Infectious bacteria are developing and spreading resistance to conventional treatments at a rapid pace. To provide novel potent antimicrobials, we must develop new bioprospecting strategies. Here, we combined in silico and phenotypic approaches to explore the bioactive potential of the marine bacterial genus Pseudoalteromonas. We found that pigmented strains in particular represent an untapped resource of secondary metabolites and that they also harbor an elaborate chitinolytic machinery. Furthermore, our analysis showed that chitin is likely a preferred substrate for pigmented species, in contrast to nonpigmented species. Potentially, chitin could facilitate the production of new secondary metabolites in pigmented Pseudoalteromonas strains.
KEYWORDS: bioactivity, glycosyl hydrolases, Pseudoalteromonas, chitin
ABSTRACT
Chitin is the most abundant polymer in the marine environment and a nutrient-rich surface for adhering marine bacteria. We have previously shown that chitin can induce the production of antibiotic compounds in Vibrionaceae, suggesting that the discovery of novel bioactive molecules from bacteria can be facilitated by mimicking their natural habitat. The purpose of this study was to determine the glycosyl hydrolase (GH) profiles of strains of the genus Pseudoalteromonas to enable selection of presumed growth substrates and explore possible links to secondary metabolism. Genomic analyses were conducted on 62 pigmented and 95 nonpigmented strains. Analysis of the total GH profiles and multidimensional scaling suggested that the degradation of chitin is a significant trait of pigmented strains, whereas nonpigmented strains seem to be driven toward the degradation of alga-derived carbohydrates. The genomes of all pigmented strains and 40 nonpigmented strains encoded at least one conserved chitin degradation cluster, and chitinolytic activity was phenotypically confirmed. Additionally, the genomes of all pigmented and a few nonpigmented strains encoded chitinases of the rare GH family 19. Pigmented strains devote up to 15% of their genome to secondary metabolism, while for nonpigmented species it was 3% at most. Thus, pigmented Pseudoalteromonas strains have a bioactive potential similar to that of well-known antibiotic producers of the Actinobacteria phylum. Growth on chitin did not measurably enhance the antibacterial activity of the strains; however, we demonstrated a remarkable co-occurrence of chitin degradation and the potential for secondary metabolite production in pigmented Pseudoalteromonas strains. This indicates that chitin and its colonizers of the Pseudoalteromonas genus represent a so far underexplored niche for novel enzymes and bioactive compounds.
IMPORTANCE Infectious bacteria are developing and spreading resistance to conventional treatments at a rapid pace. To provide novel potent antimicrobials, we must develop new bioprospecting strategies. Here, we combined in silico and phenotypic approaches to explore the bioactive potential of the marine bacterial genus Pseudoalteromonas. We found that pigmented strains in particular represent an untapped resource of secondary metabolites and that they also harbor an elaborate chitinolytic machinery. Furthermore, our analysis showed that chitin is likely a preferred substrate for pigmented species, in contrast to nonpigmented species. Potentially, chitin could facilitate the production of new secondary metabolites in pigmented Pseudoalteromonas strains.
INTRODUCTION
The increasing development and spread of antimicrobial resistance are a serious threat to human health, and humanity is in urgent need for novel antibiotics to treat infectious diseases. In particular, soil bacteria have been explored extensively as a source of antibiotics, and bacteria belonging to the phylum Actinobacteria have been a prolific source, providing two-thirds of all known microbial antibiotics (1). Exploring new environments is one strategy for finding novel compounds that are not (yet) affected by resistance. While promising bioactive molecules have been identified from marine organisms, particularly from the family Pseudoalteromonadaceae (2–5), the marine environment still remains an underexploited resource of novel bioactive compounds (6–8).
The exclusively marine genus Pseudoalteromonas constitutes, on average, 2 to 3% of the bacterial abundance in the upper ocean waters (9). Pseudoalteromonas strains are excellent biofilm formers and are often found in association with eukaryotic hosts, such as crustaceans or algae (10). As of 2018, 47 Pseudoalteromonas species had validly published names (11). The genus is phenotypically and phylogenetically divided into two main clusters that are differentiated by the ability to produce pigments and the lack thereof (12). The pigmented species produce an array of bioactive secondary metabolites, including violacein, indolmycin, and pentabromopseudilin, produced by Pseudoalteromonas luteoviolacea; prodigiosin, produced by P. rubra; bromoalterochromides, produced by P. piscicida; and tambjamines, produced by P. tunicata, P. flavipulchra, and P. maricaloris (13–16). In contrast, nonpigmented species have generally been explored as producers of unusual enzymatic activities (10).
On a global marine research expedition, we isolated strains of both pigmented and nonpigmented Pseudoalteromonas based on their antimicrobial activity (17); however, the nonpigmented strains did not retain antimicrobial activity following frozen storage (13). The genomes of four pigmented and three nonpigmented strains were sequenced and mined for biosynthetic gene clusters (BGCs) of secondary metabolites, revealing a large untapped potential in the pigmented strains (18). For most of the BGCs, the associated chemistry has not been elucidated, potentially because the BGCs are not expressed (e.g., are silent or cryptic) or are expressed at low levels under growth conditions hitherto used (13). Mimicking the natural growth substrate to induce bioactivity has been successful in the Vibrionaceae. Providing vibrios with chitin, the most abundant polymer in the marine environment, can enhance their antibacterial activity (19, 20) and can induce expression of their BGCs (21). Some Pseudoalteromonas species can also degrade chitin (18, 22), but little is known about their chitinolytic machinery and a possible influence on secondary metabolism. In the Vibrionaceae, specifically, Vibrio cholerae (23, 24), chitin degradation relies on the secretion of extracellular chitinases. In bacteria, the majority of chitinases belong to glycosyl hydrolase (GH) family 18 (25). Recently, chitinases belonging to GH family 19 have been discovered in a few groups of prokaryotes (26–30), and we have found that the genomes of 10 marine chitinolytic bacteria, including Pseudoalteromonas, all contain at least one GH19 chitinase (31).
The purpose of this study was to determine the chitinolytic abilities of species of the genus Pseudoalteromonas and explore possible links to their potential for secondary metabolite production. We used a genome sequence-guided approach combined with phenotypic assays to assess chitin degradation and antibacterial activity.
RESULTS AND DISCUSSION
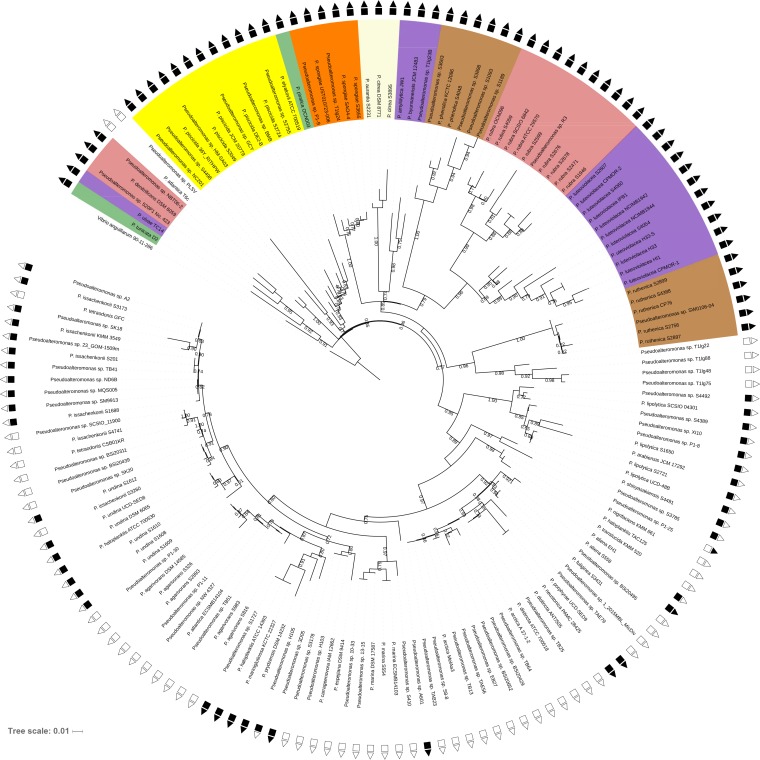
The average nucleotide identity (ANI) of 253 Pseudoalteromonas genomes obtained from isolates collected on a global marine expedition (17) and from the NCBI database was determined. The data set had a strong bias toward certain taxonomic subgroups being overrepresented among the total number of genomes. Therefore, in the case of an ANI of more than 98.3% (see Fig. S1 and S2 in the supplemental material), a representative for this phylogenetic subgroup was selected for further genomic analyses on the basis of sequence quality (Table S1). The resulting data set consisted of 157 strains covering 37 of the 47 species with standing in the prokaryote nomenclature (List of Prokaryotic Names with Standing in Nomenclature [LPSN] [11]). Single nucleotide polymorphism (SNP)-based phylogenetic analysis (Fig. 1) confirmed the previous clustering based on 16S rRNA gene analysis and separated the Pseudoalteromonas strains into two phylogenetic groups that correlated with the pigmented and nonpigmented phenotypes (13), with the exception of two nonpigmented strains (P. atlantica T6c and Pseudoalteromonas sp. strain PLSV), which clustered in the pigmented group. Sixty-two strains belonged to pigmented species, and 95 strains belonged to nonpigmented species. Pigmented species had a genome size of 5.2 ± 0.6 Mb, while nonpigmented species had smaller genomes of 4.4 ± 0.4 Mb (Table S1). In general, the phylogenetic diversity was higher in the pigmented species, whereas nonpigmented species were phylogenetically more similar (Fig. 1; Fig. S2). This could indicate a higher selection pressure for pigmented strains or a more recent evolution of nonpigmented strains. Based on the ANI analysis (Fig. S2), several of the strains, particularly those within the pigmented group, could belong to novel species. This is in agreement with the findings of a recent study by Busch et al. (5), in which the authors noted that considerable species-level diversity has yet to be described in the Pseudoalteromonas genus. Furthermore, the ANI analysis also enabled some unclassified strains to become assigned to species. The whole genomes of 10 taxonomically classified species have not yet been sequenced, and therefore, these species were not included in the analysis and some unassigned strains could belong to these species. The ANI analysis also demonstrated that the ANI of some strains were <95% and, thus, that these strains should be reclassified as other species (32).
FIG 1.
SNP-based phylogeny of 157 Pseudoalteromonas strains consisting of 50 of our isolates and 107 genomes downloaded from the NCBI database. Symbols are color coded, where black symbols represent that the feature is present in the genome and white symbols represent that the feature is absent (squares, chitinolytic genotype; triangles, GH19 chitinases are present in the genome). The pigmented strains are colored according to their pigmentation. The phylogenetic tree was constructed using the CSI Phylogeny web server, and V. anguillarum 90-11-286 (GenBank accession no. CP011460 and CP011461) was used as a root. Bootstrap values of >0.5 are included.
Correlation of percent ANI to the percentage of isolates used for inferring the cutoff percent ANI value of identical isolates. Download FIG S1, PDF file, 0.1 MB (138.6KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat map of average nucleotide identity (ANI) analysis of 157 Pseudoalteromonas strains. The color-coded bar on the left indicates the pigmentation status. Download FIG S2, PDF file, 0.4 MB (375KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of 50 Pseudoalteromonas strains isolated during the Galathea 3 expedition and 107 Pseudoalteromonas genomes obtained from NCBI, the species, their pigmentation, the genome size (in megabases), the percentage of the genome allocated to BGCs, and the NCBI accession number. Download Table S1, PDF file, 0.4 MB (447.4KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The high level of dissimilarity between some of the species could even suggest that the Pseudoalteromonas genus potentially represents multiple genera, as was the case for the genus Algicola, consisting of the two species Algicola bacteriolytica and A. sagamiensis, which belonged to the Pseudoalteromonas genus until 2007 (33). Since ANI serves as a genomic measurement only for species delineation and not for genus delineation, this is only speculative. Nonetheless, the ANI analysis clearly visualized a problem with species naming within the Pseudoalteromonas genus, as some strains were not correctly assigned to a species. For example, P. undina DSM 6065 and P. haloplanktis ATCC 700530 share 98% ANI similarity, thus exceeding the species cutoff for ANI of ≥ 95% (32). Also, P. mariniglutinosa KCTC 22327 and P. haloplanktis ATCC 700530 have 99% ANI similarity and P. neustonica PAMC 28425 and P. porphyrae UCD-SED9 share 99% ANI similarity, indicating that these are the same species. Similar issues with species delineation have been reported in the Pseudomonas genus (34), as well as on a much larger scale for prokaryotes (35).
Genes encoding chitin degradation machinery are common in pigmented Pseudoalteromonas strains.
Of the 157 strains, 102 strains had the genomic capacity to degrade chitin (Fig. 1). The chitin-degrading genotype matched the chitin-degrading phenotype (Table S2). All 62 genomes of pigmented strains encoded chitinase genes from both the GH18 and GH19 families. Forty-five percent (40 of 95) of the genomes of the nonpigmented strains encoded chitinase genes of the GH18 family. Of those, only 10 genomes also encoded chitinases from the GH19 family.
Chitin degradation on agar plates containing colloidal chitin and inhibition of Vibrio anguillarum by Pseudoalteromonas strains on media supplemented with four different carbon sources: glucose, N-acetylglucosamine, colloidal chitin, or crystalline chitin. +, inhibition; −, no inhibition. Download Table S2, PDF file, 0.3 MB (350.1KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The genomes of pigmented strains encoded, on average, 5.2 GH18 chitinases, while the nonpigmented chitinolytic strains encoded, on average, 2.1 GH18 chitinases. Hence, the pigmented strains encoded significantly more GH18 chitinases than nonpigmented chitinolytic strains, with an average of 2.4 times more GH18 chitinases (P < 0.0001) being found in pigmented strains. A three-gene chitin degradation cluster (CDC) (Fig. 2), encoding a GH18 chitinase of the ChiC type, a lytic polysaccharide monooxygenase (LPMO), and a GH18 chitinase of the ChiA type, was found in all chitinolytic strains and is likely a conserved feature in chitin-degrading Pseudoalteromonas strains independent of pigmentation. The CDC was initially described in 2002 by Tsujibo et al. at a time when LPMOs had not been described (36). Today, it is known that chitin-associated LPMOs are abundant in prokaryotes (37) and facilitate the breakdown of chitin by catalysis of the oxidative cleavage of glycosidic bonds (38–40). The genomes of the majority of pigmented strains belonging to the species P. luteoviolacea, P. rubra, P. phenolica, and P. citrea encoded two CDCs.
FIG 2.

The chitin degradation cluster (CDC) present in all chitinolytic Pseudoalteromonas strains. The CDC consists of a GH18 chitinase of the chiC type, a lytic polysaccharide monooxygenase (lpmo), and a GH18 chitinase of the chiA type.
The conservation of GH19 chitinases in all pigmented strains indicates an important role of this enzyme in this group. GH19 chitinases are mostly known from plants and are rare and thus understudied in bacteria and fungi (37, 41–43). In comparison, a recent analysis of 40 Micromonospora strains whose whole genomes have been sequenced found that all genomes carried the chiC gene, encoding the GH18 chitinase, but none contained the GH19-encoding genes (44). In plants, chitinases mainly serve as antifungal agents, and some studies have suggested that this could be one of the functions of bacterial GH19 chitinases (27, 30, 41, 45). Antifungal activity is, however, not limited to GH19 chitinases, as some GH18 chitinases also display antifungal activities (46, 47). The first GH19 chitinase described in Pseudoalteromonas was antifungal, but it also hydrolyzed colloidal and crystalline chitin (26). Thus, the functional differences between the GH18 and GH19 chitinases remain unresolved.
Pigmented and nonpigmented Pseudoalteromonas strains are distinct in their GH profiles.
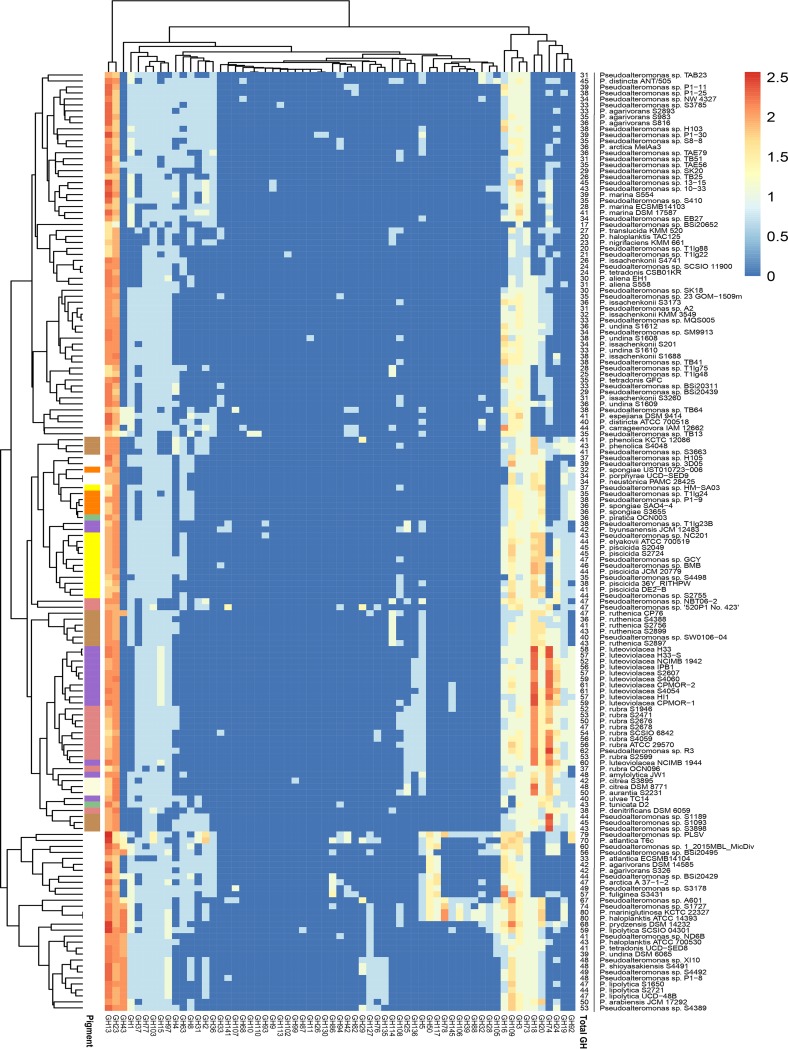
A clustered heat map was generated based on the predicted GH profiles of the strains (Fig. 3). Pigmented and nonpigmented strains contained, on average, 47 ± 8 and 40 ± 13 GHs, respectively. The functional clustering divided the strains into two groups, with one consisting solely of nonpigmented strains and the other consisting of both pigmented and nonpigmented strains, and subclustering correlated with the phylogeny and pigmentation. In total, 63 out of 156 entries of GHs described in the CAZy database by the start of 2019 (http://www.cazy.org/) were represented in the strains. Of the 63 different GH families identified, 60 GH families were represented in nonpigmented strains, whereas only 38 GH families were found in pigmented strains. The Shannon index was higher in the pigmented strains (P < 0.001), but the variation was 8-fold larger in the nonpigmented strains. This suggests that even though the average pigmented strain genome based on gene counts contains more GHs than the average nonpigmented strain genome, the GH profiles of the nonpigmented strains are more heterogeneous than those of the pigmented strains.
FIG 3.
Functionally clustered heat map based on the predicted glycosyl hydrolases (GH) of the 157 Pseudoalteromonas strains. The key displays the log-transformed gene counts of GHs.
Interestingly, the functional clustering detected a pattern of co-occurrence between the GH19 chitinase and the GH92 mannosidase, meaning that of the 71 genomes containing at least one GH19 gene, 71.8% also contained a GH92 gene (P < 0.00001). In the mycoparasitic fungus Trichoderma harzianum, chitinases and a GH92 mannosidase were significantly upregulated in response to pathogenic fungi (48). Since chitin and mannose are components of the fungal cell wall, this could indicate that the natural role of bacterial GH19s is indeed antifungal, possibly in combination with GH92.
To evaluate if the GH profiles differed on the basis of pigmentation status, multidimensional scaling (MDS) was conducted. MDS clearly separated pigmented and nonpigmented strains into two groups on the basis of their GH profiles (Fig. 4). This is in accordance with analysis of similarities (ANOSIM) showing a significant effect of pigmentation in multivariate space (P < 0.001) and suggesting that pigmentation status explains ∼50% of the GH profile (r2 = 0.50). Listed in order of the most significant contribution, GH18, GH74, GH20, GH24, GH19, GH92, and GH23 had the major impact of this divergence for pigmented strains (loadings < 0.5). For nonpigmented strains, GH13, GH43, GH50, GH16, GH78, and GH109 had a major divergent impact on the GH profile (loadings > −0.5) (Table S3). Thus, the GH18 chitinase was the strongest contributor to the GH profiles of pigmented strains, as it was present in all genomes and was present at a higher abundance than it was in nonpigmented strains. Also, the chitinolytic GH19s and GH20s contributed prominently to the separation of the GH profiles of pigmented strains, proposing that chitin degradation is a key physiological trait in pigmented Pseudoalteromonas strains. The loading coordinates of GH19 and GH92 were very similar, confirming their co-occurrence, as discussed above. For nonpigmented strains, GH13 was the strongest contributor to the diversification from pigmented species. The contribution of GH13 in nonpigmented strains is reflected by a higher gene count compared to that in pigmented species. GH43, GH50, GH16, GH78, and GH109 also contributed to the intergroup diversity of the nonpigmented GH profiles, and all are enzymes that are active on marine algal polysaccharides, such as agar, porphyrin, carrageenan, starch, and glucan. The building blocks of algal polysaccharides are relatively few, but they have a high structural complexity, as they can be linked in almost infinite ways and are also often subject to acetylation, methylation, or sulfation (49). The high diversity of GH families in nonpigmented species could indicate that this group favors the utilization of algal polysaccharides. Pigmented strains also have the genetic capacity to utilize algal components, and many nonpigmented strains can degrade chitin; however, the conserved chitinolytic machinery in pigmented strains indicates a specialized adaptation to chitin utilization.
FIG 4.
Multidimensional scaling (MDS) plot of pigmented (red dots) and nonpigmented (gray dots) Pseudoalteromonas strains based on their glycosyl hydrolase (GH) profile. GHs with loadings of more than ±0.5 are shown in the graph.
Loadings associated with multidimensional scaling plot in Fig. 4. Loadings plotted in Fig. 4 are represented in bold. Download Table S3, PDF file, 0.2 MB (232.7KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The general perception of the genus Pseudoalteromonas is that pigmented species produce many bioactive secondary metabolites, whereas nonpigmented species produce a larger amount of hydrolytic enzymes (10). The results presented herein partially support this assumption, as the diversity of GHs was higher in nonpigmented strains, but not on a gene count basis. While having less diverse GH profiles, the genomes of pigmented strains contained, on average, at least the same number of GHs as nonpigmented strains or higher number of GHs than nonpigmented strains.
Pseudoalteromonas strains devote up to 15% of their genome to the biosynthesis of secondary metabolites.
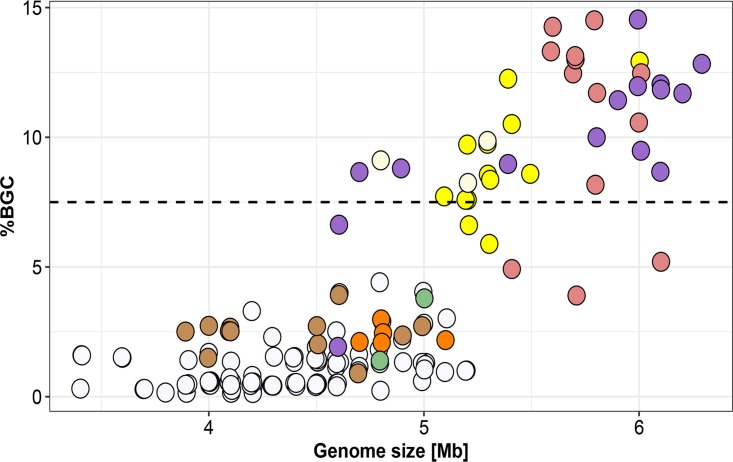
The genomic bioactive potential of all Pseudoalteromonas strains was compared to that of well-known secondary metabolite producers (50) by calculating the proportion of the genome dedicated to the biosynthesis of secondary metabolites (percent BGC) (Fig. 5; Table S1). Mining the genomes using the antiSMASH tool, we found that the genomes of pigmented strains dedicated, on average, 7.6% ± 4.2% to BGCs and that those of nonpigmented strains dedicated, on average, 1.1% ± 0.9%. In comparison, the average prokaryote devotes 3.7% ± 3.1% of its genome to BGCs (50). There was a strong linear correlation between genome size and percent BGC, which was especially pronounced in the pigmented strains (r2 = 0.60, P < 0.00001, slope = 5.18) compared to the nonpigmented strains (r2 = 0.13, P < 0.001, slope = 0.81), meaning that for every gigabase increase in genome size, a further 5.2% of the genome is dedicated to BGCs for pigmented species. The genomes of pigmented species encoded, on average, 6.8 times more BGCs (P < 0.00001) than those of nonpigmented species did. Thirty-six strains devoted more than 7.5% (50) of their genomes to BGCs, with the highest percentage of BGCs being found in P. luteoviolacea and P. rubra strains (12% to 15%). Thus, their genetic potential for the production of secondary metabolites is equal to that of species generally well-known for the production of secondary metabolites, including those in the genera Streptomyces, Myxococcus, Sorangium, and Burkholderia (50). It is also comparable to that in species from the marine actinomycete genus Salinispora, devoting approximately 10% of the investigated genomes to secondary metabolism (51, 52).
FIG 5.
Proportion of biosynthetic gene clusters (BGCs) in the Pseudoalteromonas genomes according to genome size. Each circle represents one genome and is color coded based on pigmentation. The data have been jittered to account for overlying points.
Genomic analysis of representatives of each pigmented species and three unclassified pigmented strains using antiSMASH found that most of the predicted BGCs showed little to no homology to known and characterized bioactive compounds (Table 1). Many of these BGCs may represent a novel chemistry or may encode novel homologs of the compound classes (13). Bacteria from the phylum Actinobacteria have been the cornerstone of the antibiotic era, as two-thirds of all known microbial antibiotics originate from these Gram-positive bacteria (1, 53). They remain a focal point of antibiotic research, since genome mining has revealed the presence of many potentially silent or cryptic BGCs in their genomes (54). Likewise, our study demonstrates that pigmented Pseudoaltermonas species could be a potent and promising source of new antimicrobials.
TABLE 1.
Total number of predicted BGCs in a representative selection of pigmented species, including the most similar known BGCs according to percent gene similaritya
| Species and strain | Total no. of predicted BGCs |
Most similar known BGC (% gene similarity) |
|---|---|---|
| P. luteoviolacea S2607 | 23 | Violacein (80%), kalimantacin (10%), vibriobactin (18%), griseobactin (17%), cystobactamide (8%) |
| P. spongiae S3655 | 5 | Arylpolyene (35%), flexirubin (5%), desferrioxamine B (60%) |
| P. phenolica S1189 | 6 | Xenocyloins (25%), vulnibactin (25%) |
| P. rubra S4059 | 25 | Indigoidine (40%), turnerbactin (15%), kalimantacin/batumin (10%) |
|
Pseudoalteromonas sp. strain S4498 |
9 | Serobactins (15%) |
| P. piscicida S2049 | 18 | Griseobactin (23%), feglymycin (10%), bromoalterochromides (14%), alterochromides (90%) |
| P. ruthenica S2897 | 4 | Arylpolyene (40%), desferrioxamine B (60%) |
| P. aurantia S2231 | 17 | Kalimantacin/batumin (10%), pyoverdine (1%) |
| P. citrea S3895 | 17 | Pyoverdine (1%), turnerbactin (15%), pyoverdine (1%), zeamine (17%) |
| P. ulvae TC14 | 4 | APE_Vf (40%), violacein (80%) |
| P. piratica OCN0013 | 3 | APE_Vf (40%), desferrioxamine B (60%) |
|
Pseudoalteromonas sp. strain 520P1 |
7 | Violacein (80%), pyoverdine (2%), taxlllaid (6%), eicosapentaenoic acid-like compound (18%) |
|
Pseudoalteromonas sp. strain HM SA03 |
11 | Bromoalterochromides (14%), hectochlorin (37%), alterochromides (100%), turnerbactin (15%) |
| P. amylolytica JW1 | 16 | Violacein (80%), staphylobactin (12%) |
| P. denitrificans DSM 6059 | 10 | APE_Vf (45%), eicosapentaenoic acid-like (18%), bacillibactin (23%) |
| P. tunicata D2 | 5 | Violacein (80%), prodigiosin (12%), desferrioxamine B (50%) |
| P. byunsanensis JCM 12483 | 14 | Taxlllaid (4%), staphylobactin (12%), bromoalterochromides (14%), violacein (80%) |
| P. elyakovii ATCC 700519 | 18 | Bacillibactin (60%), alterochromides (95%) |
BGCs were predicted and percent gene similarity was defined by use of the antiSMASH tool.
Enhanced antibacterial activity could not be detected on chitin.
In some Vibrionaceae, BGCs are upregulated when the bacteria are grown on chitin, and the finding of an elaborate chitinolytic machinery in Pseudoalteromonas led us to determine the antibacterial activity of the strains when grown on four different carbon sources (glucose, N-acetylglucosamine [NAG], colloidal chitin, and crystalline chitin) mimicking a natural chitinous habitat (55). Of the 50 strains, all 24 pigmented strains and one nonpigmented strain (strain S558) were inhibitory toward the target pathogen, in agreement with previous results (13); however, inhibition was independent of the carbon source (Table S2). Thus, the overall antibacterial activity was not enhanced on chitinous media compared to glucose media, as has been seen for strains of the Vibrionaceae family (20). The assay was not sensitive enough to quantify the antibacterial activity, and chitin could potentially still enhance the response of a metabolite cluster that is also expressed without addition of chitin. This was seen for the antibiotic andrimid in Vibrio coralliilyticus S2052 when grown on chitin compared to glucose (19). Also, growth on chitin may alter the expression of BGCs encoding bioactivities other than antibacterial activity.
Conclusion.
Here, we demonstrate that chitin is likely an important carbon source for pigmented Pseudoalteromonas species, as chitin hydrolysis is a key trait in this group of bacteria. Pigmented Pseudoalteromonas strains and a few nonpigmented strains contain GH19 chitinases, which are not common in bacteria. In plants, the main role of GH19 chitinases is antifungal, and the finding of a co-occurring mannosidase of the GH92 family suggests that the fungal cell wall could be the target of GH19 chitinases in bacteria as well. Nonpigmented Pseudoalteromonas strains have an extended algal polysaccharide-degrading GH profile compared to pigmented strains; in contrast, their genomic potential for producing bioactive compounds was much lower. The fact that pigmented species devote up to 15% of their genome to BGCs qualifies this genus to be in the same antibiotic-producing league as the well-known Actinobacteria, with the benefit of being less explored. We hypothesized that growth on chitin would enhance the bioactivity of the strains, but, when measured as antibiotic activity, this was not confirmed. However, this study serves as an extensive basis for ecology-based bioprospecting of secondary metabolites and hydrolytic enzymes and has uncovered a promising genomic potential for producing novel antimicrobial compounds in pigmented Pseudoalteromonas.
MATERIALS AND METHODS
Pseudoalteromonas genomes.
One hundred sixty-five Pseudoalteromonas genomes available in the NCBI database in May 2018 and 88 Pseudoalteromonas genomes (see below) from our global Galathea collection (17) were included. To avoid bias in the data set due to an uneven distribution of the number of strains per species and to exclude very similar isolates, an average nucleotide identity (ANI) analysis using the Python module pyani (56) was conducted using all 253 genomes to eliminate those sharing an ANI of greater than 98.3%. This cutoff was inferred by visually inspecting a plot of the number of isolates versus the ANI value, in which there was a notable elbow at 98.3% identity (see Fig. S1 in the supplemental material). Subsequently, 107 genomes from NCBI and 50 genomes sequenced in the present study were included in the bioinformatic analyses (Table S1). The data set included 15 pigmented species (62 strains) and 23 nonpigmented species (95 strains). These numbers are approximate, as some strains were not assigned to any species but clustered within either the pigmented or the nonpigmented group.
Isolation of genomic DNA and genome sequencing.
Ninety-eight Pseudoalteromonas strains were isolated during the Galathea 3 global marine research expedition (17). They were isolated from seawater or swab samples from biotic or abiotic surfaces and selected on the basis of their antibacterial activity against the fish pathogen Vibrio anguillarum. The origins and coordinates of isolation can be found in the work of Gram et al. (2010) (17). The genomes of 12 strains were sequenced as part of a previous study (18, 57), and we were unable to purify DNA from 10 strains. Thus, 76 new genomes were sequenced as part of the present study. High-purity genomic DNA was extracted using a NucleoSpin tissue kit (catalog number 740952; Macherey-Nagel) including an RNase treatment step. For half of the strains, we had difficulties extracting high-purity DNA; therefore, a Qiagen Genomic-tip 20/G kit (catalog number 10223; Qiagen) and a genomic DNA buffer set (catalog number 19060; Qiagen) were used for extraction instead. Quantification was done using a DeNovix DS-11+ spectrometer (DeNovix, USA) and Qubit (v2.0) analyzer (Invitrogen, United Kingdom). Genomes were sequenced at the Novo Nordisk Center for Biosustainability (Technical University of Denmark, Lyngby, Denmark) using 150-bp paired-end sequencing on an Illumina NextSeq platform. Genomes were assembled using CLC Genomics Workbench software (v8; CLC bio, Aarhus, Denmark), and contig-based draft genomes were obtained; all had genome coverage of 105-fold or higher.
Phylogenetic analysis.
A phylogenetic tree of the 157 genomes was generated with the SNP-based procedure CSI Phylogeny (http://cge.cbs.dtu.dk/services/CSIPhylogeny/) (58). Pseudoalteromonas luteoviolacea DSM 6061T (WGS accession no. GCF_001625655.1) was used as a reference genome, and Vibrio anguillarum 90-11-286 (GenBank accession no. CP011460 and CP011461) was used as the outgroup. The maximum likelihood phylogenetic tree was visualized using iTOL (https://itol.embl.de/) (59).
Bioinformatic analyses.
The genomes sequenced in the present study were annotated using the Rapid Annotation using Subsystem Technology (RAST) (60). The presence of a conserved chitin degradation cluster (CDC) was investigated using MultiGeneBlast (61) with the CDC query from (36). The lytic polysaccharide monooxygenase (LPMO) of the CDC was identified using Pfam (https://pfam.xfam.org/).
The total numbers of GHs encoded in each genome was predicted using Hidden Markov Model (HMM) searches against local versions of the dbCAN (62). A heat map of the GH profiles was constructed in R (v3.4.2) using the package pheatmap (63), in which the profiles were clustered by complete linkage on the log-transformed values for ease of visualization. The average GH18 gene content in pigmented versus nonpigmented chitinolytic strains was tested with a t test. A co-occurrence pattern between GH19 and GH92 was tested with a Fisher exact test. To visualize the differences in GH profiles based on pigmentation, a metric multidimensional scaling (MDS) analysis on a count matrix with the total number of predicted protein sequences per GH family was conducted in R using Bray-Curtis distances. Loadings with values of more than ±0.5 were plotted in the MDS plot. Analysis of similarities (ANOSIM) was used to test whether the pigmentation status was predictive of the GH gene profile in a multivariate context. To test if pigmentation status was associated with GH profile diversity, the Shannon index was calculated and tested with a t test.
All 157 genomes were analyzed for secondary metabolite gene clusters using the antiSMASH (v4.0) tool (64). The sizes of the predicted BGCs relative to the genome size were calculated for each strain (in nucleobases) and compared to the total size of the genome using linear regression. The percentage of the genome dedicated to BGCs in pigmented versus nonpigmented strains was tested with a t test.
Chitin degradation.
To determine if the chitin-degrading genotype resulted in a phenotype, the 50 strains from the Galathea collection were tested for chitin degradation. Strains were grown with aeration at 25°C and 200 rpm overnight in marine broth (MB 2216; Difco BD). Two microliters of each culture was inoculated in duplicate on chitin agar, consisting of 1.5% agar, 2% sea salt (catalog number S9883; Sigma), 0.3% Casamino Acids, and 0.2% colloidal chitin (catalog number C7170; Sigma). Colloidal chitin was prepared as described previously (20). Plates were inspected daily for 10 days, and chitin degradation was determined by the appearance of a clearing zone around the colonies.
Screening for antibacterial activity.
The antibacterial activity of the 50 Galathea strains against the fish pathogen V. anguillarum 90-11-286 (65) was tested using the agar-based assay described previously (20) with either 0.2% glucose, 0.2% N-acetylglucosamine (NAG), 0.2% colloidal chitin, or 0.2% crystalline chitin as the carbon source, in addition to 0.3% Casamino Acids. Bacterial strains were grown at 25°C and 200 rpm overnight in MB. Two microliters of each culture was inoculated in duplicate on the four media. On each plate, bacteria were inoculated in rows of four strains with a horizontal and a vertical distance of 25 mm from each other. Strains were allowed 2 days of growth before 2 μl of an overnight culture of the target strain, V. anguillarum 90-11-286, grown in MB was inoculated at a distance of 5 mm from the Pseudoalteromonas strains. The plates were incubated at 25°C, and the colony growth of the target strain was assessed 48 h after the target strain had been inoculated.
Data availability.
The 76 new draft genomes are available at the National Center for Biotechnology Information (NCBI) database under accession numbers PNBS01 to PNEL01. All accession numbers are listed in Table S1. The strains are available upon request.
ACKNOWLEDGMENTS
The study was funded by the Villum Kann Rasmussen Annual Award 2016 to Lone Gram.
We thank Kai Blin, Christopher Workman, and Magnus Hallas-Møller for many helpful comments on our analyses. We thank Jette Melchiorsen and Henrique Machado for assistance on DNA extractions and genome assembly.
The present work was carried out as a part of the Galathea 3 expedition under the auspices of the Danish Expedition Foundation. This is Galathea 3 contribution no. P129.
The study was designed by S.S.P., L.G., and E.C.S. S.S.P. conducted the experiments and analyzed the data with the help of L.G. and E.C.S. P.K.B. and M.L.S. conducted some of the genomic analyses and analyzed the data with the help of S.S.P. S.S.P., L.G., and E.C.S. wrote the manuscript. We all reviewed and approved the final version of the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.Van Der Meij A, Worsley SF, Hutchings MI, Van Wezel GP. 2017. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol Rev 41:392–416. doi: 10.1093/femsre/fux005. [DOI] [PubMed] [Google Scholar]
- 2.Kozuma S, Hirota-Takahata Y, Fukuda D, Kuraya N, Nakajima M, Ando O. 2017. Identification and biological activity of ogipeptins, novel LPS inhibitors produced by marine bacterium. J Antibiot (Tokyo) 70:79–83. doi: 10.1038/ja.2016.81. [DOI] [PubMed] [Google Scholar]
- 3.Sannino F, Parrilli E, Apuzzo GA, de Pascale D, Tedesco P, Maida I, Perrin E, Fondi M, Fani R, Marino G, Tutino ML. 2017. Pseudoalteromonas haloplanktis produces methylamine, a volatile compound active against Burkholderia cepacia complex strains. N Biotechnol 35:13–18. doi: 10.1016/j.nbt.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Whalen KE, Poulson-Ellestad KL, Deering RW, Rowley DC, Mincer TJ. 2015. Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3,4-dibromopyrrole-2,5-dione isolated from a Pseudoalteromonas sp. J Nat Prod 78:402–412. doi: 10.1021/np500775e. [DOI] [PubMed] [Google Scholar]
- 5.Busch J, Agarwal V, Schorn M, Machado H, Moore BS, Rouse GW, Gram L, Jensen PR. 2019. Diversity and distribution of the bmp gene cluster and its products in the genus Pseudoalteromonas. Environ Microbiol 21:1575–1585. doi: 10.1111/1462-2920.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong Z-Q, Wang J-F, Hao Y-Y, Wang Y. 2013. Recent advances in the discovery and development of marine microbial natural products. Mar Drugs 11:700–717. doi: 10.3390/md11030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naughton LM, Romano S, Gara FO, Dobson ADW. 2017. Identification of secondary metabolite gene clusters in the Pseudovibrio genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front Microbiol 8:1494. doi: 10.3389/fmicb.2017.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giordano D, Coppola D, Russo R, Denaro R, Giuliano L, Lauro FM, di Prisco G, Verde C. 2015. Marine microbial secondary metabolites: pathways, evolution and physiological roles. Adv Microb Physiol 66:357–428. doi: 10.1016/bs.ampbs.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Wietz M, Gram L, Jørgensen B, Schramm A. 2010. Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat Microb Ecol 61:179–189. doi: 10.3354/ame01443. [DOI] [Google Scholar]
- 10.Bowman JP. 2007. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar Drugs 5:220–241. doi: 10.3390/md504220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parte AC. 2014. LPSN—List of Prokaryotic Names with Standing in Nomenclature. Nucleic Acids Res 42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Offret C, Desriac F, Le Chevalier P, Mounier J, Jégou C. 2016. Spotlight on antimicrobial metabolites from the marine bacteria Pseudoalteromonas: chemodiversity and ecological significance. Mar Drugs 14:E129. doi: 10.3390/md14070129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vynne NG, Månsson M, Nielsen KF, Gram L. 2011. Bioactivity, chemical profiling, and 16S rRNA-based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar Biotechnol (NY) 13:1062–1073. doi: 10.1007/s10126-011-9369-4. [DOI] [PubMed] [Google Scholar]
- 14.Speitling M, Smetanina OF, Kuznetsova TA, Laatsch H. 2007. Bromoalterochromides A and A′, unprecedented chromopeptides from a marine Pseudoalteromonas maricaloris strain KMM 636 T. J Antibiot 60:36–42. doi: 10.1038/ja.2007.5. [DOI] [PubMed] [Google Scholar]
- 15.Pinkerton DM, Banwell MG, Willis AC. 2007. Total syntheses of tambjamines C, E, F, G, H, I and J, BE-18591, and a related alkaloid from the marine bacterium Pseudoalteromonas tunicata. Org Lett 9:5127–5130. doi: 10.1021/ol7024313. [DOI] [PubMed] [Google Scholar]
- 16.Fehér D, Barlow RS, Lorenzo PS, Hemscheidt TK. 2008. A 2-substituted prodiginine, 2-(p-hydroxybenzyl) prodigiosin, from Pseudoalteromonas rubra. J Nat Prod 71:1970–1972. doi: 10.1021/np800493p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gram L, Melchiorsen J, Bruhn JB. 2010. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar Biotechnol (NY) 12:439–451. doi: 10.1007/s10126-009-9233-y. [DOI] [PubMed] [Google Scholar]
- 18.Machado H, Sonnenschein EC, Melchiorsen J, Gram L. 2015. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genomics 16:158. doi: 10.1186/s12864-015-1365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wietz M, Månsson M, Gram L. 2011. Chitin stimulates production of the antibiotic andrimid in a Vibrio coralliilyticus strain. Environ Microbiol Rep 3:559–564. doi: 10.1111/j.1758-2229.2011.00259.x. [DOI] [PubMed] [Google Scholar]
- 20.Giubergia S, Phippen C, Gotfredsen CH, Nielsen KF, Gram L. 2016. Influence of niche-specific nutrients on secondary metabolism in Vibrionaceae. Appl Environ Microbiol 82:4035–4044. doi: 10.1128/AEM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giubergia S, Phippen C, Nielsen KF, Gram L. 2017. Growth on chitin impacts the transcriptome and metabolite profiles of antibiotic-producing Vibrio coralliilyticus S2052 and Photobacterium galathea S2753. mSystems 2:e00141-16. doi: 10.1128/mSystems.00141-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delpin MW, Goodman AE. 2009. Nutrient regime regulates complex transcriptional start site usage within a Pseudoalteromonas chitinase gene cluster. ISME J 3:1053–1063. doi: 10.1038/ismej.2009.54. [DOI] [PubMed] [Google Scholar]
- 23.Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Roseman S. 2004. The chitinolytic cascade in vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc Natl Acad Sci U S A 101:627–631. doi: 10.1073/pnas.0307645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beier S, Bertilsson S. 2013. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front Microbiol 4:149. doi: 10.3389/fmicb.2013.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García-Fraga B, Da Silva AF, López-Seijas J, Sieiro C. 2015. A novel family 19 chitinase from the marine-derived Pseudoalteromonas tunicata CCUG 44952T: heterologous expression, characterization and antifungal activity. Biochem Eng J 93:84–93. doi: 10.1016/j.bej.2014.09.014. [DOI] [Google Scholar]
- 27.Watanabe T, Kanai R, Kawase T, Tanabe T, Mitsutomi M, Sakuda S, Miyashita K. 1999. Family 19 chitinases of Streptomyces species: characterization and distribution. Microbiology 145:3353–3363. doi: 10.1099/00221287-145-12-3353. [DOI] [PubMed] [Google Scholar]
- 28.Kong H, Shimosaka M, Ando Y, Nishiyama K, Fujii T, Miyashita K. 2001. Species-specific distribution of a modular family 19 chitinase gene in Burkholderia gladioli. FEMS Microbiol Ecol 37:135–141. doi: 10.1111/j.1574-6941.2001.tb00861.x. [DOI] [Google Scholar]
- 29.Kawase T, Saito A, Sato T, Kanai R, Fujii T, Nikaidou N, Miyashita K, Watanabe T. 2004. Distribution and phylogenetic analysis of family 19 chitinases in Actinobacteria. Appl Environ Microbiol 70:1135–1144. doi: 10.1128/aem.70.2.1135-1144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda Y, Taniguchi H, Kitaoka M. 2008. A reducing-end-acting chitinase from Vibrio proteolyticus belonging to glycoside hydrolase family 19. Appl Microbiol Biotechnol 78:627–634. doi: 10.1007/s00253-008-1352-2. [DOI] [PubMed] [Google Scholar]
- 31.Paulsen SS, Andersen B, Gram L, Machado H. 2016. Biological potential of chitinolytic marine bacteria. Mar Drugs 14:E230. doi: 10.3390/md14120230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 33.Nam Y-D, Chang H-W, Park JR, Kwon H-Y, Quan Z-X, Park Y-H, Lee J-S, Yoon J-H, Bae J-W. 2007. Pseudoalteromonas marina sp. nov., a marine bacterium isolated from tidal flats of the Yellow Sea, and reclassification of Pseudoalteromonas sagamiensis as Algicola sagamiensis comb. nov. Int J Syst Evol Microbiol 57:12–18. doi: 10.1099/ijs.0.64523-0. [DOI] [PubMed] [Google Scholar]
- 34.Tran PN, Savka MA, Gan HM. 2017. In-silico taxonomic classification of 373 genomes reveals species misidentification and new genospecies within the genus Pseudomonas. Front Microbiol 8:1296. doi: 10.3389/fmicb.2017.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A. 2015. Microbial species delineation using whole genome sequences. Nucleic Acids Res 43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsujibo H, Orikoshi H, Baba N, Miyahara M, Miyamoto K, Yasuda M, Inamori Y. 2002. Identification and characterization of the gene cluster involved in chitin degradation in a marine bacterium, Alteromonas sp. strain O-7. Appl Environ Microbiol 68:263–270. doi: 10.1128/AEM.68.1.263-270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai Y, Eijsink VGH, Kielak AM, Van Veen JA, De Boer W. 2016. Genomic comparison of chitinolytic enzyme systems from terrestrial and aquatic bacteria. Environ Microbiol 18:38–49. doi: 10.1111/1462-2920.12545. [DOI] [PubMed] [Google Scholar]
- 38.Hamre AG, Eide KB, Wold HH, Sørlie M. 2015. Activation of enzymatic chitin degradation by a lytic polysaccharide monooxygenase. Carbohydr Res 407:166–169. doi: 10.1016/j.carres.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Walton PH, Davies GJ. 2016. On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr Opin Chem Biol 31:195–207. doi: 10.1016/j.cbpa.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, Eijsink VGH. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 41.Kawase T, Yokokawa S, Saito A, Fujii T, Nikaidou N, Miyashita K, Watanabe T. 2006. Comparison of enzymatic and antifungal properties between family 18 and 19 chitinases from S. coelicolor A3(2). Biosci Biotechnol Biochem 70:988–998. doi: 10.1271/bbb.70.988. [DOI] [PubMed] [Google Scholar]
- 42.Berlemont R, Martiny C. 2015. Genomic potential for polysaccharide deconstruction in bacteria. Appl Environ Microbiol 81:1513–1519. doi: 10.1128/AEM.03718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han B, Zhou K, Li Z, Sun B, Ni Q, Meng X, Pan G, Li C, Long M, Li T, Zhou C, Li W, Zhou Z. 2016. Characterization of the first fungal glycosyl hydrolase family 19 chitinase (NbchiA) from Nosema bombycis (Nb). J Eukaryot Microbiol 63:37–45. doi: 10.1111/jeu.12246. [DOI] [PubMed] [Google Scholar]
- 44.Carro L, Nouioui I, Sangal V, Meier-Kolthoff JP, Trujillo ME, Mo C, Sahin N, Smith DL, Kim KE, Peluso P, Deshpande S, Woyke T, Shapiro N, Kyrpides NC, Klenk H, Göker M, Goodfellow M. 2018. Genome-based classification of Micromonosporae with a focus on their biotechnological and ecological potential. Sci Rep 8:525. doi: 10.1038/s41598-017-17392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang L, Garbulewska E, Sato K, Kato Y, Nogawa M, Taguchi G, Shimosaka M. 2012. Isolation of genes coding for chitin-degrading enzymes in the novel chitinolytic bacterium, Chitiniphilus shinanonensis, and characterization of a gene coding for a family 19 chitinase. J Biosci Bioeng 113:293–299. doi: 10.1016/j.jbiosc.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Suma K, Podile AR. 2013. Chitinase A from Stenotrophomonas maltophilia shows transglycosylation and antifungal activities. Bioresour Technol 133:213–220. doi: 10.1016/j.biortech.2013.01.103. [DOI] [PubMed] [Google Scholar]
- 47.Babashpour S, Aminzadeh S, Farrokhi N, Karkhane A, Haghbeen K. 2012. Characterization of a chitinase (Chit62) from Serratia marcescens B4A and its efficacy as a bioshield against plant fungal pathogens. Biochem Genet 50:722–735. doi: 10.1007/s10528-012-9515-3. [DOI] [PubMed] [Google Scholar]
- 48.da Mota PR, Ribeiro MS, de Castro Georg R, Silva GR, de Paula RG, Silva RDN, Ulhoa CJ. 2016. Expression analysis of the α-1,2-mannosidase from the mycoparasitic fungus Trichoderma harzianum. Biol Control 95:1–4. doi: 10.1016/j.biocontrol.2015.12.013. [DOI] [Google Scholar]
- 49.Stengel DB, Connan S, Popper ZA. 2011. Algal chemodiversity and bioactivity: sources of natural variability and implications for commercial application. Biotechnol Adv 29:483–501. doi: 10.1016/j.biotechadv.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Cimermancic P, Medema MH, Claesen J, Kurita K, Brown LCW, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci U S A 104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, Mcglinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Moore BS, Jensen PR. 2009. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J 3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aminov RI. 2010. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol 1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niu G. 2017. Genomics-driven natural product discovery in actinomycetes. Trends Biotechnol 36:238–241. doi: 10.1016/j.tibtech.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Cordero OX, Datta MS, Sliwerska E, Gore J, Polz MF. 2016. Microbial interactions lead to rapid successions on model marine particles. Nat Commun 7:11965. doi: 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 57.Machado H, Vynne NG, Christiansen G, Gram L. 2016. Reclassification of Alteromonas fuliginea (Romanenko et al. 1995) as Pseudoalteromonas fuliginea comb. nov. and an emended description. Int J Syst Evol Microbiol 66:3737–3742. doi: 10.1099/ijsem.0.001259. [DOI] [PubMed] [Google Scholar]
- 58.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medema MH, Takano E, Breitling R. 2013. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol 30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolde R. 2018. R package pheatmap 1.0.10. https://github.com/raivokolde/pheatmap.
- 64.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Duran HGS, Santos E, Kim U, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema H. 2017. antiSMASH 4. 0––improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45(W1):W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skov MN, Pedersen K, Larsen JL. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl Environ Microbiol 61:1540–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation of percent ANI to the percentage of isolates used for inferring the cutoff percent ANI value of identical isolates. Download FIG S1, PDF file, 0.1 MB (138.6KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat map of average nucleotide identity (ANI) analysis of 157 Pseudoalteromonas strains. The color-coded bar on the left indicates the pigmentation status. Download FIG S2, PDF file, 0.4 MB (375KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of 50 Pseudoalteromonas strains isolated during the Galathea 3 expedition and 107 Pseudoalteromonas genomes obtained from NCBI, the species, their pigmentation, the genome size (in megabases), the percentage of the genome allocated to BGCs, and the NCBI accession number. Download Table S1, PDF file, 0.4 MB (447.4KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chitin degradation on agar plates containing colloidal chitin and inhibition of Vibrio anguillarum by Pseudoalteromonas strains on media supplemented with four different carbon sources: glucose, N-acetylglucosamine, colloidal chitin, or crystalline chitin. +, inhibition; −, no inhibition. Download Table S2, PDF file, 0.3 MB (350.1KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Loadings associated with multidimensional scaling plot in Fig. 4. Loadings plotted in Fig. 4 are represented in bold. Download Table S3, PDF file, 0.2 MB (232.7KB, pdf) .
Copyright © 2019 Paulsen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The 76 new draft genomes are available at the National Center for Biotechnology Information (NCBI) database under accession numbers PNBS01 to PNEL01. All accession numbers are listed in Table S1. The strains are available upon request.