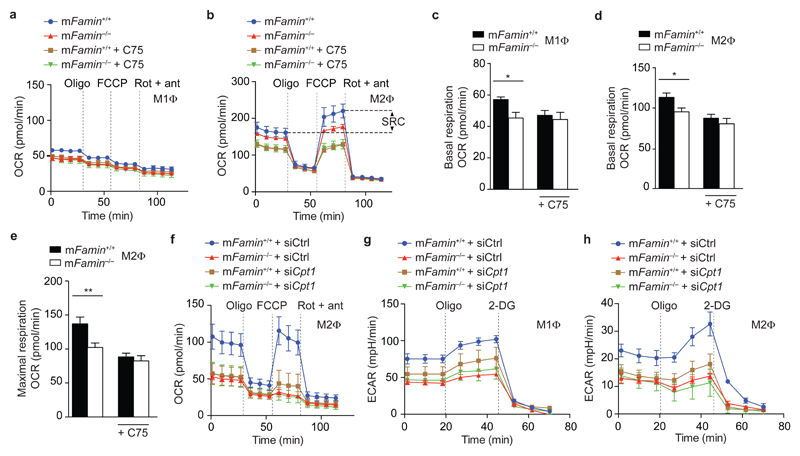
Since we had observed that the extent of carbon flux through DNL into acyl-Cn was determined by FAMIN, we hypothesized that FAMIN may control mitochondrial FAO. As expected, wild-type M2 macrophages had high basal OXPHOS, as reflected by high oxygen consumption rate (OCR), whereas M1 macrophages had low OCR that was refractory to oligomycin treatment, an inhibitor of ATP synthase (Fig. 3a–d). Impaired glycolysis usually leads to a compensatory increase in OXPHOS in order to maintain ATP intracellular concentrations22. However, in addition to defective glycolysis, mFamin–/– M1 and M2 macrophages also concomitantly exhibited significantly lower basal OCR compared to wild-type cells (Fig. 3a–d). Maximal uncontrolled OCR (MU-OCR) is elicited by dissipating the mitochondrial electrochemical proton gradient with the ‘uncoupler’ carbonyl-cyanide p-tri-fluoromethoxy-phenyl-hydrazone (FCCP) and unmasks spare respiratory capacity (SRC)12. MU-OCR was reduced and SRC could not be elicited following uncoupling via FCCP in mFamin–/– M2 macrophages (Fig. 3b,e). Complete inhibition of mitochondrial respiration via rotenone and antimycin A, complex I and III inhibitors, respectively, abolished differences in OCR between mFamin–/– and mFamin+/+ macrophages (Fig. 3a,b). Altogether these data demonstrate that mFamin–/– M1 and M2 macrophages exhibited reduced oxidative capacity in basal, but also uncoupled, respiratory states.
Figure 3. FAMIN augments fatty acid oxidation.
(a,b) Oxygen consumption rate (OCR) of mFamin–/– and mFamin+/+ M1 and M2 macrophages (MΦ) ± 20 μM C75 followed by sequential treatment (dotted vertical lines) with oligomycin (Oligo), FCCP, and rotenone plus antimycin (Rot + ant). Spare respiratory capacity (SRC) as depicted and refers to difference between maximal and basal respiratory capacities. (c–e) Basal respiratory and maximal respiratory capacity in M1 macrophages (n = 6) and M2 macrophages ± C75 (n = 14/13/9/7). (f–h) OCR and extracellular acidification rate (ECAR) of M1 and M2 macrophages silenced for Cpt1a (Cpt1a siRNA) or scrambled siRNA (Ctrl siRNA). *P < 0.05, **P < 0.01 (Unpaired, two-tailed Student’s t-test). Data are from one experiment with three mice representative of three independent experiments (a,b,f–h; mean ± S.E.M.) or pooled from two or more independent experiments (c–e; mean ± S.E.M.).
High basal OCR and SRC in M2 macrophages has been reported to be secondary to FAO12. We therefore considered defective FAO as the cause for impaired oxidation, particularly apparent in mFamin–/– M2 macrophages. To test the hypothesis that FAMIN specifically affects mitochondrial FAO and not OXPHOS fuelled by other substrates, we inhibited CPT1a activity. Genetic silencing of Cpt1a, or blocking CPT1a with the irreversible inhibitor etomoxir23, decreased the OCR in mFamin+/+ cells to that observed in mFamin–/– M2 macrophages (Fig. 3f and Supplementary Fig. 5d). Importantly, silencing Cpt1a reduced basal ECAR in mFamin+/+ M1 and M2 macrophages to the low levels seen in mFamin–/– macrophages, abrogating genotype-dependent difference in glycolysis (Fig. 3g,h). In summary, our findings demonstrate that loss of FAMIN leads to decreased FAO that is needed to sustain high rates of glycolysis.
FAMIN controls FAO of endogenously synthesized lipids
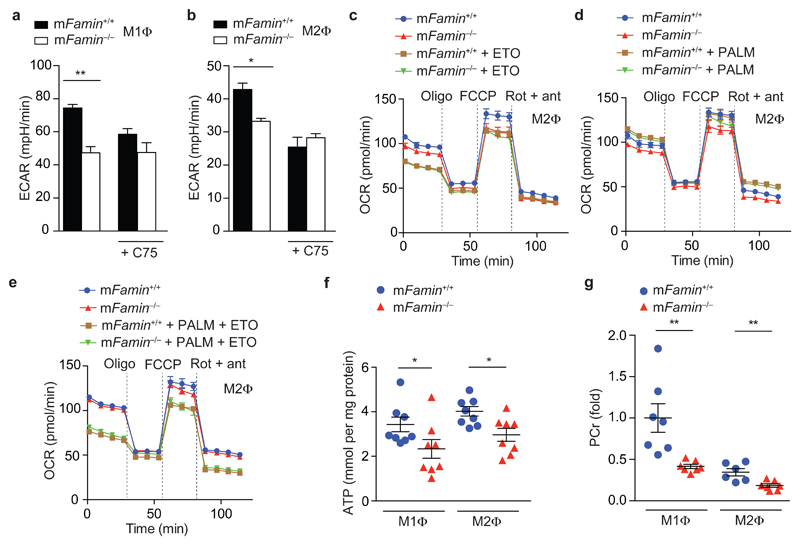
Given that FAMIN controls carbon flux from glucose into acyl-CoA, we hypothesized that FAMIN may facilitate FAO of de novo synthesized fatty acids. Blockade of FASN function with the small molecule synthetic inhibitor C7524 decreased basal OCR and SRC in mFamin+/+ M2 macrophages and abolished differences between mFamin+/+ and mFamin–/– cells (Fig. 3a–e). Importantly, blockade of FASN with C75 similarly decreased ECAR in M1 and M2 macrophages and abrogated mFamin genotype-related differences (Fig. 4a,b). To support this further, we measured OCR under conditions devoid of exogenous lipids, where FAO is thus entirely dependent upon de novo synthesized lipids by FASN. As before, baseline OCR and MU-OCR were decreased in mFamin–/– compared with mFamin+/+ M2 macrophages, with differences abrogated by CPT1a inhibition with etomoxir (Fig. 4c). Supplementation of exogenous C16:0 increased basal and MU-OCR to equivalent levels in mFamin+/+ and mFamin–/– M2 macrophages (Fig. 4d), demonstrating that exogenous C16:0 can rescue depressed basal and maximum OCR in starved mFamin–/– M2 macrophages. Finally, blockade of CPT1a in M2 macrophages supplemented with exogenous C16:0 decreased basal and MU-OCR to levels identical between mFamin+/+ and mFamin–/– cells (Fig. 4e). These data demonstrate that FAMIN supports FAO and glycolysis via a mechanism that involves DNL.
Figure 4. FAMIN controls FAO of endogenously synthesized lipids.
(a,b) Basal ECAR of mFamin–/– and mFamin+/+ M1 and M2 macrophages (MΦ) ± 20 μM C75. (c–e) OCR of M2 macrophages cultured in serum free medium and treated as indicated with 40 μM etomoxir (ETO) for 1 h and supplemented with bovine serum albumin (BSA) conjugated palmitate (PALM) or BSA alone prior to OCR measurement. (f) ATP quantification in M1 and M2 macrophage lysates (n = 8). (g) Phosphocreatine (PCr) levels in M1 and M2 macrophages. Each symbol (f,g) represents an individual mouse; small horizontal lines and error bars indicate the mean ± S.E.M. *P < 0.05, **P < 0.01 (Unpaired, two-tailed Student’s t-test). Data are from one experiment with three mice representative of three independent experiments (a–e; mean ± S.E.M.) or from one experiment with eight (f) or seven mice (g).
FAMIN controls macrophage energy stores
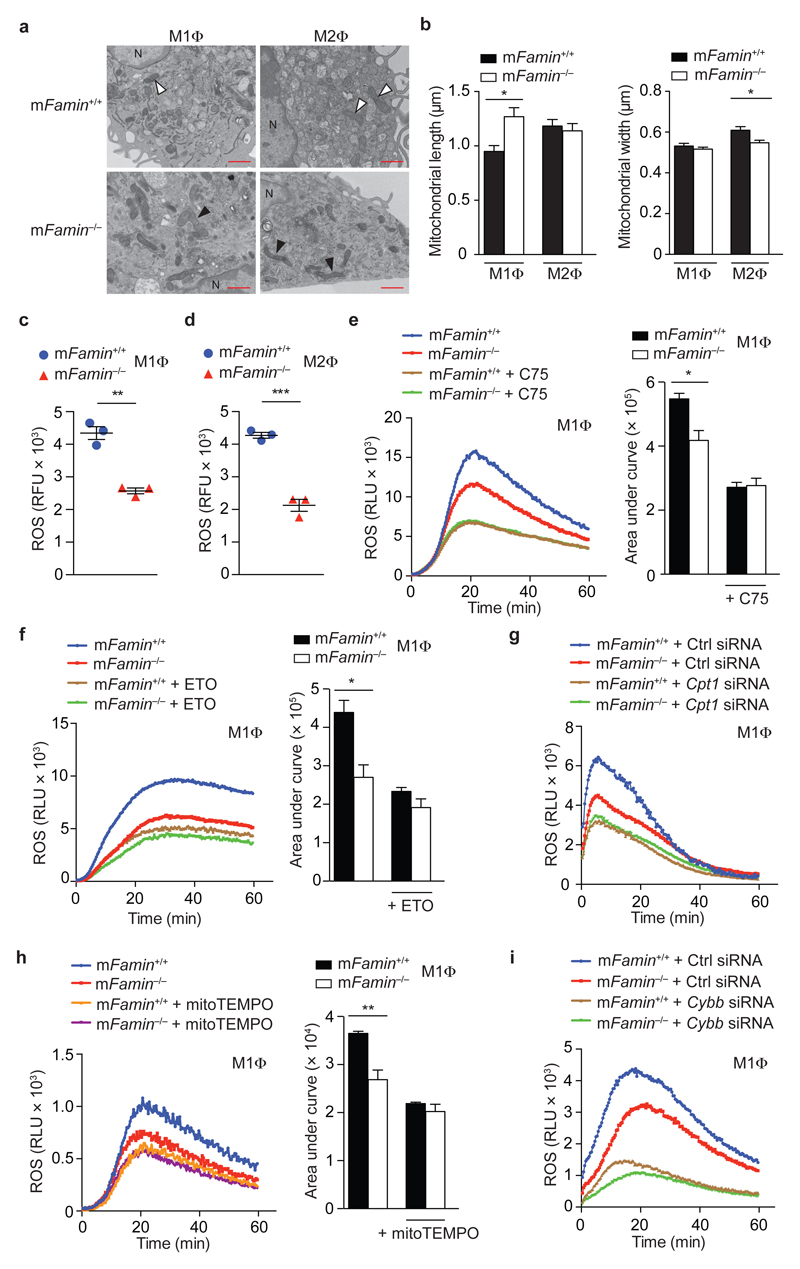
Considering that FAMIN controlled the extent of FAO and glycolysis in parallel and, remarkably, both processes did not cross-compensate for each other’s reduced function, we hypothesized that FAMIN function would impact on macrophage energy stores. Total cellular ATP was profoundly reduced by 30±12 % and 26±6 % in bone marrow-derived mFamin–/– compared to mFamin+/+ M1 and M2 macrophages (Fig. 4f). Concentrations of cellular phosphocreatine, a rapidly mobilizable energy store, were similarly reduced as ATP in mFamin–/– compared to mFamin+/+ M1 and M2 macrophages (Fig. 4g). Compared to mFamin+/+ macrophages, transmission electron microscopy (TEM) of mFamin–/– macrophages demonstrated morphological changes in mitochondria, characterized by elongation and narrowing, which are consistent with adaptation in response to ATP depletion (Fig. 5a,b)25. Hence FAMIN promotes glycolysis, FAO, and consequently cellular energy stores in macrophages.
Figure 5. FAMIN-deficient macrophages have impaired mitochondrial ROS production and exhibit features of mitochondrial injury and remodeling.
(a) Transmission electron microscopy of M1 and M2 macrophages (MΦ) showing mitochondria; black arrowheads show mitochondria that are elongated and amorphous in mFamin–/– M1 macrophages and narrower in mFamin–/– M2 macrophages compared to mFamin+/+ controls (white arrowheads). N = nucleus, scale bar = 1 μm and (b) quantification of mitochondrial size. (c,d) Mitochondrial ROS in unstimulated mFamin–/– and mFamin+/+ M1 and M2 macrophages stained with a red fluorescent mitochondrial superoxide indicator and measured in relative fluorescence units, RFU. (e–i) Zymosan induced ROS production in mFamin–/– and mFamin+/+ M1 macrophages treated as indicated for 16 h with 20 μM C75; or 1 h with 40 μM etomoxir (ETO) or 500 μM mitoTEMPO prior to stimulation; or silenced for Cpt1a (Cpt1a siRNA) or Cybb (Cybb siRNA) or scrambled siRNA (Ctrl siRNA); Left, ROS kinetic plots measured in relative light units, RLU and right, area under curve, AUC. *P < 0.05, **P < 0.01 (Mann–Whitney U-test (b) or unpaired, two-tailed Student’s t-test (c–f,h)). Data are representative of one experiment with three mice and six cells imaged per sample (a–b; mean ± S.E.M.) or from one experiment with three mice representative of at least two independent experiments (c–i; mean ± S.E.M.).
FAMIN determines macrophage ROS production
Mitochondrial respiration generates reactive oxygen species (ROS) when electrons prematurely escape electron transport chain complexes I and III and react with molecular oxygen to generate superoxide anions26. Consistent with decreased mitochondrial oxidation in the absence of FAMIN, mitochondrial ROS (mROS) determined via a red-fluorescence mitochondrial superoxide indicator was profoundly reduced in mFamin–/– compared to mFamin+/+ M1 and M2 macrophages (Fig. 5c,d). Cytoplasmic ROS measured with the cytosolic ROS indicator, CM-H2DCFDA, was also lower in mFamin–/– M1, though unchanged between genotypes in M2 macrophages (Supplementary Fig. 6a,b). Stimulation with the TLR2 agonist zymosan resulted in significantly lower extracellular ROS (eROS) production in mFamin–/– compared to mFamin+/+ M1 and M2 macophages (Fig. 5e and Supplementary Fig. 6c). FASN inhibition with C75, or silencing of Fasn with siRNA, approximately halved eROS production in macrophages polarized under both M1 and M2 conditions, and abrogated the mFamin-associated differences (Fig. 5e and Supplementary Fig. 6c–e). CPT1a inhibition with etomoxir reduced zymosan-elicited eROS generation in mFamin+/+ macrophages to levels observed in mFamin–/– cells, irrespective of whether the latter were treated with etomoxir (Fig. 5f and Supplementary Fig. 6f). A similar pattern of eROS production was observed upon Cpt1a silencing (Fig. 5g and Supplementary Fig. 6g) and, importantly, upon scavenging of mROS via mitoTEMPO (Fig. 5h and Supplementary Fig. 6h). As expected, eROS production was dependent on gp91phox NADPH oxidase27, since silencing the gene encoding gp91phox, Cybb, largely abrogated mFamin genotype-related differences (Fig. 5i and Supplementary Fig. 6i). Remarkably, mFamin–/– macrophages exhibited decreased eROS generation despite increased NADPH being available (Supplementary Fig. 6j). In contrast to macrophages, PMA-elicited eROS production in murine neutrophils was independent of FAMIN (Supplementary Fig. 6k), consistent with absent mFamin expression (6 and data not shown). In summary, FAMIN controls mROS production via a DNL- and FAO-dependent mechanism, and mROS, in turn, determines the NOX/DUOX system’s capacity to elicit eROS in macrophages.
FAMIN.pI254V and pC284R exhibit impaired function
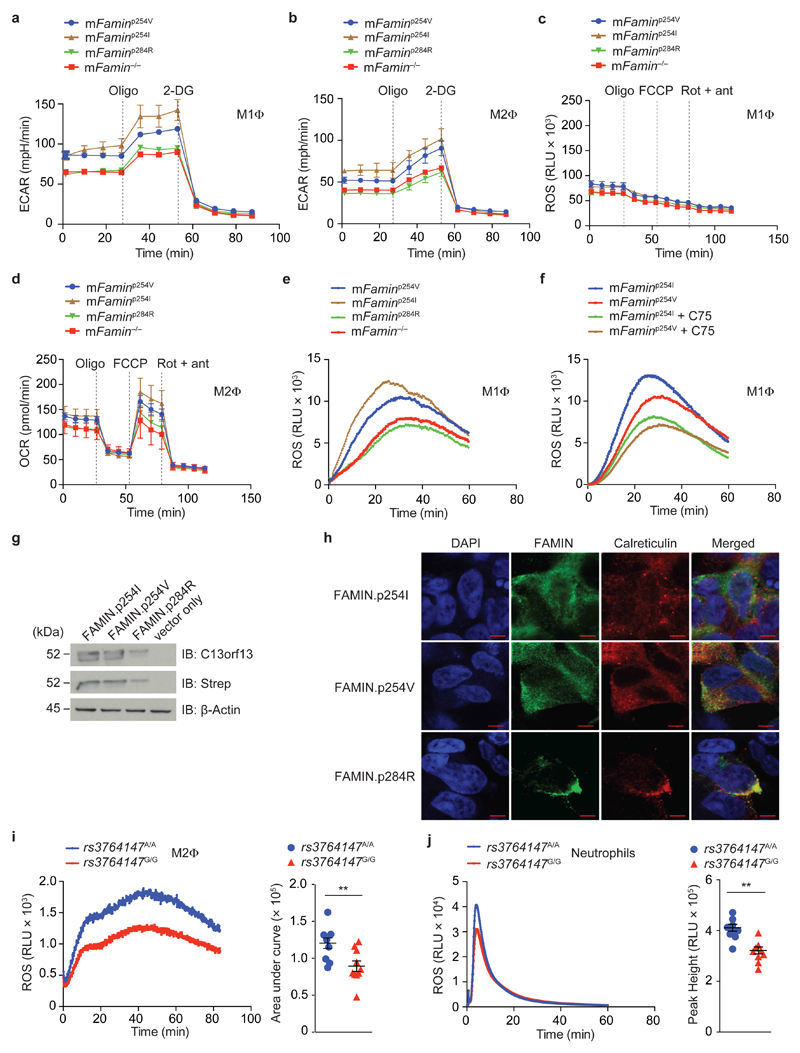
We next determined whether the metabolic mechanisms elucidated above were affected by genetic variation in C13orf31. Wild-type C57BL/6N mice encode the human ‘risk’ G allele at the corresponding aa254 position, which we refer to as ‘mFaminp254V’. Using CRISPR/Cas9, we introduced single nucleotide exchanges, generating mice homozygous for the leprosy/CD-protective [A] allele at aa254 (‘mFaminp254I’, Supplementary Fig. 3)28. We also generated mice homozygous for the sJIA/EOCD-causing [C] allele at g.43,883,879, encoding aa284 (‘mFaminp284R’) instead of the normal [T] (‘mFaminp284C’, Supplementary Fig. 3). Metabolic flux analyses were equivalent between mFaminp284R and mFamin–/– macrophages, while mFaminp254I exhibited the highest ECAR, basal OCR and MU-OCR, and mFaminp254V macrophages intermediate levels (Fig. 6a–d). Consistent with such decreased and loss of function of FAMIN.pI254V and FAMIN.pI284R, respectively, zymosan elicited higher eROS from mFaminp254I macrophages compared to mFaminp254V cells, while eROS production in mFaminp284R was similarly diminished as in mFamin–/– macrophages (Fig. 6e and Supplementary Fig 7a). Genotype-related differences between mFaminp254I and mFaminp254V in zymosan-elicited eROS production were abrogated upon inhibition of FASN with C75 (Fig. 6f and Supplementary Fig. 7b). Overexpression of human FAMIN.p254I under a CMV promoter in HEK293T cells yielded levels of protein indistinguishable from FAMIN.p254V (Fig. 6g), while expression of FAMIN.p284R was low and co-localized with calreticulin, indicating retention in the ER (Fig. 6g, h). Altogether, our results demonstrate that p.C284R causes complete loss of function, while the single methyl group change p.I254V exhibits decreased function.
Figure 6. FAMIN.pI254V is hypomorphic and FAMIN.pC284R leads to complete loss of function.
(a–d) Extracellular rate (ECAR) and Oxygen consumption rate (OCR) in murine M1 and M2 macrophages (MΦ) from mFamin–/– and C57BL/6N mice natively homozygous for the human ‘risk’ rs3764147 [G] allele (‘mFaminp254V’) or ‘non-risk’ [A] allele (‘mFaminp254I’) or Mendelian ‘risk’ variant (‘mFaminp284R’) at corresponding nucleotides of the murine gene. (e) Zymosan-stimulated ROS production in mFaminp254I, mFaminp254V, mFaminp284R and mFamin–/– M1 macrophages measured in relative light units, RLU. (f) Zymosan induced ROS production in mFaminp254I and mFamin p254V M1 macrophages treated as indicated for 16 h with 20 μM C75. (g) Immunoblot (IB) for FAMIN and Strep-tag in HEK293 cells with stable expression of N-terminal Strep-tagged FAMIN.p254I, FAMIN.p254V, FAMIN.p284R or vector control; β-actin loading control. (h) Co-localization by immunofluorescence of FAMIN (green) and calreticulin (red). DAPI, blue; scale bar = 5 μm. (i) Zymosan-stimulated ROS production in PBMC-derived M2 macrophages from healthy donors homozygous for the Crohn’s disease and leprosy risk (‘rs3764147G/G’) and non-risk (‘rs3764147A/A’) haplotypes; n = 10. (j) PMA-stimulated ROS production in neutrophils; Left, ROS kinetic plots and right, area under curve/peak height. Each symbol (i,j) represents an individual donor; small horizontal lines and error bars indicate the mean ± S.E.M. *P < 0.05, **P < 0.01 (Unpaired, two-tailed Student’s t-test). Data are from one experiment with three mice representative of two independent experiments (a–f; mean ± S.E.M), representative of three independent experiments (g,h) or pooled from 10 independent experiments (i,j).
We next obtained peripheral blood from healthy humans homozygous for the CD/leprosy risk (G/G; encoding FAMIN.p254V) and protective (A/A; encoding FAMIN.p254I) haplotypes identified by rs3764147. We isolated primary neutrophils and monocytes, and differentiated the latter in vitro into macrophages under M2 conditions. C13orf31 mRNA expression in M2 macrophages was indistinguishable between the two haplotype groups (Supplementary Fig. 7c). Human M2 macrophages homozygous for the risk haplotype exhibited decreased zymosan-elicited eROS production compared to those homozygous for the protective haplotype (Fig. 6i). Human, in contrast to murine, neutrophils express FAMIN (5 and data not shown). PMA triggered lower ROS production in neutrophils with the risk- compared to the protective haplotype (Fig. 6j), extending decreased function of the risk haplotype beyond macrophages. These data demonstrate that the CD/leprosy risk haplotype is associated with decreased eROS production, hence consistent with results obtained with macrophages from SNP-edited mFamin.pI254V mice. Together, this identifies rs3764147 encoding FAMIN.pI254V as the causal genetic variation within this risk haplotype.
FAMIN controls bactericidal and inflammasome activity
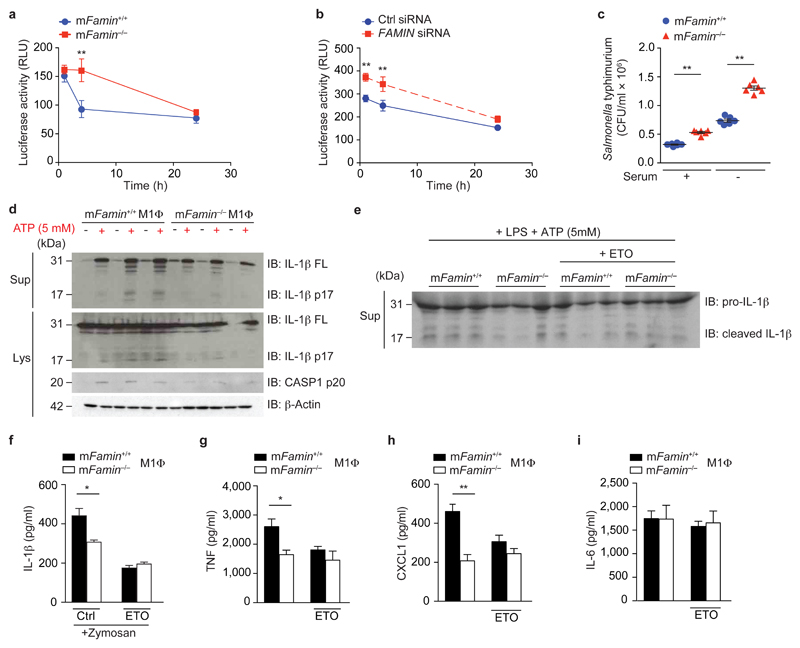
Finally, we turned our attention to the consequences of this profound metabolic derangement on immune function. Mitochondrial ROS can directly augment the bactericidal activity of macrophages toward intracellular pathogens29. Compared to mFamin+/+ cells, mFamin–/– M0 macrophages exhibited significantly decreased intracellular killing of a luminescent Mycobacterium bovis BCG strain30 (Fig. 7a). Silencing of C13orf31 in human macrophages resulted in reduced bactericidal activity towards luminescent M. bovis BCG (Fig. 7b), similar to murine macrophages. Finally, increased colony forming units (c.f.u.) of Salmonella enterica serovar Typhimurium were recovered from infected mFamin–/– compared to mFamin+/+ M0 macrophages, irrespective of whether exogenous lipids were provided in serum or not (Fig. 7c), altogether demonstrating that FAMIN determines the capacity of macrophages to kill intracellular pathogens.
Figure 7. FAMIN deficiency causes defective bacterial clearance and inflammasome activation, in vitro.
(a) mFamin–/– and mFamin+/+ M0 macrophages (MΦ) infected with Bacillus Calmette Guérin (BCG), containing a luciferase reporter gene; relative light units (RLU) measured at 0, 4, or 24 h. (b) Infection with BCG of human PBMC-derived M0 macrophages, silenced for FAMIN or scrambled siRNA control. (c) mFamin–/– and mFamin+/+ M0 macrophages infected with S. Typhimurium SL1344 in either serum containing or serum free medium; colony forming units, CFU. (d) Immunoblot (IB) of IL-1β and caspase 1 from mFamin–/– and mFamin+/+ M1 macrophages cell lysates (Lys) and supernatants (Sup); β-actin loading control. M1 macrophages were pretreated with 500 ng/mL of LPS for 2 h in serum free media and stimulated for 15 min with ATP. (e) IB of IL-1β in M1 macrophage cell supernatants; macrophages were pretreated as in (d) ± 100 μM etomoxir (ETO). (f) IL-1β levels in supernatants of M1 macrophages stimulated with 200 μg/mL of zymosan, pre-treated for 2 h with 100 μM etomoxir (ETO). (g–i) TNF, CXCL1 and IL-6 levels in supernatants of M1 macrophages stimulated with 1μg/mL of LPS pre-treated for 2 h with 100 μM etomoxir (ETO). *P < 0.05, **P < 0.01 (One-way ANOVA with post-hoc Bonferroni (a,b) or unpaired, two-tailed Student’s t-test (c,f–i)). Data are from one experiment with three mice representative of three independent experiments (a,d–i; mean ± S.E.M.) or from one experiment with six independent samples per group (b,c; mean ± S.E.M.).
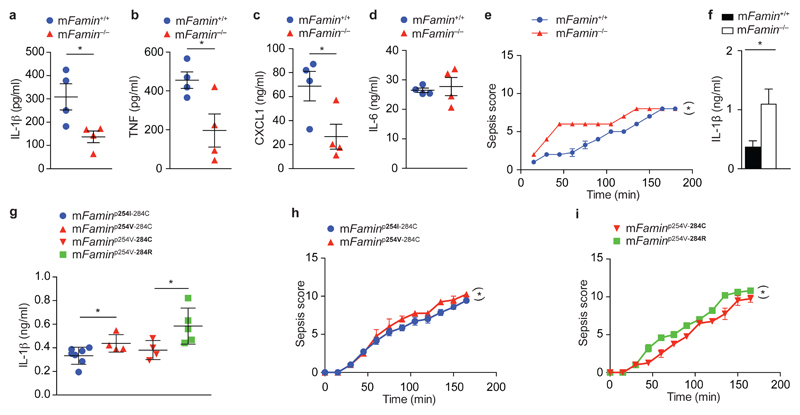
Mitochondrial ROS also plays a critical role in caspase-1-mediated maturation of IL-1β by the NLRP3 inflammasome31. This assembles on mitochondria-associated membranes (MAMs) upon triggering by pathogen-associated molecular patterns (PAMPs)31. NLRP3 inflammasome activation of mFamin–/– M1 macrophages with LPS + ATP resulted in lower levels of cleaved, active caspase-1p20 compared to mFamin+/+ cells, and consequently barely detectable cleaved IL-1βp17 in lysates and supernatants of mFamin–/– compared to mFamin+/+ M1 macrophages (Fig. 7d). Together, this demonstrates that FAMIN determines the capacity to trigger the NLRP3 inflammasome in M1 macrophages for the secretion of the host innate danger signal IL-1β. Moreover, FAMIN-dependent inflammasome activation was reliant upon intact FAO, as inhibition of FAO with etomoxir in LPS + ATP-stimulated wild-type M1 macrophages resulted in lower levels of cleaved IL-1βp17 secretion (Fig. 7e), as similarly observed in zymosan-stimulated M1 macrophages (Fig. 7f); no additive effect of FAO inhibition was observed in mFamin–/– M1 macrophages (Fig. 7e). A similar pattern of FAO-dependent cytokine regulation was observed in LPS-stimulated M1 macrophages for TNF and CXCL1, while IL-6 secretion remained unaffected (Fig. 7g–i). Importantly, intraperitoneal injection of 2 mg/kg LPS resulted in diminished IL-1β, CXCL1 and TNF serum levels in mFamin–/– compared to mFamin+/+ mice, while IL-6 levels remained unaffected (Fig. 8a–d). Notably, this pattern of LPS-induced serum cytokine regulation is reminiscent of inhibition of glycolysis in vivo with 2-DG11.
Figure 8. FAMIN deficiency causes dysregulated response to endotoxin in vivo.
(a–d) IL-1β, TNF, CXCL1 and IL-6 serum levels 2 h after intraperitoneal (i.p.) injection of LPS (2 mg/kg) of mFamin–/– and mFamin+/+ mice. (e) Clinical scoring of sepsis severity (maximum score 15) and (f) IL-1β serum levels 3 h after i.p. injection of LPS (10 mg/kg) of mFamin–/– and mFamin+/+ mice. (g) IL-1β serum levels 3 h after i.p. injection of LPS in 5 week-old mice homozygous for mFaminp254I-284C and their homozygous littermate mFaminp254V-284C controls; and homozygous mFaminp254V-284R mice and their homozygous mFaminp254V-284C littermate controls; (h,i) clinical scores of sepsis severity (maximum score 15). Each symbol (a–d,g) represents an individual mouse; small horizontal lines and error bars indicate the mean ± S.E.M. *P < 0.05, **P < 0.01 (Unpaired, two-tailed Student’s t-test (a–d,f,g) or linear mixed model (e,h,i)). Data are from one experiment with four mice per group (a–d), three mice per group (e,f; mean ± S.E.M.) or seven (mFaminp254I-284C), four (mFaminp254V-284C, mFaminp254V-284C) or five (mFaminp254V-284R) mice (g–i; mean ± S.E.M.).
Upon stimulation with LPS or LPS + ATP in vitro, mononuclear cells from patients with sJIA exhibit profoundly diminished IL-1β secretion compared to cells from healthy individuals, although it had remained unclear whether this is cause or consequence of active disease or is related to therapeutics32. Considering that FAMIN.C284R is the only firmly established genetic cause of sJIA to date1, 33, hypomorphic IL-1β induction upon stimulation with LPS and ATP in vitro is hence notably consistent with our results in mFamin–/– M1 macrophages. Nonetheless, blocking of IL-1 is therapeutic in the vast majority of sJIA patients, leading to the swift normalization of fever, rash, and acute phase reactants32, 34, 35. We therefore hypothesized that the profound impairment in energetic reserve in the absence of FAMIN function (e.g. low ATP and phosphocreatine, mitochondrial injury, reduced FAO and glycolysis) might limit the organism’s capacity to withstand serious injury. We injected a high dose of LPS (10 mg/kg) intraperitoneally, and continuously surveyed mFamin–/– and mFamin+/+ mice over a 3h period for clinical signs of sepsis. mFamin–/– mice exhibited higher sepsis scores (not covered by these scores, 2 out of 7 mFamin–/– mice also showed signs of epistaxis consistent with bleeding disorder, not observed in mFamin+/+ mice), particularly apparent during the first 90 minutes (Fig. 8e). This was associated with a more than 2-fold increase in IL-1β serum levels at the conclusion of the experiment (Fig. 8f). Turning to SNP-mutant mice, 10 mg/kg LPS resulted in highest IL-1β serum levels in mFaminp254V-284R mice, intermediate levels in mFaminp254V-284C mice, and lowest in mFaminp254I-284C mice as depicted in Fig. 8g. Correspondingly, mFaminp254V-284R mice exhibited higher clinical sepsis scores compared to mFaminp254V-284C, and mFaminp254V-284C higher than mFaminp254I-284C mice (Fig. 8h,i). Altogether, this demonstrates that in the absence of FAMIN function, high-dose LPS leads to catastrophic IL-1β activation. It can be speculated that the impaired bioenergetic reserve of mFamin–/– mice might tip the balance of sepsis-associated inflammasome activation towards a pyroptotic, pro-death response as has been reported in other contexts36.
Discussion
At an organismal level, fatty acid and glucose oxidation are reciprocal metabolic pathways, referred to as the Randle cycle37. Similarly, coupling of DNL and FAO is not energetically efficient in terms of ATP production, but nonetheless engaged in certain immune and non-immune cells38, 39, 40. In macrophages, FAMIN promotes flux through DNL to concomitantly drive high levels of both FAO and glycolysis. FAMIN thereby profoundly determines the maximum bioenergetic capacity of macrophages. Belonging to a new class of proteins (domain of unknown function 152 [DUF152] family), the closest orthologues of FAMIN with a reported function are bacterial proteins that exhibited atypical laccase (phenol oxidoreductase) activity41, 42. We purified recombinant eukaryotically expressed human FAMIN.p254I and FAMIN.p254V, and did not observe any laccase activity toward a series of prototypical substrates (data not shown). At this stage, it remains unclear how precisely FAMIN enhances DNL-dependent FAO.
Intriguingly, FAMIN-dependent FAO occurs even when uncoupled from ATP synthesis as in M1 macrophages, and may be of greatest importance in this setting for supporting glycolysis. ‘Uncoupled’ FAO has been suggested to have a critical role in cancer cells engaged in aerobic glycolysis43. Moreover, inhibition of DNL at the level of Acetyl-CoA carboxylase 1 (ACC1), the enzyme synthesising malonyl-CoA required for FASN function, has been reported to impair glycolysis in T helper cells44. As a consequence, we uncover a critical role of FAO in M1 macrophages that is required for IL-1β activation and ROS production. Glycolysis in M1 macrophages is well known to be critically required for IL-1β production11. By contrast, the importance of β-oxidation has received less attention even though FAO, not glycolysis, has been reported as the sole driver of bactericidal mROS production in Salmonella-infected macrophages45. Moreover, FAMIN-dependent mROS production also determines the maximum capacity of the NOX/DUOX oxidase system, the major bactericidal ROS source in M1 macrophages.
While metabolic pathways in bona fide immune cells are emerging as important determinants of immune function46, this is the first demonstration that a core metabolic regulator in immune cells is affected by genetic variation that causes, or predisposes for, immune-related disease and infection. Systemic JIA, which we demonstrate arises from a defect at the peroxisome – mitochondria interface, presents with a characteristic daily spiking (‘quotidian’) fever33. Deficiency in mevalonate kinase, a peroxisomal enzyme involved in cholesterol biosynthesis, causes a related juvenile periodic fever, hyper-IgD syndrome47. Remarkably, the mevalonate pathway has a common precursor with DNL, cytosolic acetyl-CoA, raising the possibility of commonality in mechanism. Intriguingly, both sJIA and hyper-IgD syndrome appear to be triggered by childhood infections or vaccinations33, 47. Diversity in such environmental triggers may also explain how homozygosity for p.C284R loss-of-function can cause two very different diseases, sJIA and EOCD1, 2, an aspect even more pertinent for the hypomorphic p.I254V, which predisposes for CD.
In summary, we report that FAMIN controls acyl-CoA synthesis through DNL at peroxisomes, thereby feeding FAO and regulating glycolysis. This has profound implications for cellular energy homeostasis and organismal immune function, with hypomorphy promoting autoinflammation.
Online Methods
Mice
Ear biopsy genomic DNA was used for routine genotyping of all mice. Primer sequences are shown in Supplementary table 3. C57BL/6N mice were bred and maintained in specific pathogen-free conditions at the Central Biomedical Services (CBS) facility, University of Cambridge and at the Wellcome Trust Sanger Institute. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted and conducted with approval of the UK home office. mFamin–/–, mFaminp284R and mFaminp254I mice generated as described below littered at Mendelian ratios, developed normally, and no spontaneous disease emerged under specific pathogen free (SPF) conditions. Six to twelve week-old mice that were age and sex-matched as described in the relevant methods sections were used for all experiments unless otherwise stated.
mFamin–/– allele
mFamin–/– mice were generated by disruption of the mouse homologue of human C13orf31 (9030625A04Rik) by homologous recombination in C57BL/6N-A/a embryonic stem (ES) cells (Supplementary Fig. 3) to generate mice on a pure C57BL/6NTac background; ES cell clone ID EPD0538_1_D06 (International Knockout Mouse Consortium)13.
CRISPR/Cas9 target sites and vector construction
CRISPR target sites were identified using http://crispr.mit.edu/ as described28 and are shown in Supplementary Table 4. The strategy to generate mouse lines carrying the p.254V and p.284R FAMIN disease associated variants (corresponding residue numbers are identical between human C13orf31 and murine 9030625A04Rik) is shown in Supplementary Fig. 3. In brief, wild-type C57BL/6NTac mice carry p.254V (human ‘risk’ variant) and p.284C. For introduction of the ‘non-risk’ p.254I allele, guide RNA (gRNA) ‘line7’ along ‘oligo 7’ or ‘oligo 7 wobble’ were used, the latter introducing two additional synonymous nucleotide changes (Supplementary Table 4 and Supplementary Fig. 3). The corresponding strategy for introduction of the ‘Mendelian variant’ p.284R with gRNA ‘line9’ along ‘oligo 9’ or ‘oligo 9 wobble’ is shown in Supplementary Fig. 3. Oligonucleotide tyrex2R (Supplementary Table 4) was co-injected with each of the four lines to target the Tyr gene, allowing for color-based selection. The selected gRNA sequences were cloned into a gRNA vector containing the gRNA backbone and a T7 promoter to facilitate RNA production (BsaI sites). Cas9 mRNA was produced using a previously described vector modified to contain a T7 promoter48. The integrity of all plasmids was confirmed by DNA sequencing. The targeting oligonucleotides were synthesized as desalted ssDNA oligonucleotides from Integrated DNA Technologies (IDT).
Cas9 and gRNA production
For Cas9 RNA production the T7/Cas9 plasmid was linearized (EcoRI), purified with PCR purification kit (Qiagen) and in vitro transcribed using mMessage mMachine T7 Ultra kit (Life Technologies). For gRNA production the plasmid was linearized (DraI), purified with PCR purification kit (Qiagen) and in vitro transcribed using the MEGAshortscript T7 kit (Life Technologies). Both, Cas9 mRNA and gRNA, were purified using the MEGAclear kit (Life Technologies) and eluted in RNase-free water. The quality of the RNA was analyzed using Agilent RNA 6000 Nano kit (Agilent Technologies, 2100 Bioanalyzer) and Qubit RNA HS assay kit (Life Technologies).
One-cell embryo injection
Twelve 4-5 week old C57BL/6NTac females were super-ovulated by intraperitoneal (i.p.) injection of 5 IU of pregnant mare’s serum (PMSG) at 12:00-13:00 h (on a 12 h light/dark cycle, on at 07:00/off at 19:00) followed 48 h later by an i.p. injection of 5 IU human chorionic gonadotrophin (hCG) and mated overnight with C57BL/6NTac stud males. The next morning the females were checked for the presence of a vaginal copulation plug as evidence of successful mating and oviducts were dissected at approximately 21-22 h post hCG. Cumulus oocyte complexes from these were released and treated with hyaluronidase as previously described and the fertilized 1-cell embryos were left in KSOM media ready for cytoplasmic injection49. 50 ng/μl Cas9 mRNA, 25 ng/μl gRNA and 100 ng/μl oligonucleotide were mixed in RNase-free water and injected into the cytoplasm of fertilized 1-cell embryos in FHM medium. 1-cell injected embryos were transferred the same day by oviductal embryo transfer into 0.5 days post-coital pseudo-pregnant female BJCB strain (CBA<Wtsi>;C57BL/6J-Jax F1) recipients, produced by estrus selecting and overnight mating to vasectomized males49.
Cytoplasmic injections
Injections were performed using a new cytoplasmic injection technique. An injector using a positive balance pressure such as an Eppendorf Femtojet was connected to a microinjection tip filled with the CRISPR materials. The microinjection tip was steadily advanced towards the opposite side of the oocyte, which was anchored by the holding pipette until the micropipette passed through the plasma membrane to break it. The pipette was then drawn back into the cytoplasm, the CRISPR mix delivered and the micropipette immediately withdrawn. A successful injection was indicated by visible movement in the cytoplasm.
SNP mutant mice breeding strategy
Illumina sequencing of the mFamin gene was used in founder and F1 generation mice, confirmed by Sanger sequencing, to assess for successful gene editing. Founder mice were crossed with wild-type C57BL/6NTac mice to generate mFaminp254I/p254V and mFaminp284R/p284C heterozygous mice, which were then intercrossed, and homozygous littermates used for experiments. Mice generated with and without wobble bases in targeting oligonucleotides were maintained separately, and used in experiments alongside their littermate controls irrespective of the presence or absence of wobble bases. Offspring from several founders were used to populate genotype categories in experiments. Routine genotyping was performed via Sanger sequencing.
Genotype-selected human samples
Peripheral blood for human neutrophil and monocyte isolation was collected from healthy volunteers from a genotype-selectable bioresource of ~10,000 individuals (Cambridge BioResource [CBR], http://www.cambridgebioresource.org.uk). Healthy age- and sex-matched individuals identified as homozygous for the risk (G/G) and non-risk (A/A) haplotype at the C13orf31 nsSNP rs3764147, encoding p.254V and p.254I respectively, were recruited to participate in the study. None of the volunteers had received corticosteroid or immunosuppressant therapy within 6 months of enrollment. Investigators were kept blinded to the genotype of samples until study completion, with all individual experimental procedures performed on (blinded) genotype pairs. The study protocol has been reviewed and approved by the Berkshire B Research Ethics Committee (15/SC/0068), and written informed consent was obtained from each subject.
Antibodies and reagents
The following antibodies were used for immunoblotting. Abcam: anti-PMP70 (1:1,000 dilution; ab3421), anti-FASN (1μg/ml dilution; ab22759); Cell Signaling: anti-cytochrome oxidase IV (1:250 dilution; 4850), anti-catalase (1:800 dilution; 12980), anti-centromere protein A (1:400 dilution; 2186), anti-calreticulin (1:200 dilution; 12238), anti-FASN (1:1,000 dilution; 3180); Santa Cruz Biotechnology: anti-C13orf31 (1:200 dilution; sc-374553; E7 and 1:500 dilution; sc-376231; E12), anti-caspase 1 p20 (1:250 dilution; sc-1218-R); R&D Systems: anti-IL-1β (1:250 dilution; AF-401-NA); Sigma: anti-β-actin (1:10,000 dilution; A5060). All antibodies used have validation profiles on either Antibodypedia or 1DegreeBio.
The following reagents were used: M-CSF (Peprotech, 300-25), LPS (from Escherichia coli K12, InvivoGen, tlrl-peklps), human IFN-γ (Peprotech, 300-02), murine IFN-γ (Peprotech, 315-05), human IL-4 (Peprotech, 200-04), murine IL-4 (Peprotech, 214-14), ATP (Sigma Aldrich, A2383), C75 (Sigma Aldrich, C5490), etomoxir (Sigma Aldrich, E1905), MitoTEMPO (Sigma Aldrich, SML0737), oligomycin (Sigma Aldrich, O4876), FCCP (Sigma Aldrich, C2920), rotenone (Sigma Aldrich, R8875), antimycin A (Sigma Aldrich, A8674), palmitate-BSA (Seahorse Bioscience, 102720-100), zymosan A (Sigma Aldrich, Z4250), PMA (Sigma Aldrich, P1585), HRP (Sigma Aldrich, P8375), luminol (Sigma Aldrich, A8511).
Murine bone marrow-derived macrophages and neutrophils
Bone marrow was flushed from mouse femurs and tibias, filtered through a 70 μm cell strainer and incubated in complete medium (RPMI-1640 containing 100 U/ml of penicillin-streptomycin, 1 mM HEPES pH 7.4) and 10% FBS. Bone marrow derived macrophages (BMDMs) were prepared by culturing cells for 6 days in the presence of 100 ng/ml of M-CSF. After harvesting and re-seeding, Macrophages were polarized for 16 h toward M1 and M2 with IFN-γ (50 ng/ml) plus LPS (20 ng/ml) and IL-4 (20 ng/ml), respectively, and where indicated, in the presence or absence of C75 (20 μM) and etomoxir (100 μM). Bone marrow neutrophils were prepared using a neutrophil isolation kit (Miltenyi Biotec) according to manufacturer’s instructions.
Human peripheral blood-derived macrophages
For studies with human cells, blood received from healthy donors was used to isolate peripheral blood mononuclear cells and neutrophils by Lymphoprep (Axis-Shield) gradient centrifugation. Monocyte-derived macrophages were generated by resuspending cells in complete RPMI-1640 and plated on Nunclon surface tissue culture dishes (Nunc). After 2 h, non-adherent cells were discarded and the remaining monocytes cultured for 4 days in complete medium with 10% FBS and 40 ng/ml M-CSF. At day 4, macrophages were harvested and polarized into M1 and M2 as described for murine macrophages. Neutrophils were concurrently isolated during Lymphoprep gradient centrifugation (Axis-Shield) by sedimentation of the red blood cell/neutrophil layer with 10% dextran, followed by hypotonic lysis of contaminating erythrocytes.
Cell lines
THP-1 and U937 cell lines were maintained in complete RPMI-1640 with 10% FBS. To polarize into M1 macrophages, cells were incubated overnight with 50 ng/ml IFN-γ plus 20 ng/ml LPS and for M2 macrophages, cells were treated with 1 μM of PMA for 6 h and then polarized with 50 ng/ml IL-4 and 50 ng/ml IL-13. HEK293 and HEK293T cells were maintained in DMEM with 10% FBS. All cell lines were purchase from ATCC, which guarantee cell line authentication.
Plasmids
FAMIN was amplified from an IMAGE clone (MHS1010-7508636) using a forward primer containing a KpnI site followed by a Kozak sequence and start codon and reverse primer containing a BamHI site, excluding the stop codon of FAMIN and was cloned into KpnI and BamHI digested and dephosphorylated pEXPR-IBA103 (IBA Life Sciences) to generate C-terminal Strep III tag FAMIN. The same IMAGE clone was amplified with a forward primer containing a SacII site and a reverse primer containing an XhoI site and cloned into pEXPR-IBA105 to generate N-terminal Strep III tag FAMIN. Strep-tag III is a sequential arrangement of two Strep-tag II sequences (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) separated by a short linker. Constructs were sequence verified, with Strep-tag III in-frame withFAMIN. FAMIN containing a stop codon was also cloned into pEXPR-IBA103 for use as an untagged FAMIN control. FAMIN variants were introduced by site-directed mutagenesis (Agilent Quick change II XL SDM kit) using the above constructs as template. To introduce the p.254V mutation the following primers were used: Forward, 5′-AGACTCATCATTCCAATGACGTCTGGATTATGGGAAGAAAG-3′; Reverse, 5′-TAAAGACTCATCATTCCAATGACATCTGGATTATGGGAAGAAAGGAG-3′ and to introduce the p.284R mutation the following primers were used: Forward, 5′-CAAAAACTATCGGTATACGGTCTGCACCAAGAGCTGC-3′; Reverse 5′-GCAGCTCTTGGTGCAGACCGTATACCGATAGTTTTTG-3′. All constructs were sequence verified in both forward and reverse orientations.
Protein affinity-purification mass spectrometry
HEK293T cells were transfected with pEXPR-IBA103 vector only, untagged FAMIN, N-terminal Strep-tag FAMIN or C-terminal Strep-tag FAMIN plasmid constructs using Lipofectamine 2000 (Invitrogen) following manufacturer's instructions. 48 h following transfection, cells were washed twice in ice-cold PBS and harvested by gently scraping into 5 ml ice-cold lysis buffer containing 20 mM HEPES-KOH, pH 7.5, 150 mM KCl, 1% Nonidet P-40, 0.1 mM phenylmethylsulfonyl fluoride, and protease inhibitor mixture. Cells were incubated on ice for 10 min then homogenized by shearing 20 times through a 21-gauge needle. The protein lysate was centrifuged for 15 min at 4 °C at 3,000g and protein supernatant collected into a freshly chilled tube to remove cellular debris. 1 U/ml of avidin was added to block endogenous biotin and incubated on ice for 15 min and then re-centrifuged for a further 10 min at 4 °C. Supernatant containing strep-tag labeled FAMIN protein was pulled down using a Strep-Tactin column (IBA Life Sciences). Strep-tagged FAMIN protein complexes were eluted using 2.5 mM desthiobiotin and eluted fractions were subjected to SDS-PAGE separation followed by silver stain or immunoblot. Gels were submitted to the Cambridge Centre for Proteomics, Department of Biochemistry and were excised manually. Proteins were reduced, carboxyamide methylated, and then digested to peptides using trypsin on a MassPrepStation (Micromass). The resulting peptides were applied to liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) on a QTof (Micromass). Fragmentation data were used to search the National Center for Biotechnology Information database using the MASCOT search engine (http://www.matrixscience.com)
Lipid mass spectrometry
Day-6 macrophages were seeded at 5 × 106 cells per 60 mm dish and polarized under M1 or M2 conditions. Cells were harvested, washed twice in ice-cold PBS and then flash frozen in liquid nitrogen. For 1, 2-13C-glucose–tracing experiments, macrophages were polarized overnight and then cultured for 24 h in the presence of 10 mM 1,2-13C-glucose (Sigma). Cell pellets were washed twice with cold PBS and re-suspended in 1.5 ml methanol. After spiking with 40 μl lipid standards cocktail mix of 50 ng 17:0-FaCN, 100 ng 17:0-FaCoA and 400 ng 17:0-FFA, 1.5 ml of LCMS-grade water and 3 ml chloroform were added in. The mixture was subjected to Folch extraction. After collection of the lower phase, the upper phase was re-extracted with 3 ml synthetic lower phase (chloroform/methanol/water at volume ratio of 2:1:1, using the lower phase for re-extraction of lipid). The lower phase from both extractions was combined and dried under vacuum at 20 °C with SpeedVac (Thermo) and re-dissolved in 100 μl chloroform. 7 μl were injected for LC/MS/MS analysis. A Thermo Orbitrap Elite system (Thermo Fisher) hyphenated with a five-channel online degasser, four-pump, column oven, and autosampler with cooler Shimadzu Prominence HPLC system (Shimadzu) was used for lipid analysis. In detail, lipid classes were separated on a normal phase silica gel column (150 × 2.1 mm, 4 μm, MicoSolv Technology) with hexane/dichloromethane/chloroform/methanol/acetanitrile/water/ethylamine solvent gradient based on the polarity of head group. High resolution (240k at m/z 400) / accurate mass (with mass accuracy <5 ppm) were used for molecular species identification and quantification. The identity of lipid was further confirmed by reference to appropriate lipids standards. Orbitrap Elite mass spectrometer operation conditions: A. For positive ion analysis, Heated ESI Source in positive ESI mode, Heater Temperature: 325 C, Sheath Gas Flow Rate (arb): 35, Aux Gas Flow Rate (arb): 5, Sweep Gas Flow Rate (arb): 0, I Spray Voltage: 3.5 kV, Capillary Temperature: 325 C, S-Lens RF Level: 60%. Orbitrap mass analyzer was operated as SIM scan mode with two events. Event 1, mass range: m/z 238-663, mass resolution: 240 k at m/z 400. Event 2, mass range: m/z 663-1088, mass resolution: 240 k at m/z 400. B. For negative ion analysis, Heated ESI Source in negative ESI mode, Heater Temperature: 325 C, Sheath Gas Flow Rate (arb): 45, Aux Gas Flow Rate (arb): 10, Sweep Gas Flow Rate (arb): 0, I Spray Voltage: 3.0 kV, Capillary Temperature: 375 C, S-Lens RF Level: 70%. Orbitrap mass analyzer was operated as SIM scan mode with two events. Event 1, mass range: m/z 218-628, mass resolution: 240 k at m/z 400. Event 2, mass range: m/z 628-1038, mass resolution: 240 k at m/z 400. All the solvents used for lipids extraction and LC/MS/MS analysis are LC-MS grade from FisherScientific.
Aqueous metabolites mass spectrometry
Metabolites were extracted from 5 × 106 macrophages using methanol/chloroform. The organic and aqueous fractions were separated by the addition of water and chloroform. The aqueous layer was analyzed subsequently for metabolomics. Mass spectrometry was performed on an AB Sciex 5500 (Warrington) coupled to an Acquity ultra performance liquid chromatography (UPLC) system from Waters Ltd. For chromatography of Krebs cycle intermediates metabolites were separated on a BEH amide HILIC column (100 × 2.1 mm, 1.7 μm; Waters Ltd) in negative ion mode. After drying, the samples were analyzed on a HSS T3 column (100 × 2.1 mm, 1.7 μm; Waters Ltd). Cone voltage, collision energy and mass transitions were optimized for each metabolite for quantification. Data are expressed in arbitrary units as the area ratio of metabolite peak relative to the internal standard.
ATP assay
ATP content in total cell lysates was measured using an ATP determination kit (Molecular probes, A22066). Briefly, M1 and M2 macrophages were trypsinized after an overnight polarization, counted and 2 × 105 cells were lysed in 100 mM Tris pH 7.75, 4 mM EDTA and 1% Triton-X. Protein concentration was determined by BCA (Pierce, 23225). Remaining lysates were boiled at 96 °C, spun down for 1 min at 1,000g and the assay was performed according to the manufacturer's protocol. The amount of ATP was determined from the standard curve and expressed as nM of ATP per mg of protein in the sample.
NADPH quantification
NADPH concentration was determined using the NADP/NADPH Assay kit (Abcam, ab65349) according to the manufacturer’s protocol. Briefly, 4 × 106 macrophages were harvested in extraction buffer and filtered through a 10-kDa Spin column (Abcam, ab93349). The NADPH cycling reaction was performed for 2 h and NADPH amount was determined from the standard curve and normalized to protein concentration.
Oxygen consumption rate and extracellular acidification rate
Macrophages were plated in XF-96 cell culture plates (7.5 × 105 cells/well) and polarized to M1 and M2 in the presence or absence of 20 μM C75 for 16 h. Macrophages were then washed and incubated for 1 h in XF assay medium (unbuffered DMEM pH 7.4 with 10 mM glucose and 2 mM L-glutamine, with or without 2 mM sodium pyruvate) in a non-CO2 incubator at 37 °C as per manufacturer’s instructions (Seahorse Bioscience). Real time measurements of macrophage extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were performed using an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). Three or more consecutive measurements were obtained under basal conditions and after the sequential addition of 1 μM oligomycin, to inhibit mitochondrial ATP synthase; 1.5 μM FCCP (fluoro-carbonyl cyanide phenylhydrazone), a protonophore that uncouples ATP synthesis from oxygen consumption by the electron-transport chain; and 100 nM rotenone plus 1 μM antimycin A, which inhibits the electron transport chain. SRC is calculated as the difference between basal OCR and maximal OCR after the addition of FCCP. To assess glycolysis, three or more consecutive ECAR measurements were obtained under basal conditions and after the sequential addition of 1 μM oligomycin, to elicit maximal glycolytic capacity and 100 mM 2-DG (2-deoxyglucose) to inhibit glycolytic dependent ECAR. Measurement of endogenous and exogenous fatty acid oxidation (FAO) was performed according to manufacturer’s instructions (Seahorse Bioscience). In brief, macrophages were seeded and polarized toward M2 as above. After 16 h, macrophages were cultured in substrate limited DMEM medium containing 0.5 mM glucose, 1 mM GlutaMAX, 0.5 mM carnitine and 1% FBS for 24 h. Macrophages were then washed and incubated for 1 h in FAO assay medium (111 mM NaCl, 4.7 mM KCl, 1.25 mM CaCl2, 2 mM MgSO4, 1.2 mM NaH2PO4) supplemented with 2.5 mM glucose, 0.5 mM carnitine, and 5 mM HEPES, pH 7.4 in the presence or absence of 40 μM etomoxir in a non-CO2 incubator at 37 °C. 1 mM palmitate conjugated to 0.17 mM BSA or BSA control was added to respective wells immediately prior to OCR measurement on XF-96 analyzer (Seahorse Bioscience).
ROS measurement
For mitochondrial and intracellular ROS measurements, macrophages were seeded at 1 × 105 cells per well in a 96-well plate and polarized toward M1 and M2 as described above. Macrophages were then washed with warm PBS and incubated at 37°C with 5 μM MitoSox (Invitrogen), mitochondrial superoxide indicator, or 10 μM CM-H2DCFDA (Invitrogen), cytosolic ROS indicator, in serum-free phenol red-free RPMI 1640 (Life Technologies) for 15 or 30 min respectively. Following washing, the fluorescence intensity of MitoSox and CM-H2DCFDA were measured on a ClarioStar plate reader at Excitation/Emission spectra of 510/580 nm or 495/520 nm respectively. Extracellular ROS kinetics was assessed using a horseradish peroxidase (HRP)-luminol-dependent chemiluminescence method. 2 × 106 cells per well were plated in a 6-well plate and polarized toward M1 and M2 and treated prior to assay with 20 μM C75, 100 μM etomoxir or 500 μM mitoTEMPO as indicated in the figure legends. Macrophages were harvested, washed in PBS then re-suspended in PBS (with added CaCl2 and MgCl2) using 5 × 105 cells per assay sample; for human macrophages 2.5 × 105 cells were used. 1 μM Luminol and 62.5 U/ml HRP were added to the cell suspension for 3 min and cells were then transferred to a white 96-well luminometer plate (Berthold Technologies). 200 ng/ml PMA, 200 μg/ml serum-opsonized zymosan or PBS control was added directly to each well and light emission recorded by a Berthold Centro LB 960 luminometer (Berthold Technologies). Experiments with murine and human neutrophils were similarly performed.
In vitro infection assays
Macrophages were infected with S. Typhimurium SL1344 for 45 min at MOI 10 in media containing 10% FCS or serum free media (Opti-MEM 1). Cells were then washed and maintained for a further 45 min with media containing 50 μg/ml gentamicin before replacing for media without antibiotics. After 2 h, cells were washed with PBS and lysed with 0.1% Triton X-100 before plating serial dilutions in triplicate onto selective agar. For Mycobacterium bovis Bacillus Calmette Guérin (BCG) strain infection, macrophages were infected with BCG containing a luciferase reporter for 0, 4, and 24 h30. Cells were then washed and lysed and relative light units (RLU), as direct correlates of colony forming units (c.f.u.), were measured as described30.
In vitro macrophage cytokine stimulation assays
1.5 × 105 macrophages per well were seeded in a 96-well plate. Cells were then washed and pre-treated with 20 μM C75, 100 μM etomoxir or vehicle only for 2 h and then stimulated with 200 μg/ml zymosan or 1 μg/ml of LPS. Supernatants were collected after 24 h and measured for IL-1β, IL-6, CXCL1 and TNF with ELISA according to manufacturer’s instruction (BD Bioscience).
In vivo endotoxin administration
E. coli LPS (2 mg/kg or 10 mg/kg) was injected intraperitoneally (i.p) into mice. All mice were male and aged between 5 and 8 weeks. At indicated time-points following i.p. LPS, serum was obtained by intracardiac puncture under terminal isoflurane anesthesia and mice were sacrificed. A graded scoring system, adapted from50, was employed to monitor the clinical severity of sepsis every 15 min following LPS injection, which consisted of scoring of respiration rate / breathing pattern (0-5), activity and movement (0-5) and appearance including changes to coat and eyes (0-5) conducted by two scientists independently.
Immunoblot
Cells were washed once in ice-cold PBS and lysed in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Igepal, 0.5% sodium deoxycholate, 0.1% SDS) with protease (Thermo Fisher Scientific, 78410) and phosphatase inhibitors (Thermo Fisher Scientific, 78426). Protein concentration was determined by BCA assay (Thermo Fisher Scientific, 23225) and equal amounts of proteins loaded onto SDS polyacrylamide gels. The proteins were transferred onto nitrocellulose membrane using a Trans-Blot Turbo transfer system (Bio-Rad). Membranes were incubated overnight with primary antibody, followed by corresponding secondary antibody and the membranes were developed using 20X LumiGLO® Reagent (Cell Signaling, 7003S).
Immunoprecipitation (IP)
Cells were washed three times with ice-cold PBS and then directly lysed with Tris-Triton buffer (10 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton-X100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate) with added protease (Thermo Scientific, 78410) and phosphatase (Thermo Scientific, 78426) inhibitor cocktails. Lysed cells were centrifuged at 9,300g for 5 min at 4 °C to remove cell debris. Lysates were pre-cleared with either rabbit or mouse IgG bound to protein A sepharose beads (GE Healthcare, 17-5138-01) suspended in a 10% BSA-PBS solution rotated for 30 min at 4 °C. Pre-cleared lysates were added to protein A sepharose beads coated with a pull-down antibody (1:100) and rotated at 4 °C for 1 h. Beads were washed with lysis buffer and the bound proteins analyzed by immunoblot.
Precipitation of IL-1β from supernatants
Equal volumes of supernatants were precipitated using the TCA-DOC method. Briefly, 1/100 of 2% sodium deoxycholate was added to one volume of the supernatant, vortexed and left for 30 min at 4 °C. Next, 1/10 of trichloroacetic acid was added, samples vortexed and left overnight at 4° C. Samples were then spun down for 15 min at 4 °C at 15,000g. Pellets were washed once with one volume of ice cold acetone, air dried, resuspended in 2x loading buffer (4% SDS, 120 mM Tris HCl pH 6.8, 10% glycerol, 100 mM DTT and dash of bromphenol blue), boiled for 5 min and ran on the SDS-Page gel for further analysis.
Silver stain
Polyacrylamide gels were fixed for 1 h at 20 °C in 40% ethanol with 10% acetic acid. Gels were washed overnight with distilled water and then sensitized with 0.02% sodium thiosulphate for 1 min, washed and then stained for 20 min at 4 °C with 0.1% silver nitrate with 0.02% formaldehyde. Gels were developed with 3% sodium carbonate with 0.05% formaldehyde and staining terminated with 5% acetic acid for 5 min.
Immunofluorescence staining
Cells were seeded on glass coverslips and differentiated overnight as required. Cells were fixed with 4% paraformaldehyde containing either 0.2% or 1% Triton X-100 for 15 min at 20 °C or 100% methanol at -20 °C for 15 min. Fixed cells were washed with PBS and non-specific binding blocked with either 5% normal serum or 0.5% milk before incubation with primary antibodies for 1 h at 20 °C. Unbound antibody was removed by washing with PBS and secondary antibody bound for 30 min at 20 °C. Coverslips were then mounted with either Prolong or Mowiol mountant containing DAPI. Fluorescence was visualized with a Zeiss LSM 510 or a DeltaVision Spectris Deconvolution Microscope.
Proximity-Ligation Assay
Cells were fixed and stained with primary antibody as described above for immunofluorescence. Proximity-ligation assay (PLA) was performed as per manufacturer’s instructions (Duolink® In Situ, Olink Bioscience). In brief, unbound antibody was removed by washing twice for 5 min with 1x wash buffer A and PLA probes bound for 1 h at 37 °C. Unbound PLA probes were removed by washing twice for 5 min with 1x wash buffer A. Coverslips were incubated with Ligation-Ligase solution for 30 min at 37 °C. Ligation-Ligase solution was removed by washing twice for 2 min with 1x wash buffer A and coverslips were incubated with Amplification-Polymerase solution for 100 min at 37 °C. Amplification-Polymerase solution was removed first by washing twice for 10 min with 1x wash buffer B and second by washing 1 min with 0.01% Wash buffer B. Coverslips were then mounted and fluorescence visualized as described in immunofluorescence staining.
Oil Red O staining
MΦ were seeded onto glass coverslips and allowed to differentiate overnight. Cells were washed twice with PBS and then incubated for 3 min in 100% propylene glycol (VWR) before staining with Oil Red O solution (0.5% Oil Red O [Amresco] in propylene glycol) for 10 min at 20 °C. Excess stain was removed by washing with 85% propylene glycol and washing with distilled water. Cells were counterstained with hematoxylin and mounted with Kaiser’s glycerol gelatine (Merck Millipore).
Transmission electron microscopy
Cells were fixed in 4% glutaraldehyde in 0.1M HEPES (pH 7.4) for 12 h at 4 °C, washed with 0.1M HEPES 5 times and then treated with 1% osmium ferricyanide at 20 °C for 2 h. Cells were rinsed in water 5 times and treated with 2% uranyl acetate in 0.05M maleate buffer (pH 5.5) for 2 h at 20 °C, rinsed and dehydrated in an ascending series of ethanol solutions (70% to 100%) and then treated with 2 changes of dry acetonitrile prior to infiltration with Quetol epoxy resin. Images were taken in an FEI Tecnai G2 operated at 120Kv using an AMT XR60B digital camera running Deben software.
Quantitative real-time PCR
Total RNA was isolated from cultured cells and tissues using RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. For RT-PCR, cDNA was synthesized using oligo(dT) primers and M-MLV reverse transcriptase followed by quantitative PCR analysis. The primers are shown in Supplementary Table 5.
Statistical analysis
Statistical significance was calculated as appropriate using unpaired, two-tailed Student’s t-test, Mann-Whitney U-test or ANOVA/Bonferroni as described in the figure legends. Data is represented as mean and standard error of the mean (S.E.M.). A P-value of <0.05 was considered significant. Data were analyzed with Graphpad Prism software (version 6).
Supplementary Material
Note: Any supplementary information and source data files are available in the online version of the paper.
Acknowledgments
We are indebted to G. Brown for help with laccase assays, J. Murkin and M. Deery for proteomics, L. Porter for metabolic flux assays, J. Skepper for electron microscopy, I. Purvis for help with in vivo procedures, R. Rodrigues, M. Md-Ibrahim and J. Jones for cellular assays, S. Dhillon for generation of constructs, and to T. Lawley, M. Pardo, J. Choudhary, K. Smith, J. Lee, D. Thomas, G. Schneditz, L. Haag, M. Parkes and R. Blumberg for helpful discussions. We gratefully acknowledge the participation of all NIHR Cambridge BioResource volunteers. We thank the Cambridge BioResource staff for their help with volunteer recruitment. We thank members of the Cambridge BioResource SAB and Management Committee for their support of our study and the National Institute for Health Research Cambridge Biomedical Research Centre for funding. Access to Cambridge BioResource volunteers and their data and samples is governed by the Cambridge BioResource SAB. Documents describing access arrangements and contact details are available at http://www.cambridgebioresource.org.uk/. Expertise and help from the NIHR Cambridge BRC Cell Phenotyping Hub is greatly acknowledged. This work was supported by the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement no. 260961 (A.K.), the Wellcome Trust (investigator award 106260/Z/14/Z to A.K., a PhD fellowship for clinicians to M.Z.C., and a Career Re-Entry Fellowship to N.C.K.), the Wellcome Trust Sanger Institute (G.D., A.B., S.M., S.C. and K.B.), the National Institutes of Health (NIH) grants 5U420D011174 and 5U54HG006348 (A.B. and K.B.), the Biotechnology and Biological Sciences Research Council BBSRC (M.J.W.), the Cambridge Biomedical Research Centre (A.K.), fellowship from the European Crohn’s and Colitis Organisation (M.T.), and grants from the Swedish Medical Research Council and the Olle Engkvist foundation (M.D’A.).
Footnotes
Author Contributions
M.Z.C., together with S.L.K., G.W.S., S.S., J.W.A., M.T., T.R. and N.C.K. designed and performed most experiments. K.B., B.D. and A.B. designed, generated and confirmed CRISPR/Cas9 mouse lines, Q.Z. and M.J.W. contributed lipidomics experimentation and analysis, J.L.G. contributed metabolomics experimentation and analysis, S.C., S.M. and G.D. contributed Salmonella and part of in vivo experimentation, K.P.B. and R.A.F. contributed mycobacterial experiments, G.A. and M.D’A. identified cellular localization of C13orf31, E.R.C. helped with metabolic flux assays and ROS experimentation, and A.K. devised and coordinated the project, and together with M.Z.C. and G.D., with contributions from all authors, designed experiments, interpreted data and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Wakil SM, et al. Association of a mutation in LACC1 with a monogenic form of systemic juvenile idiopathic arthritis. Arthritis & rheumatology. 2015;67:288–295. doi: 10.1002/art.38877. [DOI] [PubMed] [Google Scholar]
- 2.Patel N, et al. Study of Mendelian forms of Crohn's disease in Saudi Arabia reveals novel risk loci and alleles. Gut. 2014;63:1831–1832. doi: 10.1136/gutjnl-2014-307859. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nature genetics. 2015;47:267–271. doi: 10.1038/ng.3212. [DOI] [PubMed] [Google Scholar]
- 4.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hruz T, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in bioinformatics. 2008;2008 doi: 10.1155/2008/420747. 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heng TS, Painter MW, Immunological Genome Project, C The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 7.Jensen-Urstad AP, Semenkovich CF. Fatty acid synthase and liver triglyceride metabolism: housekeeper or messenger? Biochim Biophys Acta. 2012;1821:747–753. doi: 10.1016/j.bbalip.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen-Urstad AP, et al. Nutrient-dependent phosphorylation channels lipid synthesis to regulate PPARalpha. Journal of lipid research. 2013;54:1848–1859. doi: 10.1194/jlr.M036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell metabolism. 2014;19:380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang SC, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nature immunology. 2014;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillebrand M, et al. Identification of a new fatty acid synthesis-transport machinery at the peroxisomal membrane. The Journal of biological chemistry. 2012;287:210–221. doi: 10.1074/jbc.M111.272732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenkovich CF. Regulation of fatty acid synthase (FAS) Progress in lipid research. 1997;36:43–53. doi: 10.1016/s0163-7827(97)00003-9. [DOI] [PubMed] [Google Scholar]
- 16.Jha AK, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Grevengoed TJ, Klett EL, Coleman RA. Acyl-CoA metabolism and partitioning. Annual review of nutrition. 2014;34:1–30. doi: 10.1146/annurev-nutr-071813-105541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nature reviews. Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. American journal of physiology. Cell physiology. 2007;292:C125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 20.Hao W, Chang CP, Tsao CC, Xu J. Oligomycin-induced bioenergetic adaptation in cancer cells with heterogeneous bioenergetic organization. J Biol Chem. 2010;285:12647–12654. doi: 10.1074/jbc.M109.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newsholme EA, Sugden PH, Williams T. Effect of citrate on the activities of 6-phosphofructokinase from nervous and muscle tissues from different animals and its relationships to the regulation of glycolysis. The Biochemical journal. 1977;166:123–129. doi: 10.1042/bj1660123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Vats D, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell metabolism. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landree LE, et al. C75, a fatty acid synthase inhibitor, modulates AMP-activated protein kinase to alter neuronal energy metabolism. The Journal of biological chemistry. 2004;279:3817–3827. doi: 10.1074/jbc.M310991200. [DOI] [PubMed] [Google Scholar]
- 25.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell metabolism. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy MP. How mitochondria produce reactive oxygen species. The Biochemical journal. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu L, Quinn MT, Cross AR, Dinauer MC. Gp91(phox) is the heme binding subunit of the superoxide-generating NADPH oxidase. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7993–7998. doi: 10.1073/pnas.95.14.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West AP, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kampmann B, et al. Evaluation of human antimycobacterial immunity using recombinant reporter mycobacteria. The Journal of infectious diseases. 2000;182:895–901. doi: 10.1086/315766. [DOI] [PubMed] [Google Scholar]
- 31.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 32.Gattorno M, et al. The pattern of response to anti-interleukin-1 treatment distinguishes two subsets of patients with systemic-onset juvenile idiopathic arthritis. Arthritis and rheumatism. 2008;58:1505–1515. doi: 10.1002/art.23437. [DOI] [PubMed] [Google Scholar]
- 33.Mellins ED, Macaubas C, Grom AA. Pathogenesis of systemic juvenile idiopathic arthritis: some answers, more questions. Nature reviews. Rheumatology. 2011;7:416–426. doi: 10.1038/nrrheum.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quartier P, et al. A multicentre, randomised, double-blind, placebo-controlled trial with the interleukin-1 receptor antagonist anakinra in patients with systemic-onset juvenile idiopathic arthritis (ANAJIS trial) Annals of the rheumatic diseases. 2011;70:747–754. doi: 10.1136/ard.2010.134254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. The Journal of experimental medicine. 2005;201:1479–1486. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. The Journal of experimental medicine. 1997;185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. American journal of physiology. Endocrinology and metabolism. 2009;297:E578–591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan HP, et al. A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nat Med. 2002;8:1122–1128. doi: 10.1038/nm780. [DOI] [PubMed] [Google Scholar]
- 39.O'Sullivan D, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Windt GJ, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beloqui A, et al. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, structural analysis, and phylogenetic relationships. The Journal of biological chemistry. 2006;281:22933–22942. doi: 10.1074/jbc.M600577200. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, et al. Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution. Proteins. 2006;63:1097–1101. doi: 10.1002/prot.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samudio I, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berod L, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nature medicine. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 45.Hall CJ, et al. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating beta-oxidation-dependent mitochondrial ROS production. Cell metabolism. 2013;18:265–278. doi: 10.1016/j.cmet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 46.O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulders-Manders CM, Simon A. Hyper-IgD syndrome/mevalonate kinase deficiency: what is new? Seminars in immunopathology. 2015;37:371–376. doi: 10.1007/s00281-015-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behringer R, Gertsenstein M, Vintersten Nagy K, Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual (Fourth Edition) Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2014. [Google Scholar]
- 50.Bougaki M, et al. Nos3 protects against systemic inflammation and myocardial dysfunction in murine polymicrobial sepsis. Shock. 2010;34:281–290. doi: 10.1097/SHK.0b013e3181cdc327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.