Abstract
We found that protease-activated receptor 1 (PAR1) was transiently induced in cultured osteoclast (OC) precursor cells. Therefore, we examined the bone phenotype and response to resorptive stimuli of PAR1 deficient (KO) mice. Bones and bone marrow (BM)–derived cells from PAR1 KO and wild type (WT) mice were assessed using micro-computed tomography, histomorphometry, in vitro cultures, and reverse transcription–polymerase chain reaction. Osteoclastic responses to tumor necrosis factor-α (TNF) challenge in calvaria were analyzed with and without a specific neutralizing antibody to Notch2 signaling (N2-NRR Ab). In vivo under homeostatic conditions, there were minimal differences in bone mass or bone cells between PAR1 KO and WT mice. However, PAR1 KO myeloid cells demonstrated enhanced osteoclastogenesis in response to receptor activator of NF-kB ligand (RANKL) or the combination of RANKL and TNF. Strikingly, in vivo osteoclastogenic responses of PAR1 KO mice to TNF were markedly enhanced. We found that N2-NRR Ab reduced TNF-induced osteoclastogenesis in PAR1 KO mice to WT levels without affecting WT responses. Similarly, in vitro N2-NRR Ab reduced RANKL-induced osteoclastogenesis in PAR1 KO cells to WT levels without altering WT responses. We conclude that PAR1 functions to limit Notch2 signaling in responses to RANKL and TNF and moderates osteoclastogenic response to these cytokines. This effect appears, at least in part, to be cell autonomous since enhanced osteoclastogenesis was seen in highly purified PAR1 KO OC precursor cells. It is likely that this pathway is involved in regulating the response of bone to diseases associated with inflammatory signals.
INTRODUCTION
Osteoclasts (OC) are multinuclear giant cells with the unique ability to resorb bone (1). They form from a mononuclear precursor cell of hematopoietic origin. Two cytokines, macrophage colony stimulating factor (M-CSF) (2) and receptor activator of NF-κB ligand (RANKL) (3), are critical regulators of OC formation. In addition, proinflammatory cytokines like interleukin 1 (IL-1), tumor necrosis factor-α (TNF) and interleukin 6 (IL-6) can contribute to the increased osteoclastogenesis and bone loss of inflammatory diseases (4).
We previously characterized a highly enriched population of OC precursor cells in murine bone marrow (5, 6). In the current study, we used gene array expression profiling to examine highly purified OC precursor cells and identified protease-activated receptor 1 (PAR1), the product of the F2r gene, to be transiently induced by RANKL during OC differentiation.
PAR1 is a member of a four-protein family of cell surface, G-protein coupled receptors (GPCR) (PAR1–4) that are activated by proteolytic cleavage (7) and form both homo- and heterodimers (8). Activation occurs with a variety of serine proteases. Among these, thrombin is the most studied (9). However, activation also has been reported by coagulation factor Xa (10), plasmin (11), matrix metalloproteinase 1 (12), matrix metalloproteinase 13 (13), elastase (14), proteinase-3 (14), activated protein C (15) and granzyme K (16). The extracellular N-terminus of PAR1 contains multiple tethered-ligand domains that are prevented from interacting with ligand-binding domains in PAR1 by peptide sequences in the distal N-terminal extracellular domain (9). Cleavage of a section of the PAR1 distal N-terminal extracellular domain by proteases “unmasks” specific ligand domains. Once cleaved, the remaining tethered N-terminal extracellular domain (containing the unmasked ligand domain) alters its conformation and binds to a specific sequence in the extracellular region of PAR1. Depending on the protease, different ligand domains can be unmasked, producing a variety of responses (9).
In developing rat bones, PAR1 was identified by immunohistochemistry in osteoblasts, macrophages, muscle cells and endothelial cells (17). Significantly, no expression of PAR1 was observed in mature OC (17).
To determine whether PAR1 has a role in bone homeostasis, we examined the bone phenotype of mice with a global PAR1 deletion (PAR1 KO) (18). Mice were studied under basal conditions or after inducing inflammation as modeled in vitro by treating osteoclast precursor cell cultures with RANKL and TNF or in vivo by injecting mice with TNF over the calvariae.
MATERIALS AND METHODS
Experimental Animals
Mice in a C57BL/6 background were used for all experiments. Mice deficient in PAR1 (PAR1 KO) (18) were purchased from Jackson Laboratory, Bar Harbor, ME (Stock No: 002862) and housed in the Center for Comparative Medicine at UConn Health under standard housing conditions. In some experiments, recombinant mouse TNF (2.0 μg) was injected subcutaneously above the calvariae daily for 4 days and mice were sacrificed 24 hours later to analyze osteoclasts in their calvariae by histomorphometry.
Recombinant murine TNF was prepared in our laboratory as follows: A murine TNF-α cDNA fragment encoding amino acid residues 83–235 was cloned by PCR, using primers 5’-CCCCATATGCTCAGATCATCTTCTCAA-3’ and 5’-CCCCTCGAGTCACAGAGCAATGACTCC-3’. The PCR product was digested with NdeI and XhoI, and cloned into a pET28a expression vector (EMD Biosciences, Billerica, MA) to generate a HIS-fusion protein. HIS-TNF was expressed in Escherichia coli BL21 cells (Stratagene, La Jolla, CA).
The Institutional Animal Care and Use Committee (IACUC) of UConn Health approved all animal studies.
Bone Marrow Cell Cultures
Mouse bone marrow cells were isolated from the femur and tibia by a modification of published methods (19–21). Cells were then cultured (5 × 104 cells/well in 96-well plates) with complete α-MEM medium (10% heat-inactivated fetal bovine serum [HIFBS] (GE Healthcare - HyClone Laboratories, Logan, UT), 2 mM L-glutamine (Sigma-Aldrich, Saint Louis, MO), 100 U/mL penicillin-streptomycin, (Sigma-Aldrich)) in the presence of recombinant human M-CSF and/or human RANKL, which were purchased (Connstem, Cheshire CT). Bone marrow macrophage/monocyte cells (BMM) were prepared by incubating total bone marrow cells overnight in complete α-MEM on 100 mm tissue culture plastic dishes. Non-adherent cells were collected and mononuclear cells were prepared using Ficoll-Hypaque (Ficoll-Paque PLUS, GE Healthcare, Piscataway, NJ) density gradient centrifugation. The interface between Ficoll-Hypaque and medium was collected and used for BMM cultures (22).
In Vitro OC Formation Assay
Mouse BMMs were cultured at an initial plating density of 5000 cells per well in 96 well culture plates with M-CSF (30ng/ml) and RANKL (30ng/ml). In some experiments, we isolated the OC precursor population from fresh bone marrow cells, as described (5), for in vitro OC formation. The medium was replenished every 3 days and cells were fixed with 2.5% glutaraldehyde in phosphate-buffered saline (PBS) for 15 minutes at room temperature prior to TRAP enzyme histochemistry using a commercial kit (Sigma-Aldrich). TRAP-positive cells that contained 3 or more nuclei were considered to be OC. In some experiments, cultures were stained with DAPI, in addition to TRAP, to identify nuclei so that a frequency distribution of osteoclasts with different numbers of nuclei could be determined.
Tartrate Resistant Acid Phosphates Solution Assay
At the conclusion of an experiment, BMM cells were fixed in 4% paraformaldehyde for 10 minutes at room temperature. Cells were then permeabilized with a 1:1 mixture of ethanol and acetone and incubated with TRAP substrate solution (0.1 M sodium acetate (pH 5.2), 1 mM ascorbic acid, 0.15 M KCl, 10 mM disodium tartrate, and 10 mM p-nitrophenyl phosphate) for 30 minutes at room temperature. The reaction was stopped with 0.3 N NaOH, and absorbance was measured at 405 nm using a microplate reader (BioTek).
Inhibition of Notch2 Signaling
In vitro studies:
BMM cultures from wild type (WT) and PAR1 KO mice (8 to 10 weeks old) were treated with M-CSF and RANKL (30 ng/ml for both) and either a control anti-ragweed antibody (Ab) or anti-Notch2-NRR Ab (10 μg/ml each) (both Ab were a gift from Dr. Chris Siebel, Genentech, South San Francisco, CA) (23). After 5 days of culture, we determined the number of multi-nucleated osteoclasts that formed.
In vivo studies:
WT and PAR1 KO male mice (8 to 14 weeks old) were injected with recombinant mouse TNF (2.0 μg/injection), above the calvaria daily for 4 days. Mice were also treated intraperitoneally with either anti-ragweed control antibody (Ab) or anti Notch2-NRR Ab (10 mg/kg, twice per week). Ab injections were started 3 days prior to TNF injection and repeated every 3 days for a total of 3 injections per experiment. Mice were sacrificed 24 hours after the last TNF injection to analyze osteoclasts in the calvariae by histomorphometry.
Pit Formation Assay
Pit formation assays to analyze resorptive activity were performed by culturing WT or PAR1 KO BMM on UV-sterilized devitalized bovine cortical bone slices that were placed in 96-well plates. BMMs were treated with M-CSF and RANKL (both at 30 ng/mL) for 14 days (24). Pit perimeter and area per OC were measured using a light microscope (BX53, Olympus Scientific, Waltham, MA) and image analysis software (Olympus CellSens).
Flow Cytometry and Fluorescence Activated Cell Sorting (FACS)
The antibodies used for flow cytometric analysis are all commercially available. These include: anti-mouse CD45R (B220) for B-cell lineage cells; anti-mouse CD3 for T cell lineage cells; anti-mouse CD11b (Mac-1) for macrophage lineage cells; anti-mouse CD117 (c-kit) and anti-mouse CD115 (c-fms). Unless indicated, all antibodies and secondary step reagents were obtained directly conjugated to fluorochromes or biotinylated from either of 2 commercial sources (BD Biosciences, San Jose, CA, USA or eBiosciences, San Diego, CA, USA). Labeling of bone marrow cells for flow cytometric analysis was performed by standard staining procedures (6). Flow cytometric analysis was performed on a FACSCalibur (BD Biosciences) and data analysis was done using FlowJo software (Tree Star, Ashland, OR). The OC precursor population (CD45R− CD3− CD11b−/lo CD115+) was sorted in a BD-FACS Aria II (BD Biosciences) equipped with 5 lasers and 18 fluorescence detectors.
mRNA Expression Profiling
Microarray expression profiling was performed on FACS-isolated C57BL/6 murine bone marrow OC precursor cells (CD45R−, CD3−, CD11b−/lo, CD115+) that were cultured with M-CSF and RANKL (30 ng/ml for both) for 3 days. At the conclusion of the culture, RNA was extracted and analyzed by cDNA microarray hybridization using a mouse whole genome chip (Illumina Whole-Genome Expression assay), which detects the binding of fluorescent-tagged RNA from the cells to immobilized 50-mer cDNA oligonucleotides.
Data from the gene expression profiles of highly purified WT osteoclast precursor cells that were cultured with M-CSF or M-CSF + RANKL for 3 days have been deposited in the Gene Expression Omnibus of The National Center for Biotechnology Information, accession number: GSE129334 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129334).
Microcomputed Tomography Analysis (μCT)
The femurs from WT and PAR1 KO mice were removed and fixed in 70% ethanol at 4°C. Metaphyseal trabecular and mid-diaphysis cortical morphometry within the femur were quantified using μCT (μCT40; Scanco Medical AG,). Three-dimensional images were reconstructed using standard convolution back-projection algorithms with Shepp and Logan filtering, and rendered at a discrete density of 578, 704 voxels/mm3 (isòmetric 12-μm voxels). Trabecular morphometry was characterized by measuring the bone volume fraction (BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular spacing (Tb.Sp). Cortical morphometry was analyzed within a 600-μm-long section at mid-diaphysis of the femur and included measurements of average thickness and cross-sectional area. The measurement terminology and units used for μCT analysis were those recommended by the Journal of Bone and Mineral Research (25).
Histomorphometric Analysis
Bone histomorphometry was performed on mouse femurs and calvariae from WT and PAR1 KO male mice in a blinded, non-biased manner using a computerized semiautomated system (Osteomeasure, OsteoMetrics, Decatur, GA) with light microscopy. Static histomorphometry and quantification of OC were performed on paraffin embedded tissues that were stained for TRAP. OC were identified as multinucleated TRAP-positive cells adjacent to bone. A separate analysis for dynamic histomorphometry was performed on the contralateral femur, embedded in methylmethacrylate. The measurement terminology and units used for histomorphometric analysis were those recommended by the Nomenclature Committee of the American Society for Bone and Mineral Research (26). All measurements in the femoral trabecular bone were confined to the secondary spongiosa and restricted to an area between 200 and 1000 μm distal to the growth plate-metaphyseal junction of the distal femur.
For the analysis of the resorptive activity in calvaria, we measured: eroded surface per bone perimeter, osteoclast number per bone perimeter, osteoclast surface per bone perimeter and periosteum area using a computerized semiautomated system (Osteomeasure).
RNA Extraction and RT-PCR
Total RNA was extracted from both WT and PAR1 KO BMM cells at the indicated time with TRI reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s recommendation (27). Total RNA was extracted from femurs and calvaria using a tissue homogenizer (PowerGen Model 1000; Fisher Scientific) and TRI reagent. For all samples, mRNA was converted to cDNA by reverse transcriptase (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems) using random hexamer. Real-time PCR amplification was performed with multiple samples using gene-specific PCR primers and gene-specific TaqMan probes (Applied Biosystems, Foster City, CA). The PCR mixture (including TaqMan primer) was run in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Reagents used for the PCR reaction were purchased from Applied Biosystems. The relative quantification of target gene expression was normalized to the expression of a housekeeping gene, glyceraldehyde 3‐phosphate dehydrogenase (Gapdh), for each sample using the ΔΔCT method.
Western blot analysis.
For experiments that examine whole cell lysates, BMMs were cultured with M-CSF and RANKL for various periods of time and then processed for western analysis. For all western blot analysis, equal amounts of lysates were loaded and electrophoresed on sodium dodecyl sulfate–polyacrylamide electrophoresis gels using a 10% running gel under reducing conditions. Separated proteins were transferred to nitrocellulose membranes, and the membranes were probed with the indicated antibodies, which except for the antibody to PAR1 from LifeSpan Biosciences (Seattle, WA), were all from Cell Signaling Technology (Danvers, MA).
Reactive bands were detected by enhanced chemiluminescence using Chemiluminescence LumiGLO (CellSignaling Technology). Detection was by autoradiography. Quantification was achieved by digitalizing the blots using an imaging station (BioRad, Hercules, CA) and then measuring band density
To detect p65 and phospho-p65, BMMs (5×105cells/well in 6-well plates) were treated with M-CSF + RANKL (30 ng/ml M-CSF and 60 ng/ml RANKL) for 3 days to assure induction of PAR1, transferred to serum free medium for 3 hours and then treated with RANKL (60 ng/ml) for 15 minutes. Cells were lysed in cold lysis buffer and separated into cytoplasmic and nuclear fractions using a commercial kit (Cell Signaling Technology).
Overexpression of PAR1 in Murine Bone Marrow Macrophage (BMM) cells
A retroviral vector encoding the cDNA for PAR1 was purchased from Addgene (Watertown, MA) and inserted into the pMX-Puro retroviral vector (Cell Biolabs, San Diego, CA). Retroviral vectors (vector with PAR1 or empty vector) were transfected into Plat E packaging cells (Cell Biolabs) using Lipofectamine 2000 (Thermo Fisher Scientific) and retroviruses were collected 48h after transfection. BMM cells were cultured with M-CSF (150 ng/ml) for 2 days, transduced in the presence of 8 μg/ml polybrene (Millipore, Burlington, MA) for 6h and cultured overnight with M-CSF. Cells were subcultured further with 30 ng/ml M-CSF and 2 μg/ml puromycin (Millipore) for 2 days. Puromycin-resistant BMM cells were used for experiments.
Statistical Analysis
All experiments were conducted at least twice. Statistical analysis was performed by Student’s t test when comparing two groups or one-way analysis of variance (ANOVA) and the Tukey’s multiple comparison test when comparing multiple groups. Significant differences in the percentage of osteoclasts with different numbers of nuclei were calculated by chi-squared analysis.
RESULTS
Expression of PAR1 during Osteoclastogenesis
To examine genes that were induced by RANKL in purified OC precursor cells (5, 6), we isolated CD45R− CD3− CD11b−/lo CD115+ murine bone marrow cells from male mice by FACS and cultured them with either M-CSF alone or M-CSF + RANKL (30 ng/ml for both) for 3 days. Cells were then analyzed by cDNA microarray hybridization using a mouse whole genome chip for genes that were differentially expressed in cells cultured with M-CSF + RANKL compared to cells cultured with M-CSF alone. This analysis identified known RANKL-induced genes including Mmp9 (matrix metalloproteinase 9) (28) (285 fold induction), Ctsk (cathepsin K) (29) (224 fold induction) and Oscar, (osteoclast associated receptor) (30) (63 fold induction). In addition, we found that F2r (protease-activated receptor 1, PAR1) was induced by RANKL (151-fold induction).
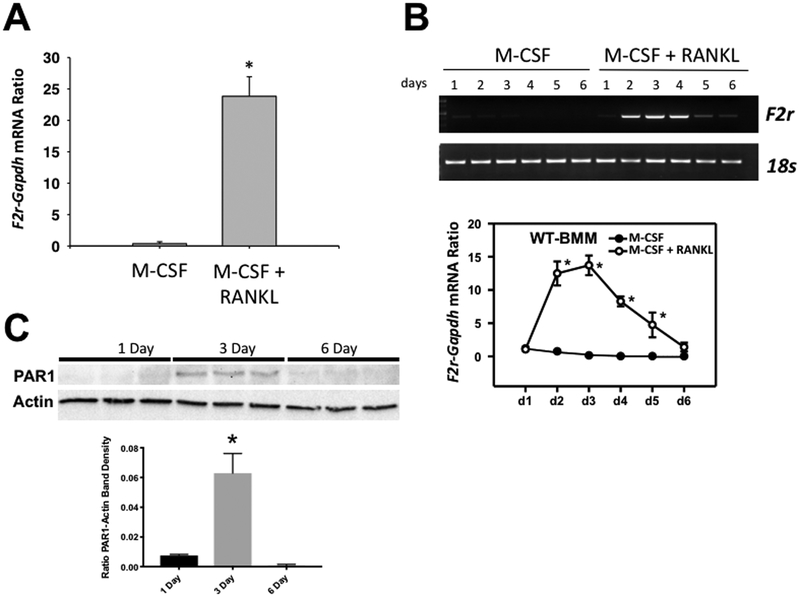
To confirm that PAR1 mRNA was induced in these cultures by RANKL, we performed qRT-PCR for F2r mRNA on the same samples that were used for gene-chip expression analysis (Figure 1A). This demonstrated an approximate 25-fold induction of F2r mRNA with RANKL.
Figure 1.
A) qRT-PCR expression of F2r mRNA in highly purified myeloid precursor cells from male mice that were cultured with M-CSF or M-CSF + RANKL (30 ng/ml for both). B) Upper Panel: Time course for expression of F2r mRNA as detected by RT-PCR relative to 18s mRNA in male BMM cultures treated with M-CSF or M-CSF + RANKL (30 ng/ml for both) for 1 to 6 days. Lower Panel: Time course for expression of F2r mRNA as detected by qRT-PCR in male BMM cultures treated with M-CSF or M-CSF + RANKL (30 ng/ml for both) for 1 to 6 days. C) Expression of PAR1 protein as detected by western blot assay in 3 replicate male BMM cultures treated with M-CSF + RANKL (30 ng/ml for both) for 1, 3 or 6 days. Upper panel is images of the blot probed for PAR1 or Actin. Lower panel is a graph of the ratio of the PAR1 to actin band densities.
N = 3 to 6 for each group, * Significantly different from M-CSF alone, p<0.01.
In this and all subsequent figures “N” is the number of biologic replicates. Values for all groups are mean ± SEM.
A time course study of BMMs from male mice treated with either M-CSF alone or M-CSF and RANKL (Figure 1B) found that at day 1 F2r mRNA levels in the two groups were similar. However, expression in RANKL + M-CSF-treated cultures peaked between days 2 to 3 and returned to that of M-CSF-treated cultures by day 6.
To confirm that PAR1 protein was upregulated in OC precursor cells, we examined its expression by western blot analysis in BMM cultures of 1-, 3- and 6-days duration that were treated with M-CSF and RANKL (Figure 1C). Consistent with the mRNA expression level (Figure 1B), we found that PAR1 protein were easily detected in 3-day cultures and minimal to undetectable in 1-day or 6-day cultures.
BMM Cultures from PAR1 Deficient Mice Form More OC In Vitro
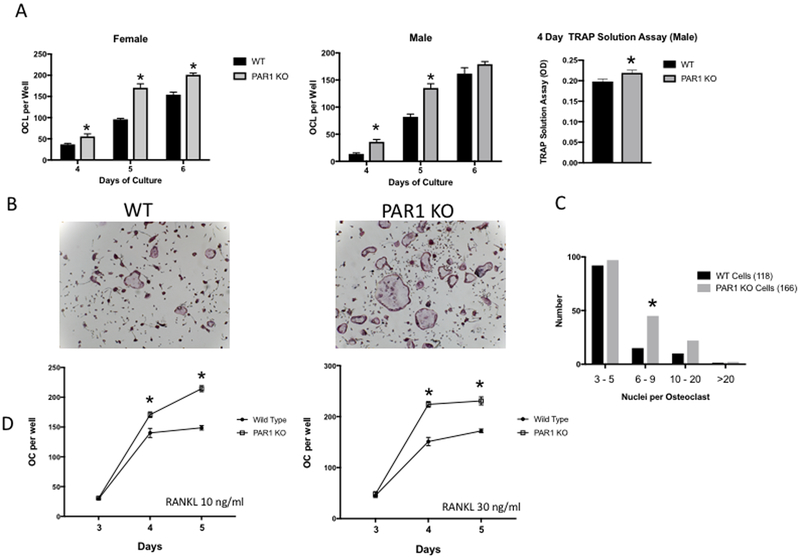
We examined in vitro osteoclastogenesis in BMMs from PAR1 KO mice (18) (Figures 2). Male and female WT and PAR1 KO BMMs were cultured with M-CSF and RANKL (30 ng/ml for both) for 4, 5 or 6 days (Figures 2A and 2B). Osteoclastogenesis was accelerated in PAR1 KO cells from both male and female mice. At days 4 and 5, all PAR1 KO cultures contained more OC than did WT cultures. In 6-day cultures OC formation in male PAR1 KO and WT BMM cultures were similar, whereas cultures of female PAR1 KO cells contained 30% more OC.
Figure 2.
A) Left two figures: OC formed in cultures of BMM cells treated with M-CSF + RANKL (30 ng/ml for both) from WT and PAR1 KO female or male mice that were cultured 4, 5 or 6 days. Far right figure: TRAP solution assay for male BMM cultures from WT or PAR1 KO mice treated with M-CSF + RANKL (30 ng/ml) for 4 days. N = 6 for each group, * Significantly different from WT, p<0.05. B) Representative images of male BMM cultures from A) stained for TRAP activity (purple color). C) Frequency distribution of nuclei in osteoclasts from cultures of male WT and PAR1 KO BMMs that were treated with RANKL and M-CSF (30 ng/ml for both) for 5 days. 118 WT cells and 166 PAR1 KO cells were analyzed. Groups were cells with 3–5 nuclei, 6–9 nuclei, 10–20 nuclei and greater than 20 nuclei. Differences were analyzed by chi-squared analysis. * Significantly different from WT, p < 0.05.
D) OC formed in cultures of highly purified osteoclast precursor cells (OCP) from male WT and PAR1 KO male mice that were treated with M-CSF (30 ng/ml) and either 10 ng/ml or 30 ng/ml of RANKL for 3, 4 or 5 days. N = 3 for each group. * Significantly different from WT, p<0.05
We also examined total tartrate-resistant acid phosphatase (TRAP) activity in 4-day BMM cultures from male mice, using a TRAP solution assay (Figure 2A far right). This demonstrated that in contrast to the approximate doubling of the number of multinucleated, TRAP-positive cells in male PAR1 KO BMM cultures at day 4, total TRAP activity in the PAR1 KO cells at this time point was only minimally elevated. This suggested that PAR1 deletion predominantly enhanced mononuclear cell fusion. To further explore this hypothesis, we examined the expression of genes associated with mononuclear osteoclast precursor cell fusion: v-ATPase V0 subunit d2 (Atp6v0d2) (31), dendritic cell-specific transmembrane protein (Dcstamp) (32) and OC-stimulatory transmembrane protein (Ocstamp) (33), in 3-day wild type and PAR1 KO BMM cultures that were treated with M-CSF + RANKL (Supplemental Figure 1). We found that expression of Atp6v0d2 and Ocstamp was significantly increased in PAR1 KO cultures. In addition, there was a trend (p=0.08) for Dcstamp to be greater in PAR1 KO cells.
We next analyzed the frequency distribution of osteoclasts with a different number of nuclei in male WT and PAR1 KO BMMs, treated with RANKL and M-CSF (30 ng/ml for both) for 5 days (Figure 2C). This demonstrated that PAR1 KO BMMs formed more osteoclasts with 6 to 9 nuclei (12.7% in WT and 27.1% in PAR1 KO, p < 0.02), a finding which further argues that PAR1 KO cells have an increased fusion capacity.
BMM cultures contain a mixture of cell types. To determine if the effect of PAR1 deletion on osteoclastogenesis was cell autonomous, we generated highly purified WT and PAR1 KO OC precursors by FACS (CD45R−, CD3−, CD11b−/lo, CD115+) (5, 6), and treated them with 10 or 30 ng/ml of RANKL together with 30 ng/ml of M-CSF (Figure 2D). At both 4 and 5 days, PAR1 KO cells generated more osteoclasts than WT cells.
To measure resorptive activity of the osteoclasts that formed in PAR1 KO and WT BMM cultures, cells were plated on bovine cortical bone slices and cultured for 14 days with M-CSF and RANKL (30 ng/ml for each). PAR1 KO cultures produced significantly more pits per slice (Supplemental Figure 2). However, the resorptive activity of individual osteoclasts as measured by the mean perimeter per pit or mean area per pit did not differ between groups.
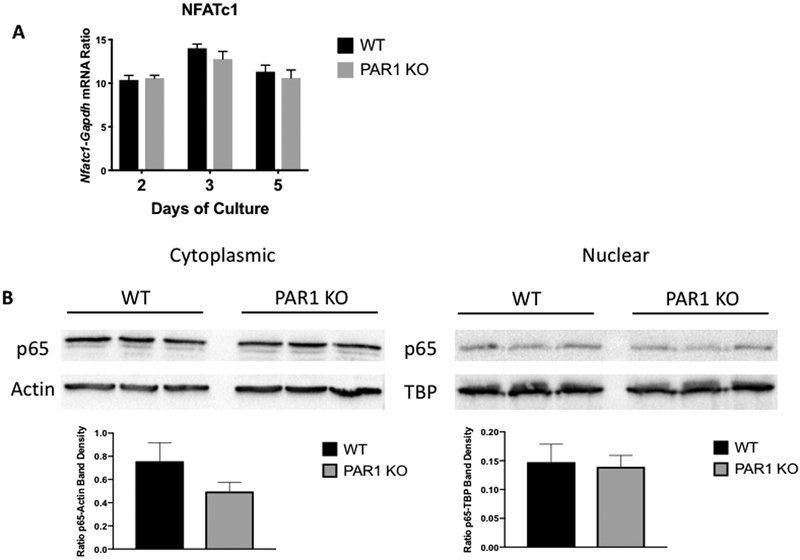
PAR1 deletion did not enhance RANKL-induced NFATc1 or NF-κB signaling in BMM cultures
To better understand the mechanism regulating the enhanced osteoclastogenesis in PAR1 KO BMM cultures, we examined the mRNA levels of nuclear factor of activated T cells c1 (NFATc1) in WT and PAR1 BMM cultures at 2, 3 and 5 days of treatment with M-CSF + RANKL (both at 30 ng/ml) (Figure 3A) and found no differences. We also examined nuclear localization of p65, a component of NF-κB signaling and found no differences in cultures of WT and PAR1 KO BMM cells that were stimulated with M-CSF + RANKL for 3 days to induce PAR1 protein, rested, without cytokines, for 3 hours and then re-challenged with RANKL for 15 minutes (Figure 3B).
Figure 3.
A) mRNA expression of Nfatc1-Gapdh ratios in male BMM cultures from WT and PAR1 KO mice treated with M-CSF and RANKL (30 ng/ml for both) for 2, 3 or 5 days. N = 3 for each group. B) Cytoplasmic and nuclear p65 protein in 3 replicate male BMM cultures from WT and PAR1 KO mice. Cells were cultured with M-CSF and RANKL (30 ng/ml for both) for 3 days to stimulate PAR1 expression in WT cells and then transferred to serum free media for 3 hours before being challenged with RANKL for 15 minutes. Cytoplasmic and nuclear fractions were isolated and run in western blot assay. Control protein blots were for actin in the cytoplasmic fraction and TATA box binding protein (TBP) in the nuclear fraction. Lower graphs are the ratio of p65 to actin band density in the cytoplasmic fraction and p65 to TBP band density in the nuclear fraction.
Parathyroid Hormone Increases the Number of OC in PAR1 KO Whole Bone Marrow Cultures without Differences between WT and PAR1 KO Cells in RANKL or Osteoprotegerin (OPG) mRNA Levels or their Ratio
It was previously found that PAR1 KO mice have lower RANKL and higher OPG levels in serum than WT mice, resulting in a lower RANKL/OPG ratio (34). Therefore, we examined the ability of bovine parathyroid hormone 1–34 (b-PTH 1–34, 10 and 100 ng/ml) to stimulate OC formation in male whole bone marrow cultures and alter RANKL (Tnfsf11) and osteoprotegerin (OPG, Tnfrsf11b) mRNA levels (Supplemental Figure 3). Unlike the BMM assay, b-PTH stimulates RANKL and inhibits (OPG) production in bone marrow stromal/mesenchymal cells, which also produce M-CSF (19, 35, 36). OC formation was increased in cultures of PAR1 KO cells at both doses of bPTH1–34 after 5 and 6 days compared to WT cultures (Supplemental Figure 3A). In cultures of cells from male mice after 3 days, both concentrations of PTH significantly increased RANKL mRNA, significantly inhibited OPG mRNA and significantly increased their ratio. However, there were no differences in these responses between PAR1 KO and WT cells (Supplemental Figure 3B).
We also examined expression levels of RANKL and OPG mRNA in freshly extracted femurs, which had their bone marrow flushed, from PAR1 KO and WT mice. These studies demonstrated similar levels of RANKL and OPG mRNA and their ratio in bones from both genotypes (Supplemental Figure 4). In addition, we found no difference in the level of M-CSF (Csf1) mRNA between WT and PAR1 KO bones.
PAR1 Deletion had Little Effect on Bone Mass or Bone Cells under Homeostatic Conditions
Using μCT, we found a trend for trabecular bone volume (BV/TV) to be decreased (p = 0.08) and trabecular spacing to be increased (p = 0.08) in the distal femurs of male 8-week-old PAR1 KO mice relative to male WT mice (Table 1). In addition, trabecular number and connectivity density were decreased. There was no effect of PAR1 deletion on cortical thickness or area in femurs. Measurement of histomorphometric indices by static and dynamic histomorphometry using 8-week-old male femurs revealed no difference in any parameter between PAR1 KO and WT mice (Table 2).
Table 1.
Micro-computed Tomography
| Trabecular Bone | BV/TV (%) |
Trabecular Thickness (μm) |
Trabecular Number (1/mm) |
Trabecular Spacing (μm) |
Connectivity Density (1/mm3) |
Structure Model Index (SMI) |
|---|---|---|---|---|---|---|
| WT | 12.7 ± 1.2 | 42 ± 2 | 5.40 ± 0.15 | 166 ± 6 | 164 ± 16 | 2.5 ± 0.1 |
| PAR1 KO | 9.9 ± 0.9 | 40 ± 2 | 5.06 ± 0.06 | 197 ± 3 | 111 ± 8 | 2.7 ± 0.1 |
| Significance | p=0.08 | NS | p<0.05 | p=0.08 | p<0.01 | NS |
| N=16 bones per group | ||||||
| Cortical Bone | Total Bone Length (mm) |
Cortical Bone Area (μm2) |
Cortical Porosity (%) |
Cortical Thickness (mm) |
|---|---|---|---|---|
| WT | 14.2 ± 0.1 | 749 ± 29 | 0.69 ± 0.09 | 0.17 ± 0.01 |
| PAR1 KO | 14.3 ± 0.1 | 703 ± 28 | 0.52 ± 0.04 | 0.16 ± 0.01 |
| Significance | NS | NS | NS | NS |
N=16 bones per group
Table 2.
Histomorphometry
| Static Parameters | BV/TV (%) |
Number OCL/ Bone Perimeter (1/mm) |
Osteoclast Surface/Bone Surface (%) |
Osteoblast Surface/ Bone Surface (%) |
Trabecular Thickness (μm) |
Trabecular Separation (μm) |
Trabecular Number (1/mm) |
|---|---|---|---|---|---|---|---|
| WT | 12.0 ± 2.2 | 2.47 ± 0.71 | 5.19 ± 0.15 | 15.18 ± 2.13 | 28.96 ± 2.78 | 228.9 ± 21.8 | 4.0 ± 0.3 |
| PAR1 KO | 10.4 ± 1.8 | 3.11 ± 0.36 | 5.06 ± 0.06 | 14.49 ± 2.27 | 26.97 ± 2.60 | 252.4 ± 26.7 | 3.8 ± 0.3 |
| Significance | NS | NS | NS | NS | NS | NS | NS |
| N=6–7 bones per group | |||||||
| Dynamic Parameters | MAR μm/day |
BFR/Bone Surface μm3/μm2/day |
|---|---|---|
| WT | 1.97 ± 0.11 | 0.068 ± 0.010 |
| PAR1 KO | 2.20 ± 0.13 | 0.092 ± 0.018 |
| Significance | NS |
N=6 bones per group
PAR1 KO Cells have an Enhanced Osteoclastogenic Response to TNF in vitro
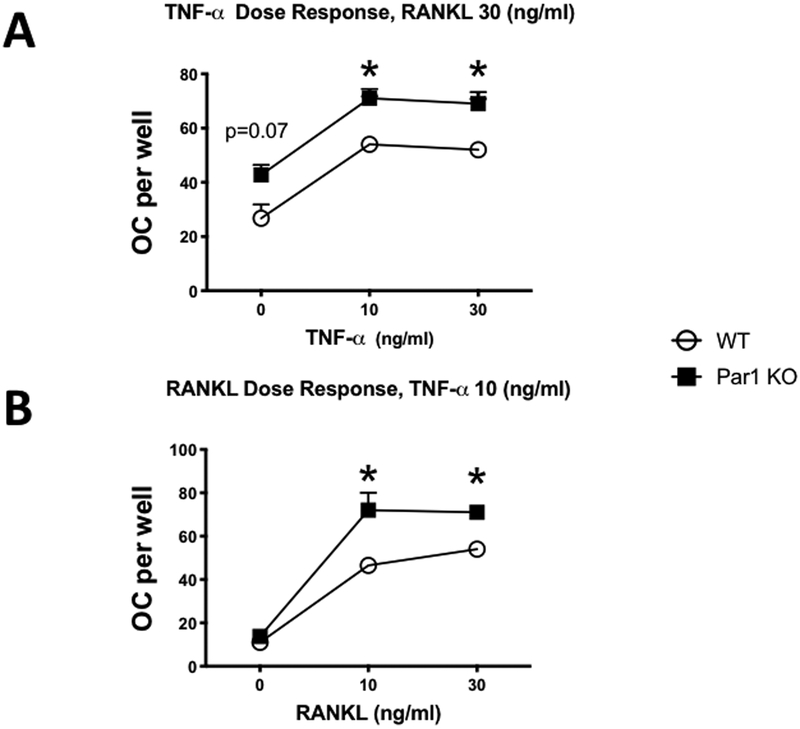
Because PAR1 KO OC precursor cells produced more osteoclasts in response to RANKL, a tumor necrosis factor superfamily member, we examined whether the response of PAR1 KO BMMs to TNF was also enhanced. Treatment of BMMs with TNF + RANKL (10 or 30 ng/ml for each) produced an enhanced osteoclastogenic response in PAR1 KO cultures compared to WT (Figures 4A and B).
Figure 4.
A) OC formed in cultures of male BMMs treated with M-CSF + RANKL (30 ng/ml for both) and 0, 10 or 30 ng/ml of TNF. Cells were from WT or PAR1 KO male mice that were cultured for 4 days. N = 3 – 4 for each group. * Significantly different from WT, p < 0.05.
B) OC formed in cultures of male BMMs treated with M-CSF (30 ng/ml) + TNF (10 ng/ml) and 0, 10 or 30 ng/ml of RANKL. Cells were from WT or PAR1 KO male mice that were cultured for 4 days. N = 3 – 4 for each group. * Significantly different from WT, p < 0.05.
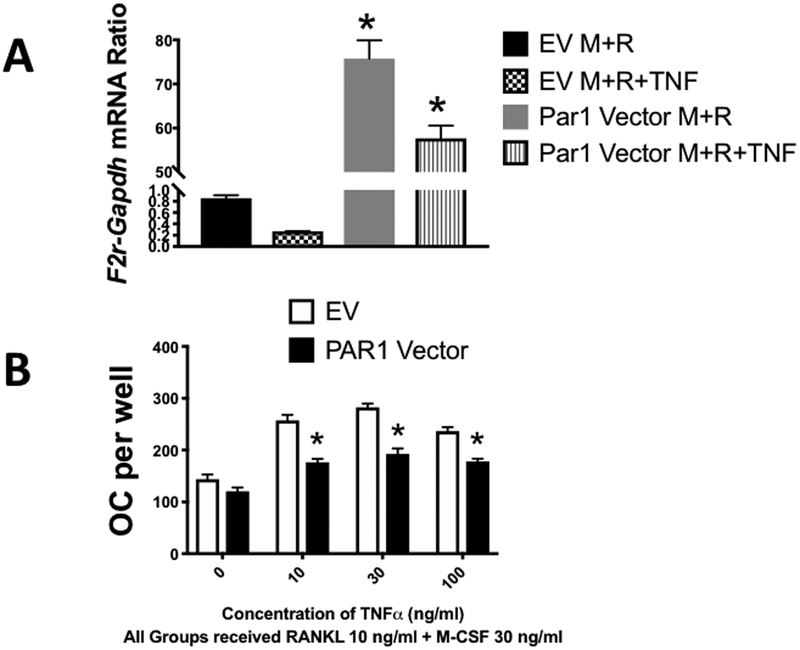
Overexpression of Par1 in BMM cultures inhibited the response to TNF +low dose RANKL but not low dose RANKL alone.
We generated a retrovirus that overexpressed PAR1 in BMM cultures and compared osteoclastogenic responses of cells cultured with M-CSF + low dose RANKL (10 ng/ml) to cells cultured with M-CSF + low dose RANKL and increasing doses of TNF (Figure 5A). In these experiments cells transduced with an empty vector retrovirus served as a control. We found that overexpression of PAR1 had little effect on the osteoclastogenic response to the low dose of RANKL but significantly inhibited the ability of TNF at 10 to 100 ng/ml to enhance the osteoclastogenic response (Figure 5B). These results argue that PAR1 is a more potent inhibitor of the in vitro osteoclastogenic response to TNF than to RANKL.
Figure 5.
A) mRNA expression of PAR1 (F2r) in WT male BMM cells measured by qRT-PCR. Cells were transduced with a retrovirus expressing either an empty vector (EV) or a vector expressing PAR1 (PAR1 Vector). Cells were then treated with M-CSF + RANKL (M+R) or M-CSF + RANKL +TNF (M+R+TNF) (30 ng/ml for each) for 3 days and extracted to analyze PAR1 (F2r) mRNA expression by qRT-PCR. N = 3 per group. * Significantly different from respective vehicle-treated group, p < 0.01.
B) OC formed in cultures of male BMM cells, which were transduced with either an empty vector (EV) or a PAR1-expressing vector (PAR1 Vector) and then treated with the indicated concentrations of M-CSF + RANKL and varying concentrations of TNF for 3 days. N = 3 per group. * Significantly different from respective EV group, p < 0.01.
PAR1 KO Cells have an Enhanced Osteoclastogenic Response to TNF in vivo
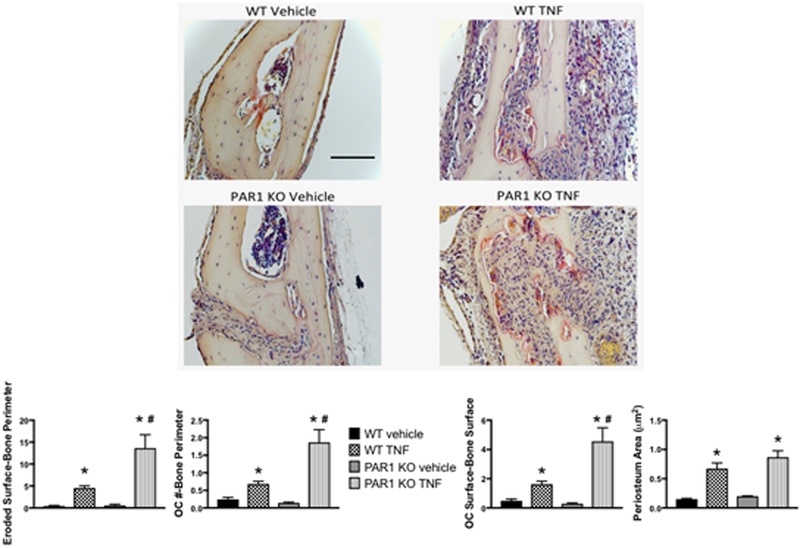
We examined if the in vivo osteoclastogenic response of PAR1 KO mice to TNF was also enhanced. We treated WT and PAR1 KO mice with daily subcutaneous injections of TNF (2 μg,) over their calvariae for 4 days (Figure 6) and examined three parameters of the osteoclastogenic response (eroded surface per bone perimeter, osteoclast number per bone perimeter and osteoclast surface per bone surface) 1 day after the last TNF injection. In WT mice TNF induced a 10.6-fold increase in eroded surface per bone perimeter, a 2.8-fold increase in osteoclast number per bone perimeter and a 3.3-fold increase in osteoclast surface per bone surface. Strikingly, in PAR1 KO mice the osteoclastogenic response to TNF was significantly greater than in WT mice (p<0.01 for all three parameters). Eroded surface per bone perimeter was increased by 23.8-fold, osteoclast number per bone perimeter was increased by 12.9-fold and osteoclast surface per bone surface was increased by 15.5-fold. In contrast, periosteal area, a measure of the size of the total inflammatory response, was not different between WT and PAR1 KO mice (a 4.4-fold increase with TNF in WT mice and a 4.3-fold increase with TNF in PAR1 KO mice).
Figure 6.
WT or PAR1 KO male mice were injected once daily with either vehicle or TNF (2 μg) subcutaneously over the calvariae for 4 days and sacrificed 1 day later. Images are representative histology slides stained for TRAP. The graphs below the images show the ratio of eroded surfaces-bone perimeter, OC number (#)-bone perimeter, OC surface-bone surface and periosteum area (μm2). N = 6 – 9 per group. * Significantly different from respective vehicle-treated group, p < 0.01. # Significantly different from WT vehicle-treated group, p < 0.01. Scale bar = 100 μm. All images are the same magnification.
PAR1 Deletion Causes Enhanced Osteoclastogenesis in Response to Inflammatory Signals through a Notch2-dependent Mechanism
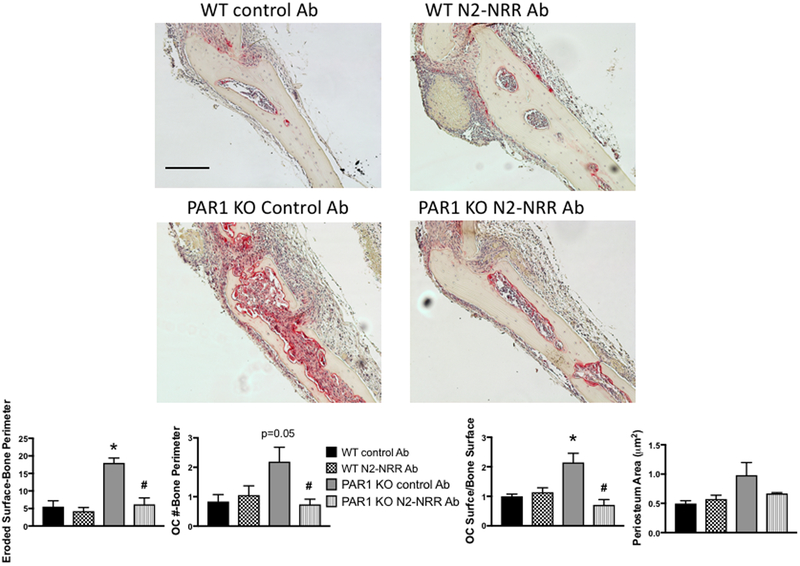
TNF-mediated inflammatory osteolysis occurs, in part, through increased Notch signaling at sites of inflammation (37) and Notch2 signaling was previously found to stimulate osteoclastogenesis (47). Therefore, we examined whether inhibiting Notch2 activation in cells from PAR1 KO mice abrogated their enhanced OC response to TNF (Figure 7). WT and PAR1 KO mice were treated with TNF over the calvariae and either a control Ab or an Ab to the Notch2 negative regulatory region (N2-NRR Ab), which specifically blocks Notch2 signaling (23). In mice treated with the control antibody, TNF treatment of PAR1 KO mice produced significantly greater levels of eroded surface per bone surface and osteoclast surface per bone surface than did TNF treatment of WT mice (P<0.01). Osteoclast number per bone perimeter was borderline significantly greater in control antibody-treated PAR1 KO mice compared to WT mice (p=0.05). We found that treatment of PAR1 KO mice with N2-NRR Ab reduced TNF-induced osteoclastogenesis to WT levels without affecting TNF responses in WT mice as measured by all three parameters of the osteoclastogenic response (eroded surface per bone perimeter, osteoclast number per bone perimeter and osteoclast surface per bone surface) (p < 0.01 for all three). In addition, Notch2 inhibition had non-significant effect on periosteum area, implying that the overall inflammatory response to TNF was less affected than the osteoclastogenic response.
Figure 7.
WT or PAR1 KO male mice were injected once daily with TNF (2 μg) subcutaneously over the calvariae for 4 days and sacrificed 1 day later. Mice were also treated with control or N2 NRR Ab starting 3 days prior to the first TNF injection. Images are representative histology slides stained for TRAP. The graphs below the images show the ratio of eroded surfaces-bone perimeter, OC number (#)-bone perimeter, OC surface-bone surface and periosteum area (μm2). N = 6 – 9 per group. N = 3 – 4 per group. * Significantly different from respective WT control Ab-treated group, p < 0.01. # Significantly different from PAR1 KO control Ab-treated group, p < 0.01. Scale bar = 100 μm. All images are the same magnification.
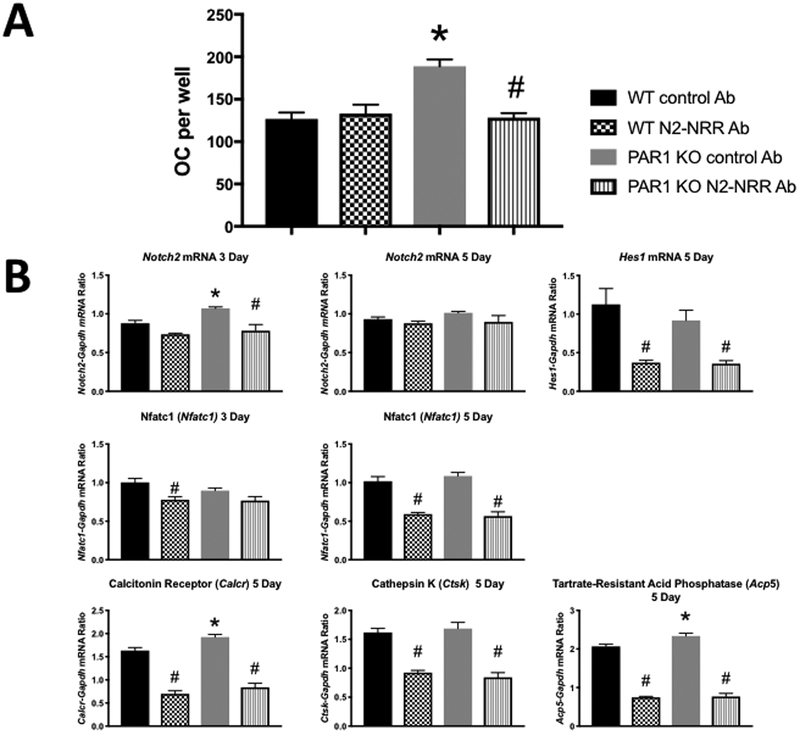
We also examined the effect of the N2-NRR antibody on osteoclastogenesis in WT and PAR1 KO cells (Figure 8). We found that it blocked the enhanced M-CSF + RANKL-stimulated osteoclastogenesis of 5-day PAR1 KO cultures without affecting responses in WT cells. However, when we examined a variety of gene that are critical for Notch2 signaling and osteoclast formation and function in these cultures, we found that the effect of PAR1 deletion and the antibody varied between 3-day and 5-day cultures. Notch2 mRNA levels were enhanced at day 3, when PAR1 (F2r) is also expressed in these cultures (Figure1), but not at day 5 when PAR1 message and protein are extinguished. Interestingly, Hes1 mRNA a principal downstream target of canonical Notch signaling in osteoclasts (38), was not increased in the cultures at 5 days, a time when its expression is greatest (38). Significantly, its levels were equally inhibited by the N2-NRR antibody in 5 day cultures of WT and PAR1 KO cells. Expression of Nfatc1, the master regulator of osteoclastogenesis (39) was not significantly different between WT and PAR1 KO cells treated with control antibody at days 3 or 5 of culture. However, Nfatc1 expression was inhibited equally in WT and PAR1 KO cells at day 5 (but not day 3) by the N2-NRR antibody. Expression levels of calcitonin receptor and tartrate -resistant acid phosphatase mRNA in day 5 cultures, treated with the control antibody, were greater in PAR1 KO cells. In contrast, there was no difference in expression levels of cathepsin K in the control antibody-treated WT and PAR1 KO cultures. Calcitonin receptor, tartrate -resistant acid phosphatase and cathepsin K are markers of the mature osteoclast phenotype (1, 40, 41). Consistent with the downregulation of Nfatc1 by the N2-NRR antibody at 5 but not 3 days of culture, we found that the N2-NRR antibody inhibited all three osteoclast-associated genes equally in WT and PAR1 KO cells at day 5. The effect of the N2-NRR antibody on mRNA levels of Nfatc1 and the three osteoclast-associated genes suggest that at longer time points, Notch2 signaling may inhibit resorptive responses equally in WT and PAR1 KO to M-CSF + RANKL. Unfortunately, this is difficult to examine in osteoclasts in vitro that form on tissue culture plastic as these cells typically undergo apoptosis at around day 7 of culture (42).
Figure 8.
A) OC formed in cultures of male BMM cells treated with M-CSF + RANKL (30 ng/ml for both) from WT and PAR1 KO male mice that were cultured for 5 days with either control Ab or N2-NRR Ab. N = 3 – 4 per group. * Significantly different from respective WT control Ab-treated group, p < 0.01. # Significantly different from PAR1 KO control Ab-treated group, p < 0.01
B) mRNA expression in BMM cultures from WT and PAR1 KO mice that treated with either control or N2-NRR Ab. Cultures were continued for either 3 or 5 days as indicated and analyzed for 3 or 5 days and analyzed for expression of Notch2, Hes1, Nfatc1, calcitonin receptor (Calcr), cathepsin K (Ctsk) or tartrate resistant acid phosphatase (Acp5) by qRT-PCR. N = 3 – 4 per group. * Significantly different from respective WT control Ab-treated group, p < 0.01. # Significantly different from PAR1 KO control Ab-treated group, p < 0.01
DISCUSSION
We found that PAR1 mRNA and protein were transiently expressed in OC precursor cells during their differentiation into mature OC. This result is consistent with a previous study (17) that found PAR1 protein was not expressed in mature OC in vivo. Recently, another group identified PAR1 mRNA and protein expression in osteoclast precursor cells (43).
OC precursors are not the only bone cells that express PAR1. Osteoblast expression has been shown to have functional consequences (17, 44). For example, the ability of thrombin to enhance cell proliferation in osteoblasts and inhibit alkaline phosphatase activity is PAR1-dependent (45), as is thrombin’s ability to increase growth factor and cytokine expression (46, 47). Transforming growth factor-β is known to stimulate PAR1 expression in osteoblasts (44) and PAR1 KO mice have a decreased rate that tibial drill-hole lesions heal (48).
To further study the role that PAR1 has in bone cells, we examined OC development and function in vitro and in vivo and the bone phenotype of PAR1 KO mice. BMM and highly purified osteoclast precursor cell cultures from PAR1 KO mice as well as whole bone marrow cultures from PAR1 KO mice formed significantly more osteoclasts than cultured cells from WT mice. Our finding of an increase in osteoclastogenesis in cultures of highly purified PAR1 KO OC precursor cells (Figure 2D) argues that this response is cell autonomous.
It is unlikely that the increased osteoclastogenic potential of PAR1 KO whole bone marrow cultures was related to changes in RANKL, OPG or their ratio since we saw little difference between WT and PAR1 KO cells in the ability of PTH to regulate the mRNA levels of RANKL and OPG or their ratio. Furthermore, we found no significant differences between PAR1 KO and WT mice in the mRNA levels of RANKL or OPG or their ratio in femurs. It also does not appear that PAR1 deletion affected either RANKL-induced p65 NF-κB signaling or NFATc1 activation as neither of these parameters differed between WT and PAR1 KO BMM cultures that were stimulated by RANKL for 3 day (Figure 3), a time when PAR1 is known to be induced (Figure 1). Rather, our result argue more that the predominant effect of PAR1 deletion on osteoclastogenesis is to enhance the rate of mononuclear cell fusion into mature OC.
Our results contrast to those of Tudpor (34), who found a decrease in serum levels of RANKL and an increase in serum levels of OPG in PAR1 KO mice, leading to a decreased RANKL/OPG ratio. However, how serum levels of RANKL and OPG, which are produced in a variety of tissues (49), relate to production of RANKL and OPG in the bone microenvironment where they act to regulate OC formation, is unknown.
Tudpor (34) also found that PAR1 KO mice had increased bone volume as determined by μCT analysis with no change in OC or osteoblast numbers. In contrast, Aronovich (50) found that PAR1 KO mice had decreased bone volume. We found a trend for PAR1 KO bones to have a small decrease in trabecular bone volume with a decrease in trabecular number and connectivity density. Our data, which are more consistent with those of Aronovich (50), argue that under basal conditions in eight-week old mice, PAR1 deletion produces a weak phenotype of decreased trabecular bone mass. It is unknown exactly why there are differences in the bone phenotype of PAR1 KO mice among different investigators since all used the same strain (18). However, our finding that challenging PAR1 KO mice with the proinflammatory cytokine, TNF, produced an enhanced resorptive response relative to WT raises the possibility that differences in the inflammatory state of mice among different investigators contributed to the variations in bone phenotype that have been reported.
The role of PAR1 in inflammation is variable. Deficiency of PAR1 in mice resulted in a reduced inflammatory response to influenza infection (51). It was also shown that PAR1 is part of a signaling cascade in osteoclasts that inhibits lipopolysaccharide (LPS)-induced osteoclastogenesis through a mechanism involving Ca++, CaMKK and AMPK (52).
We found that the enhanced osteoclastogenic response both to RANKL in vitro and TNF in vivo was mediated by mechanisms that were dependent on Notch2 signaling since they were abrogated by a specific neutralizing antibody to the Notch2 NRR (23). These results identify a previously unknown interaction of Notch2 signaling and PAR1 in osteoclastic responses to RANKL and TNF. There are only a few previous studies that demonstrate interactions of Notch signaling with PAR1. Activation of PAR1 on human neuronal cells was associated with increased expression and translocation of the voltage gated potassium channel, Kv1.3, to the neuronal cell membrane and activation of Notch1 (53). It has also been shown that overexpression of the Notch ligand, Jagged1, in PAR1 KO cells results in a rapid thrombin-induced export of FGF1 (54).
In bone, Notch signaling has been found to critically regulate both osteoblast and osteoclast function (55). In osteoclasts, Notch signaling can have variable effects (56). However, in general, Notch1 signaling inhibits osteoclastogenesis (57), while Notch2 signaling is stimulatory (58).
RBP-Jκ has a variety of functions, among which is being an intermediate in Notch signaling (55). Zhao (59) showed that deletion of RBP-Jκ in myeloid lineage cells in mice produced a phenotype that was similar to that of the PAR1 KO mouse (i.e., a minimal bone phenotype under basal conditions and an exaggerated osteoclastogenic response to TNF in vivo). Given our current findings, it is possible that some of the effects that myeloid-specific deletion of RBP-Jκ has in mice result from alterations of Notch2 signaling in OC precursor cells. However, the exact mechanism by which PAR1 and Notch influence osteoclastogenesis and the response to inflammation remains to be determined.
In summary, we found that PAR1 is transiently expressed in maturing OC precursors and its global deletion in mice has minimal effects on bone mass under homeostatic conditions. Our data are consistent with the hypothesis that PAR1 expression during osteoclast differentiation is anti-osteoclastogenic. This is supported by our data demonstrating enhanced osteoclast formation in cultured cells from PAR1 KO mice that were stimulated with RANKL alone or RANKL + TNF in vitro and TNF in vivo and by the decreased osteoclastogenic potential of WT cells that overexpress PAR1. Our results suggest that PAR1 is an inhibitor of a pro-osteoclastogenic Notch2 signaling pathway that is activated by inflammatory cytokines. This pathway is likely involved in regulating the response of bone to diseases associated with an abnormal immune system such as rheumatoid arthritis or osteomyelitis.
Supplementary Material
Key Points:
PAR1 is transiently expressed in osteoclast precursor cell during differentiation.
PAR1 deletion enhances osteoclastogenesis while PAR1 overexpression inhibits it.
The enhanced osteoclastogenesis of PAR1-deletion is mediated Notch2.
ACKNOWLEDGMENTS
We thank Dr. Douglas Adams and David Bridgewater for performing μCT analysis.
Authors’ roles: SJ, JK, SM, BS, NSA and CEJG performed experiments and data collection. FM, HLA, EC, AS, SKL and JL designed experiments and analyzed data. JL drafted the manuscript. CEJG, HLA, AS, EC, HD, SKL and JL provided comments that revised the manuscript content. JL approved the final version of manuscript. All authors accept responsibility for the integrity of data analysis.
Supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS): R01 AR048714 and RO1 AR068160 and the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK): R01 DK045227 of the National Institutes of Health (NIH)
References Cited
- 1.Teitelbaum SL 2000. Bone resorption by osteoclasts. Science 289: 1504–1508. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, and Nishikawa S. 1990. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 345: 442–444. [DOI] [PubMed] [Google Scholar]
- 3.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, and Boyle WJ. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93: 165–176. [DOI] [PubMed] [Google Scholar]
- 4.Schett G, and Smolen JS. 2005. New insights in the mechanism of bone loss in arthritis. Curr.Pharm.Des 11: 3039–3049. [DOI] [PubMed] [Google Scholar]
- 5.Jacquin C, Gran DE, Lee SK, Lorenzo JA, and Aguila HL. 2006. Identification of multiple osteoclast precursor populations in murine bone marrow. J.Bone Miner.Res. 21: 67–77. [DOI] [PubMed] [Google Scholar]
- 6.Jacome-Galarza CE, Lee SK, Lorenzo JA, and Aguila HL. 2013. Identification, characterization, and isolation of a common progenitor for osteoclasts, macrophages, and dendritic cells from murine bone marrow and periphery. J Bone Miner Res 28: 1203–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackie EJ, Loh LH, Sivagurunathan S, Uaesoontrachoon K, Yoo HJ, Wong D, Georgy SR, and Pagel CN. 2008. Protease-activated receptors in the musculoskeletal system. The international journal of biochemistry & cell biology 40: 1169–1184. [DOI] [PubMed] [Google Scholar]
- 8.Lin H, Liu AP, Smith TH, and Trejo J. 2013. Cofactoring and dimerization of proteinase-activated receptors. Pharmacol Rev 65: 1198–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao P, Metcalf M, and Bunnett NW. 2014. Biased signaling of protease-activated receptors. Front Endocrinol (Lausanne) 5: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanc-Brude OP, Archer F, Leoni P, Derian C, Bolsover S, Laurent GJ, and Chambers RC. 2005. Factor Xa stimulates fibroblast procollagen production, proliferation, and calcium signaling via PAR1 activation. Exp Cell Res 304: 16–27. [DOI] [PubMed] [Google Scholar]
- 11.Kuliopulos A, Covic L, Seeley SK, Sheridan PJ, Helin J, and Costello CE. 1999. Plasmin desensitization of the PAR1 thrombin receptor: kinetics, sites of truncation, and implications for thrombolytic therapy. Biochemistry 38: 4572–4585. [DOI] [PubMed] [Google Scholar]
- 12.Trivedi V, Boire A, Tchernychev B, Kaneider NC, Leger AJ, O’Callaghan K, Covic L, and Kuliopulos A. 2009. Platelet matrix metalloprotease-1 mediates thrombogenesis by activating PAR1 at a cryptic ligand site. Cell 137: 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffre F, Friedman AE, Hu Z, Mackman N, and Blaxall BC. 2012. beta-adrenergic receptor stimulation transactivates protease-activated receptor 1 via matrix metalloproteinase 13 in cardiac cells. Circulation 125: 2993–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihara K, Ramachandran R, Renaux B, Saifeddine M, and Hollenberg MD. 2013. Neutrophil elastase and proteinase-3 trigger G protein-biased signaling through proteinase-activated receptor-1 (PAR1). J Biol Chem 288: 32979–32990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosnier LO, Sinha RK, Burnier L, Bouwens EA, and Griffin JH. 2012. Biased agonism of protease-activated receptor 1 by activated protein C caused by noncanonical cleavage at Arg46. Blood 120: 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper DM, Pechkovsky DV, Hackett TL, Knight DA, and Granville DJ. 2011. Granzyme K activates protease-activated receptor-1. PLoS One 6: e21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham LA, Jenkins AL, Stone SR, and Mackie EJ. 1998. Expression of the thrombin receptor in developing bone and associated tissues. J Bone Miner Res 13: 818–827. [DOI] [PubMed] [Google Scholar]
- 18.Connolly AJ, Ishihara H, Kahn ML, Farese RV Jr., and Coughlin SR. 1996. Role of the thrombin receptor in development and evidence for a second receptor. Nature 381: 516–519. [DOI] [PubMed] [Google Scholar]
- 19.Lee SK, and Lorenzo JA. 1999. Parathyroid hormone stimulates TRANCE and inhibits osteoprotegerin messenger ribonucleic acid expression in murine bone marrow cultures: correlation with osteoclast-like cell formation. Endocrinology 140: 3552–3561. [DOI] [PubMed] [Google Scholar]
- 20.Katavic V, Grcevic D, Lee SK, Kalinowski J, Jastrzebski S, Dougall W, Anderson D, Puddington L, Aguila HL, and Lorenzo JA. 2003. The surface antigen CD45R identifies a population of estrogen-regulated murine marrow cells that contain osteoclast precursors. Bone 32: 581–590. [DOI] [PubMed] [Google Scholar]
- 21.Aguila HL, Mun SH, Kalinowski J, Adams DJ, Lorenzo JA, and Lee SK. 2012. Osteoblast-specific overexpression of human interleukin-7 rescues the bone mass phenotype of interleukin-7-deficient female mice. J Bone Miner Res 27: 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SK, Gardner AE, Kalinowski JF, Jastrzebski SL, and Lorenzo JA. 2006. RANKL-stimulated osteoclast-like cell formation in vitro is partially dependent on endogenous interleukin-1 production. Bone 38: 678–685. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, and Siebel CW. 2010. Therapeutic antibody targeting of individual Notch receptors. Nature 464: 1052–1057. [DOI] [PubMed] [Google Scholar]
- 24.Jacome-Galarza C, Lee S, Lorenzo J, and Aguila H. 2013. Identification, characterization and isolation of a common progenitor for osteoclasts, macrophages and dendritic cells from murine bone marrow and periphery. J Bone and Mineral Research 28: 1203–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, and Muller R. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 25: 1468–1486. [DOI] [PubMed] [Google Scholar]
- 26.Parfitt AM, Drezner MK, Glorieux F, Kanis JA, Malluche HH, Meunier PJ, Ott SM, and Recker RR. 1987. Bone histomorphometry: Standardization of nomenclature, symbols and units. J.Bone Min.Res 2: 595–610. [DOI] [PubMed] [Google Scholar]
- 27.Chirgwin JM, Przbyla AE, MacDonald RJ, and Rutter WJ. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochem. 18: 5294–5289. [DOI] [PubMed] [Google Scholar]
- 28.Wucherpfennig AL, Li YP, Stetler-Stevenson WG, Rosenberg AE, and Stashenko P. 1994. Expression of 92 kD type IV collagenase/gelatinase B in human osteoclasts. Journal of Bone and Mineral Research 9: 549–556. [DOI] [PubMed] [Google Scholar]
- 29.Saneshige S, Mano H, Tezuka K, Kakudo S, Mori Y, Honda Y, Itabashi A, Yamada T, Miyata K, Hakeda Y, Ishii J, and Kumegawa M. 1995. Retinoic acid directly stimulates osteoclastic bone resorption and gene expression of cathepsin K/OC-2. Biochemical Journal 309: 721–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim N, Takami M, Rho J, Josien R, and Choi Y. 2002. A Novel Member of the Leukocyte Receptor Complex Regulates Osteoclast Differentiation. J Exp.Med. 195: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, Pignolo RJ, Koczon-Jaremko B, Lorenzo J, and Choi Y. 2006. v-ATPase V(0) subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat.Med. . [DOI] [PubMed] [Google Scholar]
- 32.Kukita T, Wada N, Kukita A, Kakimoto T, Sandra F, Toh K, Nagata K, Iijima T, Horiuchi M, Matsusaki H, Hieshima K, Yoshie O, and Nomiyama H. 2004. RANKL-induced DC-STAMP is essential for osteoclastogenesis. J.Exp.Med. 200: 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M, Birnbaum MJ, MacKay CA, Mason-Savas A, Thompson B, and Odgren PR. 2008. Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation. J Cell Physiol 215: 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tudpor K, van der Eerden BC, Jongwattanapisan P, Roelofs JJ, van Leeuwen JP, Bindels RJ, and Hoenderop JG. 2015. Thrombin receptor deficiency leads to a high bone mass phenotype by decreasing the RANKL/OPG ratio. Bone 72: 14–22. [DOI] [PubMed] [Google Scholar]
- 35.Lee SK, Kalinowski J, Jastrzebski S, and Lorenzo JA. 2002. 1,25 (OH)(2) Vitamin D(3)-Stimulated Osteoclast Formation in Spleen-Osteoblast Cocultures Is Mediated in Part by Enhanced IL-1alpha and Receptor Activator of NF-kappaB Ligand Production in Osteoblasts. J Immunol. 169: 2374–2380. [DOI] [PubMed] [Google Scholar]
- 36.Kodama H, Nose M, Niida S, and Yamasaki A. 1991. Essential role of macrophage colony-stimulating factor in the osteoclast differentiation supported by stromal cells. J.Exp.Med. 173: 1291–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun W, Zhang H, Wang H, Chiu YG, Wang M, Ritchlin CT, Kiernan A, Boyce BF, and Xing L. 2017. Targeting Notch-Activated M1 Macrophages Attenuates Joint Tissue Damage in a Mouse Model of Inflammatory Arthritis. J Bone Miner Res 32: 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukushima H, Shimizu K, Watahiki A, Hoshikawa S, Kosho T, Oba D, Sakano S, Arakaki M, Yamada A, Nagashima K, Okabe K, Fukumoto S, Jimi E, Bigas A, Nakayama KI, Nakayama K, Aoki Y, Wei W, and Inuzuka H. 2017. NOTCH2 Hajdu-Cheney Mutations Escape SCF(FBW7)-Dependent Proteolysis to Promote Osteoporosis. Mol Cell 68: 645–658.e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, and Taniguchi T. 2002. Induction and Activation of the Transcription Factor NFATc1 (NFAT2) Integrate RANKL Signaling in Terminal Differentiation of Osteoclasts. Dev.Cell 3: 889–901. [DOI] [PubMed] [Google Scholar]
- 40.Feng X, and Teitelbaum SL. 2013. Osteoclasts: New Insights. Bone research 1: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner AG, Tjahyono F, Chiu WS, Skinner J, Sawyer R, Moore AJ, Morris HA, Findlay DM, Zajac JD, and Davey RA. 2010. The role of the calcitonin receptor in protecting against induced hypercalcemia is mediated via its actions in osteoclasts to inhibit bone resorption. Bone 17: 17. [DOI] [PubMed] [Google Scholar]
- 42.Lee SE, Chung WJ, Kwak HB, Chung CH, Kwack K, Lee ZH, and Kim HH. 2001. Tumor necrosis factor-alpha supports the survival of osteoclasts through the activation of Akt and ERK. J Biol.Chem [DOI] [PubMed] [Google Scholar]
- 43.Yoshida K, Akita N, Okamoto T, Asanuma K, Uchida A, Sudo A, Shimaoka M, Suzuki K, and Hayashi T. 2018. Activated protein C suppresses osteoclast differentiation via endothelial protein C receptor, protease-activated receptor-1, sphingosine 1-phosphate receptor, and apolipoprotein E receptor 2. Thrombosis research 163: 30–40. [DOI] [PubMed] [Google Scholar]
- 44.Abraham LA, and MacKie EJ. 1999. Modulation of osteoblast-like cell behavior by activation of protease-activated receptor-1. J Bone Miner Res 14: 1320–1329. [DOI] [PubMed] [Google Scholar]
- 45.Song SJ, Pagel CN, Pike RN, and Mackie EJ. 2005. Studies on the receptors mediating responses of osteoblasts to thrombin. The international journal of biochemistry & cell biology 37: 206–213. [DOI] [PubMed] [Google Scholar]
- 46.Pagel CN, Song SJ, Loh LH, Tudor EM, Murray-Rust TA, Pike RN, and Mackie EJ. 2009. Thrombin-stimulated growth factor and cytokine expression in osteoblasts is mediated by protease-activated receptor-1 and prostanoids. Bone 44: 813–821. [DOI] [PubMed] [Google Scholar]
- 47.Sato N, Ichikawa J, Wako M, Ohba T, Saito M, Sato H, Koyama K, Hagino T, Schoenecker JG, Ando T, and Haro H. 2016. Thrombin induced by the extrinsic pathway and PAR-1 regulated inflammation at the site of fracture repair. Bone 83: 23–34. [DOI] [PubMed] [Google Scholar]
- 48.Song SJ, Pagel CN, Campbell TM, Pike RN, and Mackie EJ. 2005. The role of protease-activated receptor-1 in bone healing. The American journal of pathology 166: 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson DM, Maraskovsky E, Billingsley WL, Dougall WC, Tometsko ME, Roux ER, Teepe MC, DuBose RF, Cosman D, and Galibert L. 1997. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390: 175–179. [DOI] [PubMed] [Google Scholar]
- 50.Aronovich A, Nur Y, Shezen E, Rosen C, Zlotnikov Klionsky Y, Milman I, Yarimi L, Hagin D, Rechavi G, Martinowitz U, Nagasawa T, Frenette PS, Tchorsh-Yutsis D, and Reisner Y. 2013. A novel role for factor VIII and thrombin/PAR1 in regulating hematopoiesis and its interplay with the bone structure. Blood 122: 2562–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khoufache K, Berri F, Nacken W, Vogel AB, Delenne M, Camerer E, Coughlin SR, Carmeliet P, Lina B, Rimmelzwaan GF, Planz O, Ludwig S, and Riteau B. 2013. PAR1 contributes to influenza A virus pathogenicity in mice. J Clin Invest 123: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanno Y, Ishisaki A, Kawashita E, Kuretake H, Ikeda K, and Matsuo O. 2016. uPA Attenuated LPS-induced Inflammatory Osteoclastogenesis through the Plasmin/PAR-1/Ca(2+)/CaMKK/AMPK Axis. International journal of biological sciences 12: 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang T, Lee MH, Choi E, Pardo-Villamizar CA, Lee SB, Yang IH, Calabresi PA, and Nath A. 2012. Granzyme B-induced neurotoxicity is mediated via activation of PAR-1 receptor and Kv1.3 channel. PLoS One 7: e43950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duarte M, Kolev V, Kacer D, Mouta-Bellum C, Soldi R, Graziani I, Kirov A, Friesel R, Liaw L, Small D, Verdi J, Maciag T, and Prudovsky I. 2008. Novel cross-talk between three cardiovascular regulators: thrombin cleavage fragment of Jagged1 induces fibroblast growth factor 1 expression and release. Molecular biology of the cell 19: 4863–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zanotti S, and Canalis E. 2016. Notch Signaling and the Skeleton. Endocrine reviews 37: 223–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashley JW, Ahn J, and Hankenson KD. 2015. Notch signaling promotes osteoclast maturation and resorptive activity. J Cell Biochem 116: 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, and Teitelbaum SL. 2008. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem 283: 6509–6518. [DOI] [PubMed] [Google Scholar]
- 58.Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, Bigas A, Jimi E, and Okabe K. 2008. The association of Notch2 and NF-{kappa}B accelerates RANKL-induced osteoclastogenesis. Mol.Cell Biol. 28: 6402–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao B, Grimes SN, Li S, Hu X, and Ivashkiv LB. 2012. TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med 209: 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.