Abstract
Human mesenchymal stem cells (MSCs) are expected to have utility as a cell source in regenerative medicine. Because we previously reported that suppression of the Wnt/β-catenin signal enhances hepatic differentiation of human MSCs, we synthesized twenty-three derivatives of small molecule compounds originally reported to suppress the Wnt/β-catenin signal in human colorectal cancer cells. We then screened these compounds for their ability to induce hepatic differentiation of human UE7T-13 MSCs. After screening using WST assay, TCF reporter assay, and albumin mRNA expression, IC-2, a derivative of ICG-001, was identified as a potent inducer of hepatic differentiation of human MSCs. IC-2 potently induced the expression of albumin, complement C3, tryptophan 2,3-dioxygenase (TDO2), EpCAM, C/EBPα, glycogen storage, and urea production. Furthermore, we examined the effects of IC-2 on human bone marrow mononuclear cell fractions sorted according to CD90 and CD271 expression. Consequently, CD90+ CD271+ cells were found to induce the highest production of urea and glycogen, important hepatocyte functions, in response to IC-2 treatment. CD90+ CD271+ cells also highly expressed albumin mRNA. As the CD90+ CD271+ population has been reported to contain a rich fraction of MSCs, IC-2 apparently represents a potent inducer of hepatic differentiation of human MSCs.
Keywords: Mesenchymal stem cells, Hepatic differentiation, Small molecule compound, Wnt/β-catenin signal inhibitor
Highlights
-
•
We screened newly synthesized derivatives of small molecule compounds generated from known Wnt/β-catenin signal inhibitors.
-
•
IC-2 was identified as an inducer of the differentiation of human mesenchymal stem cells into hepatocytes.
-
•
IC-2 potently induces hepatic differentiation of human bone marrow mononuclear CD90+ CD271+ cells.
1. Introduction
Wnt signaling plays essential roles in balancing the self-renewal and differentiation of adult stem cells in conjunction with the Notch, Hedgehog, JAK-STAT, BMP, Hippo, and FGF-MAPK signaling pathways [1]. Wnt signaling is important for the proliferation and maintenance of pluripotency [2], [3], [4] and differentiation of stem cells [5], [6]. Wnt/β-catenin signaling is required for fate decision in neural crest stem cells [7], and Wnt3A promotes differentiation into the neural and astrocytic lineages by inhibiting neural stem cell maintenance [8], clearly indicating the crucial role of the Wnt/β-catenin signaling pathway in the lineage restriction of stem cells [1].
Our previous report demonstrated that Wnt/β-catenin signaling is suppressed during hepatic differentiation of human MSCs [9], [10]. Suppression of Wnt/β-catenin signaling molecules or target genes induced the hepatic differentiation of human MSCs [9]. Dickkopf WNT signaling pathway inhibitor 1 (Dkk-1), an antagonistic inhibitor of the WNT signaling pathway, promoted hepatic differentiation of bone marrow-derived MSCs [11]. Taken together, these findings indicate that the suppression of Wnt/β-catenin signals plays an important role in the hepatic differentiation of MSCs.
Small molecules capable of modulating stem cell fate have significant potential as therapeutic agents [12]. Small molecules have been identified that modulate key developmental pathways, including Wnt, FGF, Hedgehog, Notch, and BMP/TGF-β, during stem cell differentiation [12]. Chemical screening approaches have demonstrated utility in identifying an IDE (inducer of definitive endoderm) compounds capable of inducing definitive endoderm differentiation of ES cells [13]. In contrast, the use of small molecule compounds has an advantage of being safer than the use of cytokines, nucleic acids, or protein therapies. Thus, the identification of small molecule compounds has enhanced the development of stem cell biology. In the present study, we identified a small molecule compound that efficiently induces hepatic differentiation of human MSCs.
2. Materials and methods
2.1. Compounds
Twenty-three derivatives of Wnt/β-catenin signal inhibitors formerly reported in colorectal cancer cells [15], [16], [17] were newly synthesized inhouse [18]. Each inhibitor used was dissolved in 0.1% DMSO.
2.2. Cells
To screen synthesized small molecule compounds, the human bone marrow-derived mesenchymal stem cell (BM-MSC) line, UE7T-13, was used [19]. To confirm the effects of small molecule compounds, primary BM-MSCs were prepared as follows: human bone marrow mononuclear cells purchased from Lonza Walkersville, Inc. (Walkersville, MD, USA) were plated onto culture dishes. Adherent cells were expanded as a whole cell fraction in DMEM (Life Technologies Corp., Carlsbad, CA) containing 20% fetal bovine serum (FBS; GE Healthcare UK Ltd, Little Chalfont, UK), 20 ng/ml basic FGF (bFGF; TRANS GENIC INC., Ltd. Kumamoto, Japan), 100 U/ml penicillin, and 100 μg/ml streptomycin. Cell fractionation was performed as follows: cells were stained with APC mouse anti-human CD90 antibodies (BD Biosciences, San Jose, CA, USA) and PE mouse anti-human CD271 antibodies (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) for 30 min on ice. CD90+ CD271+ cells, CD90− CD271+ cells, and CD90− CD271− cells were sorted by MoFlo XDP (Beckman Coulter Inc. Fullerton, CA, USA). Each population was cultured and expanded in DMEM containing 20% FBS, bFGF, and penicillin/streptomycin.
2.3. WST assay
UE7T-13 cells were seeded at a cell density of 9.0 × 103 cells/cm2 before the addition of various concentrations of compounds 1 day after seeding. After 4 days of treatment, media was replaced with fresh media containing each compound. Cell viability was assessed using a cell counting kit (Dojindo Laboratories, Kumamoto, Japan) at 0, 2, 4, and 8 days after treatment.
2.4. Reporter assay
To establish cell lines for the assessment of Wnt/β-catenin activity in UE7T-13 cells, a TCF-4 motif-responsive luciferase reporter gene was developed. In brief, three copies of the optimal TCF-4 motif CCTTTGATC and cytomegalovirus (CMV) promoter were cloned from the Tcf4-CMVpro-Luc plasmid [9] and inserted into the multiple cloning site of the pGL4.20 luciferase reporter plasmid (Promega Corp., Fitchburg, WI), subsequently designated pTCF4-CMVpro-GL4.2 (Fig. 1). UE7T-13 cells were stably transfected with pTCF4-CMVpro-GL4.2 by electroporation and were selected using puromycin resulting in the establishment of E7-TCF4 cells. At 24 h after seeding of E7-TCF4 cells at a density of 9.0 × 103 cells/cm2, various concentrations of small molecule compounds were added. At 4 days after treatment, fresh media containing each compound was replaced. On day 1, 4, and 8, a Luciferase reporter assay system (Promega Corp.) was performed using a plate reader (PerkinElmer, Inc., MA).
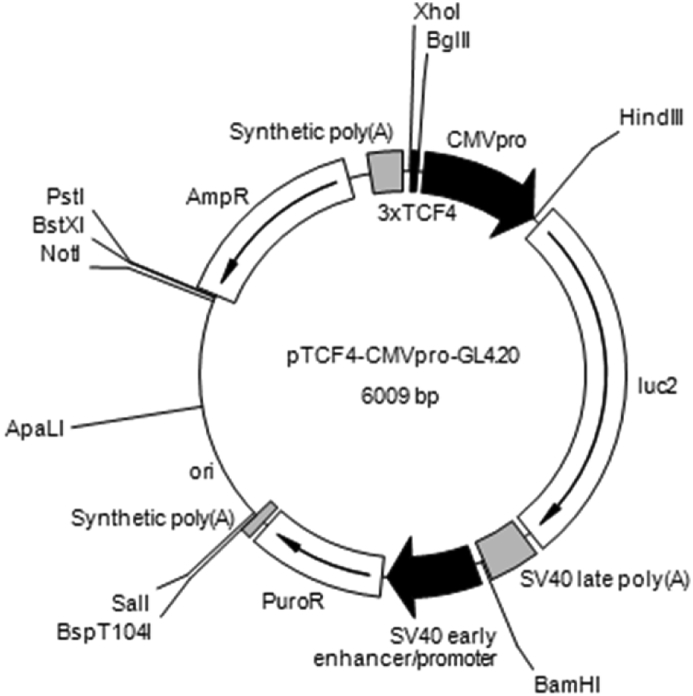
Fig. 1.
Map of the pTCF4-CMVpro-GL4.20 plasmid. Three copies of the optimal TCF-4 motif CCTTTGATC and cytomegalovirus (CMV) promoter were cloned from the Tcf4-CMVpro-Luc plasmid [9] and inserted at BglII and HindIII restriction enzyme sites in the multiple cloning sites of pGL4.20 luciferase reporter plasmid (Promega Corp., Fitchburg, WI), designated pTCF4-CMVpro-GL4.2. pTCF-CMVpro-GL4.20 was linearized by Pst1 and electroporated into UE7T-13 cells to establish cell lines for the assessment of Wnt/β-catenin activity in UE7T-13 cells.
To confirm Wnt/β-catenin signal transcriptional activity in human bone marrow mononuclear cells purchased from Lonza, cells were cultured at a density of 1.8 × 104 cells/cm2 for 1 day before treatment with various concentrations of IC-2. On day 1, the TCF/LEF reporter lentivirus (Cignal Lenti TCF/LEF Reporter [luc], SABiosciences, Qiagen N.V., Frederick, MD, USA) and control renilla lentivirus (Cignal Lenti TK Renilla Control [luc], SABiosciences, Qiagen N.V.) were transduced into cells. The Luciferase reporter assay system (Promega) was performed on day 4 using a Luminometer (Berthold Japan K.K., Tokyo, Japan). Similarly, lentiviruses were transduced on day 5 and luciferase reporter assay performed on day 8.
2.5. Hepatic differentiation
UE7T-13 cells were seeded at a density of 9.0 × 103 cells/cm2, and each compound was added at 24 h after seeding. Hepatic differentiation was assessed 8 days after treatment with each compound. Media was changed on day 4. Hepatic differentiation of primary BM-MSCs was performed using identical conditions except for a starting cell density of 1.8 × 104 cells/cm2.
2.6. RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted from UE7T-13 cells using TRIzol reagent (Invitrogen, Life Technologies Corp., Carlsbad, CA). Reverse transcription was performed using SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen) and oligo (dT)18 primers according to manufacturer's instructions. Real-time PCR analysis was performed using EXPRESS qPCR Supermix with Premixed ROX (Invitrogen) and appropriate combinations of Universal Probe Library Probes (Roche Diagnostics GmbH, Mannheim, Germany). Primer sets are listed in Table 1. For each assay, PCR was performed using a 7900 HT Fast Real-Time PCR System (Applied Biosystems, Life Technologies Corp., Carlsbad, CA, USA). Following the extraction of total RNA from primary BM-MSCs using TaqMan Gene Expression Cells-to-CT Kits (Life Technologies Corp.), qRT-PCR was performed using TaqMan® Universal PCR Master Mix (Applied biosystems, Life Technologies Corp., Carlsbad, CA) and appropriate combinations of Universal Probe Library Probes (Roche Diagnostics GmbH, Mannheim, Germany). PCR was performed using a LightCycler System (Roche Applied Science, Mannheim, Germany).
Table 1.
Primer and probe number information for qRT-PCR.
| Primer | Sequences (5′–3′) | Product (bp) | Probe no. | |
|---|---|---|---|---|
| ALB | Forward: | CAAAGATGACAACCCAAACCTC | 126 | 54 |
| Reverse: | GGATGTCTTCTGGCAATTTCA | |||
| C3 | Forward: | CAGCACCATGGGACCCACCTCAG | 120 | 40 |
| Reverse: | CTCTCCAGCCGCAAGATGTTGGG | |||
| TDO2 | Forward: | CGATGACAGCCTTGGACTTC | 76 | 67 |
| Reverse: | CGGAATTGCAAACTCTGGA | |||
| EpCAM | Forward: | GATGGCTCTCATCCCAGACTT | 111 | 3 |
| Reverse: | AGTCCATGTGAATGGGTTCC | |||
| GAPDH | Forward: | AGCCACATCGCTCAGACAC | 66 | 60 |
| Reverse: | GCCCAATACGACCAAATCC | |||
2.7. Immunofluorescence, PAS staining, and urea assay
After 8 days of incubation with each compound, cells were fixed and immunofluorescence analysis was performed as previously described [20]. Urea assays and periodic acid–Schiff (PAS) staining were performed as previously described [20].
2.8. Statistical analyses
All the values are expressed as means ± SD. Groups were compared using two-way ANOVA followed by Bonferroni's post-hoc test and one-way ANOVA followed by Dunnett's post-hoc test using predictive analytics software (SPSS Inc., Chicago, IL, USA). Differences between two groups were analyzed using the Mann–Whitney U test. P-values less than 0.05 were considered statistically significant.
3. Results
3.1. Newly synthesized derivatives of known small molecule inhibitors of Wnt/β-catenin signaling
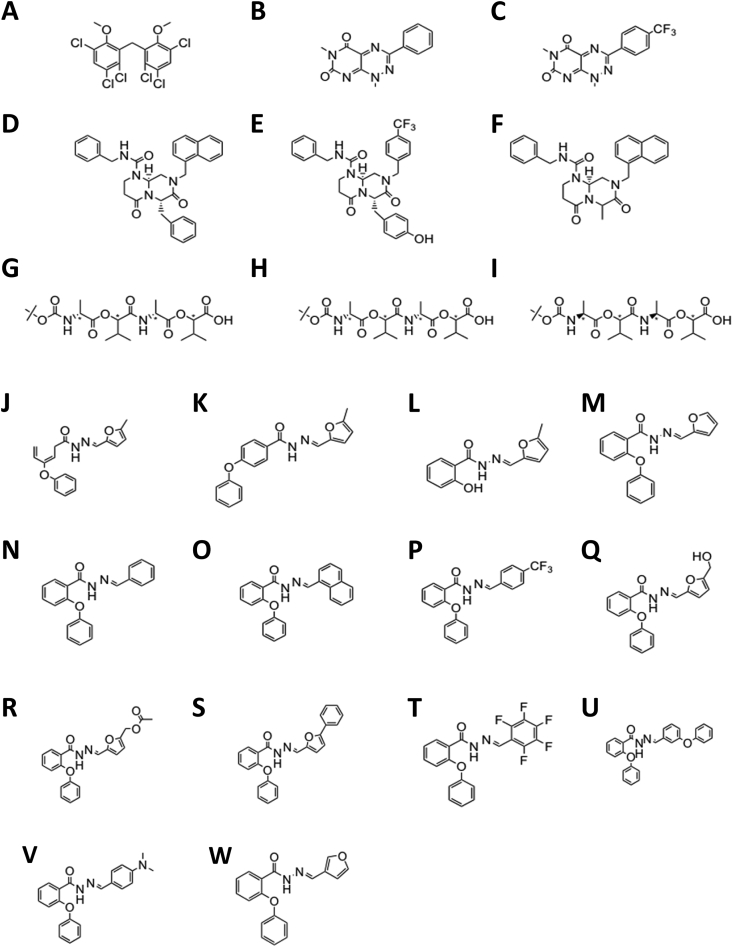
The following twenty-three compounds were newly-synthesized inhouse based on known Wnt/β-catenin signaling inhibitors [18]: HC-1, a derivative of hexachlorophene; PK-2 and PK-3, derivatives of PKF118-310; IC-2, IC-3, and IC-4, derivatives of ICG-001; RRRR, RSRS, and SRSR, derivatives of NSC668036; and PN-1-2, PN-1-3, PN-2, PN-3, PN-3-3, PN-3-4, PN-3-5, PN-3-8, PN-3-9, PN-3-12, PN-3-13, PN-3-16, PN-3-17, and PN-3-19, derivatives of PNU-74654 (Fig. 2).
Fig. 2.
Chemical structures of newly synthesized derivatives from known small molecule inhibitors of Wnt/β-catenin signaling. A, HC-1; B, PK-2; C, PK-3; D, IC-2; E, IC-3; F, IC-4; G, RRRR; H, RSRS; I, SRSR; J, PN-1-2; K, PN-3-1; L, PN-3-2; M, PN-3; N, PN-3-3; O, PN-3-4; P, PN-3-5; Q, PN-3-8; R, PN-3-9; S, PN-3-12; T, PN-3-13; U, PN-3-16; V, PN-3-17; W, PN-3-19. Inhibitors were dissolved in 0.1% DMSO for further use.
3.2. Effects on the viability of human MSCs
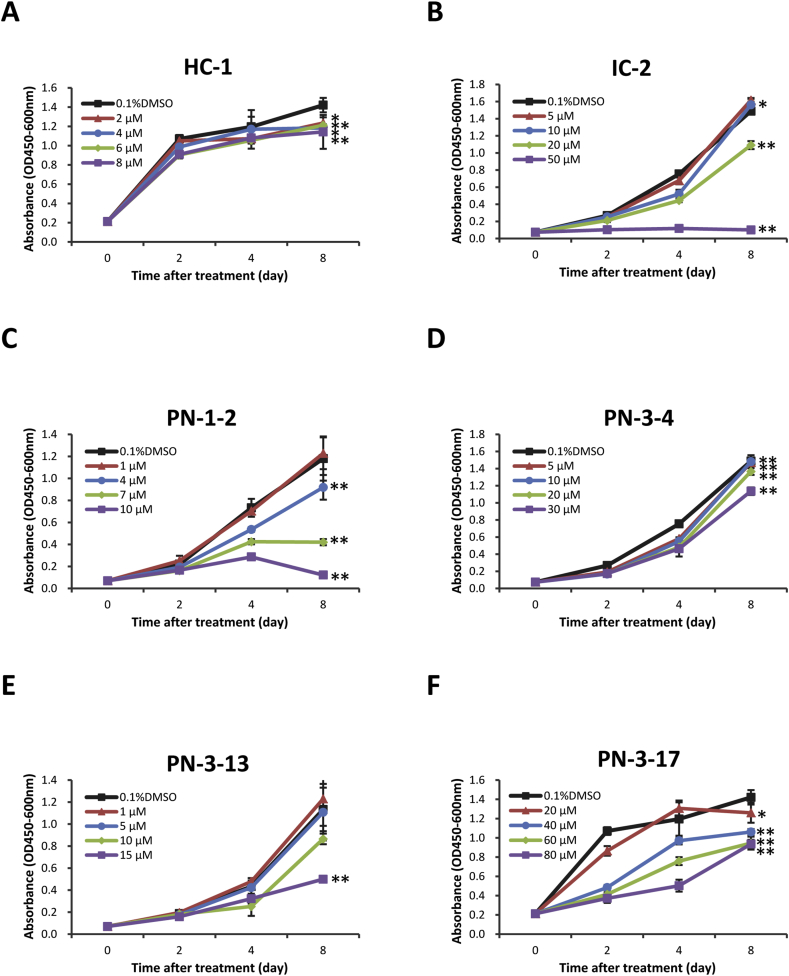
To identify compounds with low toxicity during hepatic differentiation, cell viability of human MSCs was assessed using WST assays. Not more than approximately 8 μM of HC-1 was regarded as safe over 8 days of treatment (Fig. 3A). Correspondingly, approximately 20 μM or less of IC-2, approximately 7 μM or less of PN-1-2, approximately 30 μM or less of PN-3-4, approximately 10 μM or less of PN-3-13, and approximately 40 μM or less of PN-3-17 were considered safe concentrations for use during hepatic differentiation (Fig. 3B–F).
Fig. 3.
Cell viability assays with increasing concentrations of small molecule inhibitors. The effects of each small molecule compound on cell viability were determined by WST assays. WST assays were performed on days 0, 2, 4, and 8. A, HC-1; B, IC-2; C, PN-1-2; D, PN-3-4; E, PN-3-13; F, PN-3-17. Data are expressed as the mean ± SD of 3 separate wells. Two-way ANOVA followed by Bonferroni's post-hoc test was used to compare the viability of UE7T-13 cells following treatment with different concentrations at different time points. *P < 0.05, and **P < 0.01 compared with 0.1% DMSO.
3.3. Small molecule compounds inhibiting TCF4/β-catenin transcriptional activity in E7-TCF4 cells
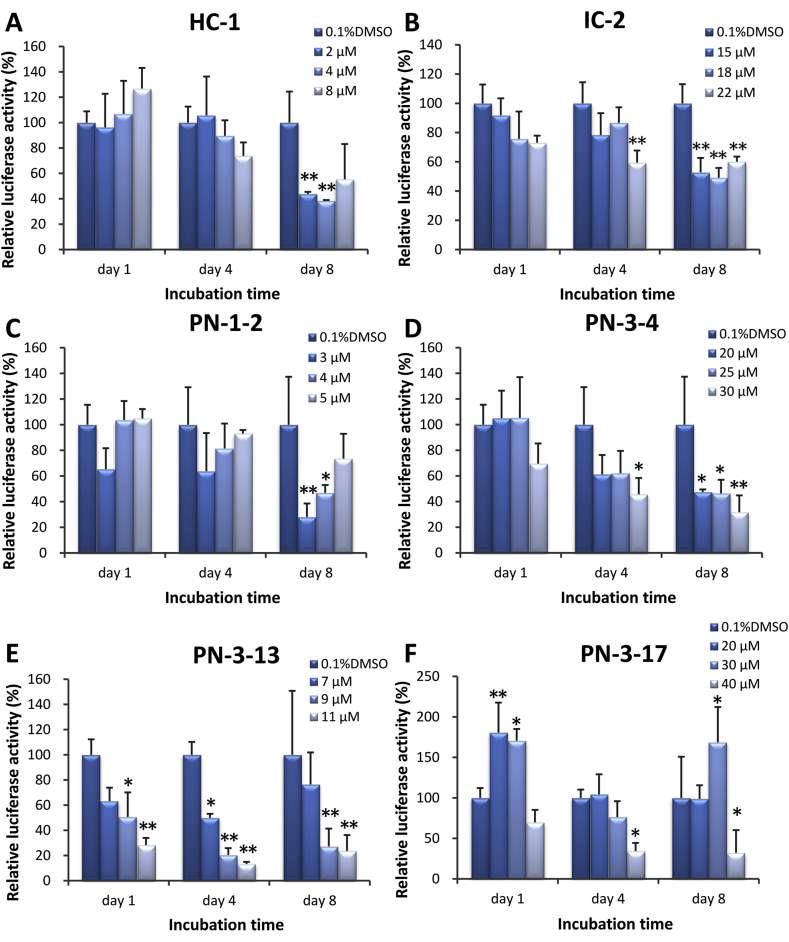
TCF4/β-catenin transcriptional activity was measured using a luciferase reporter assay. Of the twenty-three small molecule compounds, six compounds were found to suppress TCF4/β-catenin transcriptional activity in E7-TCF4 cells after 1, 4, and 8 days treatment. HC-1 suppressed luciferase activity at 2 and 4 μM on day 8 only (Fig. 4A). IC-2 clearly suppressed luciferase activity in a time- and dose-dependent manner (Fig. 4A). After 8 days of treatment, IC-2 was found to suppress rates of luciferase activity by approximately 50%. Four derivatives of PNU-74654, including PN-1-2, PN-3-4, PN-3-13, and PN-3-17, demonstrated modest inhibitory effects on luciferase activity (Fig. 4C–F, respectively). Of the four derivatives of PNU-74654, PN-3-13 was found to be the most potent inhibitor of TCF4/β-catenin transcriptional activity.
Fig. 4.
Small molecule compounds inhibiting TCF4/β-catenin transcriptional activity in E7-TCF4 cells. TCF4/β-catenin transcriptional activity was examined by luciferase reporter assay. Of the twenty-three small molecule compounds, six compounds suppressed TCF4/β-catenin transcriptional activity in E7-TCF4 cells. A, HC-1; B, IC-2; C, PN-1-2; D, PN-3-4; E, PN-3-13; F, PN-3-17. Data are expressed as mean ± SD (n = 3). Relative luciferase activity was expressed as a ratio of 0.1% DMSO. Two-way ANOVA followed by Bonferroni's post hoc-test was used to compare the relative luciferase activities of UE7T-13 cells following treatment with different concentrations at different time points. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO at each corresponding time point.
3.4. Effects on hepatic differentiation of human MSCs
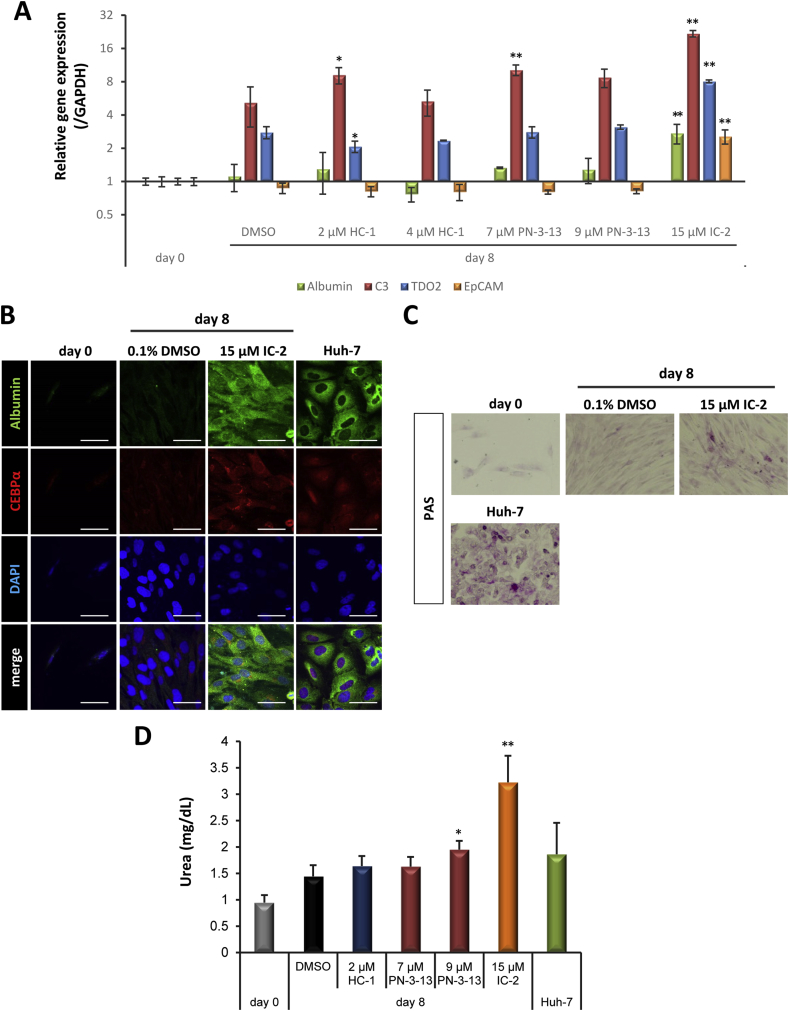
Of the six compounds that exhibited suppressive effects on TCF4/β-catenin transcriptional activity, we evaluated the ability of HC-1, PN-3-13, and IC-2 to induce hepatic differentiation of human MSCs (Fig. 5). First, we examined mRNA expression of albumin, C3, tryptophan 2,3-dioxygenase (TDO2), and EpCAM by UE7T-13 cells treated with each compound for 8 days (Fig. 5A). Increased expression of albumin was observed in cells treated with 15 μM IC-2. Treatment with 7 μM PN-3-13 and 15 μM IC-2 induced complement C3 expression. TDO2 expression was induced by 2 μM HC-1 and 15 μM IC-2, respectively. Interestingly, 15 μM IC-2 significantly increased the expression of TDO2, a key modulator of hepatocyte function, compared with treatment with 0.1% DMSO. Elevated expression of EpCAM was observed following treatment with 15 μM IC-2 only. According to the expression of albumin, C3, TDO2, and EpCAM, IC-2 is apparently a potent inducer of the hepatic differentiation of human MSCs. Moreover, immunofluorescence analysis revealed that the expression of albumin was observed in mainly in the cytoplasm and faintly in the nucleus although it was restricted to the cytoplasm in Huh-7 cells (Fig. 5B). C/EBPα was also induced in cells treated with 15 μM IC-2, however, its localization was not confined in the nucleus (Fig. 5B). These phenomena may be supported by the reports that localization of albumin and C/EBPα in the transdifferentiated hepatocytes differs from in mature hepatocytes [21], [22]. PAS staining demonstrated increased glycogen storage in response to treatment with 15 μM IC-2 compared with treatment with 0.1% DMSO (Fig. 5C). Moreover, urea production, one of the most important hepatocyte functions, was increased by approximately 3-fold in response to treatment with IC-2 compared with day 0 and was higher than in Huh-7 cells (Fig. 5D). These data indicate that IC-2 could induce hepatic specification of MSCs through inhibiting Wnt/β-catenin signal.
Fig. 5.
IC-2 potently induced hepatic differentiation of UE7T-13 cells. A. Liver-specific gene expression was confirmed by qRT-PCR analysis. Hepatocyte markers (albumin, C3, and TDO2) and a hepatic stem cell marker (EpCAM) were measured. Data are expressed as the means ± SD of three experiments. Analysis by one-way ANOVA, followed by Dunnett's post-hoc test, was used to compare the relative gene expression of UE7T-13 cells following treatment with different compounds at day 8. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO. B. Immunofluorescence microscope image of hepatocyte-specific protein expression. Expression of albumin and C/EBPα was confirmed on day 0 and on day 8. Four different images including albumin and C/EBPα, DAPI and their merge were shown in four different conditions. Scale bar represents 50 μm. C. Glycogen storage in UE7T-13 cells treated with small molecule compounds. PAS staining was performed to demonstrate glycogen storage on day 0 and 8. Huh-7 cells were used as a positive control. D. Urea production in cells treated with small molecule compounds. After incubation of UE7T-13 cells with small molecule compounds for eight days, culture media were fleshly replaced by media including 5 mM ammonium chloride and cells were further incubated for 96 h. Huh-7 cells were used as a positive control. Data are expressed as the means ± SD of six experiments. One-way ANOVA, followed by Dunnett's post-hoc test, was used to compare urea production in UE7T-13 cells following treatment with different compounds at day 8. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO.
3.5. Suppressive effects on TCF4/β-catenin transcriptional activity in primary human bone marrow mononuclear cells fractionated according to CD90 and CD271 expression
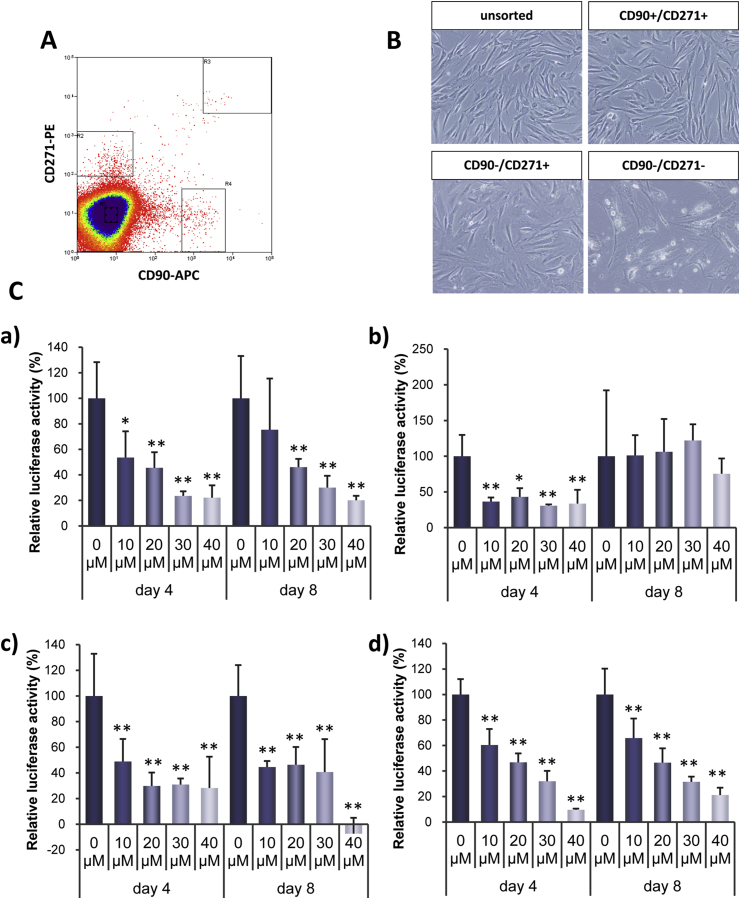
The CD90+ CD271+ population of primary human bone marrow mononuclear cells has been reported to be enriched for CFU-Fs with the potential to differentiate into osteoblasts, chondrocytes, and adipocytes, indicating the presence of MSCs in this fraction [23]. Primary human bone marrow mononuclear cells were fractionated according to expression of CD90 and CD271 (Fig. 6A). The appearances of the unsorted whole bone marrow nuclear cells, CD90+ CD271+ cells, CD90− CD271+ cells, and CD90− CD721− cells are shown in Fig. 6B. All sorted cells had fibroblastic morphology. CD90+ CD271− cells grew very slowly and poorly expanded. Accordingly, this fraction of cells was excluded from further analysis. In unsorted whole bone marrow mononuclear cells and CD90− CD721− cells, IC-2 suppressed TCF4/β-catenin transcriptional activity in a dose-dependent manner after both 4 and 8 days treatment (Fig. 6Ca and Cd). Suppressive effects of IC-2 on luciferase activity in CD90+ CD721+ cells were clearly observed on day 4 but not on day 8 (Fig. 6Cb). In CD90− CD271+ cells, IC-2 suppressed luciferase activity on day 4 and day 8 (Fig. 6Cc). These data demonstrate that IC-2 is capable of suppressing TCF4/β-catenin transcriptional activity in unsorted whole bone marrow nuclear cells, CD90+ CD271+ cells, CD90− CD271+ cells, and CD90− CD721− cells.
Fig. 6.
IC-2 suppressed TCF/β-catenin transcriptional activities in human bone marrow mononuclear cells. A. Cell populations of primary BM-MSCs were isolated according to CD90 and CD271 expression. In addition to whole bone marrow mononuclear cells, CD90+ CD271+, CD90− CD271+, and CD90− CD271− cell populations were fractionated by cell sorting. B. Bright field images of unsorted whole BM-MSCs, CD90+ CD271+, CD90− CD271+, and CD90− CD271− cells expanded after cell sorting. C. TCF4/β-catenin transcriptional activities in human bone marrow mononuclear cells. a) unsorted whole mononuclear cells, b) CD90+ CD271+ cells, c) CD90− CD271+ cells, and d) CD90− CD271− cells. Data are expressed as the means ± SD of five experiments. Two-way ANOVA followed by Bonferroni's post-hoc test was used to compare the relative luciferase activities of human bone marrow mononuclear cells following treatment with different concentrations at different time points. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO at each corresponding time point.
3.6. IC-2 potently induced hepatic differentiation of CD90+ CD271+ human bone marrow mononuclear cells
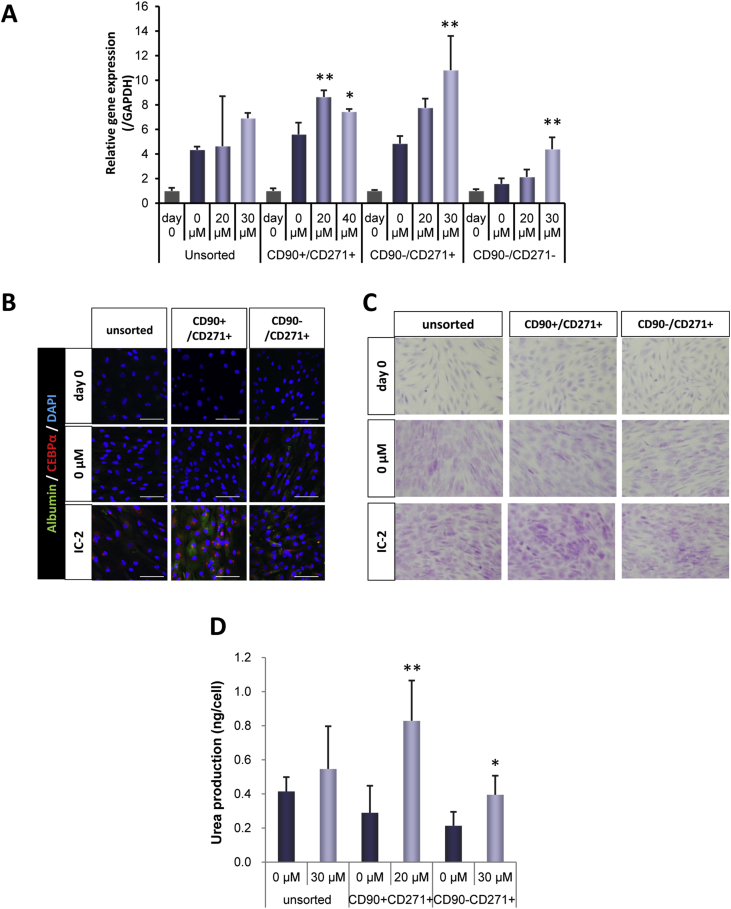
Albumin mRNA expression after IC-2 treatment was assessed in four populations, including the unsorted whole bone marrow nuclear cells, CD90+ CD271+ cells, CD90− CD271+ cells, and CD90− CD721− cells (Fig. 7A). Unsorted whole bone marrow nuclear cells, CD90+ CD271+ cells, and CD90− CD721+ cells demonstrated similar expression profiles in response to IC-2 treatment in a dose-dependent manner. However, the effects of IC-2 on CD90− CD721− cells were minimal. Immunohistochemical analysis revealed that the expression of albumin and C/EBPα in CD90+ CD271+ cells was higher than that in unsorted whole bone marrow nuclear cells and CD90− CD721+ cells (Fig. 7B). PAS staining demonstrated greater glycogen contents in CD90+ CD271+ cells than in unsorted whole bone marrow nuclear cells and CD90− CD721+ cells (Fig. 7C). Finally, urea production was found to be the highest in CD90+ CD271+ cells of the three assessed cell populations (Fig. 7D). These data indicate that IC-2 induces hepatic differentiation of human bone marrow mononuclear cells, particularly CD90+ CD271+ cells.
Fig. 7.
IC-2 induced hepatic differentiation of primary human bone marrow mononuclear cells. A. Albumin mRNA expression in IC-2-treated primary human bone marrow mononuclear cells. Data are expressed as the means ± SD of three experiments. One-way ANOVA followed by Dunnett's post-hoc test was used to compare the relative gene expression of human bone marrow mononuclear cells following treatment with different concentrations of IC-2 at day 8. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO. B. Immunofluorescence analysis of albumin and C/EBPα expression was conducted on day 8. Scale bar, 100 μm. CD90+ CD271+ cells were treated with 0 and 20 μM IC-2. Unsorted whole cells and CD90− CD271+ cells were treated with 0 and 30 μM IC-2, respectively. C. PAS staining of IC-2 treated primary human BM-MSCs to demonstrate glycogen storage on day 8. CD90+ CD271+ cells were treated with 0 and 20 μM IC-2. Unsorted whole cells and CD90− CD271+ cells were treated with 0 and 30 μM IC-2, respectively. D. Urea synthesis in IC-2-treated human bone marrow mononuclear cells. Urea contents were measured by urea assay. Human bone marrow mononuclear cells were treated with IC-2 for 8 days followed by further incubation in the presence of 5 mM ammonium chloride for 96 h. Data are expressed as the means ± SD of five experiments and analyzed by Mann–Whitney U test. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO.
4. Discussion
Human MSCs have been reported to be capable of differentiating into hepatocytes [20], [24], [25], [26], [27], [28]. Differentiating protocols predominantly involve incubation with combinations of several cytokines. However, the overexpression of hepatocyte-specific transcription factors, such as HNF-3β and HNF-4α, determines and enhances hepatic specification [20], [28]. The findings of the present study demonstrated the use of a single compound, rather than a combination of cytokines or hepatocyte nuclear factor gene expression, was able to induce the hepatocytic differentiation of human MSCs. Our and other groups have reported that the downregulation of the Wnt/β-catenin pathway induces hepatic differentiation of MSCs [9], [10], [11]. Although the mechanisms regulating hepatic differentiation of human MSCs have yet to be fully elucidated, β-catenin expression has been shown to be suppressed during the competence and specification stages of normal liver development [29], [30]. Wnt/β-catenin signaling has been reported to maintain stemness [31]. Differentiation toward endoderm is induced by activin/Nodal signals. Endodermal progenitor cells, including hepatic progenitors, can be generated by the culture of ES cells with activin, BMP-4, and FGF-4 [32]. Inhibition of Wnt/β-catenin pathways suppresses expression of BAMBI, BMP, and activin membrane-bound inhibitor, indicating that the suppression of Wnt/β-catenin signals may induce activin/Nodal signaling in response to BAMBI inhibition [33]. In the present study, HC-1, IC-2, and PN-3-13 similarly suppressed Wnt/β-catenin signaling, however, IC-2 was the most potent inducer of hepatic differentiation. These data indicate that other factors are involved in hepatic differentiation, although the suppression of Wnt/β-catenin signals is required for hepatic differentiation of human MSCs. Therefore, further studies are required to identify other factors involved in regulating the hepatic specification of human MSCs.
As small molecule compounds are more safe and stable than nucleic acids and protein products, they have greater applicability in clinical settings [12], [13], [14]. Small molecule compounds have been reported to have diverse functions, including the modulation and maintenance of pluripotent stem cells, self-renewal of tissue-specific stem and progenitor cells, and induction of stem cell differentiation and reprogramming [12]. Molecules that interact with IC-2 were not determined in the present study. It has been reported that ICG-001 suppresses Wnt/β-catenin signals by binding CREB-binding protein (CBP) [34], [35]. As IC-2 is a derivative of ICG-001 [18], IC-2 may interact with CBP. Therefore, further studies aimed at identifying proteins that interact with IC-2 and determining the precise mechanisms underlying the hepatic differentiation of human MSCs are required in the near future.
5. Conclusion
We screened twenty-three newly synthesized derivatives of small molecule compounds generated from several known Wnt/β-catenin signal inhibitors. IC-2 was identified as a potent small molecule inhibitor capable of inducing the differentiation of human mesenchymal stem cells into hepatocytes. IC-2 potently induced hepatic differentiation of human bone marrow mononuclear cells which express CD90 and CD271.
Author contributions
NI performed small compound screening, examined liver functions of human bone marrow mononuclear cells treated with IC-2, and wrote the manuscript. TS, KK, JA, HS, YK, and KA examined liver functions of human bone marrow mononuclear cells treated with IC-2. YM screened small molecule compounds. TT, HO, TS, HS, and MM designed and produced small molecule compounds. YM and YM performed the fractionation of human bone marrow mononuclear cells. GS designed the project and wrote the manuscript.
Funding sources
This study was supported by a project for the realization of regenerative medicine from the Ministry of Education, Culture, Sports, Science and Technology in Japan.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank Y. Arakaki, T. Yasui, and M. Dozono for technical assistance. We are grateful to Y. Hoshikawa for helpful discussion.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Contributor Information
Noriko Itaba, Email: itaba@med.tottori-u.ac.jp.
Tomohiko Sakabe, Email: saka0924@med.tottori-u.ac.jp.
Keita Kanki, Email: kkanki@bme.ous.ac.jp.
Junya Azumi, Email: jazumi@med.tottori-u.ac.jp.
Hiroki Shimizu, Email: h.shimizu9005@med.tottori-u.ac.jp.
Yohei Kono, Email: ykono@med.tottori-u.ac.jp.
Yoshiaki Matsumi, Email: guard610@med.tottori-u.ac.jp.
Ken-ichiro Abe, Email: ken.abe@med.tottori-u.ac.jp.
Takayuki Tonoi, Email: tonoi@rs.tus.ac.jp.
Hiroyuki Oka, Email: hoka@chem.tottori-u.ac.jp.
Toshihiko Sakurai, Email: sakurai@bio.tottori-u.ac.jp.
Hiroyuki Saimoto, Email: saimoto@chem.tottori-u.ac.jp.
Minoru Morimoto, Email: morimoto@chem.tottori-u.ac.jp.
Yo Mabuchi, Email: yomabuchi.bb@tmd.ac.jp.
Yumi Matsuzaki, Email: ymatsuzak@gmail.com.
Goshi Shiota, Email: gshiota@med.tottori-u.ac.jp.
References
- 1.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–532. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginis I., Luo Y., Miura T., Thies S., Brandenberger R., Gerecht-Nir S. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 3.Sato N., Meijer L., Skalsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 4.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and caner stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Otero J.J., Fu W., Kan L., Cuadra A.E., Kessler J.A. β-catenin signaling is required for normal differentiation of embryonic stem cells. Development. 2004;131:1171–1183. doi: 10.1242/dev.01218. [DOI] [PubMed] [Google Scholar]
- 6.Bertrand F.E., Angus C.W., Partis W.J., Sigounas G. Developmental pathways in colon cancer: crosstalk between WNT, BMP, Hedgehog and Notch. Cell Cycle. 2012;11:4344–4351. doi: 10.4161/cc.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hari L., Brault V., Kléber M., Lee H.Y., Ille F., Leimeroth R. Lineage-specific requirement of β-catenin in neural crest development. J Cell Biol. 2001;159:867–880. doi: 10.1083/jcb.200209039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muroyama Y., Kondoh H., Takada S. Wnt proteins promote neuronal differentiation in neural stem cell culture. Biochem Biophys Res Commun. 2004;313:915–921. doi: 10.1016/j.bbrc.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida Y., Shimomura T., Sakabe T., Ishii K., Gonda K., Matsuoka S. A role of Wnt/beta-catenin signals in hepatic fate specification of human umbilical cord blood-derived mesenchymal stem cells. Am J Physiol Gastrointest Liver Physiol. 2007;293:1089–1098. doi: 10.1152/ajpgi.00187.2007. [DOI] [PubMed] [Google Scholar]
- 10.Shimomura T., Yoshida Y., Sakabe T., Ishii K., Gonda K., Murai R. Hepatic differentiation of human bone marrow-derived UE7T-13 cells: effects of cytokines and CCN family gene expression. Hepatol Res. 2007;37:1068–1079. doi: 10.1111/j.1872-034X.2007.00162.x. [DOI] [PubMed] [Google Scholar]
- 11.Ke Z., Zhou F., Wang L., Chen S., Liu F., Fan X. Down-regulation of Wnt signaling could promote bone marrow-derived mesenchymal stem cells to differentiate into hepatocytes. Biochem Biophys Res Commun. 2008;367:342–348. doi: 10.1016/j.bbrc.2007.12.134. [DOI] [PubMed] [Google Scholar]
- 12.Li W., Li K., Wei W., Ding S. Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell. 2013;13:270–283. doi: 10.1016/j.stem.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borowiak M., Maehr R., Chen S., Chen A.E., Tang W., Fox J.L. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schugar R.C., Robbins P.D., Deasy B.M. Small molecules in stem cell self-renewal and differentiation. Gene Ther. 2008;15:126–135. doi: 10.1038/sj.gt.3303062. [DOI] [PubMed] [Google Scholar]
- 15.Lepourcelet M., Chen Y.N., France D.S., Wang H., Crews P., Petersen F. Small-molecule antagonists of the oncogenic Tcf/β-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 16.Voronkov A., Krauss S. Wnt/beta-catenin signaling and small molecule inhibitors. Curr Pharm Des. 2013;19:634–664. doi: 10.2174/138161213804581837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S., Gwak J., Cho M., Song T., Won J., Kim D.E. Hexachlorophene inhibits Wnt/beta-catenin pathway by promoting Siah-mediated beta-catenin degradation. Mol Pharmacol. 2006;70:960–966. doi: 10.1124/mol.106.024729. [DOI] [PubMed] [Google Scholar]
- 18.Shiota G, Hoshikawa Y, Matsumoto N, Matsumi Y, Morimoto M, Tonoi T, et al. Synthesis and analysis of novel compound capable of inducing differentiation of human mesenchymal stem cell into hepatocyte. WO2012/141038A1, PCT/JP2012/059021.
- 19.Mori T., Kiyono T., Imabayashi H., Takeda Y., Tsuchiya K., Miyoshi S. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol Cell Biol. 2005;25:5183–5195. doi: 10.1128/MCB.25.12.5183-5195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii K., Yoshida Y., Akechi Y., Sakabe T., Nishio R., Ikeda R. Hepatic differentiation of human bone marrow-derived mesenchymal stem cells by tetracycline-regulated hepatocyte nuclear factor 3beta. Hepatology. 2008;48:597–606. doi: 10.1002/hep.22362. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz R.E., Reyes M., Koodie L., Jiang Y., Blackstad M., Lund T. Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. J Clin Investig. 2002;109:1291–2302. doi: 10.1172/JCI15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burke Z.D., Shen C.-N., Ralphs K.L., Tosh D. Characterization of liver function in transdifferentiated hepatocytes. J Cell Physiol. 2006;206:147–159. doi: 10.1002/jcp.20438. [DOI] [PubMed] [Google Scholar]
- 23.Mabuchi Y., Morikawa S., Harada S., Niibe K., Suzuki S., Renault-Mihara F. LNGFR+THY-1+VCAM-1hi+ cells reveal functionally distinct subpopulations in mesenchymal stem cells. Stem Cell Rep. 2013;1:152–165. doi: 10.1016/j.stemcr.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee K.D., Kuo T.K., Whang-Peng J., Chung Y.F., Lin C.T., Chou S.H. In vitro hepatic differentiation of human mesenchymal stem cells. Hepatology. 2004;40:1275–1284. doi: 10.1002/hep.20469. [DOI] [PubMed] [Google Scholar]
- 25.Wei X., Wang C.Y., Liu Q.P., Li J., Li D., Zhao F.T. In vitro hepatic differentiation of mesenchymal stem cells from human fetal bone marrow. J Int Med Res. 2008;36:721–727. doi: 10.1177/147323000803600414. [DOI] [PubMed] [Google Scholar]
- 26.Aurich H., Sgodda M., Kaltwasser P., Vetter M., Liehr T., Brulport M. Hepatocyte differentiation of mesenchymal stem cells from human adipose tissue in vitro promotes hepatic integration in vivo. Gut. 2009;58:570–581. doi: 10.1136/gut.2008.154880. [DOI] [PubMed] [Google Scholar]
- 27.Dong X., Pan R., Zhang H., Yang C., Shao J., Xiang L. Modification of histone acetylation facilitates hepatic differentiation of human bone marrow mesenchymal stem cells. PLoS One. 2013;8:e63405. doi: 10.1371/journal.pone.0063405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M.L., Lee K.D., Huang H.C., Tsai Y.L., Wu Y.C., Kuo T.M. HNF-4α determines hepatic differentiation of human mesenchymal stem cells from bone marrow. World J Gastroenterol. 2010;16:5092–5103. doi: 10.3748/wjg.v16.i40.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nejak-Bowen K., Monga S.P. Wnt/beta-catenin signaling in hepatic organogenesis. Organogenesis. 2008;4:92–99. doi: 10.4161/org.4.2.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson M.D., Monga S.P. Wnt/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 31.Willert K., Brown J.D., Danenberg E., Duncan A.W., Weissman I.L., Reya T. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 32.Gouon-Evans V., Boussemart L., Gadue P., Nierhoff D., Koehler C.I., Kubo A. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- 33.Sekiya T., Adachi S., Kohu K., Yamada T., Higuchi O., Furukawa Y. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. J Biol Chem. 2004;20:6840–6846. doi: 10.1074/jbc.M310876200. [DOI] [PubMed] [Google Scholar]
- 34.Emami K.H., Nguyen C., Ma H., Kim D.H., Jeong K.W., Eguchi M. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription. Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ring A., Kim Y.-M., Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev Rep. 2014;10:512–525. doi: 10.1007/s12015-014-9515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]