Abstract
The potential applications of human embryonic stem cells (hESCs) in regenerative medicine and developmental research have made stem cell biology one of the most fascinating and rapidly expanding fields of biomedicine. The first clinical trial of hESCs in humans has begun, and the field of stem cell therapy has just entered a new era. Here, we report seven hESC lines (SEES-1, -2, -3, -4, -5, -6, and -7). Four of them were derived and maintained on irradiated human mesenchymal stem cells (hMSCs) grown in xenogeneic-free defined media and substrate. Xenogeneic-free hMSCs isolated from the subcutaneous tissue of extra fingers from individuals with polydactyly showed appropriate potentials as feeder layers in the pluripotency and growth of hESCs. In this report, we describe a comprehensive characterization of these newly derived SEES cell lines. In addition, we developed a scalable culture system for hESCs having high biological safety by using gamma-irradiated serum replacement and pharmaceutical-grade recombinant basic fibroblast growth factor (bFGF, also known as trafermin). This is first report describing the maintenance of hESC pluripotency using pharmaceutical-grade human recombinant bFGF (trafermin) and gamma-irradiated serum replacement. Our defined medium system provides a path to scalability in Good Manufacturing Practice (GMP) settings for the generation of clinically relevant cell types from pluripotent cells for therapeutic applications.
Keywords: Human embryonic stem cells, Xenogeneic-free medium, Stem cell expansion, Human feeder layer, Mesenchymal stem cells, Gamma irradiation
1. Introduction
Human pluripotent stem cells such as human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) have remarkable developmental plasticity and therefore possess great potential for drug screening and the development of cellular models to study diseases. hESCs also have potential applications in regenerative medicine as source for cell-based therapy. Since the initial derivation of pluripotent hESCs by Thomson and colleagues in 1998 [1], more than 1000 hESC lines have been established and are now utilized in basic and clinical research worldwide [2]. Several clinical trials using hESC lines approved by the United States Food and Drug Administration (FDA) are currently underway. However, further research is still needed to facilitate the development of safer, reproducible, and scalable culture systems for the generation of hESCs for clinical and industrial purposes.
A key consideration with a cell therapy-compliant culture system, including safe expansion of hESCs, is the choice of culture media, matrix (including feeder layers), and passage procedures. Early hESC culture systems typically include the use of mouse embryonic fibroblast (MEF) feeders or medium conditioned on MEFs in the presence of serum or serum substitutes such as knockout serum replacement [1], [3], [4], [5], [6]. Human feeders and serum have also been used for hESC culture [7]; however, the use of serum or serum replacement, which contains undefined xenogeneic factors, is still an issue for potential clinical applications of these cells. Transplantation of human cells exposed to animal-derived products can potentially transfer immunogenic sugars such as N-glycolylneuraminic acid (Neu5Gc) into the human body and may trigger chronic inflammation and immune reactions [8], [9], [10]. Recent studies have identified multiple factors that play a role in sustaining pluripotency and have led to the development of several defined medium systems for hESC culture and derivation [11], [12], [13]. Because no reports have described the successful establishment of hESCs in xenogeneic-free (XF) medium, it seems realistic to assume that the most reliable strategies for the establishment of clinical-grade hESCs include the use of a human feeder layer in XF medium, followed by expansion in a feeder-free culture system [14]. There are two strategies for developing hESC culture systems suitable for generation of clinically applicable cells. One approach is to establish XF, defined culture systems especially for future applications, and the other approach is to develop safe conditions for hESC culture systems while continuing to use xenogeneic products. In fact, ongoing clinical trials using hESCs employ conventional hESC culture systems, including mouse feeder layers, under certain conditions [15].
In this study, we developed a novel derivation/cultivation system of hESCs for potential application in translational and clinical research. To completely avoid exposure of hESCs to culture media with animal products, we established an XF culture system containing XF, defined culture medium with inactivated human mesenchymal stem cell (hMSC) feeders. The XF culture system with the hMSC feeder layers proved stable maintenance of self-renewal and pluripotency of newly established hESCs for more than 50 passages. Here, we report the successful derivation of four new hESC lines in this novel XF culture system. Furthermore, we evaluated replacement of a conventional hESC culture system with high-dose gamma-irradiated serum and pharmaceutical-grade recombinant basic fibroblast growth factor (bFGF) to provide insights into the development of a reproducible hESC cultivation system. We examined the pluripotency of SEES cell lines cultivated under the modified conventional culture system. hESCs were able to proliferate under the modified conventional conditions while retaining their pluripotency. Thus, our data provided clinically relevant alternative platforms for clinically and industrially applicable hESC culture systems.
2. Results
Since the first report describing the generation of hESCs by Thomson and coworkers [1], more than 1000 different hESC lines have been established worldwide [2]. Nevertheless, there is still a need for establishment of new hESC lines, particularly from certain HLA types and ethnic groups [16]. In this study, we derived seven new hESC lines using different culture conditions and developed a new hESC culture system using gamma-irradiated KO-SR and pharmaceutical-grade recombinant human bFGF (trafermin). Since our organization name is “Sei-iku” in Japanese, the hESC lines were designated as “SEES (Sei-iku embryonic stem)” in combination with numbers.
2.1. Stable derivation and culture of hESCs under serum replacement conditions
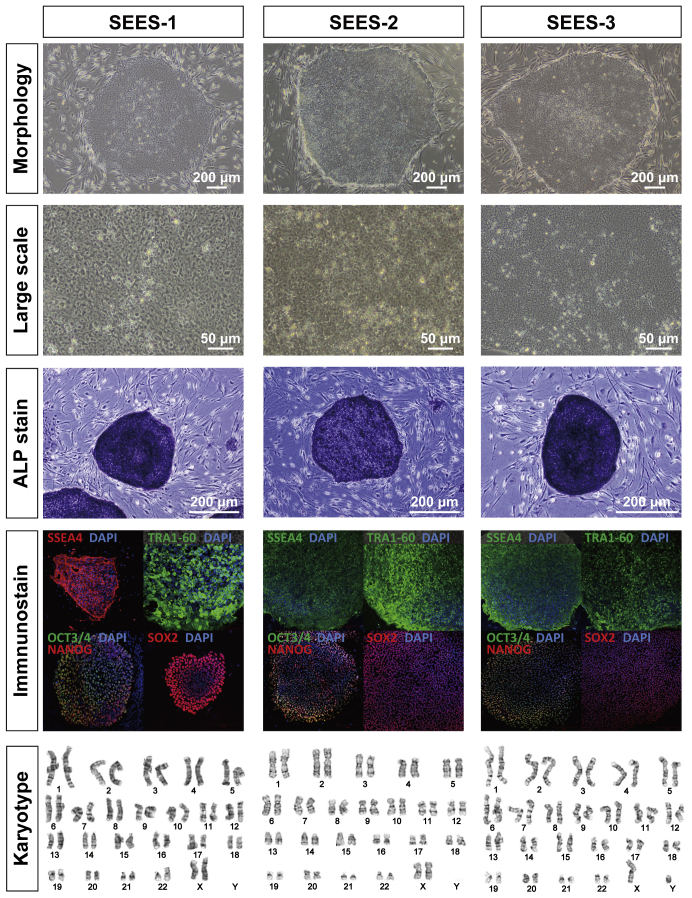
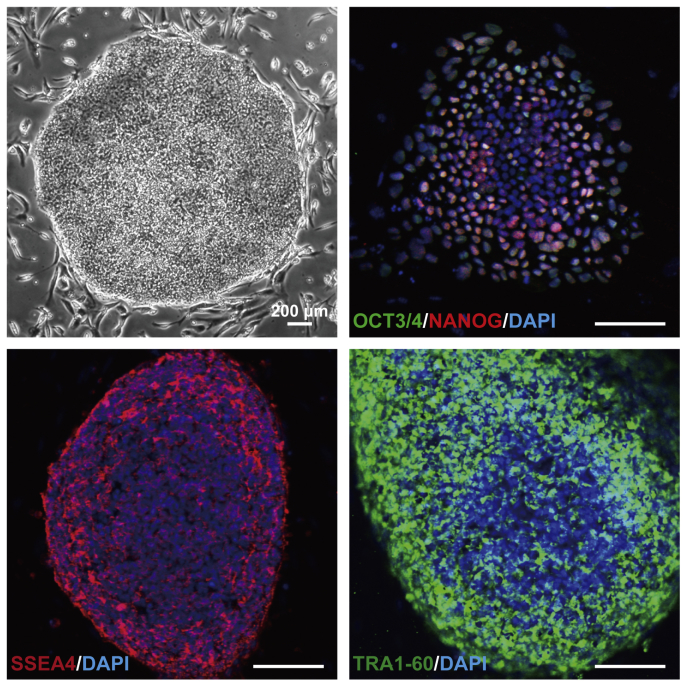
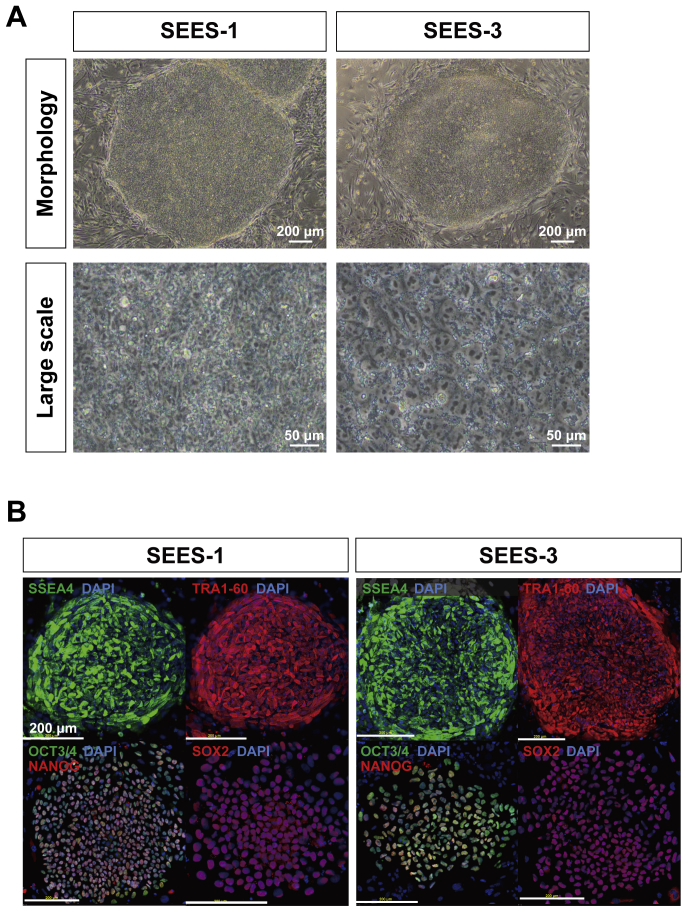
Out of five blastocysts, three hESC lines (SEES-1, SEES-2, and SEES-3) were derived from ICMs on MEF layers using the immunosurgery technique; conventional medium conditions, composed of HUES medium without human plasma protein fraction [4] (Fig. 1A), were used in this derivation. The pluripotency of each of these three SEES cell lines was confirmed by observation of typical morphology and positive immunostaining of stemness markers and differentiated derivatives comprising three embryonic germ layers (Supplemental Figs. 1 and 2). G-banding showed that SEES-1, SEES-2, and SEES-3 cells had normal karyotypes, i.e., 46,XX, 46,XX, and 46,XY, respectively (Supplemental Fig. 1).
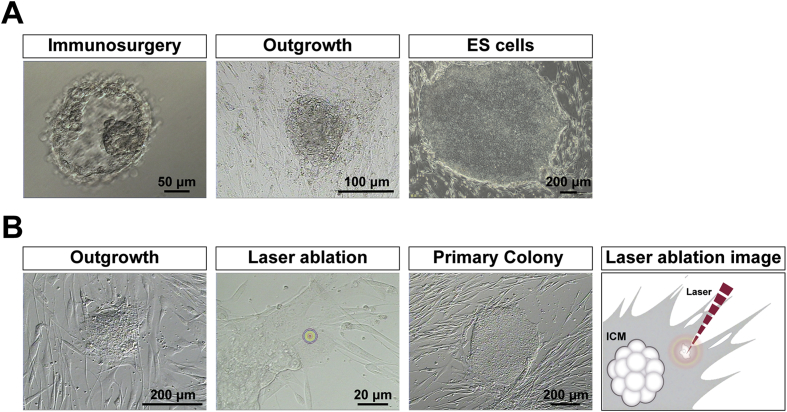
Fig. 1.
Derivation of human ES cells by immunosurgery and laser ablation A) The intact inner cell mass (ICM) was isolated from blastocysts by immunosurgery. Cells of the trophectoderm are destroyed by brief exposure to antibodies directed against human cells in tandem with complement activity. Attachment and outgrowth of the ICM grew into an ESC (SEES-1) colony. B) The intact ICM was isolated by laser ablation without any animal products. Outgrowth of the blastocyst grown on human MSC feeder layers. Trophectoderm cells were targeted with multiple pulses of laser ablation. Typical morphology of ESC colonies was readily visible at high magnification. The image shows a schematic of ICM isolation by laser ablation, which is commonly used in artificial reproductive technology (ART).
2.2. Isolation and characterization of the XF hMSC feeder layer
Primary hMSCs isolated from subcutaneous tissue samples of juvenile donors undergoing surgical procedures for polydactyly were derived and expanded in a complete XF media system that contained an XF supplement, LipoMax, on a humanized substrate, CELLstart (Fig. 2A). Proliferation analysis was performed, and cell morphology was observed by light microscopy to confirm the proliferation assay results. Our data showed that cells grown in MSC-XF medium exhibited high proliferation rates and were self-renewing (Fig. 2B). Flow cytometric characterization was performed to compare surface marker expression characteristics of the cells expanded in the MSC-XF medium. Positive CD29, CD44, and CD90 expression was observed, while CD117 and CD133 were not detected (Fig. 2C). Cells differentiated into both adipogenic and osteogenic cells (Fig. 2D and E). Taken together, our data demonstrated that cells isolated from polydactyly tissues could be classified as MSCs by flow cytometry analysis of their surface markers and evaluation of their multilineage differentiation potential.
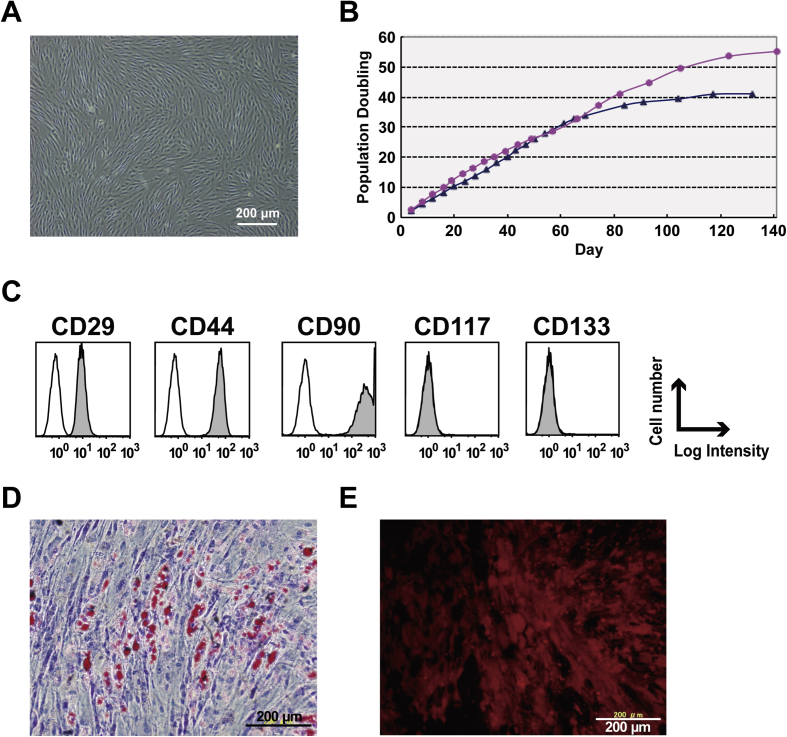
Fig. 2.
Isolation and characterization of the xenogeneic-free hMSC feeder layer A) Primary human mesenchymal cells isolated from subcutaneous tissue of the polydactyly were expanded in xenogeneic-free media. B) Cells grew well over PD 30 under xenogeneic-free conditions. Two differentially isolated cell lines (red circle and blue triangle) showed similar cellular proliferation characteristics. C) Mesenchymal markers such as CD29, CD44, and CD90 were observed. D,E) The cells differentiated into adipogenic and osteogenic cells.
2.3. Derivation of new hESC lines in XF culture conditions
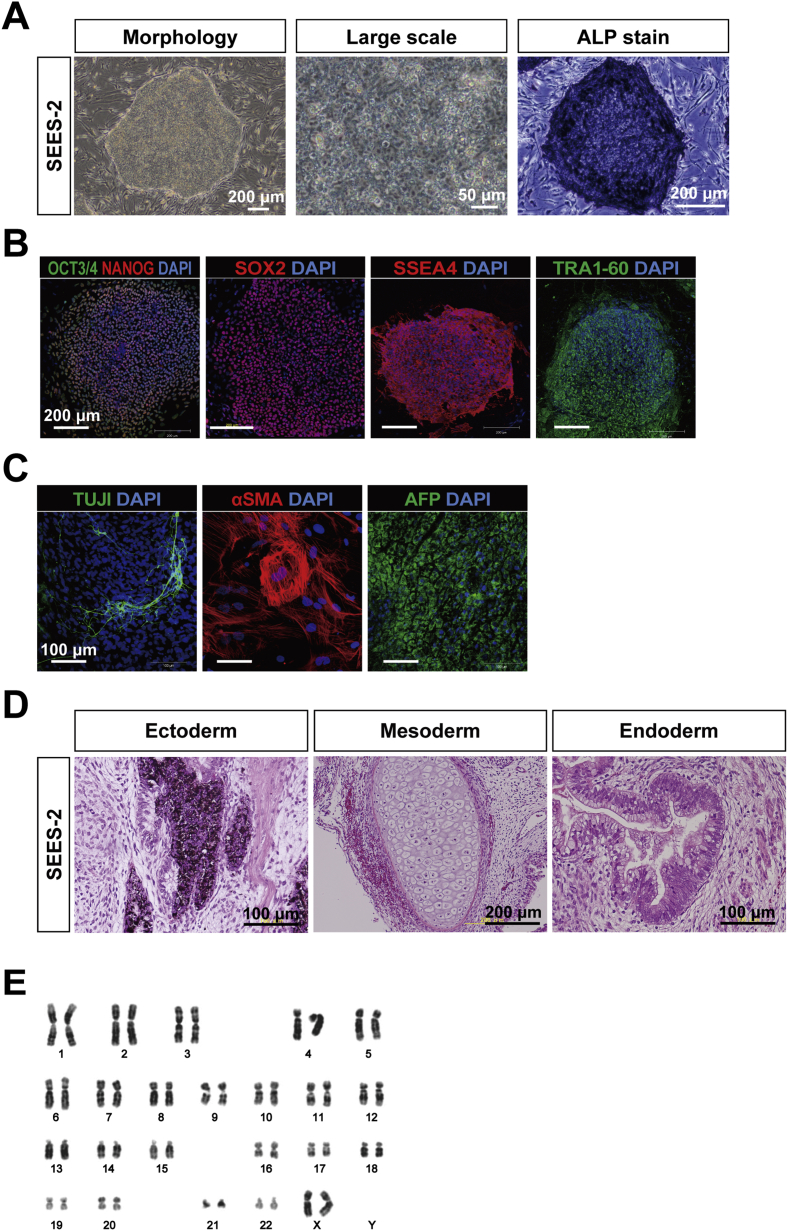
To derive an XF hESC line, we first developed an XF culture system using XF hMSC feeder layers. We optimized an XF hESC medium composed of KO-DMEM supplemented with KO-SR XF, amino acids, vitamin C, bFGF, and two other growth factors (IGF1 and heregulin) [17]. All components of the medium were synthetic, recombinant, or of human origin. As a preliminary step, we confirmed the stable cultivation of SEES-1, SEES-2, and SEES-3 cells on XF hMSC feeder layers in XF hESC medium (data not shown). Twelve frozen human embryos were thawed and cultured to the blastocyst stage, and of these, eight blastocysts were used to derive XF hESC lines. Intact blastocysts, without immunosurgery, were plated onto irradiated XF hMSC layers in XF hESC medium. ICM isolation was carried out by exposing TE cells to cell-lethal laser pulses from an XYClone laser system (Fig. 1B). Finally, four XF hESC lines (SEES-4, SEES-5, SEES-6, and SEES-7) were generated and maintained stably (Fig. 3). All of these newly derived XF hESC lines were karyotyped regularly and exhibited a normal diploid karyotype, i.e., 46,XX, 46,XX, 46,XY, and 46,XX, respectively (Fig. 3). To assess the expression of a subset of stemness markers, these cell lines were analyzed by immunocytochemical staining; all XF SEES cell lines expressed the hESC markers NANOG, OCT3/4, SOX2, SSEA4, and TRA1-60 (Fig. 4). We also generated human iPSCs from XF Yub cells by over-expressing three reprogramming factors under hXF culture conditions using XF hESC medium and XF hMSC feeder layers. The XF human iPSCs were stably maintained and expressed the hESC markers such as OCT3/4, NANOG, SSEA4 and TRA1-60 (Supplemental Fig. 3).
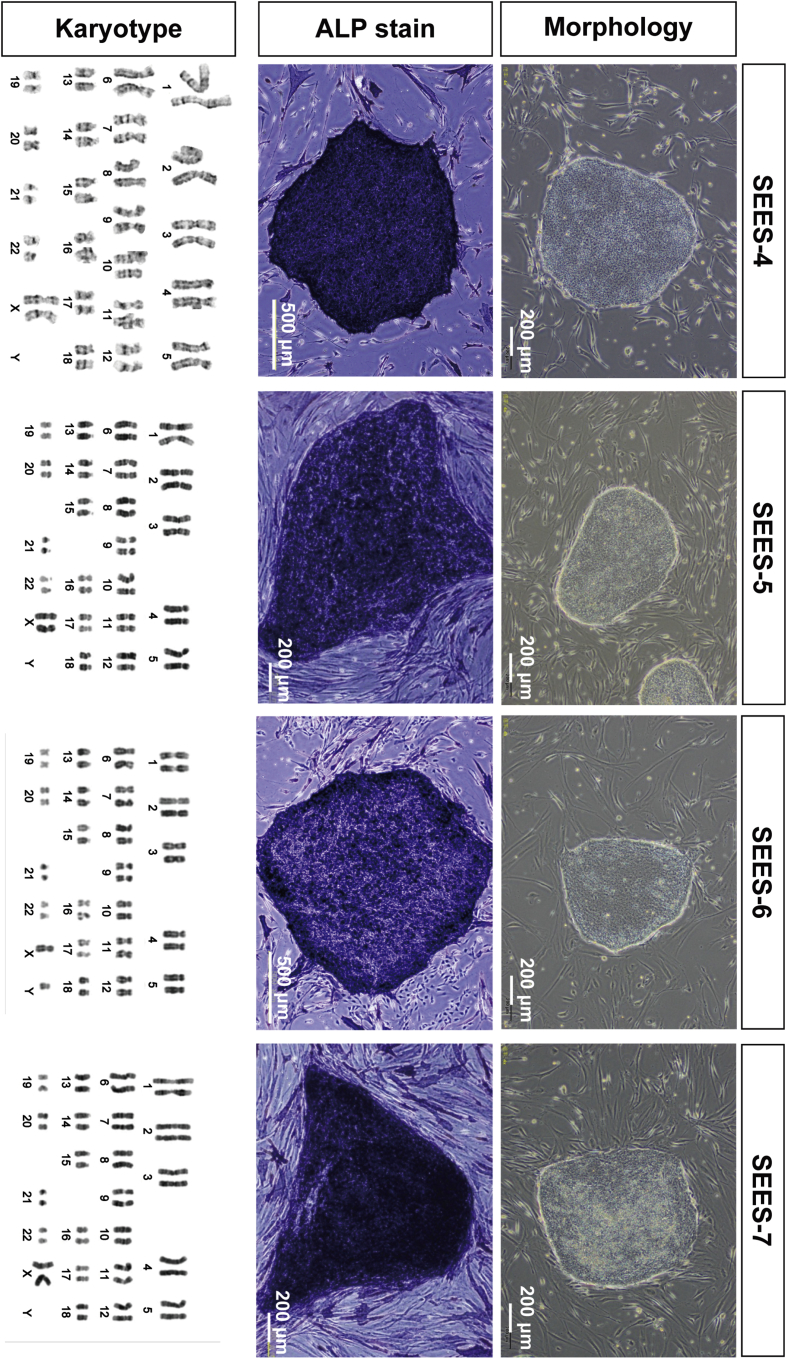
Fig. 3.
Derivation of xenogeneic-free hESCs on the hMSC feeder layer Under completely xenogeneic-free conditions, four hESC lines were derived from the inactivated hMSC feeder layer using the laser ablation system. Typical ESC morphology was readily visible. Alkaline phosphatase (ALP) activity was detected in each SEES cell line. Chromosome analysis of SEES-4, SEES-5, SEES-6, and SEES-7 cells showed normal karyotypes: 46,XX, 46,XX, 46,XY, and 46,XX, respectively.
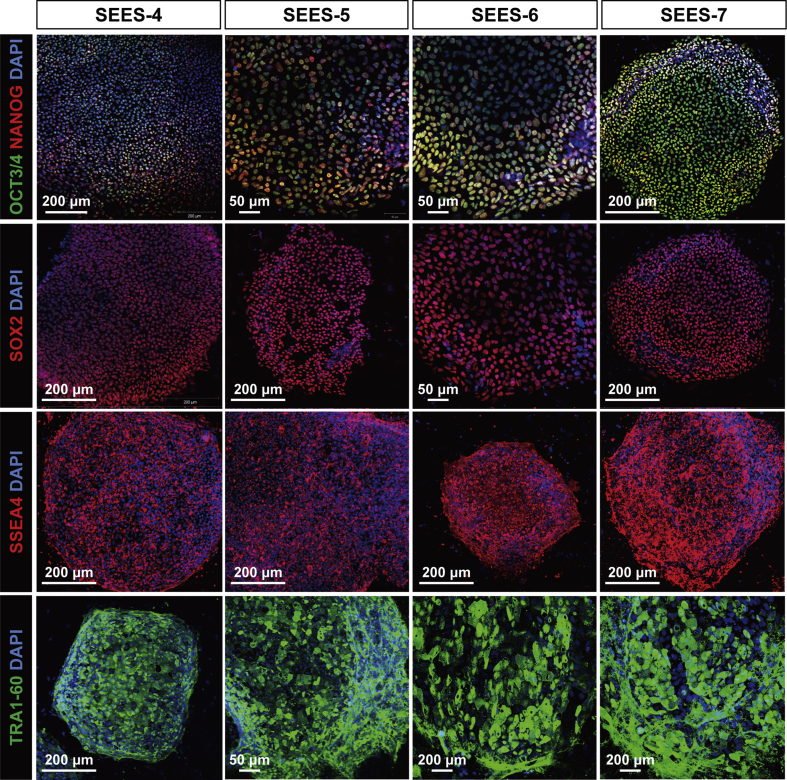
Fig. 4.
Pluripotent marker expression of xenogeneic-free SEES cell lines In the undifferentiated state, xenogeneic-free SEES cell lines expressed markers characteristic of pluripotent hESCs, including OCT4, NANOG, SOX2, SSEA4, and TRA1-60. SEES-4: scale bars are 200 μm; SEES-5: scale bars are 50 μm in OCT3/4 and TRA1-60 and 200 μm in SOX2 and SSEA4; SEES-6: scale bars are 50 μm in OCT3/4 and SOX2 and 200 μm in SSEA4; SEES-7: scale bars are 200 μm.
2.4. hESCs cultured for prolonged periods in XF culture medium maintained their pluripotency and differentiation characteristics
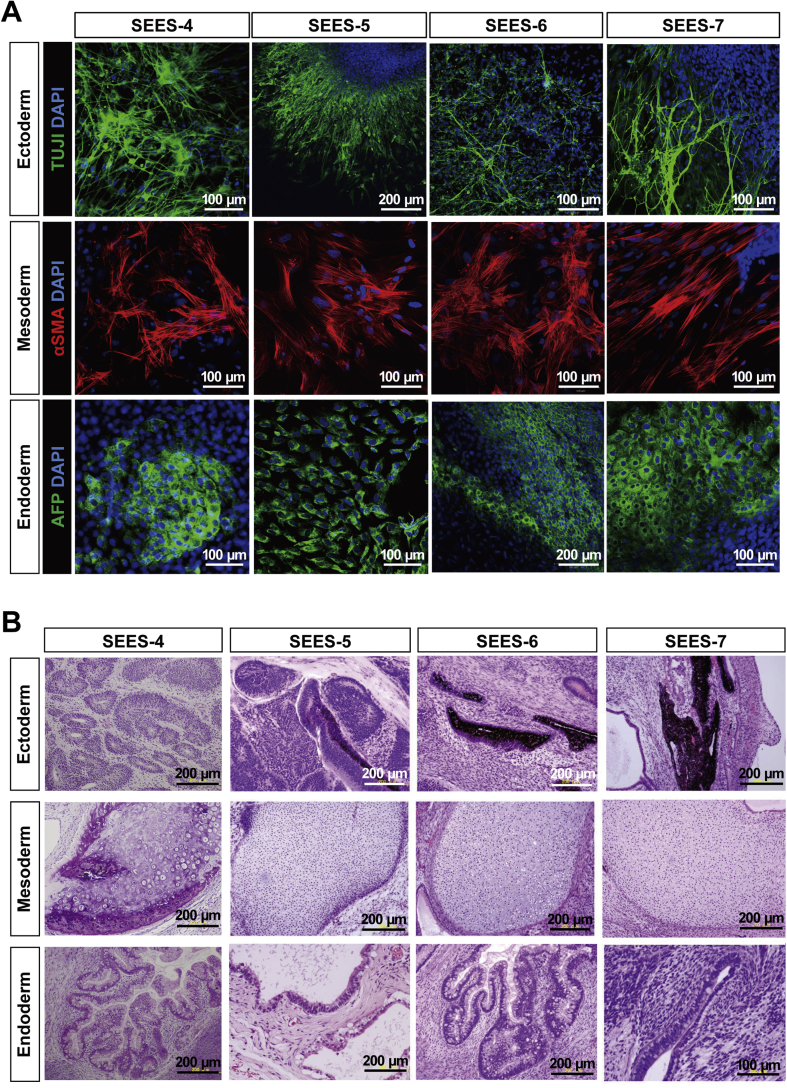
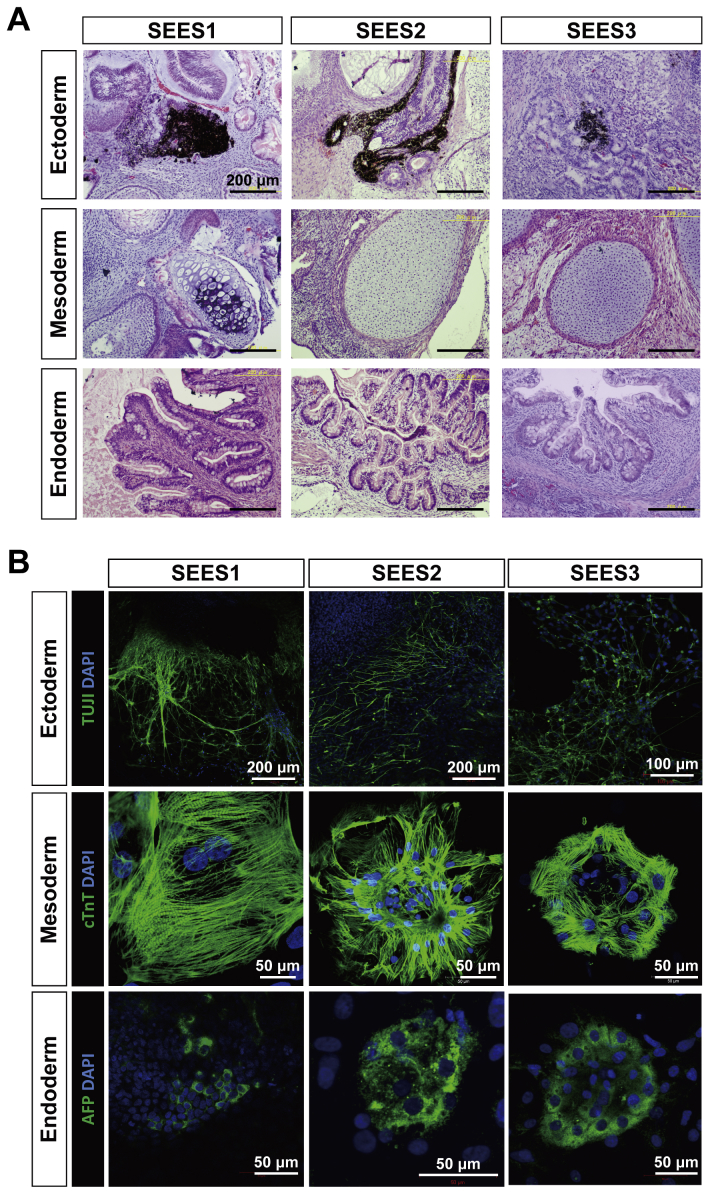
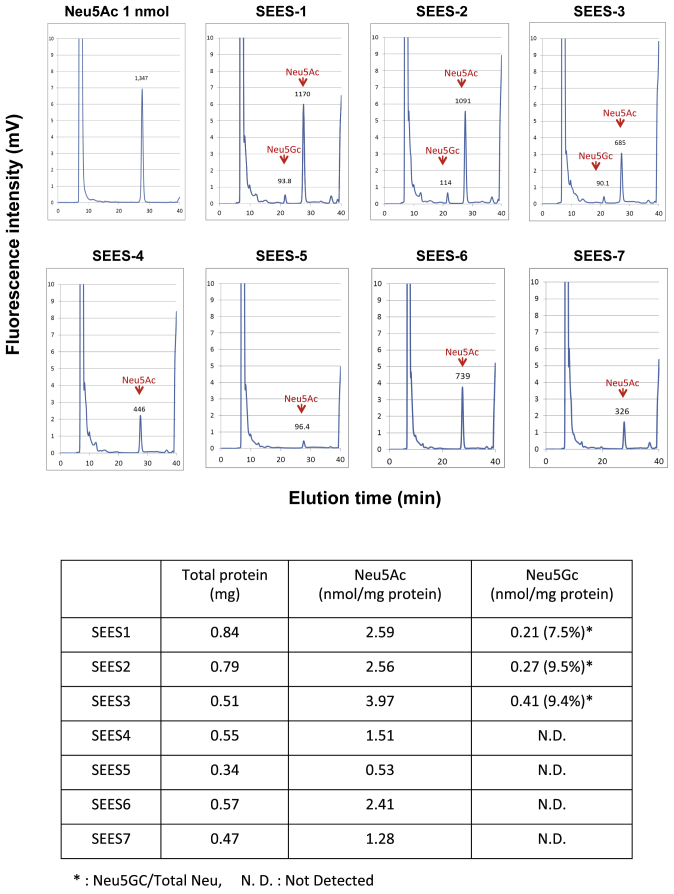
To evaluate whether the newly derived XF SEES cell lines maintained their pluripotency in vitro, we performed EBs assays. EBs differentiated from the cells of SEES-4, SEES-5, SEES-6, and SEES-7 cells expressed markers associated with the three major germ layers: TUJ1 (ectoderm), αSMA (mesoderm), and AFP (endoderm; Fig. 5A). Additionally, in an in vivo pluripotency assay, structures from all three germ layers were detected, including neural tissues and pigmented epithelium (ectoderm), cartilage (mesoderm), and gut epithelial tissues (endoderm; Fig. 5B). Sialic acid was released from seven hESCs by acid hydrolysis and quantified by DMB-HPLC. Neu5Gc levels were increased in SEES-1, SEES-2, and SEES-3 cells, while the four XF SEES cell lines (SEES-4, SEES-5, SEES-6, and SEES-7) either did not express Neu5GC at all or had negligible levels of Neu5GC (Supplemental Fig. 4).
Fig. 5.
Differentiation of three germ layers of xenogeneic-free SEES cell lines A) SEES cells differentiated in vitro via EBs expressed markers of the primary germ layers. Immunohistochemical analyses of markers of the ectoderm (TUJ1), mesoderm (αSMA), and endoderm (AFP) layers are shown. SEES-4: scale bars are 100 μm; SEES-5: scale bars are 200 μm for TUJ1 and 100 μm for αSMA and AFP; SEES-6: scale bars are 100 μm for TUJ1 and αSMA and 200 μm for AFP; SEES-7: scale bars are 100 μm. B) SEES cells differentiated in vivo via teratoma formation. Hematoxylin and eosin staining revealed germ layer derivatives, such as neural tissues, pigmented epithelium (ectoderm), cartilage (mesoderm), and gut epithelial tissues (endoderm). Scale bars are 200 μm.
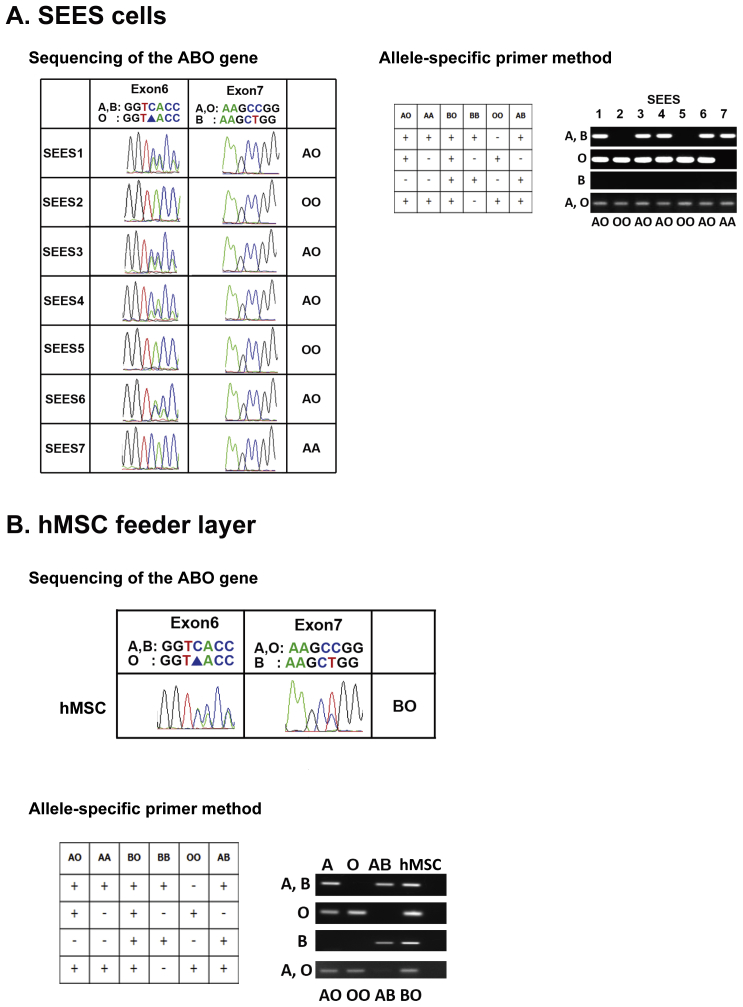
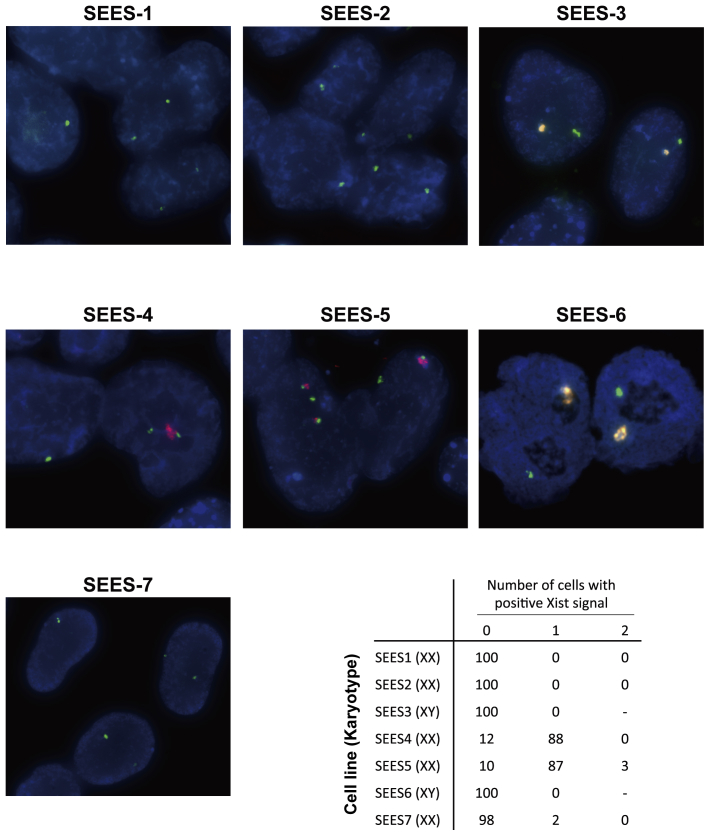
The seven SEES cell lines were further characterized by short tandem repeat (STR) analysis (Supplemental Table 1), HLA-DNA typing (Supplemental Table 2), ABO typing (Supplemental Fig. 5), and cytogenetic X-chromosome inactivation status analysis (Supplemental Fig. 6). The distinct features of SEES cell lines could be observed by STR and HLA profiling. SEES-2 and SEES-5 were blood type OO, which is most suitable for cellular transplantation.
2.5. Stable expansion of SEES cell lines under modified conventional hESC culture conditions
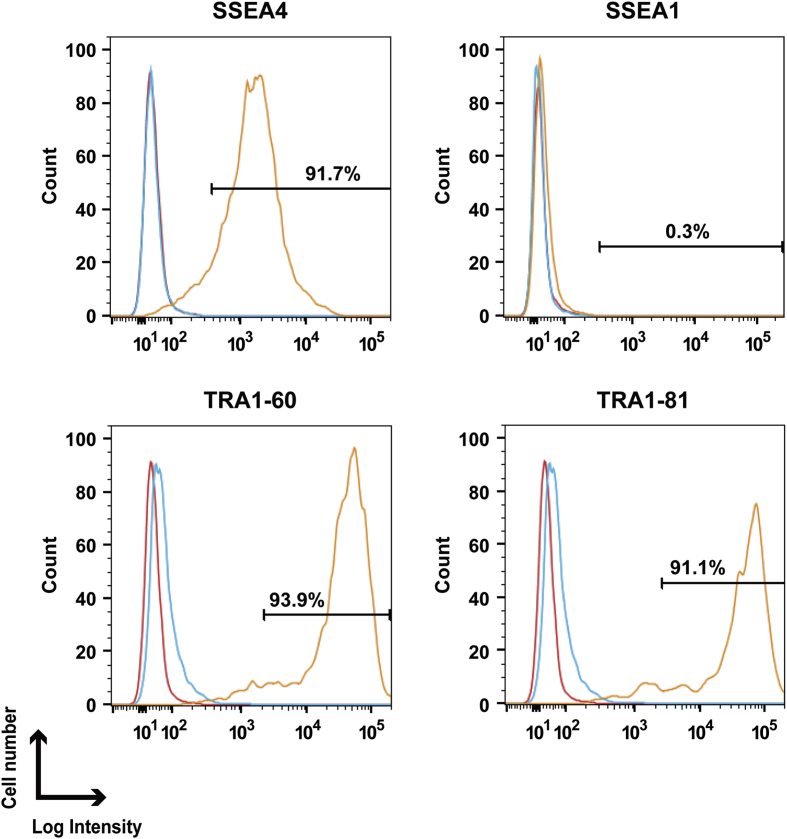
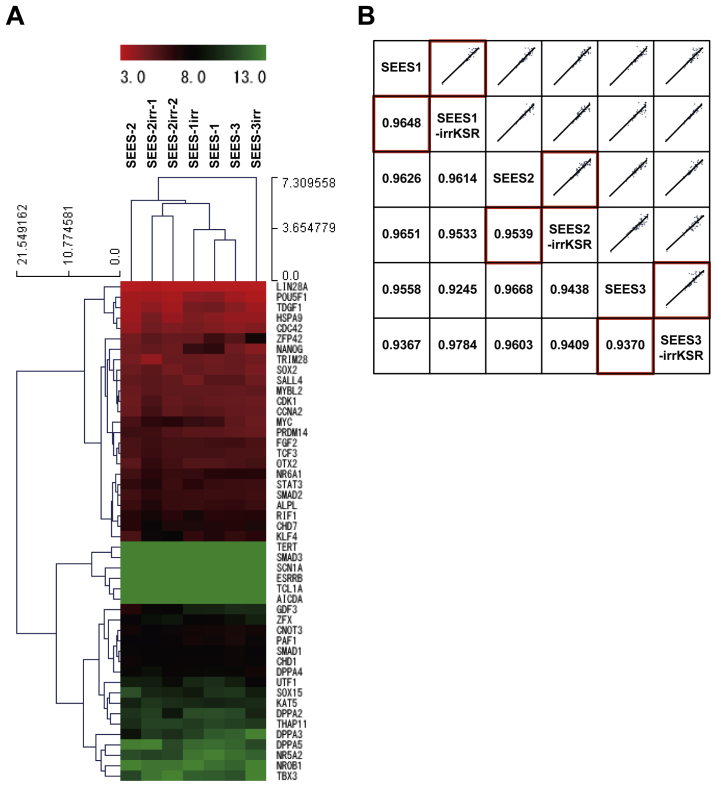
To overcome the shortcomings of conventional approaches, we determined which elements of conventional culture systems were necessary and sufficient for maintaining the pluripotency of hESCs. In our modified conventional hESC culture medium, human recombinant bFGF and KO-SR were replaced by pharmaceutical-grade recombinant human bFGF (trafermin) and high-dose gamma-irradiated KO-SR, respectively. SEES-2 cells were stably maintained on the qualified MEF layer in the modified medium without antibiotics (Fig. 6A). Under these conditions, the colonies expressed multiple pluripotency markers, including OCT3/4, NANOG, SOX2, SSEA4, and TRA1-60, as demonstrated by immunostaining (Fig. 6B). Interestingly, SEES-2 cells could differentiate into derivatives of all three embryonic germ layers in vitro and in vivo. The EB formation assay showed detection of TUJ1, αSMA, and AFP by immunostaining (Fig. 6C). Additionally, differentiation of the three germ layer in vivo was confirmed by teratoma analysis (Fig. 6D), and SEES-2 cells retained normal karyotypes following extensive passaging in culture (Fig. 6E). Under these conditions, cells were able to maintain pluripotency for over 20 passages, as confirmed by positive expression of SSEA4, TRA1-60, and TRA1-80 and absence of SSEA1 expression (Fig. 7). Taken together, our data demonstrated that the modified conventional hESC culture system, based on replacement with high-dose gamma-irradiated KO-SR and trafermin, maintained the pluripotency of hESCs. SEES-1 and SEES-3 also retaining their pluripotency, as shown by morphological analysis and immunostaining (Supplemental Fig. 7). Analysis of the expression of several genes by qRT-PCR showed that hESCs grown under conventional conditions and our modified conditions exhibited highly similar gene expression patterns (Supplemental Fig. 8). Additionally, FISH analysis of SEES cell lines showed that three out of the five SEES cell lines were X-chromosome active.
Fig. 6.
Characterization of the pluripotency of SEES-2 maintained using a modified conventional hESC culture medium SEES-2 cells were stably maintained over 20 passages on the qualified MEF feeder layer in the modified medium, which contained pharmaceutical-grade recombinant human bFGF (trafermin) and high-dose (35-K) gamma-irradiated KO-SR without antibiotics. A) Typical hESC colony morphology was readily visible. ALP activity was detected. B) SEES-2 cells expressed undifferentiated hESC markers, including OCT4, NANOG, SOX2, SSEA4, and TRA1-60. SEES-2 cells could differentiate into three embryonic germ layers in vitro and in vivo. Scale bars are 200 μm. C) SEES cells that had been differentiated in vitro via EBs expressed markers of the primary germ layers, ectoderm (TUJ1), mesoderm (αSMA), and endoderm (AFP). Scale bars are 100 μm. D) Histological analysis of teratomas containing multidifferentiated tissues derived from SEES-2 cells. Pigmented epithelium (ectoderm), cartilage (mesoderm), and gut epithelial tissues (endoderm). E) Chromosomal analysis of SEES-2 cells cultivated through 16 passages using a modified conventional hESC culture medium showed a normal 46,XX karyotype.
Fig. 7.
Expression of pluripotency markers in SEES-2 cells was maintained using a modified conventional hESC culture medium Flow cytometric analysis of hESC-specific marker expression in SEES-2 cells. The isotype control is indicated by the blue line, and the unlabeled sample, which was used as a control, is indicated by the red line. Surface staining is shown by the yellow line for SSEA4, SSEA1, TRA-1-60, and TRA-1-81.
3. Discussion
Traditional methods for isolating and expanding hESCs include using MEFs as a feeder layer and supplementing medium with FBS replacement. Conventional hESC culture systems are widely used for both basic research and clinical trials under appropriate conditions for ensuring safety [15]. Currently, there are at least two options for further development of hESC cultivation systems for use in regenerative medicine; these methods seek to achieve safe culture conditions within a stable, conventional hESC cultivation system. Future potential uses of hESCs in clinical and industrial applications will require a reproducible, XF culture system. In this study, we evaluated the replacement of a conventional hESC culture system with high-dose, gamma-irradiated serum and pharmaceutical-grade recombinant bFGF (trafermin) in order to address the reproducibility of hESC cultivation systems. hESCs could be successfully maintained using modified conventional medium supplemented with 35 KGy-irradiated KO-SR and trafermin. Our data provided evidence supporting clinically relevant alternative platforms of hESC culture for use in clinical and industrial applications. This study also demonstrated the development of a defined, novel, efficient culture system for the derivation of hESCs from blastocyst ICMs and described the expansion and maintenance of hESCs in completely XF conditions on human allogeneic MSC feeder layers. The XF culture system with XF hMSC feeder layers can stably expand several hiPSC lines (data not shown) and successfully allow the derivation of human iPSCs (Supplemental Fig. 3). These conditions are also quite applicable to iPSC derivation and cultivation as well. Importantly, the development of such an XF culture system primarily enables hESCs to culture and expand in conditions that are totally devoid of forming Neu5GC, a sialic acid glycan and xenogeneic antigen that can potentially transform cells to obtain cancer phenotypes [18], [19]. In this culture system, we developed and successfully generated completely Neu5GC-free hESC lines. Our four XF hESC lines (SEES-4, SEES-5, SEES-6, and SEES-7) can be also stably cultivated under alternative xenogeneic-free conditions. Nakagawa et al. reported a novel culture system for derivation and expansion of hiPSCs with the recombinant laminin-511 E8 fragment matrices and the xenogeneic-free culture medium (StemFit™) [20]. Four XF SEES cell lines were stably grown using the StemFit™ medium under feeder-free conditions (data not shown). The XF SEES cell lines would be accustomed to being maintained in xenogeneic-free conditions with ease, as these cell lines have been derived and expanded under animal derived components-free conditions.
Pluripotent hESCs are isolated from preimplantation embryos [1] and general characteristics of hESCs include flat morphology, dependence on FGF2 signaling, differentiation into three germ layers in vitro and in vivo, and pluripotent markers expression [21]. Numerous hESC lines have been derived [3]. It is indicated that many of hESC lines differ in the manner in which they were derived and maintained in culture, and such differences may have significant effects on the characteristics of the cell lines [22], [23]. In female hESCs, culture conditions may have significant effects on the X chromosome inactivation status and contribute to the cellular characteristics [24]. Three of five female SEES cell lines showed none of XIST expression in this study. However, it has demonstrated that there are not notable differences in gross hESCs characters including pluripotent markers and differentiation in vitro and in vivo, despite the significant differences in XIST expression status among SEES 1, 2, 4, 5 and 7. Notably, SEES cell lines are characterized not only the biological properties of human ESCs, also the genomic signatures including ABO blood typing, STR genotyping and HLA isotyping. The distinct properties of the SEES cell lines offer a scalable cell resource for clinical application.
The culture medium we described in this study consisted of a basal culture medium with well-known growth factors that define the maintenance of pluripotency. Since these ingredients are well known and are added to a simple culture medium formulation, the cells derived/expanded in such a system have the potential for reduced contamination, better kinetics of growth, good ability to differentiate into all the three germ layer derivative lineages, maintain a normal karyotype, and, most importantly, will not contribute to tumorigenicity. This defined culture system serves as a better and safer alternative to derive/culture/expand hESCs/iPSCs for larger cell therapy purposes.
4. Materials and methods
4.1. Derivation of hESC lines on inactivated mouse embryonic fibroblast (MEF) feeder cultures
All derivations and cultures of hESC lines in this study were performed in full compliance with the Guidelines on the Derivation and Distribution of Human Embryonic Stem Cells of the Ministry of Education, Culture, Sports, Science and Technology, Japan (Notification No. 156 of 2009), after approval of the Institutional Review Board regarding hESC research at the National Center for Child Health and Development (NCCHD; “Sei-iku” in Japanese title of the affiliation), Japan. Surplus frozen human embryos, donated by consenting couples, were thawed using a Cryotop Safety Thawing Kit (Kitazato BioPharma, Shizuoka, Japan; #VT602) according to the manufacturer's instructions and cultured in BlastAssist System medium (MediCult, Jyllinge, Denmark; #12150010) until they reached the blastocyst stage. The derivation of three hESC lines, i.e., SEES-1, SEES-2, and SEES-3, was performed using modified HUES derivation methods, as described previously [4], [5], [25]. Briefly, the inner cell mass (ICM) was isolated by immunosurgery by using rabbit antiserum (Rockland Immunochemicals, PA, USA; #109-4139) and guinea pig serum complement (Sigma–Aldrich, MO, USA; #S-1639) and then seeded onto a feeder layer of freshly plated gamma-irradiated MEFs, isolated from ICR embryos at 12.5 gestations and passaged two times before gamma irradiation (30 Gy), in hESC conventional derivation media. The hESC conventional derivation media consisted of Knockout Dulbecco's modified Eagle's medium (KO-DMEM; Life Technologies, CA, USA; #10829-018) supplemented with 20% Knockout Serum Replacement (KO-SR; #10828-028), 2 mM GlutaMAX-I (#35050-079), 0.1 mM nonessential amino acids (NEAAs; #11140-076), 50 U/mL penicillin/50 μg/mL streptomycin (Pen-Strep; #15070-063), 0.055 mM beta-mercaptoethanol (#21985-023), and recombinant human full-length bFGF (#PHG0261) at 10 ng/mL (all reagents were from Life Technologies). Seven to 14 days after ICMs were plated, expanded ICMs were dissected mechanically into small clumps using a finely drawn glass Pasteur pipette and transferred onto a new MEF feeder layer as previously described [4], [25]. Secondary colonies were similarly dispersed and plated onto new feeder layers of MEFs until passages 2–4. Cells were then further expanded manually using a Stem Cell Cutting Tool (Vitrolife, Kungsbacka, Sweden; #14601) and Dispase II (Eidia, Ibaraki, Japan; #GD81070).
4.2. hMSCs as feeder layers
4.2.1. Production and culture of hMSCs under XF conditions
To derive and expand hMSC feeder layers under XF conditions, we eliminated the use of media with all animal-derived components during the derivation, propagation, and passaging process in the preparation of new hMSC feeders from human subjects. Parental written informed consent was obtained from all families, and the study was approved by the Institutional Review Board (IRB #88) of the NCCHD. hMSCs were isolated from human dermal tissue samples collected from juvenile donors undergoing surgical procedures for polydactyly in the Division of Orthopedics of the NCCHD. Human dermal tissues were first washed in Dulbecco's phosphate-buffered saline (DPBS) without calcium and magnesium (#14190-250) containing penicillin/streptomycin and then minced into small pieces by using a sterile scalpel in a laminar flow cabinet. The minced tissue was centrifuged and filtered in sequential steps to separate the tissue debris. The isolated cells were then expanded in StemPro MSC SFM XenoFree (MSC-XF; #A10675-01) supplemented with StemPro LipoMax Defined XenoFree Lipid Supplement (#A10850-01) on culture dishes coated with a recombinant humanized matrix (CELLstart CTS; #A10142-01). The isolated cells were expanded in MSC-XF medium with CELLstart using animal-component free TrypLE Select (#12563-011; all reagents from Life Technologies). After approximately 20 days, a confluent monolayer of primary cells was established (passage 0).
4.2.2. Proliferation assay
To assess the proliferative capacity of the isolated cells, primary cultures from two donors were analyzed through 22 serial passages using MSC-XF medium with CELLstart. Cells were counted using a cell viability analyzer (Vi-CELL Cell Viability Analyzer; Beckman Coulter, CA, USA), and cells were subcultured at 105 cells/100-mm dish every 4 days for approximately 15 days. At each passage, the population doubling (PD) rate was calculated based on the total cell number using the following formula: [log10(Nh) – log10(N1)]/log10(2), where N1 was the number of cells plated and Nh was the number of cells harvested [26]. Growth curves were generated in triplicate using two independent cell lines.
4.2.3. Flow cytometric analysis and in vitro multilineage differentiation assay
Flow cytometric analysis was performed as described previously [27] in order to characterize the cells. Cells were incubated with primary antibodies or isotype-matched control antibodies, followed by immunofluorescence secondary antibody staining and analysis using an EPICS ALTRA analyzer (Beckman Coulter). The following cell surface epitopes were detected with anti-human fluorescein isothiocyanate (FITC)-conjugated or phycoerythrin (PE)-conjugated antibodies: CD29 (Beckman Coulter; #6604105), CD44 (Beckman Coulter; #IM1219), CD90 (BD Pharmingen, CA, USA; #555596), CD117 (Beckman Coulter; #IM1360), and CD133 (Miltenyi Biotec, Bergisch Gladbach, Germany; #130-080-801). The potential of the isolated cells to differentiate into osteogenic and adipogenic lineages was examined according to the manufacturer's instructions. Adipogenesis and osteogenesis were induced by culturing cells in medium from an hMSC Adipogenic BulletKit (Lonza, PA, USA; #PT-3004) or an hMSC Osteogenic BulletKit (Lonza; #PT-3002), respectively. After 8 weeks under differentiation conditions, cells were processed for lineage-specific staining. Oil red staining was used for the detection of accumulated oil droplets in the cytoplasm of cells maintained with adipogenic differentiation media. Alkaline phosphatase activity was used to determine the extent of osteogenesis in cells grown in osteogenic differentiation medium.
4.2.4. Preparation of hMSCs for the feeder layer
At passages 5–20, XF cells were detached from the culture plate with TrypLE Select, washed with MSC-XF medium, and counted. The cells were resuspended in culture medium at a concentration of 1 × 107 cells per tube and then gamma-irradiated with 30 Gy. The irradiated cells were washed in medium and frozen at a concentration of 2 × 106 cells per vial in freezing medium (STEM-CELLBANKER, ZENOAQ, Fukushima, Japan; #CB043).
4.3. XF hESCs
4.3.1. Derivation and expansion of new hESC lines under XF conditions
The procedure for derivation of XF hESCs was approved by the Institutional Review Board at the NCCHD, and embryos were collected from donors undergoing fertility treatment after obtaining informed consent. Frozen embryos were thawed and cultured to the blastocyst stage under the same methods as used for conventional hESC derivation. Blastocysts without immunosurgery were plated on inactivated XF hMSC feeder layers (Yub-1896 cells) in XF hESC culture medium composed of 15% Knockout SR XenoFree CTS (KO-SR XF; Life Technologies; #12618-013), 85% KO-DMED, 2 mM GlutaMAX-I, 0.1 mM NEAAs, penicillin/streptomycin, 50 μg/mL l-ascorbic acid 2-phosphate (Sigma–Aldrich; #A4544), 10 ng/mL heregulin-1β, recombinant human NRG-beta 1/HRG-beta 1 EGF domain (R&D Systems, MN, USA; #396-HB-050/CF), 200 ng/mL LONG R3-IGF1, recombinant human insulin-like growth factor-1 (Sigma–Aldrich; #85580C), and recombinant human full-length bFGF (Life Technologies; #PHG0261) at 20 ng/mL. Cells were initially maintained in a drawer-type incubator (IVF CUBE, ASTEC, Fukuoka, Japan; #AR-3100) at 37 °C under the appropriate humidified gas mixture (usually 3%–5% O2/5% CO2/90%–92% N2). Within 7 days after whole blastocysts were plated on the feeder layers, ICMs were isolated by laser-mediated ablation of trophectoderm (TE) cells by using a XYClone laser system (Hamilton Thorne Biosciences, MA, USA) with 80% pulse strength and a pulse length of 300 μs [5]. After 2 weeks, the ICM outgrowth began to resemble morphologically distinct hESCs, and cells could be handpicked and transferred to fresh plates. After the first splitting, new colonies were disaggregated with a recombinant trypsin (Roche Applied Science, Basel, Switzerland; #06369880103) and transferred to fresh mitotically inactivated XF hMSC feeder plates every 5–7 days. Undifferentiated cells, as judged by morphology, were chosen for each further passage. A total of four XF hESC lines were derived and stably maintained (SEES-4, SEES-5, SEES-6, and SEES-7). All four SEES cell lines were maintained in a standard tissue culture incubator at 37 °C under the appropriate humidified gas mixture (3%–5% O2/5% CO2/90%–92% N2) in multigas incubator (Sanyo, Osaka, Japan; #MCO-18M) in a separate culture room to avoid any contamination. SEES cells were cryopreserved using a conventional slow-rate cooling/thawing method with STEM-CELLBANKER. Validation of the cryopreservation by subsequent thawing of individual tubes resulted in efficient recovery of viable, undifferentiated hESCs.
4.3.2. Immunohistochemical analyses of stem cell and differentiated markers
Immunohistochemistry was performed as previously described [28], [29]. The primary antibodies used for hESCs were specific for Nanog (1:300; ReproCELL, Kanagawa, Japan; #RCAB0003P), Oct3/4 (1:300; Santa Cruz Biotechnology, CA, USA; #sc-5279), Sox2 (1:300; Merck Millipore, Darmstadt, Germany; #AB5603), TRA1-60 (1:300; Merck Millipore; #MAB4360), and SSEA4 (1:300; Merck Millipore; #MAB4304). To assess the differentiation of the three germ layers, the primary antibodies used for embryoid bodies (EBs) were anti-α-fetoprotein (AFP; 1:200; R&D Systems; MAB1368), mouse (ascites) anti-β-tubulin III (TUJ1; 1:1000; Promega; #G712A), and anti-α-smooth muscle actin (αSMA; 1:400; Sigma; #A2547). Cells were fixed in 4% paraformaldehyde and incubated with primary antibodies overnight at 4 °C. Cells were then probed with Alexa Fluor 546 Goat Anti-Mouse IgG- or Alexa Fluor 488 Goat Anti-Rabbit IgG-conjugated secondary antibodies (1:300 each; Life Technologies), counterstained with 1 μg/mL DAPI, for 1 h in the dark at room temperature. After incubation, cells were mounted in Vectashield mounting medium containing 4′, 6-diamidino-2-phenylindole (Vector Laboratories, CA, USA). The labeled cells were visualized using a laser-scanning confocal microscope (LSM 510 META; Carl Zeiss, Oberkochen, Germany). Alkaline phosphatase (ALP) was detected with a Vector Red kit (Vector Laboratories; #SK-5100) according to manufacturer's instructions.
4.3.3. Flow cytometry analysis of pluripotency markers
SEES cells cultured in appropriate conditions were stained for 30 min at 4 °C with primary antibodies and immunofluorescence secondary antibodies. Cells were analyzed with a Cytomics FC 500 Cytometer (Beckman Coulter), and data were analyzed with FC 500 CXP Software ver. 2.0 (Beckman Coulter). Antibodies against human SSEA-1 (R&D Systems; #FAB2155C), SSEA4 (R&D Systems; #FAB1435F), TRA1-60 (Merck Millipore; #MAB4360), and TRA1-81 (Merck Millipore; #MAB4381) were used as primary antibodies in hESCs. PE-conjugated anti-mouse IgG antibodies (BD Pharmingen; #555578) and PE-conjugated anti-mouse IgM antibodies (BD Pharmingen; #553472) were used as secondary antibodies. X-Mean, the sum of intensity divided by the total cell number, was automatically calculated and was utilized for our evaluation.
4.3.4. Differentiation assays in vitro and in vivo
For induction of differentiation in vitro, the cells were dissociated using either StemPro Accutase (Life Technologies; #A11105-01) or TrypLE Select for 5 min at 37 °C, plated into 96-well plates (low attachment surface; Lipidure; NOF Corp., Tokyo, Japan), and cultured in differentiation medium containing KO-DMEM supplemented with 20% fetal bovine serum (FBS), 2 mM GlutaMAX-I, 0.1 mM NEAAs, and penicillin/streptomycin, to generate EBs. After 7 days in suspension, EBs were transferred onto poly-l-ornithine-coated chamber slides and cultured for an additional 10–14 days. The cultures were fixed with 4% paraformaldehyde for 20 min before immunohistochemical analysis.
In vivo pluripotency was assessed by teratoma formation in severe combined immunodeficient nude mice (BALB/cAJcl-nu/nu) purchased from CLEA Japan. A 60-mm plate of undifferentiated hESCs was washed with DPBS, and the cells were harvested with a cell scraper. The cell suspension was collected into a 15-mL conical tube and centrifuged at 1000 rpm for 4 min. The cell pellet was resuspended in hESC culture medium and Matrigel (BD Biosciences, NJ, USA; #356234) to a final total volume of 400 μL. Approximately 2–5 × 106 cells in 200 μL were injected subcutaneously into the dorsolateral area on both sides. Mice were sacrificed after 8–10 weeks. Tumors were then excised surgically, fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Hematoxylin and eosin-stained paraffin-embedded sections were histologically examined for the presence of differentiated human tissue derived from all three embryonic germ layers.
The animal use protocol was approved by the Institutional Animal Care and Use Committee of the National Research Institute for Child Health and Development (NRICHD, Permit Number: A2003-002). All experiments with mice were subject to the 3 R consideration (refine, reduce, and replace), and all efforts were made to minimize animal suffering and to reduce the number of animals used.
4.3.5. Sialic acid analysis
Sialic acid was released from cell homogenate samples by acid hydrolysis with 0.05 M HCl at 80 °C for 3 h. Sialic acid samples were ultrafiltered using Amicon Ultra-0.5 mL filters (cut-off, 10 kDa; Millipore; #UFC503008), and filtrates were dried in a vacuum concentrator. Sialic acid was derivatized with 1,2-diamino-4,5-methylenedioxybenzene (DMB; Sialic Acid Fluorescence Labeling Kit; Takara Bio, Shiga, Japan) and analyzed by reverse-phase fluorometric HPLC1 using a PALPAK Type R column (Takara Bio). The excitation and emission wavelengths were 310 and 448 nm, respectively. The DMB-derivatized sialic acid was identified by comparing retention times with those of known standards (Glyko Sialic acid reference panel; ProZyme, CA, USA; #GKRP-2503) that were similarly treated. This method can evaluate sialic acids at a minimum concentration of 0.01 nmol/mg protein.
4.3.6. Karyotype analysis
Chromosomal G-band analyses were performed at the Nihon Gene Research Laboratories, Sendai, Japan. The chromosomes were classified according to the International System for Human Cytogenetic Nomenclature. At least 20 metaphase chromosomes were analyzed per cell line.
4.4. Generation of human iPSCs
Human iPSCs were generated from XF Yub-1896 cells by transduction with of a lentiviral vector carrying three reprogramming factors (OCT3/4, SOX2 and KLF4) under XF culture conditions. The vector was a generous gift of Konrad Hochedlinger (Harvard University). XF iPSCs were successfully maintained on inactivated XF hMSC feeder layers in XF hESC culture medium over 20 passages. We confirmed the XF iPSCs expressed pluripotent markers including OCT3/4, NANOG, SSEA4, and TRA1-60.
4.5. Modified conventional hESC cultivation
4.5.1. Production of MEF feeder stock
Pregnant ICR mice were obtained from a breeding colony under specific pathogen-free (SPF) conditions in a barrier room with extensive health monitoring at CLEA Japan (Atsugi facility, Kanagawa, Japan). MEFs were generated from ICR embryos at 12.5 gestations as previously described [5], [25] and expanded using MEF feeder sock medium composed of 90% KO-DMEM, 30 KGy gamma-irradiated FBS (HyClone; Thermo Fisher Scientific, MA, USA), and 2 mM GlutaMAX-I without any antibiotics. At passage 2, cells were mitotically inactivated by gamma irradiation (30 Gy) and frozen at a concentration of 4 × 106 cells per vial. To minimize the risk of introducing murine viruses and other pathogens, MEF feeder stock was tested in GMP/GLP studies by Vitology Limited (Glasgow, UK). The specifications and results for the testing of lot MEF-0001 are presented in Supplemental Table 3.
4.5.2. hESC expansion using gamma-irradiated KO-SR and pharmaceutical-grade recombinant human bFGF without antibiotics
To develop a safer culture system for clinical application of hESCs, we replaced KO-SR and recombinant human full-length bFGF with pharmaceutical recombinant human bFGF and high-dose gamma--irradiated KO-SR. Frozen KO-SR products ware gamma irradiated at 35 KGy (KOGA ISOTOPE, Ltd., Shiga, Japan). Pharmaceutical-grade recombinant human bFGF (generic name: trafermin), supplied by Kaken Pharmaceutical (Tokyo, Japan) as Fiblast Spray, which is prepared using the powdered form of trafermin, was applied for hESC culture. hESC lines were cultivated in a modified conventional culture medium composed of 20% gamma-irradiated KO-SR, 80% KO-DMEM, 2 mM GlutaMAX-I, 0.1 mM NEAAs, and 50 ng/mL trafermin without any antibiotics on plates coated with 0.1% type I collagen (Nippon Ham, Ibaraki, Japan; #307-31611) and a mitotically inactivated layer of MEFs.
Competing financial interests
The authors declare no competing financial interests.
Acknowledgments
We are grateful to Hideki Tsumura and the staff of the Animal Care Facility at NRICHD for mouse husbandry. We thank Kaken Pharmaceutical for providing us with the trafermin used in this study. We thank Kahori Minami and Nobuyuki Watanabe for technical assistance and Tomoyuki Kawasaki for help with editing the figures. The authors would like to thank members of the Umezawa laboratory for helpful suggestions.
This research was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; by Ministry of Health, Labor and Welfare (MHLW) Sciences research grants; by a Research Grant on Health Science focusing on Drug Innovation from the Japan Health Science Foundation; by the program for the promotion of Fundamental Studies in Health Science of the Pharmaceuticals and Medical Devices Agency; by the Grant of National Center for Child Health and Development; by the Takeda Science Foundation to HA and AU. This research was also by a grant from JST-CREST to HA. AU acknowledges the International High Cited Research Group (IHCRG #14-104), Deanship of Scientific Research, King Saudi University, Riyadh, Kingdom of Saudi Arabia. AU also thanks King Saud University, Riyadh, Kingdom of Saudi Arabia, for the Visiting Professorship. The funders had no control over the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.reth.2014.12.004.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Short tandem repeat (STR) profiles of SEES cell lines and hMSC feeders. Short tandem repeat (STR) profiling was performed by BEX CO., LTD, Tokyo, Japan. The 16 loci analyzed by the PowerPlex 1.2 system (Promega, Madison, WI, USA) comprised D3S1358, TH01, D21S11, D18S51, Penta E, D5S818, D13S317, D7S820, D16S539, CSF1PO, Penta D, AMEL, vWA, D8S1179, TPOX, and FGA. The unique STR profiles of seven SEES cell lines and the hMSC feeder line are shown. SEES-3.1 was a subclone of the SEES-3 cell line. Yub-1896 XF MSC was a xenogeneic-free hMSC feeder line. P indicates the passage number, and Lot. indicates the number of our stock cell lines.
HLA profiles of SEES cell lines. HLA DNA typing was performed by ReproCELL, Kanagawa, Japan.
Characterization of MEF feeder stock. MEF feeder stock was tested in GMP/GLP studies by Vitology Limited (Glasgow, UK).
Supplemental Fig. 1.
Characterization of SEES-1, SEES-2, and SEES-3 cells Typical ESC morphology is shown for SEES-1, SEES-2, and SEES-3 cells. Alkaline phosphatase (ALP) activity was detected in each SEES cell line. Three SEES cell lines, grown in conventional hESC culture medium, expressed markers characteristic of pluripotent hESCs, including OCT4, NANOG, SOX2, SSEA4, and TRA1-60. Chromosomal analysis of SEES-1, SEES-2, and SEES-3 cells showed normal karyotype, 46,XX, 46,XX, and 46,XY, respectively.
Supplemental Fig. 2.
Differentiation potential of SEES-1, SEES-2, and SEES-3 cells A) Histological analysis of teratomas containing multidifferentiated tissues derived from SEES-1, SEES-2, and SEES-3 cells. Pigmented epithelium (ectoderm), cartilage (mesoderm), and gut epithelial tissues (endoderm). Scale bars are 200 μm. B) SEES-1, SEES-2, and SEES-3 cells that were differentiated in vivo via EB formation expressed markers of the primary germ layers, ectoderm (TUJ1), mesoderm (cTnT∗), and endoderm (AFP). ∗cTnT: cardiac troponin T, a cardiac marker. SEES-1 and SEES-2 cells: scale bars are 200 μm for TUJ1 and 50 μm for cTnT and AFP. SEES-3 cells: scale bars are 100 μm for TUJ1 and 50 μm for cTnT and AFP.
Supplemental Fig. 3.
Establishment of human iPSCs under xenogeneic-free conditions Human iPSCs were generated from XF MSCs (Yub-1896 cells) by transduction of three reprogramming factors (OCT3/4, SOX2 and KLF4) under hXF culture conditions using XF hESC medium and XF hMSC feeder layers. Derived human XF iPSCs express pluripotent markers including OCT4, NANOG, SSEA4, and TRA1-60. Scale bars are 100 μm
Supplemental Fig. 4.
Detection of Neu5Gc and Neu5Ac on SEES cells N-glycolylneuraminic acid (Neu5Gc) was not detected in xenogeneic-free SEES cells, but was observed in SEES-1, SEES-2, and SEES-3 cells. N-acetylneuraminic acid (Neu5Ac) was found in all examined SEES cell lines. The detection limit of sialic acids is 0.01 nmol/mg protein.
Supplemental Fig. 5.
Blood group ABO antigen typing of SEES cell lines and hMSCs ABO blood typing was performed by sequencing of ABO gene polymorphisms as described previously [S1] with some modifications and confirmed by exome sequencing. ABO genotyping was also performed by the PCR restriction-fragment length polymorphism method [S2] and allele-specific primers method [S3]. SEES-1, SEES-3, SEES-4, and SEES-6 cells were AO type; SEES-2 and SEES-5 cells were O type; and SEES-7 cells were AA type; hMSCs feeder layer (Yub-1896 cells) was BO type.
Supplemental Fig. 6.
Xist expression in SEES cells Fluorescent in situ hybridization (FISH) of Xist in SEES cells was used to characterize the epigenetic status of X-chromosome inactivation. FISH was performed at the Chromosome Science Laboratory, Hokkaido, Japan. A total 100 cells were analyzed for Xist expression in each cell line. FISH analysis showed that SEES-1, SEES-2 and SEES-7 cells would have activated X chromosomes. RNA and DNA FISH staining with probes detecting XIST RNA (red), the X (green) and Y (yellow) chromosome, and DAPI counterstain.
Supplemental Fig. 7.
Expression of pluripotency markers in SEES-1 and SEES-3 cells was maintained using a modified conventional hESC culture medium SEES-1 and SEES-3 cells were stably maintained in the modified medium containing pharmaceutical-grade recombinant human bFGF, trafermin, and 35-K gamma-irradiated KO-SR without antibiotics. A) Typical hESC colony morphology was readily visible. B) SEES-1 and SEES-3 cells expressed undifferentiated hESC markers, including SEES-4, TRA1-60, OCT4, NANOG, and SOX2. Scale bars are 200 μm.
Supplemental Fig. 8.
Hierarchical clustering analysis of expression data from the PCR array across the 48 pluripotency-related genes We used a Human Embryonic Stem Cell PCR Array and RT2 qPCR Mastermix (Qiagen, Germany) for quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Transcript levels were determined using the QuantStudio 12K Flex real-time PCR system (Life Technologies). Relative quantification was carried out using ACTB, GAPDH, and RPL13A as endogenous control genes. Hierarchical clustering analyses across the 48 pluripotent marker genes was performed using delta Ct values for gene expression data with MEV v4.8 statistical analysis software. A) Hierarchical clustering analysis of SEES cell lines was performed using a modified conventional hESC culture medium. Gene expression levels in each sample, relative to the median level of expression of that gene across all the samples, is represented using a red-black-green color scale, as shown in the key (green: below median; black: equal to median; red: above median). B) Correlation analysis for gene expression. Scatter plots (upper diagonal) of the gene expression of 48 pluripotency markers across all pair-wise comparisons. Correlation coefficients are shown in corresponding squares below the diagonal.
References
- 1.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Schuldt B.M., Guhr A., Lenz M., Kobold S., MacArthur B.D., Schuppert A. Power-laws and the use of pluripotent stem cell lines. PLoS One. 2013;8:e52068. doi: 10.1371/journal.pone.0052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amit M., Carpenter M.K., Inokuma M.S., Chiu C.P., Harris C.P., Waknitz M.A. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 4.Cowan C.A., Klimanskaya I., McMahon J., Atienza J., Witmyer J., Zucker J.P. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 5.Chen A.E., Egli D., Niakan K., Deng J., Akutsu H., Yamaki M. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell. 2009;4:103–106. doi: 10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu C., Inokuma M.S., Denham J., Golds K., Kundu P., Gold J.D. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 7.Richards M., Fong C.Y., Chan W.K., Wong P.C., Bongso A. Human feeders support prolonged undifferentiated growth of human inner cell masses and embryonic stem cells. Nat Biotechnol. 2002;20:933–936. doi: 10.1038/nbt726. [DOI] [PubMed] [Google Scholar]
- 8.Martin M.J., Muotri A., Gage F., Varki A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat Med. 2005;11:228–232. doi: 10.1038/nm1181. [DOI] [PubMed] [Google Scholar]
- 9.Pham T., Gregg C.J., Karp F., Chow R., Padler-Karavani V., Cao H. Evidence for a novel human-specific xeno-auto-antibody response against vascular endothelium. Blood. 2009;114:5225–5235. doi: 10.1182/blood-2009-05-220400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varki A. Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci U S A. 2010;107(Suppl. 2):8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludwig T.E., Levenstein M.E., Jones J.M., Berggren W.T., Mitchen E.R., Frane J.L. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 12.Lu J., Hou R., Booth C.J., Yang S.H., Snyder M. Defined culture conditions of human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:5688–5693. doi: 10.1073/pnas.0601383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao S., Chen S., Clark J., Hao E., Beattie G.M., Hayek A. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci U S A. 2006;103:6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa K., Pomeroy J.E., Pera M.F. Current technology for the derivation of pluripotent stem cell lines from human embryos. Cell Stem Cell. 2010;6:521–531. doi: 10.1016/j.stem.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz S.D., Hubschman J.P., Heilwell G., Franco-Cardenas V., Pan C.K., Ostrick R.M. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 16.Fraga A.M., Souza de Araújo É., Stabellini R., Vergani N., Pereira L.V. A survey of parameters involved in the establishment of new lines of human embryonic stem cells. Stem Cell Rev. 2011;7:775–781. doi: 10.1007/s12015-011-9250-x. [DOI] [PubMed] [Google Scholar]
- 17.Wang L., Schulz T.C., Sherrer E.S., Dauphin D.S., Shin S., Nelson A.M. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110:4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedlund M., Padler-Karavani V., Varki N.M., Varki A. Evidence for a human-specific mechanism for diet and antibody-mediated inflammation in carcinoma progression. Proc Natl Acad Sci U S A. 2008;105:18936–18941. doi: 10.1073/pnas.0803943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearce O.M., Läubli H., Verhagen A., Secrest P., Zhang J., Varki N.M. Inverse hormesis of cancer growth mediated by narrow ranges of tumor-directed antibodies. Proc Natl Acad Sci U S A. 2014;111:5998–6003. doi: 10.1073/pnas.1209067111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa M., Taniguchi Y., Senda S., Takizawa N., Ichisaka T., Asano K. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep. 2014;4:3594. doi: 10.1038/srep03594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna J.H., Saha K., Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman L.M., Carpenter M.K. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman L.M., Hall L., Batten J.L., Young H., Pardasani D., Baetge E.E. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen H.T., Geens M., Spits C. Genetic and epigenetic instability in human pluripotent stem cells. Hum Reprod Update. 2013;19:187–205. doi: 10.1093/humupd/dms048. [DOI] [PubMed] [Google Scholar]
- 25.Akutsu H., Cowan C.A., Melton D. Human embryonic stem cells. Methods Enzymol. 2006;418:78–92. doi: 10.1016/S0076-6879(06)18005-2. [DOI] [PubMed] [Google Scholar]
- 26.Escobedo-Lucea C., Bellver C., Gandia C., Sanz-Garcia A., Esteban F.J., Mirabet V. A xenogeneic-free protocol for isolation and expansion of human adipose stem cells for clinical uses. PLoS One. 2013;8:e67870. doi: 10.1371/journal.pone.0067870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui C.H., Miyoshi S., Tsuji H., Makino H., Kanzaki S., Kami D. Dystrophin conferral using human endothelium expressing HLA-E in the non-immunosuppressive murine model of Duchenne muscular dystrophy. Hum Mol Genet. 2011;20:235–244. doi: 10.1093/hmg/ddq458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagata S., Toyoda M., Yamaguchi S., Hirano K., Makino H., Nishino K. Efficient reprogramming of human and mouse primary extra-embryonic cells to pluripotent stem cells. Genes Cells. 2009;14:1395–1404. doi: 10.1111/j.1365-2443.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- 29.Makino H., Toyoda M., Matsumoto K., Saito H., Nishino K., Fukawatase Y. Mesenchymal to embryonic incomplete transition of human cells by chimeric OCT4/3 (POU5F1) with physiological co-activator EWS. Exp Cell Res. 2009;315:2727–2740. doi: 10.1016/j.yexcr.2009.06.016. [DOI] [PubMed] [Google Scholar]
References in supplemental figures
- 1.Hosoi E. Biological and clinical aspects of ABO blood group system. J Med Invest. 2008;55:174–182. doi: 10.2152/jmi.55.174. (For Supplemental Figure S1) [DOI] [PubMed] [Google Scholar]
- 2.Ota M., Fukushima H., Kulski J.K., Inoko H. Single nucleotide polymorphism detection by polymerase chain reaction-restriction fragment length polymorphism. Nat Protoc. 2007;2:2857–2864. doi: 10.1038/nprot.2007.407. (For Supplemental Figure S2) [DOI] [PubMed] [Google Scholar]
- 3.Muro T., Fujihara J., Imamura S., Nakamura H., Kimura-Kataoka K., Toga T. Determination of ABO genotypes by real-time PCR using allele-specific primers. Leg Med Tokyo. 2012;14:47–50. doi: 10.1016/j.legalmed.2011.10.002. (For Supplemental Figure S3) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Short tandem repeat (STR) profiles of SEES cell lines and hMSC feeders. Short tandem repeat (STR) profiling was performed by BEX CO., LTD, Tokyo, Japan. The 16 loci analyzed by the PowerPlex 1.2 system (Promega, Madison, WI, USA) comprised D3S1358, TH01, D21S11, D18S51, Penta E, D5S818, D13S317, D7S820, D16S539, CSF1PO, Penta D, AMEL, vWA, D8S1179, TPOX, and FGA. The unique STR profiles of seven SEES cell lines and the hMSC feeder line are shown. SEES-3.1 was a subclone of the SEES-3 cell line. Yub-1896 XF MSC was a xenogeneic-free hMSC feeder line. P indicates the passage number, and Lot. indicates the number of our stock cell lines.
HLA profiles of SEES cell lines. HLA DNA typing was performed by ReproCELL, Kanagawa, Japan.
Characterization of MEF feeder stock. MEF feeder stock was tested in GMP/GLP studies by Vitology Limited (Glasgow, UK).