Interest in the role of intestinal microbiota in determining FE in pigs has increased in recent years. However, it is not known if the same FE-associated bacteria are found across different rearing environments. In this study, geographic location and intestinal sampling site had a greater influence on the pig gut microbiome than FE. This presents challenges when aiming to identify consistent reliable microbial biomarkers for FE. Nonetheless, seven FE-associated microbial taxa were common across two geographic locations and/or two batches within one location, and these indicated a potentially “healthier” and metabolically more capable microbiota in more-feed-efficient pigs. These taxa could potentially be employed as biomarkers for FE, although bacterial consortia, rather than individual taxa, may be more likely to predict FE. They may also merit consideration for use as probiotics or could be targeted by dietary means as a strategy for improving FE in pigs in the future.
KEYWORDS: feed efficiency, geographic location, intestinal microbiota, pigs
ABSTRACT
Optimal feed efficiency (FE) in pigs is important for economic and environmental reasons. Previous research identified FE-associated bacterial taxa within the intestinal microbiota of growing pigs. This study investigated whether FE-associated bacteria and selected FE-associated physiological traits were consistent across geographic locations (Republic of Ireland [ROI] [two batches of pigs, ROI1 and ROI2], Northern Ireland [NI], and Austria [AT]), where differences in genetic, dietary, and management factors were minimized. Pigs (n = 369) were ranked, within litter, on divergence in residual feed intake (RFI), and 100 extremes were selected (50 with high RFI and 50 with low RFI) across geographic locations for intestinal microbiota analysis using 16S rRNA amplicon sequencing and examination of FE-associated physiological parameters. Microbial diversity varied by geographic location and intestinal sampling site but not by RFI rank, except in ROI2, where more-feed-efficient pigs had greater ileal and cecal diversity. Although none of the 188 RFI-associated taxonomic differences found were common to all locations/batches, Lentisphaerae, Ruminococcaceae, RF16, Mucispirillum, Methanobrevibacter, and two uncultured genera were more abundant within the fecal or cecal microbiota of low-RFI pigs in two geographic locations and/or in both ROI batches. These are major contributors to carbohydrate metabolism, which was reflected in functional predictions. Fecal volatile fatty acids and salivary cortisol were the only physiological parameters that differed between RFI ranks. Despite controlling genetics, diet specification, dietary phases, and management practices in each rearing environment, the rearing environment, encompassing maternal influence, herd health status, as well as other factors, appears to impact intestinal microbiota more than FE.
IMPORTANCE Interest in the role of intestinal microbiota in determining FE in pigs has increased in recent years. However, it is not known if the same FE-associated bacteria are found across different rearing environments. In this study, geographic location and intestinal sampling site had a greater influence on the pig gut microbiome than FE. This presents challenges when aiming to identify consistent reliable microbial biomarkers for FE. Nonetheless, seven FE-associated microbial taxa were common across two geographic locations and/or two batches within one location, and these indicated a potentially “healthier” and metabolically more capable microbiota in more-feed-efficient pigs. These taxa could potentially be employed as biomarkers for FE, although bacterial consortia, rather than individual taxa, may be more likely to predict FE. They may also merit consideration for use as probiotics or could be targeted by dietary means as a strategy for improving FE in pigs in the future.
INTRODUCTION
Feed efficiency (FE) is critical in pig production, as feed accounts for ∼70% of production costs (1). As a result, a considerable amount of research has focused on improving FE and finding reliable biomarkers for FE in pigs. Those that have been suggested include cortisol, a hormone which is secreted in response to stress and low blood glucose levels, affecting metabolism of carbohydrates, lipids, and protein (2). Animals with a higher level of blood plasma cortisol are more likely to divert energy away from lean-meat deposition, resulting in poorer FE (3). Blood cell profile and metabolic markers are functional indicators of metabolic pathways that ultimately denote homeostasis or dysfunction (4), and some of these have also been linked with FE in pigs (e.g., hemoglobin concentration; red blood cell count; white blood cell profile; and protein, triglyceride, and cholesterol concentrations) (5–7). Immune status is another important factor associated with FE; it has been suggested that pigs with enhanced FE might have a more effective immune response without affecting growth and lean-meat deposition (8, 9). More-feed-efficient pigs were previously shown to produce lower but sufficient levels of white blood cells, i.e., lymphocytes and monocytes (5), and had increased expression of antigen-processing-related genes (10).
The resident intestinal microbiota, specifically its diversity, composition, and function, is likely to influence FE in pigs, considering its role in host metabolism and immunity (9, 11–13). An increasing number of studies aimed at characterizing the swine intestinal microbiome have identified major populations of bacteria associated with intestinal site, age of the pig, and diet (14–19). Studies have also uncovered a major role for the porcine intestinal microbiota and microbial metabolites in regulation of the immune system (20). Recently, a number of studies, including one from our group, have demonstrated an association between porcine intestinal microbiota composition and FE (12, 13, 19, 21). However, each study was limited to one batch of pigs reared in the same environment at a single geographic location.
Humans living in different environments have dramatically different intestinal microbial profiles (22), whereas in cattle, management practices appear to be more influential than geographic location (23). Previous work by this group investigated the fecal and intestinal microbiota of pigs divergent in RFI (residual feed intake; a metric for FE) within one rearing environment (12). Feed efficiency-associated bacteria were identified throughout the lifetime of the pig albeit mostly low-relative-abundance taxa. In general, the abundances of microbes related to metabolism and disease were higher and lower, respectively, in low-RFI (more-feed-efficient) pigs. Following on from this work, where a first set of FE-associated microbial taxa was identified, the present study sought to determine if these findings can be extrapolated to pigs raised in different rearing environments, even other countries, when genetics, diet, and management are controlled.
The objective was to investigate the intestinal microbiota of pigs ranked on RFI, reared at three geographic locations, each using the same animal genetics, diet specifications, dietary phases, and management protocols, with a view to determining if similar FE-associated microbiomes are found across different rearing environments. Other physiological parameters were also assessed and correlated with intestinal microbial composition, in an attempt to elucidate their role in influencing FE in pigs.
RESULTS
Growth performance of pigs ranked as having low or high RFI at different geographic locations.
Growth performance parameters, including residual feed intake (RFI), were determined between days 70 and 120 of age and are presented in Table 1. Across geographic locations (Republic of Ireland [ROI], Northern Ireland [NI], and Austria [AT]), there was a distinct separation between high- and low-RFI pigs (P < 0.001). The average daily gain (ADG) (P = 0.25) and weight at day 70 (P = 0.22) did not differ between RFI ranks across locations, but weight at day 120 (P < 0.001), average daily feed intake (ADFI) (P < 0.005), and feed conversion efficiency (FCE) (P < 0.03) did.
TABLE 1.
Growth performance and salivary cortisola of pigs ranked on residual feed intake at three geographic locationsb
| Parameter | Value for groupc
|
SEMd |
P value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High RFI |
Low RFI |
|||||||||||
| ROI1 | ROI2 | NI | AT | ROI1 | ROI2 | NI | AT | RFI × location |
RFI | Location | ||
| RFI (g/day) | 1,237 A | 1,108 A | 207 B | 1,171 AB | −1,030 C | −737 C | −200 C | −956 C | 227.6 | <0.001 | <0.001 | 0.86 |
| wt (kg), day 70 | 32.2 | 33.3 | 27.4 | 30.2 | 33.6 | 31.9 | 28.7 | 28.6 | 1.93 | 0.22 | 0.97 | 0.27 |
| wt (kg), day 134 | 83.1 BC | 82.3 BC | 91.3 AC | 85.2 ABC | 87.2 ABC | 79.5 B | 95.2 B | 82.8 BC | 1.94 | <0.001 | 0.83 | 0.35 |
| ADFI (g/day) | 2,025 BD | 2,194 AD | 2,033 AB | 2,380 A | 1,988 BD | 1,851 BC | 1,586 C | 1,942 BCD | 74.1 | 0.005 | <0.001 | 0.005 |
| ADG (g/day) | 1,028 | 902 | 891 | 1,131 | 1,065 | 847 | 894 | 1,077 | 30.0 | 0.25 | 0.44 | <0.001 |
| FCE (g/g) | 2.05 B | 2.62 A | 2.40 A | 2.11 B | 1.91 B | 2.39 A | 1.80 B | 1.84 B | 0.088 | 0.03 | <0.001 | <0.001 |
| Salivary cortisol concn (ng/ml) |
1.76 | 1.34 | 0.176 | 0.06 | ||||||||
Cortisol was measured only in ROI1 pigs on the day prior to slaughter.
ROI1, Republic of Ireland batch 1; ROI2, Republic of Ireland batch 2; NI, Northern Ireland; AT, Austria; wt, weight; ADFI, average daily feed intake; ADG, average daily gain; FCE, feed conversion efficiency.
Within each row, values that do not share a common letter are different (P ≤ 0.05).
Least-squares means and the pooled standard errors of the means (SEM) are presented.
Microbial diversity and composition in the feces and digesta of pigs ranked on RFI at different geographic locations.
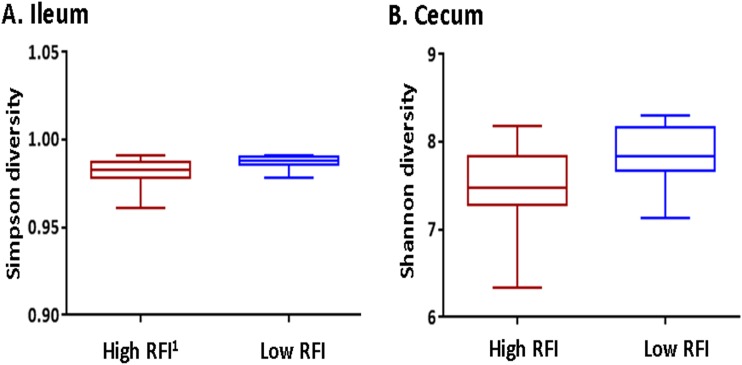
Microbial richness and diversity within the feces of pigs (at both days 70 and 134 of age) were not associated with RFI rank (see Fig. S1 in the supplemental material). However, in the ileal and cecal digesta from ROI batch 2 (ROI2), operational taxonomic unit (OTU) diversity, depicted by Shannon and Simpson α-diversity indices, was higher for low-RFI pigs (P < 0.05) (Fig. 1).
FIG 1.
Alpha diversity of the microbiota within the ileum (A) and cecum (B) of pigs ranked on residual feed intake (RFI1) from Republic of Ireland batch 2 (ROI2).
Alpha diversity of the fecal microbiota of pigs ranked on residual feed intake (RFI) at 70 days of age (A) and 134 days of age (B) across all three geographic locations. ROI, Republic of Ireland; NI, Northern Ireland; AT, Austria. Download FIG S1, DOCX file, 0.3 MB (271.4KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
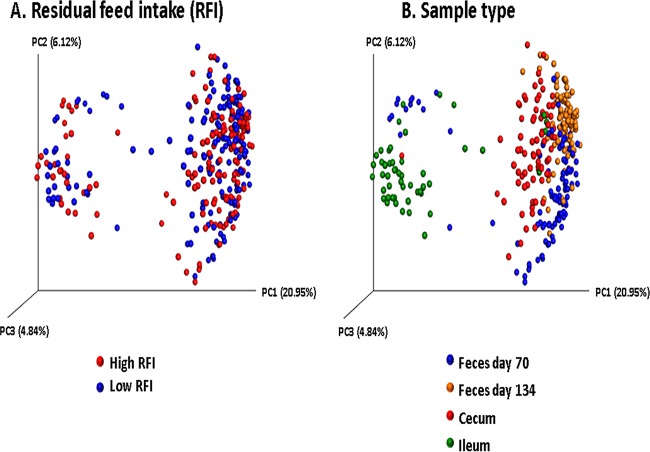
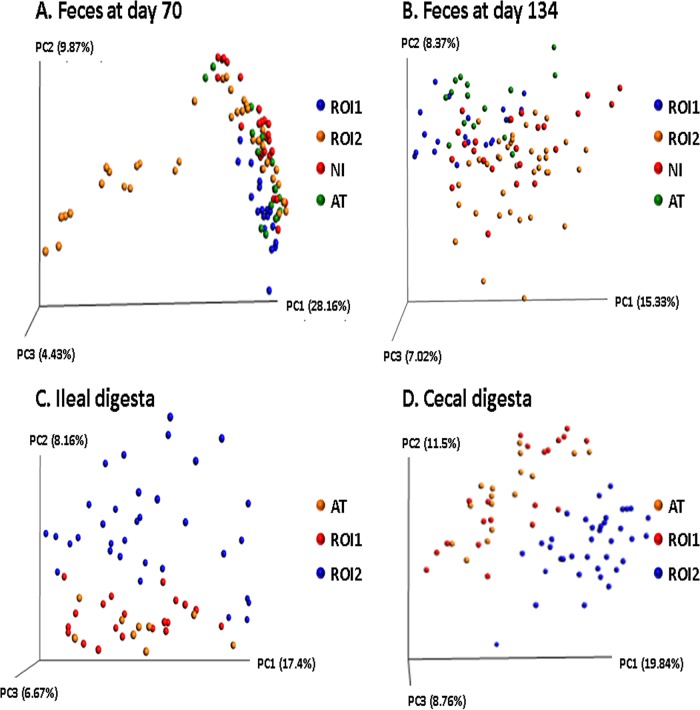
Overall, β-diversity of the intestinal microbiota was affected by sample type (i.e., feces and/or digesta) and age at sampling but not by RFI rank (Fig. 2). At each fecal time point and for each digesta type, location-specific effects were observed, with samples from the same geographic location/digesta type generally clustering together in the principal-coordinate analysis (PCoA) plots (P < 0.001) (Fig. 3). However, one exception was that samples from ROI1 and ROI2 pigs did not cluster together, even though they originated from the same location. In fact, the microbial diversity of samples from ROI1 pigs was closer to that of samples from AT pigs, and the samples from pigs from ROI2 were more similar to samples from NI pigs.
FIG 2.
Principal-coordinate analysis (PCoA) plots (based on OTUs) by RFI rank (A) and sample type (B).
FIG 3.
Principal-coordinate analysis plots (based on OTUs) for feces collected from pigs on day 70 (A) and day 134 (B) of age and in ileal (C) and cecal (D) digesta. ROI, Republic of Ireland; AT, Austria; NI, Northern Ireland.
Microbial composition was examined, at the phylum, family, and genus levels. The phylum Firmicutes was the most abundant phylum in the feces and ileal digesta in pigs from all geographic locations (Fig. S2). Pigs from ROI1 had a greater abundance of Tenericutes in the ileum than did pigs in ROI2 and AT (P < 0.05) (Fig. S2).
Median relative abundance (percent) of bacterial phyla present in pigs according to residual feed intake (RFI) rank across geographic locations for all fecal time points and intestinal sites. 1Other (no BLAST hits/uncultured). * indicates a significant difference observed for that phylum between high- and low-RFI pigs within each sample type and geographic location (P ≤ 0.05). These RFI-associated differences are as follows: for feces on day 70, ↑ Deferribacteres (AT), ↑ SHA.109 (ROI2), and ↓ Verrucomicrobia (ROI1); for feces on day 134, ↓ candidate division TM7 (ROI2), ↓ Firmicutes (ROI1), ↑ Euryarchaeota (ROI1), and ↑ Lentisphaerae (ROI1 and ROI2) and Verrucomicrobia (ROI1); for ileum, ↓ Cyanobacteria (ROI2), ↓ Firmicutes (ROI2), and ↑ Planctomycetes (ROI2); and for cecum, ↑ Cyanobacteria (AT), ↓ Firmicutes (ROI1), ↑ Proteobacteria (ROI1), and ↑ Verrucomicrobia (ROI2). Arrows indicate higher (↑) or lower (↓) relative abundances in low-RFI pigs. Download FIG S2, DOCX file, 0.2 MB (249.9KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
When the intestinal microbiota of high- versus low-RFI pigs was compared at each geographic location at the phylum, family, and genus levels, a total of 188 compositional differences, mostly for taxa at a low relative abundance, were found across all locations (P < 0.05) (Tables 2 and 3). Most of these RFI-associated differences were observed in ROI2 pigs. Phylum-level differences between high- and low-RFI pigs are shown in Fig. S2 and included the high-abundance phylum Firmicutes. A number of low-relative-abundance phyla also differed significantly between RFI ranks, including Proteobacteria, Verrucomicrobia, SHA.109, Deferribacteres, Lentisphaerae, Euryarchaeota, candidate division TM7, Cyanobacteria, and Planctomycetes. For the majority of phyla, a higher relative abundance was observed in the low-RFI (i.e., more-feed-efficient) pigs than in their high-RFI counterparts, except for Firmicutes and Verrucomicrobia in ROI1 at day 70 of age.
TABLE 2.
RFI-associated statistically significant microbial composition differences in the feces of pigs at three geographic locationsa
| Time point | Phylumb | Family | Genus | Location(s)c | Significanced | Differencee |
|---|---|---|---|---|---|---|
| Day 70 of age | Bacteroidetes | S24.7 | Uncultured | NI | F, G | ↓ |
| AT | G | ↑ | ||||
| Deferribacteres | Deferribacteraceae | Mucispirillum schaedleri | AT | P, G | ↑ | |
| Mucispirillum | NI | F, G | ↑ | |||
| AT | G | ↑ | ||||
| Day 134 of age | Firmicutes | Family XIII | Mogibacterium | NI | G | ↓ |
| Lachnospiraceae | Roseburia | NI | G | ↑ | ||
| Ruminococcaceae | Incertae sedis | ROI1 | G | ↓ | ||
| Uncultured | NI | G | ↓ | |||
| Erysipelotrichaceae | Uncultured | NI | G | ↓ | ||
| Planctomycetes | Planctomycetaceae | ROI2 | F | ↑ | ||
| SHA.109 | Uncultured | Uncultured | ROI2 | P, F, G | ↑ | |
| Spirochaetes | Spirochaetaceae | Treponema brennaborense | AT | G | ↑ | |
| Verrucomicrobia | ROI1 | P | ↓ | |||
| ROI1 | P | ↑ | ||||
| Euryarchaeota | Methanobacteriaceae | AT | F | ↑ | ||
| Methanobrevibacter | ROI1, ROI2 | G | ↑ | |||
| BS11.gut.group | Uncultured | ROI2 | F, G | ↑ | ||
| Prevotellaceae | Alloprevotella | ROI2 | G | ↓ | ||
| Bacteroidetes | RF16 | Uncultured | ROI1 | F, G | ↑ | |
| AT | G | ↑ | ||||
| Rikenellaceae | Alistipes | ROI2 | G | ↑ | ||
| Uncultured | Uncultured | ROI1 | F, G | ↓ | ||
| Candidate division TM7 | Unknown family | “Candidatus Saccharimonas” | ROI2 | P, F, G | ↓ | |
| Cyanobacteria | Uncultured | Uncultured | ROI1 | F, G | ↑ | |
| Firmicutes | Defluviitaleaceae | Uncultured | ROI1 | P, F, G | ↓ | |
| Family XI | Sedimentibacter | ROI2 | F, G | ↑ | ||
| Family XIII | Uncultured | ROI2 | F, G | ↑ | ||
| Lachnospiraceae | Lachnospira | ROI1 | G | ↓ | ||
| Marvinbryantia | ROI2 | G | ↓ | |||
| Oribacterium | NI | G | ↑ | |||
| ROI1 | G | ↓ | ||||
| Pseudobutyrivibrio | NI | G | ↑ | |||
| Ruminococcaceae | Faecalibacterium | ROI2 | G | ↓ | ||
| Papillibacter | ROI2 | G | ↑ | |||
| Ruminococcus | NI | G | ↓ | |||
| Subdoligranulum | ROI2 | G | ↓ | |||
| vadinBB60 | Unidentified | ROI2 | F, G | ↑ | ||
| Uncultured | Unidentified | AT | G | ↑ | ||
| Erysipelotrichaceae | Asteroleplasma | ROI2 | G | ↓ | ||
| Incertae sedis | ROI1 | G | ↓ | |||
| Veillonellaceae | Allisonella | ROI1 | G | ↓ | ||
| Mitsuokella | ROI1 | G | ↓ | |||
| Selenomonas | ROI2 | G | ↓ | |||
| Uncultured | ROI2 | G | ↓ | |||
| ROI1, ROI2 | P | ↑ | ||||
| Lentisphaerae | RF12 gut group, uncultured | Uncultured | ROI1 | F, G | ↑ | |
| Uncultured rumen bacterium | ROI1 | F, G | ↑ | |||
| RF12 gut group, unidentified | Uncultured | ROI1 | F, G | ↑ | ||
| WCHB1.25, uncultured | Uncultured | ROI1 | F, G | ↑ | ||
| Proteobacteria | Neisseriaceae | Leeia | ROI1 | F, G | ↑ | |
| Succinivibrionaceae | Uncultured | ROI1 | G | ↑ | ||
| TA18, uncultured | Uncultured | ROI2 | F, G | ↑ | ||
| Desulfovibrionaceae | Desulfovibrio | AT | F, G | ↑ | ||
| Tenericutes | RF9, uncultured | Uncultured | AT | F, G | ↑ | |
| Verrucomicrobia | Verrucomicrobiaceae | Akkermansia | ROI1 | P, F, G | ↑ | |
A total of 84 RFI-associated microbial composition differences were observed in the feces across the three geographic locations: 9 phyla, 23 families, and 52 genera.
All phylum-level differences between high- and low-RFI pigs are shown in Fig. S2 in the supplemental material.
AT, Austria; NI, Northern Ireland; ROI, Republic of Ireland (batches 1 and 2).
Significantly different (P < 0.05) at the phylum (P), family (F), and/or genus (G) level.
Arrows indicate relative-abundance differences in low-RFI pigs compared to high-RFI pigs at the same geographic location (↓, lower relative abundance; ↑, higher relative abundance).
TABLE 3.
RFI-associated statistically significant microbial composition differences in the ileal and cecal digesta of pigs slaughtered at 134 days of age at three geographic locationsa
| Sample type | Phylumb | Family | Genus | Location(s)c | Significanced | Differencee |
|---|---|---|---|---|---|---|
| Ileum | Actinobacteria | Dietziaceae | Dietzia | ROI2 | F, G | ↑ |
| Microbacteriaceae | Leucobacter | ROI2 | G | ↑ | ||
| Micrococcaceae | Rothia | ROI2 | G | ↑ | ||
| Pseudonocardiaceae | Saccharopolyspora | ROI2 | F, G | ↑ | ||
| Bacteroidetes | S24.7 | Uncultured | ROI2 | G | ↑ | |
| Cyanobacteria | Gastranaerophilales, uncultured | Uncultured | ROI2 | P, F, G | ↑ | |
| Firmicutes | ROI2 | P | ↓ | |||
| Bacillaceae | Oceanobacillus | ROI2 | G | ↑ | ||
| Paucisalibacillus | ROI2 | G | ↑ | |||
| Paenibacillaceae | ROI2 | F | ↑ | |||
| Aerococcaceae | ROI2 | F | ↑ | |||
| Dolosicoccus | ROI2 | G | ↑ | |||
| Globicatella | ROI2 | G | ↑ | |||
| Enterococcaceae | Enterococcus | ROI2 | F, G | ↑ | ||
| Clostridiaceae 1 | Clostridium sensu stricto 1 | ROI1 | F, G | ↑ | ||
| Lachnospiraceae | Blautia | ROI2 | G | ↑ | ||
| Pseudobutyrivibrio | ROI2 | G | ↑ | |||
| Subdoligranulum | ROI2 | G | ↑ | |||
| vadinBB60 | Uncultured | ROI1 | G | ↓ | ||
| ROI2 | G | ↑ | ||||
| Erysipelotrichaceae | Turicibacter | ROI1 | F, G | ↑ | ||
| Solobacterium | ROI2 | G | ↑ | |||
| Veillonellaceae | Dialister | ROI2 | G | ↑ | ||
| Mitsuokella | ROI2 | G | ↑ | |||
| Planctomycetes | Planctomycetaceae | p1088 a5 gut group | ROI2 | P, F, G | ↑ | |
| Proteobacteria | Desulfovibrionaceae | Desulfovibrio | ROI2 | F, G | ↑ | |
| Helicobacteraceae | Helicobacter | ROI1 | F, G | ↓ | ||
| Pasteurellaceae | Actinobacillus | ROI2 | F, G | ↑ | ||
| Spirochaetae | Spirochaetaceae | Spirochaeta | ROI2 | G | ↑ | |
| Cecum | Actinobacteria | Microbacteriaceae | Microbacterium | ROI2 | F, G | ↑ |
| Coriobacteriaceae | Uncultured | ROI2 | F, G | ↑ | ||
| Bacteroidetes | Porphyromonadaceae | Paludibacter | ROI2 | G | ↑ | |
| RF16 | Uncultured | ROI1, ROI2 | F, G | ↑ | ||
| Rikenellaceae | ROI2 | F | ↑ | |||
| dgA 11 gut group | ROI2 | G | ↑ | |||
| RC9 gut group | ROI2 | G | ↑ | |||
| Cyanobacteria | 4c0d, uncultured | Uncultured | AT | P, F, G | ↑ | |
| Gastranaerophilales, uncultured | Uncultured | ROI2 | F, G | ↑ | ||
| Firmicutes | ROI1 | P | ↓ | |||
| Erysipelotrichaceae | Uncultured | ROI2 | F, G | ↑ | ||
| Asteroleplasma | ROI2 | G | ↑ | |||
| Family XIII | Uncultured | ROI2 | G | ↑ | ||
| Lachnospiraceae | Butyrivibrio | ROI2 | G | ↓ | ||
| Peptococcaceae | Peptococcus | ROI2 | F, G | ↑ | ||
| Ruminococcaceae | ROI2, AT | F | ↑ | |||
| Incertae sedis | ROI2 | G | ↑ | |||
| Oscillospira | ROI1 | G | ↓ | |||
| Papillibacter | ROI2 | G | ↑ | |||
| vadinBB60 | Uncultured | ROI2 | G | ↑ | ||
| Streptococcaceae | Streptococcus | ROI2 | F, G | ↓ | ||
| Veillonellaceae | Uncultured | ROI1 | F, G | ↓ | ||
| Lentisphaerae | RF12 gut group, uncultured | Uncultured | ROI2 | F, G | ↑ | |
| Proteobacteria | ROI1 | P | ↑ | |||
| Rhodospirillaceae | ROI1 | F | ↑ | |||
| Alcaligenaceae | Sutterella | ROI1 | F, G | ↑ | ||
| Oxalobacteraceae | Noviherbaspirillum | ROI2 | F, G | ↑ | ||
| GR WP33 58 | Uncultured | ROI2 | F, G | ↑ | ||
| Helicobacteraceae | Helicobacter | ROI2 | F, G | ↑ | ||
| Succinivibrionaceae | Ruminobacter | ROI1 | F, G | ↑ | ||
| Spirochaetes | Spirochaetaceae | Treponema | AT | G | ↓ | |
| Synergistetes | Synergistaceae | Pyramidobacter | ROI2 | F, G | ↑ | |
| Tenericutes | Anaeroplasmataceae | Anaeroplasma | ROI1 | F, G | ↑ | |
| NB1 n, uncultured | Uncultured | ROI2 | F, G | ↑ | ||
| Verrucomicrobia | Puniceicoccaceae | Puniceicoccus | ROI2 | P, F, G | ↑ | |
| vadinHA64, uncultured | Uncultured | ROI2 | F, G | ↑ | ||
| Verrucomicrobiaceae | Akkermansia | ROI1 | F, G | ↑ |
A total of 104 RFI-associated microbial composition differences were observed across the three geographic locations.
All phylum-level differences between high- and low-RFI pigs are shown in Fig. S2 in the supplemental material.
AT, Austria; ROI, Republic of Ireland (batches 1 and 2).
Significantly different (P < 0.05) at the phylum (P), family (F), and/or genus (G) level.
Arrows indicate relative-abundance differences in low-RFI pigs compared to high-RFI pigs at the same geographic location (↓, lower relative abundance; ↑, higher relative abundance).
RFI-associated bacterial taxa in the fecal and intestinal microbiota of pigs ranked on RFI at different geographic locations.
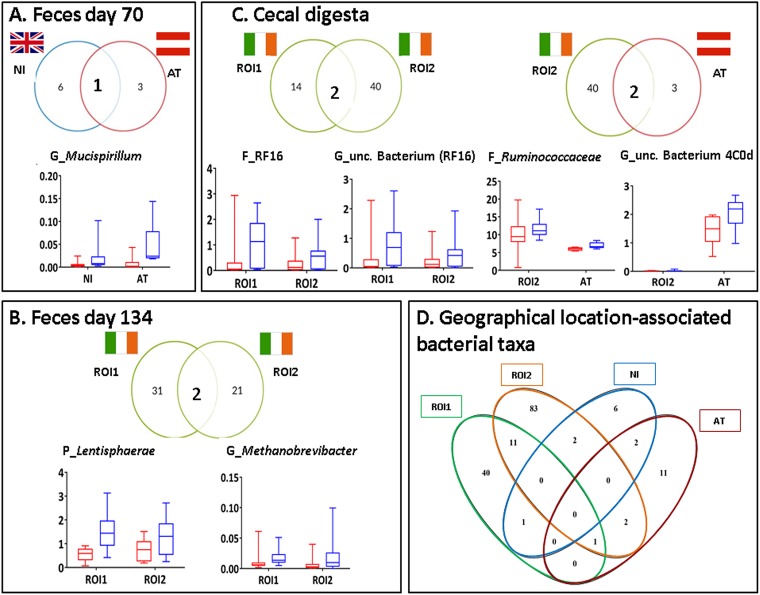
Although none of the 188 RFI-associated taxonomic differences within the fecal and/or intestinal microbiota were common to all four groups of pigs (Fig. 4D), seven taxa were found to be enriched in low-RFI (more-feed-efficient) pigs at more than one geographic location or in the two different batches reared in ROI (P < 0.05) (Fig. 4). In the feces collected on day 70 of age, Mucispirillum (from Deferribacteres) was more abundant in low- than in high-RFI pigs from both NI (2-fold) and AT (15-fold) (P < 0.05). On day 134 of age, the phylum Lentisphaerae was enriched in the feces of low-RFI pigs in both ROI1 and ROI2 (ROI1, 2.4-fold; ROI2, 1.7-fold), as was the genus Methanobrevibacter (from Euryarchaeota) (ROI1, 2.3-fold; ROI2, 3.3-fold [P < 0.05]). In the cecal digesta, four cross-locational RFI-associated taxa were observed. In both ROI1 and ROI2, the bacterial family RF16 and an uncultured bacterium from this family were enriched in low-RFI pigs (22-fold for ROI1 and 5-fold for ROI2, and 14-fold for ROI and 4-fold for ROI2, respectively [P < 0.05]). The RF16 family was also 20 times more abundant in the feces of low-RFI pigs than in the feces of high-RFI pigs on day 134 of age in ROI1 (P < 0.05) (data not shown). In addition, two similar OTUs belonging to an uncultured bacterium from RF16 were found at a higher abundance (∼20-fold) in the feces on day 134 of age in low-RFI pigs from both ROI1 and AT than their high-RFI counterparts (P < 0.05). Furthermore, in the cecum, low-RFI pigs in ROI2 and AT had higher abundances of the family Ruminococcaceae (1.2-fold and 1.1-fold, respectively) and an uncultured bacterium from the Cyanobacteria (10-fold and 1.5-fold, respectively). However, the Ruminococcaceae family was the only cross-locational RFI-associated taxon with a median relative abundance of >5%. In contrast, in the ileum, no common RFI-associated differences were observed across geographic locations.
FIG 4.
(A to C) RFI-associated bacterial taxa common to more than one geographic location in feces from pigs at day 70 (A) and day 134 (B) and in cecal digesta (C). Median relative abundances (percent) of these taxa in low-RFI (blue boxes) and high-RFI (red boxes) pigs are also shown. (D) RFI-associated bacterial taxa shared across geographic locations, irrespective of sample type. Only taxa that were significantly different between high- and low-RFI ranks across locations are depicted (P ≤ 0.05). Digesta was not sampled from NI pigs. P, phylum; F, family; G, genus.
When data from the four rearing environments were combined, a number of bacterial genera with a relative abundance of >0.1% were exclusively found in low-RFI pigs at each time point (Fig. S3). In the feces of pigs at day 70 of age, there were eight low-RFI-specific genera; in the feces at slaughter, there were two; in the ileal digesta, there were nine; and in cecal digesta, there were seven genera exclusive to low-RFI pigs.
Genera found exclusively in all low-residual-feed-intake (RFI)-ranked pigs across all geographic locations in feces at 70 days (A) and 134 days (B) of age and in ileal (C) and cecal (D) digesta. Data from all geographic locations were taken together. unc, uncultured; und, unidentified. Taxa present at a <0.1% relative abundance were not included in the analysis. Download FIG S3, DOCX file, 0.8 MB (796.3KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted microbial pathways in the feces and digesta of pigs ranked on RFI reared at different geographic locations.
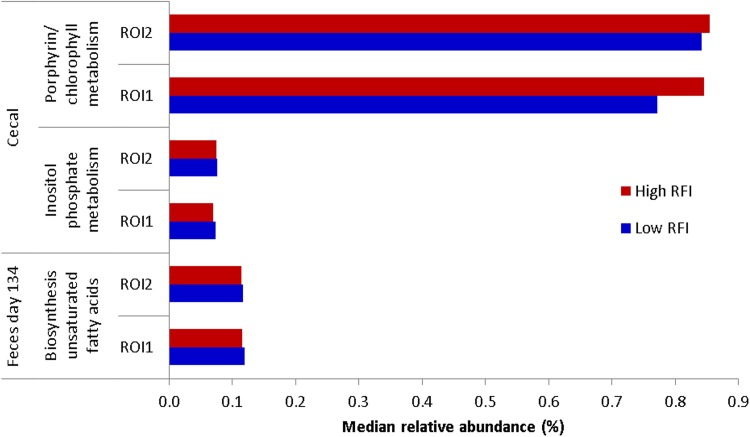
Potential functionality of the intestinal microbiota was inferred using the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) package. One hundred and two predicted microbial pathways differed significantly in relative abundance between high- and low-RFI pigs across geographic locations and by sample type and were subsequently grouped into major functional categories (Fig. S4). These pathways were present at a very low relative abundance (≤2.1%) and were mainly related to metabolism, especially carbohydrate metabolism in the feces at day 70 of age, energy metabolism in the feces of pigs at day 134 of age and in the cecum, and nucleotide metabolism in the ileum (Fig. S4). Depending on the age of the pig, there was also a relatively high representation of pathways related to genetic information processing (e.g., replication and repair or transcription). There were differences in the abundances of Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) functions between high- and low-RFI pigs across locations. In the cecal and ileal digesta, for most of the locations, most pathways were at a higher relative abundance in the low-RFI pigs than in their high-RFI counterparts. In the feces of pigs at day 70 of age, most of the differentially abundant pathways were found in the pigs from NI, whereas for the rest of the time points, most of the differences were assigned to ROI1 and ROI2 pigs. However, none of the differentially abundant predicted microbial pathways were common to all geographic locations, and only three followed the same trend in both ROI batches: biosynthesis of fatty acids in the feces at day 134 of age and inositol phosphate metabolism and porphyrin/chlorophyll metabolism in the cecal digesta (P < 0.05) (Fig. 5). The latter was most abundant in the high-RFI (less-feed-efficient) pigs, while the abundances of the other two pathways were higher in the low-RFI pigs (P < 0.05). The inositol phosphate metabolism pathway, belonging to the core carbohydrate metabolism function, was also differentially abundant in both the ileal and cecal digesta of pigs from both ROI batches.
FIG 5.
Common RFI-associated predicted microbial pathways in low- and high-RFI pigs across geographic locations. Only the predicted pathways that were significantly different between high- and low-RFI ranks across locations are depicted.
Predicted functionality of intestinal microbiota for high- and low-RFI pigs across geographic locations and by sample type (feces [A] and cecal digesta [B]). Pathways shown are those found to be differentially abundant between high- and low-RFI pigs (P < 0.05). ROI, Republic of Ireland; NI, Northern Ireland; AT, Austria. Pathways were grouped into major functional categories as follows: M1, carbohydrate metabolism; M2, metabolism of cofactors and vitamins; M3, energy metabolism; M4, lipid metabolism; M5, amino acid metabolism; M6, glycan biosynthesis and metabolism; M7, biosynthesis of other secondary metabolites; M8, metabolism of terpenoids and polyketides; M9, xenobiotic biodegradation and metabolism; M10, metabolism of other amino acids; M11, nucleotide metabolism; E, environmental information processing; G, genetic information processing; C, cell motility. Download FIG S4, DOCX file, 0.1 MB (126.2KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Volatile fatty acid concentrations in pigs ranked on RFI reared at different geographic locations.
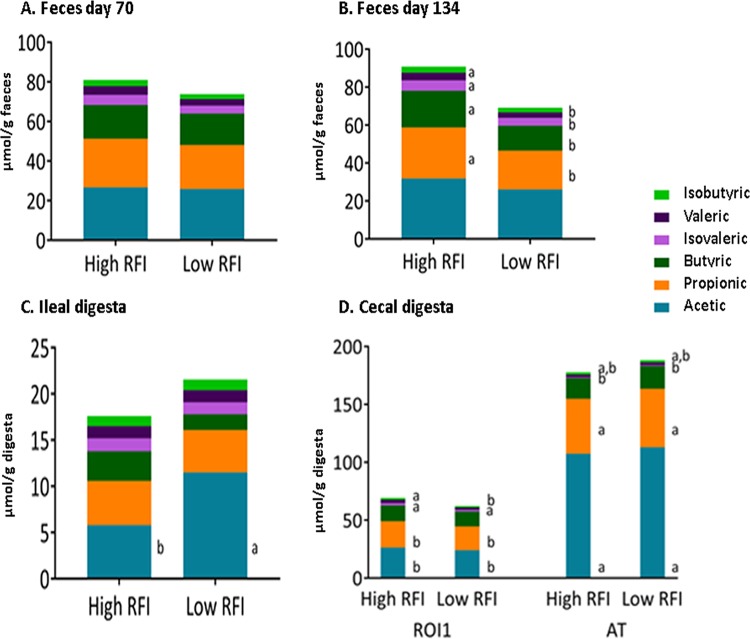
Concentrations of volatile fatty acids (VFAs) were determined in feces collected from pigs on days 70 and 134 of age, in the ileal and cecal digesta from pigs in ROI1, as well as in the cecal digesta from pigs in AT (Fig. 6). No differences were observed between RFI ranks in feces collected from pigs on day 70 of age. However, low-RFI pigs had lower concentrations of total VFAs as well as butyric and propionic acids (P < 0.05) and a tendency toward lower valeric acid (P = 0.07) and isovaleric acid (P = 0.09) concentrations in the feces collected at day 134 of age. In the ileal digesta, low-RFI pigs had higher concentrations of total VFAs and acetic acid (P < 0.05). In the cecum, a strong influence of geographic location was observed for all VFAs measured, apart from butyric and isobutyric acids. Pigs in AT had higher concentrations of total VFAs, as well as higher acetic and propionic acid concentrations, compared to ROI1 pigs (P < 0.05). The pigs in AT had lower concentrations of isovaleric acid than did pigs from ROI1 (P < 0.05). The concentration of valeric acid was lower in low-RFI pigs from ROI1 than in their high-RFI counterparts and pigs from AT (P < 0.05).
FIG 6.
Effect of ranking pigs on RFI on volatile fatty acid (VFA) concentrations (millimoles per gram) in feces of ROI1 pigs at day 70 (A) and day 134 (B) of age and in ileal digesta (C) and cecal digesta (D) of ROI1 and AT pigs. VFAs that do not share a superscript letter (a and b) are significantly different from each other (P < 0.05).
Bacterial taxa correlated with RFI values and volatile fatty acid concentrations in pigs ranked on RFI reared at different geographic locations.
A correlation analysis was performed between the intestinal microbiota composition, at the phylum and genus levels, and RFI values and VFAs that differed significantly between RFI ranks. Those correlations that were significant are shown as a heat map in Fig. S5. None of the phyla or genera that were RFI associated in this study were significantly correlated with RFI value. At the phylum level, only the phylum Verrucomicrobia in the cecum was negatively correlated with a low RFI value (R = −0.56). At the genus level, Megasphaera (R = −0.57) and an uncultured genus from the Rhodospirillaceae (R = −0.54) in the ileum were correlated with a low RFI value.
Heat map showing Spearman correlations between bacterial phyla and genera and physiological parameters measured. * indicates significance at a P value of ≤0.05. Red indicates a negative correlation, and blue indicates a positive correlation. 1Volatile fatty acid concentrations were measured in Republic of Ireland batch 1 (ROI1) (feces at day 134 and ileal and cecal digesta) and Austria (AT) (cecal digesta) pigs only. 2Residual feed intake (RFI), calculated between days 70 and 120 of age for all pigs. Download FIG S5, DOCX file, 0.6 MB (571.4KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In relation to correlations between microbes and VFA concentrations, butyric acid in the feces collected from ROI1 pigs at day 134 of age was positively correlated with Mycoplasma (R = 0.69) but negatively correlated with Pyramidobacter (R = −0.59). Valeric acid was negatively correlated with an uncultured genus from Verrucomicrobia subdivision 5b (R = −0.70), but isovaleric acid was positively correlated with Collinsella (R = 0.67). In the ileum of ROI1 pigs, there was a negative correlation between acetic acid and Clostridium sensu stricto 1 (R = −0.70).
Salivary cortisol concentrations and hematological and biochemical parameters in pigs ranked on RFI reared at different geographic locations.
The salivary cortisol concentration, measured only in pigs from ROI1 just prior to slaughter (day 130 of age), tended to be lower in low-RFI (more-feed-efficient) pigs (P = 0.06) (Table 1). In pigs from ROI1, ROI2, and AT, no significant differences were observed according to RFI rank for serum biochemistry measures or for any of the hematological parameters measured (P > 0.05) (Table S1), and most values were within the normal ranges previously reported in growing pigs (24).
Hematological and serum biochemical1 parameters in pigs ranked on residual feed intake (RFI) in ROI2 and AT3 pigs. Download Table S1, DOCX file, 0.02 MB (23.3KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ileal immunological capacity and serum haptoglobin and cecal lipopolysaccharide levels in pigs ranked on RFI reared at different geographic locations.
No significant differences were observed between high- and low-RFI pigs from ROI1 for any of the ileal lymphocyte populations or cytokine concentrations measured from lamina propria lymphocytes (LPL) and intraepithelial lymphocytes (IEL), either control or mitogen stimulated (Table S2). Most of the lymphocyte populations in pigs from both RFI ranks were within the ranges previously reported for younger, healthy/control pigs (25, 26). Similarly, no significant differences were observed between RFI ranks across geographic locations for serum haptoglobin or cecal lipopolysaccharide (LPS) concentrations (P > 0.05) (Table S3).
Pooled ileal intraepithelial lymphocyte (IEL) and lamina propria lymphocyte (LPL) populations (percent),1 with or without mitogen stimulation, from ROI1 pigs ranked based on residual feed intake (RFI) and cytokine production (picograms per milliliter) from these cells. Download Table S2, DOCX file, 0.01 MB (14.3KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cecal lipopolysaccharide1 and serum haptoglobin2 concentrations in pigs ranked on residual feed intake (RFI) across geographic locations. Download Table S3, DOCX file, 0.01 MB (13.2KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Recently, a number of studies have demonstrated an association between FE and intestinal microbiota composition and function in pigs (12, 13, 16, 19, 21). However, very few of the FE-associated microbial taxa identified are common across studies. This is most likely due to the differences in diet, genetics, management strategies, and FE metrics used across rearing environments. Following on from this, the intestinal microbial profile of pigs ranked on RFI from three geographic locations was assessed in the present study but with these factors controlled in an attempt to find reliable cross-locational microbial biomarkers for FE in pigs. However, across geographic locations, few potential microbial biomarkers that were common to more than one site were identified, and most of these were different from those pinpointed in our previous study (12). The fact that ranking based on RFI was performed during different stages of the production cycle, i.e., days 70 to 120 of age in the present study compared to between weaning (∼28 days old) and day 154 of age in the previous study, could help to explain the discrepancies. Moreover, in the present study, none of the RFI-associated differences in intestinal microbiota composition found were common across all geographic locations. Similarly, studies in chickens have highlighted the challenge of finding common FE-associated microbes across locations (27) and even across batches from within the same location (28).
The limited cross-locational RFI-associated differences in the intestinal microbiota were perhaps due to variation in the core microbiome in pigs at each location, as illustrated by the location-specific diversity found in the PCoA plots. Similarly, Metzler-Zebeli et al. found that geographic location had more of an influence on intestinal size, structure, and functionality than RFI in the same pigs (29). It has previously been shown that poultry raised in different environments have different microbiota (28). However, to date, this has not been shown for pigs, although it is likely to occur considering that even the pen in which a pig is housed impacts the intestinal microbiota (13). This also seems to be the case in the present study, despite the fact that external factors, including diet specification, diet phases, genetics, and management protocols, were the same. It is also well known that the microbiome of the sow influences the microbiota composition of her progeny (30). In the present study, we attempted to account for this when looking at RFI-associated microbiota differences by selecting pigs of both high and low RFI from within the same litter. Nonetheless, litter origin is also likely to have contributed to the microbiota differences observed across geographic locations. In addition, it is likely that the housing environment and health status differences (31), however subtle, between locations also played a part in the differences observed. Overall, these maternal and environmental influences, as well as the interindividual variability in intestinal microbiota highlighted by Yang et al. (13), make it difficult to find reliable universal biomarkers for FE.
The overall intestinal microbiota composition showed a high abundance of Firmicutes and Bacteroidetes, which mirrors the fact that these are commonly identified as major constituents of the core pig microbiome (20). The lower relative abundance of the phylum TM7 and its genus “Candidatus Saccharimonas” observed in the low-RFI pigs from ROI2 at day 134 resembles previous findings from our group (12). This phylum is diverse, comprising ubiquitous members with potential proinflammatory activity previously found to be enriched in humans with inflammatory bowel disease (32).
Although none of the RFI-associated differences in microbiota composition were common to all geographic locations, seven taxa were found to differ in abundance between high- and low-RFI pigs at more than one geographic location or from the two different batches reared at the same location in ROI. However, within RFI rank, variation between pigs was observed; these taxa differences appear to explain, at least in part, differences in FE and could potentially be used as microbial biomarkers for FE. For example, the low-RFI-associated microbial taxa have a major role in core metabolism of carbohydrates. In feces collected from pigs at day 70 of age, the abundance of Mucispirillum, which is a mucin degrader, was found to be higher in low-RFI pigs from NI and AT. However, too much of a shift toward this genus could be harmful, causing disruption of the mucus layer (33). In feces collected from pigs at day 134 of age, Methanobrevibacter was more abundant in the highly efficient than in the poorly efficient pigs in both ROI batches. This genus has previously been positively correlated with fiber digestibility in pigs (34) and also plays an important role in methane production (35). Recently, it was found to be enriched in healthy humans compared to those with irritable bowel syndrome (36), and a link with leaner phenotypes in humans is also noteworthy (37). The phylum Lentisphaerae, enriched in low-RFI pigs, has previously been associated with improved health in humans, which might suggest a healthier gut microbiome here (38). This phylum has also been associated with improved FE and, specifically, weight gain, albeit in cattle (39), which would suggest a benefit in further investigating its role in improving FE in pigs. In the cecal digesta, the family Ruminococcaceae, which has a central role in the fermentation of carbohydrates, including cellulose (40, 41), and in the production of butyrate, was identified as an RFI-associated taxon in pigs from both AT and ROI2. Moreover, this family represents a core taxon with a relative abundance of 5 to 10%, and it has previously been linked with improved FE in pigs (13, 19). Finally, an uncultured bacterium from the RF16 family along with the RF16 family itself (from Bacteroidetes) were present at higher relative abundances in low-RFI pigs and exclusively found in low-RFI pigs. Although there is little information regarding the role of OTUs from this family within the intestinal community, they were enriched in pigs fed a diet high in complex carbohydrates (42), which could suggest an enriched biofunction of carbohydrate catabolism. In work previously conducted by this group, species from Bifidobacterium found exclusively in low-RFI pigs (12) were again found exclusively in low-RFI pigs in the present study. Additionally, two members of the Clostridiales, an uncultured member of the vadinBB60 family and the genus Cellulosilyticum, were found to be present only in low-RFI pigs here, and this was also found in previous work by our group (12). Moreover, Bacteroides in the feces at slaughter may be a potential microbial marker for RFI, as low-RFI pigs had a high relative abundance compared to high-RFI pigs in both this and our previous study (12).
The predicted RFI-associated microbial function data agreed with the differential compositional data, showing that most predicted pathways enriched in low-RFI pigs were related to core metabolism, including carbohydrate, energy, and nucleotide metabolism. Perhaps not surprisingly, this is in agreement with the findings of other recent studies on FE-associated microbiota in pigs (13, 19). The findings from a hepatic gene expression study with FE-divergent pigs, suggesting that carbohydrate biofunction is enriched in highly-feed-efficient pigs (7), also complement the microbial functionality found here, with the phosphorylation of inositol pathway being FE associated in both studies.
Microbial metabolites such as VFAs have a key role in modulation of host cells and also constitute an extra source of energy in the hindgut of the host. The concentration of VFAs present impacts the host phenotype, with, for example, higher concentrations of VFAs in the feces associated with obesity (43). The fact that fecal VFA concentrations were lower in low-RFI pigs could suggest increased colonic absorption, indicating better utilization of bacterial fermentation end products in the large intestine (44). However, the contribution of colonic VFAs to energy supply in young pigs, as was the case here, is low relative to that in adult pigs (e.g., sows or breeding boars). There appeared to be a strong influence of rearing environment on the cecal VFA concentrations measured in the present study, with ROI1 pigs having lower concentrations of most VFAs than AT pigs.
Apart from intestinal microbiota profiles and associated microbial metabolites, the level of salivary cortisol was the only other measure found to be RFI associated (lower in low-RFI pigs). This suggests that cortisol could be useful as a biomarker for improved FE in pigs, as has been previously suggested for cattle (3). Despite the absence of a link between ileal immune competence and FE in our study, an adequate number of ileal immune cells (i.e., within normal ranges previously found for healthy, control pigs [25, 26]) indicates an ability to fight off disease while maintaining optimum growth performance. Likewise, no RFI-associated differences were found for cecal concentrations of the bacterial endotoxin LPS, a potent immunogen, high serum concentrations of which have previously been linked to poor FE (45). Serum concentrations of the acute-phase protein haptoglobin also did not differ between RFI ranks in the present study, although they were previously found to be lower in low-RFI pigs (46). In addition, while previous work has shown that low-RFI pigs were more efficient in terms of producing lower but sufficient levels of blood lymphocytes, monocytes, and white blood cells than high-RFI pigs (5), no RFI-associated hematological differences were found in the present study.
Conclusions.
In conclusion, the FE-associated bacterial taxa consistently found across rearing environments may have a role to play in improving FE in pigs, mainly because of their importance in relation to carbohydrate metabolism. In addition, methanogenic members of the Archaea (Methanobrevibacter) are also likely to shape FE in pigs. In the future, these FE-associated taxa could potentially be used as probiotics or targeted by dietary means as a strategy for improving FE in pigs. Alternatively, they could be exploited as potential predictive biomarkers for porcine FE. However, the unculturable nature of some of these taxa together with location-specific findings highlight the challenges associated with the translation of our data into a set of reliable usable biomarkers and/or potential probiotics for pigs. Furthermore, the complex interplay between microbes within the gut ecosystem, in particular in the cecum, makes the cause-effect relationship intricate. Therefore, the microbial taxa identified in the present study cannot be interpreted as definitive determinants for FE in pigs, without additional studies. Moreover, consortia of bacterial species, rather than individual taxa, may be more likely to predict FE.
MATERIALS AND METHODS
Ethical approval.
The trials in ROI were approved by the animal ethics committees of Teagasc (TAEC9/2013) and Waterford Institute of Technology (13/CLS/02), and an experimental license (number AE1932/P004) was obtained from the Irish Health Products Regulatory Authority (HPRA). The trial in NI was conducted under project licenses PPL 2751 and PPL 2781, obtained from the Department of Health, Social Services and Public Safety (DHSSPS), which adhered to the Animals (Scientific Procedures) Act of 1986. The trial in AT was approved by the institutional ethics committee and the national authority according to paragraph 26 of the Law for Animal Experiments, Tierversuchsgesetz 2012 (TVG 2012) (GZ 68.205/0058-WF/II/3b/2014). All animal procedures were performed according to European Union Directive 2010/63/EU on the protection of animals used for scientific purposes (47).
Animal management, performance records, and sampling.
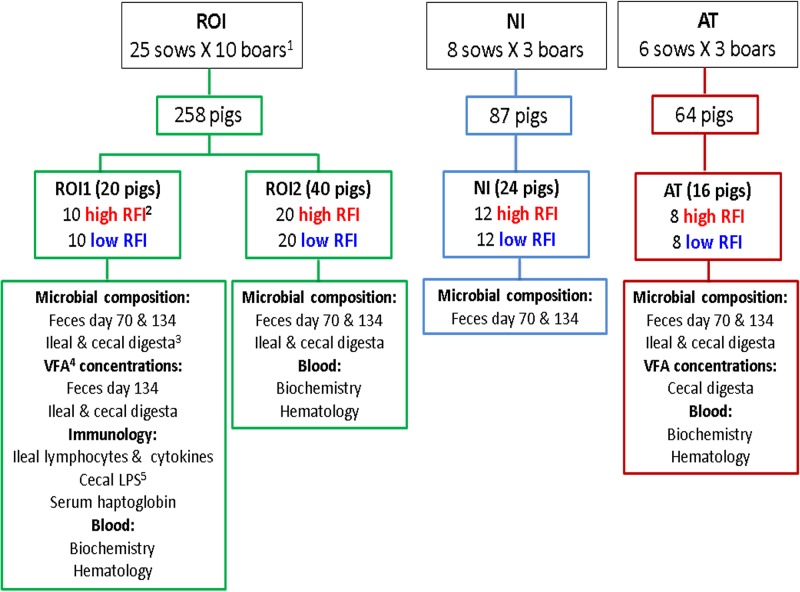
A schematic illustration depicting animal management, selection, and sample collection is shown in Fig. 7. Four trials were conducted across three geographic locations: ROI, NI, and AT. A total of 39 sows (Large White × Landrace), across all three locations (25 in ROI, 8 in NI, and 6 in AT), were blocked by body weight and inseminated with semen from individual boars (Maxgro; Hermitage Genetics Co., Kilkenny, Ireland). One common boar was used across the three locations, with an additional 10 boars specific to ROI, 3 specific to NI, and another 3 specific to AT.
FIG 7.
Schematic showing pig selection based on RFI and sampling procedure across geographic locations. 1one common boar was used across the three locations to minimize genetic variation; 2pigs were ranked on RFI at between 70 and 120 days of age; 3pigs were slaughtered at ∼134 days of age, and ileal and cecal digesta were collected; 4volatile fatty acids; 5lipopolysaccharide.
Subsequent offspring comprised 369 piglets: 218 in ROI (two batches, ROI1 [n = 80] and ROI2 [n = 138]), 87 in NI, and 64 in AT. All pigs were weaned at 28 ± 3 days of age, housed in groups of intact litters, and fed via feed intake recording equipment (FIRE) feeders (Schauer Agrotronic, Wels, Austria) (ROI1, 8 feeders with 7 to 10 pigs/pen; ROI2, 12 feeders with 10 to 13 pigs/pen; NI, 8 feeders with 10 to 12 pigs/pen; AT, 6 feeders with 9 to 12 pigs/pen). Pigs were fed the same sequence of diets (starter, link, weaner, and finisher), with the same diet specifications across all three geographic locations. Water and feed were provided on an ad libitum basis. The ingredients and chemical compositions of all experimental diets are shown in Table S4 in the supplemental material.
Composition and chemical analysis of diets used in the study (as-fed basis) (in grams per kilogram). Download Table S4, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual body weight was recorded every week, and voluntary feed intake was recorded daily between day 70 and day 120 of age, to calculate growth performance: ADFI, ADG, and FCE. Ultrasound measurements of back fat (BF) depth and muscle depth (MD) were taken at the 3rd and 4th last rib every week, using a Piglog 105 instrument (Carometec, Herlev, Denmark) for ROI, a Sonoscope A5 instrument (Keebomed, Mount Prospect, IL) for NI, and a Renco lean meater (Renco Corporation, Minneapolis, MN) for AT. All pigs were checked at least twice daily; any pigs showing signs of illness were treated as appropriate, and the details were recorded.
After day 120 of age, RFI was calculated for each pig (between days 70 and 120 of age), and extremes for RFI were selected within litter and gender. The RFI is a metric of FE that assesses the difference between the actual and predicted feed intake, with low-RFI animals being the most feed efficient. At each geographic location, RFI was calculated as the residuals from a least-squares regression model of ADFI on ADG, metabolic live weight (body weight0.75), gender, and all relevant two-way interactions as well as the effects of BF depth and MD. Pigs were ranked as having either high or low RFI, with a minimum spread of 2 standard deviations from the mean (within location and batch) between RFI ranks. A total of 100 pigs (60 from ROI [ROI1, 20; ROI2, 40], 24 from NI, and 16 from AT) were selected for sampling. A schematic depicting the selection of pigs is shown in Fig. 7.
Individual fecal samples were collected from all selected pigs following rectal stimulation on days 70 and 120 of age, immediately snap-frozen in liquid nitrogen, and stored at −80°C for subsequent microbiota and VFA analyses. Two weeks after selection of RFI extremes (approximately day 134 of age), pigs were slaughtered by CO2 stunning followed by exsanguination. Hot-carcass weight was recorded immediately following slaughter and was multiplied by 0.98 to obtain cold-carcass weight. The kill-out percentage was calculated as (cold carcass weight/body weight at slaughter) × 100. Back fat depth and MD at slaughter were measured 6 cm from the edge of the split back at the 3rd and 4th last ribs using a Hennessy grading probe (Hennessy and Chong, Auckland, New Zealand). Lean-meat yield was estimated according to the following formula: lean meat yield = 60.30 − 0.847 X1 + 0.147 X2 (where X1 is back fat depth [millimeters] and X2 is muscle depth [millimeters]). Digesta samples were collected from the terminal ileum (15 cm proximal to the ileocecal junction) and the cecum (terminal tip) from the selected pigs in ROI and AT. Samples were immediately snap-frozen in liquid nitrogen and stored at −80°C for subsequent microbiota and VFA analyses.
Salivary cortisol analysis.
On day 130 of age, saliva samples were collected from ROI1 pigs by allowing them to chew on a cotton bud (Salivette; Sarstedt Co., Wexford, Ireland). Cortisol concentrations were determined in duplicate using a high-sensitivity enzyme-linked immunosorbent assay (ELISA) kit (Salimetrics Europe Ltd., Suffolk, UK) according to the manufacturer’s instructions.
Hematology and serum biochemistry analyses.
Blood was collected from ROI and AT pigs during exsanguination at the slaughter plants for hematology and biochemistry analyses. For hematological analysis, blood was collected in vacuette tubes (from Labstock, Dublin, Ireland, for ROI and from Sarstedt, Nürnbrecht, Germany, for AT) containing EDTA to prevent clotting and analyzed within 4 h using a Beckman Coulter Ac T Diff analyzer (Beckman Coulter Ltd., High Wycombe, UK) for ROI pigs and a ProCyte dx hematology analyzer (Idexx Laboratories, Inc., Westbrook, ME, USA) for AT pigs.
For biochemical analysis, blood was collected from ROI pigs in vacuette tubes (Labstock) and allowed to clot at room temperature prior to centrifugation at 1,500 × g for 10 min. The serum was then collected and stored at −80°C for subsequent analysis. Serum samples were analyzed using an ABS Pentra 400 clinical chemistry analyzer (Horiba, ABX, North Hampton, UK) for total protein, blood urea nitrogen, cholesterol, glucose, triglycerides, creatinine, and creatine kinase. The analyzer was calibrated according to the manufacturer’s instructions, and every fifth sample was run in duplicate to determine analyzer accuracy. For AT pigs, blood was collected in serum collection tubes (Sarstedt, Nürnbrecht, Germany) and centrifuged at 1,811 × g for 10 min. Serum was then collected and stored at −80°C for subsequent analysis of levels of blood urea nitrogen, glucose, triglycerides, and cholesterol, which were determined by standard enzymatic colorimetric analysis using a clinical chemistry autoanalyzer, as outlined previously (48).
Immunological analyses.
Ileal tissue (2 to 3 cm) was collected 15 cm proximal to the ileocecal junction from ROI1 pigs at slaughter, placed in Hanks’ balanced salt solution (HBSS; Sigma-Aldrich Co., Wicklow, Ireland), and put on ice. The LPL and IEL were isolated from the ileal tissue as previously described (25) and pooled. Briefly, cells were isolated from the mucosa and submucosa of the porcine ileal tissue samples, suspended at 106 cells/ml in complete medium (Iscove’s modified Dulbecco’s medium [IMDM] plus GlutaMAX [Invitrogen], 20% heat-inactivated fetal bovine serum [FBS], 100 U/ml penicillin, and 100 mg/ml streptomycin), and transferred into 24-well plates (Sarstedt, Nürnbrecht, Germany) in quadruplicate. Cells were then pooled, treated with phosphate-buffered saline (PBS) (control) or stimulated with phorbol myristate acetate (PMA) (25 ng/ml; Sigma-Aldrich) plus ionomycin (I) (1 μg/ml; Sigma-Aldrich) (PMA+I) (mitogen stimulated) at 37°C in a 5% (vol/vol) CO2-humidified atmosphere for 4 to 5 h, and incubated for 18 h. Cells were centrifuged at 1,230 × g for 20 min, and supernatants were stored at −80°C until cytokine analysis as outlined below.
Immunophenotyping was then performed on the pooled LPL and IEL that were washed and resuspended in 2% FBS–PBS using a BD FACSCanto II flow cytometer (BD Biosciences, Devon, UK), with at least 50,000 events acquired and analyzed. Data were analyzed using FACSDiva software (BD Biosciences). Primary and secondary antibodies were added at concentrations determined by previous titration, and incubations were performed in the dark at room temperature for 15 min. Antibodies used were CD45 fluorescein isothiocyanate (FITC) (lymphocyte marker) (AbD Serotec/Bio-Rad, Kidlington, UK) to verify the white blood cell population identified by light scatter, anti-porcine B cell marker phycoerythrin (PE) (Abcam, Cambridge, UK), CD3 phycoerythrin/cyanine 5 (Cy5) (T cell marker) (Abcam), anti-porcine CD14 PE/Cy7 (monocyte marker) (Abcam), mouse anti-porcine CD4a FITC (BD Biosciences), mouse anti-porcine CD8a (BD Biosciences), purified rat anti-pig γδ T lymphocytes (BD Biosciences), and goat anti-rat FITC (AbD Serotec/Bio-Rad). Proportions of B cells, total T cells, and monocytes were calculated as percentages of the total white blood cells that were identified by light scatter and verified with CD45 antibody to be 63.51% ± 24.49% positive in high-RFI pigs and 51.53% ± 16.45% positive in low-RFI pigs. The T cell subsets were calculated based on the percentage of CD3-positive cells.
Concentrations of interleukin-4 (IL-4), IL-6, IL-8, and tumor necrosis factor alpha (TNF-α) were subsequently determined in the supernatants from pelleted immune cells treated with PBS and PMA+I using a multiplex ELISA (R&D Systems, Minneapolis, MN, USA) in triplicate according to the manufacturer’s instructions.
Haptoglobin in serum.
The concentration of haptoglobin was determined in serum samples collected from ROI1, AT, and NI pigs using a porcine-specific commercial ELISA kit (GenWay, San Diego, CA, USA) according to the manufacturer’s instructions. Serum samples were diluted 7,000- to 12,500-fold, depending on the actual haptoglobin concentrations in samples. Haptoglobin concentrations were determined in duplicate, and the intra-assay coefficient of variation was below 10%.
Lipopolysaccharides in cecal digesta.
Concentrations of cell-free LPS in cecal digesta collected from all pigs were determined using the pyrochrome Limulus amebocyte lysate (LAL) assay (Associates of Cape Cod, Inc., East Falmouth, MA, USA) as previously described (49). After dilution and deproteinization by heating, the supernatants were used in the assay. Changes in the optical density of samples at 405 nm were measured against calibration curves using Pyros EQS software (Associates of Cape Cod, Inc.) after the addition of pyrochrome LAL reagent and incubation at 37°C. Reactions were run in duplicate, and the intra-assay coefficient of variation was <10%.
Microbiota profiling.
Total DNA was extracted from fecal, ileal, and cecal samples using the QIAamp DNA stool minikit (Qiagen, Crawley, United Kingdom) according to the manufacturer’s instructions, apart from adding a beat beating step and increasing the lysis temperature to 95°C, to increase DNA yield (14).
Microbial profiling was performed using high-throughput sequencing of the V3-V4 region of the 16S rRNA gene (paired-end reads of 2 by 250 bp) on an Illumina MiSeq platform. The Illumina-recommended 16S metagenomic library preparation (Nextera) protocol was followed, except that the PCR mix volume was doubled in the first PCR step and the number of amplification cycles was increased to 30 instead of 25. Any samples with fewer than 40,000 post-quality reads on the MiSeq platform were removed from the analysis. Raw sequences were merged using Flash (with a minimum overlap of 30 bp and a minimum read length of 460 bp) and quality checked using the split libraries script (with default parameters) from the QIIME package, version 1.9.1. Reads with 97% sequence homology were clustered into OTUs by de novo OTU picking, and chimeras were removed with the 64-bit version of USEARCH (50). Subsequently, OTUs were aligned to the SILVA rRNA-specific database (version 111), and a phylogenetic tree was generated within QIIME. Alpha and beta diversity analyses were also performed using QIIME. Principal-coordinate analysis plots based on unweighted UniFrac distances were visualized using EMPeror v0.9.3-dev. Further downstream images were generated using the R package Phyloseq.
Microbial function prediction.
The predicted functionality of the microbiota for each sample based on 16S rRNA data was determined using PICRUSt according to RFI rank, geographic location, and sample type. PICRUSt is a software tool which uses the 16S rRNA gene sequence to predict the functionality of microorganisms (51). Prediction of functions was inferred based on KEGG (52) and Clusters of Orthologous Groups of Proteins (COG) annotations, which assign annotations according to the database. The KO functions that were not bacterium related or for which the relative abundance was <0.01% for all of the RFI ranks were dismissed.
Volatile fatty acid concentrations in feces and digesta.
Concentrations of VFAs (acetic, butyric, isobutyric, propionic, valeric, and isovaleric acids) were measured in triplicate in feces collected from pigs on days 70 and 134 of age and in ileal and cecal digesta from the ROI1 pigs, and in cecal digesta from AT pigs using gas chromatography (GC) (Agilent 5890 gas chromatograph for ROI1 and Fisons gas chromatograph model 8060 MS DPFC for AT). For samples analyzed in ROI, ∼8 g of sample was weighed, diluted with 5% trichloroacetic acid (TCA) (2.5× the weight of the sample), and centrifuged at 1,800 × g at 4°C for 10 min. One and a half milliliters of the resultant supernatant and 1.5 ml of the internal standard (0.043 M 3-methylvaleric acid in 0.15 M oxalic acid; Sigma-Aldrich) were mixed gently, and the mixture was filtered through a 0.45-μm filter (VWR International Ltd., Dublin, Ireland) into a labeled 8-mm amber GC vial (Antech Solutions Ltd., Waterford, Ireland) and stored at −80°C until analyzed, as previously described (12, 53).
For cecal digesta analyzed in AT, aliquots of 1 g were thawed on ice and mixed with 0.2 ml metaphosphoric acid (4.3 M; Sigma-Aldrich), 1 ml of double-distilled water, and 200 μl of the internal standard (0.024 M 4-methylvaleric acid in 4.3 M phosphoric acid; Sigma-Aldrich), and the mixture was centrifuged at 3,148 × g for 10 min. The clear supernatant was filtered through a 0.45-μm filter (VWR International Ltd.) into a labeled 8-mm GC vial and analyzed as previously described (54).
Statistical analyses.
Growth performance parameters (weight, ADG, ADFI, and FCE) were analyzed using a mixed linear model in SAS 9.4. Fixed effects included in the model were RFI rank, geographic location, gender, and time period as well as their interactions. While adjusting for gender, sow was included as a random effect, and a repeated-measures model was used to describe correlations between time periods (weekly weight gain and feed intake recordings). Physiological parameters measured at only one time point (i.e., ileal lymphocyte populations, cytokine data, salivary cortisol concentrations, biochemical and hematological parameters, and haptoglobin and VFA concentrations) were also analyzed using a mixed linear model; the above-mentioned fixed effects were included in the model. Comparisons of means were undertaken using a Tukey correction for multiple testing. Residual diagnostics were made to ensure that the assumptions of the analysis were met.
Statistical differences for microbiota abundance at the phylum, family, and genus levels were calculated in R using the SILVA 16S-specific database (version 111) and estimated using the Kruskal-Wallis test for independent samples and the Wilcoxon rank test for paired samples. Corrections for multiple comparisons were made using the Benjamini-Hochberg method (55). Each geographic location was statistically analyzed separately.
The RFI values and VFA concentrations found to differ between low- and high-RFI pigs were correlated with the taxonomic relative abundance at the phylum and genus levels (at all time points for each geographic location where appropriate). Spearman correlation values were calculated in SAS, using the PROC CORR procedure, and P values were adjusted using the stepdown Bonferroni test.
For all statistical analyses conducted, significance was set at a P value of ≤0.05.
Data availability.
The raw 16S rRNA gene sequence data generated from this study are available in the European Nucleotide Archive under accession number PRJEB22209.
ACKNOWLEDGMENTS
We thank Tomas Ryan and the farm staff in the Pig Development Department at Teagasc Moorepark (ROI) for assistance with pig management. We also acknowledge work placement students assisting with the pig trials at Teagasc as well as Orla O’Donovan (Waterford Institute of Technology), John Butler (Meso Scale Diagnostics), Vicki Murray, and Gwynneth Halley for assistance with laboratory work. We gratefully acknowledge staff of the research pig farm Medau, Suchitra Sharma and Anita Dockner (Institute of Animal Nutrition and Functional Plant Compounds), and colleagues from the University Clinic for Swine, all at the University of Veterinary Medicine, Vienna, Austria (AT), for assisting with animal and laboratory work. We acknowledge research farm staff at the Agri-Food and Biosciences Institute (AFBI), Hillsborough (NI), for assistance with animal work conducted there.
P.G.L., B.U.M.-Z., E.M., G.E.G., and D.P.B. conceived the study and designed the experiments. U.M.M., S.G.B., P.G.L., B.U.M.-Z., and E.M. conducted the animal study and collected intestinal samples. S.G.B. and D.P.B. analyzed and ranked the pigs on RFI. U.M.M., B.U.M.-Z., M.L.P., H.R., F.C., and S.G.B. performed laboratory analysis. M.H. provided assistance regarding hematological and immunological laboratory work. N.R. analyzed the FACS data. Bioinformatics and microbiota statistical analyses were performed by O.O. U.M.M. statistically analyzed animal growth performance and physiological data and, together with T.C., G.E.G., and P.G.L., interpreted the microbial data. U.M.M. and T.C. wrote the manuscript, and G.E.G., D.P.B., B.U.M.-Z., E.M., P.D.C., F.C., O.O., and P.G.L. revised the manuscript. All authors read and approved the final version of the manuscript.
We declare that we have no competing interests.
The research leading to these results received funding from the European Union’s Seventh Framework Programme (ECO-FCE project number 311794) for research, technological development, and demonstration independently of any commercial input, financial or otherwise. U.M.M. was funded by the Teagasc Walsh Fellowship program. Gwynneth Halley, who assisted with laboratory work, was funded by a Society for Applied Microbiology Students into Work grant.
REFERENCES
- 1.Teagasc. 2017. Pig herd performance report. Teagasc Moorepark, Fermoy, Ireland. [Google Scholar]
- 2.Hillmann E, Schrader L, Mayer C, Gygax L. 2008. Effects of weight, temperature and behaviour on the circadian rhythm of salivary cortisol in growing pigs. Animal 2:405–409. doi: 10.1017/S1751731107001279. [DOI] [PubMed] [Google Scholar]
- 3.Richardson E, Herd R, Archer J, Arthur P. 2004. Metabolic differences in Angus steers divergently selected for residual feed intake. Aust J Exp Agric 44:441–452. doi: 10.1071/EA02219. [DOI] [Google Scholar]
- 4.Pond WG, Mersmann HJ. 2001. Biology of the domestic pig, 2nd ed Cornell University Press, Ithaca, NY. [Google Scholar]
- 5.Mpetile Z, Young J, Gabler N, Dekkers J, Tuggle C. 2015. Assessing peripheral blood cell profile of Yorkshire pigs divergently selected for residual feed intake. J Anim Sci 93:892–899. doi: 10.2527/jas.2014-8132. [DOI] [PubMed] [Google Scholar]
- 6.Grubbs J, Dekkers J, Huff-Lonergan E, Tuggle C, Lonergan S. 2016. Identification of potential serum biomarkers to predict feed efficiency in young pigs. J Anim Sci 94:1482–1492. doi: 10.2527/jas.2015-9692. [DOI] [PubMed] [Google Scholar]
- 7.Reyer H, Oster M, Magowan E, Dannenberger D, Ponsuksili S, Wimmers K. 2017. Strategies towards improved feed efficiency in pigs comprise molecular shifts in hepatic lipid and carbohydrate metabolism. Int J Mol Sci 18:E1674. doi: 10.3390/ijms18081674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mpetile Z. 2014. Effect of divergent selection for residual feed intake on immune system of Yorkshire pigs. MS thesis. Iowa State University, Ames, IA. [Google Scholar]
- 9.Vigors S, O’Doherty JV, Kelly AK, O’Shea CJ, Sweeney T. 2016. The effect of divergence in feed efficiency on the intestinal microbiota and the intestinal immune response in both unchallenged and lipopolysaccharide challenged ileal and colonic explants. PLoS One 11:e0148145. doi: 10.1371/journal.pone.0148145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Nguyen YT, Nettleton D, Dekkers JC, Tuggle CK. 2016. Post-weaning blood transcriptomic differences between Yorkshire pigs divergently selected for residual feed intake. BMC Genomics 17:73. doi: 10.1186/s12864-016-2395-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramayo-Caldas Y, Mach N, Lepage P, Levenez F, Denis C, Lemonnier G, Leplat J-J, Billon Y, Berri M, Doré J, Rogel-Gaillard C, Estellé J. 2016. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J 10:2973. doi: 10.1038/ismej.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormack UM, Curião T, Buzoianu SG, Prieto ML, Ryan T, Varley P, Crispie F, Magowan E, Metzler-Zebeli BU, Berry D, O’Sullivan O, Cotter PD, Gardiner GE, Lawlor PG. 2017. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl Environ Microbiol 83:e00380-17. doi: 10.1128/AEM.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Huang X, Fang S, He M, Zhao Y, Wu Z, Yang M, Zhang Z, Chen C, Huang L. 2017. Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front Microbiol 8:1555. doi: 10.3389/fmicb.2017.01555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buzoianu SG, Walsh MC, Rea MC, O’Sullivan O, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. 2012. High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt maize) for 31 days. Appl Environ Microbiol 78:4217–4224. doi: 10.1128/AEM.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HB, Isaacson RE. 2015. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet Microbiol 177:242–251. doi: 10.1016/j.vetmic.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Mach N, Berri M, Estellé J, Levenez F, Lemonnier G, Denis C, Leplat J-J, Chevaleyre C, Billon Y, Doré J, Rogel-Gaillard C, Lepage P. 2015. Early life establishment of the swine gut microbiome and impact on host phenotypes. Environ Microbiol Rep 7:554–569. doi: 10.1111/1758-2229.12285. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W, Wang Y, Liu S, Huang J, Zhai Z, He C, Ding J, Wang J, Wang H, Fan W, Zhao J, Meng H. 2015. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One 10:e0117441. doi: 10.1371/journal.pone.0117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, Huang X, Fang S, Xin W, Huang L, Chen C. 2016. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci Rep 6:27427. doi: 10.1038/srep27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quan J, Cai G, Ye J, Yang M, Ding R, Wang X, Zheng E, Fu D, Li S, Zhou S, Liu D, Yang J, Wu Z. 2018. A global comparison of the microbiome compositions of three gut locations in commercial pigs with extreme feed conversion ratios. Sci Rep 8:4536. doi: 10.1038/s41598-018-22692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao L, Estellé J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, Liang S, Pedersen AØ, Kjeldsen NJ, Liu C, Maguin E, Doré J, Pons N, Le Chatelier E, Prifti E, Li J, Jia H, Liu X, Xu X, Ehrlich SD, Madsen L, Kristiansen K, Rogel-Gaillard C, Wang J. 2016. A reference gene catalogue of the pig gut microbiome. Nat Microbiol 1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 21.Tan Z, Yang T, Wang Y, Xing K, Zhang F, Zhao X, Ao H, Chen S, Liu J, Wang C. 2017. Metagenomic analysis of cecal microbiome identified microbiota and functional capacities associated with feed efficiency in Landrace finishing pigs. Front Microbiol 8:1546. doi: 10.3389/fmicb.2017.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klem TB, Bleken E, Morberg H, Thoresen SI, Framstad T. 2010. Hematologic and biochemical reference intervals for Norwegian crossbreed grower pigs. Vet Clin Pathol 39:221–226. doi: 10.1111/j.1939-165X.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 25.Walsh MC, Buzoianu SG, Gardiner GE, Rea MC, Gelencsér E, Jánosi A, Epstein MM, Ross RP, Lawlor PG. 2011. Fate of transgenic DNA from orally administered Bt MON810 maize and effects on immune response and growth in pigs. PLoS One 6:e27177. doi: 10.1371/journal.pone.0027177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nofrarias M, Manzanilla E, Pujols J, Gibert X, Majo N, Segalés J, Gasa J. 2006. Effects of spray-dried porcine plasma and plant extracts on intestinal morphology and on leukocyte cell subsets of weaned pigs. J Anim Sci 84:2735–2742. doi: 10.2527/jas.2005-414. [DOI] [PubMed] [Google Scholar]
- 27.Siegerstetter S-C, Schmitz-Esser S, Magowan E, Wetzels SU, Zebeli Q, Lawlor PG, O’Connell NE, Metzler-Zebeli BU. 2017. Intestinal microbiota profiles associated with low and high residual feed intake in chickens across two geographical locations. PLoS One 12:e0187766. doi: 10.1371/journal.pone.0187766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley D, Hughes RJ, Geier MS, Moore RJ. 2016. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol 7:187. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzler-Zebeli BU, Lawlor PG, Magowan E, McCormack UM, Curiao T, Hollmann M, Ertl R, Aschenbach JR, Zebeli Q. 2017. Finishing pigs that are divergent in feed efficiency show small differences in intestinal functionality and structure. PLoS One 12:e0174917. doi: 10.1371/journal.pone.0174917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson CL, Wang B, Holmes AJ. 2008. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. ISME J 2:739–748. doi: 10.1038/ismej.2008.29. [DOI] [PubMed] [Google Scholar]
- 31.Patience JF, Rossoni-Serão MC, Gutiérrez NA. 2015. A review of feed efficiency in swine: biology and application. J Anim Sci Biotechnol 6:33. doi: 10.1186/s40104-015-0031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuehbacher T, Rehman A, Lepage P, Hellmig S, Fölsch UR, Schreiber S, Ott SJ. 2008. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol 57:1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- 33.Belzer C, Gerber GK, Roeselers G, Delaney M, DuBois A, Liu Q, Belavusava V, Yeliseyev V, Houseman A, Onderdonk A, Cavanaugh C, Bry L. 2014. Dynamics of the microbiota in response to host infection. PLoS One 9:e95534. doi: 10.1371/journal.pone.0095534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu Q, Li P, Hao S, Zhang Y, Kim SW, Li H, Ma X, Gao S, He L, Wu W, Huang X, Hua J, Zhou B, Huang R. 2015. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci Rep 5:9938. doi: 10.1038/srep09938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pimentel M, Gunsalus RP, Rao SS, Zhang H. 2012. Methanogens in human health and disease. Am J Gastroenterol Suppl 1:28. doi: 10.1038/ajgsup.2012.6. [DOI] [Google Scholar]
- 36.Pozuelo M, Panda S, Santiago A, Mendez S, Accarino A, Santos J, Guarner F, Azpiroz F, Manichanh C. 2015. Reduction of butyrate- and methane-producing microorganisms in patients with irritable bowel syndrome. Sci Rep 5:12693. doi: 10.1038/srep12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. 2012. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes (Lond) 36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, Hu Y, Li J, Liu Y. 2015. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep 5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myer PR, Smith TP, Wells JE, Kuehn LA, Freetly HC. 2015. Rumen microbiome from steers differing in feed efficiency. PLoS One 10:e0129174. doi: 10.1371/journal.pone.0129174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE, Louis P, Flint HJ, de Vos WM. 2014. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J 8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J, Monagas M, Jang S, Molokin A, Harnly JM, Urban JF, Solano-Aguilar G, Chen P. 2015. A high fat, high cholesterol diet leads to changes in metabolite patterns in pigs—a metabolomic study. Food Chem 173:171–178. doi: 10.1016/j.foodchem.2014.09.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandes J, Su W, Rahat-Rozenbloom S, Wolever T, Comelli E. 2014. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes 4:e121. doi: 10.1038/nutd.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams BA, Verstegen MW, Tamminga S. 2001. Fermentation in the large intestine of single-stomached animals and its relationship to animal health. Nutr Res Rev 14:207–228. doi: 10.1079/NRR200127. [DOI] [PubMed] [Google Scholar]
- 45.Mani V, Weber TE, Baumgard LH, Gabler NK. 2012. Growth and development symposium: endotoxin, inflammation, and intestinal function in livestock. J Anim Sci 90:1452–1465. doi: 10.2527/jas.2011-4627. [DOI] [PubMed] [Google Scholar]
- 46.Mani V, Harris A, Keating AF, Weber TE, Dekkers JC, Gabler NK. 2013. Intestinal integrity, endotoxin transport and detoxification in pigs divergently selected for residual feed intake. J Anim Sci 91:2141–2150. doi: 10.2527/jas.2012-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.European Union. 2010. Directive 2010/63/EU of the European Parliament and of the council of 22 September 2010 on the protection of animals used for scientific purposes.
- 48.Metzler-Zebeli BU, Eberspächer E, Grüll D, Kowalczyk L, Molnar T, Zebeli Q. 2015. Enzymatically modified starch ameliorates postprandial serum triglycerides and lipid metabolome in growing pigs. PLoS One 10:e0130553. doi: 10.1371/journal.pone.0130553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzler-Zebeli BU, Mann E, Schmitz-Esser S, Wagner M, Ritzmann M, Zebeli Q. 2013. Changing dietary calcium-phosphorus level and cereal source selectively alters abundance of bacteria and metabolites in the upper gastrointestinal tracts of weaned pigs. Appl Environ Microbiol 79:7264–7272. doi: 10.1128/AEM.02691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. 2010. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prieto ML, O’Sullivan L, Tan SP, McLoughlin P, Hughes H, O’Donovan O, Rea MC, Kent RM, Cassidy JP, Gardiner GE, Lawlor PG. 2014. Evaluation of the efficacy and safety of a marine-derived Bacillus strain for use as an in-feed probiotic for newly weaned pigs. PLoS One 9:e88599. doi: 10.1371/journal.pone.0088599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metzler-Zebeli BU, Schmitz-Esser S, Mann E, Grüll D, Molnar T, Zebeli Q. 2015. Adaptation of the cecal bacterial microbiome of growing pigs in response to resistant starch type 4. Appl Environ Microbiol 81:8489–8499. doi: 10.1128/AEM.02756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alpha diversity of the fecal microbiota of pigs ranked on residual feed intake (RFI) at 70 days of age (A) and 134 days of age (B) across all three geographic locations. ROI, Republic of Ireland; NI, Northern Ireland; AT, Austria. Download FIG S1, DOCX file, 0.3 MB (271.4KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Median relative abundance (percent) of bacterial phyla present in pigs according to residual feed intake (RFI) rank across geographic locations for all fecal time points and intestinal sites. 1Other (no BLAST hits/uncultured). * indicates a significant difference observed for that phylum between high- and low-RFI pigs within each sample type and geographic location (P ≤ 0.05). These RFI-associated differences are as follows: for feces on day 70, ↑ Deferribacteres (AT), ↑ SHA.109 (ROI2), and ↓ Verrucomicrobia (ROI1); for feces on day 134, ↓ candidate division TM7 (ROI2), ↓ Firmicutes (ROI1), ↑ Euryarchaeota (ROI1), and ↑ Lentisphaerae (ROI1 and ROI2) and Verrucomicrobia (ROI1); for ileum, ↓ Cyanobacteria (ROI2), ↓ Firmicutes (ROI2), and ↑ Planctomycetes (ROI2); and for cecum, ↑ Cyanobacteria (AT), ↓ Firmicutes (ROI1), ↑ Proteobacteria (ROI1), and ↑ Verrucomicrobia (ROI2). Arrows indicate higher (↑) or lower (↓) relative abundances in low-RFI pigs. Download FIG S2, DOCX file, 0.2 MB (249.9KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genera found exclusively in all low-residual-feed-intake (RFI)-ranked pigs across all geographic locations in feces at 70 days (A) and 134 days (B) of age and in ileal (C) and cecal (D) digesta. Data from all geographic locations were taken together. unc, uncultured; und, unidentified. Taxa present at a <0.1% relative abundance were not included in the analysis. Download FIG S3, DOCX file, 0.8 MB (796.3KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Predicted functionality of intestinal microbiota for high- and low-RFI pigs across geographic locations and by sample type (feces [A] and cecal digesta [B]). Pathways shown are those found to be differentially abundant between high- and low-RFI pigs (P < 0.05). ROI, Republic of Ireland; NI, Northern Ireland; AT, Austria. Pathways were grouped into major functional categories as follows: M1, carbohydrate metabolism; M2, metabolism of cofactors and vitamins; M3, energy metabolism; M4, lipid metabolism; M5, amino acid metabolism; M6, glycan biosynthesis and metabolism; M7, biosynthesis of other secondary metabolites; M8, metabolism of terpenoids and polyketides; M9, xenobiotic biodegradation and metabolism; M10, metabolism of other amino acids; M11, nucleotide metabolism; E, environmental information processing; G, genetic information processing; C, cell motility. Download FIG S4, DOCX file, 0.1 MB (126.2KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat map showing Spearman correlations between bacterial phyla and genera and physiological parameters measured. * indicates significance at a P value of ≤0.05. Red indicates a negative correlation, and blue indicates a positive correlation. 1Volatile fatty acid concentrations were measured in Republic of Ireland batch 1 (ROI1) (feces at day 134 and ileal and cecal digesta) and Austria (AT) (cecal digesta) pigs only. 2Residual feed intake (RFI), calculated between days 70 and 120 of age for all pigs. Download FIG S5, DOCX file, 0.6 MB (571.4KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hematological and serum biochemical1 parameters in pigs ranked on residual feed intake (RFI) in ROI2 and AT3 pigs. Download Table S1, DOCX file, 0.02 MB (23.3KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pooled ileal intraepithelial lymphocyte (IEL) and lamina propria lymphocyte (LPL) populations (percent),1 with or without mitogen stimulation, from ROI1 pigs ranked based on residual feed intake (RFI) and cytokine production (picograms per milliliter) from these cells. Download Table S2, DOCX file, 0.01 MB (14.3KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cecal lipopolysaccharide1 and serum haptoglobin2 concentrations in pigs ranked on residual feed intake (RFI) across geographic locations. Download Table S3, DOCX file, 0.01 MB (13.2KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Composition and chemical analysis of diets used in the study (as-fed basis) (in grams per kilogram). Download Table S4, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2019 McCormack et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The raw 16S rRNA gene sequence data generated from this study are available in the European Nucleotide Archive under accession number PRJEB22209.