Summary
Lymphatic vessels (LVs) are important in the regulation of tissue fluid homeostasis and the pathogenesis of tumor progression. We investigated the innervation of LVs and the response to agonists and antagonists of the autonomic nervous system in vivo. While skin-draining collecting LVs express muscarinic, α1- and β2-adrenergic receptors on lymphatic endothelial cells and smooth muscle cells, intestinal lacteals express only β-adrenergic receptors and muscarinic receptors on their smooth muscle cells. Quantitative in vivo near-infrared imaging of the exposed flank-collecting LV revealed that muscarinic and α1-adrenergic agonists increased LV contractility, whereas activation of β2-adrenergic receptors inhibited contractility and initiated nitric oxide (NO)-dependent vasodilation. Tumor-draining LVs were expanded and showed a higher innervation density and contractility that was reduced by treatment with atropine, phentolamine, and, most potently, isoproterenol. These findings likely have clinical implications given the impact of lymphatic fluid drainage on intratumoral fluid pressure and thus drug delivery.
Keywords: lymphatic vessels, contractility, autonomic nervous system, Ca2+ imaging, tumor-draining lymphatics
Graphical Abstract
Highlights
-
•
Murine lymphatic vessels are innervated in an organ-specific manner
-
•
α1-adrenergic and muscarinic agents enhance lymphatic contractility in vivo
-
•
β2-adrenergic agents reduce lymphatic contractility
-
•
Tumor-draining lymphatic vessels have increased innervation and contractility
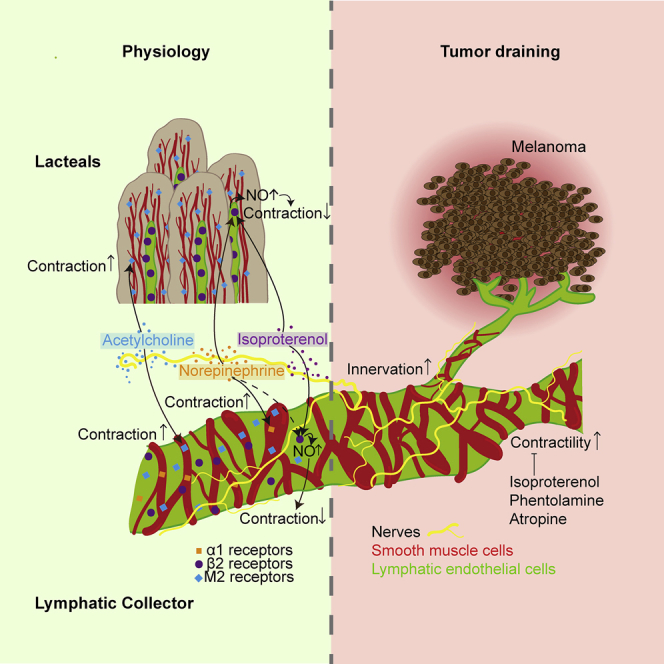
Bachmann et al. use in vivo imaging to demonstrate that muscarinic and α1-adrenergic agonists increase lymphatic contractility, whereas activation of β2-adrenergic receptors inhibits contractility and mediates NO-dependent vasodilation. Tumor-draining lymphatics have a denser innervation and increased contractility, which can be diminished by β2-adrenergic agonists.
Introduction
The lymphatic vascular system is responsible for tissue fluid homeostasis, lipid absorption, and mediation of immune responses. It initiates as a network of blind-ended capillaries throughout most organs of the body. These capillaries have discontinuous tight junctions in order to take up free fluid and solutes from the interstitium or lipids from the intestinal tract. Antigen-presenting cells, as well as antigen itself, enter the lymphatic system at the level of the capillaries before being conducted into collecting lymphatic vessels (LVs). Concurrent with an increasing diameter, collecting LVs are covered by a thin layer of smooth muscle cells that are required for the intrinsic contractility of lymphatic collectors (Gashev, 2008, Zawieja et al., 1993). Together with intraluminal valves, their active contractions ensure unidirectional lymph flow against an increasing pressure gradient. Collecting LVs conduct lymph through one or more lymph nodes until it ultimately reaches the bloodstream at the subclavian veins.
The mechanisms regulating collecting LV contractility are not entirely understood. Generally, there are both extrinsic and intrinsic factors that influence lymph flow. Extrinsic factors include the passive movement of lymph by contractions of striated muscles (e.g., during walking), as well as by nearby arteries, which through vasomotion can conduct their pulsation to LVs (Gashev, 2008, Zawieja et al., 1993). With regard to intrinsic mechanisms, mechanical parameters such as shear stress and intraluminal pressure gradients within the LVs exert important influences on smooth muscle cell behavior (reviewed in Scallan et al., 2016). Vasoregulatory factors, including nitric oxide (NO), also have a strong influence on the contractility. NO is an effective vasodilator and has negative inotropic effects on LVs (Scallan and Davis, 2013). The release of NO is induced by high shear stress within the vessel (Kornuta et al., 2015, Kunert et al., 2015) or by a variety of signaling molecules such as vascular endothelial growth factor A (VEGF-A) (Lahdenranta et al., 2009). Surprisingly, the potential regulation of LV contractility by the autonomic nervous system has not received much attention thus far.
Nerves in close proximity to LVs have been described in humans (D’Andrea et al., 2013, D’Andrea et al., 2015) and large animals, including cows and dogs (Ohhashi et al., 1982, Todd and Bernard, 1973), and ex vivo studies have reported responses of the contraction pattern of isolated LVs to several neurotransmitters (McHale et al., 1980, McHale, 1990, Ohhashi and Azuma, 1986). However, the results of these ex vivo studies are somewhat contradictory. While previous studies consistently found activation of lymphatic contractility by the α-adrenergic nervous system, some attributed these effects solely to the activation of α1 receptors (Benoit, 1997), whereas others reported that only α2 receptors, but not α1 receptors, are needed (Hashimoto et al., 1994). Similarly, conflicting results have been reported regarding the role of the muscarinic nervous system, since its agonist acetylcholine (ACh) induced LV relaxation (Ferguson, 1992, Hashimoto et al., 1994, Ohhashi and Takahashi, 1991) as well as contraction (Ohhashi et al., 1978). Recently, ex vivo studies using human tissue found a strong induction of LV contracting activity by the adrenergic agonist norepinephrine (NE) and an inhibitory effect of the muscarinic antagonist atropine (Telinius et al., 2010, Telinius et al., 2014). Overall, most of the current knowledge about the interaction between the autonomic nervous system and LV contractility stems from ex vivo studies, while in vivo studies are still lacking (Choe et al., 2015, Ono et al., 2000). Importantly, virtually nothing is known about potentially different responses of LVs to neurotransmitters in pathological conditions such as chronic inflammation and cancer, where LVs play major pathogenetic roles (Christiansen and Detmar, 2011, Stacker et al., 2014).
In this study, we investigated in detail the role of the autonomic nervous system in the regulation of LV function under physiological and pathological conditions. To this end, we first studied the presence of different subtypes of nerves on murine LVs and analyzed the expression profile of neurotransmitter receptors on lymphatic endothelial cells (LECs) in vitro and in situ. These studies revealed organ-specific expression patterns that might explain previously reported diverging effects of neurotransmitters in different organs. We then applied quantitative in vivo near-infrared imaging techniques to directly investigate the effects of a broad spectrum of agonists and antagonists of the autonomic nervous system on the contraction frequency and amplitude of collecting LVs under physiological conditions. These investigations were accompanied by live Ca2+ imaging studies in vitro and in vivo. Finally, we investigated the effects of selected neurotransmitters on tumor-draining LVs. Our results reveal that under physiological conditions, contractions of collecting LVs are activated via α-adrenergic and muscarinic receptors, while they are inhibited via activation of β2-adrenergic receptors. Importantly, we found that tumor-draining LVs exhibited more active contractility and that this contraction activity can be inhibited by distinct agonists and antagonists of the autonomic nervous system.
Results
Neurotransmitter Receptors Are Expressed on LVs in an Organ-Specific Manner
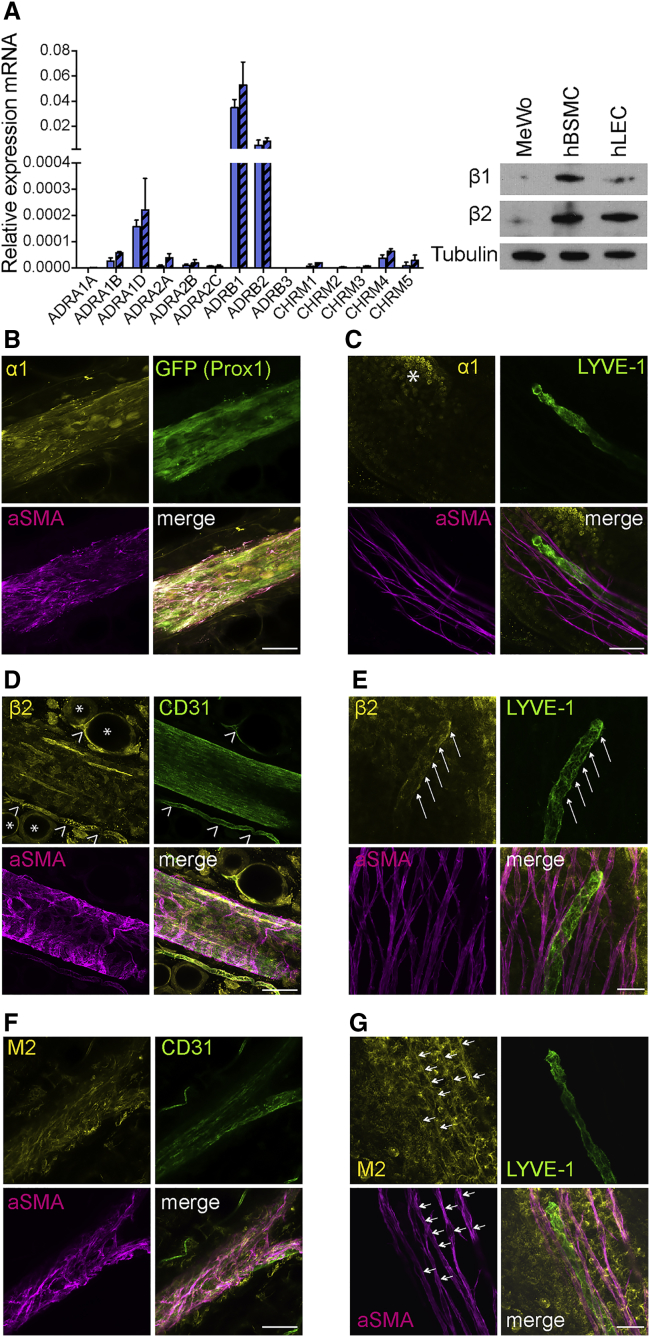
We first investigated the mRNA expression levels of all known adrenergic and muscarinic neurotransmitter receptors in five different primary human dermal LEC lines. LECs specifically expressed β1- and β2-adrenergic receptors (Figure 1A shows two cell lines, and Figure S1A the additional three lines). These findings were confirmed at the protein level by western blot (Figure 1A), with human bronchial smooth muscle cells (hBSMCs) serving as a positive control and the tumor cell line MeWo as a negative control. In a next step, we investigated the neurotransmitter receptors expressed in mouse tissue. Whole-mount staining of the flank collecting LVs and lacteal villi in the small intestine was performed. Adrenergic α1 receptors were present on CD31+ LECs as well as on the αSMA+ smooth muscle cells of the flank collector (Figure 1B). In contrast to the collecting LVs, neither the intestinal LYVE-1+ lacteals nor their associated smooth muscle cells (αSMA+) demonstrated expression of α1 receptors (Figure 1C). Interestingly, the mesenteric collecting LVs lacked expression of α1 receptors (Figure S1F). Whole-mount staining revealed no detectable expression of β1 receptors on either type of LVs (Figures S1B and S1C), whereas the β2 receptors were expressed on LECs of all vessel types, lacteals, the flank, and mesenteric collectors (Figures 1D, 1E, and S1G). β2-Receptor expression was also detected on smooth muscle cells of the flank and mesenteric collecting LVs, but not on the smooth muscle cells surrounding the lacteal.
Figure 1.
Neurotransmitter Receptors Are Expressed on LVs in an Organ-Specific Manner
(A) qPCR-based detection of adrenergic β1 and β2 receptor (ADRB1 and ADRB2) mRNA in two primary human LEC lines (shown as blue and blue-striped columns). Expression was confirmed at the protein level using western blots with human bronchial smooth muscle cells (hBSMCs) as a positive control and the tumor cell line MeWo as a negative control.
(B, D, and F) Whole-mount staining of flank collector showing α1 (B), β2 (D), and M2 (F) receptors expressed on CD31+ LECs and αSMA+ smooth muscle cells. Asterisks indicate β2-positive CD31+ blood vessels, and open arrowheads indicate β2-positive adipocytes.
(C, E, and G) LYVE-1+ lacteals and their associated αSMA+ cells do not express α1 receptors (C), but the lacteals express β2 receptors (E, arrows), and the associated smooth muscle cells express M2 receptors (G, arrows). The α1 receptor expressing enterocytes (Baglole et al., 2006) are clearly visible (asterisk in C). Images were obtained using confocal microscopy.
Scale bars represent 50 μm (B, D, and F) and 25 μm (C, E, and G). Data in (A) are presented as mean ± SD. See also Figure S1.
Even though it was not detected at the RNA level in cultured human LECs, we next performed whole-mount staining for the muscarinic receptor M2. We found expression of M2 receptors by smooth muscle cells of the flank and mesenteric collector and of the lacteal villi, whereas the LECs themselves only showed expression of M2 receptors in the collecting vessels (Figures 1F, 1G, and S1H). As a second method, flow-cytometry studies on freshly isolated flank collector LECs were performed (Figures S1I and S1J). Thereby, we were able to confirm the expression of α1 and β2 receptors. Additionally, but to a lower extent than β2 receptors, we also detected positive expression of β1 receptors. Due to the lack of a suitable antibody, we were unable to assess M2 receptor expression using this approach. Taken together, these findings reveal that LVs specifically express receptors in an organ-site-dependent manner that may enable them to respond to neurotransmitters released from the autonomic nervous system.
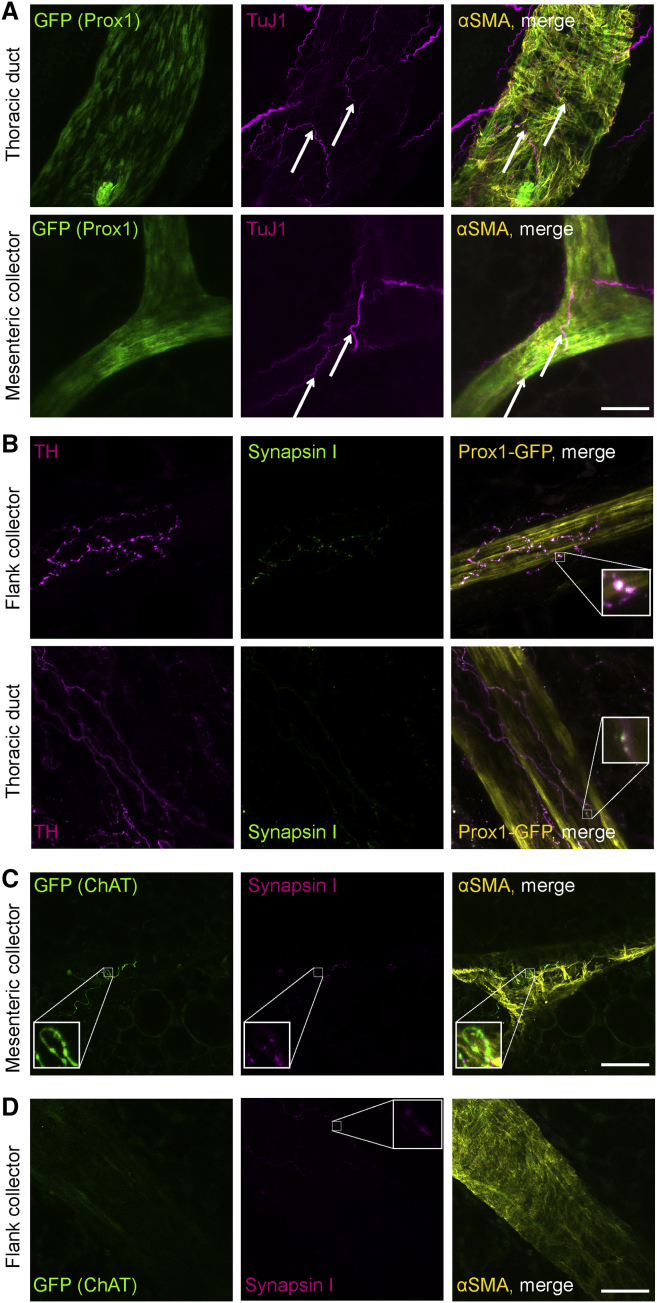
Nerves Are Closely Associated with LVs
In order to exert neural effects on LVs, the presence of synapses formed by nerves is required, as these represent the source of neurotransmitters. Whole-mount staining of collecting LVs from different organ sites of Prox1-GFP mice for the pan-neural marker TuJ1 detected nerves running along LVs (Figure 2A). Tyrosine hydroxylase (TH)-positive sympathetic nerves were detected, forming synapsin-I-positive synapses on lymphatic collectors (Figure 2B). Parasympathetic nerves are characterized by their expression of choline acetyltransferase (ChAT). We therefore performed whole-mount staining of ChAT-GFP mice in combination with synapsin I staining to detect synapses. Mesenteric lymphatic collectors demonstrated innervation with parasympathetic nerves (Figure 2C, zoom-in), whereas no ChAT+ neural structures were detected on collectors at other organ sites, such as the flank (Figure 2D).
Figure 2.
Nerves Are Closely Associated with LVs
(A) Lymphatic collectors of Prox1-GFP mice are closely associated with pan-neural marker TuJ1+ nerves (arrows).
(B) Tyrosine hydroxylase (TH)+ sympathetic nerves run along lymphatic collectors of Prox1-GFP mice and form synapses (synapsin I, zoom-in).
(C) Choline acetyltransferase (ChAT) GFP+ parasympathetic nerves, which form synapses (synapsin I), accompany mesenteric collectors (zoom-in).
(D) Collectors other than mesenteric collectors show no ChAT+ innervation. Synapsin I staining shows the presence of other types of nerves (zoom-in). Images were obtained using confocal microscopy.
Scale bars, 50 μm. See also Figure S2.
Whole-mount staining showed a close proximity of nerves (TuJ1+), distinguishable as either sympathetic (TH+) or parasympathetic (ChAT+), with intestinal lacteals (Figures S2A–S2C). Synapsin I staining revealed that the nerves adjacent to lacteals also formed synapses (Figures S2D and S2E).
Overall, these results demonstrate that autonomic nerves of different subtypes are present near LVs, indicating the possibility of a direct influence on LV function.
LECs Are Direct Targets of Neurotransmitters
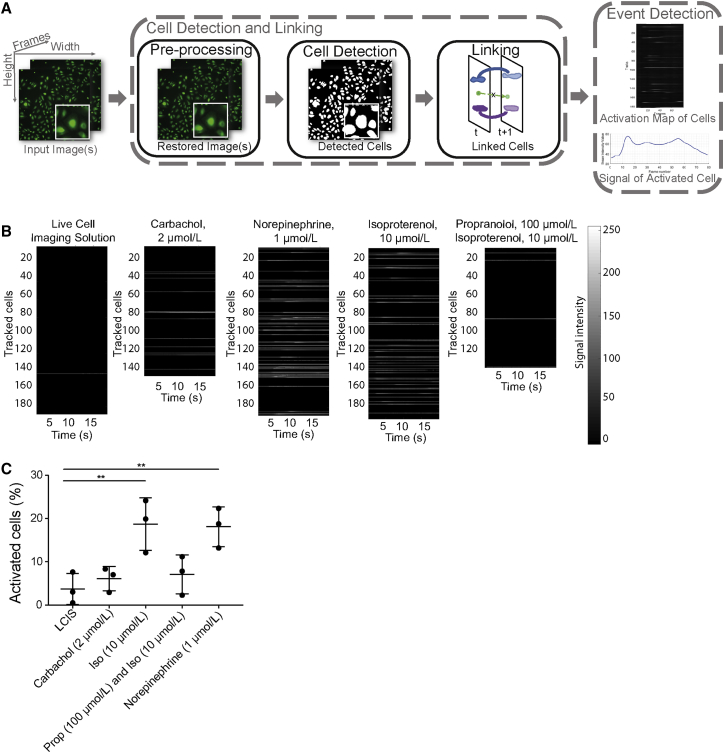
To evaluate if LECs might demonstrate downstream signaling in response to neurotransmitters, we next investigated their Ca2+ response in vitro as well as in vivo. This approach was chosen because activation of most muscarinic receptors and some adrenergic receptors leads to an increase of intracellular calcium as a second messenger. In vitro imaging of human LECs using Fluo8H was quantified with a self-written algorithm that is explained in STAR Methods and Figure 3A. We found that intracellular Ca2+ was increased upon treatment with NE (1 μmol/L) or isoproterenol (10 μmol/L) (Figures 3B and S3). To evaluate the specificity of this experiment, pretreatment of cells with an antagonist was performed. Propranolol (100 μmol/L) strongly inhibited the increase of intracellular Ca2+ induced by isoproterenol (Figure 3B). The quantification of the percentage of activated cells was in line with the visual quantification of Ca2+ activation (Figure 3C).
Figure 3.
LECs Are Direct Targets of Neurotransmitters In Vitro
(A) Visualization of the algorithm made for Ca2+ quantification. After enhancing the quality of each frame (image pre-processing), each cell is detected separately and linked between consecutive frames in order to track it over the whole duration of the video (Cell Detection and Linking). For each tracked cell (Track), the fluorescence intensity over time is computed. If the signal changes notoriously over time, an activation event is recorded (Event Detection). A detailed description can be found in STAR Methods.
(B) Quantification of in vitro Ca2+ imaging of human primary LECs using Fluo8H in response to different neural agonists. Each horizontal line of the bar represents a tracked cell; only activated cells are shown in gray. Fluo8H intensity is shown in grayscale, proportional to intracellular Ca2+.
(C) Quantification of the percentage of cells reacting with an increase of Ca2+ upon treatment with different agonists. LCIS, live cell imaging solution; Iso, isoproterenol; Prop, propranolol. ∗∗p ≤ 0.01. Data in are presented as mean ± SD; n = average per well of cells. Individual cells can be seen Figure S3.
See also Figure S3.
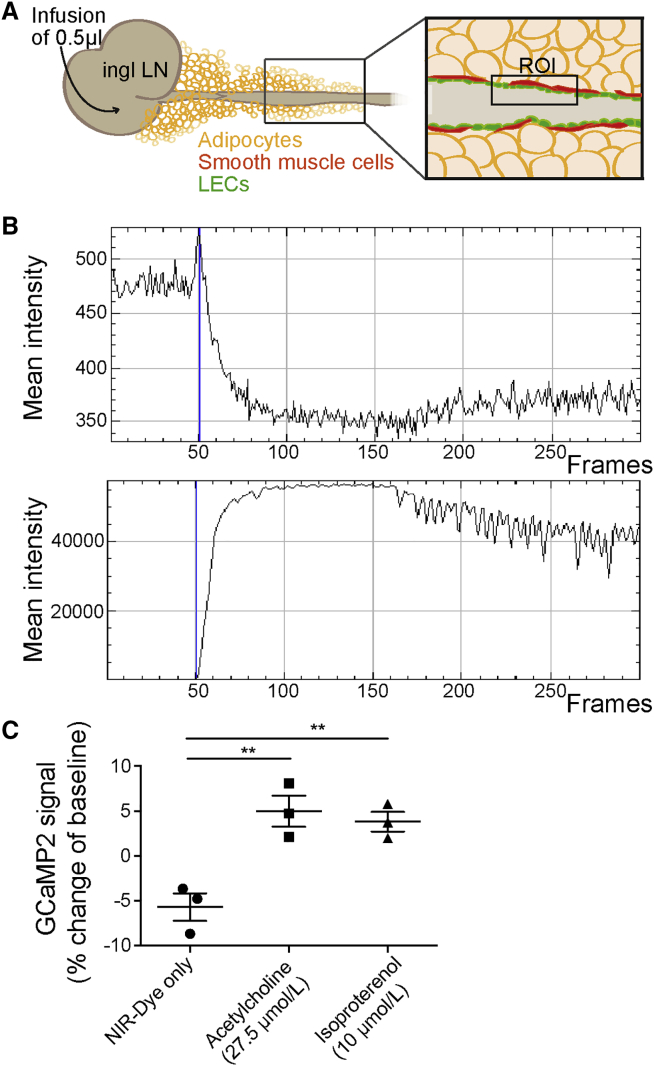
For in vivo imaging, Cx40 GCaMP2 transgenic mice were used (Tallini et al., 2007). These mice express the calcium indicator GCaMP2 under control of the connexin 40 promoter and provide a dynamic readout of GFP signal intensity that is proportional to the intracellular calcium concentration. Connexin 40 is expressed in arterial endothelial cells and to a lesser degree in LECs. In these mice, we infused 0.5 μL of 10 μmol/L 20 kDa PEG-IRDye680 (P20D680) near-infrared (NIR) tracer (Proulx et al., 2013) either alone or mixed with adrenergic or muscarinic agonists into the inguinal lymph node (Figure 4A) and evaluated a region of interest over the efferent collecting LV. The tracer was used to define the time point when the infused agonist arrives in the region of interest. Ca2+ fluxes in the LECs were analyzed by measuring changes in the GFP intensity before arrival of the tracer compared to the time point of arrival (Figure 4B). When tracer alone was infused, the GFP intensity dropped due to the volume-induced dilation of the collecting vessel (Figure 4C). When the muscarinic agonist ACh (27.5 μmol/L) was infused with the tracer, a clear transient spike in GFP intensity was observed (Figure 4C). The same was observed for the β adrenergic agonist isoproterenol (10 μmol/L).
Figure 4.
LECs Are Direct Targets of Neurotransmitters In Vivo
(A) Schematic overview of in vivo Ca2+ imaging using Cx40 GCaMP2 mice. The flank collector was used for imaging. Ingl LN, inguinal lymph node; ROI, region of interest.
(B) Representative example of GCaMP2 intensity (top) measurement while a mixture of pegylated NIR dye and acetylcholine was infused into the inguinal lymph node. At time of arrival of the dye (bottom), there is a visible peak of the GCaMP2 expression (frame 50, blue line).
(C) The GCaMP2 signal intensity at time point of arrival of the dye in the imaged section was normalized to the signal intensity before arrival of the dye. Differences are shown as percent change of the baseline signal intensity. ∗∗p ≤ 0.01. Data are presented as mean ± SD; n = 3 mice.
Together, these results indicate that LECs might represent direct targets of neurotransmitters.
The Autonomic Nervous System Regulates Lymphatic Contractility In Vivo
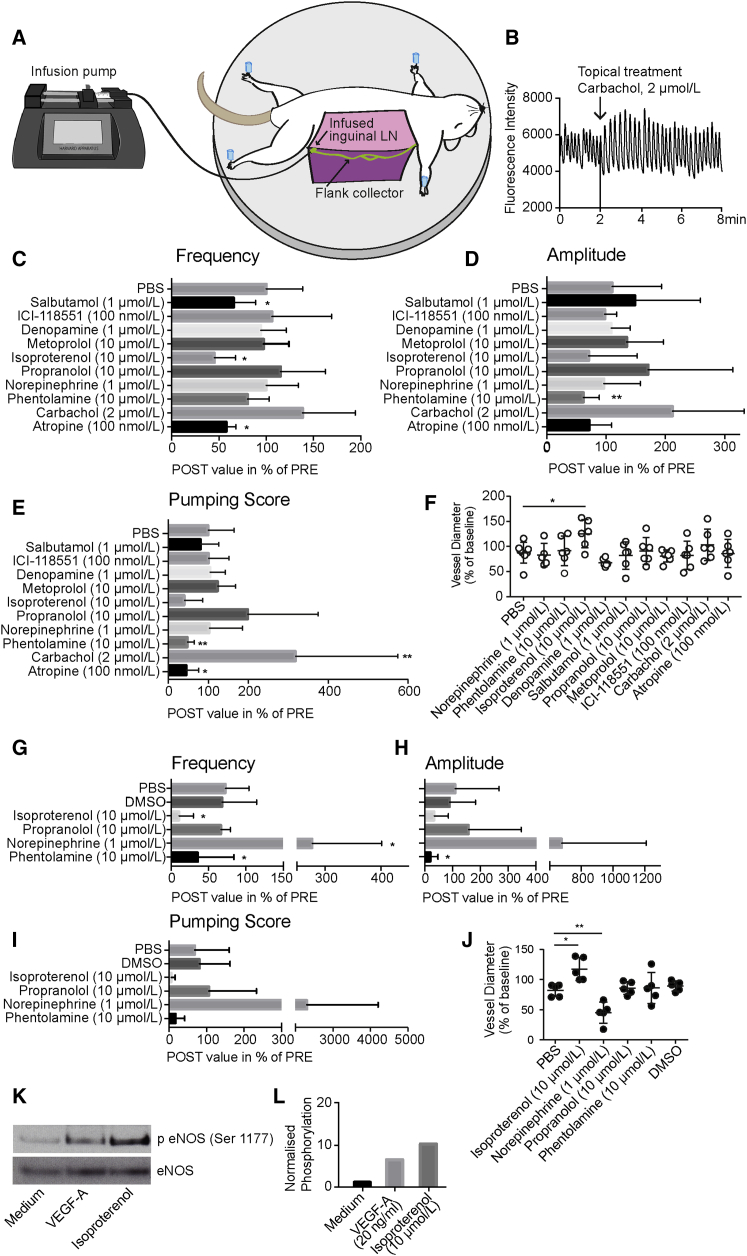
To investigate the regulation of LV contractility by the autonomic nervous system in vivo, we used a previously described method (Chong et al., 2016). In brief, after the infusion of 0.5 μL of the P20D680 NIR dye into the inguinal lymph node, the flank collector becomes visible and contractile activity can be imaged (Figure 5A). Over a period of 8 min, the fluorescent intensity of the tracer in the vessel is imaged, which corresponds to diameter changes of the vessel (Figure 5B) and enables quantification of the contractility using a custom-made algorithm (Chong et al., 2016). After the first 2 min, a topical treatment was applied. We quantified contraction frequency, amplitude, and pumping score, which is the product of frequency and amplitude, after treatment with a range of muscarinic, α-adrenergic, and β-adrenergic agonists and antagonists.
Figure 5.
The Autonomic Nervous System Regulates Lymphatic Contractility In Vivo
(A) Schematic overview of in vivo imaging of flank collector contractility.
(B) Representative example of measured NIR-dye intensity in the flank collector over time; topical treatment was done at time point 2 min.
(C–E) Effects of topical treatment with neural agonists and antagonists on lymphatic contraction frequency (C), amplitude (D), and pumping score (E). Shown are the post-treatment values as a percentage of pretreatment values for mice under injection anesthesia (ketamine, xylazine, and acepromazine).
(G–I) Same is shown for mice under isoflurane: changes in frequency (G), amplitude (H) and pumping score (I).
(F and J) Quantification of the tonic contraction of the flank collector upon treatment with neural agonists and antagonists under injection anesthesia (F; ketamine, xylazine, acepromazine) and isoflurane (J).
(K and L) Isoproterenol-mediated phosphorylation of eNOS, as assessed (K) and quantified (L) by western blot. VEGF-A was used as positive control. The western blot shown is a representative example of at least three independent experiments.
∗p < 0.05, ∗∗p ≤ 0.01. Data are presented as mean ± SD. (C–F) n = 8 vessels for PBS, and n = 6 vessels for all other treatments. (G–J) n = 4 vessels per treatment. (K and L) n = 1. See also Figure S5.
Comparing the post-treatment period (minutes 4–7) with the pretreatment period (first 2 min), potent effects of several agonist and antagonists were found (Figures 5C–5E, S5C, and S5D). There was a strong increase of contractile activity upon treatment with the muscarinic agonist carbachol (2 μmol/L; amplitude 212.62% of pre-value; pumping score 317.67% of pre-value). Conversely, antagonization of the muscarinic system by atropine (100 nmol/L) reduced the contraction activity significantly (frequency 57.63% of pre-value; pumping score 44.26% of pre-value). For the adrenergic system, we detected reduced contractility upon treatment with the unspecific β-adrenergic agonist isoproterenol (10 μmol/L; frequency 45.1% of pre-value). To determine if this effect was mediated by β1 or β2 receptors, the specific β1 agonist denopamine and the specific β2 agonist salbutamol (both at 1 μmol/L) were studied. Only salbutamol induced an inhibitory effect (frequency 65.29% of pre-value), indicating that the effect is predominantly mediated by β2 receptors. Treatment with agonists at two additional concentrations (0.1 and 10 μM) also showed no significant effect of the β1 agonist, while the higher concentration of the β2 agonist showed a strong trend of reduced frequency (62.88% of pre-value) (Figure S5B). The unspecific β adrenergic antagonist propranolol (10 μmol/L) exhibited a trend to activate the contraction activity (frequency 115.07% of pre-value, amplitude 171.55% of pre-value, pumping score 197.86% of pre-value). To ensure specificity of the β2 agonist, competitive blocking studies were performed. After the exposed vessels were treated at t = 2 min, either PBS or the β2 antagonist were added at t = 4 min, and the vessels’ post-treatment was assessed at t = 6–9 min. While PBS treatment had no effect on the reduction of contractility, treatment with the β2 antagonist neutralized the effect of the initial β2 agonist, resulting in no change in contractility (Figure S5A).
The α-adrenergic system has been reported to induce contractility in isolated LVs (Ono et al., 2000, Telinius et al., 2014). In line with these results, we found a reduced contraction activity upon antagonization of the α adrenergic system using phentolamine (10 μmol/L; amplitude 62.42% of pre-value; pumping score 46.79% of pre-value; Figures 5D and 5E). Phentolamine was dissolved in DMSO, reaching a final DMSO concentration of 0.01%. In previous studies, we confirmed that this concentration of DMSO had no effect on the contraction activity (Chong et al., 2016). Surprisingly, we could not detect any effect of the α-adrenergic (and, with much lower affinity, β-adrenergic) agonist NE. This lack of response was most probably due to the usage of the injection anesthesia (ketamine, xylazine, and acepromazine) that contained α agonists itself. To investigate this hypothesis, we repeated the adrenergic treatments with mice under isoflurane anesthesia (Figures 5G–5I and S6). We confirmed the inhibiting effect of the α antagonist phentolamine (10 μmol/L; amplitude 19.87% of pre-value), while the DMSO control showed no significant effect. More importantly, we found a strong activating effect of NE (1 μmol/L; frequency 277.29% of pre-value), indicating that the lack of effect in the previous experiments was due to the choice of anesthesia. To investigate whether the results observed after treatment with other mediators might also have been affected by the injection anesthesia, we next treated collecting LVs in mice anesthetized with isoflurane with β adrenergic compounds. Isoproterenol (10 μmol/L) induced comparable effects (frequency 10.32% of pre-value) and propranolol (10 μmol/L) had only minor effects on the contractility (frequency 66.63% of pre-value) (Figures 5G–5I).
Overall, these in vivo experiments identified a strong impact of neurotransmitters on the contraction activity of lymphatic collectors in mice.
β-Adrenergic Stimulation Regulates LV Tone by eNOS Phosphorylation
Besides the effects on phasic contractions, we next investigated whether treatment with neurotransmitters might also mediate changes in vessel tone, since previous studies indicated that NE increases the vessel tone of lymphatic collectors from various organ sites by inducing vasoconstriction (Dobbins, 1992, Ono et al., 2000, Takahashi et al., 1990). While mice under injection anesthesia did not show any effect of NE on the vessel tone (Figure 5F), in line with the lack of any response in phasic contractions, the vessel tone was increased in mice under isoflurane anesthesia (Figure 5J). We observed a decrease in vessel tone (vasodilation) upon treatment with isoproterenol, independent of the anesthesia regimen used. As a potential mediator of vasodilation, we next investigated the phosphorylation state of eNOS in LECs upon treatment with β-adrenergic agonists by western blot and found an increased phosphorylation after treatment with isoproterenol (Figures 5K and 5L). Thus, treatment with isoproterenol may lead to increased NO production in LECs. The released NO acts on the nearby smooth muscle cells, induces their relaxation, and, therefore, leads to a dilation of the collecting LV.
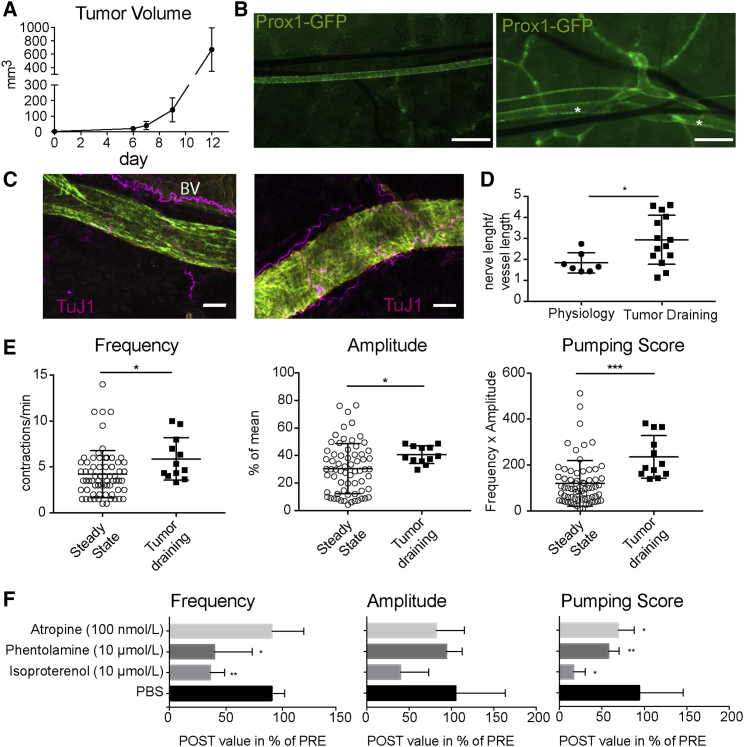
Tumor-Draining Lymphatic Collectors Contract More Actively
B16F10 melanomas were grown in the flank skin of Prox-1 GFP mice (Figure 6A). When tumors reached 12 mm in one dimension, tumor-draining LVs were imaged in vivo. As reported by many studies (Padera et al., 2002, Skobe et al., 2001, Stacker et al., 2014), we found that several LVs were expanded in the vicinity of the flank collector, demonstrating the direct activating effect of the tumor on the lymphatic vasculature (Figure 6B). TuJ1 staining of the affected vessels showed a denser innervation (Figures 6C and 6D). In line with this finding, the tumor-draining flank collectors had a higher baseline contraction activity (Figure 6E). Based on the results obtained under physiological conditions, we next investigated whether treatment with distinct neurotransmitters might inhibit the contractility of tumor-draining LVs. Indeed, atropine, phentolamine, and, most potently, isoproterenol reduced the pumping score significantly (Figures 6F and S6).
Figure 6.
Tumor-Draining Lymphatic Collectors Contract More Actively
(A) Growth of intradermally injected B16F10 tumors was measured over time using a caliper (n = 7 mice).
(B) Overview picture of the flank area in a naive (left) and a tumor-bearing Prox1-GFP mouse (right). In the vicinity of the flank collector (asterisk), additional lymphatic collectors were formed draining the tumor.
(C) Compared to physiological LVs (left), tumor-draining vessels show denser innervation, visualized with the pan-neural marker TuJ1 (Prox1-GFP staining, green; αSMA, yellow).
(D) Quantification of innervation density (nerve length/vessel length).
(E) Tumor-draining LVs show higher baseline activity than the respective vessels in non-tumor-bearing mice with regard to frequency, amplitude, and pumping score.
(F) Contractility of tumor-draining vessels can be inhibited by atropine, phentolamine, and isoproterenol.
∗p < 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001. Scale bars represent 500 μm (B) and 25 μm (C). Data represent mean ± SD, and dots in (D) and (E) represent vessels. (F) n = 4 vessels per treatment. BV, blood vessel. See also Figure S6.
Discussion
In this study, we provide the first comprehensive in vivo characterization of the regulation of LV contractility by agonists and antagonists of the autonomic nervous system. Our findings reveal that both smooth muscle cells and endothelial cells of LVs express distinct neurotransmitter receptors and that nerves and synapses are in contact with these vessels. We identified specific functions of the sympathetic and the parasympathetic nervous system with regard to LV contraction and confirmed the distinct activation of signaling in LECs by Ca2+ imaging in vitro and in vivo. The studies of tumor-draining LVs revealed their increased innervation and contraction activity and also identified neurotransmitters that may inhibit the activity of tumor-draining LVs.
In our investigation of the expression of neurotransmitter receptors in LECs, we found that cultured human LECs only express β-adrenergic receptors, whereas, in addition, both α1-adrenergic and M2-muscarinic receptors were found on murine collecting LVs by whole-mount staining and flow-cytometry analysis. The lack of expression of several of these receptors in vitro might indicate a change of the expression profile of LECs under the in vitro culture conditions, in agreement with previous studies (Amatschek et al., 2007, Wick et al., 2007). In addition, these differences might also represent vessel-type-specific expression patterns, since the cultured human LECs were isolated from the skin and might be more representative of LECs originating from capillaries and not from collecting LVs. Consistent with this concept, murine lacteal LECs, representing cells of specialized intestinal lymphatic capillaries, also only express β-adrenergic receptors. However, species-associated differences between humans and mice cannot be ruled out as well. Confirmation for the differently expressed receptors in LECs in vitro compared to the in vivo situation was obtained by the Ca2+ imaging studies, since cultured cells only responded to β agonists, whereas there was also a response to the muscarinic agonist ACh in vivo.
Our finding of an organ-specific expression pattern of neurotransmitter receptors may provide an explanation for the previously reported discrepancies of NE effects on LV function. While contractility was increased in ex vivo preparations of peripheral collecting LVs from different species (including human) after exposure to NE (Dobbins, 1992, Ono et al., 2000, Takahashi et al., 1990, Telinius et al., 2014), in agreement with our in vivo studies in mice, a recent study reported a diminished contraction activity of intestinal lacteals after NE treatment (Choe et al., 2015). Our findings indicate that the opposing effects of the same adrenergic agonist can be explained by the absence of α-adrenergic receptors on the lacteals. Thus, NE (being an α-adrenergic agonist and, with much lower affinity, also a β-adrenergic agonist) likely acts as a β agonist in lacteals, leading to reduced contractility, whereas it promotes contractions in lymphatic collectors via the α-adrenergic receptors. This organ-specific response to NE makes sense physiologically, since under systemic stress conditions, enhanced return of peripheral lymph fluid together with reduced energy expense for the gastrointestinal lipid transport might be advantageous.
A major conclusion from our studies is that muscarinic and α-adrenergic agonists activate the contractility of collecting LVs, whereas β2-adrenergic agonists reduce the contraction frequency. The activating effect mediated by NE is in agreement with previous ex vivo studies (Hashimoto et al., 1994, Igarashi et al., 1998, McHale, 1992, McHale et al., 1980, Telinius et al., 2014). However, whereas some of these studies indicated that this effect is mediated via α2 receptors (Hashimoto et al., 1994), others suggested that both α1 and α2 receptors are involved (Igarashi et al., 1998). Our findings that there are profound differences in the expression patterns of neurotransmitter receptors on different types of LVs indicate that some of the reported differences might relate to organ-specific responses to neurotransmitters. In support of this concept, LVs isolated from the liver and lungs of pigs showed decreased contraction and were dilated upon ACh treatment (Ferguson, 1992, Hashimoto et al., 1994), similarly to isolated thoracic ducts of dogs (Ohhashi and Takahashi, 1991). By contrast, in rat mesenteric vessels (Fang et al., 2007), operation-induced vagotomy resulted in a reduced contraction rate, indicating an activating role of the parasympathetic nervous system with its main neurotransmitter, ACh, in line with the contraction-inducing effects of ACh in bovine mesenteric LVs (Ohhashi et al., 1978). Another possible explanation for these opposing effects of ACh might be the ratio of muscarinic receptors expressed on LECs versus smooth muscle cells. If muscarinic receptors are expressed predominantly on smooth muscle cells, then their activation likely induces LV contraction, whereas binding of ACh on muscarinic receptors on LECs induces NO release (Scallan and Davis, 2013), resulting in vessel dilation and reduced contractility. Thus, a detailed characterization of the neurotransmitter receptor expression profiles of human LVs from different organs, as well as from model organisms such as the mouse, is needed to better understand the organ- and species-specific effects of the autonomic nervous system.
Our data indicate that the choice of anesthesia is of great importance for the design and interpretation of functional studies of LV contractility. We were unable to detect the activating effects of the α-agonist NE when using an injection anesthesia regimen that contained the α-agonist xylazine, thus masking the effects of NE on LV contractility. By contrast, NE induced an increased LV contraction frequency under isoflurane anesthesia. However, this anesthesia regimen turned out to be associated with a general depression of LV contractions. Thus, a thorough investigation of different anesthesia regimens for functional in vivo studies of LVs would be helpful to ensure a better cross comparison of published work.
The results of our in vivo studies are in agreement with the improvement of secondary lymphedema after therapeutic inhibition of the sympathetic nervous system in patients after breast cancer surgery (E. Choi et al., 2015) and gynecologic-cancer-related surgery (Woo et al., 2013). In these clinical settings, the treatment blocks the complete sympathetic nervous system locally. Thus, one might speculate that the parasympathetic nervous system, which is left intact after the treatment, promotes LV contractions, resulting in improvement of the lymphedema.
One should keep in mind that the autonomic nervous system influences both lymphatic and blood vessels, indicating that therapies aimed at modulating blood vessel functions might also impact the function of the lymphatic vascular system. As an example, the long-term usage of β blockers for hypertension might negatively influence the balanced system of physiological lymphatic contractions. Similarly, while β2 agonists are successfully used to induce bronchodilation in asthma patients, they might reduce the contractility of lymphatics of the lung and increase NO release by LECs, potentially leading to a reduced fluid clearance of the lung tissue.
To our knowledge, our studies identified for the first time that the expanded tumor-draining LVs have a higher innervation density, which might contribute to the observed increase in contractility of the preexisting and newly formed LVs. The increased contractility of tumor-draining LVs is in agreement with a previous study in mice where increased smooth muscle cell coverage was also detected (Gogineni et al., 2013). Together, these findings reveal that tumors not only induce LEC proliferation and lymphangiogenesis but also profoundly change the anatomical structure and function of tumor-draining LVs. Our findings that specific agonists and antagonists of the autonomic nervous system reduce the contractility of tumor-draining LVs might be of potential relevance for clinical cancer therapy. In the future, targeted long-term treatment of tumor-draining LVs would be needed to investigate whether the inhibition of lymphatic contractility ultimately results in reduced tumor growth and metastasis. At the same time, these studies could also address possible negative effects of an increased interstitial pressure induced by such long-term treatments, including potentially reduced tumor tissue penetration of therapeutic compounds.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| alpha-1 adrenergic receptor | Santa Cruz | cat# sc-1477; RRID:AB_630854 |
| beta-1 adrenergic receptor | abcam | cat# ab3442; RRID:AB_10890808 |
| beta-2 adrenergic receptor | abcam | cat# ab13989; RRID:AB_300816 |
| muscarinic acetylcholine receptor 2 | abcam | cat# ab109226; RRID:AB_10858602 |
| muscarinic acetylcholine receptor 3 | Santa Cruz | cat# sc-9108; RRID:AB_2291779 |
| Synapsin-1 | Invitrogen | cat# 51-5200; RRID:AB_87668 |
| Tyrosine Hydroxylase | R&D | cat# AF7566 |
| Tyrosine Hydroxylase | Invitrogen | cat# P21962; RRID:AB_2539844 |
| III beta Tubulin | BioLegend | cat# 801201; RRID:AB_2313773 |
| GFP | Aves | cat# GFP-1010; RRID:AB_2307313 |
| CD31 | BD Bioscience | cat# 550274; RRID:AB_393571 |
| alpha smooth muscle actin | Sigma-Aldrich | cat# C6198; RRID:AB_476856 |
| LYVE-1 | AngioBio | cat# 11-034 |
| LYVE-1 | ReliaTech | cat# 103-M130 |
| eNOS | ThermoFisher | cat# PA1-037; RRID:AB_325774 |
| Phospho-eNOS (Ser1177) | CellSignaling | cat# 9571; RRID:AB_329837 |
| Tubulin | Sigma-Aldrich | cat# T9026V; RRID:AB_477593 |
| goat Alexa 594 | Invitrogen | cat# A-11058; RRID:AB_2534105 |
| rabbit Alexa 488 | Invitrogen | cat# A-21206; RRID:AB_2535792 |
| rabbit Alexa 647 | Invitrogen | cat# A-31573; RRID:AB_2536183 |
| chicken Alexa 488 | Jackson ImmunoResearch | cat# 703-545-1551 |
| sheep Alexa 594 | Invitrogen | cat# A-11016; RRID:AB_2534083 |
| rat Alexa 488 | Invitrogen | cat# A-21208; RRID:AB_2535794 |
| rabbit HRP | GE Healthcare | cat# NA9340V; RRID:AB_772191 |
| mouse HRP | GE Healthcare | cat# NA931; RRID:AB_772210 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DL-Norepinephrine hydrochloride | Sigma-aldrich | cat# A7256 |
| Phentolamine hydrochloride | Sigma-aldrich | cat# P7547 |
| Atropine sulfate salt monohydrate | Sigma-aldrich | cat# A0257 |
| Carbachol | Millipore | cat# 212385 |
| (−)-Isoproterenol hydrochloride | Sigma-aldrich | cat# I6504 |
| (±)-Propranolol hydrochloride | Sigma-aldrich | cat# P0884 |
| R(-)-Denopamine | Sigma-aldrich | cat# D7815 |
| (±)-Metoprolol (+)-tartrate salt | Sigma-aldrich | cat# M5391 |
| Salbutamol | Sigma-aldrich | cat# S8260 |
| ICI-118,551 hydrochloride | Sigma-aldrich | cat# I127 |
| Experimental Models: Cell Lines | ||
| MeWo | Sigma-Aldrich | cat# 93082609 |
| human bronchial smooth muscle cells | ScienCell | cat#3400 |
| B16F10 | ATCC | CRL-6475, RRID:CVCL_0159 |
| human dermal lymphatic endothelial cells | Group Prof. Detmar | N/A |
| Experimental Models: Organisms/Strains | ||
| Prox-1 GFP | Dr. Young-Kwon Hong, University of Southern California | I. Choi et al., 2011 |
| ChAT GFP | Jackson Laboratories | Strain No 007902 |
| Cx40BAC-GCaMP2 | Dr. Michael Kotlikoff, Cornell University and the CHROMus mouse resource | R24HL120847 |
| C57BL/6J | Jackson Laboratories | Strain No 000664 |
| Oligonucleotides | ||
| See Table S4 for used primers | N/A | |
| Software and Algorithms | ||
| Quantification algorithms | For full MATLAB codes please see STAR Methods. | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Detmar (michael.detmar@pharma.ethz.ch).
Experimental Models and Subject Details
Cell culture
Primary male human LECs (Hirakawa et al., 2003) were cultured under standard culture conditions (37°C and 5% CO2) on collagen (Advanced BioMatrix) coated dishes (50 μg/mL) in EBM medium (Lonza) containing 20% FBS (GIBCO), 1% penicillin/streptomycin (GIBCO), 2 mmol/L L-glutamine (GIBCO), 25 μmol/mL cAMP (Sigma-Aldrich) and 10 μg/mL hydrocortisone (Sigma-Aldrich).
Human bronchial smooth muscle cells were purchased from ScienCell and cultured under standard conditions in Smooth Muscle Cell Medium 2 containing the Supplemental Mix (both PromoCell). MeWo cells (Sigma Aldrich) were cultured under standard culture conditions in DMEM containing pyruvate (GIBCO) and 10% FBS.
Mice
Mice were group housed in IVC cages containing a mouse house and environmental enrichment under pathogen-free conditions. Mice were not involved in previous procedures. Mice of both genders were used at the age of 7-20 weeks for organ collection and 7-12 weeks for in vivo imaging. Prox1-GFP mice on the C57BL/6J background, used for clear visualization of LVs in whole mounts and tumor studies, were a kind gift of Dr. Young-Kwon Hong, University of Southern California (Choi et al., 2011). B6.Cg-Tg(RP23-268L19-EGFP)2Mik/J (ChAT-GFP, strain number 007902) mice and C57BL/6J mice were purchased from Jackson Laboratories, Bar Harbor, ME. Cx40BAC-GCaMP2 mice (Tallini et al., 2007) were obtained from Dr. Michael Kotlikoff, Cornell University and the CHROMus mouse resource (R24HL120847).
For organ collection at the end of imaging, mice were euthanized with an overdose of anesthesia (160 mg/kg ketamine; 0.4 mg/kg medetomidine) followed by cervical dislocation and opening of the chest cavity. All experiments were approved by Kantonales Veterinaeramt Zurich (protocols: 11/2012, 12/2015).
Method Details
LEC treatment in vitro
LECs were incubated in EBM containing 1% FBS overnight (starvation medium). Compounds of interest (Table S1) were added in starvation medium. Antagonists were added 30 min before treatment of the cells with the corresponding agonist. 15 min after agonist addition, total protein was extracted and protein concentration was measured using a Pierce BCA assay (ThermoFisher).
Western blotting
Equal amounts of denaturized protein were loaded per slot, using pre-cast NuPage gels (ThermoFisher). A Mini Gel Tank system was used to run and blot the gels (ThermoFisher) according to manufacturer’s instructions. After blocking the membranes, protein was detected using specific primary antibodies (Table S2) and HRP-conjugated secondary antibodies (Table S3). ECL was used for visualization using an Agfa Curix Developer. Quantification of phosphorylation was performed using FIJI (Schindelin et al., 2012).
RNA isolation and qPCR
For RNA extraction, the NucleoSpin RNA kit from Macherey-Nagel was used according to the manufacturer’s instructions. RNA was reverse transcribed with the High Capacity cDNA kit (Applied Biosystems/ThermoFisher). qPCR analyses were done in triplicate using SYBRGreen (Roche) on a 7900 HT Fast Real-Time PCR System (Applied Biosystems). The following genes were analyzed: ADRA1A, ADRA1B, ADRA1d, ADRA2A, ADRA2B, ADRA2C, ADRB1, ADRB2, ADRB3, ChRM1, ChRM2, ChRM3, ChRM4 and ChRM5 (for primers see Table S4). For normalization, GAPDH expression was analyzed.
Tissue digestion and FACS analysis
For FACS analysis of LECs, the flank collectors were dissected, minced, and digested in a collagenase solution [530 u/mL Collagenase II (Worthington), 0.744 u/mL Elastase (Worthington),5 u/mL DNase I (Roche)] for 70 min at 37°C. The digested tissue was passed through a cell strainer, washed and labeled with antibodies (see Tables S2 and S3) for 30 min at 4°C, followed by incubation of secondary antibody for 30 min at 4°C. Cells were stained for Zombie-NIR used for life/dead discrimination. After washing, cells were resuspended in FACS buffer for acquisition. Data were acquired on a FACS ARIA (BD) and analyzed using FlowJo v10.2 software (Treestar Inc.).
Imaging of flank collector contractions
The contractility of the flank collecting LV was analyzed as described (Chong et al., 2016). In brief, mice were anesthetized using a combination of xylazine (20 mg/ kg, Rompun® 2%, Provet), ketamine (100 mg/kg, Ketasol®, Graeub) and acepromazine (3 mg/kg, Prequillan®, Arovet) or with isoflurane (2.5%, Attane®, Provet). After loss of a detectable response to toe pinches, mice were fixed on a silicone dish and the flank collector was exposed. Tissue was kept moist by applying warm PBS and mice were kept on a heating pad during the imaging. Cannulation of the inguinal lymph node enabled a 0.5 μL infusion of 10 μmol/L P20D680 dye into the node and thereby visualization of the flank collector. For infusion, a pump (PHD2000, Harvard Apparatus, Cambridge, MA) was used, connected to a custom-made catheter using polyethylene PE-10 tubing (SCI, Lake Havasu City, AZ) and a 30-g needle. After stabilization of the contractility, videos of 8 min (9 min for the specificity testing) duration were acquired. Local topical treatment of the flank collector was done at time point 2 min (compound information see Table S1). For the specificity testing, the second treatment was topically added at time point 4 min. Videos were acquired using either a Zeiss StereoLumar.V12 stereomicroscope with AxioVision (Carl Zeiss, Feldbach, Switzerland) software and a Photometrics Evolve 512 camera or a Zeiss AxioZoom.V16 fluorescence zoom microscope with Zen Pro (Carl Zeiss) software and a QImaging OptiMOS sCMOS camera (QImaging, Surrey, Canada) in combination with a light-emitting diode illumination system pE-4000 (CoolLED Ltd, Andover, UK) and filters for Cy5 (Carl Zeiss). The contractility was quantified using a previously described algorithm (Chong et al., 2016).
Mice were allocated randomly to a specific treatment. Group size was evaluated after first pilot studies using power-analysis. Exclusion criteria for an infused vessel were: complete coverage with fat or visible luminal obstruction of the vessel induced by the infusion by e.g., blood coagulates.
In vivo Ca2+ imaging
Flank collectors of Cx40BAC-GCaMP2 mice were exposed. Infusion of either 0.5 μL of 10 μmol/L P20D680 tracer or a mixture of tracer with agonists was performed into the inguinal lymph node. During infusion, a video of the flank collector was acquired using a custom built 2-photon laser-scanning microscope. For quantification of a possible Ca2+ peak, mean intensity of the GCaMP2 signal was measured over the vessel wall during the video (Figure 5A). Specific compounds were randomly allocated to mice.
In vitro Ca2+ imaging
The Fluo8H, AM (AAT Bioquest, cat-no. 21091) was used according to the manufacturer’s instructions. In brief, Fluo8H stock solution (1.14 mol/L) was prepared by adding 1.5% Pluronic and 2.4% DMSO in PBS. Cells were incubated in EBM containing 1% FBS for 4h, Fluo8H 1:500 in starvation medium was added and cells were incubated for 30 min at 37°C, followed by 10 min at room temperature. Cells were washed twice with Live Cell Imaging Solution (Molecular Probes). Imaging of cells was performed in Live Cell Imaging Solution; compounds were added in the same volume of live cell imaging solution as the cells were cultured in, using 2x treatment solutions. Imaging was performed using a fluorescent microscope (Nikon Eclipse Ti-E, Tokyo, Japan) in combination with a Hamamatsu ORCA-Flash4.0 CCD camera (Hamamatsu, Japan). FITC fluorescence videos were acquired and quantified as stated below.
Whole mount stainings
Vessels or pieces of small intestine were dissected out of 7- to 20-week-old mice of either gender. The small intestine samples were cut open longitudinally to give access to villi. Samples were fixed with 4% PFA for 2h at 4°C or for 1h in 100% methanol at 4°C. After washing in PBS, tissues were incubated in 0.2% BSA, 5% donkey serum, 0.1% Triton-X in PBS for 3h at 4°C. Primary antibodies were incubated over night at 4°C. After washing in 0.1% Triton X in PBS, secondary antibodies were applied in 0.1% Triton X in PBS for 2h at room temperature or at 4°C for the small intestine. After several washes in PBS, tissues were mounted using Vectashield (Vector) for confocal imaging. The antibodies used are listed in Tables S2 and S3. Whole mount pictures were acquired using a LSM 780 FCS confocal microscope and ZEN software (Zeiss, Jena, Germany) and processed with FIJI. Positive control stainings for the neurotransmitter receptors are as follows: α1-positive enterocytes are visible in Figure 1C, β1-positive heart tissue is provided in Figure S1D, β2-positive blood vessels are visible in Figure 1D and M2-positive liver tissue is provided in Figure S1E.
B16F10 melanoma studies
Male C57BL/6J mice of 7 weeks were bilaterally injected intradermally with 105 B16F10 tumor cells in the flank skin. Tumor growth was monitored by size measurements using a caliper. When tumors reached 12 mm in one dimension, tumor-draining LVs were imaged in vivo. Mice were randomly assigned to compound treatments.
Quantification and Statistical Analysis
Quantification of innervation
Overall length of nerves covering vessels was measured manually in FIJI and divided by the length of the LV.
Statistical analyses
All statistical analyses were performed using the GraphPad Prism 5 software. Data are shown as means ± SD, information on exact value and representation of n can be found in the respective figure legends. For comparison of more than two groups, one-way ANOVA with Bonferroni post-test was used for normally distributed data. For quantifications from in vivo flank imaging, a paired two-tailed Student’s t test was used for normal distributed data. A Mann-Whitney test was used for the comparison of two groups if the data was non-normal distributed, tested by a Shapiro-Wilk test. A p value of p < 0.05 was considered significant (p < 0.05 ∗, p ≤ 0.01 ∗∗, p ≤ 0.001∗∗∗).
Data and Software Availability
Quantification of low frequency contractions
Experiments performed with isoflurane resulted in a much lower contraction activity which could not be analyzed with the previously developed algorithm (Chong et al., 2016). Therefore, a new algorithm was developed (Figure S4). In order to determine the characteristics of the peaks (i.e., amplitudes and periodicity) of the measured fluorescence signal, a MATLAB (MathWorks Inc., Natick MA, USA) function with a graphical user interface was developed that performs the signal processing in a semi-automatically manner. The user selects the peaks of the fluorescence signal manually and based on these selected values, the absolute amplitudes, relative amplitudes and the frequency are then calculated automatically. These parameters were calculated according to the formulas
| (1) |
| (2) |
| (3) |
with the baseline value of each peak according to in case of or in case of , and the time-difference between the two peaks and with the first peak assigned to and the second to (and so on). For a visualization of the mathematical nomenclature used, see Figure S4. The formulation ensures that the amplitude and periodicity values are also calculated correctly in case the signal contains negative peaks, or a mixture of positive and negative peaks.
MATLAB Code
function [Amplitude_abs, Amplitude_rel, Period, Frequency] = AnalyzePeaks_Ampl_Freq_v2(x,f_s)
% Function to analyze a time-series regarding the characteritics of the
% peaks (i.e., amplitudes, period)
% _________________________________________________________________________
% For Steven Proulx and Samia Bachmann, ETH.
% May 2017 | Felix Scholkmann | Version 2
%__________________________________________________________________________
% INPUT
% x: signal to analyze
% f_s: sampling frequency in Hz
% OUTPUT
% Amplitude_abs: absolute amplitude values (first value: median, second value: inter quantile range, IQR)
% Amplitude_rel: relative amplitude values (first value: median, second value: inter quantile range, IQR)
% Period: period values [in seconds] (first value: median, second value: inter quantile range, IQR)
% Frequency: frequency values [in 1/min] (first value: median, second value: inter quantile range, IQR)
% important:
% The input signal (x) must contain at least 3 peask (i.e., at least 9
% points needs to be selected from the signal). This is necessary to be
% able to calculate the period values.
%% ________________________________________________________________________
close all
scrsz = get(0,’ScreenSize’); figure(‘Position’,[100 50 1300 850]);
set(gcf., ‘color’, ‘w’); set(gcf.,’defaultaxesfontsize’,12)
subplot(2,20,[1,3])
% Please insert here the path to the file “Points.png”
currentFolder = pwd;
Path = strcat(currentFolder,’\Points.png’)
img = imread(Path);
image(img);
set(gca, ‘XTickLabelMode’, ‘Manual’)
set(gca, ‘XTick’, [])
set(gca, ‘YTickLabelMode’, ‘Manual’)
set(gca, ‘YTick’, [])
title (‘Definition’,’fontsize’,14)
fig = subplot(2,20,[6,20]);
plot(t,x,’linewidth’,2,’Color’,[0.3, 0.5, 1])
axis tight
ylabel(‘Intensity’); xlabel(‘Time [s]’)
title (‘- > Please select the points (\alpha_{i}, \alpha_{i+1}, \alpha_{i+2}, i = 1, 2, …, \it{N})’,’fontsize’,14)
% Get the points from the figure
disp(‘→ Please select the points in the figure. The press “Enter”’)
[x, y] = getpts(fig);
hold on
vline_red(x(2:3:end))
vline_black(x(1:3:end))
vline_black(x(3:3:end))
n = length(x);
% Calculate the amplitudes
j = 1;
- for i = 1:3:n-2;
- if y(i+2) > y(i)
- M(j) = (y(i+2)-y(i))/2 + y(i);
- A_abs(j) = abs(M(j) - y(i+1));
- A_rel(j) = 100 ∗ (A_abs(j)/M(j));
- j = j+1;
- else
- M(j) = (y(i)-y(i+2))/2 + y(i+2);
- A_abs(j) = abs(M(j) - y(i+1));
- A_rel(j) = 100 ∗ (A_abs(j)/M(j));
- j = j+1;
- end
end
plot(x(2:3:end),M,’ob’)
plot(x(1:3:end),y(1:3:end),’ok’)
plot(x(3:3:end),y(3:3:end),’ok’)
plot(x(2:3:end),y(2:3:end),’or’)
% Calculate the periods
j = 1;
- for i = 1:3:n-4;
- P(j) = x(i+4)-x(i+1);
- j = j+1;
end
F = (1./(P))∗60; % Frequency [1/min]
subplot(2,20,[21:23])
bar(A_abs,’b’);
ylabel(‘Amplitude (A_{abs}) [absolute]’); xlabel(‘Peak number’)
title ({’Amplitude values’,’[absolute]’},’fontsize’,14)
xlim([0,length(A_abs)+1])
subplot(2,20,[25:27])
bar(A_rel,’b’);
ylabel(‘Amplitude (A_{rel}) [%]’); xlabel(‘Peak number’)
title ({’Amplitude values’,’[%]’},’fontsize’,14)
xlim([0,length(A_rel)+1])
subplot(2,20,[29:31])
bar(F,’r’);
ylabel(‘Frequency [1/min]’); xlabel(‘Peak number’)
title ({’Frequency values’,’[1/min]’},’fontsize’,14)
xlim([0,length(F)+1])
subplot(2,20,35:36)
errorbar(median(A_rel),iqr(A_rel)/2,’k’);
hold on
plot(median(A_rel),’k.’,’markersize’,30)
ylabel(‘Amplitude [%] (median +- IQR)’)
set(gca, ‘XTickLabelMode’, ‘Manual’)
set(gca, ‘XTick’, [])
title([‘A_{rel} = ’ num2str(median(A_rel)),’ (‘ num2str(iqr(A_rel)),’) %’]);
subplot(2,20,39:40)
errorbar(median(F),iqr(F)/2,’k’);
hold on
plot(median(F),’k.’,’markersize’,30)
ylabel(‘Frequency [1/min] (median +- IQR)’)
set(gca, ‘XTickLabelMode’, ‘Manual’)
set(gca, ‘XTick’, [])
title([‘F = ’ num2str(median(F)),’ (‘ num2str(iqr(F)),’) 1/min’]);
% Generate output data
Amplitude_abs = [median(A_abs), iqr(A_abs)];
Amplitude_rel = [median(A_rel), iqr(A_rel)];
Period = [median(P), iqr(P)];
Frequency = [median(F), iqr(F)];
Quantification of in vitro Ca2+ imaging
Pre-processing
An important step to restore images is denoising, which consists of reducing noise present in images but at the same time preserving objects’ features such as edges, texture, etc. In this context, we have opted to use the Block-Matching and 3D filtering (BM3D) method which has shown its denoising capabilities in different datasets by preserving finer image structures (Dabov et al., 2007, Danielyan et al., 2014).
Cell Detection
Once the image has been pre-processed, the next step is to detect the foreground objects, i.e., cells. To do this, we use the Otsu thresholding method (Otsu, 1979), however instead of using a single threshold value, we adopt a two-level threshold to detect the nucleus and the cytoplasm as joint single objects.
After thresholding, small objects are further removed since they usually correspond to non-relevant objects, such as air-bubbles. In addition, since some cells may spatially be close to each other, they can end up being merged after thresholding. To cope with this issue, we use the Watershed Transform. More precisely, each segmented object is represented as a basin and the detected Watershed Lines with the distance transform are used to separate the objects.
Cell Linking
Cells are detected at each single frame separately. In order to analyze how a cell activates over time, the detected cells are first linked together over time using the Kalman filter (Kalman, 1960).
The Kalman filter comprises two steps: prediction and update. In the prediction step, the Kalman filter predicts the current state of the system, based on the past, through a dynamic model. In the update step, the status of the system is corrected based on the observed variables. In our case, the assignment/linking of a given cell to a specific track is based on the area of overlap of the segmented cells between consecutive frames. The Kalman Filter therefore seeks to maximize the area of overlap between cell detections over time. To obtain more robust predictions at time t, our system averages the bounding-box of the detected cells over the last four frames. To remove false positives such as air-bubbles, our system discards objects with large location changes over time. Also, short tracks are discarded for further processing. In our case, we impose the constraint that tracks shall cover at least 75% of the total length of the input video.
Event Detection
Our event detection module is versatile and besides detecting which cells got activated, it can also find out when exactly a given activation starts and ends over time. The module starts first by smoothing the cell’s intensity signal (the cell-intensity track) to reduce noise. Smoothing is done with a moving average filter. Note that the smoothing step is optional and the raw signal can be directly used if so desired.
Next, the cell signals are normalized to compensate for effects such as photobleaching and also to facilitate the detection of cell activations in the subsequent steps. The normalization of a cells’ signal is done by computing the average intensity , of the detected cell at the frames before the injection time (see Equation 1).
| (Equation 1) |
where is the mean intensity value of the collection of pixels belonging to the detected cell i at time t.
The computed mean value of cell i is used to normalize the signal by dividing it into the cell signal values at all later times (see Equation 2).
| (Equation 2) |
where is the normalized cells’ signal and N corresponds to the total number of frames. Figure S3B shows an example of cell signals before (upper graph) and after normalization (lower graph). Detected activations above the threshold value (displayed as a black dashed line) are shaded, see next section of details of the detection algorithm.
To detect cell-activations, the normalized cell signals are thresholded. More precisely, signal values larger than a threshold value are considered as activations and values lower than the threshold are set to zero. We set this threshold value empirically to be 1.25. Note that this value can be tuned, if more detections are targeted, one can increase this value. It is important though to set this value in a conservative manner. An example of cell activation is displayed in yellow in the left graph of Figure S3C. The thresholding step is applied for all cell signals, in such a way that all activations can be mapped as an activation map. In Figure S3C, each row represents a single cell signal and its mean-intensity pixel values are displayed for each frame along the x axis. After thresholding the cell signals on the normalized cell signals (middle graph in Figure S3C), a binary activation map is obtained in which cell activations are displayed as white stripes (right graph, Figure S3C). The signal values where the cell did not activate are set to zero. In addition, to remove potential false negatives, our method first fills short gaps inside an event and also prunes short cell activations to remove false positives. In our experiments, both values were empirically set to five frames.
One important property of our method is that it detects where a cell activation starts and ends. This is done by computing the first order derivative over the binary activated signal and by looking respectively for the indexes where the derivative is respectively positive or negative (i.e., +1 or −1).
MATLAB Code (2 parts)
function kalmanMultiObjectTracking(videoName)
% KalmanMultiObjectTracking: this function detects a set of cells and
% tracks them over time. Cell detection involves image pre-processing where
% the original video frames are enhanced, e.g., denoising. The cell
% detections are mapped into cell-tracks, i.e., a set of temporal
% bounding-boxes for each detected cell.
%
% Input:
% videoName, e.g., videoName = ‘hu_NE1uM_2.avi’;
%
% Output:
- % tracksHist.mat MATLAB file containing the cell-tracks.
- opts = getDefaultParameters(); %# reading configuration parameters.
- pathInputVideo = fullfile(opts.videoPath, videoName);
- digitStr = ‘%d-’;
- strAllMotionDigits = repmat(digitStr, 1, numel(opts.motionNoise));
- motionNoiseStr = sprintf(strAllMotionDigits(1:end-1),opts.motionNoise);
- strAllInitErrorDigits = repmat(digitStr, 1, numel(opts.initialEstimateError));
- initErrorStr = sprintf(strAllInitErrorDigits(1:end-1),opts.initialEstimateError);
- paramsDir = sprintf(‘%s_LRate%2.2f_visi%2.2f_age%d_invib%d_initErr%s_motNoise%s_measNoise%d_%s_cSze%d’, …
- opts.motionModel, opts.learningRate, opts.trackVisibility, opts.ageThreshold, …
- opts.invisibleForTooLong, initErrorStr, motionNoiseStr, opts.measurementNoise, opts.thresh.type, opts.cell.size);
- [videosPath, inVideoname, ∼] = fileparts(pathInputVideo);
- videoNameOutputTrackRGB = sprintf(‘%s_track_rgb.avi’, inVideoname);
- videoNameOutputTrackProc = sprintf(‘%s_track_proc.avi’, inVideoname);
- videoNameOutputTrackBW = sprintf(‘%s_track_bw.avi’, inVideoname);
- videoNameOutputBW = sprintf(‘%s_bw.avi’, inVideoname);
- tracksDir = fullfile(videosPath, ‘tracks_kalmann_sel’);
- tracksDirVideo = fullfile(tracksDir, inVideoname, paramsDir);
- opts.framesOrigDir = fullfile(tracksDir, inVideoname, ‘frames’);
- opts.framesProcDir = fullfile(tracksDirVideo, ‘frameProc’);
- opts.maskDir = fullfile(tracksDirVideo, ‘frameMasks’);
- opts.maskDirTracks = fullfile(tracksDirVideo, ‘frameMasksTracks’);
- opts.framesOrigTracksDir = fullfile(tracksDirVideo, ‘framesOrigTracks’);
- opts.framesProcTracksDir = fullfile(tracksDirVideo, ‘framesProcTracks’);
- opts.maskDirTracksAll = fullfile(tracksDirVideo, ‘frameMasksTracksAll’);
- opts.framesOrigTracksDirAll = fullfile(tracksDirVideo, ‘framesOrigTracksAll’);
- createVideoDirs(tracksDirVideo, opts); %# creating output directories
- videoObjTrackRGB = VideoWriter(fullfile(tracksDirVideo, videoNameOutputTrackRGB), ‘Uncompressed AVI’);
- videoObjTrackProc = VideoWriter(fullfile(tracksDirVideo, videoNameOutputTrackProc), ‘Uncompressed AVI’);
- videoObjTrackBW = VideoWriter(fullfile(tracksDirVideo, videoNameOutputTrackBW), ‘Uncompressed AVI’);
- videoObjBW = VideoWriter(fullfile(tracksDirVideo, videoNameOutputBW), ‘Uncompressed AVI’);
- fprintf(‘video: %s\n’,(fullfile(tracksDirVideo, videoNameOutputTrackRGB)));
- tracksObj = fullfile(tracksDirVideo, sprintf(‘%s.mat’, inVideoname));
- obj = setupSystemObjects(pathInputVideo, opts);
- tracks = initializeTracks(); %# Create an empty array of tracks.
- tracksHist = initializeTracksHistory();
- nextId = 1; % ID of the next track
- noFrames = 0;
- if ∼exist(tracksObj, ‘file’)
- %# Detect moving objects, and track them across video frames.
- while ∼isDone(obj.reader)
- noFrames = noFrames + 1;
- fprintf(‘# Processing frame: %d\n’, noFrames);
- frame = readFrame(obj);
- %# image pre-processing and per-frame cell detection.
- [centroids, bboxes, mask, frameDen] = detectObjects(frame, opts);
- %# predict cell location.
- [tracks, tracksHist] = predictNewLocationsOfTracks(tracks, tracksHist, noFrames);
- %# link cells over time.
- [assignments, unassignedTracks, unassignedDetections] = …
- detectionToTrackAssignment(tracks, centroids, bboxes, opts);
- %# update existing cell-tracks.
- [tracks, tracksHist] = updateAssignedTracks(tracks, tracksHist, assignments, centroids, bboxes, noFrames);
- [tracks, tracksHist] = updateUnassignedTracks(tracks, tracksHist, unassignedTracks);
- %# delete short tracks.
- [tracks, tracksHist] = deleteLostTracks(tracks, tracksHist, opts);
- %# create new tracks.
- [tracks, tracksHist, nextId] = createNewTracks(tracks, tracksHist, centroids, bboxes, unassignedDetections, nextId, noFrames);
- %# saving tracks to disk.
- dumpTrackingResults(frame, mask, tracks, tracksHist, tracksDirVideo, videoObjTrackRGB, videoObjTrackBW, videoObjBW, noFrames, opts);
- end
- opts.totNoFrames = noFrames;
- tracksHist = updateEmptyTracks(tracksHist, opts); %# check empty tracks frames and reduce its totalVisibleCount
- save(tracksObj, ‘tracksHist’, ‘-v7.3′); %# save to disk the detected tracks.
- else
- fprintf(‘# Loading tracks\n’);
- tracksHist = load(tracksObj);
- tracksHist = tracksHist.tracksHist;
- reset(obj.reader);
- while ∼isDone(obj.reader)
- frame = readFrame(obj);
- noFrames = noFrames + 1;
- end
- fprintf(‘# No of. frames: %d\n’, noFrames);
- end
- opts.totNoFrames = noFrames;
- if ∼isfield(tracksHist, ‘numEmptyMask’)
- tracksHist = updateEmptyTracks(tracksHist, opts);
- end
- open(videoObjTrackRGB); %# create RGB video with cell tracks.
- open(videoObjTrackProc); %# create RGB video for cell tracks (segmented/cells + bounding boxes).
- open(videoObjTrackBW); %# create BW video for cell tracks (segmented/detected cells + bounding boxes).
- open(videoObjBW);%# create BW video file for cell detections (segmented/detected cells as binarys masks).
- noFrames = 0;
- for ff = 1:opts.totNoFrames
- noFrames = noFrames + 1;
- frameOrigFile = fullfile(opts.framesOrigDir, sprintf(‘frame_%05d.tif’, ff));
- frameDenFile = fullfile(opts.framesProcDir, sprintf(‘frame_%05d.tif’, ff));
- frameMaskFile = fullfile(opts.maskDir, sprintf(‘frame_%05d.tif’, ff));
- fprintf(‘#Processing frame: %d\n’, noFrames);
- if exist(frameOrigFile, ‘file’) && exist(frameDenFile, ‘file’) && exist(frameMaskFile, ‘file’)
- frameOrig = imread(frameOrigFile);
- frameDen = imread(frameDenFile);
- frameMask = imread(frameMaskFile);
- %# storing into disk each frame overlayed with the cell-tracks.
- saveTrackingResults(frameOrig, frameDen, frameMask, tracksHist, videoObjTrackRGB, videoObjTrackProc, videoObjTrackBW, videoObjBW, ff, opts);
- end
- end
- close(videoObjTrackRGB); %# closing videoFile.
- close(videoObjTrackProc); %# closing videoFile.
- close(videoObjTrackBW); %# closing videoFile.
- close(videoObjBW); %# closing videoFile.
end
function tracksThresh2Activations(videoName)
% tracksThresh2Activations: given the extracted cell-tracks of a
% given video (linked cells over time), this function detects which of
% the cells got activated. For that purposes, the mean pixel intensity
% of a segmented cell is computed at each frame. If the mean pixel
% intensity value is higher than a threshold, then an activation is fired.
% The original intensity values can also be smoothed to remove noise and if
% the interval of the activation is too short, then a false-positive activation
% is detected.
%
% Input:
% videoName, e.g., videoName = ‘hu_NE1uM_2.avi’;
%
% Output:
% a set of figures are stored into disk, containing a
- % matrix-representation of the activated cell-signals.
- addpath(genpath(‘./3rd/overlay/’));
- %# reading default parameters, e.g., location of detected cells frames.
- opts = getDefaultParameters();
- if ∼isfield(opts, ‘endThreatment’)
- opts.endThreatment = 10;
- end
- if ∼isfield(opts, ‘signal’)
- opts.signal = [];
- end
- if ∼isfield(opts.signal, ‘Type’)
- opts.signal.Type = ‘orig’;
- end
- %# smoothing type for the cell signal.
- if ∼isfield(opts.signal, ‘smoothType’)
- opts.signal.smoothType = ‘box’; % ‘golay’, ‘gauss’, ‘box’
- end
- %# threshold value
- if ∼isfield(opts.signal, ‘threshVal’)
- opts.signal.threshVal = 1.25;
- end
- %# minimum duration of cell-activation
- if ∼isfield(opts.signal, ‘pruneEventDuration’)
- opts.signal.pruneEventDuration = 5;
- end
- if ∼isfield(opts.signal, ‘fillGaps’)
- opts.signal.fillGaps = true;
- end
- if ∼isfield(opts.signal, ‘fillGapsSze’)
- opts.signal.fillGapsSze = 5;
- end
- [videosPath, videoNameInput, ∼] = fileparts(videoName);
- digitStr = ‘%d-’;
- strAllMotionDigits = repmat(digitStr, 1, numel(opts.motionNoise));
- motionNoiseStr = sprintf(strAllMotionDigits(1:end-1),opts.motionNoise);
- strAllInitErrorDigits = repmat(digitStr, 1, numel(opts.initialEstimateError));
- initErrorStr = sprintf(strAllInitErrorDigits(1:end-1),opts.initialEstimateError);
- paramsDir = sprintf(‘%s_LRate%2.2f_visi%2.2f_age%d_invib%d_initErr%s_motNoise%s_measNoise%d_%s_cSze%d’, …
- opts.motionModel, opts.learningRate, opts.trackVisibility, opts.ageThreshold, …
- opts.invisibleForTooLong, initErrorStr, motionNoiseStr, opts.measurementNoise, opts.thresh.type, opts.cell.size);
- %# creating subdirectories where resuls are going to be stored.
- tracksDir = fullfile(videosPath, ‘tracks_paper’);
- tracksDirVideo = fullfile(tracksDir, videoNameInput, paramsDir);
- signalDir = fullfile(tracksDirVideo, ‘signal’);
- signalThreshDir = fullfile(signalDir, ‘thresh’);
- signalEventsDir = fullfile(signalDir, ‘events’);
- if ∼exist(signalEventsDir, ‘dir’), mkdir(signalEventsDir); end
- tracksFile = fullfile(tracksDirVideo, ‘validTracksIdx.mat’);
- validTracksIdx = load(tracksFile);
- validTracksIdx = validTracksIdx.validTracksIdx;
- %# loading detected cell-tracks
- if strcmpi(opts.signal.Type, ‘orig’)
- threshSubDir = fullfile(signalThreshDir, ‘origdata’);
- signalMeanFile = fullfile(tracksDirVideo, ‘signalMean.mat’);
- signalMean = load(signalMeanFile);
- signalMean = signalMean.signalMean;
- signalMean(∼validTracksIdx,:) = [];
- signalArray = signalMean;
- if ∼exist(threshSubDir, ‘dir’), error(‘Unknown directory: %s\n’, threshSubDir); end
- signalEventsDir = fullfile(signalEventsDir, ‘origdata’);
- if ∼exist(signalEventsDir, ‘dir’), mkdir(signalEventsDir); end
- signalTypeStr = opts.signal.Type;
- tracksIdx = find(validTracksIdx);
- elseif strcmpi(opts.signal.Type, ‘smooth’)
- fprintf(‘# Loading %s data\n’, opts.signal.smoothType);
- signalSmoothFile = fullfile(signalDir, ‘smooth’, sprintf(‘tracksSummary_%s.mat’, opts.signal.smoothType));
- signalMeanSmooth = load(signalSmoothFile);
- signalMeanSmooth = signalMeanSmooth.signalMeanSmooth;
- signalMeanSmooth(∼validTracksIdx,:) = [];
- signalArray = signalMeanSmooth;
- threshSubDir = fullfile(signalThreshDir, opts.signal.smoothType);
- if ∼exist(threshSubDir, ‘dir’), error(‘Unknown directory: %s\n’, threshSubDir); end
- signalEventsDir = fullfile(signalEventsDir, opts.signal.smoothType);
- if ∼exist(signalEventsDir, ‘dir’), mkdir(signalEventsDir); end
- signalTypeStr = sprintf(‘%s-%s’, opts.signal.Type, opts.signal.smoothType);
- tracksIdx = 1:size(signalArray,1);
- else
- error(‘Unkown type: %s’, opts.signal.Type);
- end
- signalArrayNormFilename = fullfile(threshSubDir, ‘signalNorm.mat’);
- if ∼exist(signalArrayNormFilename, ‘file’), error(‘Unknown file: %s\n’, signalArrayNormFilename); end
- signalNorm = load(signalArrayNormFilename);
- signalNorm = signalNorm.signalNorm;
- %# thresholding for activation.
- signalBinaryActivation = signalNorm > opts.signal.threshVal;
- [noTracks, ∼] = size(signalBinaryActivation);
- %# fill possible gaps in the cell-activations
- if opts.signal.fillGaps > 0
- filledSignal = signalBinaryActivation;
- for ii = 1:noTracks
- filledSignal(ii,:) = ∼bwareaopen(∼filledSignal(ii,:), opts.signal.fillGapsSze);
- end
- signalBinaryActivation = filledSignal;
- strFillGap = sprintf(‘fillSze%d’, opts.signal.fillGapsSze);
- else
- strFillGap = sprintf(‘nofill’);
- end
- signalActivation = zeros(size(signalBinaryActivation));
- if opts.signal.pruneEventDuration > 0
- strPruneEvent = sprintf(‘pruned%d’, opts.signal.pruneEventDuration);
- else
- strPruneEvent = sprintf(‘noprune’);
- end
- %# analyzing cell-track duration.
- stats = repmat(struct(‘startIdx’,[], ‘endIdx’, [], ‘duration’, [], ‘eventIdx’, [], ‘activation’, 0), noTracks, 1);
- for tt = 1:noTracks
- trackActivation = signalBinaryActivation(tt,:);
- trackActivation = diff([0 trackActivation 0]);
- eventStartIdx = find(trackActivation > 0);
- eventEndIdx = find(trackActivation < 0)-1;
- eventDuration = eventEndIdx - eventStartIdx + 1;
- if opts.signal.pruneEventDuration
- stringIdx = (eventDuration > = opts.signal.pruneEventDuration);
- eventStartIdx = eventStartIdx(stringIdx);
- eventEndIdx = eventEndIdx(stringIdx);
- end
- eventIdx = zeros(1,max(eventEndIdx)+1);
- eventIdx(eventStartIdx) = 1;
- eventIdx(eventEndIdx+1) = eventIdx(eventEndIdx+1)-1;
- eventIdx = find(cumsum(eventIdx));
- signalActivation(tt, eventIdx) = 2;
- signalActivation(tt,eventStartIdx) = +1;
- signalActivation(tt,eventEndIdx) = −1;
- stats(tt).startIdx = eventStartIdx;
- stats(tt).endIdx = eventEndIdx;
- stats(tt).duration = eventDuration;
- stats(tt).eventIdx = eventIdx;
- stats(tt).activation = isempty(eventIdx) = = 0;
- end
- videoName4Plot = strrep(videoNameInput,’_’,’\_’);
- signalImFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_%s_label.tif’, videoNameInput, signalTypeStr));
- normSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_norm.tif’, videoNameInput));
- activationSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_activ_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- binActivationSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_bin_activ_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- signalEventsDirBox = strrep(signalEventsDir, ‘origdata’, ‘box’);
- activationBoxSignalFilename = fullfile(signalEventsDirBox, sprintf(‘%s_signal_activ_%s_%s_%s.mat’, videoNameInput, signalTypeStr, strPruneEvent, strFillGap));
- activationBoxSignalFilename = strrep(activationBoxSignalFilename, ‘orig’, ‘smooth-box’);
- signalActivationBox = load(activationBoxSignalFilename);
- signalActivationBox = signalActivationBox.signalActivation;
- signal2disk(signalArray, videoName4Plot, signalImFilename, opts);
- signal2disk(signalNorm, videoName4Plot, normSignalFilename, opts);
- signal2disk(signalActivation, videoName4Plot, activationSignalFilename, opts);
- signal2disk(signalActivation∼ = 0, videoName4Plot, binActivationSignalFilename, opts);
- overlaySignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlay_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- overlayNormSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlaynorm_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- signal2diskOverlay(signalArray, signalActivation∼ = 0, videoName4Plot, overlaySignalFilename, opts);
- signal2diskOverlay(signalNorm, signalActivation∼ = 0, videoName4Plot, overlayNormSignalFilename, opts);
- overlaySignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlay_%s_%s_%s_box.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- overlayNormSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlaynorm_%s_%s_%s_box.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- signal2diskOverlay(signalArray, signalActivationBox∼ = 0, videoName4Plot, overlaySignalFilename, opts);
- signal2diskOverlay(signalNorm, signalActivationBox∼ = 0, videoName4Plot, overlayNormSignalFilename, opts);
- signalFilenameMat = fullfile(signalEventsDir, sprintf(‘%s_signal_activ_%s_%s_%s.mat’, videoNameInput, signalTypeStr, strPruneEvent, strFillGap));
- save(signalFilenameMat, ‘signalActivation’);
- signalStatsFilenameMat = fullfile(signalEventsDir, sprintf(‘%s_stats_%s_%s_%s.mat’, videoNameInput, signalTypeStr, strPruneEvent, strFillGap));
- save(signalStatsFilenameMat, ‘stats’);
end
Acknowledgments
The authors thank Carlos Ochoa for excellent technical assistance, Dr. Markus Rudin for kindly providing Cx40BAC-GCaMP2 mice, and the Scientific Center for Optical and Electron Microscopy (ScopeM) of ETH Zurich for assistance with confocal imaging. This work was supported by Swiss National Science Foundation grants 310030B_147087 and 310030_166490, European Research Council grant LYVICAM, and Oncosuisse, Krebsliga Zurich, and Leducq Foundation Transatlantic Network of Excellence grant Lymph Vessels in Obesity and Cardiovascular Disease (11CVD03) (all to M.D.).
Author Contributions
Conceptualization, S.B.B. and M.D.; Software, J.M., F.S., and S.F.N.; Investigation, S.B.B., D.G., M.S., S.T.P., and C.T.; Resources, M.D.; Writing – Original draft, S.B.B.; Writing – Review & Editing, all authors; Supervision, M.D. and S.F.N.; Funding Acquisition, M.D.
Declaration of Interests
The authors declare no competing interests.
Published: June 11, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.05.050.
Supplemental Information
References
- Amatschek S., Kriehuber E., Bauer W., Reininger B., Meraner P., Wolpl A., Schweifer N., Haslinger C., Stingl G., Maurer D. Blood and lymphatic endothelial cell-specific differentiation programs are stringently controlled by the tissue environment. Blood. 2007;109:4777–4785. doi: 10.1182/blood-2006-10-053280. [DOI] [PubMed] [Google Scholar]; Amatschek, S., Kriehuber, E., Bauer, W., Reininger, B., Meraner, P., Wolpl, A., Schweifer, N., Haslinger, C., Stingl, G., and Maurer, D. (2007). Blood and lymphatic endothelial cell-specific differentiation programs are stringently controlled by the tissue environment. Blood. 109, 4777-4785. [DOI] [PubMed]
- Baglole C.J., Sigalet D.L., Martin G.R., Yao S., Meddings J.B. Acute denervation alters the epithelial response to adrenoceptor activation through an increase in α1-adrenoceptor expression on villus enterocytes. Br. J. Pharmacol. 2006;147:101–108. doi: 10.1038/sj.bjp.0706424. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baglole, C.J., Sigalet, D.L., Martin, G.R., Yao, S., and Meddings, J.B. (2006). Acute denervation alters the epithelial response to adrenoceptor activation through an increase in α1-adrenoceptor expression on villus enterocytes. Br. J. Pharmacol. 147, 101-108. [DOI] [PMC free article] [PubMed]
- Benoit J.N. Effects of alpha-adrenergic stimuli on mesenteric collecting lymphatics in the rat. Am. J. Physiol. 1997;273:R331–R336. doi: 10.1152/ajpregu.1997.273.1.R331. [DOI] [PubMed] [Google Scholar]; Benoit, J.N. (1997). Effects of alpha-adrenergic stimuli on mesenteric collecting lymphatics in the rat. Am. J. Physiol. 273, R331-R336. [DOI] [PubMed]
- Choe K., Jang J.Y., Park I., Kim Y., Ahn S., Park D.-Y., Hong Y.-K., Alitalo K., Koh G.Y., Kim P. Intravital imaging of intestinal lacteals unveils lipid drainage through contractility. J. Clin. Invest. 2015;125:4042–4052. doi: 10.1172/JCI76509. [DOI] [PMC free article] [PubMed] [Google Scholar]; Choe, K., Jang, J.Y., Park, I., Kim, Y., Ahn, S., Park, D.-Y., Hong, Y.-K., Alitalo, K., Koh, G.Y., and Kim, P. (2015). Intravital imaging of intestinal lacteals unveils lipid drainage through contractility. J. Clin. Invest. 125, 4042-4052. [DOI] [PMC free article] [PubMed]
- Choi I., Chung H.K., Ramu S., Lee H.N., Kim K.E., Lee S., Yoo J., Choi D., Lee Y.S., Aguilar B., Hong Y.-K. Visualization of lymphatic vessels by Prox1-promoter directed GFP reporter in a bacterial artificial chromosome-based transgenic mouse. Blood. 2011;117:362–365. doi: 10.1182/blood-2010-07-298562. [DOI] [PMC free article] [PubMed] [Google Scholar]; Choi, I., Chung, H.K., Ramu, S., Lee, H.N., Kim, K.E., Lee, S., Yoo, J., Choi, D., Lee, Y.S., Aguilar, B., and Hong, Y.-K. (2011). Visualization of lymphatic vessels by Prox1-promoter directed GFP reporter in a bacterial artificial chromosome-based transgenic mouse. Blood 117, 362-365. [DOI] [PMC free article] [PubMed]
- Choi E., Nahm F.S., Lee P.B. Sympathetic block as a new treatment for lymphedema. Pain Physician. 2015;18:365–372. [PubMed] [Google Scholar]; Choi, E., Nahm, F.S., and Lee, P.B. (2015). Sympathetic block as a new treatment for lymphedema. Pain Physician 18, 365-372. [PubMed]
- Chong C., Scholkmann F., Bachmann S.B., Luciani P., Leroux J.-C., Detmar M., Proulx S.T. In vivo visualization and quantification of collecting lymphatic vessel contractility using near-infrared imaging. Sci. Rep. 2016;6:22930. doi: 10.1038/srep22930. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chong, C., Scholkmann, F., Bachmann, S.B., Luciani, P., Leroux, J.-C., Detmar, M., and Proulx, S.T. (2016). In vivo visualization and quantification of collecting lymphatic vessel contractility using near-infrared imaging. Sci. Rep. 6, 22930. [DOI] [PMC free article] [PubMed]
- Christiansen A., Detmar M. Lymphangiogenesis and cancer. Genes Cancer. 2011;2:1146–1158. doi: 10.1177/1947601911423028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Christiansen, A., and Detmar, M. (2011). Lymphangiogenesis and cancer. Genes Cancer 2, 1146-1158. [DOI] [PMC free article] [PubMed]
- Dabov K., Foi A., Katkovnik V., Egiazarian K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans Image Process. 2007;16:2080–2095. doi: 10.1109/tip.2007.901238. [DOI] [PubMed] [Google Scholar]; Dabov, K., Foi, A., Katkovnik, V., and Egiazarian, K. (2007) Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans Image Process 16, 2080-2095. [DOI] [PubMed]
- D’Andrea V., Bianchi E., Taurone S., Mignini F., Cavallotti C., Artico M. Cholinergic innervation of human mesenteric lymphatic vessels. Folia Morphol. (Warsz) 2013;72:322–327. doi: 10.5603/fm.2013.0054. [DOI] [PubMed] [Google Scholar]; D’Andrea, V., Bianchi, E., Taurone, S., Mignini, F., Cavallotti, C., and Artico, M. (2013). Cholinergic innervation of human mesenteric lymphatic vessels. Folia Morphol. (Warsz) 72, 322-327. [DOI] [PubMed]
- D’Andrea V., Panarese A., Taurone S., Coppola L., Cavallotti C., Artico M. Human lymphatic mesenteric vessels: morphology and possible function of aminergic and NPY-ergic nerve fibers. Lymphat. Res. Biol. 2015;13:170–175. doi: 10.1089/lrb.2015.0018. [DOI] [PubMed] [Google Scholar]; D’Andrea, V., Panarese, A., Taurone, S., Coppola, L., Cavallotti, C., and Artico, M. (2015). Human lymphatic mesenteric vessels: morphology and possible function of aminergic and NPY-ergic nerve fibers. Lymphat. Res. Biol. 13, 170-175. [DOI] [PubMed]
- Danielyan A., Wu Y.W., Shih P.Y., Dembitskaya Y., Semyanov A. Denoising of two-photon fluorescence images with block-matching 3D filtering. Methods. 2014;68:308–316. doi: 10.1016/j.ymeth.2014.03.010. [DOI] [PubMed] [Google Scholar]; Danielyan, A., Wu, Y.W., Shih, P.Y., Dembitskaya, Y., and Semyanov, A. (2014) Denoising of two-photon fluorescence images with block-matching 3D filtering. Methods 68, 308-316. [DOI] [PubMed]
- Dobbins D.E. Catecholamine-mediated lymphatic constriction: involvement of both alpha 1- and alpha 2-adrenoreceptors. Am. J. Physiol. 1992;263:H473–H478. doi: 10.1152/ajpheart.1992.263.2.H473. [DOI] [PubMed] [Google Scholar]; Dobbins, D.E. (1992). Catecholamine-mediated lymphatic constriction: involvement of both alpha 1- and alpha 2-adrenoreceptors. Am. J. Physiol. 263, H473-H478. [DOI] [PubMed]
- Fang Y., Ding Z., Bi Y., Gong N., Liu Y., Wei L., Liu Z. Effect of vagotomy on dynamics of mesenteric lymphatic vessels in the rat. Chin. J. Physiol. 2007;50:89–92. [PubMed] [Google Scholar]; Fang, Y., Ding, Z., Bi, Y., Gong, N., Liu, Y., Wei, L., and Liu, Z. (2007). Effect of vagotomy on dynamics of mesenteric lymphatic vessels in the rat. Chin. J. Physiol. 50, 89-92. [PubMed]
- Ferguson M.K. Modulation of lymphatic smooth muscle contractile responses by the endothelium. J. Surg. Res. 1992;52:359–363. doi: 10.1016/0022-4804(92)90116-h. [DOI] [PubMed] [Google Scholar]; Ferguson, M.K. (1992). Modulation of lymphatic smooth muscle contractile responses by the endothelium. J. Surg. Res. 52, 359-363. [DOI] [PubMed]
- Gashev A.A. Lymphatic vessels: pressure- and flow-dependent regulatory reactions. Ann. N Y Acad. Sci. 2008;1131:100–109. doi: 10.1196/annals.1413.009. [DOI] [PubMed] [Google Scholar]; Gashev, A.A. (2008). Lymphatic vessels: pressure- and flow-dependent regulatory reactions. Ann. N Y Acad. Sci. 1131, 100-109. [DOI] [PubMed]
- Gogineni A., Caunt M., Crow A., Lee C.V., Fuh G., van Bruggen N., Ye W., Weimer R.M. Inhibition of VEGF-C modulates distal lymphatic remodeling and secondary metastasis. PLoS ONE. 2013;8:e68755. doi: 10.1371/journal.pone.0068755. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gogineni, A., Caunt, M., Crow, A., Lee, C.V., Fuh, G., van Bruggen, N., Ye, W., and Weimer, R.M. (2013). Inhibition of VEGF-C modulates distal lymphatic remodeling and secondary metastasis. PLoS ONE 8, e68755. [DOI] [PMC free article] [PubMed]
- Hashimoto S., Kawai Y., Ohhashi T. Effects of vasoactive substances on the pig isolated hepatic lymph vessels. J. Pharmacol. Exp. Ther. 1994;269:482–488. [PubMed] [Google Scholar]; Hashimoto, S., Kawai, Y., and Ohhashi, T. (1994). Effects of vasoactive substances on the pig isolated hepatic lymph vessels. J. Pharmacol. Exp. Ther. 269, 482-488. [PubMed]
- Hirakawa S., Hong Y.-K., Harvey N., Schacht V., Matsuda K., Libermann T., Detmar M. Identification of vascular lineage-specific genes by transcriptional profiling of isolated blood vascular and lymphatic endothelial cells. Am. J. Pathol. 2003;162:575–586. doi: 10.1016/S0002-9440(10)63851-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hirakawa, S., Hong, Y.-K., Harvey, N., Schacht, V., Matsuda, K., Libermann, T., and Detmar, M. (2003). Identification of vascular lineage-specific genes by transcriptional profiling of isolated blood vascular and lymphatic endothelial cells. Am. J. Pathol. 162, 575-586. [DOI] [PMC free article] [PubMed]
- Igarashi T., Ikomi F., Ohhashi T. Electrical stimulation-induced α1- and α2-adrenoceptors-mediated contraction in isolated dog thoracic ducts. J. Auton. Nerv. Syst. 1998;71:18–24. doi: 10.1016/s0165-1838(98)00057-5. [DOI] [PubMed] [Google Scholar]; Igarashi, T., Ikomi, F., and Ohhashi, T. (1998). Electrical stimulation-induced α1- and α2-adrenoceptors-mediated contraction in isolated dog thoracic ducts. J. Auton. Nerv. Syst. 71, 18-24. [DOI] [PubMed]
- Kalman R.E. A new approach to linear filtering and prediction problems. J. Basic Eng. 1960;82:35–45. [Google Scholar]; Kalman, R.E. (1960). A new approach to linear filtering and prediction problems. J. Basic Eng. 82, 35-45.
- Kornuta J.A., Nepiyushchikh Z., Gasheva O.Y., Mukherjee A., Zawieja D.C., Dixon J.B. Effects of dynamic shear and transmural pressure on wall shear stress sensitivity in collecting lymphatic vessels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;309:R1122–R1134. doi: 10.1152/ajpregu.00342.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kornuta, J.A., Nepiyushchikh, Z., Gasheva, O.Y., Mukherjee, A., Zawieja, D.C., and Dixon, J.B. (2015). Effects of dynamic shear and transmural pressure on wall shear stress sensitivity in collecting lymphatic vessels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 309, R1122-R1134. [DOI] [PMC free article] [PubMed]
- Kunert C., Baish J.W., Liao S., Padera T.P., Munn L.L. Mechanobiological oscillators control lymph flow. Proc. Natl. Acad. Sci. USA. 2015;112:10938–10943. doi: 10.1073/pnas.1508330112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kunert, C., Baish, J.W., Liao, S., Padera, T.P., and Munn, L.L. (2015). Mechanobiological oscillators control lymph flow. Proc. Natl. Acad. Sci. USA 112, 10938-10943. [DOI] [PMC free article] [PubMed]
- Lahdenranta J., Hagendoorn J., Padera T.P., Hoshida T., Nelson G., Kashiwagi S., Jain R.K., Fukumura D. Endothelial nitric oxide synthase mediates lymphangiogenesis and lymphatic metastasis. Cancer Res. 2009;69:2801–2808. doi: 10.1158/0008-5472.CAN-08-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lahdenranta, J., Hagendoorn, J., Padera, T.P., Hoshida, T., Nelson, G., Kashiwagi, S., Jain, R.K., and Fukumura, D. (2009). Endothelial nitric oxide synthase mediates lymphangiogenesis and lymphatic metastasis. Cancer Res. 69, 2801-2808. [DOI] [PMC free article] [PubMed]
- McHale N.G. Lymphatic innervation. Blood Vessels. 1990;27:127–136. doi: 10.1159/000158803. [DOI] [PubMed] [Google Scholar]; McHale, N.G. (1990). Lymphatic innervation. Blood Vessels 27, 127-136. [DOI] [PubMed]
- McHale N.G. Noradrenergic control of pumping in lymphatic vessels. Phlebology. 1992;7:26–29. [Google Scholar]; McHale, N.G. (1992). Noradrenergic control of pumping in lymphatic vessels. Phlebology 7, 26-29.
- McHale N.G., Roddie I.C., Thornbury K.D. Nervous modulation of spontaneous contractions in bovine mesenteric lymphatics. J. Physiol. 1980;309:461–472. doi: 10.1113/jphysiol.1980.sp013520. [DOI] [PMC free article] [PubMed] [Google Scholar]; McHale, N.G., Roddie, I.C., and Thornbury, K.D. (1980). Nervous modulation of spontaneous contractions in bovine mesenteric lymphatics. J. Physiol. 309, 461-472. [DOI] [PMC free article] [PubMed]
- Ohhashi T., Azuma T. Sympathetic effects on spontaneous activity in bovine mesenteric lymphatics. Retraction. Am. J. Physiol. 1986;247:H610–H615. doi: 10.1152/ajpheart.1984.247.4.H610. [DOI] [PubMed] [Google Scholar]; Ohhashi, T., and Azuma, T. (1986). Sympathetic effects on spontaneous activity in bovine mesenteric lymphatics. Retraction. Am. J. Physiol. 247, H610-H615. [DOI] [PubMed]
- Ohhashi T., Takahashi N. Acetylcholine-induced release of endothelium-derived relaxing factor from lymphatic endothelial cells. Am. J. Physiol. 1991;260:H1172–H1178. doi: 10.1152/ajpheart.1991.260.4.H1172. [DOI] [PubMed] [Google Scholar]; Ohhashi, T., and Takahashi, N. (1991). Acetylcholine-induced release of endothelium-derived relaxing factor from lymphatic endothelial cells. Am. J. Physiol. 260, H1172-H1178. [DOI] [PubMed]
- Ohhashi T., Kawai Y., Azuma T. The response of lymphatic smooth muscles to vasoactive substances. Pflugers Arch. 1978;375:183–188. doi: 10.1007/BF00584242. [DOI] [PubMed] [Google Scholar]; Ohhashi, T., Kawai, Y., and Azuma, T. (1978). The response of lymphatic smooth muscles to vasoactive substances. Pflugers Arch. 375, 183-188. [DOI] [PubMed]
- Ohhashi T., Kobayashi S., Tsukahara S., Azuma T. Innervation of bovine mesenteric lymphatics: from the histochemical point of view. Microvasc. Res. 1982;24:377–385. doi: 10.1016/0026-2862(82)90024-3. [DOI] [PubMed] [Google Scholar]; Ohhashi, T., Kobayashi, S., Tsukahara, S., and Azuma, T. (1982). Innervation of bovine mesenteric lymphatics: from the histochemical point of view. Microvasc. Res. 24, 377-385. [DOI] [PubMed]
- Ono N., Mizuno R., Nojiri H., Ohhashi T. Development of an experimental apparatus for investigating lymphatic pumping activity of murine mesentery in vivo. Jpn. J. Physiol. 2000;50:25–31. doi: 10.2170/jjphysiol.50.25. [DOI] [PubMed] [Google Scholar]; Ono, N., Mizuno, R., Nojiri, H., and Ohhashi, T. (2000). Development of an experimental apparatus for investigating lymphatic pumping activity of murine mesentery in vivo. Jpn. J. Physiol. 50, 25-31. [DOI] [PubMed]
- Otsu N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. Syst. 1979;SMC-9:62–66. [Google Scholar]; Otsu, N. (1979). A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. Syst. SMC-9, 62-66.
- Padera T.P., Kadambi A., di Tomaso E., Carreira C.M., Brown E.B., Boucher Y., Choi N.C., Mathisen D., Wain J., Mark E.J. Lymphatic metastasis in the absence of functional intratumor lymphatics. Science. 2002;296:1883–1886. doi: 10.1126/science.1071420. [DOI] [PubMed] [Google Scholar]; Padera, T.P., Kadambi, A., di Tomaso, E., Carreira, C.M., Brown, E.B., Boucher, Y., Choi, N.C., Mathisen, D., Wain, J., Mark, E.J., et al. (2002). Lymphatic metastasis in the absence of functional intratumor lymphatics. Science 296, 1883-1886. [DOI] [PubMed]
- Proulx S.T., Luciani P., Dieterich L.C., Karaman S., Leroux J.-C., Detmar M. Expansion of the lymphatic vasculature in cancer and inflammation: new opportunities for in vivo imaging and drug delivery. J. Control. Release. 2013;172:550–557. doi: 10.1016/j.jconrel.2013.04.027. [DOI] [PubMed] [Google Scholar]; Proulx, S.T., Luciani, P., Dieterich, L.C., Karaman, S., Leroux, J.-C., and Detmar, M. (2013). Expansion of the lymphatic vasculature in cancer and inflammation: new opportunities for in vivo imaging and drug delivery. J. Control. Release 172, 550-557. [DOI] [PubMed]
- Scallan J.P., Davis M.J. Genetic removal of basal nitric oxide enhances contractile activity in isolated murine collecting lymphatic vessels. J. Physiol. 2013;591:2139–2156. doi: 10.1113/jphysiol.2012.250662. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scallan, J.P., and Davis, M.J. (2013). Genetic removal of basal nitric oxide enhances contractile activity in isolated murine collecting lymphatic vessels. J. Physiol. 591, 2139-2156. [DOI] [PMC free article] [PubMed]
- Scallan J.P., Zawieja S.D., Castorena-Gonzalez J.A., Davis M.J. Lymphatic pumping: mechanics, mechanisms and malfunction. J. Physiol. 2016;594:5749–5768. doi: 10.1113/JP272088. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scallan, J.P., Zawieja, S.D., Castorena-Gonzalez, J.A., and Davis, M.J. (2016). Lymphatic pumping: mechanics, mechanisms and malfunction. J. Physiol. 594, 5749-5768. [DOI] [PMC free article] [PubMed]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. [DOI] [PMC free article] [PubMed]
- Skobe M., Hamberg L.M., Hawighorst T., Schirner M., Wolf G.L., Alitalo K., Detmar M. Concurrent induction of lymphangiogenesis, angiogenesis, and macrophage recruitment by vascular endothelial growth factor-C in melanoma. Am. J. Pathol. 2001;159:893–903. doi: 10.1016/S0002-9440(10)61765-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Skobe, M., Hamberg, L.M., Hawighorst, T., Schirner, M., Wolf, G.L., Alitalo, K., and Detmar, M. (2001). Concurrent induction of lymphangiogenesis, angiogenesis, and macrophage recruitment by vascular endothelial growth factor-C in melanoma. Am. J. Pathol. 159, 893-903. [DOI] [PMC free article] [PubMed]
- Stacker S.A., Williams S.P., Karnezis T., Shayan R., Fox S.B., Achen M.G. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer. 2014;14:159–172. doi: 10.1038/nrc3677. [DOI] [PubMed] [Google Scholar]; Stacker, S.A., Williams, S.P., Karnezis, T., Shayan, R., Fox, S.B., and Achen, M.G. (2014). Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer 14, 159-172. [DOI] [PubMed]
- Takahashi N., Kawai Y., Ohhashi T. Effects of vasoconstrictive and vasodilative agents on lymphatic smooth muscles in isolated canine thoracic ducts. J. Pharmacol. Exp. Ther. 1990;254:165–170. [PubMed] [Google Scholar]; Takahashi, N., Kawai, Y., and Ohhashi, T. (1990). Effects of vasoconstrictive and vasodilative agents on lymphatic smooth muscles in isolated canine thoracic ducts. J. Pharmacol. Exp. Ther. 254, 165-170. [PubMed]
- Tallini Y.N., Brekke J.F., Shui B., Doran R., Hwang S.M., Nakai J., Salama G., Segal S.S., Kotlikoff M.I. Propagated endothelial Ca2+ waves and arteriolar dilation in vivo: measurements in Cx40BAC GCaMP2 transgenic mice. Circ. Res. 2007;101:1300–1309. doi: 10.1161/CIRCRESAHA.107.149484. [DOI] [PubMed] [Google Scholar]; Tallini, Y.N., Brekke, J.F., Shui, B., Doran, R., Hwang, S.M., Nakai, J., Salama, G., Segal, S.S., and Kotlikoff, M.I. (2007). Propagated endothelial Ca2+ waves and arteriolar dilation in vivo: measurements in Cx40BAC GCaMP2 transgenic mice. Circ. Res. 101, 1300-1309. [DOI] [PubMed]
- Telinius N., Drewsen N., Pilegaard H., Kold-Petersen H., de Leval M., Aalkjaer C., Hjortdal V., Boedtkjer D.B. Human thoracic duct in vitro: diameter-tension properties, spontaneous and evoked contractile activity. Am. J. Physiol. Heart Circ. Physiol. 2010;299:H811–H818. doi: 10.1152/ajpheart.01089.2009. [DOI] [PubMed] [Google Scholar]; Telinius, N., Drewsen, N., Pilegaard, H., Kold-Petersen, H., de Leval, M., Aalkjaer, C., Hjortdal, V., and Boedtkjer, D.B. (2010). Human thoracic duct in vitro: diameter-tension properties, spontaneous and evoked contractile activity. Am. J. Physiol. Heart Circ. Physiol. 299, H811-H818. [DOI] [PubMed]
- Telinius N., Baandrup U., Rumessen J., Pilegaard H., Hjortdal V., Aalkjaer C., Boedtkjer D.B. The human thoracic duct is functionally innervated by adrenergic nerves. Am. J. Physiol. Heart Circ. Physiol. 2014;306:H206–H213. doi: 10.1152/ajpheart.00517.2013. [DOI] [PubMed] [Google Scholar]; Telinius, N., Baandrup, U., Rumessen, J., Pilegaard, H., Hjortdal, V., Aalkjaer, C., and Boedtkjer, D.B. (2014). The human thoracic duct is functionally innervated by adrenergic nerves. Am. J. Physiol. Heart Circ. Physiol. 306, H206-H213. [DOI] [PubMed]
- Todd G.L., Bernard G.R. The sympathetic innervation of the cervical lymphatic duct of the dog. Anat. Rec. 1973;177:303–315. doi: 10.1002/ar.1091770210. [DOI] [PubMed] [Google Scholar]; Todd, G.L., and Bernard, G.R. (1973). The sympathetic innervation of the cervical lymphatic duct of the dog. Anat. Rec. 177, 303-315. [DOI] [PubMed]
- Wick N., Saharinen P., Saharinen J., Gurnhofer E., Steiner C.W., Raab I., Stokic D., Giovanoli P., Buchsbaum S., Burchard A. Transcriptomal comparison of human dermal lymphatic endothelial cells ex vivo and in vitro. Physiol. Genomics. 2007;28:179–192. doi: 10.1152/physiolgenomics.00037.2006. [DOI] [PubMed] [Google Scholar]; Wick, N., Saharinen, P., Saharinen, J., Gurnhofer, E., Steiner, C.W., Raab, I., Stokic, D., Giovanoli, P., Buchsbaum, S., Burchard, A., et al. (2007). Transcriptomal comparison of human dermal lymphatic endothelial cells ex vivo and in vitro. Physiol. Genomics 28, 179-192. [DOI] [PubMed]
- Woo J.H., Park H.S., Kim S.C., Kim Y.H. The effect of lumbar sympathetic ganglion block on gynecologic cancer-related lymphedema. Pain Physician. 2013;16:345–352. [PubMed] [Google Scholar]; Woo, J.H., Park, H.S., Kim, S.C., and Kim, Y.H. (2013). The effect of lumbar sympathetic ganglion block on gynecologic cancer-related lymphedema. Pain Physician 16, 345-352. [PubMed]
- Zawieja D.C., Davis K.L., Schuster R., Hinds W.M., Granger H.J. Distribution, propagation, and coordination of contractile activity in lymphatics. Am. J. Physiol. 1993;264:H1283–H1291. doi: 10.1152/ajpheart.1993.264.4.H1283. [DOI] [PubMed] [Google Scholar]; Zawieja, D.C., Davis, K.L., Schuster, R., Hinds, W.M., and Granger, H.J. (1993). Distribution, propagation, and coordination of contractile activity in lymphatics. Am. J. Physiol. 264, H1283-H1291. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Quantification of low frequency contractions
Experiments performed with isoflurane resulted in a much lower contraction activity which could not be analyzed with the previously developed algorithm (Chong et al., 2016). Therefore, a new algorithm was developed (Figure S4). In order to determine the characteristics of the peaks (i.e., amplitudes and periodicity) of the measured fluorescence signal, a MATLAB (MathWorks Inc., Natick MA, USA) function with a graphical user interface was developed that performs the signal processing in a semi-automatically manner. The user selects the peaks of the fluorescence signal manually and based on these selected values, the absolute amplitudes, relative amplitudes and the frequency are then calculated automatically. These parameters were calculated according to the formulas
| (1) |
| (2) |
| (3) |
with the baseline value of each peak according to in case of or in case of , and the time-difference between the two peaks and with the first peak assigned to and the second to (and so on). For a visualization of the mathematical nomenclature used, see Figure S4. The formulation ensures that the amplitude and periodicity values are also calculated correctly in case the signal contains negative peaks, or a mixture of positive and negative peaks.
MATLAB Code
function [Amplitude_abs, Amplitude_rel, Period, Frequency] = AnalyzePeaks_Ampl_Freq_v2(x,f_s)
% Function to analyze a time-series regarding the characteritics of the
% peaks (i.e., amplitudes, period)
% _________________________________________________________________________
% For Steven Proulx and Samia Bachmann, ETH.
% May 2017 | Felix Scholkmann | Version 2
%__________________________________________________________________________
% INPUT
% x: signal to analyze
% f_s: sampling frequency in Hz
% OUTPUT
% Amplitude_abs: absolute amplitude values (first value: median, second value: inter quantile range, IQR)
% Amplitude_rel: relative amplitude values (first value: median, second value: inter quantile range, IQR)
% Period: period values [in seconds] (first value: median, second value: inter quantile range, IQR)
% Frequency: frequency values [in 1/min] (first value: median, second value: inter quantile range, IQR)
% important:
% The input signal (x) must contain at least 3 peask (i.e., at least 9
% points needs to be selected from the signal). This is necessary to be
% able to calculate the period values.
%% ________________________________________________________________________
close all
scrsz = get(0,’ScreenSize’); figure(‘Position’,[100 50 1300 850]);
set(gcf., ‘color’, ‘w’); set(gcf.,’defaultaxesfontsize’,12)
subplot(2,20,[1,3])
% Please insert here the path to the file “Points.png”
currentFolder = pwd;
Path = strcat(currentFolder,’\Points.png’)
img = imread(Path);
image(img);
set(gca, ‘XTickLabelMode’, ‘Manual’)
set(gca, ‘XTick’, [])
set(gca, ‘YTickLabelMode’, ‘Manual’)
set(gca, ‘YTick’, [])
title (‘Definition’,’fontsize’,14)
fig = subplot(2,20,[6,20]);
plot(t,x,’linewidth’,2,’Color’,[0.3, 0.5, 1])
axis tight
ylabel(‘Intensity’); xlabel(‘Time [s]’)
title (‘- > Please select the points (\alpha_{i}, \alpha_{i+1}, \alpha_{i+2}, i = 1, 2, …, \it{N})’,’fontsize’,14)
% Get the points from the figure
disp(‘→ Please select the points in the figure. The press “Enter”’)
[x, y] = getpts(fig);
hold on
vline_red(x(2:3:end))
vline_black(x(1:3:end))
vline_black(x(3:3:end))
n = length(x);
% Calculate the amplitudes
j = 1;
- for i = 1:3:n-2;
- if y(i+2) > y(i)
- M(j) = (y(i+2)-y(i))/2 + y(i);
- A_abs(j) = abs(M(j) - y(i+1));
- A_rel(j) = 100 ∗ (A_abs(j)/M(j));
- j = j+1;
- else
- M(j) = (y(i)-y(i+2))/2 + y(i+2);
- A_abs(j) = abs(M(j) - y(i+1));
- A_rel(j) = 100 ∗ (A_abs(j)/M(j));
- j = j+1;
- end
end
plot(x(2:3:end),M,’ob’)
plot(x(1:3:end),y(1:3:end),’ok’)
plot(x(3:3:end),y(3:3:end),’ok’)
plot(x(2:3:end),y(2:3:end),’or’)
% Calculate the periods
j = 1;
- for i = 1:3:n-4;
- P(j) = x(i+4)-x(i+1);
- j = j+1;
end
F = (1./(P))∗60; % Frequency [1/min]
subplot(2,20,[21:23])
bar(A_abs,’b’);
ylabel(‘Amplitude (A_{abs}) [absolute]’); xlabel(‘Peak number’)
title ({’Amplitude values’,’[absolute]’},’fontsize’,14)
xlim([0,length(A_abs)+1])
subplot(2,20,[25:27])
bar(A_rel,’b’);
ylabel(‘Amplitude (A_{rel}) [%]’); xlabel(‘Peak number’)
title ({’Amplitude values’,’[%]’},’fontsize’,14)
xlim([0,length(A_rel)+1])
subplot(2,20,[29:31])
bar(F,’r’);
ylabel(‘Frequency [1/min]’); xlabel(‘Peak number’)
title ({’Frequency values’,’[1/min]’},’fontsize’,14)
xlim([0,length(F)+1])
subplot(2,20,35:36)
errorbar(median(A_rel),iqr(A_rel)/2,’k’);
hold on
plot(median(A_rel),’k.’,’markersize’,30)
ylabel(‘Amplitude [%] (median +- IQR)’)
set(gca, ‘XTickLabelMode’, ‘Manual’)
set(gca, ‘XTick’, [])
title([‘A_{rel} = ’ num2str(median(A_rel)),’ (‘ num2str(iqr(A_rel)),’) %’]);
subplot(2,20,39:40)
errorbar(median(F),iqr(F)/2,’k’);
hold on
plot(median(F),’k.’,’markersize’,30)
ylabel(‘Frequency [1/min] (median +- IQR)’)
set(gca, ‘XTickLabelMode’, ‘Manual’)
set(gca, ‘XTick’, [])
title([‘F = ’ num2str(median(F)),’ (‘ num2str(iqr(F)),’) 1/min’]);
% Generate output data
Amplitude_abs = [median(A_abs), iqr(A_abs)];
Amplitude_rel = [median(A_rel), iqr(A_rel)];
Period = [median(P), iqr(P)];
Frequency = [median(F), iqr(F)];
Quantification of in vitro Ca2+ imaging
Pre-processing
An important step to restore images is denoising, which consists of reducing noise present in images but at the same time preserving objects’ features such as edges, texture, etc. In this context, we have opted to use the Block-Matching and 3D filtering (BM3D) method which has shown its denoising capabilities in different datasets by preserving finer image structures (Dabov et al., 2007, Danielyan et al., 2014).
Cell Detection
Once the image has been pre-processed, the next step is to detect the foreground objects, i.e., cells. To do this, we use the Otsu thresholding method (Otsu, 1979), however instead of using a single threshold value, we adopt a two-level threshold to detect the nucleus and the cytoplasm as joint single objects.
After thresholding, small objects are further removed since they usually correspond to non-relevant objects, such as air-bubbles. In addition, since some cells may spatially be close to each other, they can end up being merged after thresholding. To cope with this issue, we use the Watershed Transform. More precisely, each segmented object is represented as a basin and the detected Watershed Lines with the distance transform are used to separate the objects.
Cell Linking
Cells are detected at each single frame separately. In order to analyze how a cell activates over time, the detected cells are first linked together over time using the Kalman filter (Kalman, 1960).
The Kalman filter comprises two steps: prediction and update. In the prediction step, the Kalman filter predicts the current state of the system, based on the past, through a dynamic model. In the update step, the status of the system is corrected based on the observed variables. In our case, the assignment/linking of a given cell to a specific track is based on the area of overlap of the segmented cells between consecutive frames. The Kalman Filter therefore seeks to maximize the area of overlap between cell detections over time. To obtain more robust predictions at time t, our system averages the bounding-box of the detected cells over the last four frames. To remove false positives such as air-bubbles, our system discards objects with large location changes over time. Also, short tracks are discarded for further processing. In our case, we impose the constraint that tracks shall cover at least 75% of the total length of the input video.
Event Detection
Our event detection module is versatile and besides detecting which cells got activated, it can also find out when exactly a given activation starts and ends over time. The module starts first by smoothing the cell’s intensity signal (the cell-intensity track) to reduce noise. Smoothing is done with a moving average filter. Note that the smoothing step is optional and the raw signal can be directly used if so desired.
Next, the cell signals are normalized to compensate for effects such as photobleaching and also to facilitate the detection of cell activations in the subsequent steps. The normalization of a cells’ signal is done by computing the average intensity , of the detected cell at the frames before the injection time (see Equation 1).
| (Equation 1) |
where is the mean intensity value of the collection of pixels belonging to the detected cell i at time t.
The computed mean value of cell i is used to normalize the signal by dividing it into the cell signal values at all later times (see Equation 2).
| (Equation 2) |
where is the normalized cells’ signal and N corresponds to the total number of frames. Figure S3B shows an example of cell signals before (upper graph) and after normalization (lower graph). Detected activations above the threshold value (displayed as a black dashed line) are shaded, see next section of details of the detection algorithm.
To detect cell-activations, the normalized cell signals are thresholded. More precisely, signal values larger than a threshold value are considered as activations and values lower than the threshold are set to zero. We set this threshold value empirically to be 1.25. Note that this value can be tuned, if more detections are targeted, one can increase this value. It is important though to set this value in a conservative manner. An example of cell activation is displayed in yellow in the left graph of Figure S3C. The thresholding step is applied for all cell signals, in such a way that all activations can be mapped as an activation map. In Figure S3C, each row represents a single cell signal and its mean-intensity pixel values are displayed for each frame along the x axis. After thresholding the cell signals on the normalized cell signals (middle graph in Figure S3C), a binary activation map is obtained in which cell activations are displayed as white stripes (right graph, Figure S3C). The signal values where the cell did not activate are set to zero. In addition, to remove potential false negatives, our method first fills short gaps inside an event and also prunes short cell activations to remove false positives. In our experiments, both values were empirically set to five frames.
One important property of our method is that it detects where a cell activation starts and ends. This is done by computing the first order derivative over the binary activated signal and by looking respectively for the indexes where the derivative is respectively positive or negative (i.e., +1 or −1).
MATLAB Code (2 parts)
function kalmanMultiObjectTracking(videoName)
% KalmanMultiObjectTracking: this function detects a set of cells and
% tracks them over time. Cell detection involves image pre-processing where
% the original video frames are enhanced, e.g., denoising. The cell
% detections are mapped into cell-tracks, i.e., a set of temporal
% bounding-boxes for each detected cell.
%
% Input:
% videoName, e.g., videoName = ‘hu_NE1uM_2.avi’;
%
% Output:
- % tracksHist.mat MATLAB file containing the cell-tracks.
- opts = getDefaultParameters(); %# reading configuration parameters.
- pathInputVideo = fullfile(opts.videoPath, videoName);
- digitStr = ‘%d-’;
- strAllMotionDigits = repmat(digitStr, 1, numel(opts.motionNoise));
- motionNoiseStr = sprintf(strAllMotionDigits(1:end-1),opts.motionNoise);
- strAllInitErrorDigits = repmat(digitStr, 1, numel(opts.initialEstimateError));
- initErrorStr = sprintf(strAllInitErrorDigits(1:end-1),opts.initialEstimateError);
- paramsDir = sprintf(‘%s_LRate%2.2f_visi%2.2f_age%d_invib%d_initErr%s_motNoise%s_measNoise%d_%s_cSze%d’, …
- opts.motionModel, opts.learningRate, opts.trackVisibility, opts.ageThreshold, …
- opts.invisibleForTooLong, initErrorStr, motionNoiseStr, opts.measurementNoise, opts.thresh.type, opts.cell.size);
- [videosPath, inVideoname, ∼] = fileparts(pathInputVideo);
- videoNameOutputTrackRGB = sprintf(‘%s_track_rgb.avi’, inVideoname);
- videoNameOutputTrackProc = sprintf(‘%s_track_proc.avi’, inVideoname);
- videoNameOutputTrackBW = sprintf(‘%s_track_bw.avi’, inVideoname);
- videoNameOutputBW = sprintf(‘%s_bw.avi’, inVideoname);
- tracksDir = fullfile(videosPath, ‘tracks_kalmann_sel’);
- tracksDirVideo = fullfile(tracksDir, inVideoname, paramsDir);
- opts.framesOrigDir = fullfile(tracksDir, inVideoname, ‘frames’);
- opts.framesProcDir = fullfile(tracksDirVideo, ‘frameProc’);
- opts.maskDir = fullfile(tracksDirVideo, ‘frameMasks’);
- opts.maskDirTracks = fullfile(tracksDirVideo, ‘frameMasksTracks’);
- opts.framesOrigTracksDir = fullfile(tracksDirVideo, ‘framesOrigTracks’);
- opts.framesProcTracksDir = fullfile(tracksDirVideo, ‘framesProcTracks’);
- opts.maskDirTracksAll = fullfile(tracksDirVideo, ‘frameMasksTracksAll’);
- opts.framesOrigTracksDirAll = fullfile(tracksDirVideo, ‘framesOrigTracksAll’);
- createVideoDirs(tracksDirVideo, opts); %# creating output directories
- videoObjTrackRGB = VideoWriter(fullfile(tracksDirVideo, videoNameOutputTrackRGB), ‘Uncompressed AVI’);
- videoObjTrackProc = VideoWriter(fullfile(tracksDirVideo, videoNameOutputTrackProc), ‘Uncompressed AVI’);
- videoObjTrackBW = VideoWriter(fullfile(tracksDirVideo, videoNameOutputTrackBW), ‘Uncompressed AVI’);
- videoObjBW = VideoWriter(fullfile(tracksDirVideo, videoNameOutputBW), ‘Uncompressed AVI’);
- fprintf(‘video: %s\n’,(fullfile(tracksDirVideo, videoNameOutputTrackRGB)));
- tracksObj = fullfile(tracksDirVideo, sprintf(‘%s.mat’, inVideoname));
- obj = setupSystemObjects(pathInputVideo, opts);
- tracks = initializeTracks(); %# Create an empty array of tracks.
- tracksHist = initializeTracksHistory();
- nextId = 1; % ID of the next track
- noFrames = 0;
- if ∼exist(tracksObj, ‘file’)
- %# Detect moving objects, and track them across video frames.
- while ∼isDone(obj.reader)
- noFrames = noFrames + 1;
- fprintf(‘# Processing frame: %d\n’, noFrames);
- frame = readFrame(obj);
- %# image pre-processing and per-frame cell detection.
- [centroids, bboxes, mask, frameDen] = detectObjects(frame, opts);
- %# predict cell location.
- [tracks, tracksHist] = predictNewLocationsOfTracks(tracks, tracksHist, noFrames);
- %# link cells over time.
- [assignments, unassignedTracks, unassignedDetections] = …
- detectionToTrackAssignment(tracks, centroids, bboxes, opts);
- %# update existing cell-tracks.
- [tracks, tracksHist] = updateAssignedTracks(tracks, tracksHist, assignments, centroids, bboxes, noFrames);
- [tracks, tracksHist] = updateUnassignedTracks(tracks, tracksHist, unassignedTracks);
- %# delete short tracks.
- [tracks, tracksHist] = deleteLostTracks(tracks, tracksHist, opts);
- %# create new tracks.
- [tracks, tracksHist, nextId] = createNewTracks(tracks, tracksHist, centroids, bboxes, unassignedDetections, nextId, noFrames);
- %# saving tracks to disk.
- dumpTrackingResults(frame, mask, tracks, tracksHist, tracksDirVideo, videoObjTrackRGB, videoObjTrackBW, videoObjBW, noFrames, opts);
- end
- opts.totNoFrames = noFrames;
- tracksHist = updateEmptyTracks(tracksHist, opts); %# check empty tracks frames and reduce its totalVisibleCount
- save(tracksObj, ‘tracksHist’, ‘-v7.3′); %# save to disk the detected tracks.
- else
- fprintf(‘# Loading tracks\n’);
- tracksHist = load(tracksObj);
- tracksHist = tracksHist.tracksHist;
- reset(obj.reader);
- while ∼isDone(obj.reader)
- frame = readFrame(obj);
- noFrames = noFrames + 1;
- end
- fprintf(‘# No of. frames: %d\n’, noFrames);
- end
- opts.totNoFrames = noFrames;
- if ∼isfield(tracksHist, ‘numEmptyMask’)
- tracksHist = updateEmptyTracks(tracksHist, opts);
- end
- open(videoObjTrackRGB); %# create RGB video with cell tracks.
- open(videoObjTrackProc); %# create RGB video for cell tracks (segmented/cells + bounding boxes).
- open(videoObjTrackBW); %# create BW video for cell tracks (segmented/detected cells + bounding boxes).
- open(videoObjBW);%# create BW video file for cell detections (segmented/detected cells as binarys masks).
- noFrames = 0;
- for ff = 1:opts.totNoFrames
- noFrames = noFrames + 1;
- frameOrigFile = fullfile(opts.framesOrigDir, sprintf(‘frame_%05d.tif’, ff));
- frameDenFile = fullfile(opts.framesProcDir, sprintf(‘frame_%05d.tif’, ff));
- frameMaskFile = fullfile(opts.maskDir, sprintf(‘frame_%05d.tif’, ff));
- fprintf(‘#Processing frame: %d\n’, noFrames);
- if exist(frameOrigFile, ‘file’) && exist(frameDenFile, ‘file’) && exist(frameMaskFile, ‘file’)
- frameOrig = imread(frameOrigFile);
- frameDen = imread(frameDenFile);
- frameMask = imread(frameMaskFile);
- %# storing into disk each frame overlayed with the cell-tracks.
- saveTrackingResults(frameOrig, frameDen, frameMask, tracksHist, videoObjTrackRGB, videoObjTrackProc, videoObjTrackBW, videoObjBW, ff, opts);
- end
- end
- close(videoObjTrackRGB); %# closing videoFile.
- close(videoObjTrackProc); %# closing videoFile.
- close(videoObjTrackBW); %# closing videoFile.
- close(videoObjBW); %# closing videoFile.
end
function tracksThresh2Activations(videoName)
% tracksThresh2Activations: given the extracted cell-tracks of a
% given video (linked cells over time), this function detects which of
% the cells got activated. For that purposes, the mean pixel intensity
% of a segmented cell is computed at each frame. If the mean pixel
% intensity value is higher than a threshold, then an activation is fired.
% The original intensity values can also be smoothed to remove noise and if
% the interval of the activation is too short, then a false-positive activation
% is detected.
%
% Input:
% videoName, e.g., videoName = ‘hu_NE1uM_2.avi’;
%
% Output:
% a set of figures are stored into disk, containing a
- % matrix-representation of the activated cell-signals.
- addpath(genpath(‘./3rd/overlay/’));
- %# reading default parameters, e.g., location of detected cells frames.
- opts = getDefaultParameters();
- if ∼isfield(opts, ‘endThreatment’)
- opts.endThreatment = 10;
- end
- if ∼isfield(opts, ‘signal’)
- opts.signal = [];
- end
- if ∼isfield(opts.signal, ‘Type’)
- opts.signal.Type = ‘orig’;
- end
- %# smoothing type for the cell signal.
- if ∼isfield(opts.signal, ‘smoothType’)
- opts.signal.smoothType = ‘box’; % ‘golay’, ‘gauss’, ‘box’
- end
- %# threshold value
- if ∼isfield(opts.signal, ‘threshVal’)
- opts.signal.threshVal = 1.25;
- end
- %# minimum duration of cell-activation
- if ∼isfield(opts.signal, ‘pruneEventDuration’)
- opts.signal.pruneEventDuration = 5;
- end
- if ∼isfield(opts.signal, ‘fillGaps’)
- opts.signal.fillGaps = true;
- end
- if ∼isfield(opts.signal, ‘fillGapsSze’)
- opts.signal.fillGapsSze = 5;
- end
- [videosPath, videoNameInput, ∼] = fileparts(videoName);
- digitStr = ‘%d-’;
- strAllMotionDigits = repmat(digitStr, 1, numel(opts.motionNoise));
- motionNoiseStr = sprintf(strAllMotionDigits(1:end-1),opts.motionNoise);
- strAllInitErrorDigits = repmat(digitStr, 1, numel(opts.initialEstimateError));
- initErrorStr = sprintf(strAllInitErrorDigits(1:end-1),opts.initialEstimateError);
- paramsDir = sprintf(‘%s_LRate%2.2f_visi%2.2f_age%d_invib%d_initErr%s_motNoise%s_measNoise%d_%s_cSze%d’, …
- opts.motionModel, opts.learningRate, opts.trackVisibility, opts.ageThreshold, …
- opts.invisibleForTooLong, initErrorStr, motionNoiseStr, opts.measurementNoise, opts.thresh.type, opts.cell.size);
- %# creating subdirectories where resuls are going to be stored.
- tracksDir = fullfile(videosPath, ‘tracks_paper’);
- tracksDirVideo = fullfile(tracksDir, videoNameInput, paramsDir);
- signalDir = fullfile(tracksDirVideo, ‘signal’);
- signalThreshDir = fullfile(signalDir, ‘thresh’);
- signalEventsDir = fullfile(signalDir, ‘events’);
- if ∼exist(signalEventsDir, ‘dir’), mkdir(signalEventsDir); end
- tracksFile = fullfile(tracksDirVideo, ‘validTracksIdx.mat’);
- validTracksIdx = load(tracksFile);
- validTracksIdx = validTracksIdx.validTracksIdx;
- %# loading detected cell-tracks
- if strcmpi(opts.signal.Type, ‘orig’)
- threshSubDir = fullfile(signalThreshDir, ‘origdata’);
- signalMeanFile = fullfile(tracksDirVideo, ‘signalMean.mat’);
- signalMean = load(signalMeanFile);
- signalMean = signalMean.signalMean;
- signalMean(∼validTracksIdx,:) = [];
- signalArray = signalMean;
- if ∼exist(threshSubDir, ‘dir’), error(‘Unknown directory: %s\n’, threshSubDir); end
- signalEventsDir = fullfile(signalEventsDir, ‘origdata’);
- if ∼exist(signalEventsDir, ‘dir’), mkdir(signalEventsDir); end
- signalTypeStr = opts.signal.Type;
- tracksIdx = find(validTracksIdx);
- elseif strcmpi(opts.signal.Type, ‘smooth’)
- fprintf(‘# Loading %s data\n’, opts.signal.smoothType);
- signalSmoothFile = fullfile(signalDir, ‘smooth’, sprintf(‘tracksSummary_%s.mat’, opts.signal.smoothType));
- signalMeanSmooth = load(signalSmoothFile);
- signalMeanSmooth = signalMeanSmooth.signalMeanSmooth;
- signalMeanSmooth(∼validTracksIdx,:) = [];
- signalArray = signalMeanSmooth;
- threshSubDir = fullfile(signalThreshDir, opts.signal.smoothType);
- if ∼exist(threshSubDir, ‘dir’), error(‘Unknown directory: %s\n’, threshSubDir); end
- signalEventsDir = fullfile(signalEventsDir, opts.signal.smoothType);
- if ∼exist(signalEventsDir, ‘dir’), mkdir(signalEventsDir); end
- signalTypeStr = sprintf(‘%s-%s’, opts.signal.Type, opts.signal.smoothType);
- tracksIdx = 1:size(signalArray,1);
- else
- error(‘Unkown type: %s’, opts.signal.Type);
- end
- signalArrayNormFilename = fullfile(threshSubDir, ‘signalNorm.mat’);
- if ∼exist(signalArrayNormFilename, ‘file’), error(‘Unknown file: %s\n’, signalArrayNormFilename); end
- signalNorm = load(signalArrayNormFilename);
- signalNorm = signalNorm.signalNorm;
- %# thresholding for activation.
- signalBinaryActivation = signalNorm > opts.signal.threshVal;
- [noTracks, ∼] = size(signalBinaryActivation);
- %# fill possible gaps in the cell-activations
- if opts.signal.fillGaps > 0
- filledSignal = signalBinaryActivation;
- for ii = 1:noTracks
- filledSignal(ii,:) = ∼bwareaopen(∼filledSignal(ii,:), opts.signal.fillGapsSze);
- end
- signalBinaryActivation = filledSignal;
- strFillGap = sprintf(‘fillSze%d’, opts.signal.fillGapsSze);
- else
- strFillGap = sprintf(‘nofill’);
- end
- signalActivation = zeros(size(signalBinaryActivation));
- if opts.signal.pruneEventDuration > 0
- strPruneEvent = sprintf(‘pruned%d’, opts.signal.pruneEventDuration);
- else
- strPruneEvent = sprintf(‘noprune’);
- end
- %# analyzing cell-track duration.
- stats = repmat(struct(‘startIdx’,[], ‘endIdx’, [], ‘duration’, [], ‘eventIdx’, [], ‘activation’, 0), noTracks, 1);
- for tt = 1:noTracks
- trackActivation = signalBinaryActivation(tt,:);
- trackActivation = diff([0 trackActivation 0]);
- eventStartIdx = find(trackActivation > 0);
- eventEndIdx = find(trackActivation < 0)-1;
- eventDuration = eventEndIdx - eventStartIdx + 1;
- if opts.signal.pruneEventDuration
- stringIdx = (eventDuration > = opts.signal.pruneEventDuration);
- eventStartIdx = eventStartIdx(stringIdx);
- eventEndIdx = eventEndIdx(stringIdx);
- end
- eventIdx = zeros(1,max(eventEndIdx)+1);
- eventIdx(eventStartIdx) = 1;
- eventIdx(eventEndIdx+1) = eventIdx(eventEndIdx+1)-1;
- eventIdx = find(cumsum(eventIdx));
- signalActivation(tt, eventIdx) = 2;
- signalActivation(tt,eventStartIdx) = +1;
- signalActivation(tt,eventEndIdx) = −1;
- stats(tt).startIdx = eventStartIdx;
- stats(tt).endIdx = eventEndIdx;
- stats(tt).duration = eventDuration;
- stats(tt).eventIdx = eventIdx;
- stats(tt).activation = isempty(eventIdx) = = 0;
- end
- videoName4Plot = strrep(videoNameInput,’_’,’\_’);
- signalImFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_%s_label.tif’, videoNameInput, signalTypeStr));
- normSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_norm.tif’, videoNameInput));
- activationSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_activ_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- binActivationSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_bin_activ_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- signalEventsDirBox = strrep(signalEventsDir, ‘origdata’, ‘box’);
- activationBoxSignalFilename = fullfile(signalEventsDirBox, sprintf(‘%s_signal_activ_%s_%s_%s.mat’, videoNameInput, signalTypeStr, strPruneEvent, strFillGap));
- activationBoxSignalFilename = strrep(activationBoxSignalFilename, ‘orig’, ‘smooth-box’);
- signalActivationBox = load(activationBoxSignalFilename);
- signalActivationBox = signalActivationBox.signalActivation;
- signal2disk(signalArray, videoName4Plot, signalImFilename, opts);
- signal2disk(signalNorm, videoName4Plot, normSignalFilename, opts);
- signal2disk(signalActivation, videoName4Plot, activationSignalFilename, opts);
- signal2disk(signalActivation∼ = 0, videoName4Plot, binActivationSignalFilename, opts);
- overlaySignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlay_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- overlayNormSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlaynorm_%s_%s_%s.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- signal2diskOverlay(signalArray, signalActivation∼ = 0, videoName4Plot, overlaySignalFilename, opts);
- signal2diskOverlay(signalNorm, signalActivation∼ = 0, videoName4Plot, overlayNormSignalFilename, opts);
- overlaySignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlay_%s_%s_%s_box.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- overlayNormSignalFilename = fullfile(signalEventsDir, sprintf(‘%s_signal_overlaynorm_%s_%s_%s_box.tif’, videoNameInput, strPruneEvent, strFillGap, signalTypeStr));
- signal2diskOverlay(signalArray, signalActivationBox∼ = 0, videoName4Plot, overlaySignalFilename, opts);
- signal2diskOverlay(signalNorm, signalActivationBox∼ = 0, videoName4Plot, overlayNormSignalFilename, opts);
- signalFilenameMat = fullfile(signalEventsDir, sprintf(‘%s_signal_activ_%s_%s_%s.mat’, videoNameInput, signalTypeStr, strPruneEvent, strFillGap));
- save(signalFilenameMat, ‘signalActivation’);
- signalStatsFilenameMat = fullfile(signalEventsDir, sprintf(‘%s_stats_%s_%s_%s.mat’, videoNameInput, signalTypeStr, strPruneEvent, strFillGap));
- save(signalStatsFilenameMat, ‘stats’);
end