Abstract
Real-time reverse transcription polymerase chain reaction (RT-PCR) is regarded as one of the most useful and powerful tools for characterizing hematopoietic stem cells (HSCs), because samples of extremely small cell numbers can be analyzed. The expression levels determined by RT-PCR are based on relative quantification; therefore, the selection of an appropriate reference gene with a relatively stable expression level under most conditions is crucial. Here, we determined that beta2-microglobulin (B2m) is an appropriate reference gene for analyzing mouse HSCs by a novel method using single-cell RT-PCR. Clonally sorted HSCs were subjected to RT reactions with exogenous RNA fragments and then to real-time PCR. Next, the relative gene expression levels of 4 well-known housekeeping genes were quantified in each single cell sample based on the threshold cycle of exogenous RNA. The analysis revealed that B2m expression was reproducibly detected in almost all HSCs and that B2m had the most stable expression level among the compared genes, even after the cells had been cultured under various conditions. Thus, our results indicate that B2m can reliably be used as a reference gene for the relative quantification of expression levels in HSCs across various conditions. Furthermore, our work proposes a novel method for the selection of appropriate reference genes.
Keywords: Hematopoietic stem cells, Reference gene, Single-cell RT-PCR, Beta2-microglobulin
Abbreviations: RT-PCR, reverse-transcription polymerase chain reaction; HSCs, hematopoietic stem cells; B2m, beta2-microglobulin; HKGs, housekeeping genes; MPPs, multi-potential progenitors; ERCC, External RNA Controls Consortium; Actb, beta-actin; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Hprt, hypoxanthine phosphoribosyl transferase; SCF, stem cell factor; TPO, Thrombopoietin; Ct, threshold cycles; MHC, major histocompatibility complex
1. Introduction
Hematopoietic stem cells (HSCs) have the ability to self-renew and to differentiate into multiple lineages, and they govern the maintenance of the hematopoietic system by giving rise to all blood cell types. Generally, to elucidate the biological mechanisms that regulate HSC functions, gene expression analyses, such as reverse-transcription polymerase chain reaction (RT-PCR), DNA microarrays and RNA sequencing, are employed [1], [2]. In addition, gene expression assays can be used with an extremely small number of cells because these assays estimate the expression level of mRNA through the quantification of amplified cDNA. Therefore, these assays greatly contribute to the characterization of HSCs, which are difficult to be prepared in large numbers. To compare the gene expression levels of target genes between samples by using RT-PCR, expression levels are generally normalized to housekeeping genes (HKGs) as internal controls. Therefore, the selection of an appropriate reference gene in real-time RT-PCR is crucial for an accurate comparison of samples based on a relative quantitative estimation of gene expression. Ideally, genes utilized as references should exhibit stable expression levels in individual cells under different conditions. Unstable expression levels of a reference gene may result in inaccurate calculations of the expression levels of target genes and incorrect interpretations of experimental results. Although an appropriate reference gene for HSCs has never been proposed, many researchers have still analyzed gene expression in these cells by using real-time RT-PCR.
It is known that HSCs typically reside in a specialized microenvironment, termed a “niche”, within the bone marrow that regulates their functions [3]. Most HSCs reportedly maintain a quiescent state in the bone marrow niche [4], [5], [6], and this non-cycling state is thought to protect HSCs from external stresses. However, under the appropriate in vitro conditions, HSCs reportedly show a greater potential for cell proliferation, compared to other hematopoietic cells containing multi-potential progenitors (MPPs) [7], [8]. During such ex vivo culture, HSCs are likely to exhibit a highly active state, in contrast to their quiescent state in vivo. This suggests that the expression levels of various genes, including some of the HKGs used as reference genes, could be altered between in vitro and in vivo conditions [9].
In the present study, we show that the expression level of beta2-microglobulin (B2m) is relatively stable compared with other reference genes, even after culturing HSCs under 3 different conditions. Using cDNA synthesized from clonally sorted HSCs together with exogenously added RNA fragments, we estimated the expression levels of genes that are frequently used as reference genes using relative quantification based on the threshold cycle of exogenous RNA in single cells. Our present study demonstrates that B2m is one of the most appropriate reference genes for RT-PCR analysis of HSCs and provides a novel method for examining appropriate reference genes for single-cell RT-PCR.
2. Materials and methods
All animal experiments were performed according to the “Guidelines of Tokyo Women's Medical University on Animal Use”, the “Principles of Laboratory Animal Care” formulated by the National Society for Medical Research, and the “Guide for the Care and Use of Laboratory Animals” prepared by the Institute of Laboratory Animal Resources and published by the National Institutes of Health (NIH Publication No. 86-23, revised 1985).
2.1. Antibodies and the 2nd reagent
Unless otherwise noted, the following monoclonal antibodies were purchased from BioLegend (San Diego, CA) and were used for cell sorting and flow cytometric analysis: anti-c-Kit (2B8), anti-CD34 (RAM34, eBiosciences, San Diego, CA), anti-CD150 (TC15-12F12.2), anti-CD48 (HM48-1), anti-Sca-1 (E13-161.7), anti-CD45.2 (104), anti-CD45.1 (A20), anti-B220/CD45R (RA3-6B2), anti-Mac-1 (M1/70), anti-Gr-1 (RB6-8C5), anti-CD4 (RM4-5) and anti-CD8 (53-6.72). PE-Texas Red-conjugated streptavidin was purchased from BD Biosciences (Pharmingen, San Jose, CA).
2.2. Cell sorting and flow cytometric analysis
Whole bone marrow cells were prepared from mice as previously described [10]. HSCs were sorted using a MoFlo™ XDP flow cytometer (Beckman Coulter, Brea, CA), as previously described [11].
2.3. Single-cell gene expression analysis
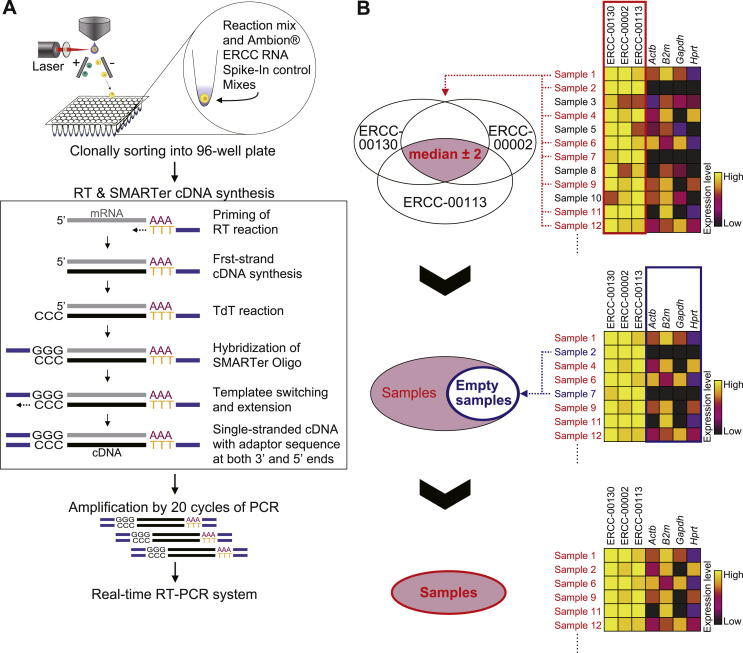
The synthesis and pre-amplification of cDNA derived from single cells were performed using a SMARTer Pico cDNA amplification kit (Clontech, Mountain View, CA) according to the manufacturer's instructions, with minor changes. Briefly, cells were clonally sorted directly into 96-well PCR plates that contained 3 μl of initial reaction mix containing 0.0067% NP40 (Roche, Basel, Switzerland), 0.53 U/μl RNaseOUT (Life Technologies, Carlsbad, CA) and Ambion® Extenal RNA Controls Consortium (ERCC) RNA Spike-In Mixes (Life Technologies) per well and treated at 72 °C for 2 min. Next, reverse transcription (RT) and SMART (Switching Mechanism at 5′ End of RNA Template) reactions were performed at 42 °C for 90 min using the original RT reaction mix supplemented with 0.53 U/μl RNaseOUT and 0.033 μg/μl T4 gene 32 protein (Roche), followed by heat-treatment at 70 °C for 10 min to inactivate the RT enzyme. The synthesized cDNA templates with universal sequences at both their 5′ and their 3′ ends were pre-amplified by 20 cycles of PCR. Real-time RT-PCR was performed using the BioMark™ HD system (Fluidigm, South San Francisco, CA) with a TaqMan® probe (Fig. 1A). To control for template quality, the threshold cycle (Ct) values of 3 selected control ERCC RNAs (ERCC-00130, ERCC-00002, ERCC-00113) were simultaneously examined, and only experimental samples that showed threshold cycles within 2 cycles of the median values of each ERCC RNA were employed in the relative quantification of the expression level of the reference genes (Fig. 1B). The following pre-made TaqMan® MGB probes and primer sets were purchased from Life Technologies: beta-actin (Actb) (Mm00607939_s1, Mm01205647_g1), B2m (Mm00437762_m1, Mm00437764_m1), glyceraldehyde-3-phosphate dehydrogenase (Gapdh) (Mm99999915_g1), hypoxanthine phosphoribosyl transferase (Hprt) (Mm01318741_m1, Mm00446968_m1), ERCC-00130 (Ac03459943_a1), ERCC-00002 (Ac03459872_a1), ERCC-00113 (Ac03459936_a1). In addition, an alternative probe and primer set named “AIFASHW” was designed using the Custom TaqMan® Assay Design Tool for the detection of Gapdh expression. Approximately 7% of the samples had no detectable signal for any of the target genes (data not shown). These samples were assumed to lack cells as a result of cell-sorting error and were removed (Fig. 1B). We confirmed that these samples had no detectable signal even after further amplification by PCR (data not shown).
Fig. 1.
Single-cell gene expression analysis strategy. (A) Experimental design of the single-cell gene expression assay. HSCs were clonally sorted into 96-well PCR plates containing initial reaction mix, including Ambion® ERCC RNA Spike-In Mixes for quality control. Using the indicated primers, an RT reaction followed by an SMART reaction was used to synthesize first-strand cDNAs with adapter sequences at both the 5′ and 3′ ends. Subsequently, cDNA was amplified by 20 cycles of PCR using a primer containing the adapter sequences, and these amplicons were used as the templates for single-cell real-time RT-PCR. (B) For quality control of these templates, the threshold cycle (Ct) values of control ERCC RNA were examined by real-time PCR simultaneously with the target genes. As the first step of quality control, only single-cell samples that exhibited Ct values within ±2 cycles of the median Ct of each of 3 control ERCC RNAs in each independent experiment were used. In addition, samples that had no detectable signal for any gene were deemed to be empty samples (lacking a cell), attributed to a cell sorting error, and were removed from the analysis. These samples repeatedly showed negative signals for all target genes even when templates were further amplified through 10 additional cycles of PCR (data not shown).
2.4. Real-time RT-PCR analysis of bulk samples
Using 10,000 sorted cells from each sample, total RNA was extracted by ISOGEN (Nippon gene, Tokyo, Japan), prior to the synthesis of cDNA by using the SMARTer Pico cDNA amplification kit, as previously described [1]. Next, cDNA templates were subjected to pre-amplification by PCR, as previously described [1]. Amplified cDNA samples were assessed by real-time RT-PCR using the BioMark™ HD system with a TaqMan MGB probe (Life Technologies).
2.5. The calculation of “expression values”
To assess the expression levels of genes in each sample, we calculated “expression values” for the target genes by normalizing their expression to the Ct values for B2m or ERCC-00130, one of the control ERCC RNAs, using the ΔΔCt method.
2.6. HSC culture
CD150+CD34−KSL HSCs were sorted and cultured for 5 days in S-Clone SF-03 medium (EIDIA, Tokyo, Japan) supplemented with 0.5% bovine serum albumin (Sigma, St. Louis, MO) and 50 ng/ml mouse stem cell factor (SCF) and/or 50 ng/ml mouse Thrombopoietin (TPO) (both from R&D Systems, Minneapolis, MN), as previously described [1].
3. Results
To date, reports on reference genes have examined threshold cycles (Ct) determined by real-time RT-PCR using equal amounts of total RNA, thereby comparing the expression levels of these reference genes between samples [12], [13], [14]. However, the possibility that an assay based on the quantity of the total RNA in a sample could incorrectly reflect changes in the expression of that RNA at the cellular level cannot be excluded, particularly when the amount of total RNA per cell differs between samples. Therefore, because the results of such pooled RT-PCR assays seem to be dependent on the changes of the amount of total RNA in cells, we focused on single-cell RT-PCR as a tool to accurately examine changes in RNA expression at the cellular level.
Initially, we clonally sorted fresh CD150+CD34−KSL cells into the wells of a 96-well PCR plate and subsequently examined the expression of frequently used reference genes, Actb, B2m, Gapdh and Hprt, using real-time RT-PCR (Fig. 1A and B). The assays showed that B2m was detected in all uncultured, fresh HSCs (Fig. 2A). However, unexpectedly, the expression of Actb, Gapdh and Hprt was undetectable in approximately 15%, 40% and 80% of HSCs, respectively (Fig. 2A). We further examined the expression of these genes by real-time RT-PCR using alternative primer and probe sets, but obtained similar results (Fig. 2A). This indicated that almost all fresh, uncultured HSCs consistently express B2m but not the other 3 commonly used reference genes. However, these 3 genes are essential factors for the maintenance of cell function [15], [16], [17], suggesting the possibility that these 3 genes were expressed but the sensitivity of our single-cell gene expression assay was insufficient to detect the transcripts. Therefore, to confirm the validity of this single-cell PCR system, we compared the expression levels of these 3 reference genes between templates prepared from 10,000 cells and single-cell samples by normalizing to the Ct values of B2m. Consequently, the bulk and the single-cell samples showed similar mean expression values for each gene (Fig. 2B–D). These results indicated that the scale-down necessary for the single-cell system had little influence on the detection sensitivity for the expression of these 3 reference genes, which strongly suggested that our single-cell RT-PCR system is comparable to RT-PCR using bulk samples in terms of detection sensitivity for gene expression. Therefore, our single-cell PCR system likely reflected the heterogeneous expression pattern of Actb, Gapdh and Hprt in fresh HSCs.
Fig. 2.
B2m expression could be stably detected in uncultured, fresh HSCs. The expression of the 4 indicated housekeeping genes was examined using single-cell RT-PCR in fresh HSCs (CD150+CD34−KSL cells). (A) Frequency of HSCs in which the expression of the indicated genes could be detected. The numbers in parentheses represent cells with positive signal/examined cells in an experiment; 3 independent experiments were performed. To investigate the influence of scaling down to the single-cell level, the expression levels of Actb (B), Gapdh (C) and Hprt (D) were compared between templates prepared from either 10,000 cells or single cells. The graphs represent the mRNA expression levels of indicated genes that were normalized to the Ct values of B2m. Data are shown as mean ± S.D. (single cell: n = 159; 10,000 cells: n = 3).
Next, we examined whether ex vivo stimulation affects the expression of these reference genes using single-cell RT-PCR. Fresh HSCs were cultured for 5 days under 3 different conditions in medium containing SCF, TPO, or both. After the culture, CD48−KSL cells, a cultured HSC-enriched population [18], were clonally sorted and subjected to real-time RT-PCR at the single-cell level (Fig. 3A). All 3 culture conditions led to an increased frequency of cells expressing Actb, Gapdh or Hprt, and the expression of B2m was detected in almost all the HSCs under the any conditions (Fig. 3B). Moreover, using the results of single-cell real-time RT-PCR assays, we calculated the expression values of the 4 examined reference genes in individual cells based on the Ct values of control ERCC RNA. Although the expression levels of each of these genes in HSCs increased regardless of culture condition, particularly in the presence of SCF and TPO, the change in B2m expression is likely to lower than any other genes (Fig. 3C). In order to quantitatively confirm this result, we compared the mean expression values between fresh and cultured HSCs. Culturing HSCs in the presence of SCF and TPO led to a more than 4-fold increase in the expression levels of Actb, Gapdh and Hprt compared with fresh, uncultured cells, whereas the expression of B2m increased approximately 1.5-fold in these conditions (Fig. 4). Furthermore, following culture under other conditions, the expression level of B2m in HSCs exhibited the least change out of the 4 reference genes (Fig. 4). Taken together, these data indicated that the expression of B2m in HSCs is relatively stable compared with the 3 other reference genes, suggesting that B2m is the most appropriate reference gene in HSCs under the examined conditions.
Fig. 3.
B2m is stably expressed in HSCs, even after ex vivo culture. (A) CD150+CD34−KSL cells were cultured for 5 days under 3 different conditions in the presence of SCF and/or TPO. After the culture, CD48−KSL cells, referred to as the cultured HSC population [18], were clonally sorted and subsequently subjected to single-cell gene expression analysis. (B) Frequency of cells in which the expression of the indicated genes could be detected before and after culture. The numbers in parentheses represent cells that had positive signals/examined cells under each condition. (C) Heat maps showing the expression values of the indicated genes in individual HSCs before and after the culture under the 3 different conditions. The expression levels of these genes were normalized to the Ct values of ERCC-00130, an exogenously added control RNA. (Fresh: n = 159, SCF + TPO: n = 148, SCF: n = 163, TPO: n = 127).
Fig. 4.
The expression level of B2m in HSCs is relatively independent of the influence of ex vivo culture. Using the results of single-cell real-time RT-PCR, changes in the expression levels of the indicated genes in HSCs were examined after ex vivo culture. The expression levels of these genes in individual single-cell samples were normalized to the Ct values of ERCC-00130, an external control RNA. The graphs show relative expression values of Actb (A), B2m (B), Gapdh (C) and Hprt (D). Data are shown as mean ± S.D. (**p < 0.05, Fresh: n = 159, SCF + TPO: n = 148, SCF: n = 163, TPO: n = 127).
4. Discussion
In this study, we demonstrated that B2m expression was relatively constant in mouse HSCs, even under 3 different culture conditions (Fig. 4B). In addition, B2m expression was detected at the single-cell level in almost all HSCs (Figs. 2A and 3B). Therefore, our results suggest that B2m is a candidate reference gene for RT-PCR analyses of mouse HSCs and that normalizing RT-PCR results to B2m expression accurately reflects changes in the expression pattern of target genes in HSCs. In addition to functioning as a reference gene for relative quantification in real-time RT-PCR, B2m may be useful as a marker to confirm the presence of cells in single-cell RT-PCR assays using HSCs. B2m is a component of major histocompatibility complex (MHC) class I molecules. MHC class I molecules not only play a key role in protection against virus-infected cells or tumor cells by presenting their antigens to killer T cells but also function as markers for self-recognition to prevent attacks by autologous killer T cells [19], [20], [21]. Our observation that B2m expression was uniformly stable in individual HSCs is consistent with these functions of MHC class I molecules in immune defense. However, the possibility cannot be excluded that B2m expression is more significantly altered under conditions that were not tested in this study. In fact, treatment with inflammatory cytokines (e.g., interferon-γ) reportedly enhances B2m expression in macrophages [22]. Therefore, it may be necessary to compare B2m expression levels between samples before examining the expression levels of target genes by relative quantification using real-time RT-PCR, particularly when the influence of inflammatory cytokines is predicted.
Typically, to compare reference gene expression levels between samples, an equivalent amount of total RNA from each sample is subjected to real-time RT-PCR and the samples' Ct values are compared. However, this method may not accurately reflect the expression levels of the reference genes, particularly if the amount of total RNA per cell is significantly different between samples. By contrast, our method, which is based on single-cell RT-PCR, always enables the accurate comparison of reference gene expression levels between samples with the same number of cells. Moreover, this method can reveal changes in expression level per cell, but not the proportion of a particular mRNA within the total RNA. There are no reports of this type of clonal analysis being employed to compare the expression levels of reference genes between samples of interest. In addition, our system utilized exogenous RNA spiked into each single-cell sample to more strictly control for template quality and to more accurately estimate reference gene expression as a standard control. In conclusion, we provide a novel method based on single-cell analysis to select an appropriate reference gene for RT-PCR based on changes in its per-cell gene expression. In addition, this assay can be used to calculate fold changes in the expression levels of reference genes between samples, resulting in a correction factor for accurately comparing target gene expression between the samples.
Although our single-cell system shows equal sensitivity to RT-PCR using bulk samples as evidenced by similar expression values for Actb, Gapdh and Hprt between single cell and bulk samples (Fig. 2B–D), expression of these 3 reference genes could not be detected in all of the fresh HSCs at the single-cell level (Fig. 2A). These results indicate that our single-cell assay reveals the heterogeneity of the expression levels of these reference genes in the HSC population, particularly in an uncultured state. However, whether cells without positive signals in real-time RT-PCR actually lack expression of these genes remains controversial. The possibility that our method is insufficient to detect very low mRNA levels of these genes cannot be excluded. However, because our assay is based on the SMART method, which uses oligo-d(T) primers, it may be sensitive to the secondary structure of mRNA. This method is useful for preparing cDNAs with adapters at both the 5′ and the 3′ ends, although the addition of an adapter at the 3′ end of the cDNA is not induced by the template switch unless the RT reaction completely finishes at the 5′ end of the RNA fragment. Therefore, the secondary structure of mRNA may negatively influence pre-amplification of the cDNA by PCR with the universal primer because the adapter at the 3′ end of the cDNA may not become incorporated due to an incomplete RT reaction, resulting in decreased signal in real-time PCR. Indeed, B2m has the shortest length of the 4 genes analyzed (B2m: 858 bp, Actb: 1889 bp, Gapdh: 1296 bp, Hprt: 1349 bp), and analyzing the secondary structures of these 4 mRNAs using CentroidFold (http://www.ncrna.org/centroidfold/) reveals that B2m has a simpler secondary structure compared with the other 3 mRNAs (data not shown). Thus, these findings support the idea that the problematic secondary structure of RNA may have a negative influence on the SMART reaction method using oligo-d(T) primers. Therefore, other methods that are more resistant to the influence of RNA secondary structure (e.g., RT reaction and subsequent pre-amplification using gene-specific primers) may improve the detection of these 3 genes at the single-cell level.
Taken together, these data demonstrate that the expression of B2m in HSCs is stable and constant in both uncultured cells and cells under several culture conditions, as detected via single-cell RT-PCR. Therefore, our results suggest that B2m is an appropriate reference gene for accurate estimation of gene expression level by RT-PCR in mouse HSCs. Furthermore, we propose a novel method for examining the expression levels of reference genes using the single-cell RT-PCR system, which can be adapted for other cell types in addition to HSCs. In conclusion, this study provides a novel method for the selection of reference genes to correctly estimate the expression levels of target genes and provides evidence that B2m is an appropriate reference gene in mouse HSCs.
Acknowledgments
The authors thank Dr. Y. Shiratsuchi and Dr. J. Ishihara for their kind assistance in providing cells and all of the members of our research group for providing discussions. This study was supported by the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems “Cell Sheet Tissue Engineering Center (CSTEC)” from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Contributor Information
Terumasa Umemoto, Email: umemoto.teruamsa@twmu.ac.jp.
Masayuki Yamato, Email: yamato.masayuki@twmu.ac.jp.
References
- 1.Umemoto T., Yamato M., Ishihara J., Shiratsuchi Y., Utsumi M., Morita Y. Integrin-alphavbeta3 regulates thrombopoietin-mediated maintenance of hematopoietic stem cells. Blood. 2012;119:83–94. doi: 10.1182/blood-2011-02-335430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkatraman A., He X.C., Thorvaldsen J.L., Sugimura R., Perry J.M., Tao F. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin T., Li L. The stem cell niches in bone. J Clin Investig. 2006;116:1195–1201. doi: 10.1172/JCI28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai F., Hirao A., Ohmura M., Sato H., Matsuoka S., Takubo K. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Yoshihara H., Arai F., Hosokawa K., Hagiwara T., Takubo K., Nakamura Y. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1:685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Qian H., Buza-Vidas N., Hyland C.D., Jensen C.T., Antonchuk J., Mansson R. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Ema H., Takano H., Sudo K., Nakauchi H. In vitro self-renewal division of hematopoietic stem cells. J Exp Med. 2000;192:1281–1288. doi: 10.1084/jem.192.9.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C.C., Lodish H.F. Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood. 2005;105:4314–4320. doi: 10.1182/blood-2004-11-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thellin O., Zorzi W., Lakaye B., De Borman B., Coumans B., Hennen G. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 10.Umemoto T., Yamato M., Shiratsuchi Y., Terasawa M., Yang J., Nishida K. CD61 enriches long-term repopulating hematopoietic stem cells. Biochem Biophys Res Commun. 2008;365:176–182. doi: 10.1016/j.bbrc.2007.10.168. [DOI] [PubMed] [Google Scholar]
- 11.Umemoto T., Yamato M., Shiratsuchi Y., Terasawa M., Yang J., Nishida K. Expression of integrin 3 is correlated to the properties of quiescent hemopoietic stem cells possessing the side population phenotype. J Immunol. 2006;177:7733–7739. doi: 10.4049/jimmunol.177.11.7733. [DOI] [PubMed] [Google Scholar]
- 12.Radonic A., Thulke S., Mackay I.M., Landt O., Siegert W., Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 13.Wan H., Zhao Z., Qian C., Sui Y., Malik A.A., Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399:257–261. doi: 10.1016/j.ab.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Tricarico C., Pinzani P., Bianchi S., Paglierani M., Distante V., Pazzagli M. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 15.Bunnell T.M., Burbach B.J., Shimizu Y., Ervasti J.M. beta-Actin specifically controls cell growth, migration, and the G-actin pool. Mol Biol Cell. 2011;22:4047–4058. doi: 10.1091/mbc.E11-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sculley D.G., Dawson P.A., Emmerson B.T., Gordon R.B. A review of the molecular basis of hypoxanthine-guanine phosphoribosyltransferase (HPRT) deficiency. Hum Genet. 1992;90:195–207. doi: 10.1007/BF00220062. [DOI] [PubMed] [Google Scholar]
- 17.Sirover M.A. Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J Cell Biochem. 1997;66:133–140. [PubMed] [Google Scholar]
- 18.Noda S., Horiguchi K., Ichikawa H., Miyoshi H. Repopulating activity of ex vivo-expanded murine hematopoietic stem cells resides in the CD48-c-Kit+Sca-1+lineage marker- cell population. Stem Cells. 2008;26:646–655. doi: 10.1634/stemcells.2007-0623. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka T., Ebata T., Tajima A., Kinoshita K., Okumura K., Yagita H. Beta2-microglobulin required for cell surface expression of blastocyst MHC. Biochem Biophys Res Commun. 2005;332:311–317. doi: 10.1016/j.bbrc.2005.03.249. [DOI] [PubMed] [Google Scholar]
- 20.Cogen A.L., Moore T.A. Beta2-microglobulin-dependent bacterial clearance and survival during murine Klebsiella pneumoniae bacteremia. Infect Immun. 2009;77:360–366. doi: 10.1128/IAI.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorfman J.R., Zerrahn J., Coles M.C., Raulet D.H. The basis for self-tolerance of natural killer cells in beta2-microglobulin- and TAP-1- mice. J Immunol (Baltim Md – 1950) 1997;159:5219–5225. [PubMed] [Google Scholar]
- 22.Keskinen P., Ronni T., Matikainen S., Lehtonen A., Julkunen I. Regulation of HLA class I and II expression by interferons and influenza A virus in human peripheral blood mononuclear cells. Immunology. 1997;91:421–429. doi: 10.1046/j.1365-2567.1997.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]