Abstract
Human pluripotent stem cells (hPSCs), such as human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), are leading candidate cells as raw materials for cell therapy products, because of their capacity for pluripotent differentiation and unlimited self-renewal. hPSC-derived products have already entered the scope of clinical application. However, the assessment and control of their tumorigenicity remains to be a critical challenge. Sensitive detection of the pluripotent cellular impurities is necessary for the safety and quality control of the hPSC-derived products. In the present study, we established a sensitive assay for detection of the residual undifferentiated hiPSCs in cardiomyocytes, using droplet digital PCR (ddPCR). The ddPCR method with a probe and primers for LIN28 significantly detected as low as 0.001% undifferentiated hiPSCs in primary cardiomyocytes, which is equivalent to the ratio of a single hiPSC to 1 × 105 cardiomyocytes. The ddPCR also showed that LIN28 expression is extremely low in human tissues including liver, heart, pancreas, kidney, spinal cord, corneal epithelium and lung. These results suggest that the ddPCR method targeting LIN28 transcripts is highly sensitive and useful for the quality assessment of various cell therapy products derived from hPSCs.
Keywords: HiPSCs, HiPSC-derived cell therapy products, Quality control, Tumorigenicity, ddPCR, LIN28
Highlights
-
•
A highly sensitive in vitro assay for detection of residual undifferentiated hiPSCs in hiPSC-derived cell therapy products.
-
•
LIN28 mRNA is highly sensitive marker of residual undifferentiated hiPSCs in hiPSC-derived cardiomyocytes.
-
•
The ddPCR assay using LIN28 as a target is able to detect 0.001% undifferentiated hiPSCs in primary cardiomyocytes.
Abbreviations
- hPSCs
human pluripotent stem cells
- hESCs
human embryonic stem cells
- hiPSCs
human induced pluripotent stem cells
- ddPCR
droplet digital PCR
- RPE
retinal pigment epithelial
- LLOD
lower limit of detection
1. Introduction
Human pluripotent stem cells (hPSCs) such as human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) have the ability to differentiate into a variety of cell types and undergo numerous cell division cycles while maintaining their cellular identity. Because of their pluripotency and self-renewal property, it has been expected that hPSCs will provide new sources for robust and continuous production of many cell and tissue types for regenerative medicine and cell therapy. As previously reported, many attempts are currently underway to differentiate hPSCs into various tissues [1], [2]. Cell therapy using hPSCs has already entered the scope of clinical application. Clinical trials using hESC-derived retinal pigment epithelial (RPE) cells have been initiated to treat patients with Stargardt's disease and the dry type of age-related macular degeneration [3]. Clinical research using autologous hiPSC-derived RPE for the wet type of age-related macular degeneration has also started in Japan.
One of the most important issues in the development of a safe cell therapy product derived from hPSCs is ensuring that the final product does not form tumors after implantation [4]. The products derived from hPSCs may contain residual undifferentiated cells that eventually proliferate and form a teratoma [5]. The required cell number of hPSC-derived products varies depending on the diseases. In the case of RPE cell transplantation, approximately 5 × 104 RPE cells is thought to be required for retinal degeneration diseases. We have previously reported that qRT-PCR successfully detects 0.002% residual undifferentiated cells in hiPSC-derived RPE cells using LIN28 as a target gene [6]. The sensitivity of this assay is sufficient for the quality control hiPSC-derived RPE cells. However, at least 109 cardiomyocytes would be needed for transplant into one patient with heart failure [7], indicating that testing hPSC-derived cardiomyocytes products using the LIN28/qRT-PCR method may lead to false-negative test results. For the quality control of hPSC-derived cardiomyocytes, the detection of pluripotent cellular impurities in the products needs to be as sensitive as possible.
Recently, a new PCR format called droplet digital PCR (ddPCR) that directly quantifies DNA/mRNA copies has been developed. The ddPCR takes advantage of water-in-oil droplets to divide a 20 μL mixture of sample and reagents into ∼20,000 droplets. For droplets that contain template, specific cleavage of TaqMan probes generates a strong fluorescence signal. An automated droplet flow-cytometer reads each set of droplets after PCR, and droplets are assigned as positive or negative based on their fluorescence amplitude. The number of positive and negative droplets in each channel is used to calculate the concentration of the target nucleic acid [8]. This assay can avoid bias that arises from inefficient target gene amplification using qRT-PCR [9], [10]. In addition, ddPCR effectively enriches template concentrations in partitions and allows for the more sensitive detection of rare targets compared with qRT-PCR [8].
In this study, we established a highly sensitive assay for detection of residual undifferentiated hiPSCs in cell therapy products. The LIN28/qRT-PCR method was shown to detect 0.01% undifferentiated hiPSCs spiked into primary cardiomyocytes. We also determined that the LIN28/ddPCR method is able to detect as low as 0.001% undifferentiated hiPSCs in primary cardiomyocytes, which is equivalent to a single hiPSC contained in 1 × 105 cardiomyocytes.
2. Results
2.1. Detection of undifferentiated hiPSCs in cardiomyocytes using the LIN28/qRT-PCR method
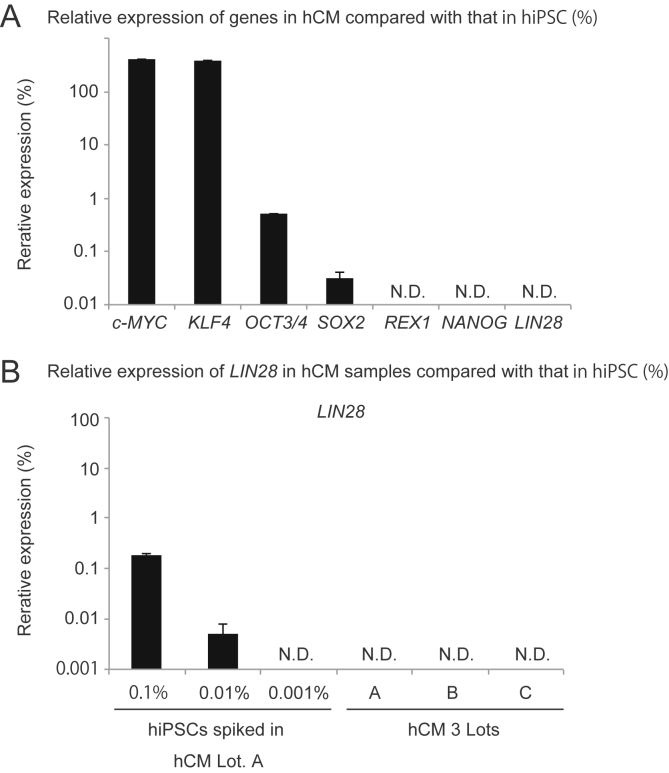
The marker genes for detecting trace amount of residual undifferentiated cells in cell therapy products should be highly expressed in hPSCs but be strongly silenced in differentiated cells. We first ascertained whether LIN28 mRNA is a superior marker for detecting residual undifferentiated cells in hiPSC-derived cardiomyocytes. To identify highly selective markers for undifferentiated hiPSCs, we compared MYC (c-MYC), KLF4, POU5F1 (OCT3/4), SOX2, REX1, NANOG and LIN28A (LIN28) mRNA levels in hiPSCs and primary cardiomyocytes. REX1, NANOG and LIN28 gene expression was not detected in primary cardiomyocytes (Fig. 1A). However, in hiPSCs, the Ct values of REX1, NANOG and LIN28 was 34.2, 25.2 and 22.6, respectively (data not shown). Thus, the LIN28 expression level was drastically changed between primary cardiomyocytes and hiPSCs. These results indicated that LIN28 was the most sensitive marker of residual undifferentiated cells in hiPSC-derived cardiomyocytes among seven candidate marker genes, as well as in hiPSC-derived RPE cells [6].
Fig. 1.
Detection of undifferentiated hiPSCs in primary cardiomyocytes using the LIN28/qRT-PCR method. (A) LIN28, NANOG, REX1, SOX2, OCT3/4, KLF4 and c-MYC relative mRNA expression in cardiomyocytes was determined using qRT-PCR analysis. (B) qRT-PCR analysis of hiPSCs spiked into three lots of primary cardiomyocytes. Single-cell hiPSCs (0.1%, 1 × 103 cells; 0.01%, 1 × 102 cells; 0.001%, 1 × 101 cells) were spiked into 1 × 106 primary cardiomyocytes, and total RNA was isolated from the mixed cells. All values are expressed as mRNA levels relative to those in undifferentiated hiPSCs. hCM: human cardiomyocyte. The expression levels of target genes were normalized to those of the GAPDH transcript. Results are presented as the mean ± standard deviation (n = 3).
To define the minimum number of hiPSCs that may be detected using the LIN28/qRT-PCR method, we spiked 1 × 104 (1%), 1 × 103 (0.1%), 1 × 102 (0.01%) and 1 × 10 (0.001%) hiPSCs into 1 × 106 primary cardiomyocytes. Total RNA isolated from these mixed cells was subjected to the LIN28/qRT-PCR method, and LIN28 mRNA signals of 0.1% and 0.01% were detected for spiked hiPSCs in a dose-dependent manner, but 0.001% spiked hiPSCs in the sample were not detectable (Fig. 1B), indicating that the LIN28/qRT-PCR method is able to detect at least 0.01% residual undifferentiated hiPSC contamination in cardiomyocytes.
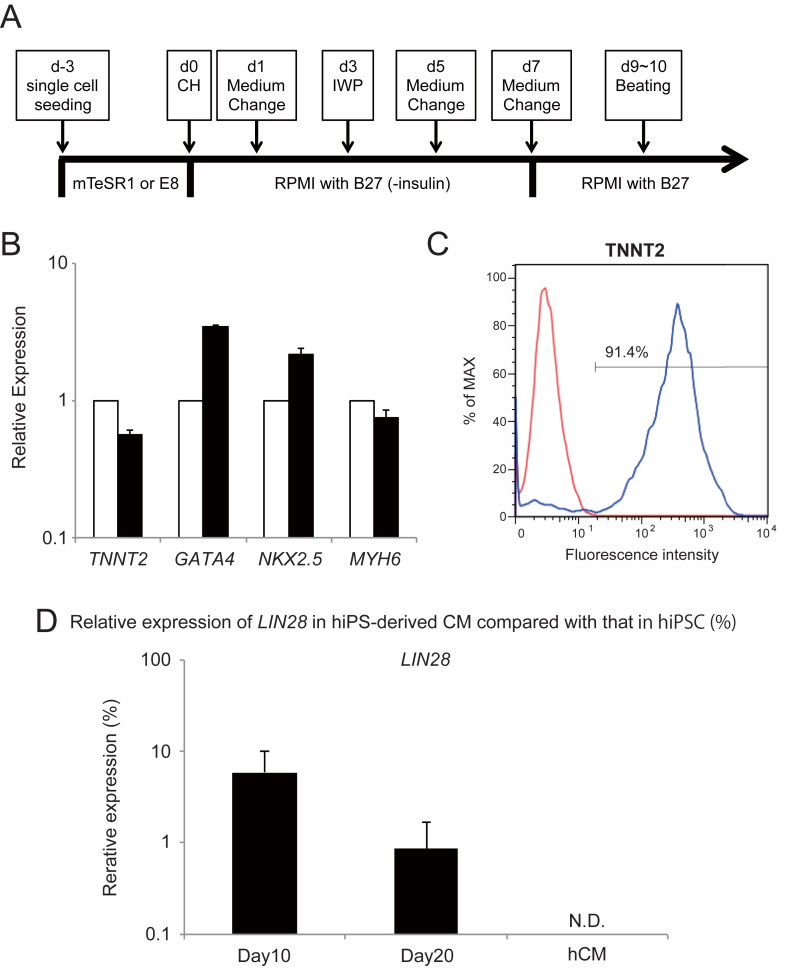
To evaluate residual undifferentiated hiPSCs in the final products, we differentiated hiPSCs into cardiomyocytes using an in vitro differentiation protocol, as previously described [11]. The GSK3 inhibitor CHIR99021 was added on day 0–1 to activate Wnt signaling, and then Wnt production-4 (IWP4) was added on day 3–5 to inhibit Wnt signaling (Fig. 2A). Beating cells were found on day 8–9 and they were broadly spread out by day 15 after cardiomyocyte induction (Movie S1). Gene-expression analysis using qRT-PCR revealed that TNNT2, GATA4, NKX2.5 and MYH6, which are specific markers for cardiomyocytes, were equally well expressed both in the hiPSC-derived cardiomyocytes and human heart (Fig. 2B). In addition, flow cytometry analysis showed that 91.4% of differentiated cells at day 20 were TNNT2-positive (Fig. 2C). These results indicate that hiPSCs were differentiated into cardiomyocytes at day 20. Total RNA extracted from differentiated cells (day 10 and 20) was subjected to the LIN28/qRT-PCR method. LIN28 mRNA levels continuously decreased during the differentiation process, to 1.2% of that of hiPSCs at day 20 (Fig. 2D).
Fig. 2.
Detection of undifferentiated hiPSCs in hiPSC-derived cardiomyocytes using the LIN28/qRT-PCR method. (A) Schematic diagram of culture procedure for cardiomyocyte. (B) qRT-PCR analysis of cardiomyocyte markers, TNNT2, GATA4, NKX2.5 and MYH6. Total RNA was isolated from hiPSC-derived cardiomyocytes (black bar) and primary cardiomyocytes (white bar). The mRNA levels are shown relative to those in primary cardiomyocytes. (C) Flow cytometry analysis of TNNT2 in hiPSCs (red), and hiPSC-derived cardiomyocytes at day 20 (blue). (D) LIN28 expression in hiPSCs differentiating into cardiomyocytes (d10 and d20). LIN28 mRNA levels are shown relative to that in undifferentiated hiPSCs. hCM: human cardiomyocyte. The expression levels of target genes were normalized to those of the GAPDH transcript. Results are presented as the mean ± standard deviation (n = 3).
Supplementary video related to this article can be found at http://dx.doi.org/10.1016/j.reth.2015.08.001.
The following is the supplementary data related to this article:
2.2. Detection of undifferentiated hiPSCs in cardiomyocytes using the LIN28/ddPCR method
ddPCR has recently been shown to enable the precise quantification of target nucleic acids in samples. To establish a highly sensitive assay for detection of residual undifferentiated hiPSCs in the final product, we employed ddPCR (the LIN28/ddPCR method) as a substitute for qRT-PCR in this study. The ddPCR system measures fluorescence intensities of droplets after completing all thermal cycling. The copy number of target genes is determined based on the number of fluorescent-positive and -negative droplets in a sample well. We first determined the optimal experimental conditions for the newly designed assay. We tested 6 different probe and primer sets to select an optimum one (Table S2) and also determined their optimum annealing temperature (Fig. S1). We then selected LIN28-1 and 64 °C as the probe and primer set and an annealing temperature, respectively, for the following experiments with ddPCR.
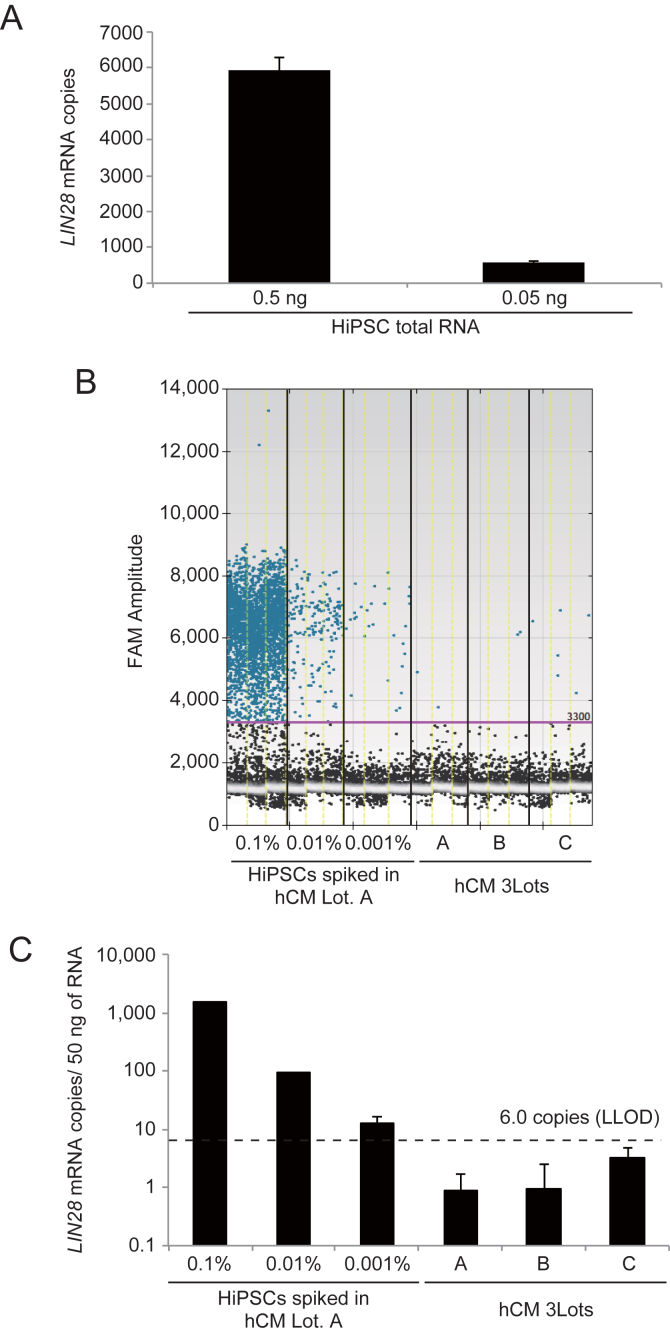
ddPCR has an advantage because it provides an absolute number of LIN28 mRNA copies present in the sample. To estimate the LIN28 mRNA copy number present in a single hiPSC, we determined the LIN28 mRNA copy number in 0.5 ng and 0.05 ng total RNA isolated from hiPSCs by the LIN28/ddPCR method. There were 6000 ± 380 copies of LIN28 mRNA obtained from 0.5 ng RNA, and 572 ± 42 copies of LIN28 mRNA obtained from 0.05 ng of RNA (Fig. 3A). Since 500 ng of RNA was obtained from 1 × 105 hiPSCs, approximately 60 copies of LIN28 mRNA were estimated to exist in one hiPSC. Therefore, 1 copy of LIN28 mRNA per 50 ng of RNA is calculated to be equivalent to 0.0002% hiPSCs contamination. The theoretical limit of detection for the LIN28/ddPCR method is estimated as 0.0002% (one cell in 500,000 cell).
Fig. 3.
Detection of undifferentiated hiPSCs in primary cardiomyocytes using the LIN28/ddPCR method. (A) The copy number of LIN28 mRNA in 0.5 and 0.05 ng total RNA of hiPSC was investigated by ddPCR. (B) The copy number of LIN28 mRNA in 50 ng total RNA of hiPSC-spiked cardiomyocytes and three lots of primary cardiomyocytes were investigated by ddPCR. Dots indicate the fluorescence intensity of the droplets. Droplets were assigned “positive” or “negative” based on their fluorescence amplitude. (C) Raw droplet data shown in (B) were quantified as LIN28 mRNA copies. These samples used in Fig. 1B and Fig. 3B/C were same ones. hCM: human cardiomyocyte. Results are presented as the mean ± standard deviation (n = 3).
To examine the detection limit of the LIN28/ddPCR method, we analyzed the LIN28 mRNA copy number of the hiPSCs-spiked primary cardiomyocytes with the LIN28/ddRT-PCR method. The 0.1%, 0.01% and 0.001% hiPSC-spiked samples contained 1495.3, 91.3 and 12.4 copies of LIN28 mRNA per sample (50 ng of total RNA), respectively. On the other hand, three lots of primary cardiomyocyte samples showed 1.7 ± 1.3 copies of LIN28 mRNA. The lower limit of detection (LLOD, LLOD = mean + 3.3 × SD [12]) was 6.0 copies (Fig. 3B, C), when primary cardiomyocyte samples were employed as the negative control. These results indicate that the LIN28/ddPCR method is able to detect 0.001% hiPSCs spiked into primary cardiomyocytes. The LLOD in Fig. 3B and C was the same as those based on the data from two other lots of cardiomyocytes (Fig. S2).
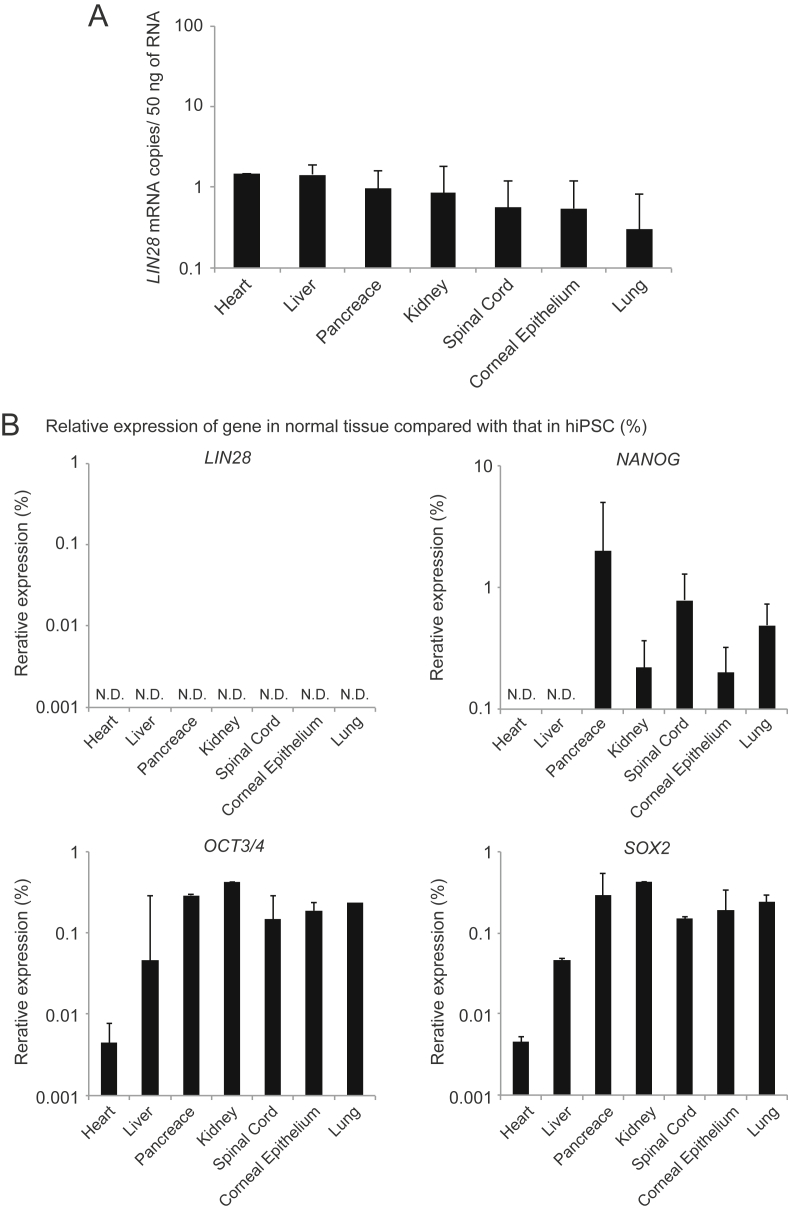
Finally, to confirm the versatility of LIN28 mRNA as a high sensitive marker of residual undifferentiated hiPSCs, we measured background expression levels of LIN28 mRNA in various types of human tissues using ddPCR. Since a high signal to noise ratio is commonly required for sensitive detection, the lower expression of LIN28 mRNA in normal tissues would be preferable as the background. As expected, LIN28 mRNA levels were extremely low in all of liver, heart, pancreas, kidney, spinal cord, corneal epithelium and lung samples detecting with LIN28/ddPCR methods. Mean copy numbers of all these tissues were less than 1.5 copies per sample (Fig. 4A), which was consistent with the results with cardiomyocytes (Fig. 3B, C). To compare the versatility of LIN28 mRNA with other genes, we further measured the NANOG, OCT3/4 and SOX2 mRNA levels in these normal human tissues using qRT-PCR. Small amounts of NANOG, OCT3/4 and SOX2 mRNA were present in the tissues, except for NANOG expression in heart and liver (Fig. 4B). These results suggest that LIN28 mRNA is a suitable marker to detect residual undifferentiated hiPSCs in products and that the LIN28/ddPCR method is applicable to the quality control of hiPSC-derived cell therapy products, i.e. hepatocytes, cardiomyocytes, pancreatic β-cells, kidney cells, spinal cord, corneal epithelium and lung cells.
Fig. 4.
Detection of LIN28 mRNA in various types of human tissues using the LIN28/ddPCR method. (A) Absolute quantification of LIN28 mRNA copy number in normal tissues using ddPCR. Total RNA (50 ng) was analyzed. (B) qRT-PCR analysis of normal tissues. Total RNA (50 ng) was analyzed. LIN28, NANOG, OCT3/4 and SOX2 mRNA levels are shown as expression relative to those in undifferentiated hiPSCs. The expression levels of target genes were normalized to those of the ribosomal RNA. Results are presented as the mean ± standard deviation of three different lots of each tissue.
3. Discussion
For the safety and quality of cell therapy products derived from hPSCs, the level of residual undifferentiated cells that have tumorigenic potential is one of the critical attributes. Therefore, it is necessary to develop highly sensitive, validated assays for the detection of the pluripotent cellular impurities. In this study, we developed a novel, highly sensitive in vitro assay to detect residual undifferentiated hiPSCs in cardiomyocytes using ddPCR. The ddPCR assay appeared to detect as few as 0.001% undifferentiated iPSCs spiked into primary cardiomyocytes using LIN28 as a target gene.
In the previous study, we reported that a qRT-PCR assay successfully detected 0.002% residual undifferentiated cells in hiPSC-derived RPE cells using LIN28 as a target gene [6]. In the present study, we modified this method to detect residual undifferentiated hiPSCs in cardiomyocytes. When detecting a trace amount of cellular impurities in hiPSC-derived products by expression analysis of pluripotency marker genes, it is important that their expression levels are sufficiently low in normal somatic cells of interest. As in the case of RPE cells, we found that the LIN28 mRNA expression was suppressed in primary cardiomyocytes (Fig. 1B). In addition, we also confirmed the expression of LIN28, NANOG, OCT3/4 and SOX2 in normal tissues (human liver, heart, pancreas, kidney, spinal cord, corneal epithelium and lung) using qRT-PCR. NANOG, OCT3/4 and SOX2 mRNA was present in most of these tissues in low amounts, whereas LIN28 expression was totally absent in all of the tissues examined (Fig. 4B). These results indicate that LIN28 is a useful marker for detecting residual hiPSCs in cell therapy products.
The ddPCR technology avoids amplification efficiency bias, which is observed with matrix-linked inhibition that occurs with qRT-PCR. Therefore, ddPCR is more sensitive than qRT-PCR and can detect rare targets, thus providing accurate data [13], [14]. In the present study, we have compared qRT-PCR with one-step ddPCR, using biological samples that have a low target copy number. ddPCR detected LIN28 signals when as little as 0.001% hiPSCs were spiked into cardiomyocytes, but this was not detected using qRT-PCR, namely the sensitivity of ddPCR is ten times higher than that of qRT-PCR in this assay (Fig. 1, Fig. 3B). However, reaction conditions with ddPCR should be carefully titrated to optimize the new platform. First, depending on the design of probes and primers, the number of positive drops and their fluorescence intensity were impacted. The fluorescence intensity was also affected by the annealing temperature, but amplicon size had no effect (Fig. S1, Table S2). Second, determination of the threshold to distinguish between positive and negative droplets is critical to accurately quantify target gene copy number using ddPCR [15]. We measured water, which was used as the negative control, in triplicate to determine the threshold between the positive and negative droplets.
The one-step ddPCR technology provides an absolute count of target mRNA copies per input sample as described. Absolute quantification provided from the LIN28/ddPCR method showed that limit of detection is theoretically 0.0002% hiPSCs contained in cell samples (Fig. 3A). This sensitivity would not be sufficient for transplant of cardiomyocytes because at least 109 cardiomyocytes is thought to be necessary for one patient with heart failure [7]. This indicates that tests of hPSC-derived cardiomyocytes products using the current in vitro detection methods may still lead to false-negative test results. However, to date and to our knowledge, the LIN28/ddPCR method in the present study is the most sensitive in vitro methods to quantitatively detect a trace amount of pluripotent cellular impurities in normal somatic cells. The LIN28/ddPCR method can be applicable at least to quality assessment and process validation of hPSC-derived products (or their intermediate products). Namely, although “Negative in the LIN28/ddPCR test” may not directly indicate the “safety”, the negative result can be one of critical “quality” attributes to demonstrate the absence a certain amount of hPSCs. The problems with sensitivity may be resolved using a combination of a new, highly efficient culture amplification method for the undifferentiated hPSCs contained in cell therapy products [16] and the LIN28/ddPCR method. It should be also noted that the theoretical limit of detection would be different in cell types, due to variation of estimated total RNA contents per cell. Moreover, the LLOD for the LIN28/ddPCR method may vary, if the copy numbers of LIN28 are different between hiPSC lines. However, the LIN28 mRNA numbers were similar across all the hiPSC lines examined (253G1, 201B7, R-1A, R-2A, R-12A) (Fig. S3), indicating that the LLOD of LIN28/ddPCR method is quite constant among hiPSC lines.
There are several in vivo and in vitro assays available for detection of pluripotent cellular impurities [17]. In vivo tumorigenicity tests using severely immunodeficient mice are suggested to detect a small amount of undifferentiated hPSCs. Kanemura et al. [18] reported that in vivo tumorigenicity tests using NOD/Shi-scid IL2rγnull (NOG) mice can detect tumors derived from as few as 10 hiPSCs. Although the in vivo tumorigenicity assay is costly and time-consuming, it can directly analyze tumor formation. On the other hand, our in vitro LIN28/ddPCR method is simple and rapid enough to obtain the results prior to the clinical application of the final product. However, in vitro LIN28/ddPCR method indirectly detects undifferentiated cells. Namely, it cannot determine whether completely undifferentiated hiPSCs or partially differentiated hiPSCs are contained in products. The hPSC-detecting assays should be chosen to assess the safety of hPSC-derived products, and consideration should be given to the limitations of each test. The overall safety of each hPSC-derived product should be estimated based on the results of appropriate the hPSC-detecting assays. Moreover, it is necessary to understand that residual undifferentiated hiPSC is not only a cause of the tumorigenicity. Tumorigenicity studies for hPSC-derived products should examine not only the existence of residual undifferentiated pluripotent cells, but also the existence of tumorigenic transformants, and whether the transplant forms tumor in microenvironments at the site of transplantation [17].
The LIN28/ddPCR method is the most sensitive in vitro assay for detecting undifferentiated hiPSCs at present, and has the potential to be widely applied to quality assessment of hPSC-derived products. We expect our findings to assist with the use of regenerative medicine to treat a wide variety of diseases with hPSC-derived products in the near future.
4. Methods
4.1. Cell culture
The hiPSC cell lines, 253G1 [19], 201B7 [2], HiPS-RIKEN-1A (R-1A), HiPS-RIKEN-2A (R-2A) and HiPS-RIKEN-12A (R-12A) were provided by the RIKEN BRC through the Project for Realization of Regenerative Medicine and the National Bio-Resource Project of the MEXT, Japan. Undifferentiated hiPSCs were maintained on Laminin-521 (BioLamina, Stockholm) in Essential 8 medium (Invitrogen) [16]. Colonies were passaged by dissociating the cells into single cells once every 3–4 days using 0.5 mM EDTA in PBS at a density of 2 × 104 cells/cm2. Adult human cardiomyocytes (Promocell, Heidelberg) were cultured on Laminin-211 (BioLamina, Stockholm) in the Promocell myocyte growth medium. Cells were grown in a humidified atmosphere of 5% CO2 and 95% air at 37 °C.
4.2. Cardiomyocyte differentiation of hiPSCs
Cardiomyocyte differentiation was induced as previously reported, with a few modifications [11]. hiPSCs were detached by incubation with 0.5 mM EDTA in PBS for 7 min and seeded onto cell culture plates coated with laminin-521 or Matrigel at a density of 50,000 cells/cm2 in Essential 8 or mTeSR1, respectively, for 3–4 days before cardiomyocyte induction. Confluent hiPSCs were treated with 12 μM CHIR99021 (Stemgent), a GSK3 inhibitor, in RPMI1640 medium (Sigma–Aldrich) supplemented with B27-insulin (Invitrogen) (RPMI/B27-insulin) for 24 h. At day 1, the medium was changed to RPMI/B27-insulin. At day 3, cells were treated with 5 μM IWP4 (Stemgent), a Wnt inhibitor, in RPMI/B27-insulin for 48 h. At day 5, the medium was changed to RPMI/B27-insulin. The cells were transferred to RPMI/B27 without insulin at day 7, and the medium was changed every 3 days. Differentiated cells were harvested at the indicated time for further analysis.
4.3. qRT-PCR
Total RNA was isolated from cells using an RNeasy Mini Kit with DNase I treatment (Qiagen), according to the manufacturer's instructions. RNA concentration was determined using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific). In the spike experiments, 253G1 cells and primary human cardiomyocytes (PromoCell) were mixed at a defined cell number before RNA isolation. qRT-PCR was performed with the QuantiTect Probe one-step RT-PCR Kit (Qiagen) on a 7300 Real-Time PCR System (Applied Biosystems). Total RNA (50 ng per sample) was used for the analysis. The expression levels of target genes were normalized to those of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcript or ribosomal RNA, which were quantified using TaqMan human GAPDH control reagents (Life Technologies) or TaqMan ribosomal RNA control reagents (Life Technologies). Probes labeled with 5′-FAM/3′-TAMRA and primers were obtained from Sigma–Aldrich. The sequences of primers and probes used in the present study are listed in Table S1. All qRT-PCR reactions were run at 45 cycles.
4.4. Flow cytometry
253G1-derived cardiomyocytes were fixed using the BD Cytofix fixation buffer (BD Biosciences) for 20 min and permeabilized using BD Perm/Wash buffer (BD Biosciences) for 10 min at room temperature. Cells were incubated for 1 h at room temperature with mouse anti-tnnt2 monoclonal antibody (1:1000) (Abcam). Indirect immunostaining was then completed with anti-mouse IgG Alexa Fluor 488-conjugated secondary antibody (1:1000) (Molecular Probes) for 1 h. Normal mouse IgG antibody was used as a negative control (Millipore). Stained cells were analyzed using a BD FACScalibur II (BD Biosciences). Data retrieved from sorting was analyzed using Flowjo software (Tree Star, Ashland, OR).
4.5. One-step digital droplet PCR
Total RNA was prepared as described above. One-step droplet digital PCR reaction mixtures (20 μl volume) were composed as follows: 1 × One-Step RT-ddPCR Supermix (Bio-Rad), 750 nM forward and reverse primers, 250 nM probe labeled with 5′-FAM/3′-BHQ-1, and 1 mM manganese acetate solution (Bio-Rad). Five μl of total RNA sample (50 ng) was added to the mixture. The sequences of primers and probes used in the present study are listed in Table S2. Droplets were generated in 8-well cartridges using the QX100 droplet generator (Bio-Rad) according to the manufacturer's instructions. Water-in-oil emulsions were transferred to a 96-well plate, and RT-PCR was performed using a T100™ thermal cycler (Bio-Rad). Thermal cycling conditions were as follows: 30 min reverse transcription at 60 °C, followed by 5 min enzyme activation at 95 °C, and 40 cycles of a thermal profile comprising 30 s denaturation at 94 °C and 60 s annealing/extension at 64 °C. After PCR amplification, products were denatured at 98 °C for 10 min and cooled at 4 °C. Fluorescence intensities of each droplet from the samples were measured using the QX100 droplet reader (Bio-Rad). Positive droplets containing amplification products were distinguished from negative droplets and counted by applying a fluorescence amplitude threshold in QuantaSoft software. The threshold was manually determined at the highest point of the sample droplet cluster of water and usually set at 3300 under our experimental conditions. QuantaSoft software provides concentration results in copies of target per microliter (copies/μl). The number of copies of target per template RNA (i.e., copies/50 ng of RNA) was calculated as concentration (copies/μl) x 20 (μl).
4.6. Tissue RNA
Total RNA isolated from human heart, liver, pancreas, kidney, spinal cord, and lung was obtained from Clontech Laboratories or BioChain. Total RNA isolated from human corneal epithelium was purchased from ScienCell Research Laboratories.
Author contributions
TK SY YSato designed the experiments. TK SM performed the experiments. TK SY KT SKu YSawa SKa YSato analyzed the data. TK SY YSato wrote the manuscript. YSawa SKa YSato acquired the funding. All authors read and approved the final manuscript.
Competing financial interests
The authors declare no competing financial interests.
Acknowledgments
This work was supported by a Strategic Fund for the promotion of Science and Technology from the Japan Science and Technology Agency, and Research Grants from the Japanese Ministry of Health, Labour and Welfare (H23-SAISEI-IPPAN-004, H23-SAISEI-IPPAN-005, H24-IYAKU-SHITEI-027, H25-JITSUYOKA(SAISEI)-IPPAN-008), and the Marketing Authorization Facilitation Program for Innovative Therapeutic Products.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.reth.2015.08.001.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Lu B., Malcuit C., Wang S., Girman S., Francis P., Lemieux L. Long-term safety and function of RPE from human embryonic stem cells in preclinical models of macular degeneration. Stem Cells. 2009;27:2126–2135. doi: 10.1002/stem.149. [DOI] [PubMed] [Google Scholar]
- 4.Benvenisty UB-DaN The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:10. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 5.Knoepfler P.S. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda T., Yasuda S., Kusakawa S., Hirata N., Kanda Y., Suzuki K. Highly sensitive in vitro methods for detection of residual undifferentiated cells in retinal pigment epithelial cells derived from human iPS cells. PloS One. 2012;7:e37342. doi: 10.1371/journal.pone.0037342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laflamme M.A., Murry C.E. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- 8.Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhat S., Herrmann J., Armishaw P., Corbisier P., Emslie K.R. Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal Bioanal Chem. 2009;394:457–467. doi: 10.1007/s00216-009-2729-5. [DOI] [PubMed] [Google Scholar]
- 10.Corbisier P., Bhat S., Partis L., Xie V.R., Emslie K.R. Absolute quantification of genetically modified MON810 maize (Zea mays L.) by digital polymerase chain reaction. Anal Bioanal Chem. 2010;396:2143–2150. doi: 10.1007/s00216-009-3200-3. [DOI] [PubMed] [Google Scholar]
- 11.Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller J.N.M.J.C. 5th ed. Person Education Limited; Harlow: 2005. Statistics and chemometrics for analytical chemistry. [Google Scholar]
- 13.Whale A.S., Cowen S., Foy C.A., Huggett J.F. Methods for applying accurate digital PCR analysis on low copy DNA samples. PloS One. 2013;8:e58177. doi: 10.1371/journal.pone.0058177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Racki N., Morisset D., Gutierrez-Aguirre I., Ravnikar M. One-step RT-droplet digital PCR: a breakthrough in the quantification of waterborne RNA viruses. Anal Bioanal Chem. 2014;406:661–667. doi: 10.1007/s00216-013-7476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones M., Williams J., Gartner K., Phillips R., Hurst J., Frater J. Low copy target detection by droplet digital PCR through application of a novel open access bioinformatic pipeline, 'definetherain'. J Virol Methods. 2014;202C:46–53. doi: 10.1016/j.jviromet.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tano K., Yasuda S., Kuroda T., Saito H., Umezawa A., Sato Y. A novel in vitro method for detecting undifferentiated human pluripotent stem cells as impurities in cell therapy products using a highly efficient culture system. PloS One. 2014;9:e110496. doi: 10.1371/journal.pone.0110496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda T., Yasuda S., Sato Y. Tumorigenicity studies for human pluripotent stem cell-derived products. Biol Pharm Bull. 2013;36:189–192. doi: 10.1248/bpb.b12-00970. [DOI] [PubMed] [Google Scholar]
- 18.Kanemura H., Go M.J., Shikamura M., Nishishita N., Sakai N., Kamao H. Tumorigenicity studies of induced pluripotent stem cell (iPSC)-derived retinal pigment epithelium (RPE) for the treatment of age-related macular degeneration. PloS One. 2014;9:e85336. doi: 10.1371/journal.pone.0085336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.