Abstract
Trypanothione (T(SH)2) is the main antioxidant metabolite for peroxide reduction in Trypanosoma cruzi; therefore, its metabolism has attracted attention for therapeutic intervention against Chagas disease. To validate drug targets within the T(SH)2 metabolism, the strategies and methods of Metabolic Control Analysis and kinetic modeling of the metabolic pathway were used here, to identify the steps that mainly control the pathway fluxes and which could be appropriate sites for therapeutic intervention. For that purpose, gamma-glutamylcysteine synthetase (γECS), trypanothione synthetase (TryS), trypanothione reductase (TryR) and the tryparedoxin cytosolic isoform 1 (TXN1) were separately overexpressed to different levels in T. cruzi epimastigotes and their degrees of control on the pathway flux as well as their effect on drug resistance and infectivity determined. Both experimental in vivo as well as in silico analyses indicated that γECS and TryS control T(SH)2 synthesis by 60–74% and 15–31%, respectively. γECS overexpression prompted up to a 3.5-fold increase in T(SH)2 concentration, whereas TryS overexpression did not render an increase in T(SH)2 levels as a consequence of high T(SH)2 degradation. The peroxide reduction flux was controlled for 64–73% by TXN1, 17–20% by TXNPx and 11–16% by TryR. TXN1 and TryR overexpression increased H2O2 resistance, whereas TXN1 overexpression increased resistance to the benznidazole plus buthionine sulfoximine combination. γECS overexpression led to an increase in infectivity capacity whereas that of TXN increased trypomastigote bursting. The present data suggested that inhibition of high controlling enzymes such as γECS and TXN1 in the T(SH)2 antioxidant pathway may compromise the parasite's viability and infectivity.
Keywords: Trypanothione, Gamma-glutamylcysteine synthetase, Trypanothione synthetase, Tryparedoxin, Trypanothione reductase, Flux control coefficient, Benznidazol
Graphical abstract
Highlights
-
•
The trypanothione synthesis flux is primarily but not exclusively controlled by γECS.
-
•
Tryparedoxin exerts high control on the peroxide reduction flux.
-
•
Kinetic metabolic modeling may reliably predict the in vivo pathway behavior.
-
•
TXN1 overexpression provides benznidazole resistance.
-
•
γECS and TXN contribute to parasite infectivity.
Abbreviations
- Cys
cysteine
- Glu
glutamate
- γEC
gamma-glutamylcysteine
- Gly
glycine
- GSH
glutathione
- Spd
spermidine
- Put
putrescine
- AdoMet
S-adenosyl methionine
- dAdoMet
decarboxylated S-adenosyl methionine
- T(SH)2
reduced trypanothione
- TS2
oxidized trypanothione
- ROOH
hydroperoxide
- γECS
gamma-glutamylcysteine synthetase
- GS
glutathione synthetase
- TryS
trypanothione synthetase
- SpdS
spermidine synthase
- AdoMetDC
S-adenosyl methionine decarboxylase
- PutT
putrescine transporter
- SpdT
spermidine transporter
- TryR
trypanothione reductase
- TXN1
tryparedoxin isoform 1
- TXNPx
tryparedoxin peroxidase
- GPx
non-selenium glutathione peroxidase-type tryparedoxin peroxidase
- OE
overexpressing
- CJai
flux control coefficient
- t-butOOH
tert-butyl hydroperoxide
- Wt
wild type
- Bnz
benznidazol
- BSO
buthionine sulfoximine
1. Introduction
Trypanosoma cruzi is the etiological agent of human Chagas disease. The World Health Organization estimates that 6–7 million people mainly in the Americas are infected with this parasitic protist, with ≈7500 annual deaths, whereas ≈70 million persons are at risk of becoming infected because of living in endemic regions [1,2,63,64]. Moreover, the disease has now also been found in non-endemic countries due to emigration of infected persons, with consequent non-vectorial transmission [2,3].
The current drugs available to treat the infection, benznidazol (Bnz) and nifurtimox have several drawbacks such as (i) high toxicity which causes severe side effects [4]; (ii) their lack of efficacy in the treatment of the chronic stage of infection [5]; and (iii) poor medical infrastructure: less than 1% of infected people have access to diagnostics and treatment [65]. Therefore, there is a need for new therapeutic strategies, safer and affordable drugs and validated drug-targets against Chagas disease [6,7]. Indeed, many T. cruzi enzymes and processes have been proposed as drug targets [7], including the trypanothione-dependent antioxidant pathway [[8], [9], [10], [11]].
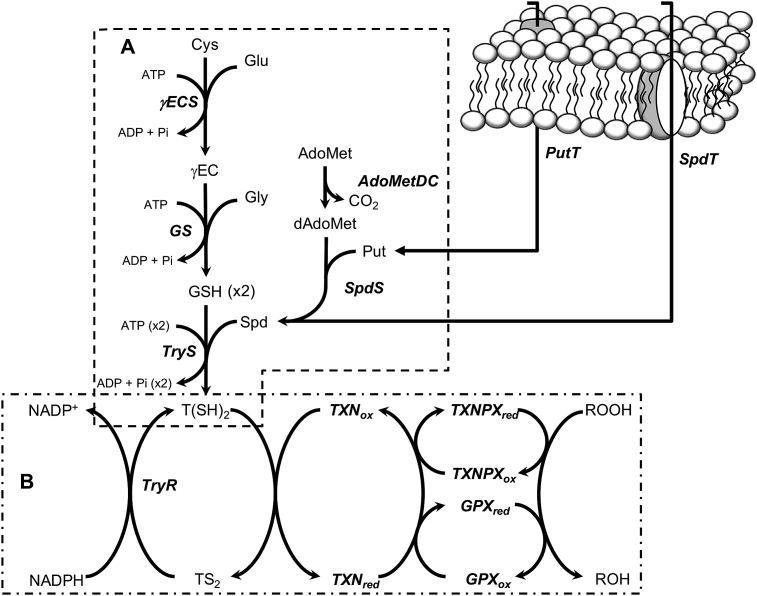
Trypanothione (T(SH)2) is a conjugate of two glutathione (GSH) and one spermidine (Spd) molecules that replaces the antioxidant functions that GSH has in most cells, including mammalian ones [12]. The antioxidant system of T. cruzi is constituted by two modules, the T(SH)2-synthesis pathway (Fig. 1A) and the T(SH)2-dependent hydroperoxide reduction pathway (Fig. 1B). In the first one, cysteine (Cys) and glutamate (Glu) are covalently linked by gamma-glutamylcysteine synthetase (γECS) to form gamma-glutamylcysteine (γEC), which then is bound to glycine (Gly) by glutathione synthetase (GS) thus producing GSH. The other precursor, Spd, can be imported from the extracellular environment or can also be synthesized from putrescine (Put) and decarboxylated S-adenosylmethionine (dAdoMet) by spermidine synthase (SpdS). Finally, trypanothione synthetase (TryS) synthesizes T(SH)2 by binding two GSH molecules to a Spd molecule [8].
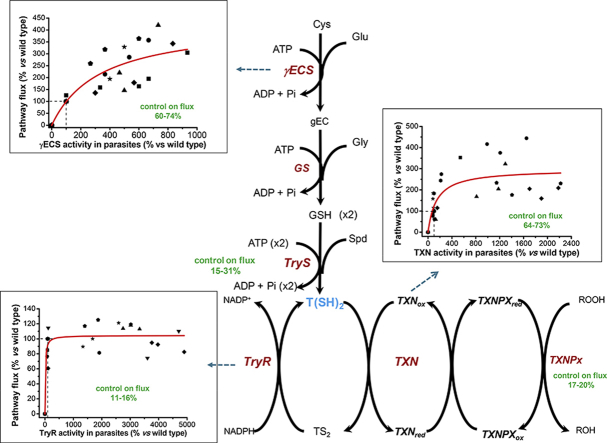
Fig. 1.
T. cruzi antioxidant pathway. (A) The trypanothione synthesis pathway starting from intracellular Cys. (B) The TXN1-dependent hydroperoxide reduction pathway. Metabolites are: Cys, cysteine; Glu, glutamate; γEC, gamma-glutamyl cysteine; Gly, glycine; GSH, glutathione; Spd, spermidine, Put, putrescine; AdoMet, S-adenosyl methionine; dAdoMet, decarboxylated S-adenosyl methionine; T(SH)2, reduced trypanothione; TS2, oxidized trypanothione; ROOH, hydroperoxide. Transporters and enzymes are: γECS, gamma glutamylcysteine synthetase; GS, glutathione synthetase; TryS, trypanothione synthetase; SpdS, spermidine synthase; AdoMetDC, S-adenosyl methionine decarboxylase; PutT putrescine transporter; SpdT, spermidine transporter; TryR, trypanothione reductase; TXN1, tryparedoxin 1; TXNPx, tryparedoxin peroxidase; GPx, glutathione peroxidase-type tryparedoxin peroxidase.
The cytosolic enzymes belonging to the main hydroperoxide reduction pathway catalyze peroxide reduction and oxidized trypanothione (TS2) reduction. Firstly, T(SH)2 reduces tryparedoxin 1 (TXN1), which then transfers its electrons to either tryparedoxin peroxidase (TXNPx), which has preference for H2O2 and short-chain alkyl/aryl hydroperoxide reduction, or to a TXN1-dependent non-selenium glutathione peroxidase-like enzyme (GPx), which has preference for long-chain alkyl peroxides, although it also uses other peroxides with one order of magnitude lower affinity [13,14]. These reactions produce oxidized trypanothione (TS2), which is regenerated by trypanothione reductase (TryR) using NADPH [8].
The arguments supporting the notion that T(SH)2 metabolism enzymes may serve as drug targets are: (i) TryS, TXN, TXNPx and TryR have no counterparts in the host; (ii) through gene expression manipulation, all the pathway enzymes (Fig. 1) have been proved to be essential in Trypanosoma brucei and Leishmania spp. (reviewed in [8,11,15]); (iii) TryR, the most intensively studied enzyme for drug-target design and screening studies, seems to be druggable [[16], [17], [18]]; (iv) TryS as well as mitochondrial and cytosolic TXNPx's have been proposed as virulence factors [[19], [20], [21]]. Nevertheless, their metabolic validation as potential sites for therapeutic intervention is still a pending experimental issue. In this regard, an approach is to determine the role that each enzyme has in controlling the T(SH)2 metabolism pathway, because inhibition of the most controlling pathway enzymes would affect more the pathway function than inhibition of enzymes exerting limited control (for a review see [22,23].
Metabolic control analysis (MCA) is a theoretical and experimental framework in the study of the control and regulation of metabolic pathways [24,25]; it can be applied to identify suitable drug targets in the intermediate metabolism of parasites [22,23]. Experimental MCA studies in several microorganisms and mammalian cells have demonstrated the non-existence of only one “rate-limiting” or “bottle neck” enzyme in metabolic pathways. Instead, they have shown that control of a metabolic pathway flux is shared among all the pathway enzymes/transporters, with only a few (2–3) steps showing the highest control [24,25]. MCA allows to quantitatively determine the degree in which a metabolic pathway flux depends on the activity of each individual pathway step, a value called flux control coefficient (CJai), where J is the pathway flux and ai is the activity of the pathway enzyme i. The sum of the positive CJai (steps that favor the pathway flux) and the negative CJai (flux-draining steps, e.g. pathway leaks) of all the pathway components should add up to one (summation theorem). An enzyme/transporter with a CJai approaching to one means that it has a predominant (but not unique) role in determining the pathway flux, whereas enzymes with CJai approaching to zero have negligible control on it. Therefore, enzymes with high CJai in essential metabolic pathways of the parasite are promising drug targets from a metabolic perspective [22,23,25].
The CJai are systemic properties, i.e. their determination requires a whole functional pathway, which is achieved only when all involved players interact with each other. Hence, the CJai cannot be determined by analyzing or manipulating the enzyme in isolation either in vitro or in vivo. The CJai can be theoretically predicted by in silico kinetic modeling of the metabolic pathway. Using this latter strategy, it was previously predicted that γECS and TryS exert most of the control on the T(SH)2 synthesis flux, with CJai values of 0.58–0.7 and 0.49–0.58, respectively, and with other steps displaying negative CJai to preserve the summation theorem [26]. On the other hand, by in vitro reconstitution of the hydroperoxide-reducing pathway using the recombinant enzymes, it was predicted that TXN1 and either TXNPx or GPx are the steps that mostly control the hydroperoxide reduction flux with CJai values of 0.9 [14]. In contrast, TryR showed negligible control on both, the T(SH)2 synthesis [26] and hydroperoxide reduction [14] fluxes.
The present work aims to expand the previous MCA studies and to authenticate the in silico and in vitro predictions of flux control distribution of the T(SH)2 metabolism in T. cruzi, by now developing in vivo experimentation. To this end, the CJai of γECS and TryS on the T(SH)2 synthesis flux, and TXN1 and TryR on the peroxide reduction pathway, were determined by modulating the expression of these enzymes in the parasites and determining its effects on the pathways’ fluxes. In addition, the previously reported metabolic model of T(SH)2 synthesis [26] was updated and expanded with additionally determined kinetic data and a new kinetic model of the peroxide reduction pathway was also constructed. Furthermore, correlations between the degree of control of the pathway enzymes with Bnz and peroxide resistance, and infectivity in human cells, were also analyzed. The results indicated that γECS and TXN1 have high control on their respective pathway fluxes and are relevantly involved in drug resistance and infectivity.
2. Methods
2.1. Cell culture
The Mexican T. cruzi Queretaro strain (DTUI) [27] was used throughout the study. Epimastigotes of non-transfected wild type (Wt), and stable clones created by transfection with empty plasmid (mock) and plasmids for overexpressing (OE) specific enzymes were grown in liver infusion-tryptose (LIT) medium {0.5% tryptose - 0.5% liver infusion (DIFCO; Detroit, MI, USA), 0.4% NaCl, 0.04% KCl, 0.42% Na2HPO4, 0.2% glucose}; supplemented with 10% fetal bovine serum FBS (Biowest; Nuaillé, France), 25 μg hemin/mL and 100 U penicillin/mL plus 100 μg streptomycin/mL, and maintained at 28 °C. Where indicated, 300 μg G418/mL (Cayman; Ann Arbor, MI, USA) were added. To determine their generation time (G), epimastigotes were cultured at an initial concentration of 0.8 × 106 parasites/mL and incubated at 28 °C for 96 h. Each 24 h the number of parasites was determined by direct counting of motile parasites in a Neubauer chamber or by absorbance at 600 nm. The G was calculated as the inverse of the growth rate constant (μ), the latter being the inverse of the slope of a Log2 (OD) vs. time curve.
2.2. Gene amplification
The TryR and TXN1 genes were ligated into the pTREXn expression vector specific for T. cruzi [28]. The genes were amplified by PCR using standard methods with the following nucleotide primers: TryR, sense 5'gcggcggcggcgaagcttatgatgtcaaagatttttg3′, antisense 5′ggcggccgcttacagagatgcttctgaagg3’; TXN1, sense 5'ggcaagcttatgtctggtttggcgaag3′, antisense 5'ggcggccgcttagtcggaccaggggaag3′, both containing HindIII and NotI restriction sites for oriented ligation. For TryR, a previously reported plasmid containing the gene from the T. cruzi Ninoa strain [26] was used as template for gene amplification. For TXN1, genomic DNA from the T. cruzi Ninoa strain was used. The PCR products were ligated into the pTREXn vector by standard methodologies. Construction of plasmids for γECS and TryS overexpression has been previously reported [29]. Genes' nucleotide sequences and in-frame plasmid constructs were verified by automated Sanger DNA sequencing.
2.3. Parasite transfection
Epimastigotes overexpressing γECS and TryS were created as reported elsewhere [29]; the same protocol was used to obtain clones overexpressing TryR and TXN1. Briefly, 3 × 108 epimastigotes resuspended in 350 μL of cold non-supplemented LIT medium were transfected with 100 μg of cesium chloride-purified plasmid DNA by electroporation with a BTX ECM 830 electroporator (Harvard Apparatus; Holiston, MA, USA) at 300 V for 70 ms in 2 mm gap BTX electroporation cuvettes. The parasites were maintained at 4 °C for 5 min and then resuspended in fully supplemented LIT medium and incubated at 28 °C. After 48 h, 500 μg G418/mL was added to the culture to select plasmid-containing parasites. After 7 days, an aliquot (500 μL) of the culture was diluted 10 times in medium lacking the antibiotic and cells were grown for 5 days; later, they were diluted in medium and exposed again to antibiotic for 7 days. This procedure was repeated twice to ensure that most of the parasites were transfectants. The selection procedure took approximately two months, after which the parasites were maintained under an antibiotic concentration of 300 μg G418/mL. As control of the transfection and selection procedure, epimastigotes were transfected with a pTREXn-GFP construct, treated likewise, and the total number of transfected parasites was monitored by direct counting using a fluorescence microscope. Whenever ≈100% of parasites were expressing GFP, it was assumed that the parasites transfected with the other constructs were also completely selected. The selected parasites grown without drug and grown back with G418, maintained the resistance and/or fluorescence (GFP control), indicating that the obtained transfected parasites were stable.
2.4. Cloning of transfected parasites
An heterogeneous population (pop) of stable-transfected parasites, all selected for antibiotic resistance indicative of harboring an expression plasmid, was obtained and processed to obtain clones with different levels of enzyme expression by the following protocol. Serial dilutions (1:2) were performed, starting with 58 parasites in 200 μL of complete LIT medium with antibiotics and G418 in a 96 well plate and the cells were grown at 28 °C for at least 4 weeks. From the last dilution of parasites yielding clones, at least three clones (numbered 1, 2 and 3 as the overexpression level increased) were selected for each overexpressed protein.
2.5. Protein content in the overexpressing clones
Soluble cell protein (0.1 mg) of selected OE-clones was prepared as for enzymatic activity (section 2.6). The proteins were separated by SDS-PAGE. For OE-γECS, OE-TryS and OE-TryR, the separating gel was prepared at 10% polyacrylamide whereas for OE-TXN1 it was at 20%. The gels were processed for Coomassie-Blue staining. As we have not specific antibodies against these enzymes, Western blot analyses were not performed.
2.6. Enzyme activities in parasites
Control (Wt or mock) and different stable OE-epimastigote clones were cultured to the late logarithmic phase, harvested by centrifugation at 4250 x g and washed with PBS. The soluble-enriched fractions were prepared as described before [29] by lysing the parasites with three cycles of freezing/thawing and the lysate was centrifuged to collect the soluble fraction and immediately used for activity determination.
Activities of γECS and TryS in the OE clones and GS in all parasites were determined as described before [29] by coupling the ADP production via pyruvate kinase/lactate dehydrogenase (PyK/LDH) to NADH oxidation and the change in absorbance was monitored in real time under initial velocity conditions at 340 nm and 37 °C in a diode array spectrophotometer (Agilent, Santa Clara, CA, USA). In all assays it was made sure that the activity was linear with respect to the amount of soluble cell protein. The 0.5 mL reaction contained 100 mM Hepes buffer pH 7.4, 1 mM EDTA, 5 mM MgSO4, 100 mM KCl, 0.2 mM NADH, 2 mM ATP, 2 mM phosphoenolpyruvate (PEP) and at least 600 mU PyK and 900 mU LDH. In addition, the following components were added (the specific thiol substrates were added to start the reactions): for γECS activity, 0.15–0.25 mg soluble cell protein, 1.3 mM Glu and 2.1 mM Cys; for GS activity, 0.005–0.040 mg soluble cell protein, 8 mM Gly and 0.4 mM γEC; and for TryS activity, 0.01–0.2 mg soluble cell protein, 11 mM Spd and 3 mM GSH. To subtract the high spurious ATPase activity present in the cell samples (accounting for 150–300 nmol/min x mg soluble cell protein in the enzymatic assay), a master mix for two reactions was prepared with all components (including the soluble cell protein) except the specific thiol substrate; the mixture was divided in two reactions and only one was supplemented with the corresponding thiol substrate and changes in the absorbance of the two reactions were followed in parallel. The activity in the absence of the thiol substrate accounted for the non-specific activity and was always subtracted. γECS and TryS basal activities in Wt and mock cells could not be determined with this protocol since no reliable increased rates above the high ATPase activity could be distinguished. TryS activity was determined in soluble cell protein fraction of Wt cells by HPLC. A mixture of reaction buffer (100 mM Hepes pH 7.4, 100 mM KCl, 1 mM EDTA, 5 mM MgSO4), 2 mM PEP, 2 mM ATP, 0.6–0.9 U PyK/LDH and 0.2–0.4 mg of soluble cell protein was prepared and then divided in three: one was supplemented with 3 mM GSH, the second with 11 mM Spd (both were control reactions) and the third was supplied with both substrates. Aliquots of 90 μL were transferred to 1.5 mL tubes and incubated at 37 °C for 0, 15, 30 or 60 min. After each time, the reaction was stopped by adding perchloric acid (PCA) at 3% v/v final concentration and T(SH)2 was determined by HPLC as described in section 2.7 [29].
TryR, TXN1 and TXNPx activities were determined as previously described [14] by monitoring the T(SH)2-dependent peroxide reduction associated to NADPH oxidation. For TryR activity the mixture contained 40 mM Hepes pH 7.4, 1 mM EDTA, 0.16 mM NADPH, 0.23 mM TS2 and the cellular sample (5–50 μg soluble cell protein from Wt clones; and 0.1–0.2 μg soluble cell protein from OE-clones) was added to start the reaction. For TXN1 and TXNPx, the mixture contained 40 mM Hepes pH 7.4, 1 mM EDTA, 0.16 mM NADPH, 0.45 mM (TSH)2, 0.5 μM TryR, > 24 μM TXNPx (for TXN1 determination) or > 22 μM TXN1 (for TXNPx determination) and 0.1 mM cumene hydroperoxide (CumOOH) was added. The reason to use CumOOH instead of H2O2 is that the inhibitory effect of the former on TXNPx occurs at a higher concentration than that of the latter [14], making it feasible to monitor the steady state of the reaction for a longer period of time. After 3 min baseline stabilization, the reaction was started by adding the soluble cell protein; when the endogenous enzyme activity levels were determined, the amounts were 0.03–0.4 mg soluble cell protein and 0.03–0.35 mg soluble cell protein for TXN1 and TXNPx, respectively; and 3–30 μg soluble cell protein from the OE-TXN1 clones.
2.7. Metabolite determination
Concentrations of thiol metabolites were determined in the parasites as previously described [29]. The cells were grown to the late exponential phase, harvested and washed twice with PBS. They were resuspended in lysis buffer (20 mM Hepes pH 7.4, 1 mM EDTA, 0.15 mM KCl) plus 20 mM dithiothreitol, disrupted by freezing and thawing, centrifuged at 17000 x g for 10 min and the soluble fraction was separated. The samples were strongly reduced with NaBH4, acidified with PCA (3% v/v final concentration) and centrifuged at 16,843 x g for 2 min. Twenty microliters of the supernatant were analyzed by HPLC. The thiol-molecules were post-column derivatized with DTNB and detected at 412 nm. It was previously demonstrated that non-significant T(SH)2 degradation occurs with this protocol [29]. To calculate the millimolar intracellular concentration, it was here determined that 1 × 106 epimastigotes have 5 ± 0.8 μg total cell protein (n = 10) and considered that 1 × 109 epimastigotes have an intracellular water volume of 30 μL [30].
2.8. Supplementation of thiol-molecules and polyamine to epimastigotes
Cultures of epimastigotes (controls and OE-clones) were initiated at a concentration of 0.8 × 106 parasites/mL and incubated at 28 °C for 48 h (reaching a concentration of ≈3 × 106 parasites/mL), after which they were separated in 25 mL aliquots. One of them was used as a control (no supplementation) and the other were supplemented with different concentrations of Cys (0.03, 0.06, 0.1 mM; ≈10–33 fmol/cell), GSH (0.3, 0.6, 1.0 mM; ≈100–333 fmol/cell), Spd (0.1 mM; ≈33 fmol/cell) or the combination of Cys plus Spd (0.1 mM each; ≈33 fmol/cell each). The parasites were incubated at 28 °C for further 24 h and processed for metabolite determination (section 2.7). The difference in metabolite concentrations before and after 24 h supplementation was calculated and normalized in percentage versus the control condition (no supplementation).
2.9. Ex vivo T(SH)2 synthesis flux
Control (Wt and mock) and OE-γECS or OE-TryS epimastigotes were cultured to the late logarithmic phase and soluble cell protein fractions were prepared as for enzyme activity determination (section 2.6). The T(SH)2 synthesis flux was determined ex vivo (using saturating concentrations of the precursors Cys, Glu, Gly, ATP and Spd) at room temperature in 0.5 mL of 100 mM Hepes pH 7.4, 5 mM MgSO4, 100 mM KCl, 1 mM EDTA, 2.1 mM Cys, 1.3 mM Glu, 8 mM Gly, 11 mM Spd, 3 mM ATP, 2 mM PEP, 20 mM DTT, at least 600 mU PyK and 900 mU LDH (the latter two to maintain a high and constant ATP concentration) and 0.8–1.2 mg of soluble cell protein were added to start the reaction. A control reaction lacking Cys was made in parallel. At 0, 1, 3, 5, 10 and 15 min, 90 μL of the reaction were withdrawn and mixed with PCA (3% final concentration) to stop the reaction. The time-dependent T(SH)2 formation was determined by HPLC as described in the metabolite determination assay (section 2.7). The T(SH)2 formation rate of the control reaction lacking Cys was subtracted from the full reaction. For these experiments it was made sure that the flux was linear with respect to the amount of protein used.
2.10. Ex vivo hydroperoxide reduction flux
A soluble cell protein fraction was prepared as for the enzyme activities determination assay (section 2.6). The peroxide reduction ex vivo flux was carried out as previously described [14]. Briefly, the 0.5 mL reaction mixture contained 40 mM Hepes pH 7.4, 1 mM EDTA, 0.2 mM NADPH, 0.45 mM T(SH)2 in-house prepared according to [31] and 0–2.5 mg of soluble cell protein. The reaction was initiated by adding 0.1 mM CumOOH and the NADPH oxidation was monitored at 340 nm and 37 °C. Also, for these experiments it was ensured that the flux was linear with respect to the amount of protein used.
2.11. Flux control coefficients
For rigorous determination of flux control coefficients, actual enzyme activities are required, regardless of the protein contents. In order to determine the CJai of the enzymes of the T(SH)2 metabolism, three clones of the OE-epimastigotes with different levels of enzyme activity were selected. The effect of changes in enzyme activity on either the ex vivo T(SH)2 synthesis flux or the ex vivo peroxide reduction flux were determined. The CJai is determined from the slope of the tangent (i.e., the derivative) to a curve of pathway flux versus enzyme activity multiplied by a scalar factor (aio/Jo) that represents the ratio of the values of flux and enzyme activity at the reference metabolic state (for further details, see [[23], [24], [25]]). To simplify the procedure due to the high variability in the biological samples, the percentage of pathway flux versus percentage of enzyme activity compared to control cells (Wt or mock) were plotted, and the CJai calculated from the derivative at the 100% (control) activity level in each individual experiment. A requisite for CJai determination is that no significant modifications in the other pathway enzymes be attained. Therefore, the activities of the other enzymes from the pathway were also determined on the OE-clones.
2.12. Pathway modeling
Kinetic models for T(SH)2 synthesis and T(SH)2-dependent peroxide reduction pathways were built using the metabolic simulator software GEPASI/COPASI [32,33]. The full information for their construction is included in Supplementary Material 2 (SM2). The models included the reactions displayed in Fig. S2.1 and Fig. S2.3. A summary of the reaction codification in the software, the kinetic parameters and reaction kinetic mechanisms [62] are shown in Tables S2.1 and S2.3. The initial and fixed metabolite concentrations used in the models are provided in Table S2.2 and Table S2.4. The full kinetic rate equations are described in SM2. The model files are available on request from the corresponding author. The characteristics of each model are outlined below.
For the T(SH)2 synthesis, the previously published kinetic model [26] was improved (Fig. S2.1) by (i) including the kinetic parameters of the Cys supply reaction (Cys transport; CysT) to simulate the effects of Cys supplementation on the T(SH)2 pool; the reaction included the Km value for external Cys previously reported [34], whereas the other required kinetic and thermodynamic parameters were parameterized to simulate the internal Cys concentration as experimentally determined. (ii) including a new TryS rate equation with substrate inhibition by GSH with kinetic parameters obtained from OE-TryS cell samples and recently reported (Fig. S2 in [29]) with KmGSH = 1.6 mM and KiGSH = 7.3 mM). (iii) removing GSH and Spd leaks to allow higher variation of T(SH)2; the reasons for these changes are described in Results section 3.7. And (iv) replacing TryR and NADPH supply reactions with a reaction of T(SH)2-demand with kinetic parameters of the peroxide reducing enzymes as previously reported [14] (see further details in SM2). This kinetic model was also parameterized to simulate the increase in the thiol pools of parasites supplemented with 0.1 mM Cys.
The kinetic model of the T(SH)2-dependent peroxide reduction pathway reported here for the first time included the reactions of TryR, TXN1, TXNPx and NADPH supply (Fig. S2.3). The rate equations were bi-bi ordered reversible for TryR; ping-pong kinetics for TXN1 and TXNPx and mass action reversible for NADPH supply (Table S2.3). The enzyme kinetic parameters Vmax, Km and Ki were those previously determined by our research group under near-physiological conditions using the recombinant enzymes [14] (Table S2.3).
The models were refined until they were able to simulate (i) the steady-state metabolite concentrations as determined within the parasites under the different experimental settings used; and (ii) the experimentally determined fluxes, i.e. the ex vivo pathway flux determined here with parasite cell samples and with the in vitro reconstituted pathway [14].
2.13. Benznidazole (±BSO) and peroxide resistance assays
Two different protocols were used for Bnz and peroxide resistance assays. The first protocol included a 24 h exposure, and direct counting of motile parasites in a Neubauer chamber or the OD600nm determination. Epimastigotes were cultured at an initial concentration of 0.8 × 106 parasites/mL and incubated at 28 °C for 48 h, after which the parasite concentration was determined by direct counting and different concentrations of Bnz (0.5–11 μM; ≈0.16–3.6 fmol/cell) or Bnz (2.0–25 μM; ≈0.66–8.3 fmol/cell) plus 0.1 mM (≈33 fmol/cell) buthionine sulfoximine (BSO) were added. The parasites were further incubated at 28 °C and then counted again 24 h later. The difference in parasite concentrations before and after 24 h drug exposure was considered as the relative growth, which then was normalized in percentage versus the control condition (no drug added). The concentration at which the relative growth was decreased by 50% (IC50) was then calculated. For the second protocol, a bolus addition of different H2O2 concentrations (60–250 μM; ≈3.2–13.3 fmol/cell) was used, and the OD600nm was determined every 24 h over a period of 96 h. The growth rate constant (μ) was calculated (i.e. the slope of a Log2 (OD) vs. time curve), and the concentration at which the μ was decreased by 50% (IC50) was then calculated.
2.14. Parasite host cell infection
Human foreskin fibroblasts (HFF-1) and Rhesus monkey kidney epithelial cells (LLC-MK2) were grown in culture-treated flasks in Dulbecco's-MEM with high glucose (GIBCO; MD, USA), supplemented with 10% FBS (Biowest; Nuaillé, France), 100 U penicillin/mL and 100 μg streptomycin/mL and incubated under 5% CO2 at 37 °C until 70% confluence was reached. The infection protocol was carried out as previously described [35] with some modifications. Primary infections of HFF-1 cells cultured in 25 cm2 flasks were initiated with Wt and newly transfected and selected populations of mock and OE-epimastigotes (2 × 106 parasites/mL) in Dulbecco's-MEM high glucose supplemented with 2% FBS; the interaction of parasites with human cells was allowed to last for 48 h. Then, the cells were daily washed with serum-free medium for epimastigote removal and replenished with fresh medium supplemented with 2% FBS. After 7–8 days post infection (dpi), trypomastigotes derived from the first burst (0.2 × 106 parasites/mL) were used for a secondary infection of LLC-MK2 cells cultured in 25 cm2 flasks and incubated for 48 h, afterwards the culture was processed as in the previous step. Trypomastigotes from the first burst were used to infect HFF-1 cells (tertiary infection)ultured over coverslips in 24 well-plates in quadruplicates; after 2 h incubation the cells were washed twice to remove non-internalized parasites and replenished with fresh medium. After 18, 48 or 66 h post-infection, the coverslips were exhaustively washed, fixed with 4% formaldehyde, stained with DAPI by standard methods and the number of infected cells as well as the number of internal parasites was analyzed by fluorescence microscopy. At least 500 cells per coverslip were analyzed.
To examine the trypomastigotes bursting, HFF-1 cells were grown in 24 well-plates (in the absence of coverslip) and infected as above. The trypomastigotes in the supernatants were resuspended and collected at the fourth and fifth day to determine by direct counting in the Neubauer chamber the number of trypomastigotes that had burst into the extracellular medium.
3. Results
3.1. Cell growth in stable transfectants
The generation time (G) in at least three independent cultures of the different OE-TryS (23.6 ± 2.5 h), OE-TryR (23.4 ± 3.1 h) and OE-TXN1 (25.6 ± 4.1 h) clones was similar in comparison to that of Wt or clones of mock cells (24 ± 2.7 h and 24.3 ± 2.1 h, respectively), except for OE-γECS, whose value (26.3 ± 3.7 h) was higher (p < 0.05), thus growing slower than Wt and mock cells. In addition, the generation time of mock cells in the absence or presence of G418 showed no difference vs. control cells (23.9 ± 1.4 h). The G values were similar whether optical density or direct cell counting methods were used.
3.2. Protein contents and enzyme activities in control and enzyme-overexpressing parasites
SDS-PAGE analysis of the soluble cell protein fractions from Wt, mock and OE-parasites showed that the targeted proteins in the OE-TryS, OE-TryR and OE-TXN1 clones were indeed overexpressed (Fig. S1.1 in supplementary material 1; SM1). However, no clear overexpression of γECS was apparent in the cell samples.
The basal γECS activity in the absence of overexpression cannot be accurately determined due to a high spurious ATPase activity in the assay (150–300 nmol/min x mg soluble cell protein). By performing the appropriate control reactions as described in the methods section and subtracting the unspecific ATPase reaction, it was established that a reliable difference for the specific activity should be at least 3 nmol/min x mg soluble cell protein above the unspecific rates; hence the basal γECS activity was below this threshold value. Despite this high ATPase activity, γECS and TryS were reliably determined above the threshold in their respective OE-clones using the PyK/LDH coupled assay.
An effort was made to determine TryS activity in Wt cells by determining the T(SH)2 production through HPLC as described in methods. A typical HPLC profile (Fig. S1.2 in SM1) shows a time-dependent increase in T(SH)2 using 3 mM GSH. A value of 0.63 ± 0.19 nmol/min x mg cell protein for TryS activity was determined in 5 independent parasite cultures. This value does not correspond to the actual Vmax, since TryS shows substrate inhibition by GSH. To circumvent this problem, it was determined from GSH saturation curves of TryS activity in OE-TryS clones and by fitting of the experimental data, that the TryS Vmax was underestimated by nearly 50% (Table S1.1 in SM1). Therefore, it was assumed that in Wt epimastigotes the TryS Vmax was ≈1.2 ± 0.4 nmol/min x mg cell protein.
The levels of increased activity were on average ≈ 8 for OE-γECS, ≈75 for OE-TryS, ≈31 for OE-TryR and ≈13 for OE-TXN1 (Table 1). This indicated that the parasite clones indeed functionally overexpressed their respective enzymes. These overexpression levels did not significantly vary among clones of the same OE-parasites, except in some cases (Table 1). Unless overexpressed, the basal activities of TryR, TXN1 and TXNPx were unaltered in the OE-clones, whereas a decreased GS activity was determined in several clones. OE-clones randomly evaluated showed that their activities did not depend on the G418 concentration used (300 or 500 μg G418/mL) or its absence for about 2 months (data not shown). Nevertheless, the parasites were always grown in the presence of G418 to prevent unexpected changes in the enzyme activities.
Table 1.
Activities of the T(SH)2 metabolism enzymes in T. cruzi epimastigotes overexpressing different enzymes.
| Specific activity (nmol/min x mg soluble cell protein) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| γECS | OE/Mock | GS | OE/Mock | TryS | OE/Mock | TryR | OE/Mock | TXN | OE/Mock | TXNPx | OE/Mock | |
| Wt | <3& | 1 | 23 ± 10 | 0.8 ± 0.3 | 1.2 ± 0.4§ | 1 | 334 ± 111 | 1.1 ± 0.4 | 33 ± 11 | 1.0 ± 0.3 | 66 ± 25 | 0.9 ± 0.3 |
| Mock | <3& | 1 | 28 ± 7 | 1 | 1.2$ | 1 | 291 ± 100 | 1 | 32 ± 11 | 1 | 73 ± 20 | 1 |
| γECS 1 | 12 ± 2* | 4 ± 1* | 23 ± 13 | 0.8 ± 0.5 | ND | ND | 341 ± 54 | 1.2 ± 0.2 | 30 ± 12 | 0.9 ± 0.4 | 85 ± 15 | 1.2 ± 0.2 |
| γECS 2 | 16 ± 7*ℵ | 5 ± 2* | 15 ± 6* | 0.5 ± 0.2* | ND | ND | 368 ± 63 | 1.3 ± 0.2 | 57 ± 33 | 1.8 ± 1.0 | 86 ± 16 | 1.2 ± 0.2 |
| γECS 3 | 22 ± 3*ℵ | 7 ± 1* | 18 ± 8 | 0.6 ± 0.3 | ND | ND | 321 ± 49 | 1.1 ± 0.2 | 27 ± 13 | 0.9 ± 0.4 | 88 ± 7 | 1.2 ± 0.1 |
| TryS 1 | ND | ND | 14 ± 6* | 0.5 ± 0.2* | 77 ± 24* | 64 ± 20* | 347 ± 120 | 1.2 ± 0.4 | 29 ± 8 | 0.9 ± 0.3 | 97 ± 37 | 1.3 ± 0.5 |
| TryS 2 | ND | ND | 11 ± 4* | 0.4 ± 0.2* | 95 ± 17* | 79 ± 14* | 283 ± 60 | 1.0 ± 0.2 | 27 ± 12 | 0.8 ± 0.4 | 67 ± 27 | 0.9 ± 0.4 |
| TryS 3 | ND | ND | 11 ± 4* | 0.4 ± 0.1* | 98 ± 45* | 81 ± 37* | 346 ± 160 | 1.2 ± 0.6 | 49 ± 26 | 1.5 ± 0.8 | 86 ± 30 | 1.2 ± 0.4 |
| TryR 1 | ND | ND | 20 ± 10 | 0.7 ± 0.3 | ND | ND | 6.1 ± 3.4 (x103)*# | 21 ± 12*# | 34 ± 13 | 1.1 ± 0.4 | 68 ± 16 | 0.9 ± 0.2 |
| TryR 2 | ND | ND | 21 ± 4 | 0.7 ± 0.5 | ND | ND | 6.8 ± 2.5 (x103)*# | 23 ± 8.5*# | 34 ± 11 | 1.1 ± 0.3 | 77 ± 27 | 1.1 ± 0.4 |
| TryR 3 | ND | ND | 14 ± 3* | 0.5 ± 0.1* | ND | ND | 14.4 ± 7.7 (x103)* | 50 ± 27* | 32 ± 12 | 1.0 ± 0.4 | 64 ± 12 | 0.9 ± 0.2 |
| TXN1-1 | ND | ND | 8 ± 3* | 0.3 ± 0.1* | ND | ND | 293 ± 71 | 1.0 ± 0.2 | 379 ± 175* | 12 ± 5* | 73 ± 23 | 1.0 ± 0.3 |
| TXN1-2 | ND | ND | 9 ± 5* | 0.3 ± 0.2* | ND | ND | 317 ± 69 | 1.1 ± 0.2 | 399 ± 164* | 12 ± 5* | 79 ± 19 | 1.1 ± 0.3 |
| TXN1-3 | ND | ND | 11 ± 6** | 0.4 ± 0.2** | ND | ND | 318 ± 51 | 1.1 ± 0.2 | 490 ± 230* | 15 ± 7* | 83 ± 22 | 1.1 ± 0.3 |
& threshold reliable specific enzymatic activity after the spurious ATPase activity was subtracted. ND: below the threshold confidence activity of the PyK/LDH coupled assay. § value determined by HPLC. $ value assumed to be similar to that of Wt. Values are the mean ± SD of at least three independent cultures. Student's t-test for non-paired samples *p < 0.01, **p < 0.05 vs. mock. ℵp <0.01 vs. γECS 1, #p < 0.05 vs. TryR3.
3.3. Metabolite pools in transfected cells
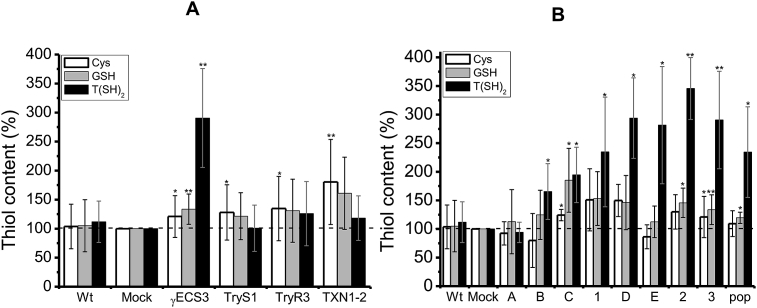
Mock and Wt cells showed similar thiol-molecule contents (Fig. 2A). OE-γECS3 cells showed 3.5-fold increased T(SH)2 concentration in comparison to mock cells (Fig. 2A). Analysis of the thiol contents in different OE-γECS clones revealed that when the γECS activity reached >10 nmol/min x mg soluble cell protein (clone C in Fig. 2B and Table S1.2 in SM1), the T(SH)2 concentration became clearly higher than in mock cells, reaching up to 4-fold more when the activity was 22 nmol/min x mg soluble cell protein. Moreover, the T(SH)2 level increased by ≈ 6-fold when OE-γECS cells were supplied with 0.1 mM Cys or 1 mM GSH, which was ≈2-fold above the increases attained in similarly supplemented mock cells (Fig. S1.3 in SM1). Higher concentrations of added Cys (0.2–1 mM) or GSH (up to 5 mM) resulted in lower T(SH)2 increases of 4.5- and 3.5-fold and even some inhibition (data not shown). On the other hand, OE-TryS1 cells showed similar T(SH)2 levels as Wt and mock cells, either supplemented with Cys, GSH or Spd or non-supplemented. This unexpected result is examined in the next section.
Fig. 2.
Thiol contents in control and overexpressing parasites. (A) Thiol metabolites in parasites correspond to: Wt (wild type); mock (cells transformed with empty plasmid); γECS3 (OE-γECS clone 3); TryS1 (OE-TryS clone 1); TryR3 (OE-TryR clone 3), TXN1-2 (OE-TXN1 clone 2). 100% levels in mock cells corresponded to: Cys, 6.6 ± 2.2 nmol/mg cell protein (1.2 ± 0.4 mM); GSH, 9.4 ± 3.5 nmol/mg cell protein (1.1 ± 0.4 mM); T(SH)2, 4.7 ± 1.8 nmol/mg cell protein (0.9 ± 0.3 mM). At least 28–45 different independent cell cultures for Wt and mock cells and at least 12–35 independent cultures for the OE-clones were used. t (B) Thiol metabolite concentrations in different γECS overexpressing clones. γECS activities for clones 1–3 are shown in Table 1; the values for clones A-D are shown in Table S1.2 in Supplementary Material 1.100% thiol concentration in mock cells correspond to: Cys = 6.8 ± 0.8 nmol/mg cell protein (1.1 ± 0.1 mM); GSH = 9.8 ± 3.8 nmol/mg cell protein (1.6 ± 0.6 mM); T(SH)2 = 4.5 ± 1.7 nmol/mg cell protein (0.7 ± 0.3 mM). Student's t-test for non-paired samples *p < 0.05, **p < 0.01 vs. mock.
The OE-TXN1-2 cells consistently showed increased Cys and GSH concentrations (≈75% and 60%, respectively) above control cell levels, but they showed no changes in T(SH)2 levels (Fig. 2A). OE-TryR3 cells showed a slight increase in thiol contents (≈30–40%) compared to control cells, although it was not statistically significant (Fig. 2A).
3.4. TryS overexpression did not induce increased T(SH)2 levels
Unexpectedly, OE-TryS1 cells showed non-significant changes in T(SH)2 vs. mock cells (Fig. 2A), despite the at least 64-fold increase in enzyme activity (Table 1). This has also been observed by others [36], where TryS overexpressing epimastigotes (but not trypomastigotes) did not increase their T(SH)2 pool. To further examine this counterintuitive result, the OE-TryS1 cells were grown with supplementation of (i) Cys, to circumvent a possible Cys deficit for endogenous γECS activity and hence low GSH synthesis; (ii) GSH, to directly increase the TryS substrate; and (iii) Spd, to determine whether this precursor was limiting. OE-TryS1 cells supplemented with 1 mM GSH showed a similar increase in T(SH)2 to that observed in control cells, and lower levels when supplemented with 0.1 mM Cys (Fig. S1.3). On the other hand, Spd seemed not to be limiting for T(SH)2 synthesis because (i) their basal concentration in Wt cells is high 0.8 ± 0.2 mM [29]; (ii) there should be no increase in T(SH)2 in the OE-γECS cells by merely supplementing with Cys or GSH (Fig. S1.3); and (iii) the OE-TryS1 parasites supplemented with Spd alone did not increase their T(SH)2 (Fig. S1.4 in SM1).
Furthermore, in experiments to determine the ex vivo T(SH)2 synthesis flux in OE-TryS clones, no net increase in T(SH)2 was attained, despite the presence of saturating Cys, Glu, Gly, ATP and Spd concentrations (see Methods section for details). Instead, a time-dependent increase in GSH was observed in the cell protein samples of OE-TryS cells, which was not evident in the ex vivo flux determinations in Wt, mock and OE-γECS cell samples. Such pattern in the OE-TryS clones suggested that (i) T(SH)2 might be simultaneously synthesized and degraded at similar rates or (ii) overexpressed TryS was not active. The latter possibility can be ruled out, because high TryS activities were indeed determined in the OE-TryS parasites (Table 1), although it is unknown whether in intact cells any type of negative metabolic regulation may occur.
To analyze the possibility of T(SH)2 degradation in the OE-TryS cells, soluble cell protein fractions from the OE-TryS1 clone were incubated in the absence or presence of 1 mM T(SH)2. Time-dependent increases in GSH (40–80 nmol/min x mg soluble cell protein) and decreases in T(SH)2 (10–30 nmol/min x mg soluble cell protein) were observed (Fig. S1.5 in SM1), indicating T(SH)2 degradation. The calculated rates of T(SH)2 degradation in the OE-TryS cell samples were 13 ± 5 and 21 ± 9 nmol/min x mg soluble cell protein in the absence or presence of added T(SH)2, respectively. In contrast, cell soluble protein fractions from Wt and mock cells did not show significant T(SH)2 degradation (Fig. S1.5 in SM1). These observations may explain the lack of net increase in T(SH)2 in the OE-TryS1 epimastigotes in comparison to control cells (Fig. 2A).
3.5. Trypanothione synthesis and peroxide reduction ex vivo fluxes
T(SH)2 is not a metabolic pathway end-product (such as lactate or ethanol for glycolysis or CO2 for the Krebs cycle) since there are still enzymes or processes using it. Hence, at a specific metabolic steady state, the T(SH)2 moiety pool in the cell is the result of the dynamic balance between its synthesis (i.e. supply) and consumption (i.e. demand). For these reasons, determination of fluxes of the T(SH)2 metabolism in intact parasites would require more sophisticated techniques such as labeling studies or fluxomics. To circumvent this limitation, the fluxes were determined ex vivo, in parasite soluble cell protein fractions. However, it has to be considered that (i) the values determined represent the maximal fluxes of synthesis with the enzymes expressed in the cell sample, because the pathway precursors Cys, Glu, Gly, Spd and ATP are saturating and thus the limiting factor should only be the content of active enzymes in the cell sample; and (ii) in the ex vivo system most likely some physiological regulatory interactions are lost.
The T(SH)2 synthesis flux was linear for up to 5 min using 0.3–0.8 mg soluble cell protein when Wt or OE-γECS3 soluble cell protein fractions were used (data not shown). If the specific pathway substrate Cys was not added to the reaction, the basal T(SH)2 content did not change over time, indicating that it was not significantly synthesized or degraded. Under such conditions, the maximal ex vivo T(SH)2 synthesis flux in Wt cells was 0.6 ± 0.2 nmol T(SH)2/min x mg soluble cell protein (n = 5) (Table 2), whereas in the OE-γECS3 clone with the highest activity (20 nmol/min x mg soluble cell protein) the flux was 2 ± 0.4 nmol T(SH)2/min x mg soluble cell protein (Table S1.2 in SM1). Since in the OE-γECS3 cells γECS activity was in excess in comparison to GS and TryS activities, these results also suggested that (i) the basal TryS activity should not exceed 2 nmol/min x mg soluble cell protein, i.e. the maximal T(SH)2 synthesis flux in OE-γECS3 cells; such a value is in the range of the TryS activity calculated from the HPLC results; and (ii) the basal γECS activity should not exceed 1.2 nmol/min x mg soluble cell protein, i.e. the pathway flux in the Wt cells multiplied by two (considering the pathway stoichiometry). In contrast, for the OE-TryS1 cells, which have a TryS activity of 77 nmol/min x mg soluble cell protein, a maximal flux output of only 0.3 nmol T(SH)2/min x mg soluble cell protein was obtained, which was even lower than in Wt cells.
Table 2.
Experimental and kinetic modeling results for the T(SH)2 synthesis pathway.
| 40 μM external Cys |
140 μM external Cys |
|||
|---|---|---|---|---|
| In vivo/ex vivo | Model | In vivo | Model | |
| Metabolite (mM) | ||||
| Cysin | 1 ± 0.2 | 0.5 | 1.8 ± 0.7 | 1.78 |
| GSH | 1.4 ± 0.4 | 1.36 | 2.7 ± 0.9 | 2.6 |
| T(SH)2 | 0.9 ± 0.4 | 0.75 | 2.6 ± 0.9 | 3.5 |
| Spd int | 0.78 ± 0.2 | 1.06 | 1.05 ± 0.3 | 0.99 |
| TS2 | ND | 0.06 | ND | 0.16 |
| Flux | ||||
| JTryS | 0.6 ± 0.2 (max)* | 0.12 | ND | 0.31 |
| CJai | ||||
| CJCysT | 0.13** | 0.09 | 0.02 | |
| CJγECS | 0.69 ± 0.15 0.87 (γECS-TryS)** |
0.74 | 0.6 | |
| CJGS | 8 e−6 | 5 e−6 | ||
| CJTryS | 0.15 | 0.26 | ||
| CJSpdT | 0.003 | 0.015 | ||
| CJTSH dem | 0.003 | 0.06 | ||
| CJTSH leak | 0.012 | 0.05 | ||
Flux in nmol/min x mg cell soluble protein. * Maximal ex vivo T(SH)2 synthesis.
** Values obtained by elasticity analysis. ND not determined. Cysin means internal Cys.
For the peroxide reduction flux, the maximal flux outputs in the Wt and mock cells were 11 ± 5 and 11 ± 4 nmol/min x mg soluble cell protein, respectively, values that were not significantly different in the OE-TryR3 cells (14 ± 4 nmol/min x mg), which showed the highest level of TryR overexpression. In contrast, in the OE-TXN1-3 with the highest TXN1 activity, the flux reached a value of 46 nmol/min x mg soluble cell protein. This latter observation suggests that the basal TXN1 levels are limiting for the peroxide reduction flux.
3.6. Flux control distribution of the T(SH)2 synthesis and peroxide reduction pathway from parasites
To determine the CJai of the pathway enzymes, the dependence of the pathway flux on the enzyme activities has to be analyzed. It is necessary to emphasize that the actual enzyme activity in the cell is required for CJai determination; the protein contents (e.g. determined by Western blotting) may provide inaccurate values since a considerable protein fraction may have low or no activity. This was the reason why in the present study the enzyme activities within the parasites were rigorously established. The levels of enzymes were specifically varied in the OE clones without substantial changes in other pathway enzymes (Table 1), which is another mandatory requisite for appropriate CJai determination. The enzyme variability shown among the different biological replicas of each cell clone allowed obtaining clustered points along the curve of pathway flux versus enzyme activity using only three clones per enzyme.
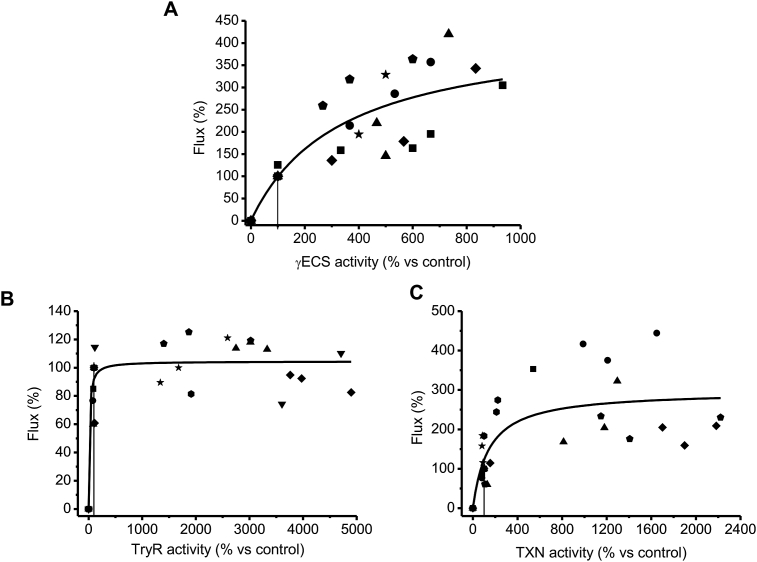
Fig. 3A shows the variations in the ex vivo T(SH)2 synthesis flux when the activity of γECS was varied using the different parasite clones. A CJai of 0.69 ± 0.15 (Table 2) was obtained for γECS at the Wt level of activity (taken to be 3 nmol/min x mg soluble cell protein, the threshold confidence value of the enzymatic assay). Unfortunately, the CJai for TryS could not be determined because of lack of variation of the T(SH)2 content in the parasites as described above. Notwithstanding this inconvenience for this ex vivo analysis, it was estimated that TryS may have a CJai of at most 0.31, by using the MCA summation theorem and assuming negligible control exerted by GS [26], the latter based on its higher activity in the parasites with respect to γECS and TryS (Table 1).
Fig. 3.
Determination of flux control coefficients. Variations in activity were achieved from clones described in Table 1. At least 5 independent titrations (indicated with different symbols) were made with three different clones. 100% pathway fluxes were: (A) T(SH)2 synthesis, 0.6 ± 0.2 nmol T(SH)2/min x mg soluble cell protein; (B) and (C) CumOOH reduction, 11 ± 4 nmol/min x mg cell soluble protein. The solid lines represent the global fitting of all data points by the Michaelis-Menten equation (without a mechanistic meaning). The vertical line shows the point at which the CJai was calculated and corresponds to the basal activity level in Wt (panel A) or mock (panels B and C) cells. Due to experimental variations between Wt and mock cells in the hydroperoxide reduction flux determination assays, some points showed values above 100% of flux (corresponding to Wt).
On the other hand, the peroxide reduction flux showed non-significant variations when TryR activity was increased up to ≈50-fold above Wt level (Fig. 3B); the calculated CJai for TryR was 0.15 ± 0.09 (n = 8). In contrast, when TXN1 was varied (Fig. 3C), the flux changed almost linearly near the Wt level, with a concomitant CJai of 0.73 ± 0.29 (n = 7). As TXNPx was not overexpressed in the parasites, the ex vivo CJai could not be obtained in a similar fashion to that for TryR and TXN1. In an attempt to determine the CJai of TXNPx, the peroxide reduction flux of soluble cell protein fractions from Wt and mock cells was titrated by adding recombinant TXNPx (Fig. S1.6 in SM1). The data showed that the flux increased while increasing TXNPx activity in a similar fashion to that of TXN1, yielding a CJai near 0.57–0.64 for TXNPx; hence, the control of the flux is shared equally by both TXN1 and TXNPx.
3.7. Flux control distribution of T(SH)2 synthesis by pathway modeling
An updated kinetic model of T(SH)2 synthesis was constructed based upon a previous version published by our group [26]. In the previous version of the model the predicted CJai of TryS (0.46–0.58) was higher than that obtained ex vivo here using parasite samples. In addition, the predicted CJai of SpdT (0.22–0.24) was also high in the previous model; however, the Spd supplementation experiments in parasites showed no increases in T(SH)2 (Fig. S1.4 in SM1), suggesting lower control than previously predicted for the polyamine supply. Simulations for the present study using the published model indicated that the high control values predicted for TryS and SpdT were due to the inclusion of GSH and Spd leak reactions. These two leak reactions decrease substrate availability to TryS, decreasing its rate, with the concomitant higher predicted CJai value for TryS. The GSH and Spd leaks also increased the CJai predicted for γECS, resulting in the sum of the CJai of γECS and TryS being higher than one, which was compensated by the negative CJai of the GSH and Spd leaks. For all these reasons the latter reactions were removed. In contrast, the supplementation experiments with Cys (Fig. S1.3 in SM1) indicated that the basal Cys level was limiting for the γECS activity and therefore, an extracellular Cys uptake (CysT reaction) was now included, for which some reported kinetic parameters are available. However, it should be noted that CysT may actually represent any other reaction or metabolic pathways providing Cys to the cell.
A refined model was here obtained, which was able to closely reproduce the metabolite concentrations found experimentally in the parasites without Cys supplementation (Cys concentration in the medium was 0.04 mM), although the simulated flux was lower than that obtained ex vivo (Table 2). The CJai of 0.74 for γECS in the model simulation (Table 2) agreed with that obtained ex vivo of CJai = 0.69 (Fig. 3A). The model predicted a CJai of 0.15 for TryS, a lower value than that previously reported, but this enzyme was still the second most flux-controlling enzyme in the T(SH)2 synthesis pathway. The CysT CJai of 0.09 was low whereas GS showed negligible control.
To further demonstrate the degree of control of Cys supply in vivo, elasticity analysis [[23], [24], [25],37] was performed (Fig. S2.2 in SM2) using data of the Cys, GSH and T(SH)2 thiol changes after Cys supplementation and BSO inhibition experiments previously published (Figs. 3A and 2 left panel, respectively in [29]). In elasticity analysis, intact cells are used and no modification of the enzymes is required, but manipulation of the intracellular steady-state levels of the pathway intermediates is performed by varying the pathway's initial substrate and by inhibiting the downstream pathway steps (see reference [23] for further details on elasticity analysis). The elasticity coefficients toward internal Cys for the Cys-supply reactions (in this case CysT) and for the Cys-consuming group of reactions (γECS, GS and TryS) were determined. Their respective CJai were determined by applying the summation and connectivity theorems of MCA [[23], [24], [25]]. CJai values of 0.13 for CysT and 0.87 for the Cys-consuming group of reactions were obtained (Table 2), which are in agreement with the sum of the CJai of γECS, GS and TryS obtained by modeling.
Afterwards, it was tested whether the model was able to simulate the increases in thiol intermediates found in parasites supplied with 0.1 mM Cys; however, merely increasing the Cys concentration did not predict the in vivo metabolite contents. The model required to simultaneously increase the Vmax values of CysT, γECS and TryS (parameterized in the model; Table S2.1 in SM2) to become able to simulate the metabolite concentrations found in Cys supplied parasites (Table 2). With these latter modifications, the pathway flux increased by 2.6-fold, which was still within the value experimentally determined, and the model predicted a high control by γECS, an increased CJai for TryS of 0.26 and again negligible control exerted by GS. Thus, the TryS control was lower than that of γECS, but higher than that of the Cys supply.
3.8. Flux control distribution of the peroxide reduction flux by pathway modeling
The kinetic model for the T(SH)2-dependent peroxide reduction flux considered TXN1 as an enzyme with ping-pong kinetics using the kinetic parameters previously determined [14]. The model closely simulated the pathway flux determined ex vivo using soluble cell protein fractions from Wt and mock cells (Table 3), as well as the flux attained in the in vitro pathway reconstitution with the recombinant enzymes previously reported [14]. Likewise, the model simulated with high accuracy the CJai determined here ex vivo by TXN1 and TryR titrations in the parasite and predicted a CJai for TXNPx of 0.17–0.2.
Table 3.
Ex vivo, in vitro and kinetic modeling results for the T(SH)2-dependent peroxide reduction pathway.
| Ex vivob | In vitro pathway reconstitution c | Predictions of the kinetic modeld | |
|---|---|---|---|
| CumOOH mM | 0.1 | 0.1** | 0.1 fixed |
| [T(SH)2] mM | 0.45 | 0.45** | 0.465 |
| [TS2] mM | ND | 0.00058 | |
| NADPH mM | 0.16 | 0.16** | 0.16 |
| NADP+ mM | ND | 0.025 | |
| [TXNox] μM | 0.022–0.078* | 0.1** | 0.083–0.089 |
| [TXNred] μM | 0.021–0.027 | ||
| JTXNPxa | 11 ± 5 | 4.9 ± 2.4 | 6.1–7.8 |
| CJTryR | 0.15 ± 0.09 | 0.2 ± 0.04 | 0.11–0.16 |
| CJTXN1 | 0.73 ± 0.29 | 1 ± 0.1 | 0.64–0.73 |
| CJTXNPx | ND | 1 ± 0.1 | 0.17–0.2 |
| CJNADPH supply | ND | ND | 3.3–6.5 e −5 |
*interval calculated amount of total TXN1 in 1 mg soluble cell protein using the formula Vmax = kcat x [total enzyme] where Vmax was that of Table 1 and kcat was 1560 min−1.
**Initial metabolite and TXN1 concentrations used in the in vitro reconstituted pathway with the recombinant enzymes. ND Not determined.
Flux of CumOOH reduction through TXNPx in nmol/min x mg soluble cell protein.
The present study.
Data from [14].
Model predictions obtained with different KmTXN1 in the TXN1 reaction as discussed in Table S2.3 in SM2.
On the other hand, it has been proposed that kinetics of redoxins, such as TXN1, should be described using a mass-action equation, since Michaelis-Menten kinetics appears to be an inaccurate descriptor of redoxin activities [38]. Thus, the model was modified with the mass action reversible equation for TXN1, which required parameterization of the mass action k values of TXN1 and NADPH supply reactions (described in SM2). With this modification, the model closely predicted pathway flux (6.4 nmol/min x mg cell protein) and CJai values for TXN1 (0.74) and TXNPx (0.2), as well as for TryR (CJai = 0.001) and NADPH supply (CJai = 0.06).
3.9. Resistance to Bnz and peroxides
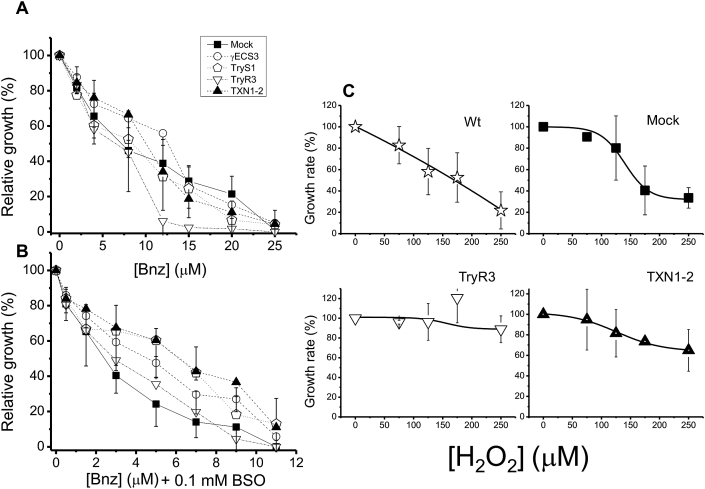
γECS overexpression led to increased T(SH)2 synthesis and T(SH)2 pool, whereas TXN1 overexpression led to an increase in the peroxide reduction flux. To determine whether these changes in the parasite anti-oxidant metabolism could affect other cellular functions, Wt, mock, and OE-clones were exposed to the antichagasic drug Bnz, alone or in combination with BSO. Statistically non-significant differences in the IC50 on growth were observed between mock and the OE-clones using Bnz alone, except for OE-TryR which was more sensitive (Fig. 4A, Table 4). For comparative purposes, the IC50 of Wt epimastigotes was determined, which was higher than that for mock cells (Table 4). A possible explanation for the difference in their IC50 values is that mock cells (and OE-clones) were simultaneously exposed to G418 and Bnz, whereas Wt cells were not treated with the antibiotic.
Fig. 4.
Benznidazole and H2O2 resistance. Percentage of relative growth of control and OE-epimastigotes after exposure for 24 h to different Bnz concentrations and in the absence (A) or presence of 0.1 mM BSO (B). (C) Percentage of rate growth monitored each 24 h for 96 h in control and OE-epimastigotes in the presence of different concentrations of H2O2. Data are mean ± SD for at least 4 independent cultures. Concentrations expressed as fmol/cell can be found in section 2.13.
Table 4.
IC50 on growth of T. cruzi epimastigotes for Bnz, Bnz plus BSO, and H2O2.
| Cellsa | IC50 (μM) |
||
|---|---|---|---|
| Bnz | Bnz + 0.1 mM BSO | H2O2 | |
| Wt | 12 ± 2 | 5 ± 1** | 145 ± 36 |
| Mock | 9 ± 3 | 3 ± 1 | 135 ± 50 |
| OE- γECS3 | 11 ± 3 | 4 ± 1 | 172 ± 48 |
| OE-TryS1 | 11 ± 5 | 5 ± 2 | 163 (2) |
| OE-TXN1-2 | 12 ± 6 | 6 ± 1** | >250 (4) |
| OE-TryR3 | 5 ± 1* | 3 ± 1 | >250 (4) |
*p < 0.05 **p < 0.01 vs. mock cells.
Mock and OE-parasites were grown in the presence of the G418 antibiotic, in addition to the other indicated compounds. Values are mean ± SD of at least three independent parasite cultures, except when indicated with the number in parentheses. Student's -t- test for non-paired samples.
The susceptibility of the cells to Bnz was increased by exposing them to a combination of this drug with 0.1 mM BSO (Fig. 4B). It was previously demonstrated that BSO not only inhibits γECS, but also TryS [29]. The reported IC50 value on growth for BSO alone in Wt and OE-clones is > 3.3 mM [29]; hence, BSO at a concentration of 0.1 mM is expected to have no effect on parasite growth; nonetheless, BSO could potentiate the Bnz effect. In the Bnz + BSO combination, only OE-TXN1 showed a significant increased resistance, compared to mock parasites (Fig. 4B and Table 4).
Next, the parasites were exposed to H2O2. OE-TryR3 and OE-TXN1-2 clones were more resistant than the other clones analyzed (Fig. 4C, Table 4) with IC50 values higher than 250 μM.
3.10. HFF-1 cell infection
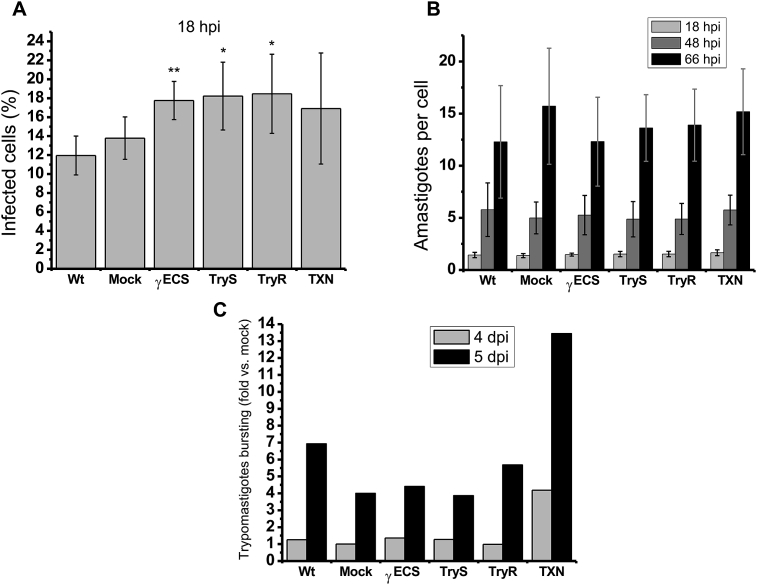
In order to investigate if these antioxidant enzymes are involved in the infection process of T. cruzi, the infectivity of OE parasites was assessed. It is known that epimastigotes have to differentiate to trypomastigotes in order to acquire the capability of invading host cells. Therefore, differences in infectivity using a primary infection with epimastigotes may be due to differences in differentiation, leading to misinterpretation. Thus, to discard any epimastigotes engagement, trypomastigotes derived from secondary infection were used to assess infectivity in tertiary infections of non-phagocytic cells, since these cells are major targets for T. cruzi infection in vivo. After 2 h of parasite and HFF-1 cells interaction, the invasion capabilities of the trypomastigotes was analyzed at 18 h post-infection (hpi) by determining the number of infected cells. The results showed that Wt and mock parasites have similar infection rates, in contrast OE-γECS trypomastigotes displayed a significantly (p < 0.01) higher infection rate of 30%. TryS trypomastigotes (32%, p < 0.05) and TryR trypomastigotes (34%, p < 0.05) only showed a tendency to a higher infection rates in comparison to controls (Fig. 5A).
Fig. 5.
In vitro infection capacity. (A) Percentage of infected cells at 18 hpi. (B) number of intracellular amastigotes per infected cell. (C) Trypomastigotes bursting compared to mock cells at 4 and 5 dpi . Results are the mean ± SD of three experiments each started from independent primary infections with epimastigotes, except for C in which n = 2 but the difference in the values from the two experiments was less than 30%. **p < 0.01, *p < 0.05 Student's t-test for non-paired samples versus mock.
Afterwards, the evolution of the infection was analyzed by determining the number of intracellular amastigotes as an indicative of transformation from trypomastigote into amastigote inside the cell (at 18 hpi) and amastigotes proliferation (at 48 and 66 hpi). There were no differences among controls and OE-parasites, showing that they were able to transform and replicate to the same extent (Fig. 5B).
Finally, to determine whether the overexpression of any enzyme confers an advantage in parasite burst, released trypomastigotes at 4 and 5 dpi were determined (Fig. 5C). Remarkably, OE-TXN parasites showed a 3-fold increase in trypomastigotes burst vs other OE- and control parasites.
4. Discussion
In the studies reported so far about T(SH)2 metabolism in T. cruzi, the pathway enzymes have been mostly individually analyzed, focusing on studying the effects of its manipulation on some metabolic or physiological functions, or characterizing the resulting phenotypes (for instance [19,36,39,40]). Therefore, the aim of the present work was to perform integral analyses of the complete pathway by modulating several enzyme activities in the parasites and performing parallel determinations of enzyme activities, metabolite concentrations and pathway fluxes. This approach allowed to identify which enzymes, and why, have most of the control on the T(SH)2 synthesis and on the peroxide reduction pathways, and to establish whether their degree of control correlate with essential physiological functions such as peroxide management, antichagasic drug resistance, and infectivity.
4.1. In silico and ex vivo flux control coefficients of the T(SH)2 synthesis pathway
The updated T(SH)2 synthesis model reported here predicted CJai values of 0.74 for γECS, 0.15 for TryS and 0.09 for CysT under control non-Cys supplied conditions. The ex vivo and in silico high CJai determined for γECS correlated with the significant increase in the T(SH)2 and GSH pools developed in the absence and presence of external Cys supplementation. Although the flux-control distribution was not determined in vivo in parasites supplied with Cys, the model predicted a decrease in the CJai of γECS and CysT and a gain in TryS control, the latter attaining CJai values of 0.15–0.31. These results indicated that γECS retained the main (but not exclusive) control of the T(SH)2 synthesis flux, sharing control with TryS. Under all conditions analyzed, GS exhibited negligible control on the T(SH)2 synthesis.
The high control attained by γECS may be explained by its very likely low activity in the parasites (<2 nmol/min x mg soluble cell protein), the latter value predicted from the ex vivo T(SH)2 synthesis fluxes using the OE-clones (section 3.5) and by pathway modeling (section 3.7). Determination of basal γECS and TryS activities in trypanosomatids is not a trivial task; the high spurious endogenous ATPase activity makes it cumbersome to discern specificity using enzymatic coupled assays. However, we were able to determine TryS activity in Wt epimastigotes by HPLC and using appropriate control reactions; the value was remarkably similar to that initially predicted by pathway modeling. Recently, a TryS basal activity in T. cruzi epimastigotes (Silvio strain) of ≈8 nmol ATP transformed to ADP/min x mg cell protein as determined by an end-point colorimetric assay of Pi release was reported [36]. Unfortunately, inclusion of control enzymatic reactions, such as whether the ATPase activity was subtracted or whether the assay was conducted under conditions of initial velocity regarding protein sample, substrate saturation and time were not described; therefore, such basal TryS activity value in the parasites may have been overestimated. To our knowledge, our study represents the first significant effort in determining the activities of the T(SH)2 synthesis enzymes within trypanosomatids. Unfortunately, γECS activity could not be determined by HPLC due to overlapping of γEC and GSH peaks.
On the other hand, OE-TryS epimastigotes did not increase the T(SH)2 content above the wild type level under any condition. A similar observation was found in [36] when TryS was also overexpressed in epimastigotes of the Silvio strain. In the latter study, the authors proposed that polyamine uptake exerted a higher metabolic control of the pathway; however, this seemed unlikely, because Spd supplementation in our OE-TryS clones did not bring about a further increase in T(SH)2. Since in the present study a high rate of T(SH)2 degradation was observed in the OE-TryS soluble cell protein extracts under conditions of ex vivo flux determination, it was hypothesized that the unaltered T(SH)2 levels in the OE-TryS1 cells could be due to an active T(SH)2 synthesis and degradation. This is supported by the previously reported in vitro TryS amidase activity [41]. It is clear that further experimental analyses in intact cells are required to assess that hypothesis, which however, are beyond the main objective of the present study. Moreover, T(SH)2 degradation was not detected in Wt and mock parasites, which have wild-type TryS levels, ruling out any physiologically relevant meaning for T(SH)2 degradation.
Finally, the predicted control exerted by CysT depended on the availability of external Cys surrounding the parasite; the flux control could be low for the intracellular parasite stages in the mammalian cells, since higher Cys concentration than in blood can be found in the cytosol of human host cells [42]. However, further experimentation is required to establish the control of the Cys supplying reactions (such as Cys transport and other metabolic pathways like de novo Cys synthesis and trans-sulfuration) on T(SH)2 synthesis.
4.2. In silico and ex vivo flux control coefficients of the T(SH)2 –dependent peroxide reduction pathway
The flux control distribution analysis by in vitro reconstitution of the pathway with the recombinant enzymes previously reported by our group [14] indicated that both TXN1 and TXNPx showed flux-control coefficients of 1.0, whereas that of TryR was 0.2. In that experimental setting, the sum of the flux control coefficients was close to 2, not one as implied by the ‘classical’ summation theorem of MCA. The explanation is that, unlike canonical metabolic pathways, this is an electron-transfer pathway in which each individual redox reaction (process) compulsorily involves two enzymes, leading to a stoichiometry of one reaction/two enzymes (and second-order dependence reaction on enzyme concentration) instead of the usual one reaction/one enzyme (and first-order reaction dependence on enzyme concentration). Therefore, the sum of the enzymes’ flux control coefficients on the transfer of groups such as in redox pathways must be added to the sum of two, whilst the sum of the flux control coefficients on the whole process of peroxide reduction remains to be one (the pathway flux maintains a first-order dependence on enzyme activities) [43]. Kinetic modeling of the redox pathway reported here for the first time, allowed to obtain the CJai on the peroxide reduction process, enabling to dissect CJai values of 0.11–0.16 for TryR, 0.64–0.73 for TXN1 and 0.17–0.2 for TXNPx, respectively, considering that the sum of all CJai must add up to 1.0. These in silico predicted values were in agreement with those obtained here by titration of the activity in the parasites, whose CJai values were 0.73 and 0.15 for TXN1 and TryR, respectively. Moreover, overexpression of TXN1 but not of TryR led to steady increases in the pathway flux, which agreed with the high control attained by TXN1. Therefore, variations in TryR activity do not change the hydroperoxide reduction flux, but variations in TXN1 do, hence, demonstrating that TXN1 has high pathway control. Regarding TXNPx overexpression in the parasites, this was not addressed in this work; however, in a mixed reconstituted system using soluble cell protein extracts of mock and WT cells the ex vivo peroxide reduction flux was titrated with recombinant TXNPx. The CJai predicted by this strategy was similarly high to that of TXN1. This result suggested that the control of the flux is shared equally by both TXN1 and TXNPx.
TXN1 and TXNPx are abundant proteins in trypanosomatids (1–6% of the total cellular protein) [20,44,45]. In T. cruzi the values calculated by activity were 0.1% and 1% for TXN1 and TXNPx, respectively, whereas the TryR protein content was 0.02% [14]. Reduction of TXN1 by T(SH)2 is the step with the lowest catalytic efficiency in the TXN-dependent peroxide reduction pathway [14,45], which may explain the high control that TXN1 exerts on the pathway flux. Certainly, TXNPx overexpression prompts increased resistance to peroxides [19,39] and infective stages overexpress this enzyme [20,21]. However, in these reports TXN1 was not evaluated and higher TXN1-TXNPx stoichiometric coupling may favor higher peroxide reduction fluxes. On the other hand, despite its lower protein content, TryR exhibited a comparatively high activity in the cells and high catalytic efficiency, as well as lack of regulatory properties, which led to a low controlling enzyme.
It should be also emphasized that the kinetic properties of the enzymes, analyzed separately, reveal little about how and by which steps the pathway is controlled. However, when all pathway enzymes (i.e. with the content of active enzyme in the cells) are allowed to interact with each other and with all ligands (in their physiological concentration ranges), the most controlling steps can be identified and the control mechanisms clearly emerge. Such controlling enzymes can be now proposed as very attractive targets for therapeutic intervention, since their low inhibition can have the greatest negative effect in the pathway flux and metabolite levels [23].
4.3. Fluxes and resistance to hydroperoxides and anti-chagasic compounds
Wt and mock cells exhibited ex vivo hydroperoxide reduction fluxes of ≈11 nmol/min x mg soluble cell protein which is similar to those reported for intact cells of the T. cruzi Y strain (3.3–12.9H2O2 nmol/min x mg cell protein) [46]. Remarkably, these maximal outputs of peroxide reduction values were one order of magnitude higher than those of the T(SH)2 synthesis (0.6 ± 0.2 nmol/min x mg soluble cell protein) in Wt cells. These results indicated that there was a 20-fold lower maximal output of de novo T(SH)2 synthesis versus its usage for peroxide reduction. In consequence, the peroxide detoxification pathway, with a highly active and efficient enzymatic machinery, seems to function during immediate and acute (short-term) responses to oxidants, whereas changes in the T(SH)2 synthesis flux are expected to have their metabolic effect after a longer period of time upon the insult (long-term response).
TXN1 down-regulation in T. brucei causes growth arrest and increased sensitivity to H2O2 [47,48]. So far, there are no reports on the effects of TXN1 overexpression in trypanosomatids. Remarkably, our results showed for the first time that overexpression of TXN1 induced higher resistance to this peroxide (Fig. 4). Furthermore, the OE-TryR cells also showed enhanced H2O2 resistance despite TryR having low control on the flux. As OE-TryR epimastigotes showed no changes in the TXN1 and TXNPx activities that could contribute to the peroxide resistance, the effect was probably due to the two orders of magnitude higher TryR activity compared to the other OE-parasites. On the other hand, no increased H2O2 resistance was observed in the OE-γECS and OE-TryS epimastigotes. The lack of resistance in our OE-TryS epimastigotes contrasts with the result obtained by others [36], where higher (70%) H2O2 resistance on epimastigotes growth was found when TryS was overexpressed. However, in that study the activities of TryR, TXN and TXNPx were not assessed to determine that they were not changed.
Furthermore, our results with overexpressing TXN1 (≈10-fold) were similar in terms of hydroperoxide resistance to those reported when TXNPx was overexpressed [19,20,39], where the authors found that 2.5-fold TXNPx overexpression increased by 50% the epimastigotes viability exposed to H2O2 and helped the cells to contend with ONOO−. A plausible explanation is that both enzymes interact tightly and function in channeling, with TXN1 being the only reducing partner for TXNPx. However, the higher activity in the parasite and higher catalytic efficiency of TXNPx will require higher levels of inhibition than TXN1 to affect the peroxide reduction pathway. Thus, TXN1 (or TXN1-TXNPx interaction) may be a better target in order to inhibit the antioxidant machinery as well as many other processes in which TXN1 is involved (e.g. DNA synthesis, polyamine metabolism, protein translation and degradation [11,49,50]). In this regard, a high throughput screening of a library of compounds identified some compounds that preferably inhibited TXN in the T. brucei peroxide reduction pathway over its human counterpart thioredoxin. The compounds showed a selectivity index >5 for bloodstream trypomastigotes versus HeLa cells [51], rendering TXN1 as a suitable target for therapeutic intervention.
Regarding TryR, it was here demonstrated that its control on the hydroperoxide reduction flux was low because it is one of the most efficient enzymes of the pathway. Consistently, its overexpression in the T cruzi Silvio strain showed no effect on the cell's hydroperoxide reduction capacity [40] and it has been demonstrated that, in order to affect the redox homeostasis in T. brucei, TryR has to be decreased by more than 90% of its wild-type level [52]. Therefore, TryR seems to be a very difficult target for therapeutic intervention.
Remarkably, TXN1 overexpression induced increased resistance to Bnz (in combination with BSO). The proposed mode of cytotoxic Bnz action is adduct formation with thiol metabolites such as GSH, T(SH)2, and other molecules, rather than ROS formation [[53], [54], [55]]. Our results suggested that a small dithiol protein such as TXN1 may also provide some protection to the parasite against Bnz (and BSO effects), a hypothesis that needs to be further experimentally analyzed. In this regard, it has been recently proposed that some small proteins such as TXN or glutaredoxins (Grx) may contribute to the reduction of glutathione disulfide (GSSG) in organisms lacking glutathione reductase [56]. The authors found that reduction of GSSG directly by T(SH)2 is a slow process, whereas it is faster when it is mediated by Grx and/or TXN. If both proteins are present at the physiological concentrations analyzed, the reduction is preferably performed by Grx (75%) rather than by TXN (25%). Whether TXN overexpression could favor GSSG reduction was not here evaluated.
4.4. Parasite infectivity
As intracellular parasite, oxidative stress is a major challenge for T. cruzi, suggesting that its antioxidant enzymes may contribute to its survival and persistence. In fact, some of the enzymes have been studied during the infection process of the parasite in phagocytic and non-phagocytic cells, showing their participation in survival, replication and differentiation of the parasite, proposing them as virulent factors [[19], [20], [21]]. Therefore, we determined the role of γECS, TryS, TryR and TXN1 in the infection process of T. cruzi in non-phagocytic cells, since only in these ones the pathogenesis of the disease is established. It has been determined for some T. cruzi strains that replication within in vitro cultured host cells occurs from 18-72 hpi [57]; for this reason and based on some preliminary observations, the infection process was analyzed over time by examining the percentage of infected cells at 18 hpi, as well as the number of internal parasites at 18, 48 and 66 hpi. Also, the trypomastigotes that burst into the extracellular medium were monitored at 4 and 5 dpi.
At 18 hpi, OE-γECS trypomastigotes were more infective than Wt or mock trypomastigotes (Fig. 5A), suggesting that this enzyme may contribute to the invasion. These findings concur with those reported for Leishmania infantum, where the inactivation of one allele of the γECS genes showed decreased GSH and T(SH)2 contents and decreased survival inside activated macrophages [58], which in turn showed an increased production of oxygen and nitrogen oxidative species. Moreover, the γECS content also increased in natural antimony-resistant isolates of L. donovani [59,60].
On the other hand, OE-TryS and OE-TryR trypomastigotes showed a tendency to higher infectivity (Fig. 5A). It has been reported that TryS (and TXNPx) are involved in infectivity in mouse models and cell cultures; hence both enzymes have been proposed as virulence factors [20,21]. These two enzymes have been also found overexpressed in (i) T. cruzi acute vs. chronic Chagas disease isolates [61]; (ii) virulent vs. attenuated strains; and (iii) in metacyclic trypomastigotes vs. epimastigotes (independently of the strain virulence) [21]. In the present work, correlation between TryS overexpression and infectivity was low, most probably because of the side effects observed in the OE-clones. On the other side, Piacenza et al. found no correlation between the TryR protein content and degree of virulence in different T. cruzi strains [21]; moreover, Kelly et al. [40] also found no correlation between TryR overexpression and peroxide resistance. Here, in contrast, OE-TryR showed an increased capacity to cope with H2O2 (Fig. 4C) which could contribute to the slight increased infectivity at 18 hpi (Fig. 5A). The different results may be related to the higher overexpression level of 50-fold attained in our OE-TryR clones in comparison to the 15-fold attained in the transfectant of Kelly et al. study. Lastly, to the best of our knowledge this is the first time that the correlation between TXN1 (the electron donor of TXNPx) and infectivity was analyzed, finding a statistically non-significant correlation between its overexpression and the capacity to infect cells at 18 hpi.
The different OE-parasites showed a similar capacity to transform into amastigotes, because at 18 hpi all parasites exhibited the same number of internal amastigotes (Fig. 5B). In turn, the internal parasites showed similar capacity to replicate within the cell, at 48 and 66 hpi (Fig. 5B). However, a remarkable finding emerged at 4 and 5 dpi, where OE-TXN displayed a notorious increase in trypomastigotes bursting (Fig. 5C). A possible explanation is that the OE-TXN amastigotes have a higher differentiation rate to trypomastigotes; however, this hypothesis has to be analyzed in more detail to understand how TXN could be involved in this process.
5. Conclusion
The results of the present investigation allowed to identify by both in silico and ex vivo experimentation that γECS and TXN1 are enzymes with high control on the T(SH)2 synthesis and peroxide reduction fluxes, respectively. TryS has a lower, but meaningful contribution to the control due to its very low activity in the parasite; thus, a slight inhibition of this enzyme may also negatively impact the pathway flux. Then, when T(SH)2 metabolism is compromised by thiol-conjugating drugs such as Bnz, high T(SH)2 or thiol-protein contents may confer drug-resistance. Hence, to prevent parasite resistance against this type of antichagasic drugs, γECS and TXN1 (and TryS) activities should be blocked. Therefore, therapeutic targeting of γECS and TXN1 will more severely affect parasite viability than intervention at other, lower-controlling pathway steps which will require much higher levels of inhibition. In addition, despite its lower control, TryS should be still considered an adequate drug target since its specific inhibition will affect both T(SH)2 synthesis and peroxide reduction pathways. Furthermore, γECS and TXN may contribute at different levels of the infection process, strengthening its proposal as drug targets.
Acknowledgments
Z. G-C was supported by CONACYT-Mexico Ph.D. fellowship No. 355168 and acknowledges to Programa de Posgrado en Ciencias Bioquímicas of the Universidad Nacional Autónoma de México for academic preparation. The research in the authors laboratories was supported by CONACYT-Mexico grants 178638, 264292, 272941 and 282663 to E.S. and 240086 to R.M.C. The University of Edinburgh’s BBSRC Global Challenge Research Fund Impact Acceleration Account Scheme is acknowledged for grants BB GC IAA 16/17-PM and BB GC IAA 17/18-PM, which enabled a collaboration resulting in the preparation of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101231.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Health Organization Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Wkly. Epidemiol. Rec. 2015;90(6):33–43. [PubMed] [Google Scholar]
- 2.Perez-Molina J.A., Molina I. Chagas disease. Lancet. 2018;391(10115):82–94. doi: 10.1016/S0140-6736(17)31612-4. [DOI] [PubMed] [Google Scholar]
- 3.Coura J.R., Viñas P.A., Junqueira A.C. Ecoepidemiology, short history and control of Chagas disease in the endemic countries and the new challenge for non-endemic countries. Mem. Inst. Oswaldo Cruz. 2014;109(7):856–862. doi: 10.1590/0074-0276140236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro J.A., de Mecca M.M., Bartel L.C. Toxic side effects of drugs used to treat Chagas' disease (American trypanosomiasis) Hum. Exp. Toxicol. 2006;25(8):471–479. doi: 10.1191/0960327106het653oa. [DOI] [PubMed] [Google Scholar]
- 5.Rassi A., Jr., Marin J.A. Neto, Rassi A. Chronic Chagas cardiomyopathy: a review of the main pathogenic mechanisms and the efficacy of aetiological treatment following the BENznidazole Evaluation for Interrupting Trypanosomiasis (BENEFIT) trial. Mem. Inst. Oswaldo Cruz. 2017;112(3):224–235. doi: 10.1590/0074-02760160334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paucar R., Moreno-Viguri E., Pérez-Silanes S. Challenges in Chagas disease drug discovery: a review. Curr. Med. Chem. 2016;23(28):3154–3170. doi: 10.2174/0929867323999160625124424. [DOI] [PubMed] [Google Scholar]
- 7.Duschak V.G. Targets and patented drugs for chemotherapy of Chagas disease in the last 15 years-period. Recent Pat. Anti-Infect. Drug Discov. 2016;11(2):74–173. doi: 10.2174/1574891X11666161024165304. [DOI] [PubMed] [Google Scholar]
- 8.Olin-Sandoval V., Moreno-Sánchez R., Saavedra E. Targeting trypanothione metabolism in trypanosomatid human parasites. Curr. Drug Targets. 2010;11(12):1614–1630. doi: 10.2174/1389450111009011614. [DOI] [PubMed] [Google Scholar]
- 9.Flohé L. The trypanothione system and the opportunities it offers to create drugs for the neglected kinetoplast diseases. Biotechnol. Adv. 2012;30(1):294–301. doi: 10.1016/j.biotechadv.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Leroux A.E., Krauth-Siegel R.L. Thiol redox biology of trypanosomatids and potential targets for chemotherapy. Mol. Biochem. Parasitol. 2016;206(1–2):67–74. doi: 10.1016/j.molbiopara.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Irigoín F., Cibilis L., Comini M., Wilkinson S., Flohé L., Radi R. Insights into the redox biology of Trypanosoma cruzi: trypanohione metabolism and oxidant detoxification. Free Radic. Biol. Med. 2008;45(6):733–742. doi: 10.1016/j.freeradbiomed.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Manta B., Comini M., Medeiros A., Hugo M., Trujillo M., Radi R. Trypanothione: a unique bis-glutathionyl derivative in trypanosomatids. Biochim. Biophys. Acta. 2013;1830(5):3199–3216. doi: 10.1016/j.bbagen.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Diechtierow M., Krauth-Siegel R.L. A tryparedoxin-dependent peroxidase protects African trypanosomes from membrane damage. Free Radic. Biol. Med. 2011;51(4):856–868. doi: 10.1016/j.freeradbiomed.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 14.González-Chávez Z., Olin-Sandoval V., Rodíguez-Zavala J.S., Moreno-Sánchez R., Saavedra E. Metabolic control analysis of the Trypanosoma cruzi peroxide detoxification pathway identifies tryparedoxin as a suitable drug target. Biochim. Biophys. Acta. 1850:263–273. doi: 10.1016/j.bbagen.2014.10.029. 2015. [DOI] [PubMed] [Google Scholar]
- 15.Krauth-Siegel R.L., Comini M.A. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim. Biophys. Acta. 2008;1780(11):1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Vázquez K., Paulino M., Salas C.O., Zarate-Ramos J.J., Vera B., Rivera G. Trypanothione reductase: a target for the development of anti- Trypanosoma cruzi drugs. Mini Rev. Med. Chem. 2017;17(11):939–946. doi: 10.2174/1389557517666170315145410. [DOI] [PubMed] [Google Scholar]
- 17.Beltran-Hortelano I., Perez-Silanes S., Galinao S. Trypanothione reductase and superoxide dismutase as current drug targets for Trypanosoma cruzi: an overview of compounds with activity against Chagas disease. Curr. Med. Chem. 2017;24(11):1066–1138. doi: 10.2174/0929867323666161227094049. [DOI] [PubMed] [Google Scholar]
- 18.Lima C.R., Carels N., Guimaraes A.C., Tufféy P., Derreumaux P. In silico structural characterization of protein targets for drug development against Trypanosoma cruzi. J. Mol. Model. 2016;22(10):244. doi: 10.1007/s00894-016-3115-9. [DOI] [PubMed] [Google Scholar]
- 19.Piacenza L., Peluffo G., Alvarez M.N., Kelly J.M., Wilkinson S.R., Radi R. Peroxiredoxins play a major role in protecting Trypanosoma cruzi against macrophage-and endogenously-derived peroxynitrate. Biochem. J. 2008;410(2):359–368. doi: 10.1042/BJ20071138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piñeyro M.D., Parodi-Talice A., Arcari T., Robello C. Peroxiredoxins from Trypanosoma cruzi: virulence factors and drug targets for treatment of Chagas disease? Gene. 2008;408:45–50. doi: 10.1016/j.gene.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Piacenza L., Zago M.P., Peluffo G., Alvarez M.N., Basombrio M.A., Radi R. Enzymes of the antioxidant network as novel determiners of Trypanosoma cruzi virulence. Int. J. Parasitol. 2009;39(13):1455–1464. doi: 10.1016/j.ijpara.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakker B.M., Westerhoff H.V., Opperdoes F.R., Michels P.A. Metabolic control analysis of glycolysis in trypanosomes as an approach to improve selectivity and effectiveness of drugs. Mol. Biochem. Parasitol. 2000;106(1):1–10. doi: 10.1016/s0166-6851(99)00197-8. [DOI] [PubMed] [Google Scholar]
- 23.Saavedra E., Gonzalez-Chávez Z., Moreno-Sanchez R., Michels P. Drug target selection for Trypanosoma cruzi metabolism by metabolic control analysis and kinetic modeling. Curr. Med. Chem. 2018 doi: 10.2174/0929867325666180917104242. [DOI] [PubMed] [Google Scholar]
- 24.Fell D. Portland Press; London: 1997. Understanding the Control of Metabolism; p. 301. [Google Scholar]
- 25.Moreno-Sanchez R., Saavedra E., Rodriguez-Enriquez S., Olin-Sandoval V. Metabolic control analysis: a tool for designing strategies to manipulate metabolic pathways. J. Biomed. Biotechnol. 2008:597913. doi: 10.1155/2008/597913. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olin-Sandoval V., González-Chávez Z., Berzunza-Cruz M., Martínez I., Jasso-Chávez R., Becker I., Espinoza B., Moreno-Sánchez R., Saavedra E. Drug target validation of the trypanothione pathway enzymes through metabolic modelling. FEBS J. 2012;279(10):1811–1833. doi: 10.1111/j.1742-4658.2012.08557.x. [DOI] [PubMed] [Google Scholar]
- 27.López-Olmos V., Pérez-Nasser N., Piñero D., Ortega E., Hernández R., Espinoza B. Biological characterization and genetic diversity of Mexican isolates of Trypanosoma cruzi. Acta Trop. 1998;69(3):239–254. doi: 10.1016/s0001-706x(97)00131-9. [DOI] [PubMed] [Google Scholar]
- 28.Vazquez M.P., Levin M.J. Functional analysis of the intergenic regions of TcP2beta gene loci allowed the construction of an improved Trypanosoma cruzi expression vector. Gene. 1999;239(2):217–225. doi: 10.1016/s0378-1119(99)00386-8. [DOI] [PubMed] [Google Scholar]
- 29.Vázquez C., Mejia-Tlachi M., González-Chávez Z., Silva A., Rodríguez-Zavala J.S., Moreno-Sánchez R., Saavedra E. Buthionine sulfoximine is a multitarget inhibitor of trypanothione synthesis in Trypanosoma cruzi. FEBS Lett. 2017;591(23):3881–3894. doi: 10.1002/1873-3468.12904. [DOI] [PubMed] [Google Scholar]
- 30.Rangel-Aldao R., Allende O., Triana F., Piras R., Henriquez D., Piras M. Possible role of cAMP in the differentiation of Trypanosoma cruzi. Mol. Biochem. Parasitol. 1987;22(1):39–43. doi: 10.1016/0166-6851(87)90067-3. [DOI] [PubMed] [Google Scholar]
- 31.Comini M.A., Dirdjaja N., Kaschel M., Krauth-Siegel R.L. Preparative enzymatic synthesis of trypanothione and trypanothione analogues. Int. J. Parasitol. 2009;39:1059–1062. doi: 10.1016/j.ijpara.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Mendes P. GEPASI: a software package for modelling the dynamics, steady states and control of biochemical and other systems. Comput. Appl. Biosci. 1993;9(5):563–571. doi: 10.1093/bioinformatics/9.5.563. [DOI] [PubMed] [Google Scholar]
- 33.Hoops S., Sahle S., Gauges R., Lee C., Pahle J., Simus N., Singhal M., Xu L., Mendes P., Kummer U. COPASI-a COmplex PAthway SImulator. Bioinformatics. 2006;22(24):3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 34.Canepa G.E., Bouvier L.A., Miranda M.R., Uttaro A.D., Pereira C.A. Characterization of Trypanosoma cruzi L-cysteine transport mechanisms and their adaptive regulation. FEMS Microbiol. Lett. 2009;292(1):27–32. doi: 10.1111/j.1574-6968.2008.01467.x. [DOI] [PubMed] [Google Scholar]
- 35.Manning-Cela R., Cortés A., González-Rey E., Van Voorhis W.C., Swindle J., González A. LYT1 protein is required for efficient in vitro infection by Trypanosoma cruzi. Infect. Immun. 2001;69(6):3916–3923. doi: 10.1128/IAI.69.6.3916-3923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesías A.C., Sasoni N., Arias D.G., Pérez Brandán C., Orban O.C.F., Kunick C., Robello C., Comini M.A., Garg N.J., Zago M.P. Trypanothione synthetase confers growth, survival advantage and resistance to anti-protozoal drugs in Trypanosoma cruzi. Free Radic. Biol. Med. 2018;130:23–34. doi: 10.1016/j.freeradbiomed.2018.10.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown G.C., Hafner R.P., Brand M.D. A ‘top-down’ approach to the determination of control coefficients in metabolic control theory. Eur. J. Biochem. 1990;188(2):321–325. doi: 10.1111/j.1432-1033.1990.tb15406.x. [DOI] [PubMed] [Google Scholar]
- 38.Pillay C.S., Hofmeyr J.H., Olivier B.G., Snoep J.L., Rohwer J.M. Enzymes or redox couples? The kinetics of thioredoxin and glutaredoxin reactions in a systems biology context. Biochem. J. 2009;417(1):269–275. doi: 10.1042/BJ20080690. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson S.R., Temperton N.J., Mondragon A., Kelly J.M. Distinct mitochondrial and cytosolic enzymes mediate trypanothione-dependent peroxide metabolism in Trypanosoma cruzi. J. Biol. Chem. 2000;275(11):8220–8225. doi: 10.1074/jbc.275.11.8220. [DOI] [PubMed] [Google Scholar]
- 40.Kelly J.M., Taylor M.C., Smith K., Hunter K.J., Fairlamb A.H. Phenotype of Leishmania donovani and Trypanosoma cruzi which over-express trypanothione reductase. Sensitivity towards agents that are thought to induce oxidative stress. Eur. J. Biochem. 1993;218(1):29–33. doi: 10.1111/j.1432-1033.1993.tb18348.x. [DOI] [PubMed] [Google Scholar]
- 41.Oza S.L., Tetaud E., Ariyanayagam M.R., Warnon S.S., Fairlamb A.H. A single enzyme catalyses formation of trypanothione from glutathione and spermidine in Trypanosoma cruzi. J. Biol. Chem. 2002;277(39):358853–358861. doi: 10.1074/jbc.M204403200. [DOI] [PubMed] [Google Scholar]
- 42.Vitvitsky V., Witcher M., Banerjee R., Thoene J. The redox status of cystinotic fibroblasts. Mol. Genet. Metab. 2010;99(4):384–388. doi: 10.1016/j.ymgme.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dam K., van der Vlag J., Kholodenko B.N., Westerhoff H.V. The sum of the control coefficients of all enzymes on the flux through a group-transfer pathway can be as high as two. Eur. J. Biochem. 1993;212(3):791–799. doi: 10.1111/j.1432-1033.1993.tb17720.x. [DOI] [PubMed] [Google Scholar]
- 44.Nogoceke E., Gommel D.U., Kiess M., Kalisz H.M., Flohé L. A unique cascade of oxidoreductases catalyses trypanothione-mediated peroxide metabolism in Crithidia fasciculata. Biol. Chem. 1997;378:827–836. doi: 10.1515/bchm.1997.378.8.827. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson S.R., Meyer D.J., Taylor M.C., Bromley E.V., Miles M.A., Kelly J.M. The Trypanosoma cruzi enzyme TcGPXI is a glycosomal peroxidase and can be linked to trypanothione reduction by glutathione or tryparedoxin. J. Biol. Chem. 2002;277(19):17062–17071. doi: 10.1074/jbc.M111126200. [DOI] [PubMed] [Google Scholar]
- 46.Carnieri E.G., Moreno S.N., Docampo R. Trypanothione-dependent peroxide metabolism in Trypanosoma cruzi different stages. Mol. Biochem. Parasitol. 1993;61(1):79–86. doi: 10.1016/0166-6851(93)90160-y. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson S.R., Horn D., Prathalingam S.R., Kelly J.M. RNA interference identifies two hydroperoxide metabolizing enzymes that are essential to the bloodstream form of the African Trypanosome. J. Biol. Chem. 2003;278(34):31640–31646. doi: 10.1074/jbc.M303035200. [DOI] [PubMed] [Google Scholar]
- 48.Comini M.A., Krauth-Siegel R.L., Flohé L. Depletion of the thioredoxin homologue tryparedoxin impairs antioxidative defence in African trypanosomes. Biochem. J. 2007;402(1):43–49. doi: 10.1042/BJ20061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machado-Silva A., Cerqueira P.G., Grazielle-Silva V., Gadhela F.R., Peloso E.F., Teixeira S.M., Machado C.R. How Trypanosoma cruzi deals with oxidative stress: antioxidant defence and DNA repair pathways. Mutat. Res. Rev. Mutat. Res. 2016;767:8–22. doi: 10.1016/j.mrrev.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Piñeyro M.D., Parodi-Talice A., Arias D.G., Guerrero S.A., Robello C. Molecular characterization and interactome analysis of Trypanosoma cruzi tryparedoxin 1. J. Proteom. 2011;74(9):1683–1692. doi: 10.1016/j.jprot.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Fueller F., Jehle B., Putzker K., Lewis J.D., Krauth-Siegel L. High throughput screening against the peroxidase cascade of African trypanosomes identifies antiparasitic compounds that inactivate tryparedoxin. J. Biol. Chem. 2012;287(12):8792–8802. doi: 10.1074/jbc.M111.338285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krieger S., Schwarz W., Ariyanayagam M.R., Fairlamb A.H., Krauth-Siegel R.L., Clayton C. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol. Microbiol. 2000;35:542–552. doi: 10.1046/j.1365-2958.2000.01721.x. [DOI] [PubMed] [Google Scholar]
- 53.Díaz de Toranzo E.G., Castro J.A., Franke de Cazzulo B.M., Cazzulo J.J. Interaction of benznidazole reactive metabolites with nuclear and kinetoplastic DNA, proteins and lipids from Trypanosoma cruzi. Experientia. 1988;44(10):880–881. doi: 10.1007/BF01941187. [DOI] [PubMed] [Google Scholar]
- 54.Maya J.D., Cassels B.K., Iturriaga-Vasquez P., Ferreira J., Faúndez M., Galanti N., Ferreira A., Morelio A. Mode of action of natural and synthetic drugs against Trypanosoma cruzi and their interaction with the mammalian host. Comp. Biochem. Physiol. Mol. Integr. Physiol. 2007;146(4):601–620. doi: 10.1016/j.cbpa.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Trochine A., Creek D.J., Faral-Tello P., Barrett M.P., Robello C. Benznidazole biotransformation and multiple targets in Trypanosoma cruzi revealed by metabolomics. PLoS Neglected Trop. Dis. 2014;8(5) doi: 10.1371/journal.pntd.0002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manta B., Möller M., Bonilla M., Deambrosi M., Grunberg K., Bellanda M., Comini M.A., Ferrer-Sueta G. Kinetics studies reveal a key role of a redox-active glutaredoxin in the evolution of the thiol-redox metabolism of trypanosomatids parasites. J. Biol. Chem. 2019;249(9):3235–3246. doi: 10.1074/jbc.RA118.006366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dumoulin, P. C. and Burleigh, B. A. Stress-induced proliferation and cell cycle plasticity of intracellular Trypanosoma cruzi amastigotes. mBio. 9:e00673-18. [DOI] [PMC free article] [PubMed]
- 58.Mukjerjee A., Roy G., Guimond C., Ouellette M. The gammaglutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol. Microbiol. 2009;74:914–927. doi: 10.1111/j.1365-2958.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 59.Mukherjee A., Padmanabhan P.K., Singh S., Roy G., Girard I., Chatterjee M., Ouellette M., Madhubala R. Role of ABC transporter MRPA, gammaglutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J. Antimicrob. Chemother. 2007;59:204–211. doi: 10.1093/jac/dkl494. [DOI] [PubMed] [Google Scholar]
- 60.Singh N., Chatterjee M., Sundar S. The overexpression of genes of thiol metabolism contribute to drug resistance in clinical isolates of visceral leishmaniasis (kala azar) in India. Parasites Vectors. 2014;7:596. doi: 10.1186/s13071-014-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Díaz M.L., Solari A., González C.I. Differential expression of Trypanosoma cruzi I associated with clinical forms of Chagas disease: overexpression of oxidative stress protein in acute patient isolate. J. Proteom. 2011;74(9):1673–1682. doi: 10.1016/j.jprot.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 62.Segel I.H. Wiley; New York, USA: 1975. Enzyme Kinetics. [Google Scholar]
- 63.What is Chagas disease? https://www.dndi.org/diseases-projects/chagas/
- 64.Chagas disease (American trypanosomiasis) http://www.who.int/en/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis)
- 65.Chagas disease. https://www.dndi.org/wp-content/uploads/2017/08/Factsheet_2016_Chagas.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.