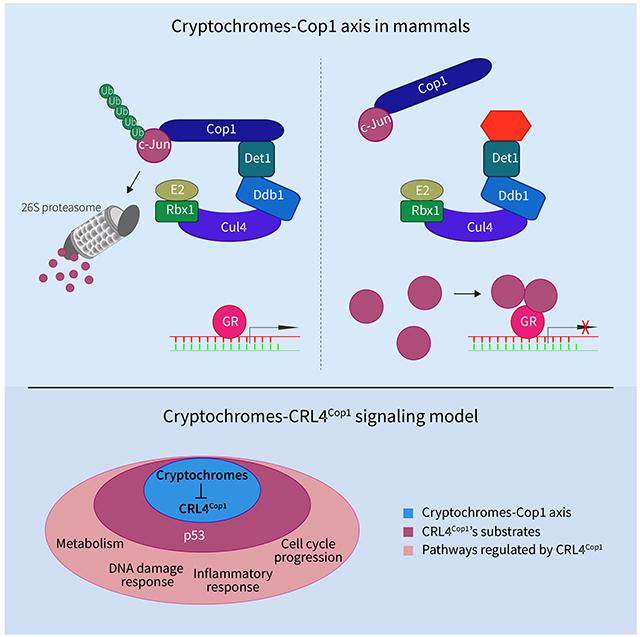
Summary
In plants, cryptochromes are photoreceptors that negatively regulate the ubiquitin ligase CRL4Cop1. In mammals, cryptochromes are core components of the circadian clock and repressors of the glucocorticoid receptor (GR). Moreover, mammalian cryptochromes lost their ability to interact with Cop1, suggesting that they are unable to inhibit CRL4Cop1. Contrary to this assumption, we found that mammalian cryptochromes are also negative regulators of CRL4Cop1, and through this mechanism they repress the GR transcriptional network both in cultured cells and in the mouse liver. Mechanistically, cryptochromes inactivate Cop1 by interacting with Det1, a subunit of the mammalian CRL4Cop1 complex that is not present in other CRL4s. Through this interaction, the ability of Cop1 to join the CRL4 complex is inhibited; therefore, its substrates accumulate. Thus, the interaction between cryptochromes and Det1 in mammals mirrors the interaction between cryptochromes and Cop1 in planta, pointing to a common ancestor in which the cryptochrome-Cop1 axis originated.
Keywords: Cryptochromes, Cop1, Det1, CRL4, Cul4, glucocorticoid receptor, c-Jun, glucagon, circadian clock
Graphical Abstract
In Brief
Rizzini et al. find that the cryptochromes-Cop1 axis is conserved in mammals. Cryptochromes inhibit CRL4Cop1 complex formation by competing with Cop1 to bind Det1. Through this molecular mechanism, the substrates of the CRL4Cop1 complex are stabilized, leading, among other effects, to repression of glucocorticoid receptor transcriptional activity.
Introduction
Plant cryptochromes are UVA and blue light photoreceptors. One of their major functions, after UVA and blue light perception, is to inactivate the ubiquitin ligase activity of Cop1 (Constitutively photomorphogenic 1) [1, 2]. Cop1 is a DCAF (Ddb1 and Cul4-Associated Factor) protein that functions as a substrate receptor subunit of the CRL4Cop1 (Cul4-RING ubiquitin ligase) complex, which is comprised of the following five subunits: Cul4, Ddb1, Rbx1, Dda1, and Cop1 [3–5]. In plants, CRL4Cop1 promotes the ubiquitylation and the consequent proteasomal-dependent degradation of downstream substrates, such as the photomorphogenesis-promoting transcription factors HY5, HYH, and LAF1 [1]. Cryptochromes-mediated inactivation of CRL4Cop1 results in the stabilization of these factors and contributes to the reprogramming of plant growth and development [1].
Mammalian cryptochromes, Cry1 and Cry2, are instead core repressors of the circadian clock [6, 7] and, in contrast to their plant orthologs, do not interact with Cop1 [8]. Cryptochromes, the Period proteins, and Rev-Erbα constitute the negative limb of the transcription-translation feedback loop (TTFL) by binding and inhibiting the Clock/Bmal1 complex within the forward limb of the clock. In addition, mammalian cryptochromes regulate the activity of the glucocorticoid receptor (GR) and modulate glucagon receptor signaling pathways important in gluconeogenesis [9, 10].
Cop1 is also present in mammals where it functions as a DCAF protein allowing the CRL4Cop1 complex to recognize specific substrates [1]. In particular, mammalian Cop1 promotes the ubiquitylation and subsequent proteasomal-dependent degradation of downstream substrates (mostly transcription factors and metabolic enzymes), such as c-Jun, p53, Mta1, Stat3, Ets1, and Atgl [3, 11–15]. Thus, Cop1 constitutes a hub that rapidly modulates the stability of key cellular regulators in response to flux of nutrients and hormones, as well as upon cellular stress.
Given the lack of physical interaction between mammalian cryptochromes and Cop1 [8], it is assumed that the cryptochromes-Cop1 axis is not evolutionary conserved. However, functional studies to investigate the presence of this regulatory axis in mammals have not been performed. Thus, we asked whether mammalian cryptochromes are also negative regulators of CRL4Cop1. The results of these studies are described herein.
Results
Cop1 mediates the cryptochromes-dependent rhythmic repression of the GR
As a first step to understand if mammalian cryptochromes are negative regulators of Cop1, we investigated the involvement of Cop1 in the well-established function of the mammalian cryptochromes in repressing the GR transcriptional network [9]. Compared to wild-type mouse embryonic fibroblasts (MEFs), Cry1−/−;Cry2−/− double knockout (DKO) MEFs display an elevated level of GR activity [9]. We expanded these findings by exogenously expressing either Cry1 or Cry2 using a doxycycline-inducible system (TET-ON), in DKO MEFs. After activation of the GR pathway with dexamethasone (a synthetic glucocorticoid), we found that the expression of Sgk1 and Gilz, two canonical GR target genes, was reduced upon induction of either Cry1 or Cry2 (Figures 1A–B). Next, we measured the dexamethasone-dependent activation of Sgk1 and Gilz upon depletion of Cop1. We found that Cop1 silencing inhibited GR-induced transcription of Sgk1 and Gilz (Figures 1C–D). Moreover, in cells depleted of Cop1, induction of Cry1 or Cry2 had no additional effect on the repression of Sgk1 and Gilz (Figures 1C–D), suggesting that cryptochromes and Cop1 operate in the same GR-repressing signaling pathway.
Figure 1. Cop1 mediates the cryptochromes-dependent repression of canonical GR target genes.
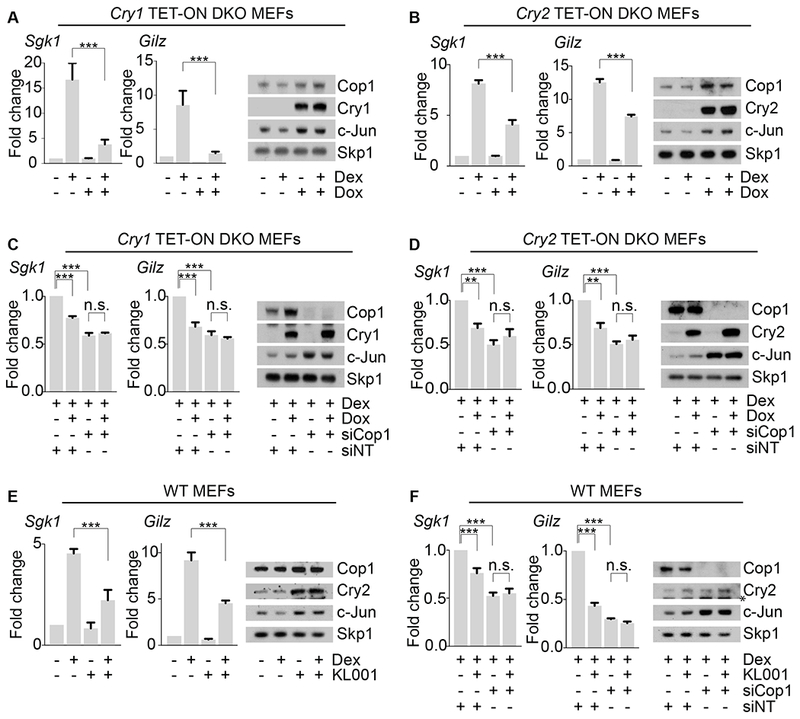
(A) Left, qPCR analysis of cDNAs prepared from DKO MEFs infected with lentiviruses expressing untagged human Cry1 under the control of a doxycycline-inducible promoter before and after dexamethasone treatment. Right, corresponding cell extracts were analyzed by immunoblotting.
(B) The experiment was performed as in (A), except that inducible human Cry2 was expressed in DKO MEFs.
(C) The experiment was performed as in (A), except that cells were treated with an siRNA oligo to Cop1 or a non-targeting siRNA oligo (NT).
(D) The experiment was performed as in (B), except that cells were treated with an siRNA oligo to Cop1 or a non-targeting siRNA oligo (NT).
(E) Left, qPCR analysis of cDNAs prepared from wild-type MEFs, before and after KL001 treatment and in presence or absence of dexamethasone treatment. Right, corresponding cell extracts were analyzed by immunoblotting.
(F) Left, qPCR analysis of cDNAs prepared from wild-type MEFs, before and after KL001 treatment, after dexamethasone treatment and in presence or absence of Cop1 knockdown. Right, corresponding cell extracts were analyzed by immunoblotting. The asterisk indicates a cross-reacting band.
To analyze the function of the cryptochromes-Cop1 axis in GR signaling in wild-type MEFs, we stabilized endogenous cryptochromes using the carbazole derivative KL001. KL001 competes with CRL1Fbxl3, a Cul1-RING ubiquitin ligase complex, for its binding to cryptochromes, thereby inhibiting their ubiquitylation and degradation [16–19]. KL001 repressed the dexamethasone-mediated induction of Sgk1 and Gilz, in agreement with the accumulation of Cry2 in KL001-treated cells (Figure 1E). Moreover, in cells depleted of Cop1, KL001 treatment had no additive effect on the expression of Sgk1 and Gilz (Figure 1F). These data agree with the results obtained with the induction of exogenous cryptochromes in DKO MEFs and, together, they suggest that, in the absence of Cop1, cryptochromes are impaired in their ability to inhibit GR. Moreover, these results indicate that KL001 inhibits GR activity through a pathway involving Cop1.
Next, we performed differential expression analysis using RNA-sequencing to determine the impact of the genetic interactions between cryptochromes, Cop1, and GR on a genomic scale. Dexamethasone-mediated activation of the GR was analyzed in wild-type MEFs treated with KL001 (to stabilize the cryptochromes) in the presence or in the absence of Cop1 knockdown. We found that out of 3,540 genes significantly differentially expressed by dexamethasone, 792 lost significant expression after KL001 treatment [see next-generation sequencing data deposited in Gene Expression Omnibus (GEO) database under the accession code GSE124388]. The scatter plots and the heatmap representations of these 792 genes shows that KL001 treatment blocked the effects of dexamethasone on both up- and down-regulated genes (Figures 2A–B). Importantly, in cells depleted of Cop1, KL001 treatment had no significant effect on these 792 GR-regulated genes (Figures 2A–B). Altogether, these results indicate that Cop1 is a regulator of glucocorticoid signaling and a downstream effector of the cryptochromes-mediated repression of the GR transcriptional activity.
Figure 2. Cop1 mediates the cryptochromes-dependent repression of the GR transcriptional network.
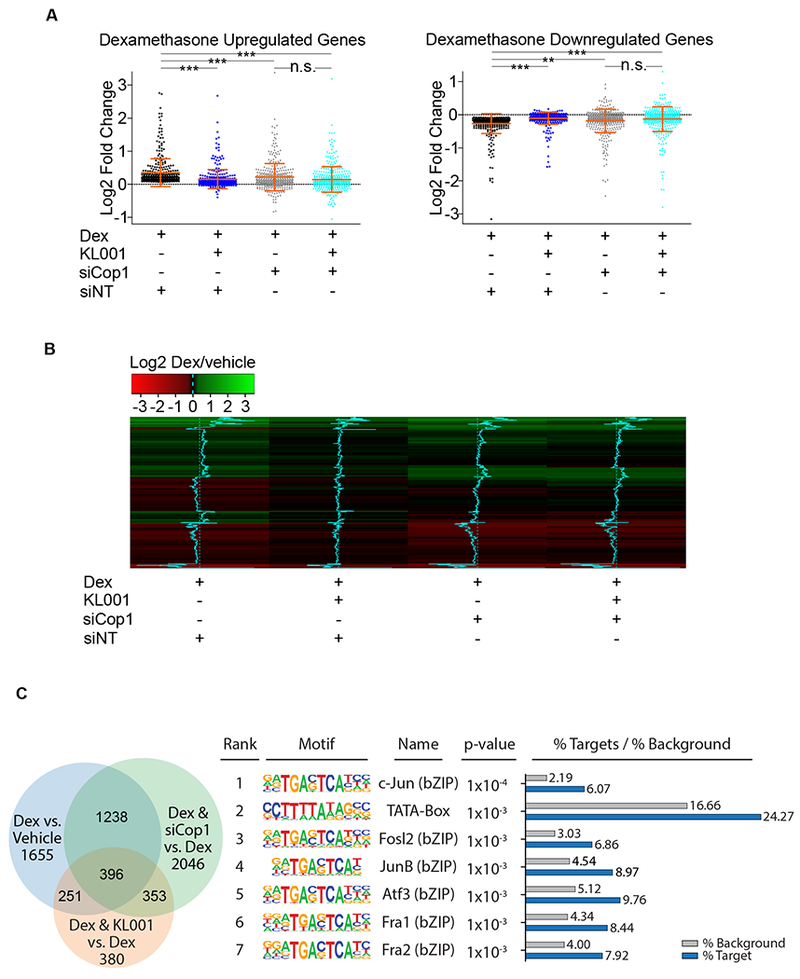
(A) RNA isolated from MEFs treated with dexamethasone, with and without KL001 in the presence or absence of Cop1 knockdown was quantified by RNA-seq. Differential expression analysis of RNA-seq data sets, [see next-generation sequencing data deposited in Gene Expression Omnibus (GEO) database under the accession code GSE124388] is represented in the scatter plots as log2 fold change for 792 genes divided in upregulated and downregulated (370 and 422, respectively). Dexamethasone-regulated genes were considered differentially expressed when the p-adjusted value was <0.05. n=3 biologically independent experiments.
(B) Heatmap as log2 fold change for 370 upregulated and 422 downregulated genes shown in (A).
(C) RNA isolated from MEFs treated with dexamethasone, with and without KL001 in the presence or absence of Cop1 knockdown was quantified by RNA-seq. Left, Venn diagram represents all genes which were significantly differentially expressed among all conditions. [see next-generation sequencing data deposited in Gene Expression Omnibus (GEO) database under the accession code GSE124388]. Right, a table showing the first 7 most highly overrepresented motifs within the promoter regions of 396 genes that were differentially expressed under all three tested conditions. See also Figure S1.
We also noticed that 396 GR-regulated genes were significantly differentially expressed when dexamethasone was provided either together with KL001 or in Cop1 knockdown cells (Figure 2C, left panel). Motif enrichment analysis for these 396 genes showed that the first 7 most highly overrepresented motifs within the promoter regions of these genes contained the core sequence TGAC/GTCA, which is the recognition site for AP-1 transcription factors (Figure 2C, right panel). The AP-1 specific signature was absent from the remaining dexamethasone-dependent gene promoters identified in this analysis (Figure S1A), suggesting that part of the transcriptional output downstream of Cop1 and KL001 is regulated by c-Jun. This is noteworthy since c-Jun is both a constitutive substrate of Cop1 and (in complex with Fos family members, such as Fra1 and Fra2) a repressor of the GR transcriptional activity [20–23].
In the mouse liver, cryptochromes mediate the rhythmic repression of GR that, as a result of this regulation, displays its maximal and minimal response to dexamethasone at the zeitgebers (ZT, which are the hours after exposing mice to light in the animal facility) ZT2 and ZT10, respectively [9]. We used these two ZTs to test the cryptochromes-Cop1 axis in vivo. We confirmed that the expression of three GR target genes (Sgk1, Gilz, and Pck1) is inhibited in the mouse liver specifically at ZT10, but not at ZT2 (Figure 3A). To test whether time-of-day dependent GR target gene activation is dependent on Cop1, we depleted Cop1 in vivo using an adenoviral-mediated shRNA injected retro-orbitally in mice. Confirming our data in cell systems, in vivo depletion of Cop1 was found to inhibit the dexamethasone-mediated activation of GR (Figures 3A and S1B). Cop1 depletion inhibited specifically the GR activation at ZT2 (Figure 3A), when the GR is not repressed by the cryptochromes [9]. Instead, Cop1 depletion at ZT10 did not additionally inhibit the activation of the GR, at a time when GR transcriptional activity is already repressed by the cryptochromes. These data confirm our results in MEFs where the cryptochromes- and Cop1-mediated transcriptional repression of GR are not synergistic (Figures 1C–D, 1F and 2A–B).
Figure 3. Cop1 mediates the cryptochromes-dependent rhythmic repression of the GR transcriptional activity in vivo and the repression of the glucagon receptor signaling.
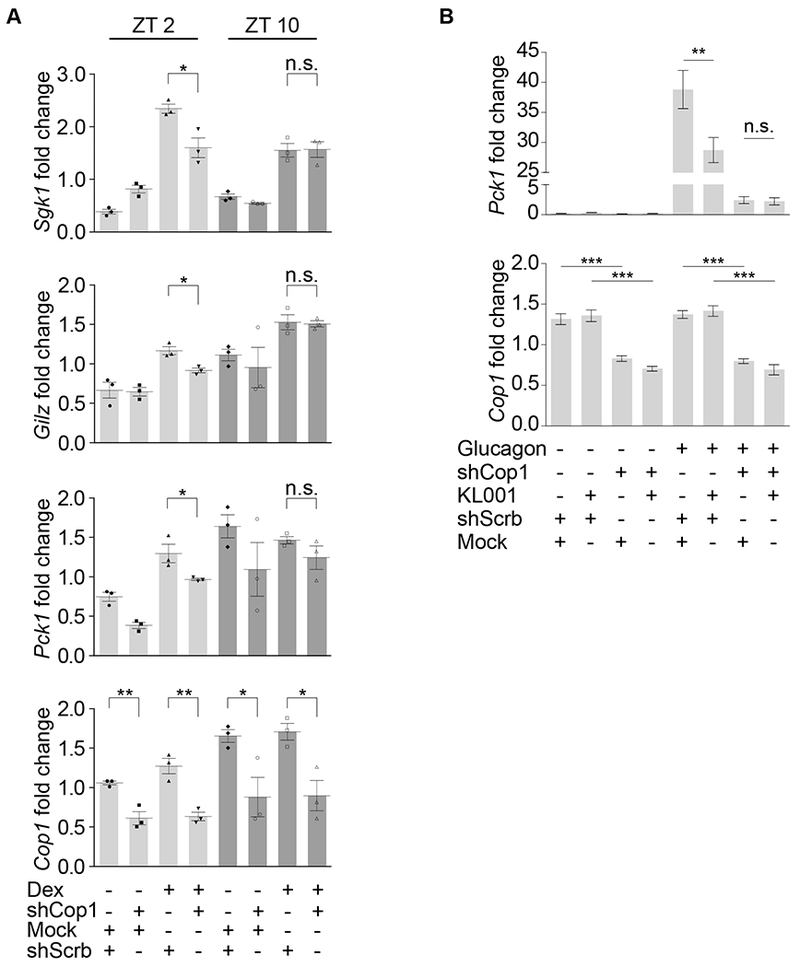
(A) Mice were retro-orbitally injected with Adeno-associated viruses expressing either scramble shRNA or Cop1 shRNA. After 4 weeks, mice were tail-injected with dexamethasone for 1 hour and then sacrificed at ZT2 and ZT10. Liver tissue was then collected, and RNA was extracted and analyzed by qPCR.
(B) Mice were retro-orbitally injected with Adeno-associated viruses expressing either scramble shRNA or Cop1 shRNA. After 4 weeks, primary hepatocytes were isolated and treated with KL001 followed by glucagon treatment. Hepatocytes were then collected, and RNA was extracted and analyzed by qPCR. See also Figure S1.
Cryptochromes are known to inhibit Pck1 transcription not only downstream of GR, but also downstream of the glucagon-activated GPCR [10]. Hence, we tested if the cryptochromes-Cop1 axis mediates also the repression of the glucagon receptor. We depleted Cop1 in vivo as described above and isolated primary hepatocytes to treat them with KL001 ex vivo as previously done [16] due to the poor pharmacokinetics of this drug [24, 25]. Hepatocytes were incubated with KL001 for 21 hours, followed by 3 hours treatment with glucagon. As expected, KL001 repressed the glucagon receptor-mediated induction of Pck1 transcription (Figure 3B). Notably, Cop1 depletion inhibited the induction of Pck1, and Cop1 depletion combined with KL001 treatment did not further reduced the transcription of Pck1 (Figure 3B). These results suggest that the cryptochromes-Cop1 axis mediates also the repression of the glucagon receptor signaling.
Cry2 controls the level and stability of Cop1’s substrates
The above results revealed the existence of a genetic interaction among cryptochromes and Cop1 that spurred us to investigate the underlying molecular mechanism. We noticed that the protein levels of c-Jun and JunB, two canonical Cop1 substrates, were lower in Cry1/2 DKO MEFs than those present in MEFs isolated from wild-type littermate mice (Figure 4A). Similarly, Cop1, which is known to induce its own degradation [26], displayed slightly lower levels in DKO MEFs compared to wild-type MEFs. Notably, doxycycline-mediated expression of Cry1 or Cry2 in DKO MEFs increased the levels of c-Jun, JunB, and Cop1 over time (Figures 1A–B and 4B). These findings mirror what was observed for the protein levels of the transcription factor HY5, a canonical substrate of Cop1 in plants, which are lower in cryptochromes knockout Arabidopsis seedlings than in wild-type seedlings [27] and increase upon expression of exogenous cryptochromes [28].
Figure 4. Cry2 controls the level and stability of Cop1’s substrates.
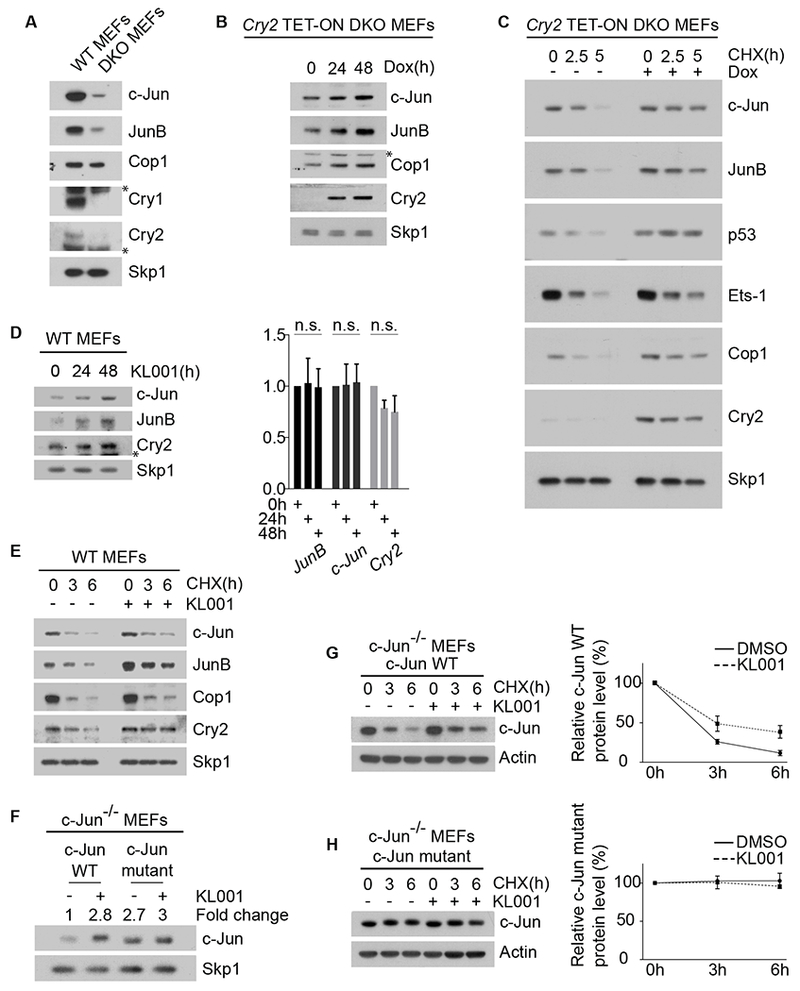
(A) Cell extracts of wild-type MEFs and DKO MEFs were analyzed by immunoblotting. The asterisks indicate cross-reacting bands.
(B) Western blot analysis of Cop1’s substrates in DKO MEFs infected with lentiviruses expressing untagged human Cry2 under the control of a doxycycline-inducible promoter. After doxycycline treatment, cells were collected, lysed and immunoblotted. The asterisk indicates a cross-reacting band.
(C)The experiment was performed as in (B), except that, one day after infection, DKO MEFs were treated with cycloheximide (CHX) for the indicated times.
(D) Left, wild-type MEFs were treated with 10 μM KL001 for the indicated times before they were collected and analyzed by immunoblotting. The asterisk indicates a cross-reacting band. Right, qPCR analysis of cDNAs prepared from the same samples.
(E) Wild-type MEFs were treated with 10 μM KL001 for 18 hours. Cells were then treated with CHX for the indicated times and cells extracts were analyzed by immunoblotting.
(F) c-Jun−/− MEFs stably expressing either wild-type c-Jun or a degron mutant of c-Jun were treated for 2 days with 10 μM KL001 and then analyzed by immunoblotting. The fold change indicates the protein levels of wild-type c-Jun and c-Jun mutant after densitometric analysis of the blot.
(G) c-Jun−/− MEFs stably expressing wild-type c-Jun were treated overnight with either vehicle or 10 μM KL001. CHX was applied for the indicated times and cells extracts were analyzed by immunoblotting. The graph shows the quantification of protein levels ±SEM (n = 3 biologically independent experiments).
(H) The experiment was performed as in (G), except that MEFs stably expressed a c-Jun mutant in its Cop1’s degron. See also Figure S2.
We also observed that the half-life of c-Jun, JunB, and Cop1 increased after induction of exogenous Cry2 (Figure 4C), showing that Cry2 modulates their protein stability. Similar protein stabilization was observed for various Cop1 substrates in all cell systems tested, including mouse Beta-TC-6 pancreatic beta cells and human HEK-293T embryonic kidney cells (Figures 4C and S2). Accordingly, KL001, which stabilizes cryptochromes, induced the stabilization of Cop1 substrates without affecting their corresponding mRNAs (Figures 4D–E and 1E–F).
To uncouple post-translational regulation of protein stability from endogenous transcriptional regulation and other RNA regulatory elements, we also analyzed in c-Jun−/− MEFs the protein level and the stability of exogenously-expressed c-Jun [either wild-type c-Jun or a c-Jun stable mutant unable to bind Cop1 [3]]. We found that KL001 increased both the levels and stability of exogenous wild-type c-Jun (Figures 4F–G). Instead, the c-Jun Cop1-insensitive stable mutant did nod accumulate and was not further stabilized by KL001 treatment (Figures 4F and 4H). As expected, induction of either Cry1 or Cry2, as well as treatment with KL001 had no additive effect on the levels of c-Jun in Cop1-depleted MEFs (Figures 1C–D and 1F). Thus, cryptochromes appear to promote the stabilization of c-Jun in a Cop1-mediated fashion.
The N-terminus of Cry2 interacts with Det1 to stabilize Cop1’s substrates and to inhibit GR transcriptional activity
The association between Cop1 and Cul4 is essential to form an active CRL4Cop1 ubiquitin ligase complex and to mediate the ubiquitylation of downstream substrates [3]. Other subunits of the CRL4Cop1 complex include Ddb1, which is also present in all CRL4 complexes, and Dda1, which is present in most but not all CRL4 complexes [4]. Moreover, CRL4Cop1 is the only mammalian CRL4 complex known to contain an additional subunit, namely Det1, which plays an essential role in linking Cop1 to Ddb1 [3]. Using a panel of eight DCAF proteins, we confirmed that Det1 specifically binds Cop1 (Figure S3A). We also confirmed that Cop1 associates more stably with Cul4A than with its paralog Cul4B, as previously observed [29]. Importantly, when we expressed increasing amounts of Det1 in HEK-293T cells, we observed that Cop1 co-immunoprecipitated increasing amounts of Cul4A, Dda1, and Ddb1 (Figure S3B), suggesting that Det1 is a rate-limiting factor for Cop1 to associate with the rest of the CRL4 complex. While in mammals Det1 is essential for Cop1 binding to Ddb1 and to form an active CRL4Cop1 complex, plant Det1 is not present in the CRL4Cop1 complex and it assembles instead in a distinct ligase complex, namely CRL4Det1 [1, 3, 5]. Moreover, plant cryptochromes physically interact and inhibit Cop1, whereas in mammals this physical interaction is lost [8]. Because of these differences, we tested the hypothesis that mammalian cryptochromes, instead of binding Cop1, could bind Det1. Immunoprecipitation of HA-tagged Det1 expressed in HEK-293T cells revealed an interaction between Det1 and wild-type Cry2, which was tagged at the C-terminus with a FLAG epitope (Figure 5A). We noticed that adding the FLAG tag to the N-terminus of the two cryptochrome proteins impaired their binding to Det1 (data not shown), suggesting that the N-terminal portion of the cryptochrome is important for the binding to Det1. Therefore, we generated C-terminal deletion mutants of Cry2, all C-terminally tagged with a FLAG epitope (see schematics in Figure 5B) and evaluated them for their binding to Det1. Immunopurification of HA-tagged Det1 co-expressed with either wild-type Cry2 or Cry2 truncation mutants revealed that the first N-terminal 130 amino acids of human Cry2 were sufficient to bind Det1 (Figures 5A–B). This is also divergent from the mechanism in planta where cryptochromes bind and inhibit Cop1 through their C-terminal domain [28, 30]. Instead, the C-terminus of mammalian cryptochromes is critical for the interaction with components of the circadian clock, such as the Period proteins and Fbxl3 [31–33]. Hence, it was not surprising that the Cry2-N130 truncation mutant, although able to interact with endogenous Det1, Dda1, and Ddb1, lost its ability to bind with the components of the circadian clock (Figure 5C).
Figure 5. The N-terminus of Cry2 interacts with Det1 to stabilize Cop1’s substrates and inhibit GR activity.
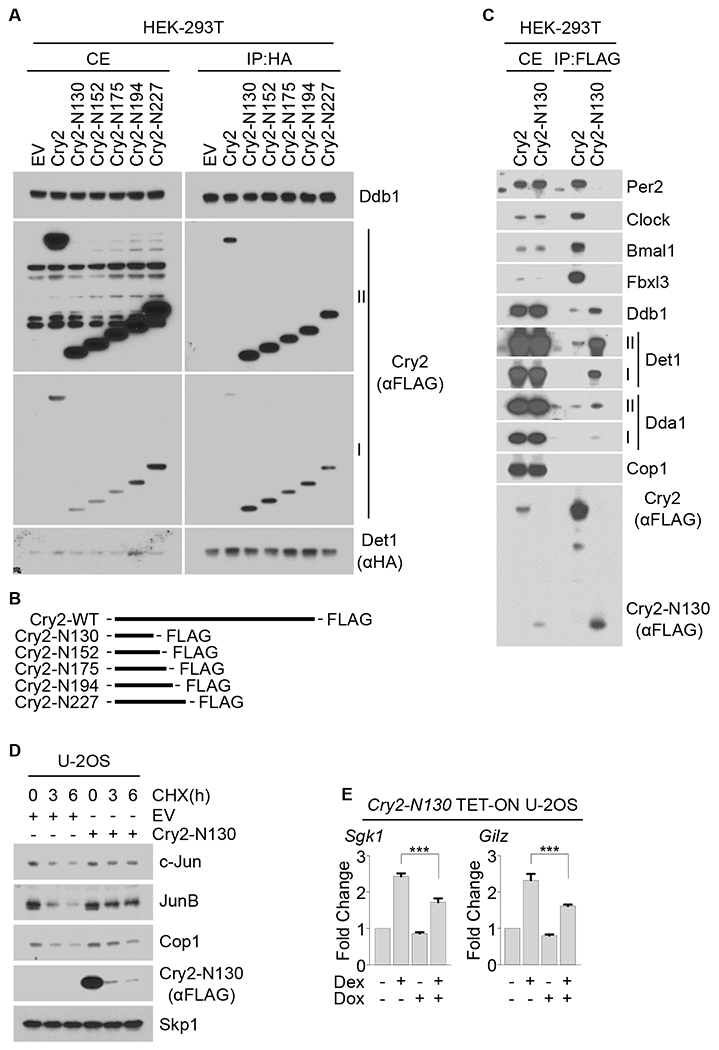
(A) HA-tagged Det1 was co-expressed in HEK-293T cells with either FLAG-tagged wild-type Cry2 or FLAG-tagged Cry2 truncation mutants followed by immunoprecipitation from cell extracts with an anti-HA resin. Cells extracts (CE) and immunoprecipitates (IP) were analyzed by immunoblotting. Roman numbers indicate different exposure times: I, short exposure; II, long exposure.
(B) Schematic representation of wild-type Cry2 and Cry2 truncation mutants.
(C) FLAG-tagged Cry2 or FLAG-tagged Cry2-N130 truncation mutant were expressed in HEK-293T cells and immunoprecipitated from cell extracts with an anti-FLAG resin. CEs and immunoprecipitates were then analyzed by immunoblotting.
(D) U-2OS cells were transfected with either an empty vector (EV) or with a vector expressing Cry2-N130. 18 hours after transfection, cells were treated with CHX for the indicated times and cell extracts were immunoblotted.
(E) qPCR analysis of cDNAs prepared from U-2OS cells infected with lentiviruses expressing Cry2-N130 under the control of a doxycycline-inducible promoter and treated with dexamethasone where indicated. See also Figures S3, S4 and S5.
Notably, expression of the truncation mutant Cry2-N130 in U-2OS cells was sufficient to increase the half-life of Cop1 substrates (Figure 5D) and to reduce the dexamethasone-dependent activation of Sgk1 and Gilz (Figure 5E), similar to what we observed with full length cryptochromes (Figures 1 and 4). These data uncover a previously unknown function of the N-terminal region of the mammalian cryptochromes that is independent from their interactions with other components of the TTFL.
Cryptochromes inhibit the binding between Cop1 and Det1-Ddb1
To gain insights into the role of cryptochromes in the assembly of the CRL4Cop1 complex, we immunopurified FLAG-tagged Cop1 expressed in HEK-293T cells either alone or in combination with Cry1 or Cry2. Mass-spectrometric analysis of the proteins present in the Cop1 complex showed that, when either Cry1 or Cry2 were co-expressed with Cop1, there was a reduction in the number of peptides corresponding to endogenous Dda1, Ddb1, and Det1 (Figure S4A). Next, we tested the interaction between Cop1 and Ddb1 in both wild-type and DKO MEFs. Strikingly, immunopurification of endogenous Cop1 revealed a higher level of binding to endogenous Det1 and Ddb1 in DKO MEFs compared to wild-type MEFs (Figure 6A). Additionally, stable DKO MEFs for inducible expression of exogenous Cry2 reduced the binding of endogenous Cop1 to endogenous Det1 and Ddb1 (Figure 6B). Similarly, transient expression of either Cry1 or Cry2 reduced the binding of exogenous Cop1 to endogenous Det1 and Ddb1 without affecting the interaction between COP1 and c-Jun (Figure 6C).
Figure 6. Cryptochromes inhibit the binding between Cop1 and Det1-Ddb1.
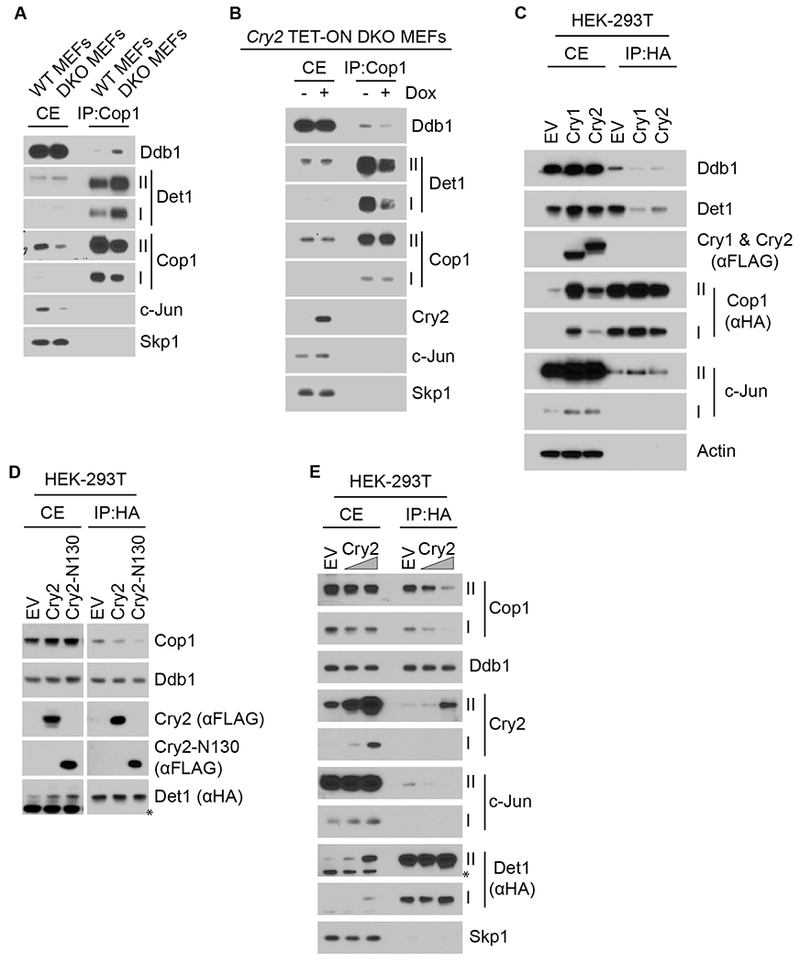
(A) Endogenous Cop1 was immunoprecipitated from wild-type MEFs and DKO MEFs. Cell extracts (CE) and immunoprecipitates (IP) were analyzed by immunoblotting. Roman numbers indicate different exposure times: I, short exposure; II, long exposure.
(B) Endogenous Cop1 was immunoprecipitated from DKO MEFs infected with lentiviruses expressing untagged human Cry2 under the control of a doxycycline-inducible promoter. CEs and immunoprecipitates were analyzed by immunoblotting.
(C) HEK-293T cells were transfected with HA-tagged Cop1 together with either an empty vector (EV) or with either Cry1 or Cry2 expressing vectors and immunoprecipitated from CEs with an anti-HA resin. CEs and immunoprecipitates were analyzed by immunoblotting.
(D) HA-tagged Det1 was co-expressed in HEK-293T cells with either EV, wild-type Cry2, or Cry2-N130 and immunoprecipitated from CEs with an anti-HA resin. CEs and immunoprecipitates were then analyzed by immunoblotting.
(E) The experiment was performed as in (D), except that increasing amounts of wild-type Cry2 expressing vector were transfected. See also Figures S4 and S5.
The physical interaction between cryptochromes and Det1 (Figures 5A and 5C), together with the increased interaction between Cop1 and Det1 in absence of the cryptochromes (Figure 6A) and the reduced interaction between Cop1 and Det1 after transient expression of either Cry1 or Cry2 (Figures 6C and S4A) led us to postulate that cryptochromes could displace Cop1 from Det1. To test this hypothesis, we immunopurified HA-tagged Det1 from cells co-transfected with either a control vector, wild-type Cry2, wild-type Cry1, Cry2-N130, or Cry1-N112 (the latter being the Cry1 truncated mutant equivalent to Cry2-N130; see Figure S5). Strikingly, co-expression of these constructs, but not the control vector, reduced the binding between Cop1 and Det1 (Figures 6D and S4B). Additionally, expression of increasing amounts of exogenous wild-type cryptochromes resulted in a dose-dependent displacement of endogenous Cop1 from Det1 (Figures 6E and S4C). In contrast, expression of Cry2, Cry2-N130, Cry1, and Cry1-N112 did not affect the binding between Det1 and Ddb1 (Figures 5A, 6D–E and S4B–C).
All together, these results strongly suggest that the interaction between cryptochromes and Det1 displaces Cop1 from Det1 and the rest of the CRL4 complex, which could explain the effect of cryptochromes on the stabilization of Cop1 substrates.
Discussion
Contrary to the current assumption, our results indicate that the cryptochromes-Cop1 axis is present in mammals. The similarity between the axis in plants and in mammals comes from the cryptochromes-mediated inactivation of the CRL4Cop1 complex and the consequent stabilization of Cop1’s substrates. However, the molecular mechanisms of this inactivation, as well as the signaling events downstream of the cryptochromes-Cop1 axis differ from plants to mammals. Whereas in plants cryptochromes bind constitutively Cop1 and inhibit its activity in a blue light-dependent manner (leading to the accumulation of its substrates), we found that in mammals cryptochromes bind Det1, instead of Cop1, blocking the ability of Cop1 to interact with the CRL4 complex, leading to the accumulation of CRL4Cop1 substrates, and altering downstream signaling pathways (see graphical abstract).
We tested the cryptochromes-Cop1 axis in the context of the well-established cryptochromes-mediated repression of the GR transcriptional network, finding that both in MEFs and in vivo, cryptochromes repress GR, at least in part, through the inactivation of Cop1. Among the numerous CRL4Cop1 substrates, we focused on c-Jun, which is both a canonical and constitutive substrate of Cop1, as well as a repressor of the GR transcriptional output [3, 20–23]. We found that cryptochromes positively control c-Jun protein levels by inhibiting Cop1. Moreover, our motif enrichment analysis shows an overrepresentation of c-Jun and Fos recognition motifs at the promoter of genes regulated by dexamethasone in combination with either cryptochromes’ stabilization or Cop1 silencing. Although it has been proposed that cryptochromes repress GR-dependent transcription via physical interaction, it has remained puzzling the fact that they have no effect on the GR-dependent repression of the NF-κB inflammatory gene network [9]. We propose that cryptochromes repress GR, at least in part, via the Cop1-c-Jun axis. Since c-Jun is known to transrepress GR but not NF-κB, our model may explain why cryptochromes do not interfere with the NF-κB-inflammatory gene network.
We studied the effect of the cryptochromes-Cop1-GR axis on the GR transcriptional activity in cell systems and mouse liver. However, Cop1 has an ever-growing number of substrates, raising the possibility that the cryptochromes-Cop1 axis controls a vast signaling network. In this respect, we found that the cryptochromes-Cop1 axis mediates also the repression of Pck1 transcription dependent on glucagon signaling.
Thus, our study opens the door to future investigations on the cryptochromes-dependent stabilization of Cop1 substrates and their physiological roles in different organs and tissues.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michele Pagano (michele.pagano@nyumc.org)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal care and use procedures were in accordance with guidelines of the Institutional Animal Care and Use Committee. All experiments were performed using male C57BL/6J (RRID: IMSR_JAX:000664) mice between 3-4 months of age, and mice were maintained in a temperature and humidity controlled vivarium (22-24 °C; ~40% humidity), on a 12:12 light dark (LD) cycle in the Northwestern University Center for Comparative Medicine and were provided with food and water ad libitum.
Cell Cultures
CRY1−/−;CRY2−/− male MEFs and male Beta TC-6 cells stably infected with pTRIPZ vectors were propagated at 37 °C in DMEM supplemented with 10% Tet system-approved FBS (Takara/Clontech Laboratories). C-Jun−/− male MEFs were stably infected with pBabe. And were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (Corning Life Sciences). Doxycycline (Sigma-Aldrich) was used at 100 ng/mL. All the other cell lines (male WT MEFs, female HEK-293T cells and female U-2OS cells) were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (Corning Life Sciences). MG132 (Peptides International) was used at 10 μM concentration. Cells were periodically screened for Mycoplasma contamination. All donated cell lines were verified by western blot analysis.
METHOD DETAILS
Plasmids
cDNAs were subcloned in pcDNA 3.1 (Life Technologies), pBabe (Cell Biolabs), and in pTRIPZ (Dharmacon).
c-Jun cDNA was amplified by PCR using a cDNA library generated from HEK293T cells and sub-cloned into pBabe (Cell Biolabs). Human CRY1 and CRY2 mutants were generated by using the QuikChange Site-directed Mutagenesis kit (Stratagene). All cDNAs were sequenced.
Gene Silencing by siRNA
ON-TARGETplus SMARTpool siRNA oligos targeting COP1 was transfected using RNAi Max (Dharmacon). ON-TARGETplus non-targeting siRNA (Dharmacon, catalog no. D-001810-01) served as a negative control.
Retro- and Lentivirus-Mediated Gene Transfer
HEK-293T cells were transiently co-transfected with retroviral (pBabe) vectors containing vesicular stomatitis virus G protein (VSV-G) and the gene of interest along with pCMV-Gag-Pol using polyethylenimine. Alternatively, lentivirus (pTRIPZ) vectors containing vesicular stomatitis virus G protein (VSV-G) and the gene of interest along with pCMV Delta R8.2 were co-transfected using polyethylenimine. Retrovirus- or lentivirus-containing medium, 48 hr after transfection, was collected and supplemented with 8 mg/ml Polybrene (Sigma).
Cells of interest were then infected by replacing the cell culture medium with the viral supernatant for 6 hours. Selection of stable clones was carried out with puromycin.
Immunoprecipitation and immunoblotting
HEK-293T cells were transiently transfected using polyethylenimine (PEI). U-2OS cells were transiently transfected using siLentFect (Bio-rad). Cell lysis was carried out with lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM EDTA, and 0.5% NP-40) supplemented with protease and phosphatase inhibitors. Immunoprecipitations were carried out after 3 hours treatment with MG132 and with either an anti-FLAG antibody conjugated to agarose resin or an anti-HA antibody conjugated to agarose resin. For immunoprecipitation of endogenous proteins, wild-type MEFs were collected and lysed with lysis buffer. Cop1 was immunoprecipitated with the listed antibody mixed with Protein A sepharose beads (Invitrogen). Elution of the immunoprecipitate was carried out with NuPAGE® LDS sample buffer (Thermo Fisher Scientific) supplemented with β-mercaptoethanol (Sigma-Aldrich) and incubation at 95°C for 3 minutes.
qRT-PCR
Total RNA was generated using RNeasy Mini Kit (Qiagen). cDNA was generated using Random Hexamers EcoDry kit (Takara Clontech). qPCR was performed using Absolute SYBR green (Thermo Fisher Scientific) on a Roche Lightcycler 480. Analysis of the qPCR experiments was conducted via absolute relative quantification with in-experiment standard curves for each primer set to control for primer efficiency. See also Table S1.
Mass spectrometry
HEK-293T cells were transfected with constructs encoding FLAG-tagged human COP1, co-transfected with either GFP control vector, or human wild-type CRY1, or human wild-type CRY2. Forty-eight hours after transfection, cells were collected and lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, plus protease and phosphatase inhibitors). Proteins were immunopurified with anti-FLAG M2 agarose beads (Sigma) and, after extensive washing, eluted by competition with 3×-FLAG peptide (Sigma). The eluate was then precipitated with TCA. Next, mass-spectrometric analysis of the immunoprecipitates was carried out according to [34].
RNA Extraction and Sequencing Analysis
Libraries for RNA-seq were prepared according to manufacturer’s instructions (Illumina). Briefly, total RNA was extracted from cultured cells using the RNeasy Plus Mini Kit (QIAGEN). Poly(A) RNA was isolated using Dynabeads Oligo(dT)25 (Invitrogen) and used as input for library construction utilizing the dUTP method. Barcodes were used for sample multiplexing. RNA libraries were sequenced on an Illumina HiSeq. RNAseq differential expression analysis was performed for three lanes of a single-read 50 Illumina HiSeq 2500 run. Per-sample FASTQ files were generated using the bcl2fastq Conversion software (v.1.8.4) to convert per-cycle BCL base call files outputted by the sequencing instrument into the FASTQ format. The alignment program, STAR (v2.4.5a), was used for mapping reads of 24 mouse samples to the mouse reference genome mm10 and the application FastQ Screen (v0.5.2) was utilized to check for contaminants. The software, featureCounts (Subread package v1.4.6-p3), was used to generate the matrix of read counts for annotated genomic features. For the differential gene statistical comparisons between groups of samples contrasted by non-targeting RNA interference and targeting Cop1 RNA interference conditions as well as KL001 and dexamethasone chemical treatments, the DESeq2 package (Bioconductor v3.0) in the R statistical programming environment was utilized. The heatmap was generated using the function ‘heatmap.2’ within the package ‘gplots’ in the R statistical programming environment.
Transcription factor motif analysis
Motifs enriched within promoters of dexamethasone-inducible genes whose expression was modified by treatment with KL001 and COP1 siRNA were identified using the Homer script “findMotifs.pl” (PMID 20513432). Specifically, we supplied the gene IDs for either the 396 dexamethasone-regulated genes that were also differentially regulated by KL001 and siRNA-COP1 treatment or the remaining 3144 dexamethasone-sensitive genes and searched for motifs among these two groups within −300 to +50 bp of the annotated transcription start site and allowing for at most two mismatched bases to known or de novo motif assignments.
In vivo delivery of shRNA
AAV-technology was utilized to delivery shRNAs to liver tissues in vivo. AAVs were cloned, packaged into serotype 8, purified by Cesium Chloride centrifugation, concentrated to ~1×1012 GC/mL, and buffer exchanged to PBS w/5% glycerol by Vector Biolabs (Malvern, PA). Pre-validated shRNAs targeting mRFWD2 (Cop1, NM_011931) or a scrambled control were expressed under the U6 promoter and GFP was co-expressed under the CMV promoter. Mice were anaesthetized using isoflurane inhalation and 1×1011 GC (100 μL) of AAV expressing either shCop1 or scrambled control RNAs were delivered by retroorbital injection. After 4 weeks, mice were injected with 100 μl saline alone or 1 μg water-soluble dexamethasone (Sigma) resuspended in saline one hour prior to being sacrificed at ZT2 and ZT10. Liver tissue was excised and snap frozen in liquid nitrogen. Total RNA was extracted from frozen liver tissue using Tri-Reagent (Molecular Research Center, Inc), and quantitative PCR was performed and analyzed using a CFX384 (Bio-Rad). PCR conditions were: 10 min at 95°C, then 35 cycles of 10 s at 95°C, 15 s at 60°C. Relative expression levels (normalized to β-actin) were determined using the comparative CT method. See also Table S2.
Hepatocytes isolation and glucagon treatment
Primary hepatocytes were isolated from mice by hepatic portal vein perfusion with buffered saline followed by 0.025% collagenase (Sigma) solution. Cells were maintained on collagen-coated plates in high glucose (4.5 g/L) DMEM medium supplemented with 10% non-heat-inactivated FBS. 24 hours after collection, hepatocytes were treated with 10 μM KL001 (Sigma) or DMSO vehicle for 24 hours. During the last 3 hours of treatment, cells were treated with or without 10 nM glucagon (Sigma) followed by RNA extraction and RT-qPCR analysis. See also Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were collected and analyzed by Prism 5 (Graphpad). Unless otherwise noted, data are representative of at least three biologically independent experiments. Two-group datasets were analyzed by student’s T-test. For three- or four-group analysis, one-way ANOVA was used. All graphs show mean values. Error bars indicate +/− S.D. except for Figures 3B and 4G–H, where error bars indicate +/− S.E.M..
DATA AND SOFTWARE AVAILABILITY
The next-generation sequencing data has been deposited in the Gene Expression Omnibus (GEO) database under ID code GSE124388. This dataset supports the findings of this study in Figure 2 and Figure S1A. The raw images of western blots have been deposited in the Mendeley Data: doi:10.17632/9ydg4zjb76.1. All other data supporting the findings of this study are available from the Lead Contact on reasonable request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Cop1, 1:2000 | Bethyl | Cat# A300-894A, RRID:AB_625290 |
| Rabbit anti-Cry1, 1:2000 | Bethyl | Cat# A302-614A, RRID:AB_10555376 |
| Rabbit anti-c-Jun, 1:5000 | Cell Signaling Technology | Cat# 9165, RRID:AB_2130165 |
| Rabbit anti-Skp1, 1:5000 | Michele Pagano laboratory | N/A |
| Rabbit anti-Cry2, 1:2000 Human-Cry2 detection |
Bethyl | Cat# A302-615A, RRID:AB_10554665 |
| Rabbit anti-Cry2, 1:1000 Endogenous mouse-Cry2 detection |
Proteintech Group | Cat# 13997-1-AP, RRID:AB_10860100 |
| Rabbit anti-JunB, 1:1000 | Bethyl | Cat# A302-704A, RRID:AB_10749029 |
| Mouse anti-β-Actin 1:5000 | Sigma-Aldrich | Cat# A5441, RRID:AB_476744 |
| Rabbit anti-Ets-1, 1:2000 | Cell signaling | Cat# 14069, N/A |
| Rabbit anti-p53, 1:2000 | Santa Cruz Biotechnology | Cat# sc-6243, RRID:AB_653753 |
| Rabbit anti-FLAG, 1:5000 | Sigma-Aldrich | Cat# F7425, RRID:AB_439687 |
| Mouse anti-Stat3, 1:2000 | Santa Cruz Biotechnology | Cat# sc-8019, RRID:AB_628293 |
| Mouse anti-Mta1, 1:1000 | Santa Cruz Biotechnology | Cat# sc-17773, RRID:AB_627969 |
| Mouse anti-Atgl, 1:2000 | Santa Cruz Biotechnology | Cat# sc-365278, RRID:AB_10859044 |
| Rabbit anti-Ddb1, 1:5000 | Bethyl | Cat# A300-462A, RRID:AB_420928 |
| Rabbit anti-HA, 1:5000 | Bethyl | Cat# A190-108A, RRID:AB_67465 |
| Rabbit anti-Per2, 1:2000 | Bethyl | Cat# A303-109A, RRID:AB_10892917 |
| Rabbit anti-Clock, 1:2000 | Bethyl | Cat# A302-618A, RRID:AB_10555233 |
| Rabbit anti-Bmal1, 1:2000 | Bethyl | Cat# A302-616A, RRID:AB_10555918 |
| Rabbit anti-Fbxl3, 1:1000 | Signalway | Cat# 21459-1, RRID:AB_10760331 |
| Mouse anti-Det1, 1:500 | Santa Cruz Biotechnology | Cat# sc-514348, N/A |
| Rabbit anti-Cul4A, 1:2000 | Bethyl | Cat# A300-739A, RRID:AB_533380 |
| Rabbit anti-Dda1, 1:2000 | Proteintech Group | Cat# 14995-1-AP, RRID:AB_10859095 |
| Secondary HRP-conjugated antibodies | Thermo Fisher Scientific | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MG-132 (10 uM) | Peptides International | Cat# IZL-3175-v |
| Doxycycline (100 ng/mL) | Sigma-Aldrich | Cat# D9891 |
| ABsolute Blue qPCR SYBR Green Mix | Thermo Fisher Scientific | Cat# AB-4166B |
| Cycloheximide (20mg/mL) | Sigma-Aldrich | Cat# C7698 |
| Polybrene (8 mg/mL) | Sigma-Aldrich | Cat# TR-1003 |
| Puromycin | Sigma-Aldrich | Cat# P9620 |
| RNAi Max (Lipofectamine) | Thermo Fisher Scientific | Cat# 13778-500 |
| SiLentFect | Biorad | Cat# 1703360 |
| KL001 (10 μM) | Sigma | Cat# SML1032 |
| Critical Commercial Assays | ||
| RNA to cDNA EcoDry Premix (Random Hexamers) | Clontech/ Takara Bio | Cat# 639546 |
| QuikChange Site-directed Mutagenesis kit | Agilent | Cat# 200523 |
| Deposited Data | ||
| RNA sequencing data | GEO | GSE124388 |
| Raw images of western blots | This paper, Mendeley database | doi:10.17632/9ydg4zjb76.1 |
| Experimental Models: Cell Lines | ||
| Human: HEK-293T | ATCC | Cat# CRL-3216, RRID:CVCL_0063 |
| Human: U-2OS | ATCC | Cat# HTB-96, RRID:CVCL_0042 |
| Mouse: Cry1−/−Cry2−/− MEFs | Gift: Dr. Choogon Lee | N/A |
| Mouse: WT MEFs | Gift: Dr. Choogon Lee | N/A |
| Mouse: Beta-TC-6 | ATCC | Cat# CRL-11506, RRID:CVCL_0605 |
| Mouse: c-Jun−/− MEFs | Gift: Dr. Michael Karin | N/A |
| Experimental Models: Organisms/Strains | ||
| Mice: C57BL/6J, male | Jackson Laboratory | Cat# JAX:000664 IMSR_JAX:000664 |
| Oligonucleotides | ||
| siRNA: RFWD2 | Dharmacon | Cat# L-043438-01 |
| siRNA: Non-Targeting | Dharmacon | Cat# D-001810-01 |
| Quantitative PCR primer sequences for cell culture experiments | Table S1 | N/A |
| Quantitative PCR primer sequences for mouse liver tissue experiments | Table S2 | N/A |
| Recombinant DNA | ||
| Plasmid: pfmh-hCry1 | Addgene | Cat# 25843, RRID: Addgene_25843 |
| Plasmid: pSO2002-Cry2 | Addgene | Cat# #25842, RRID:Addgene_25842 |
| Plasmid: pcDNA4/TO/myc-His-Det1 | Abgent | Cat# DC09517 |
| Plasmid: pcDNA3-c-Jun | Michele Pagano laboratory | N/A |
| Plasmid: pcDNA3-Rfwd2 | Michele Pagano laboratory | N/A |
| pTRIPZ | Dharmacon | N/A |
| pBabe | Cell Biolabs | N/A |
| pcDNA3.1 | Life Technologies | N/A |
| Software and Algorithms | ||
| Prism software (version 5.0.3) | Graphpad Software, Inc | www.graphpad.com |
| STAR (version 2.4.5a) | ||
| bcl2fastq Conversion software (v.1.8.4) | ||
| FastQ Screen (v0.5.2) | ||
| featureCounts (Subread package v1.4.6-p3) | ||
| DESeq2 package (Bioconductor v3.3.0) | R statistical programming environment | |
| ‘heatmap.2’ within the package ‘gplots’ | R statistical programming environment | |
| Homer script “findMotifs.pl” | PMID: 20513432 | |
| Jalview version 2 | PMID: 19151095 | |
Highlights.
Cryptochrome proteins interact with Det1 to inhibit CRL4Cop1 complex formation
Cryptochromes stabilize the substrates of the CRL4Cop1 ubiquitin ligase complex
Cop1 is a mediator of cell signaling networks downstream of the cryptochromes
The cryptochromes-Cop1 axis is evolutionarily conserved
Acknowledgments:
The authors thank the NYU Genome Technology Center (partially supported by the Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center) for expert library preparation and sequencing for RNA-seq. The mass spectrometric experiments were supported in part by NYU School of Medicine, the Laura and Isaac Perlmutter Cancer Center Support grant P30CA016087 from the National Cancer Institute, and a shared instrumentation grant from the NIH, 1S10OD010582-01A1, for the purchase of an Orbitrap Fusion Lumos. Michele Pagano is an Investigator with the Howard Hughes Medical Institute. Michele Pagano is grateful to T.M. Thor for continuous support. This work was funded by grants from the NIH (R01-GM057587 and R01-CA076584) to Michele Pagano, (R01DK100814 and 2R01DK090625-05) to Joseph Bass, (K01DK105137) to Clara Peek and (F32HL143978) to Mark Perelis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
Michele Pagano is a consultant for BeyondSpring Pharmaceuticals and a member of the scientific advisory boards of CullGen, Inc. and Kymera Therapeutics.
References
- 1.Lau OS, and Deng XW (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17, 584–593. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Zuo Z, Wang X, Liu Q, Gu L, Oka Y, and Lin C (2018). Beyond the photocycle-how cryptochromes regulate photoresponses in plants? Curr Opin Plant Biol 45, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, and Dixit VM (2004). Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371–1374. [DOI] [PubMed] [Google Scholar]
- 4.Olma MH, Roy M, Le Bihan T, Sumara I, Maerki S, Larsen B, Quadroni M, Peter M, Tyers M, and Pintard L (2009). An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. J Cell Sci 122, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22, 108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Partch CL, Green CB, and Takahashi JS (2014). Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bass J, and Lazar MA (2016). Circadian time signatures of fitness and disease. Science 354, 994–999. [DOI] [PubMed] [Google Scholar]
- 8.Yang HQ, Tang RH, and Cashmore AR (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13, 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, and Evans RM (2011). Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. (2010). Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16, 1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’ Rourke K, Koeppen H, and Dixit VM (2004). The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429, 86–92. [DOI] [PubMed] [Google Scholar]
- 12.Li DQ, Ohshiro K, Reddy SD, Pakala SB, Lee MH, Zhang Y, Rayala SK, and Kumar R (2009). E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc Natl Acad Sci U S A 106, 17493–17498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallavalle C, Thalmann G, Catapano CV, and Carbone GMR (2016). The E3 ubiquitin ligase COP1 controls STAT3 turnover and its loss leads to increased STAT3 stabilization and activation in prostate cancer. Cancer Res 76. [Google Scholar]
- 14.Lu G, Zhang Q, Huang Y, Song J, Tomaino R, Ehrenberger T, Lim E, Liu W, Bronson RT, Bowden M, et al. (2014). Phosphorylation of ETS1 by Src family kinases prevents its recognition by the COP1 tumor suppressor. Cancer Cell 26, 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh M, Niyogi S, Bhattacharyya M, Adak M, Nayak DK, Chakrabarti S, and Chakrabarti P (2016). Ubiquitin Ligase COP1 Controls Hepatic Fat Metabolism by Targeting ATGL for Degradation. Diabetes 65, 3561–3572. [DOI] [PubMed] [Google Scholar]
- 16.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, et al. (2012). Identification of small molecule activators of cryptochrome. Science 337, 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. (2007). The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900. [DOI] [PubMed] [Google Scholar]
- 18.Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, and Takahashi JS (2007). Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, and Pagano M (2007). SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904. [DOI] [PubMed] [Google Scholar]
- 20.Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, and Herrlich P (1990). Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell 62, 1189–1204. [DOI] [PubMed] [Google Scholar]
- 21.Schule R, Rangarajan P, Kliewer S, Ransone LJ, Bolado J, Yang N, Verma IM, and Evans RM (1990). Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell 62, 1217–1226. [DOI] [PubMed] [Google Scholar]
- 22.Lucibello FC, Slater EP, Jooss KU, Beato M, and Muller R (1990). Mutual transrepression of Fos and the glucocorticoid receptor: involvement of a functional domain in Fos which is absent in FosB. EMBO J 9, 2827–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, and Karin M (1990). Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell 62, 1205–1215. [DOI] [PubMed] [Google Scholar]
- 24.Humphries PS, Bersot R, Kincaid J, Mabery E, McCluskie K, Park T, Renner T, Riegler E, Steinfeld T, Turtle ED, et al. (2018). Carbazole-containing amides and ureas: Discovery of cryptochrome modulators as antihyperglycemic agents. Bioorg Med Chem Lett 28, 293–297. [DOI] [PubMed] [Google Scholar]
- 25.Humphries PS, Bersot R, Kincaid J, Mabery E, McCluskie K, Park T, Renner T, Riegler E, Steinfeld T, Turtle ED, et al. (2016). Carbazole-containing sulfonamides and sulfamides: Discovery of cryptochrome modulators as antidiabetic agents. Bioorg Med Chem Lett 26, 757–760. [DOI] [PubMed] [Google Scholar]
- 26.Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, and Chua NH (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999. [DOI] [PubMed] [Google Scholar]
- 27.Osterlund MT, Hardtke CS, Wei N, and Deng XW (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Ma LG, Li JM, Zhao HY, and Deng XW (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158. [DOI] [PubMed] [Google Scholar]
- 29.Bennett EJ, Rush J, Gygi SP, and Harper JW (2010). Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell 143, 951–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, and Cashmore AR (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103, 815–827. [DOI] [PubMed] [Google Scholar]
- 31.Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, Green CB, and Zheng N (2014). Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. Elife 3, e03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmalen I, Reischl S, Wallach T, Klemz R, Grudziecki A, Prabu JR, Benda C, Kramer A, and Wolf E (2014). Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157, 1203–1215. [DOI] [PubMed] [Google Scholar]
- 33.Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, and Zheng N (2013). SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dankert JF, Rona G, Clijsters L, Geter P, Skaar JR, Bermudez-Hernandez K, Sassani E, Fenyo D, Ueberheide B, Schneider R, et al. (2016). Cyclin F-Mediated Degradation of SLBP Limits H2A.X Accumulation and Apoptosis upon Genotoxic Stress in G2. Molecular Cell 64, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The next-generation sequencing data has been deposited in the Gene Expression Omnibus (GEO) database under ID code GSE124388. This dataset supports the findings of this study in Figure 2 and Figure S1A. The raw images of western blots have been deposited in the Mendeley Data: doi:10.17632/9ydg4zjb76.1. All other data supporting the findings of this study are available from the Lead Contact on reasonable request.


