Abstract
Background
Experimental inoculation is an important tool for common cold and asthma research. Producing rhinovirus (RV) inocula from nasal secretions has required prolonged observation of the virus donor to exclude extraneous pathogens. We produced a RV-A16 inoculum using reverse genetics and determined the dose necessary to cause moderate colds in seronegative volunteers.
Methods
The consensus sequence of RV-A16 from a previous inoculum was cloned, and inoculum virus was produced using reverse genetics techniques. After safety testing, volunteers were inoculated with either RV-A16 (n = 26) or placebo (n = 10), Jackson cold scores were recorded, and nasal secretions were tested for shedding of RV-A16 ribonucleic acid.
Results
The reverse genetics process produced infectious virus that was neutralized by specific antisera and had a mutation rate similar to conventional virus growth techniques. The 1000 median tissue culture infectious dose (TCID50) dose produced moderate colds in most individuals with effects similar to that of a previously tested conventional RV-A16 inoculum.
Conclusions
Reverse genetics techniques produced a RV-A16 inoculum that can cause clinical colds in seronegative volunteers, and they also serve as a stable source of virus for laboratory use. The recombinant production procedures eliminate the need to derive seed virus from nasal secretions, thus precluding introduction of extraneous pathogens through this route.
Keywords: common cold, inoculation, reverse genetics, rhinovirus
Reverse genetics technology was used to develop a new rhinovirus inoculum, which induced respiratory illness in experimentally inoculated volunteers. This inoculum virus was cloned instead of being grown from human nasal secretions, therefore reducing risks of contamination with extraneous pathogens.
(See the Editor Commentary by Proud on pages 181–3)
Rhinoviruses (RV) are the most frequent cause of the common cold, and they can also cause lower respiratory illnesses in susceptible populations including young children, the elderly, immunocompromised individuals, and people with chronic respiratory conditions such as asthma, chronic obstructive lung disease, or cystic fibrosis [1]. Although the morbidity associated with these respiratory illnesses is considerable, specific treatments are lacking.
The RV experimental inoculation model has been used to (1) investigate mechanisms of RV pathogenesis and transmission, (2) test the efficacy of treatments for the common cold, and (3) understand how RV infections contribute to acute exacerbation of chronic airway diseases such as asthma and chronic obstructive pulmonary disease (COPD) [2–11]. Recent advancement in safety test technologies led to the introduction of new standards of current Good Manufacturing Procedures (GMP) for production of viral inocula [12]. The traditional method of producing a virus inoculum for use in experimental infection is to isolate a “seed virus” from nasal secretions of a donor who had been infected via natural exposure. This approach is labor intensive in that the donor needs to be checked for any other infectious agents and then observed for 1 year to ensure there are no other coinfections [13].
In this report, we describe the development of a reverse genetics (RG) approach to produce an inoculum of a major group clinical isolate (RV-A16). This approach has 2 advantages compared to traditional procedures. First, several “new” respiratory viruses (eg, Middle Eastern Respiratory Syndrome coronavirus, WU and KI polyomaviruses [14, 15]) have been discovered in the past decade, and additional infectious agents will likely be discovered in the future. Therefore, it is difficult to ensure that nasal secretions that are chosen for isolation of seed virus do not contain any other pathogens. This problem is minimized through the use of a cloned viral genome to produce the inoculum virus in vitro. A second potential advantage is the ability to produce multiple inocula from the same cloned sequence. Ribonucleic acid (RNA) viruses, such as RV, have high mutation rates during genome replication because their RNA polymerases have no error-correcting function. A complementary deoxyribonucleic acid (cDNA) clone, which is amplified by the highly accurate Escherichia coli DNA polymerase, provides a stable source of virus sequence for production of future inocula. This paper describes the development of (1) an RG-RV-A16 inoculum and (2) a first-in-human, phase 1 study to assess the safety of RG-RV-A16 in humans and identify the dose needed to produce moderate-to-severe colds in 75% of RV-A16-seronegative human volunteers.
MATERIALS AND METHODS
Master Cell Bank
Passage 1 human lung fibroblasts for viral culture ([HLF-VC1] University of Wisconsin-Madison) thawed in November 2003 were used to produce a Master Cell Bank at the Waisman Clinical Biomanufacturing Facility (Madison, WI) under GMP conditions. Extensive testing for identity, quality, and safety revealed no evidence of microbial or viral contamination (Supplemental Data and Supplemental Table 1).
Safety Testing
Safety testing of the inoculum as directed by the US Food and Drug Administration and regulatory agencies [12, 16, 17] was negative for contaminants and adventitious agents (Supplemental Table 2).
Clinical Trial
This study was approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board (protocol 2012-1036-CP002). All study participants and household contacts provided written informed consent. Regulatory approvals are listed in Supplemental Table 3. Animal experiments were conducted after approval by the IIT Research Institute (Chicago, IL; IACUC Protocol 2324-2011). The data supporting this publication are available at ImmPort (immport.org) under study accession SDY1300.
Study Design
The inoculation study had a single-blind, 5 + 5 adaptive dosing design with dose escalation or de-escalation with a maximum of 4 dosing groups of up to 10 adult subjects. Inclusion criteria included otherwise healthy adults between 18 and 50 years of age who had no neutralizing antibody to the inoculum virus. Exclusion criteria included chronic respiratory disease, smoking, and subjects with household contacts deemed at-risk (eg, pregnancy, elderly, young children). Detailed inclusion and exclusion criteria are listed in Supplemental Table 4.
Subjects were inoculated on day 0 with either placebo (phosphate-buffered saline with 0.1% human serum albumin) or 100, 500, 1000, or 10000 median tissue culture infectious dose (TCID50) of RG-RV-A16 (Supplemental Table 5). The inoculum was administered as an aerosol (MAD Nasal Intranasal Mucosal Atomization Device; Teleflex, Morrisville, NC), and 100 μL was administered via each nostril. The initial dose of the RV-A16 was 100 TCID50; 5 subjects were inoculated at a given dose level, and the dose for the next group of 5 subjects was determined based on clinical symptoms of the previous 5 subjects and the dose received by the previous 5 subjects (details in Supplemental Figure 1 and Supplemental Table 4). The study was designed such that a maximum of 10 subjects would receive any of the dosing levels (placebo, 100, 500, 1000, or 10000 TCID50).
Symptom Assessments
Symptom scores (modified Jackson Cold Symptom Scores; Supplemental Figure 2) were assessed twice daily for each subject beginning on the day of inoculation and continuing for at least 7–10 days or until the symptoms resolved, and then again on the final visit. The Daily Symptom Score represents the sum of the highest score (the am or the pm score) obtained for each of 13 symptoms. The Peak Symptom Score for each subject represents the highest of the Daily Symptom Scores for the 7-day evaluation period. The severity of the induced cold for each study participant was defined by the Peak Symptom Score and was categorized as either mild (score <7), moderate (score 7–11), or severe (score ≥12). The Mean Cold Symptom Score for each dosing group is the average of the Peak Symptom Scores.
Nasal Lavage and Viral Diagnostics
Nasal lavage was performed for cell counts and diagnostic virology (details in online Supplement). Preinoculation nasal lavage was assayed by multiplex polymerase chain reaction (PCR) (RVP; Luminex, Austin TX) to detect any virus present at the time of inoculation. Nasal lavage fluid, collected after RG-RV-A16 inoculation, was tested by RV-specific quantitative reverse transcription (RT)-PCR and partial genomic sequencing to confirm infection with RG-RV-A16 and to determine viral load [18]. Viral shedding was determined by quantitative PCR (qPCR) and reported in log RNA copies/mL [19]. Serum obtained 7–10 days after inoculation was also tested by RV qPCR to assess for viremia.
Study Outcomes
The 2 primary endpoints for the study were (1) to identify the dose of RG-RV-A16 that caused colds of at least moderate intensity (peak symptom score >7) during the first week (or longer at the discretion of the principal investigator in case of a delayed peak in symptoms) after inoculation and (2) safety as determined by adverse event reporting. Secondary endpoints were (1) the Mean Cold Symptom Score per RG-RV-A16 dose, (2) Infection rate per RG-RV-A16 dose (percentage of individuals in the dosing group with detectable RV-A16 RNA in nasal secretions), and (3) Mean Cold Symptom Scores for each RG-RV-A16 dose versus placebo.
Safety Assessments
Safety laboratory tests, to include complete blood count with differential and platelets, blood urea nitrogen, creatinine, aspartate aminotransferase, alanine aminotransferase, and immunoglobulin (Ig)A and IgG serum Igs, were drawn at screening to determine eligibility and 21–28 days after inoculation to monitor for any inoculum-induced laboratory changes.
Household Contacts
Close contacts of research subjects (see online Supplement for details) were invited to join in a surveillance study to obtain information about the frequency of natural transmission of RG-RV-A16 colds and their clinical characteristics. Consenting contacts collected cold-like symptom scores using the modified Jackson Criteria beginning on the day of inoculation (Visit 1) and for 10 consecutive days. If cold symptoms were reported, the contacts were asked to collect nasal secretions using a nose-blow technique [20] for viral diagnostics.
Statistical Analysis
Sample size was estimated by examining operating characteristics of the proposed dose ranging study design in a variety of scenarios. Expected findings were based on the results of a previous dose-ranging study of RV-A16 (Lot 1086) that was grown with standard techniques and tested in 2000–2001 in which an infecting dose of 1000 TCID50/mL was associated with median (11 of 39) and mean (11.4 of 39) peak symptom scores in the moderate range and at least 75% of subjects with moderate-to-severe colds (Table 3). The expected sample size for the optimal dose or closest available dose ranged from 9 to 10 subjects. The probability that no cohort will receive the closest available dose to the target was 1.4% or less in all but 1 scenario. The relationship between dose (log-transformed) and the rate of colds of at least moderate intensity (maximal weekly symptom score of ≥7 of 39 on the modified Jackson criteria) after inoculation was assessed using logistic regression models. The logistic regression equation will be solved for the optimal dose, eg, the dose for which the estimated rate of moderate-to-severe colds is 75%.
Table 3.
Peak Symptom Scores Caused by Inoculation With a Conventional Inoculum (1086) Compared With RG-RV-A16
| Dose (TCID50/mL) | 1086 RV-A16 | RG-RV-A16 | ||||
|---|---|---|---|---|---|---|
| n | Mean (SD) | Median (25%–75%) | n | Mean (SD) | Median (25%–75%) | |
| 0 (placebo) | 9 | 3.2 (2.9) | 2 (1–5.3) | 10 | 2.0 (2.1) | 2.0 (0–3.0) |
| 100 | 8 | 4.6 (3.8) | 3 (2–7) | 9 | 7.2 (5.2)a | 6.0 (3.0–9.8) |
| 1000 | 10 | 11.4 (5.0) | 11 (9–16) | 8 | 12.3 (7.8)b | 10.5 (9.0–14) |
Abbreviations: RG, reverse genetics; RV, rhinovirus; SD, standard deviation; TCID50, median tissue culture infectious dose.
a P = .01 vs placebo.
b P = .004 vs placebo.
RESULTS
Production of Recombinant Rhinovirus-A16
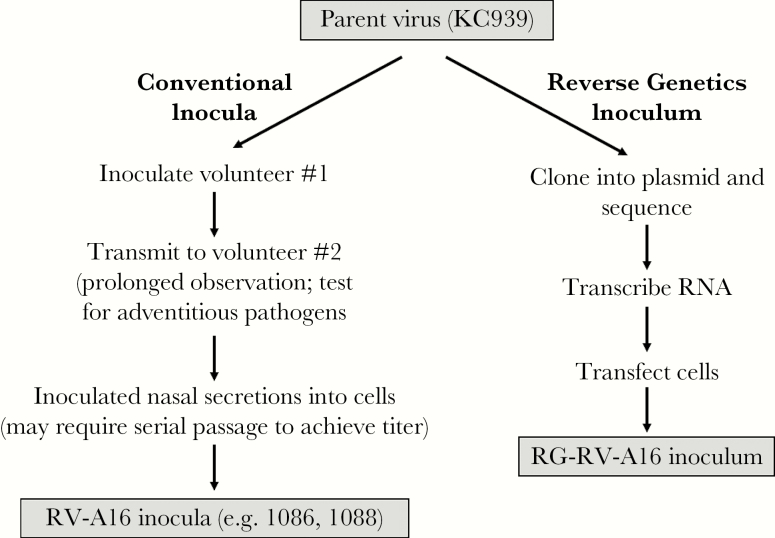
The recombinant inoculum was produced using 4 steps: (1) viral cloning, (2) transcription of viral RNA, (3) transfection into Wis.L cells, and (4) purification and resuspension of the inoculum virus (Figure 1; see online Supplement for details).
Figure 1.
Overview of conventional and reverse genetics approaches to producing a rhinovirus (RV) inoculum. RNA, ribonucleic acid.
The source of the recombinant RV-A16 genetic sequence was a previous lot of RV-A16 human inoculum (KC939) that was cryopreserved in 1985. KC939 had been used extensively for virus inoculation [5, 21–26] and as a source virus for production of 2 other viral inocula (WIS1086 and WIS1088 virus) grown using traditional culture techniques (Figure 1) that were used in additional experimental inoculation studies [19, 27, 28]. The viral RNA was extracted, and overlapping cDNA segments of the viral genome were amplified by RT-PCR, cloned, and sequenced. The cloned cDNA segments with the consensus sequences were selected and then assembled in a stepwise fashion into 1 cDNA clone with the full-length viral genome (Supplemental Figure 3). The resulting plasmid, pR16.939, was sequenced entirely and no unexpected sequence was found (GenBank accession number KX891411).
To produce infectious virus, RV-A16 RNA was synthesized and then transfected into HLF-VC1 cells. Infectious virus was released by freeze/thaw, and debris was removed by filtration and centrifugation. Safety testing of the RG-RV-A16 inoculum revealed no evidence of toxins or adventitious agents (Supplemental Table 2).
Sequence Analysis of Reverse Genetics-Rhinovirus-A16 Compared to Two Traditional Inoculum Viruses
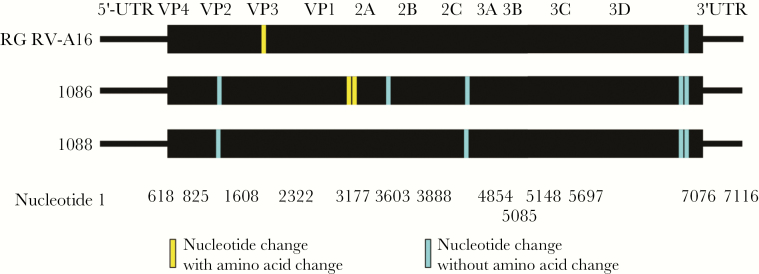
The error rate for RV 3C polymerase is high (~1 × 10–4), and RV types exist as quasispecies with consensus sequences that can rapidly adapt to different conditions in vitro and in vivo [1, 29]. Because RG-RV-A16 was produced with 1 round of replication, compared to use of 2 or more passages using traditional techniques, we hypothesized that the RG procedures would introduce fewer mutations from the reference sequence. To test this hypothesis, we compared the full-length sequence for KC939 to those of RG-RV-A16 and 2 inocula (1086 and 1088) produced from KC939 using conventional techniques; RG-RV-A16 had 2 mutations, 1086 had 7 mutations, and 1088 had 4 mutations (Figure 2).
Figure 2.
Comparisons of gene sequences of rhinovirus (RV)-A16 inocula. RG, reverse genetics; UTR, untranslated region; VP, viral protein.
Clinical Study
Study Population
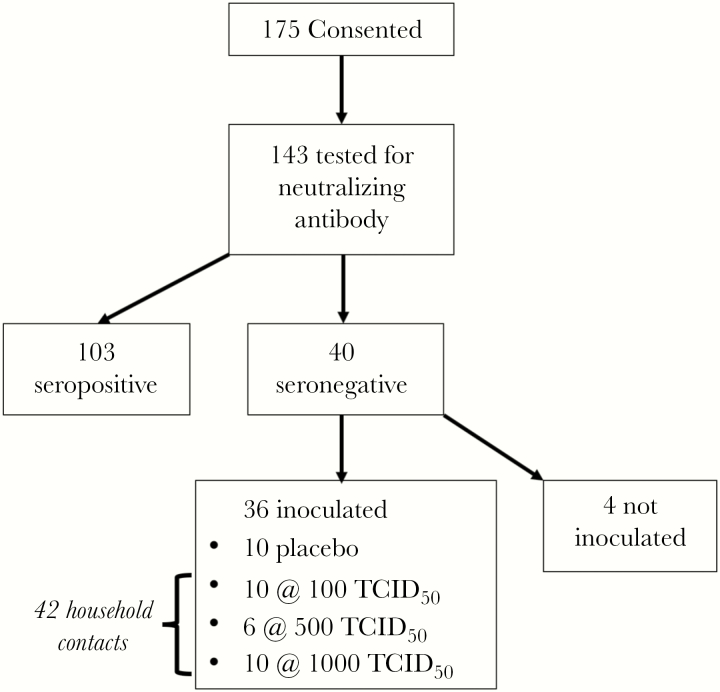
Of the 175 volunteers who were screened for the study and gave informed consent, 143 were tested for neutralizing antibody for RV-A16, and 40 (28%) had no detectable antibody (Figure 3). The 36 study subjects who were inoculated had a mean age of 25.6 years and included 10 men and 26 women (Table 1). Individuals who met study criteria and provided informed consent were similar to the screened population (Table 1).
Figure 3.
Selection of study subjects. TCID50, median tissue culture infectious dose.
Table 1.
Study Participants
| Characteristic | Not Enrolled n = 139 |
Enrolled n = 36 |
P Value |
|---|---|---|---|
| Age (years) | 26.9 ± 5.3 | 25.6 ± 3.5 | .08 |
| Gender (female, %) | 56% | 72% | .06 |
| Race (%) | |||
| White[AU: Per style, the term “Caucasian” is not used, unless you are referring to persons from the Caucasus region of eastern Europe.] | 83 | 86 | |
| Asian | 9 | 6 | |
| African American | 2 | 0 | .70 |
| Other | 2 | 3 | |
| Unknown | 4 | 6 | |
| Ethnicity (Hispanic, %) | 3 | 8 | .29 |
Concurrent Infections With Community Viruses
In total, 10 subjects were inoculated at the 100 TCID50 and 1000 TCID50 doses, 6 additional subjects (a group of 5 plus 1 replacement due to detection of a community-acquired virus) were inoculated with 500 TCID50, and 10 subjects had placebo inoculations. Although none of the subjects reported colds at the time of inoculation, 5 subjects (1, 2, and 2 subjects in the 100, 500, and 1000 TCID50 dosing groups, respectively) were found to have community-acquired RV genotypes in their nasal secretions at baseline and/or during the first week after inoculation. These subjects were excluded from the analysis of RG-RV-A16 effects on cold symptoms and viral RNA shedding. Due to the low remaining number (n = 4) of subjects inoculated with 500 TCID50/mL, this dosing level was not included in subsequent analyses. Two of 10 subjects inoculated with placebo had community-acquired RVs detected in their nasal secretions 21 days after mock-inoculation.
Primary Outcome
Inoculation with RG-RV-A16 induced clinical colds in most individuals, and the percentage of colds that were at least moderate in severity was dose related (Table 2); 0 of 10 (0%) in the placebo group, 4 of 9 (44%) in the 100 TCID50 group, and 7 of 8 (87.5%) in the 1000 TCID50 group. Thus, the 1000 TCID50 dose met the dose selection criteria for the study.
Table 2.
Number of Moderate-Severe Colds (Peak Score ≥7) at Each Dosing Level
| Dose (TCID50/mL) | Peak Scores <7 | Peak Scores ≥7 | Excluded From Analysisa |
|---|---|---|---|
| 0 (placebo) | 10 (100%) | 0 (0%) | 0 |
| 100 | 5 (56%) | 4 (44%) | 1 |
| 500 | 2 (50%) | 2 (50%) | 2 |
| 1000 | 1 (12.5%) | 7 (87.5%) | 2 |
Abbreviations: TCID50, median tissue culture infectious dose.
aSubjects who had community-acquired viruses detected in nasal secretions during the acute cold phase were excluded from the analysis.
Secondary Outcomes
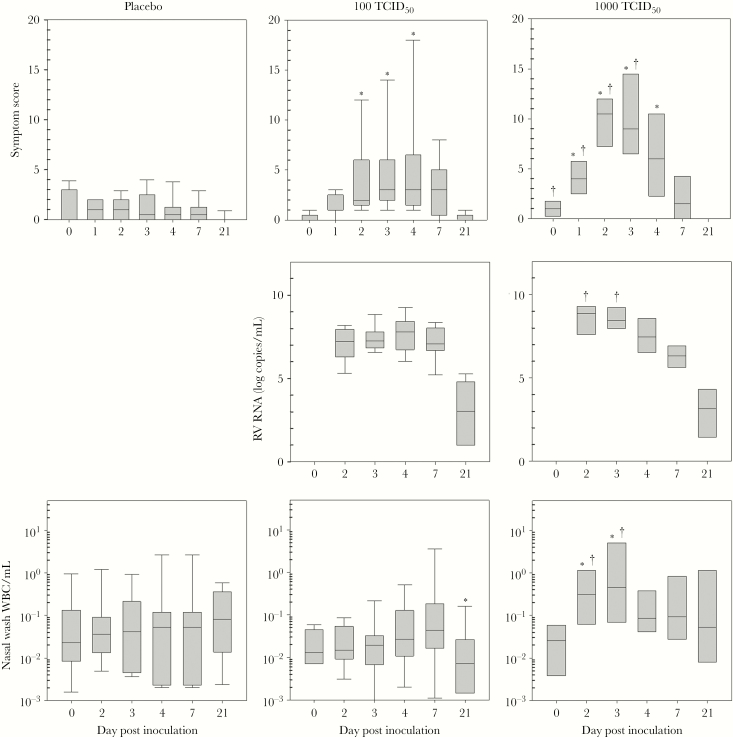
All subjects who were inoculated with RG-RV-A16 in both dose groups were infected as indicated by detection of RG-RV-A16 RNA in all samples of nasal secretions obtained within 7 days after inoculation (Figure 4). Rhinovirus inoculation significantly increased cold symptom scores 2–5 days after inoculation, and all subjects shed RG-RV-A16 in nasal secretions for at least 7 days after inoculation. Serum samples obtained at day 7 postinoculation (PI) were negative for viremia (data not shown). Neutralizing antibody responses 21–28 days PI were similar for the 100 TCID50 and 1000 TCID50 dosing groups (6 of 8 subjects in each group were positive [≥1:2.8], with 1 missing value in the 100 TCID50 group).
Figure 4.
Effects of inoculation with reverse genetics-rhinovirus (RG-RV)-A16. After inoculation with placebo or RG-RV-A16 (100 or 1000 median tissue culture infectious dose [TCID50]), clinical symptoms (daily symptom score), viral shedding (RV ribonucleic acid [RNA]), and leukocytes in nasal secretions were measured during the acute cold and during recovery. *, P ≤ .05 compared with placebo; †, P ≤ .05 compared with 100 TCID50 dose.
Several dose-related effects were noted when comparing the course of the cold after inoculation with 1000 vs 100 TCID50 (Figure 4). The 1000 TCID50 group reported slightly higher symptom scores at baseline (median 0 vs 1, P = .05). After inoculation, the 1000 TCID50 group had increased symptoms scores 1–3 days PI and greater viral RNA shedding 2–3 days PI. Inoculation with 1000 but not 100 TCID50 increased leukocyte counts in nasal wash fluid. Peak symptom scores for each group were also dose related; mean values were 2.0, 7.2, and 12.3 for the placebo, 100 and 1000 TCID50 dosing groups (Table 3). The timing of peak symptom scores was similar, and occurred 4 days after inoculation with 100 TCID50, and 3 days after inoculation with 1000 TCID50 (medians, P = nonsignificant).
Transmission of Reverse Genetics-Rhinovirus-A16 Colds to Household and Close Contacts
A total of 42 household or close contacts of study subjects consented to surveillance for natural transmission of RG-RV-A16. Three contacts reported cold symptoms during the 10-day monitoring period. One contact tested positive for a community virus (RV-A94), one tested positive for the inoculum virus (RG-RV-A16), and the third tested negative for RV and other common respiratory viruses. The contact who tested positive for the inoculum virus had mild respiratory symptoms 2 days after subject inoculation, which resolved by day 3. Cold symptoms then restarted 9 days after the index inoculation and peaked (score = 13) over the next 2–3 days. She reported symptoms (headache, congestion) consistent with sinusitis; however, the symptoms had completely resolved 1 week later (21 days after the index inoculation). Nasal secretions of the household contact tested positive for the RV-A16 inoculum strain 4 and 14 days after inoculation of the index case.
Adverse Events
Study subjects who were inoculated with RG-RV-A16 had 3 adverse events classified as related or possibly related to inoculation: a cold sore 22 days PI, 2 episodes of sinusitis (including the household contact as mentioned above), and anterior cervical lymphadenopathy during the acute illness. All were transient and judged as mild in severity.
Comparing Illnesses Caused by Reverse-Genetics Versus Conventional Rhinovirus-A16 Inocula
In 2000, we performed a dose-ranging study of a RV-A16 inoculum (1086) that was produced using conventional procedures (Figure 1). The procedures used in the 2 studies were similar, except that different devices (Devilbiss atomizer [27] vs Mucosal Atomization Device [Teleflex, Morrisville, NC] in the current study) were used to atomize and administer the viral suspension. In each case, the dose needed to produce colds of at least moderate severity in 75% of the participants was 1000 TCID50, and the mean peak symptom scores were similar (Table 3).
DISCUSSION
This study establishes that RV for use in human inoculation studies can be produced using RG technology. Advantages of this approach include increased assurance that the viral inoculum is not contaminated with extraneous pathogens, because the seed virus is derived from cloned RV sequence in pathogen-free cultured cells instead of undefined nasal secretions. In this dose-ranging inoculation study, a dose of 1000 TCID50 produced colds in 85% of those inoculated. It is notable that viral replication and cold symptoms were quite similar to those induced by a previous inoculum virus produced using traditional techniques.
Another advantage of this approach is that the cloned virus provides a genetically stable source of virus. The E coli polymerase that is used to replicate the plasmid has an error rate of ~10–9, compared with ~10–4 for picornavirus polymerases. Thus, the sequence on the plasmid is quite stable, and this enables a reliable source for virus production. This is an important feature because picornaviruses that are serially passaged in tissue culture adapt quickly to cultured cells that can change their functional characteristics [30]. We demonstrated that the RG-RV-A16 suspension acquired numerically fewer mutations compared with conventionally grown viruses, likely due to less time in tissue culture.
Because RG enables virus to be produced quickly, easily, and in large quantities, it is thus possible to use viruses that are virtually identical for inoculation studies and for in vitro studies. In contrast, many viruses now used for laboratory studies come from American Type Culture Collection isolates that are adapted to tissue culture cell lines such as HeLa cells. It is notable that the RG-RV-A16 1086 and 1088 sequences differ from those of a previously sequenced HeLa-adapted laboratory strain [31] at approximately 200 bases (data not shown). This represents a difference of approximately 3%, which is similar to the genetic discordance between a mouse and a rat. These findings underscore the utility of having a stable sequence source for RV-A16 that can be used to make virus suspensions with a high degree of sequence identity that can be used for both inoculation studies and in vitro studies.
This study was designed to identify the dose that would cause colds of moderate severity in at least 75% of study subjects. Symptoms and viral RNA shedding were both dose related. It is notable that 5 of 26 (19%) volunteers who were inoculated developed infections with community-acquired viruses. In these subjects, the community-acquired viruses were present but not yet symptomatic at the day of inoculation, and either coinfections or sequential infections were established. This demonstrates the importance of screening for other viruses during inoculation studies and determining the type of any viruses that are detected.
One challenge in conducting this study was the prolonged regulatory pathway for this first-in-human recombinant RV inoculum (Supplemental Table 3). Some of the delays were due to regulatory changes that were prompted by the 2009 H1N1 influenza epidemic, raising concerns about introducing a virus into the wild that had been synthesized in the laboratory, albeit from wild-type sequences. In addition, the detection of porcine circovirus in a live-attenuated rotavirus vaccine in 2010 [32] led to additional requirements for testing of the viral inoculum for bovine and porcine viruses that could theoretically be introduced by animal products (eg, serum) used to manufacture RG-RV-A16. Now that RG procedures for RV have been established, production of additional viral vaccines or inocula using recombinant techniques should be straightforward. Once the virus is cloned, production is a 2-day process. One significant delay in the traditional production process of inocula grown from nasal secretions is that the virus donor must undergo thorough testing for other infectious agents, and a 1-year follow-up is recommended [13]. Using a cloned virus instead of nasal secretions as the source of the seed virus obviates this 1-year delay.
Strengths of this study include the use of a novel viral inoculum, use of viral diagnostics to identify community viruses in the study subjects, and molecular characterization of all viruses detected. We also documented for the first time the low risk (1 in 42 [2.4%]) for transmission of inoculum virus to household contacts. Another potential advantage of the RG approach is that it could be used in the future to study effects of mutations of specific viral sequences. Limitations include inclusion of healthy adults as study subjects; it is possible that individuals with chronic respiratory disease (eg, asthma, COPD) could develop upper and even lower respiratory illnesses with lower doses of virus [33].
CONCLUSIONS
In conclusion, we have developed the first recombinant RV inoculum using RG techniques and demonstrated that it induces colds that are of similar intensity to those caused by traditional inocula. Using a cloned virus eliminates one potential source of contamination and provides a genetically stable source and a simplified manufacturing pathway for production of RV for human inoculation studies. Furthermore, because several RV clinical strains have been cloned [34, 35], these techniques could be used to produce additional inocula, including RV-C, for investigations into RV pathogenesis and the efficacy of antiviral drugs or vaccines.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank John Centanni (Institute for Clinical and Translational Research, University of Wisconsin-Madison) for assistance in preparing The Investigational New Drug application to the US Food and Drug Administration.
Fincancial support. This work was funded by the National Institute of Allergy and Infectious Diseases through Grant Numbers U19 AI070503 and U19 AI104317 and the Clinical and Translational Science Award program through the National Institutes of Health (NIH), National Center for Advancing Translational Sciences, Grant UL1TR000427.
Potential conflicts of interest. J. E. G. reports the following: grants from NIH during the conduct of the study; and personal fees from PREP Biopharm Inc., Regeneron, MedImmune, and Meissa Vaccines Inc. outside the submitted work. L. D. reports grants from NIH during the conduct of the study and personal fees from AstraZeneca and Sanofi outside the submitted work. R. G., M. E., and Y. A. B. report grants from NIH during the conduct of the study. J. E. G. is a consultant to Regeneron, PREP Biopharma, and Meissa Vaccines. L. D. is a consultant to AstraZeneca and Sanofi. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Gern JE, Palmenberg AC. Rhinoviruses. In: Knipe DM, Howley PM, eds. Field’s Virology. Philadelphia: Lippincott Williams & Wilkins, 2013. [Google Scholar]
- 2. Mallia P, Message SD, Gielen V, et al. . Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am J Respir Crit Care Med 2011; 183:734–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D’Alessio DJ, Meschievitz CK, Peterson JA, Dick CR, Dick EC. Short-duration exposure and the transmission of rhinoviral colds. J Infect Dis 1984; 150:189–94. [DOI] [PubMed] [Google Scholar]
- 4. Turner BW, Cail WS, Hendley JO, et al. . Physiologic abnormalities in the paranasal sinuses during experimental rhinovirus colds. J Allergy Clin Immunol 1992; 90:474–8. [DOI] [PubMed] [Google Scholar]
- 5. Lemanske RF Jr, Dick EC, Swenson CA, Vrtis RF, Busse WW. Rhinovirus upper respiratory infection increases airway hyperreactivity and late asthmatic reactions. J Clin Invest 1989; 83:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheung D, Dick EC, Timmers MC, et al. . Rhinovirus inhalation causes long-lasting excessive airway narrowing in response to methacholine in asthmatic subjects in vivo. Am J Respir Crit Care Med 1995; 152:1490–6. [DOI] [PubMed] [Google Scholar]
- 7. Proud D, Turner RB, Winther B, et al. . Gene expression profiles during in vivo human rhinovirus infection: insights into the host response. Am J Respir Crit Care Med 2008; 178:962–8. [DOI] [PubMed] [Google Scholar]
- 8. Hayden FG, Gwaltney JM Jr. Intranasal interferon-alpha 2 treatment of experimental rhinoviral colds. J Infect Dis 1984; 150:174–80. [DOI] [PubMed] [Google Scholar]
- 9. Contoli M, Message SD, Laza-Stanca V, et al. . Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med 2006; 12:1023–6. [DOI] [PubMed] [Google Scholar]
- 10. Beale J, Jayaraman A, Jackson DJ, et al. . Rhinovirus-induced IL-25 in asthma exacerbation drives type 2 immunity and allergic pulmonary inflammation. Sci Transl Med 2014; 6:256ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Toussaint M, Jackson DJ, Swieboda D, et al. . Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat Med 2017; 23:681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Group IEW. Guidance for Industry: E6 Good Clinical Practice Consolidated Guidance. In: Guidance document on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use Silver Spring, MD: US Department of Health and Human Services, Food and Drug Administration, 1996. [Google Scholar]
- 13. Gwaltney JM Jr, Hendley O, Hayden FG, et al. . Updated recommendations for safety-testing of viral inocula used in volunteer experiments on rhinovirus colds. Prog Med Virol 1992; 39:256–63. [PubMed] [Google Scholar]
- 14. Gaynor AM, Nissen MD, Whiley DM, et al. . Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 2007; 3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mackay IM, Arden KE. MERS coronavirus: diagnostics, epidemiology and transmission. Virol J 2015; 12:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. US Department of Health and Human Services, Foodand Drug Administration, Centers for Drug Evaluation and Research (CDER). Guidance for Industry, Investigators, and Reviewers: Exploratory IND Studies, Silver Spring, MD, 2006. https://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm078933.pdf. Accessed 13 November 2018. [Google Scholar]
- 17. Centers for Drug Evaluation and Research, Centers for Biologics Evaluation and Research. Guidancefor Industry: Content and Format of Investigational New Drug Applications (INDs) for Phase I Studies of Drugs, Including Well-Characterized, Therapeutic, Biotechnology-Derived Products. Silver Spring, MD: US Department of Health and Human Services, Food and Drug Administration, 1995. https://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm071597.pdf. Accessed 13 Novenber 2018. [Google Scholar]
- 18. Bochkov YA, Grindle K, Vang F, Evans MD, Gern JE. Improved molecular typing assay for rhinovirus species A, B, and C. J Clin Microbiol 2014; 52:2461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DeMore JP, Weisshaar EH, Vrtis RF, et al. . Similar colds in subjects with allergic asthma and nonatopic subjects after inoculation with rhinovirus-16. J Allergy Clin Immunol 2009; 124:245–52, 252.e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kloepfer KM, Lee WM, Pappas TE, et al. . Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol 2014; 133:1301–7, 1307.e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warshauer DM, Dick EC, Mandel AD, Flynn TC, Jerde RS. Rhinovirus infections in an isolated antarctic station. Transmission of the viruses and susceptibility of the population. Am J Epidemiol 1989; 129:319–40. [DOI] [PubMed] [Google Scholar]
- 22. Calhoun WJ, Swenson CA, Dick EC, et al. . Experimental rhinovirus 16 infection potentiates histamine release after antigen bronchoprovocation in allergic subjects. Am Rev Respir Dis 1991; 144:1267–73. [DOI] [PubMed] [Google Scholar]
- 23. Weidner TG, Cranston T, Schurr T, Kaminsky LA. The effect of exercise training on the severity and duration of a viral upper respiratory illness. Med Sci Sports Exerc 1998; 30:1578–83. [DOI] [PubMed] [Google Scholar]
- 24. Calhoun WJ, Dick EC, Schwartz LB, Busse WW. A common cold virus, rhinovirus 16, potentiates airway inflammation after segmental antigen bronchoprovocation in allergic subjects. J Clin Invest 1994; 94:2200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gern JE, Galagan DM, Jarjour NN, Dick EC, Busse WW. Detection of rhinovirus RNA in lower airway cells during experimentally induced infection. Am J Respir Crit Care Med 1997; 155:1159–61. [DOI] [PubMed] [Google Scholar]
- 26. Parry DE, Busse WW, Sukow KA, et al. . Rhinovirus-induced PBMC responses and outcome of experimental infection in allergic subjects. J Allergy Clin Immunol 2000; 105:692–8. [DOI] [PubMed] [Google Scholar]
- 27. Gern JE, Mosser AG, Swenson CA, et al. . Inhibition of rhinovirus replication in vitro and in vivo by acid-buffered saline. J Infect Dis 2007; 195:1137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gern JE, Stone CK, Nakano M, et al. . Effect of upper respiratory tract infection on AIR inhaled insulin pharmacokinetics and glucodynamics in healthy subjects. Clin Pharmacol Ther 2008; 83:307–11. [DOI] [PubMed] [Google Scholar]
- 29. Cordey S, Junier T, Gerlach D, et al. . Rhinovirus genome evolution during experimental human infection. PLoS One 2010; 5:e10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bochkov YA, Watters K, Basnet S, et al. . Mutations in VP1 and 3A proteins improve binding and replication of rhinovirus C15 in HeLa-E8 cells. Virology 2016; 499:350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee WM, Wang W, Rueckert RR. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes 1995; 9:177–81. [DOI] [PubMed] [Google Scholar]
- 32. Dubin G, Toussaint JF, Cassart JP, et al. . Investigation of a regulatory agency enquiry into potential porcine circovirus type 1 contamination of the human rotavirus vaccine, Rotarix: approach and outcome. Hum Vaccin Immunother 2013; 9:2398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mallia P, Message SD, Kebadze T, et al. . An experimental model of rhinovirus induced chronic obstructive pulmonary disease exacerbations: a pilot study. Respir Res 2006; 7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakagome K, Bochkov YA, Ashraf S, et al. . Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol 2014; 134:332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bochkov YA, Palmenberg AC, Lee WM, et al. . Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med 2011; 17:627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.