Abstract
The M2 isoform of pyruvate kinase, the final rate-limiting enzyme of aerobic glycolysis, is expressed during embryonic development. In contrast, the M1 isoform is expressed in differentiated cells due to alternative splicing. Here we investigated murine embryonic stem cells (ESCs) with Pkm1 or Pkm2 knock-in alleles. Pkm1 allele knock-in resulted in excessive oxidative phosphorylation and induced the formation of cysteine-thiol disulfide-dependent complexes of forkhead box class-O (FOXO) transcription factors, which resulted in altered endoderm differentiation. In contrast, Pkm2 knock-in induced synthesis of a methylation-donor, S-adenosylmethionine, and increased unsaturated eicosanoid groups, which contributed to the redox control and maintenance of ESC undifferentiated status. Because PKM2 is also a critical enzyme for the cancer-specific Warburg effect, our results demonstrate an important role for the Pkm2 allele in establishing intracellular redox conditions and modulating PKM1-dependent oxidative phosphorylation events to achieve an appropriate ESC differentiation program.
Keywords: Pyruvate kinase, Embryonic stem cell, Metabolism, Differentiation
1. Introduction
In mammals, there are four isoforms of pyruvate kinase (PK): PKM1, PKM2, PKL, and PKR [1], [2], [3]. PKL and PKR genes on mouse chromosome 3 are controlled by tissue-specific promoters in liver and red blood cells, respectively [4]. PKM1 and PKM2 genes on mouse chromosome 9 are preferentially expressed in various cells and are the result of alternative splicing of a single PK gene. PKM1 and PKM2 transcripts include exons 9 and 10, respectively [5]. PK expression is tightly regulated according to the particular tissues involved and developmental stages [3]. Although each PK isozyme plays a role in the metabolism in various cells [4], PKM2 is uniquely expressed in early fetal tissues, including undifferentiated embryonic stem cells (ESCs), but it is also expressed in most adult tissues, suggesting that PKM2 is a prototype enzyme during mammalian development [5], [6].
The PKM2 isoform gradually switches to the PKM1 isoform in skeletal muscle, heart, and brain in a developmental stage-dependent manner. Carcinogenesis apparently reverses this process [7]. During tumor development, tissue-specific enzyme expressions, such as those of PKM1 in brain and PKL in the liver, are decreased, whereas PKM2 expression is increased and is frequently altered in numerous tumors [4], [8]. Proliferating cells, particularly cancer cells, express the PKM2 isoform [4].
Cell metabolism plays a pivotal role in determining whether a cell would proliferate, differentiate, or remain in an undifferentiated state. A particular biochemical characteristic that distinguishes cancer cells and induced pluripotent stem cells (PSCs) from differentiated cells is their metabolic regulation, which is characterized by limited oxidative capacity and active anaerobic glycolysis [9]. Proliferative ESCs and cancer cells exhibit high glycolysis rates, which result in lactate production despite high oxygen levels. Recent studies have suggested a critical role for epigenetics during stem cell differentiation relative to differentiated cells [10], including upregulated expression of threonine dehydrogenase (TDH) in early blastocysts and ESCs and induced PSC reprogramming [11], [12]. TDH and glycine dehydrogenase regulate 5-methyltetrahydrofolate synthesis, thereby modulating histone H3 lysine 4 (H3K4) trimethylation [9]. H3K4 trimethylation is associated with open euchromatin, which is crucial for PSC epigenetic plasticity and self-renewal via their gene expression [13], [14]. This indicates that there is a close association between stem cell metabolism and differentiation.
In this study, we investigated murine ESCs with stable Pkm1 or Pkm2 knock-in for one allele at a time (here denoted Pkm1KI and Pkm2KI, respectively). Pkm2KI ESCs were difficult to differentiate into early mesoderm and endoderm lineages as compared with wild type (WT) ESCs and Pkm1KI ESCs. However, WT and Pkm1KI ESCs successfully differentiated into specific lineages, such as pancreatic cells, as compared with Pkm2 knock-in ESCs. PKM2 expression induced synthesis of a methylation-donor, S-adenosylmethionine (SAM), and oxidation that promoted the formation of cysteine-thiol disulfide-dependent complexes of FOXO transcription factors and p300/CBP. Thus, our data suggest that PKM1 plays an important role during ESC differentiation and that PKM2 may modulate oxidative phosphorylation effects to maintain ESC immature metabolism status at a branching stage for differentiation into specific cell lineages.
2. Results and discussion
2.1. Alternative Pk alleles control ESC differentiation
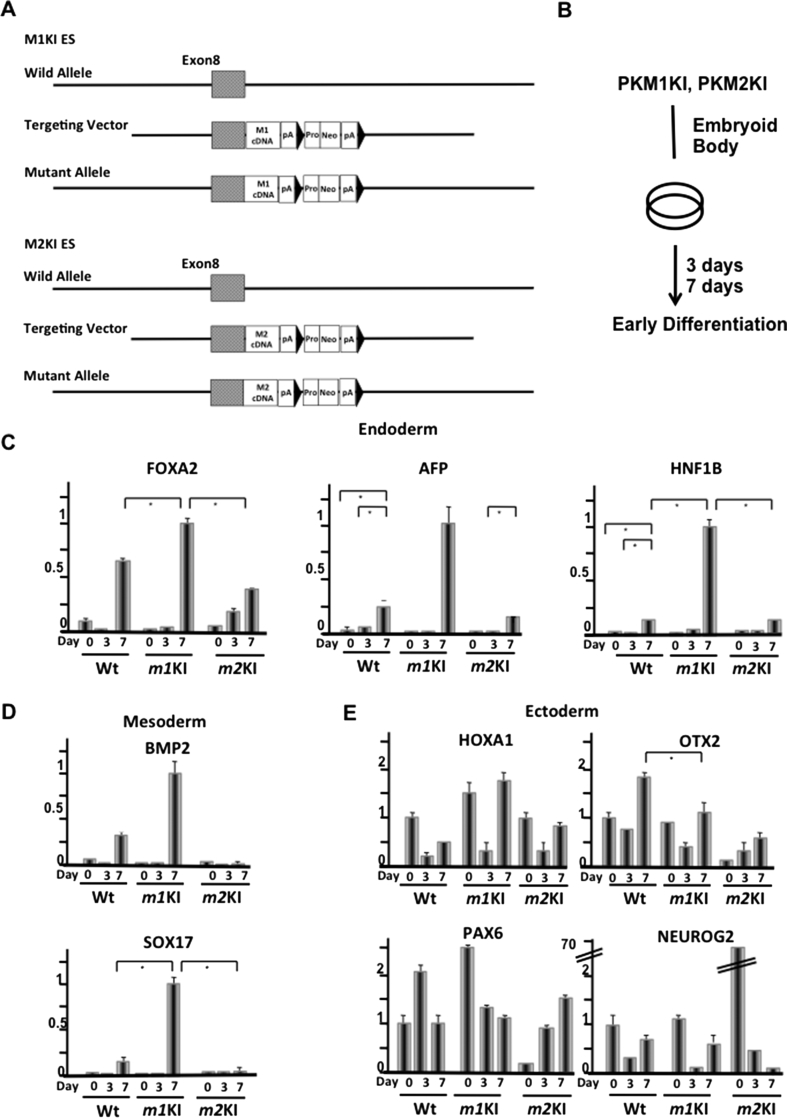
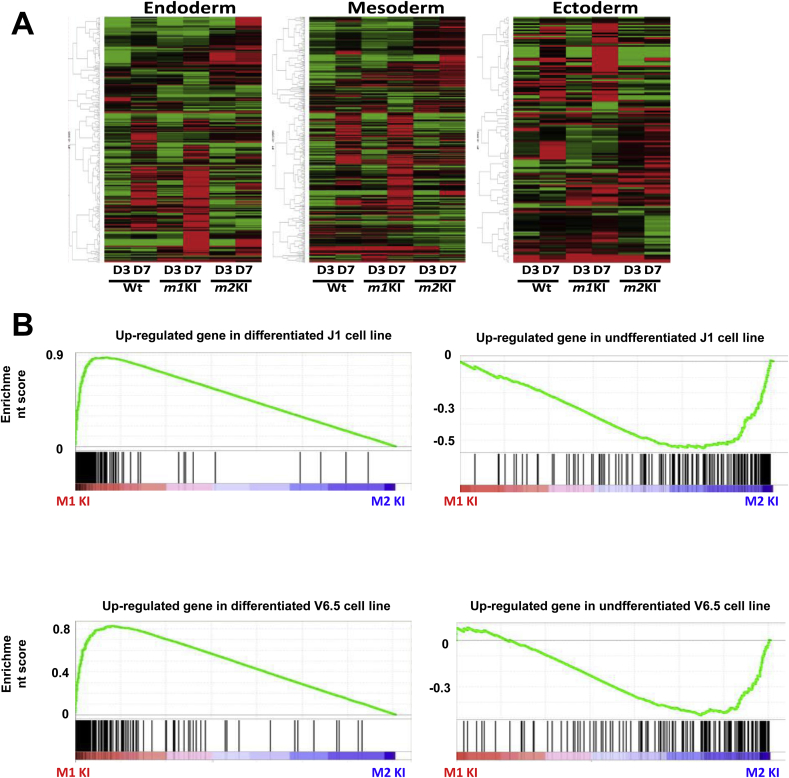
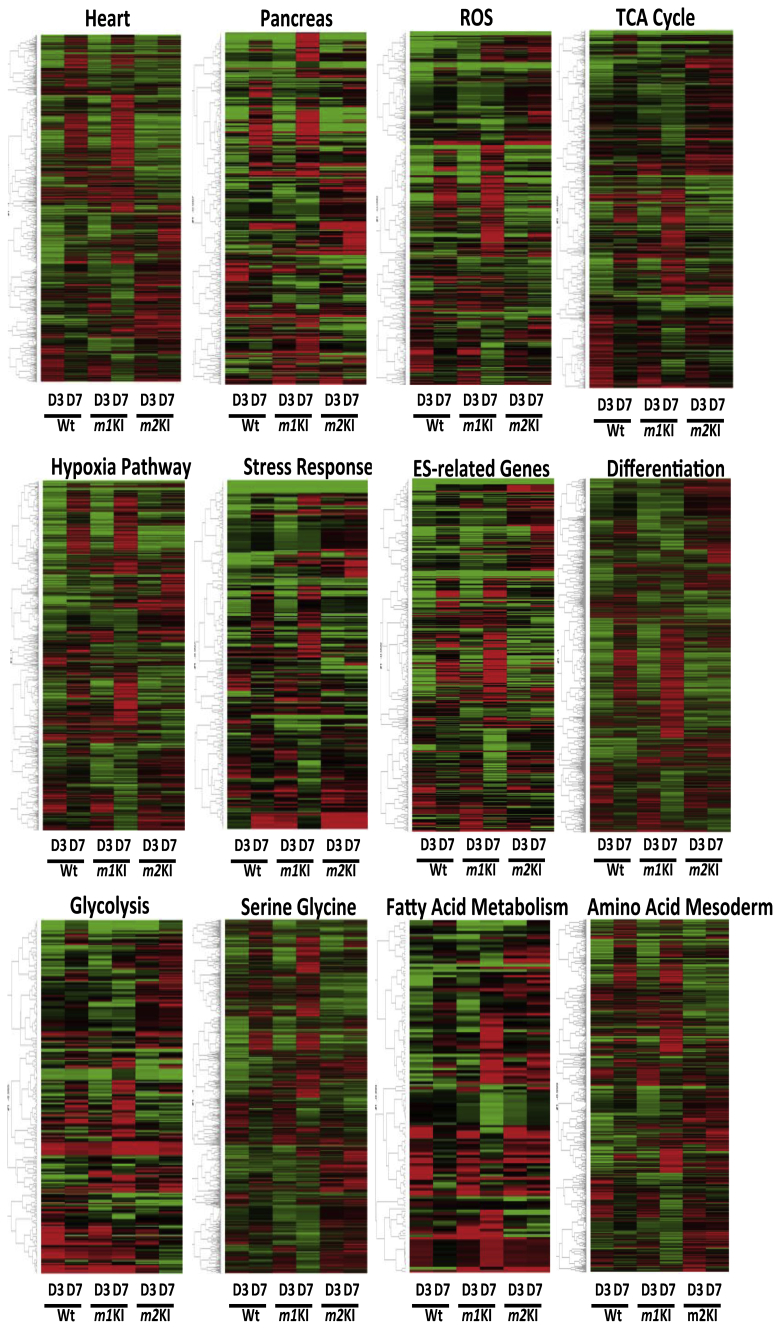
To unravel the stabilizing effect of PK on differentiation, we cultured Pkm1-knock-in (for one allele; the other allele was wild type; Pkm1KI ESCs), Pkm2-knock-in (for one allele; the other allele was wild type; Pkm2KI ESCs), and normal control ESC lines under differentiation-inducing conditions (Fig. 1AB). The expressions of FOXA2, α-fetoprotein (AFP), and HINF1B (endoderm markers), and BMP2 and SOX17 (mesoderm) were markedly increased in Pkm1KI ESCs in a time-dependent manner, whereas HOXA1, OTX2, PAX6, and Neurogenin 2 (ectoderm) expressions were context dependent (Fig. 1C–E). Microarray data (GSE57504) for cells cultured in differentiation-inducing medium for 7 days indicated that the expressions of metabolism-related genes in Pkm1KI ESCs were similar to those in WT ESCs but not those in Pkm2KI ESCs (Fig. 2A, Fig. S1). This GSEA analysis revealed that differentiation marker gene expressions [15] were enriched in Pkm1KI ESCs as compared with Pkm2KI ESCs at day 7 (Fig. 2B). This suggested that the PKM2 isoform contributed to modulating against induced ESC differentiation.
Fig. 1.
PK pathway affects cellular differentiation. Schematic representation of homologous recombination of Pkm1KI and Pkm2KI alleles in ESCs. Experimental scheme. Roles of PKM1 and PKM2 derived from alternative slicing for maintaining pluripotency and controlling early differentiation. Pkm1KI and Pkm2KI embryonic stem cells (ESCs) were studied. (C, D, E) Gene expression patterns during early differentiation of Pkm1KI and Pkm2KI ESCs that were cultured in medium without LIF. Specific differentiation markers for endoderm (B), mesoderm (C), and ectoderm (E) were examined.
Fig. 2.
PKM splicing affects ESC differentiation. Expression array analysis of Pkm1KI, Pkm2KI, and WT embryonic stem cells (ESCs) under differentiation conditions (days 3 and 7). GSEA analysis of Pkm1KI and Pkm2KI ESCs. Data for cells cultured in differentiation medium for 7 days are shown. For the J1 cell line (top two panels; Hailesellasse-Sene et al., 2007), differentiation gene set signatures were enriched in Pkm1KI cells (left), whereas undifferentiated gene set signatures were enriched in Pkm2KI cells (right). These results were confirmed using another V6.5 cell line, as shown in the bottom two panels (Hailesellasse-Sene et al., 2007).
2.2. PKM2 enhances methionine metabolism during ESC differentiation
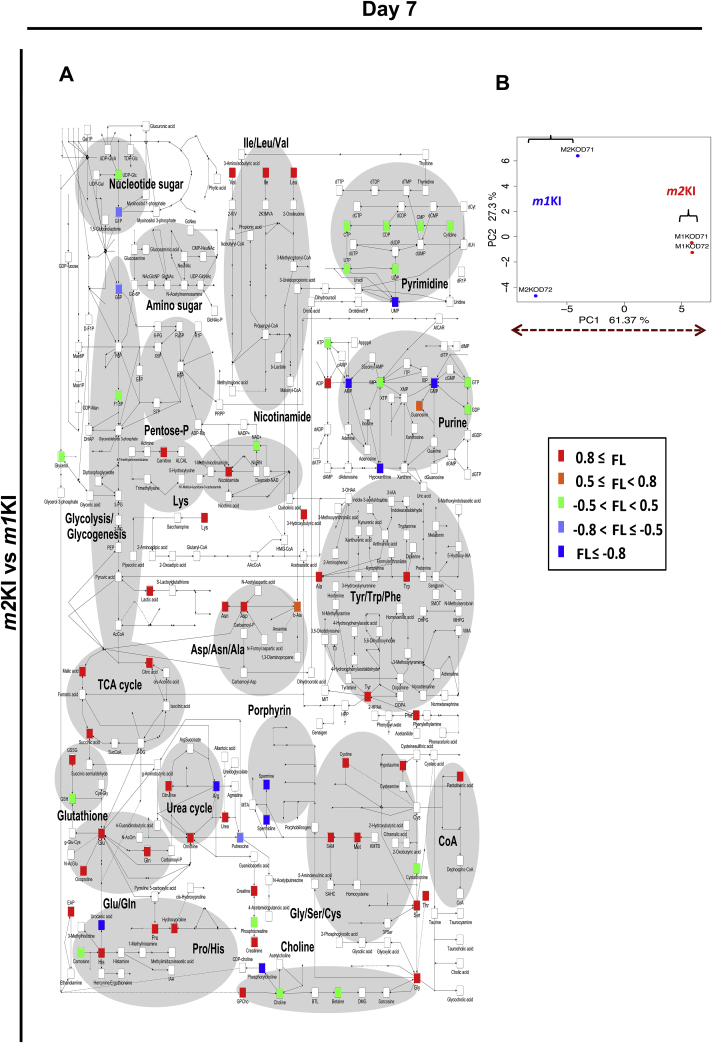
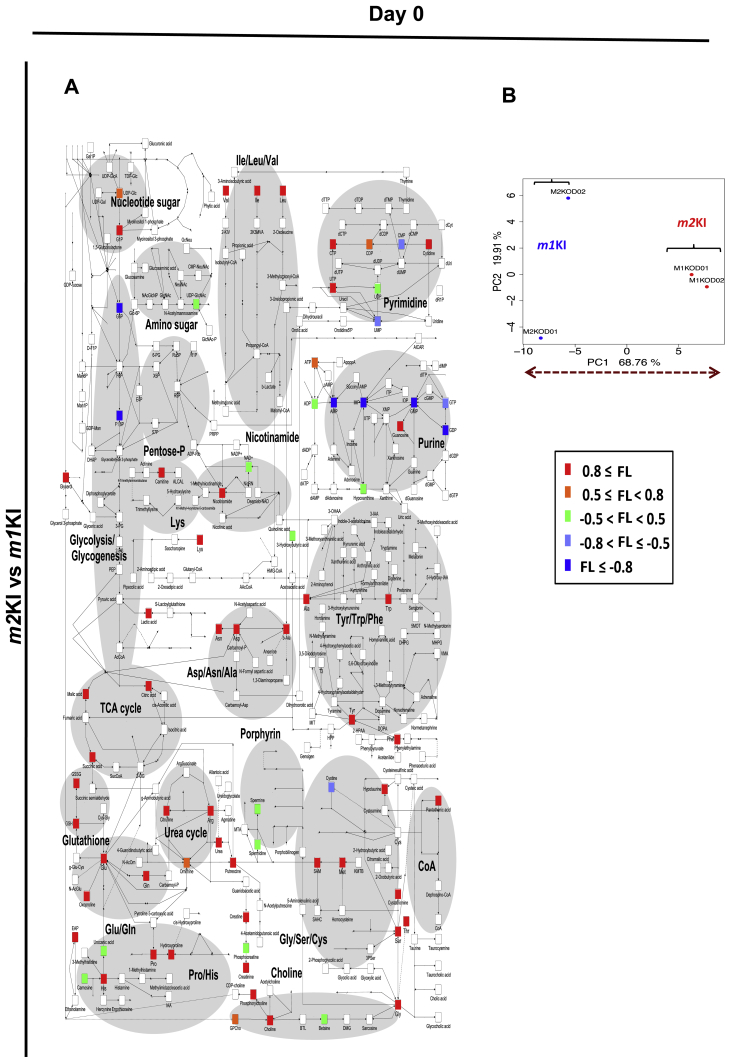
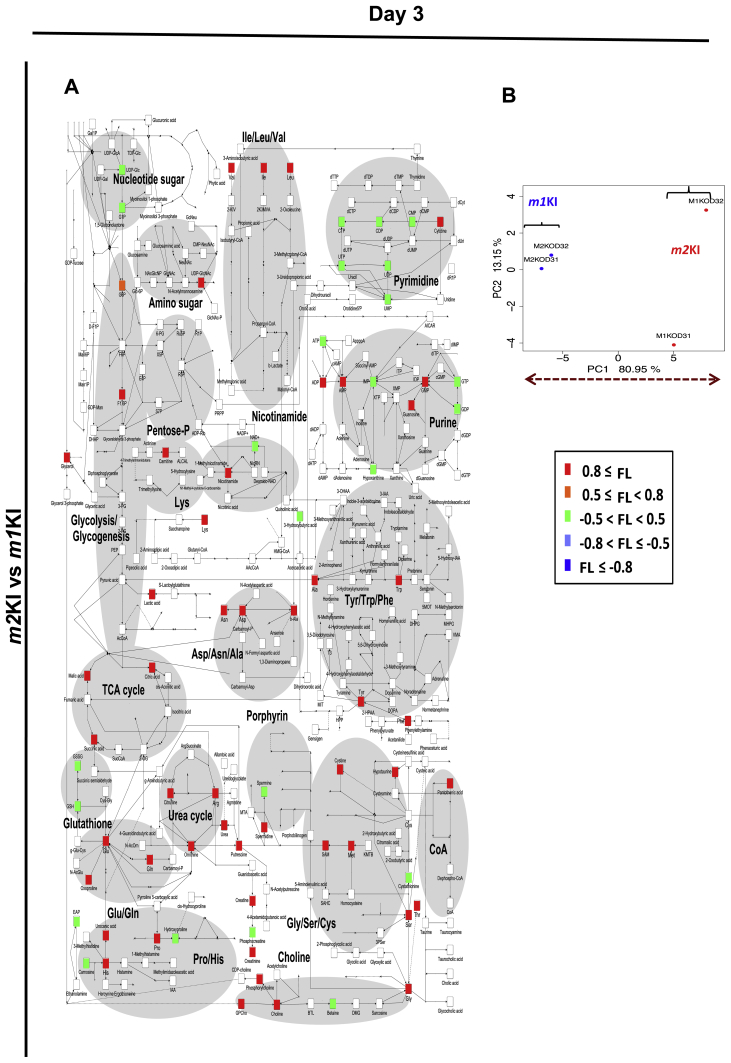
To determine the differences in metabolism between Pkm1KI and Pkm2KI ESCs during differentiation, we analyzed the metabolomes of WT, Pkm1KI, and Pkm2KI ESCs for 0, 3, and 7 days that were cultured without LIF in vitro using liquid chromatography-electrospray ionized time-of-flight MS (LC–MS). It was previously shown that oxidative stress influenced the function of transcription factor networks, such as FOXO, by altering the expression of downstream target genes, such as Pdx1 and Fbxo32[16], [17]. However, the basic metabolite dynamics in response to PKM1-dependent oxidative phosphorylation during early development remain unclear. Thus, we investigated the early-stage metabolomes of ESCs (Fig. 3, Figs.S2, S3). Principal component analysis showed that the glutathione pathway was enhanced and that SAM levels were higher in Pkm2KI ESCs at days 0, 3, and 7 than those in controls (Fig. 3, Figs.S2, S3). SAM is important for H3K4 di- or trimethylation and euchromatin formation [11]. Because the ratio of SAM:S-adenosyl homocysteine was high, SAM may have played a role in maintaining histone methylation events during the early differentiation of Pkm2KI ESCs. Thus, we hypothesized that PKM2 may have a pivotal role in modulating oxidative stress-induced mechanism.
Fig. 3.
PKM1 and PKM2 effects on cell metabolomes during early differentiation (Day 7). Embryoid bodies were prepared from Pkm1KI and Pkm2KI embryonic stem cells (ESCs) and cultured for 7 days without LIF. Principal component analysis results indicated a good separation between Pkm1KI and Pkm2KI ESCs. Comparison of all metabolites of Pkm1KI vs. Pkm2KI ESCs shown as loading factors (labeled colors). Loading (−1 < FL < 1) factors indicated the importance of each variable to account for the variability in PC1. When the FL number was small, this was regarded as an important metabolic product of Pkm1KI cells.
2.3. PKM2 controls pro-oxidative events during ESC differentiation
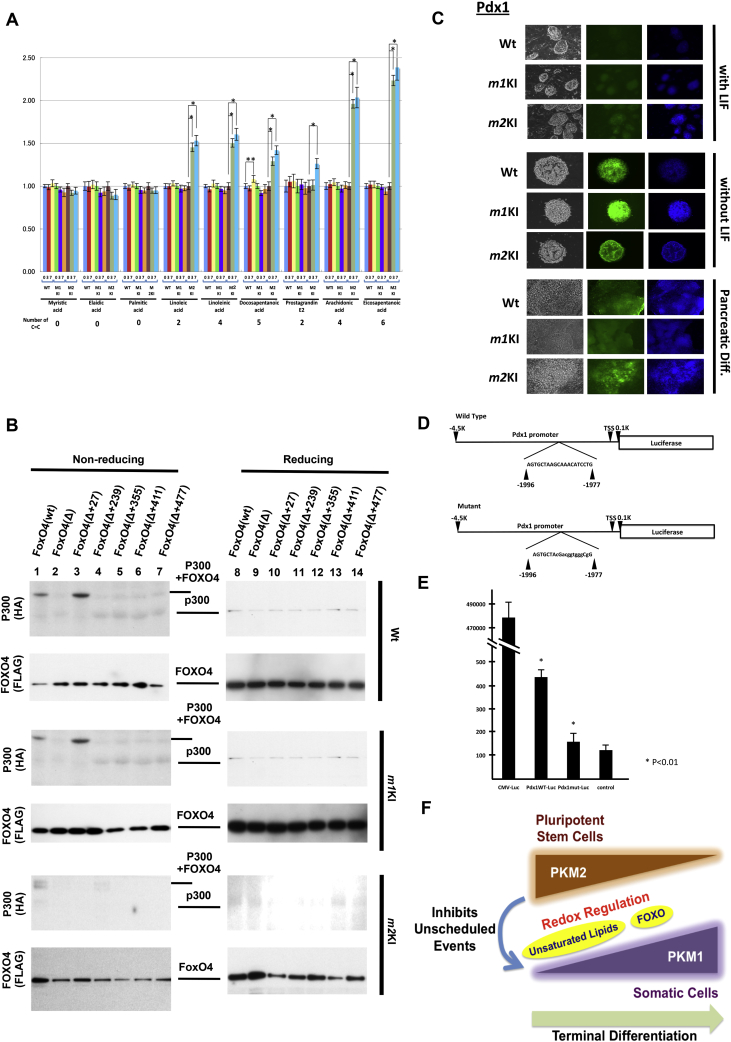
Principal component analysis also showed that lactate was the predominant metabolite in WT and Pkm2KI ESCs but not in Pkm1KI ESCs. Pluripotent ESCs are enriched in unsaturated lipids (i.e., ω3 and ω6) that contain readily oxidizable carbon–carbon double bonds [18]. Our LC–MS analysis showed that levels of unsaturated metabolites, such as linoleic acid, linolenic acid, docosapentaenoic acid, prostaglandin E2, arachidonic acid, and eicosapentaenoic acid (but not saturated acids), increased more in Pkm2KI ESCs than in Pkm1KI and WT ESCs in a time-dependent manner under differentiation conditions (Fig. 4A). These data strongly suggested that the Pkm2KI allele contributed to maintaining unsaturated lipid levels. Although a recent study indicated that highly unsaturated metabolites may be susceptible to pro-oxidative events [18], it remains unclear if this causes or results from the conditions of ESC differentiation; thus, we investigated if oxidation modulated the transcription response, which is critical for ESC differentiation.
Fig. 4.
PKM splicing is involved in redox control. Saturated and unsaturated fatty acids determined by mass spectroscopy. Pkm1KI, Pkm2KI, and WT ESCs were subjected to the early differentiation protocol for the indicated times. Numbers of unsaturated carbon bonds are shown at the bottom. (B) Redox-dependent heterodimerization of FOXO4 with p300/CBP. Wild-type (WT) and mutants (6) of Pkm1KI, Pkm2KI, and WT embryonic stem cells (ESCs) were studied under reducing and nonreducing conditions. (C) Immunocytochemical analysis of PDX1 expression with and without LIF or pancreatic differentiation of Pkm1KI, Pkm2KI, and WT ESCs. (D, E) Luciferase reporter activity. The PDX1 promoter sequence was inserted upstream of the luciferase sequence. FOXO4 binding nucleotides were substituted in a mutant vector. (F) Proposed scheme for a PKM splicing effect. PKM2 contributes to redox regulation of unsaturated metabolites in response to oxidative phosphorylation. In the pancreas, PKM2 inhibits unscheduled events by preventing Pdx1-induced excess differentiation progression. PKM1 and PKM2 orchestration may contribute to the terminal differentiation of somatic cells.
2.4. Alternative PKM1 and PKM2 splicing determines oxidation-dependent cysteine bridge FOXO transcription factors during ESC differentiation
Principal component analysis showed that the levels of cystine (sensitive to oxidation) were higher at days 3 and 7 than at day 0, which suggested that redox-sensitive modulation had occurred during ESC differentiation (Fig. 3, Figs.S2, S3). In general, cysteine oxidation of transcription factors affects DNA binding and the transcription activity of several nuclear factors, including activating-protein-1 (AP-1), nuclear factor-κB (NF-κB), p53, hypoxia-inducible factor (HIF) [19], and forkhead box class-O (FOXO) transcription factors [16]. Transcriptome pathway analysis has indicated that gene sets are enriched in the pancreatic PDX1 pathway [17] and the cardiocyte atrogin pathway [20], which are downstream of FOXO.
FOXO activity is inhibited by hydrogen peroxide (H2O2) by two independent mechanisms [21]: 1) FOXO associates with p300/CBP histone acetyltransferase via an intracellular disulfide bond, followed by protein acetylation and impaired transcriptional activity [16], which results in attenuated suppression of PDX1-dependent β-cell proliferation and failure against oxidative stress [17]; 2) An indirect mechanism of ROS-dependent phosphorylation of FOXO by Akt, which results in nuclear exclusion of this factor [22], [23] and causes in vitro alterations in cardiocyte modeling [20].
To determine if PK axis oxidation induced cysteine-thiol disulfide-dependent complexes of FOXO transcription factors and p300/CBP were involved, we tagged mutant vectors and transfected these into Pkm1KI, Pkm2KI, and WT ESCs. These cells were cultured in LIF-free medium to induce their differentiation and then subjected to immunoblot analysis. FOXO4 contains 5 cysteine residues (at positions 27, 239, 355, 411, and 477) and is involved in redox-dependent heterodimerization to p300 (1:1 ratio) via Cys477 to p300/CBP binding under exogenous H2O2 exposure [16]. We used 5 mutants, in which 4 of the 5 cysteine residues were mutated to serine residues, which left a single unaltered cysteine residue [e.g., denoted FOXO4(Δ+27), excluding Cys27].
Under nonreducing conditions, both FLAG-FOXO4 (WT) and FOXO4 (Δ+27) associated with HA-p300/CBP in Pkm1KI and WT ESCs as a slowly mobilized band. This association was reduced in FOXO4 (WT) and was undetectable in FOXO4 (Δ+27) in Pkm2KI ESCs, and it disappeared and showed only a single p300/CBP band under reducing conditions (Fig. 4B). Thus, Cys27 was probably involved in binding to p300/CBP under endogenous PKM1-derived redox regulation in ESCs, which may have been a condition distinct from exogenous peroxide-induced alteration of Cys477 in HEK293T cells [16] and suggested a unique response in PSCs. Immunohistochemistry results for embryoid bodies cultured without LIF for 7 days showed that the nonregulation of Pdx1 was considerably increased during pancreatic lineage differentiation of mutant ESCs (Fig. 4C), which suggested that early differentiation was attenuated by oxidative phosphorylation.
FOXO family inhibits unscheduled β-cell proliferation by suppressing PDX1 by competing with FOXA2 on the PDX1 promoter sequence [17]. To determine the importance of the PDX1 promoter sequence with a FOXO4 binding site, a luciferase reporter assay was performed using the mouse insulinoma cell line Min6 that expressed Pdx1. Luciferase activity was reduced due to a mutation in the FOXO4 binding site. This suggested that FOXO4 was an important factor for Pdx1 gene expression (Fig. 4D, E). Thus, the association of FOXO with p300/CBP might have impaired transcriptional function and protected against β-cell failure due to oxidative stress.
In addition, Akt was phosphorylated, and it attenuated the transcriptional activity of downstream F-box only protein 32, an important factor for cardiocyte remodeling [20] in Pkm1KI ESCs (data not shown). The role of PKM2 was confirmed by expression profiling during early differentiation, which was considerably different from those in Pkm1KI and WT ESCs. The PK axis appeared to play a pivotal role during differentiation via oxidative-sensitive FOXO transcription factors (Fig. 4F).
In conclusion, the present study results demonstrated that PKM2 contributed to fine-tuning controls of oxidative phosphorylation during the early stages of ESC differentiation of which mechanisms may be deleteriously altered in numerous cancer cells.
3. Methods
3.1. Cell culture and transfection
PKM knock-in ESCs and normal ESCs (v6.5 cell lines) were cultured in high-glucose DMEM (Nacalai Tesque) supplemented with 15% FBS (Nitirei), 1 mM sodium pyruvate solution (Gibco), 1× MEM nonessential amino acids (Gibco), 0.1 mM 2-mercaptoethanol (Nakarai), and 1000 U/ml of ESGRO (Millipore) on mitomycin treated MEF feeder cells at 37 °C in a CO2 incubator. The Min6 cell line was cultured in DMEM (high-glucose) supplemented with 10% FBS (Nitirei) and 0.1 mM 2-mercaptoethanol.
3.2. Genetic recombination in ESCs
Knock-in and knock-out Pkm1 and Pkm2 ESCs were generated by homologous recombination. Knock-in and knock-out ESCs for PKM1 and PKM2 were generated by homologous recombination of exon 9 with poly A tailed cDNA to disrupt splicing to exon 10 (Pkm1KI ESCs) and by homologous recombination of exon 10 with poly A tailed cDNA to disrupt splicing to exon 9 (Pkm2KI ESCs). ESCs were maintained in embryonic stem cell (ESC) medium supplemented with LIF and ESC certified serum (Invitrogen Life Technologies Japan, Tokyo, Japan). Our experimental protocols were approved by our Institutional Committee for Animal Use.
3.3. ESC differentiation
After culturing, ESCs were trypsinized, and embryoid bodies (EBs) were generated by the 3-D hanging drop method (Day 0). Briefly, EBs were grown in hanging drops for 2 days (Day 0 to Day 2), each of which initially included 500 cells in 19 μL of EB differentiation medium. EBs were cultured in DMEM medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% FBS and 0.08 mM 2-mercaptoethanol on an ultra-low attachment dish for 5 days (days 3–7). Pancreatic cell differentiation was performed using the methods described by Nakanishi et al. [24].
3.4. RNA analysis
Total RNA was extracted using phenol, followed by precipitation (TRIzol kit, Invitrogen Life Technologies Japan, Tokyo, Japan). cDNA was synthesized and used for quantitative real-time polymerase chain reaction (qRT-PCR) with a Light Cycler (Roche). Amplified signals were normalized against β-actin signals.
3.5. RT-PCR
The expressions of marker genes for differentiation, HOXA1, OTX2, PAX6, and NEUROG2 (ectoderm), BMP2 and SOX17 (mesoderm), AFP, HNF1B, and FOXA2 (endoderm), were determined by RT-PCR. Total RNA was extracted using Trizol (Life Technologies), according to the manufacturer's instructions and then treated with ReverTra Ace qRT-PCR Master Mix with gDNA Remover (Toyobo) to generate cDNA. qRT-PCR amplification was performed using Light Cycler FastStart DNA Master SYBR Green I (Roche). Primers used in this study were: HOXA1 (5′-CTACTCCAGCCCAACTCTGC-3′ and 5′-AATTGATGTGGACACCCGAT-3′), OTX2 (5′-ACCAGCAACCTGAAATGGAC-3′ and 5′-CCTGTTGGTCATGAAACGTG-3′), PAX6 (5′-AACAACCTGCCTATGCAACC-3′ and 5′-ACTTGGACGGGAACTGACAC-3′), NEUROG2 (5′-GCTGTGGGAATTTCACCTGT-3′ and 5′-CCTGGCCCTCTAACAAAACA-3′), BMP2 (5′-TGGAAGTGGCCCATTTAGAG-3′ and 5′-GCTTTTCTCGTTTGTGGAGC-3′), SOX17 (5′-ACCCTAAACACAAACAGCGG-3′ and 5′-GCAGGTGTCGAGATGACTGA-3′), AFP (5′-GAAGCAAGCCCTGTGAACTC-3′ and 5′-CCGAGAAATCTGCAGTGACA-3′), HNF1B (5′-GATGTCAGGAGTGCGCTACA-3′ and 5′-CTGAGATTGCTGGGGATTGT-3′), FOXA2 (5′-GGATCATGGACCTCTTCCCT-3′ and 5′-GGCACCTTGAGAAAGCAGTC-3′), GLUCAGON (5′-GCACATTCACCAGCGACTACAG-3′ and 5′-GGGAAAGGTCCCTTCAGCATGTCT-3′), and β-actin (5′-GTAGCTGTCTTCAGACACTCC-3′ and 5′-AATTAGAGCTATGCAGAGAAA-3′).
3.6. miRNA microarray experiments
For miRNA microarray experiments, after assessments for quality, 500 ng of extracted total RNA was labeled with Cyanine-3 (Cy3) using a Low Input Quick Amp Labeling Kit (Agilent). Dye incorporation and cRNA yields were checked using a NanoDrop ND-2000 Spectrophotometer. Labeled RNAs were hybridized on an Agilent Mouse GE 8×60K Microarray at 65 °C for 17 h in a rotating Agilent hybridization oven. After hybridization, microarrays were washed with GE Wash Buffer 1 (Agilent, Tokyo, Japan) at room temperature for 1 min and with GE Wash buffer 2 (Agilent) at 37 °C for 1 min, and then, dried immediately by brief centrifugation. After thorough washes with GE Wash Buffer 1 (Agilent) and GE Wash buffer 2 (Agilent) for 1 min each, fluorescent signals were scanned with an Agilent DNA Microarray Scanner (G2565CA) and analyzed using Feature Extraction Software 10.10 (Agilent).
3.7. Western blot analysis
Cells were lysed in a buffer containing EDTA-free protease inhibitors (Sigma Aldrich, Tokyo, Japan; 50 mM HEPES, pH 7.5, 150 mM NaCl, and 1% Triton X-100). Protein concentrations were determined with a protein assay kit (BioRad, Tokyo, Japan). Then, 20 μg of protein was subjected to SDS-PAGE, transferred to membranes, and antigens were detected using primary antibodies. All antibodies were from Sigma Aldrich except for isoform-specific antibodies against M1 (rabbit polyclonal; #AP7476b; Proteintech, Chicago, IL) and M2 (rabbit polyclonal; #15821-1-AP; Proteintech), which were generated using antigen-specific peptides. Immunoblot signals were quantified with an image analyzer (Multi Gauge Ver.3; FUJIFILM, Fuji, Tokyo, Japan). Results were expressed as means and standard deviations for 3 independent experiments.
3.8. Immunocytochemistry
Cells were fixed with 4% paraformaldehyde at room temperature for 30 min and then treated successively with 0.3% Triton X-100 (Wako Chemical) in PBS (Sigma Aldrich, Tokyo, Japan) for 15 min, followed by 3% bovine serum albumin (Sigma Aldrich) for 30 min to reduce nonspecific binding. Samples were reacted overnight with anti-PDX1 (#ab47308; Abcam, Tokyo, Japan) antibodies (1:200 dilution) at 4 °C, followed by reaction with Alexa Fluor 488 conjugated anti-guinea pig IgG antibody (#ab150185; Abcam) at room temperature for 1 h. Cell nuclei were stained with DAPI for 10 min. Photomicrographs were acquired with a fluorescence microscope (BZ-9000, Keyence, Osaka, Japan).
3.9. In-gel digestion and mass spectrometric protein identification
After immunoprecipitation, capillary electrophoresis (CE) and LC-MS analyses [Human Metabolome Technologies (HMT), Yamagata, Japan], in-gel digestion, and mass spectrometric protein identification were performed. Peptides were purified with ZipTip (Millipore Corporation, Billerica, MA), according to the manufacturer's instructions, and analyzed with an Ultraflex tof/tof (Bruker Daltonics, Bremen, Germany) MALDI mass spectrometer and MASCOT software (Matrix Science, Boston, MA).
3.10. Nontargeted metabolome analysis
For nontargeted metabolome analysis, cells cultured on a dish (106 cells/sample) were used for metabolite extraction. Cells were washed twice with a 5% mannitol solution and treated with 800 μL of methanol. Cell extracts were treated with 550 μL of Milli-Q water that contained internal standards (H3304-1002; Human Metabolome Technologies, Inc., Tsuruoka, Japan) and allowed to rest for another 30 s. Extracts were collected and centrifuged at 2,300 g at 4 °C for 5 min. Then, 800 μL of the upper aqueous layer was filtered by centrifugation using a Millipore 5-kDa cutoff filter at 9,100 g at 4 °C for 120 min to remove proteins before being subjected to CE–MS.
Peaks were extracted using automatic integration software MasterHands (Keio University, Tsuruoka, Japan) to obtain peak information, including m/z ratios, migration time for CE-TOFMS measurements (MT), and peak areas. Signal peaks that corresponded to isotopomers, adduct ions, and other product ions of known metabolites were excluded, and the remaining peaks were annotated with putative metabolites from the HMT metabolite database based on their MT and m/z values determined by MS. The tolerance range for peak annotation was configured at ±0.5 min for MT and ±10 parts/million for m/z ratios. Peak areas were normalized against those of the internal standards and the relative area values were further normalized by sample amounts. Hierarchical cluster analysis (HCA) and principal component analysis (PCA) were performed using our proprietary software, PeakStat and SampleStat, respectively. Detected metabolites were plotted on metabolic pathway maps using Visualization and Analysis of Networks containing Experimental Data (VANTED) software.
3.11. Luciferase assay
WT mouse Pdx1 promoter sequence and a mutant sequence with a FOXO4 binding site substitution were synthesized (Ynitech, Chiba, Japan). In the wild-type DNA fragment, the Pdx1 promoter (−4.5 kb–0.1 kb) was changed to the sequence 5′-AGTGCTACGACGGTGGGCGG-3′ at the position −1996 to −1977. Each fragment of Pdx1 promoter DNA was cloned into a pGL4.20 [luc2/Puro] vector (E6751, Promega, Tokyo, Japan). A total of 1 × 105 cells were grown in 6-well plates. After 24 h, the luciferase vector was transfected using Lipofectamine 2000 (Life Technologies, 11668027). At day 2, luciferase luminescence was measured using a Luciferase Assay System (E1500, Promega).
3.12. Statistical analysis
Results for categorical variables were compared by a Chi-square test. Results for continuous variables (medians/interquartile ranges) were compared using a Wilcoxon test. Statistical analyses were performed with JMP (JMP version 8.01, SAS Institute, Cary, NC). P-values of <0.05 were considered significant.
Acknowledgments
We thank the members of our laboratory for useful advice. We also thank Dr. Boudewijn MT Burgering (University Medical Center Utrecht, The Netherlands) for the p300 and FOXO mutant vectors. This study was financially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (#23390199, #25112708, #25134711, #30253420, #26670604) (MK, HI, MM); a Grant-in-Aid from the Ministry of Health, Labor, and Welfare (#H23-003) (MK, HI, MM); grants from National Institute of Biomedical Innovation (#12-4) (MK, HI, MM) and Osaka University Drug Discovery Funds, Japan (MK, HI, MM); grants from the Kobayashi Cancer Research Foundation (HI), Princess Takamatsu Cancer Research Fund (HI), Takeda Science and Medical Research Foundation (HI), Suzuken Memorial Foundation (MK), Yasuda Medical Foundation (NN), and Pancreas Research Foundation of Japan (KK). MK and HI received partial support from Chugai Co., Ltd., Yakult Honsha Co., Ltd., through institutional endowments. HI received partial support from Merck Co., Ltd., Unitech Co., Ltd., EBM Research Center Inc., and Taiho Therapeutic Co., Ltd. through institutional endowments; those funders had no role in main experimental equipments, supplies expenses, study design, data collection and analysis, decision to publish, or preparation of the manuscript, in this work.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.reth.2015.01.001.
Contributor Information
Hideshi Ishii, Email: hishii@gesurg.med.osaka-u.ac.jp.
Masaki Mori, Email: mmori@gesurg.med.osaka-u.ac.jp.
Conflict of interest
The authors have no conflict of interest to declare.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Fig. S1.
Expression profiling of Pkm1KI, Pkm2KI, and Wt ESCs in induced differentiation. Array analysis was performed (day 3 and day7), and gene expression was classified according to each pathway.
Fig. S2.
The effect of PKM1 and PKM2 on metabolomes of undifferentiated state. Metabolic analysis. Principal component analysis indicates good separation of Pkm1KI and Pkm2KI ESCs. Comparison of whole metabolites of Pkm1KI vs Pkm2KI ESCs is shown as loading factors (labeled colors).
Fig. S3.
The effect of PKM1 and PKM2 on metabolomes of early differentiation (Day3). Embryoid body were constructed from Pkm1KI, and Pkm2KI embryonic stem cells (ESCs) and cultured for 3 days without LIF. Principal component analysis indicates good separation of Pkm1KI and Pkm2KI ESCs. Comparison of whole metabolites of Pkm1KI vs Pkm2KI ESCs is shown as loading factors (labeled colors).
References
- 1.Nakashima K., Miwa S., Oda S., Tanaka T., Imamura K. Electrophoretic and kinetic studies of mutant erythrocyte pyruvate kinases. Blood. 1974;43:537–548. [PubMed] [Google Scholar]
- 2.Tanaka T., Harano Y., Sue F., Morimura H. Crystallization, characterization and metabolic regulation of two types of pyruvate kinase isolated from rat tissues. J Biochem. 1967;62:71–91. doi: 10.1093/oxfordjournals.jbchem.a128639. [DOI] [PubMed] [Google Scholar]
- 3.Imamura K., Tanaka T. Pyruvate kinase isozymes from rat. Methods Enzymol. 1982;90:150–165. doi: 10.1016/s0076-6879(82)90121-5. [DOI] [PubMed] [Google Scholar]
- 4.Mazurek S., Boschek C.B., Hugo F., Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Noguchi T., Inoue H., Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 6.Imamura K., Taniuchi K., Tanaka T. Multimolecular forms of pyruvate kinase. II. Purification of M2-type pyruvate kinase from Yoshida ascites hepatoma 130 cells and comparative studies on the enzymological and immunological properties of the three types of pyruvate kinases, L, M1, and M2. J Biochem. 1972;72:1001–1015. doi: 10.1093/oxfordjournals.jbchem.a129962. [DOI] [PubMed] [Google Scholar]
- 7.van der Horst A., Tertoolen L.G., de Vries-Smits L.M., Frye R.A., Medema R.H., Burgering B.M. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 8.Mazurek S., Grimm H., Boschek C.B., Vaupel P., Eigenbrodt E. Pyruvate kinase type M2: a crossroad in the tumor metabolome. Br J Nutr. 2002;87:S23–S29. doi: 10.1079/BJN2001454. [DOI] [PubMed] [Google Scholar]
- 9.Shyh-Chang N., Daley G.Q., Cantley L.C. Stem cell metabolism in tissue development and aging. Development. 2013;140:2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papp B., Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shyh-Chang N., Locasale J.W., Lyssiotis C.A., Zheng Y., Teo R.Y., Ratanasirintrawoot S. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Alexander P., Wu L., Hammer R., Cleaver O., McKnight S.L. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 14.Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hailesellasse-Sene K., Porter C.J., Palidwor G., Perez-Iratxeta C., Muro E.M., Campbell P.A. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dansen T.B., Smits L.M., van Triest M.H., de Keizer P.L., van Leenen D., Koerkamp M.G. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol. 2009;5:664–672. doi: 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- 17.Kitamura T., Ido-Kitamura Y. Role of FoxO proteins in pancreatic beta cells. Endocr J. 2007;54:507–515. doi: 10.1507/endocrj.KR-109. [DOI] [PubMed] [Google Scholar]
- 18.Yanes O., Clark J., Wong D.M., Patti G.J., Sánchez-Ruiz A., Benton H.P. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H., Colavitti R., Rovira, Finkel T. Redox-dependent transcriptional regulation. Circ Res. 2005;97:967–974. doi: 10.1161/01.RES.0000188210.72062.10. [DOI] [PubMed] [Google Scholar]
- 20.Tremblay M.L., Giguère V. Phosphatases at the heart of FoxO metabolic control. Cell Metab. 2008;7:101–103. doi: 10.1016/j.cmet.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 21.van Veelen C.W., Staal G.E., Verbiest H., Vlug A.M. Alanine inhibition of pyruvate kinase in gliomas and meningiomas. A diagnostic tool in surgery for gliomas? Lancet. 1977;2:384–385. doi: 10.1016/s0140-6736(77)90308-7. [DOI] [PubMed] [Google Scholar]
- 22.Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 23.Nemoto S., Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi M., Hamazaki S.T., Komazaki S., Okochi H., Asashima M. Pancreatic tissue formation from murine embryonic stem cells in vitro. Differentiation. 2007;75:1–11. doi: 10.1111/j.1432-0436.2006.00109.x. [DOI] [PubMed] [Google Scholar]