Abstract
The bidirectional interaction between pancreatic cancer (PanCa) and diabetes has been confirmed by epidemiological studies, but until now, the underlying molecular mechanisms for this connection is not fully understood yet. Here, we analyzed the clinical and genomic data of 26 pancreatic ductal adenocarcinoma (PDAC) patients without diabetes, and six diabetic PDAC patients, whose tumors underwent targeted next-generation sequencing (551 cancer-related genes included). Ingenuity Pathway Analysis (IPA) was performed to investigate genetic alterations and biological consequences associated with PDACs with or without diabetes. We identified 345 somatic mutations of 153 genes in test cohort and a positive association between diabetes duration and somatic mutation burden. KRAS, TP53, and SMAD4 were the top3 commonly mutated genes at a similar frequency compared to the Cancer Genome Atlas (TCGA) data. Several novel but infrequent mutations in other genes (MDC1, PRB2, and PRB4) were also found. Besides, 13 mutated genes (PIK3CD, SNCAIP, IRF4, HLA-A, NOTCH4, PIM1, ETV6, B2M, CD70, PRDM14, TGFBR1, FLT1, and PARP2) were uniquely found in the diabetic group, mainly involved in immune-related pathways. Further targeted sequencing of these genes in an independent validation cohort (n = 50) revealed significant enrichment in the diabetic group (n = 18, P = 2.6964E-08). Long-standing diabetes (≥3-year duration) may induce increasing somatic mutations with time, facilitating tumor initiation. Gene mutants associated with immune-related pathways could be used to distinguish the diabetic PDAC patients from the non-diabetic cases and allow more selective treatment.
Introduction
Pancreatic cancer (PanCa) is currently the fourth leading cause of cancer-associated mortality and predicted to be the second leading cause within the next decade in developed countries. Risk factors, including chronic pancreatitis, Type 2 diabetes mellitus (T2DM), and tobacco smoking, account for over one-quarter of cases. Pancreatic cancer therapy remains a formidable challenge due to its late diagnosis and a notorious resistance to most conventional treatments, resulting in a low 5-year survival rate of 8% [1], [2]. Actually, therapeutic options are limited and progress in drug development is hampered by the complexity of pancreatic cancers at the genomic, epigenetic and metabolic levels, with multiple aberrant pathways and crosstalk, which deserves further investigation.
Diabetes mellitus (DM) is an endocrine disease among the top 10 leading causes of death globally, which becomes one of the largest global health emergencies of the 21st century. According to the latest global estimate from the International Diabetes Federation (IDF), there were approximately 425 million people (20–79 years) worldwide with diabetes in 2017. This number will increase to 629 million by 2045 in this trend [3]. There is an obvious association between diabetes and pancreatic cancer, although it is still under debate which is the cause. Long-standing T2DM is known to be a risk factor for PanCa; however, increasing clinical and epidemiological evidence indicates pancreatic ductal adenocarcinoma (PDAC) as also a presumed cause of diabetes due to unclear mechanisms [4].
In the mutational landscape of pancreatic cancer, the most commonly mutated genes are KRAS (KRAS Proto-Oncogene, GTPase), CDKN2A (cyclin-dependent kinase inhibitor 2A), TP53 (tumor protein p53), and SMAD4 (SMAD family member 4). KRAS mutation is almost ubiquitous and present in >90% of tumors, whereas TP53, CDKN2A and SMAD4 occur in 50–80% of pancreatic cancers. However, none of them are currently druggable [5], [6], [7]. Unlike PDAC, the genetic risk of T2DM cannot be delineated as due to mutations in major driver genes. Similarly, lower-frequency and rare variants don't contribute significantly to disease risk as well [8]. Arising from the same organ, diabetes and pancreatic cancer tend to occur concurrently. Despite the well-known association of these two diseases, the differential molecular profiles in PDAC with or without diabetes remain elusive.
In this study, we explored the molecular signatures of PDAC with or without diabetes by NGS-based gene panel sequencing in a test cohort of 32 subjects and identified a putatively positive association between somatic mutation burden and diabetes duration in PDAC patients. Apart from the similar patterns of commonly mutated genes in PDAC compared to TCGA database, we found somatic mutations in 13 genes that were specifically present in PDAC patients with diabetes. Ingenuity Pathway Analysis (IPA) showed that these genes were enriched in immune-related signaling pathways. This result may help to better understand the underlying mechanism of PDAC and diabetes, providing novel insights into new therapeutic opportunities in this specific subgroup.
Material and Methods
Patients and Samples
Fresh tumor samples were collected by fine needle biopsy, and matched blood samples were collected after fine needle aspiration (FNA) procedure, and sequenced after pathological diagnosis at Changhai Hospital (Shanghai, People's Republic of China) in 2018. None of them had received therapeutic procedures, for instance, chemotherapy or radiotherapy. Diabetes was defined as two fasting glucose measurements above 7.0 mmol/l (126 mg/dl). Patients' baseline data (gender, age, BMI, etc.) and medical history were collected through inpatient inquiry. Lymph node metastasis, distant metastasis and tumor staging were determined by imageological examination (CT, MRI, etc.) and endoscopic ultrasonography after admission. Clinicopathological features of the 32 patients for NGS-based gene panel sequencing (551 genes) were listed in Table 1. And another 50 cases were subsequently recruited for validation by deep sequencing with a specific 13-gene panel, the clinicopathological features of which was summarized in Table 2. Samples were frozen immediately in liquid nitrogen and stored at −80°C until analysis.
Table 1.
Demographic and clinicopathologic characteristics of 32 PDACs with or without T2DM in test cohort
| Parameters | PDAC without T2DM | PDAC with T2DM | P value |
|---|---|---|---|
| Age (years) | |||
| >60 | 11 | 3 | |
| ≤60 | 15 | 3 | 1.000 |
| Gender | |||
| Female | 7 | 2 | |
| Male | 19 | 4 | 1.000 |
| Stage | |||
| I-II | 8 | 1 | |
| III-IV | 18 | 5 | 0.648 |
| Lymphatic metastasis | |||
| Positive | 12 | 3 | |
| Negative | 5 | 0 | 0.539 |
| Remote metastasis (Liver) | |||
| Positive | 9 | 3 | |
| Negative | 17 | 3 | 0.647 |
Table 2.
Demographic and clinicopathologic characteristics of 50 PDACs with or without T2DM in validation cohort
| Parameters | PDAC without T2DM | PDAC with T2DM | P value |
|---|---|---|---|
| Age (years, median) | 60 (39–74) | 70.5 (46–83) | 0.077 |
| Gender (Female/male) | 11/21 | 9/9 | 0.370 |
| Stage (I + II/III + IV) | 27/5 | 16/2 | 1.000 |
DNA Extraction and Quality Control
Genomic DNA (gDNA) from fresh frozen tissues was extracted by GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany). Quantity and purity of gDNA were assessed by Qubit® 3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE, USA). Fragmentation status were evaluated by the Agilent 2200 TapeStation system using the Genomic DNA ScreenTape assay (Agilent Technologies, Santa Clara, CA, USA) to produce a DNA Integrity Number (DIN). An additional quality control (QC) step to assess fresh frozen tissue DNA integrity was performed using a multiplex Polymerase Chain Reaction (PCR) approach [29]. Briefly, 30 ng of gDNA were amplified using three different-size set of primers of Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) gene (200–400 base pair), and the concentration of PCR products was determined by Agilent 2100 Bioanalyzer instrument (Agilent Technologies). Then, to estimate fresh frozen tissue gDNA fragmentation, we evaluated an Average Yield Ratio (AYR) value, calculated by yield ratio of each amplicon compared with a reference DNA (Promega Madison, WI, USA).
Library Preparation and Hybridization Capture
A total of 300 ng of each gDNA sample based on Qubit quantification was mechanically fragmented on a E220 focused ultrasonicator Covaris (Covaris, Woburn, MA, USA); 200 ng of sheared gDNA was used to perform end repair, A-tailing, and adapter ligation with either Agilent SureSelect XT (Agilent Technologies) or KAPA library preparation kits (Kapa Biosystems Inc. Wilmington, MA, USA), following the manufacturer's instructions. Subsequently, the libraries were captured using Agilent SureSelect XT custom 0.5–2.9 M (Agilent Technologies) probes, and amplified.
Illumina Sequencing
After QC and quantification by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) and Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA), libraries were sequenced on an Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA) High output mode using 2 × 150 cycles was performed with TruSeq SBS v3 chemistry. For each library preparation type, 10 samples were loaded in a single lane of a flow-cell v3.
NGS-Based Gene Panel Sequencing
Thirty-two DNA samples were analyzed for target-capture sequencing of 551 genes with the SureSelect Target Enrichment Kit on an Illumina HiSeq 2500 platform. The average coverage of the targeted region was 600×, and 95% of the target was covered at >50×. Sequencing reads were aligned to the human genome (NCBI build 37) with the BWA algorithm on default settings. Moreover, another fifty DNA samples were analyzed for target-capture sequencing of 13 genes with the same method. Finally, 82 cases passed internal quality control and quality matrix and were included in further analyses.
Bioinformatic Analysis
BCL files generated by Illumina for the panel sequencing samples were converted to FASTQ format by CASAVA software (v.1.8.1, Illumina) and aligned to the human reference genome hg19 with the Burrows-Wheeler Aligner. Local realignment, PCR duplicate marking, base-quality recalibration, and calculation of coverage metrics were performed with GATK33 and Picard tools. Putative somatic SNVs and indels were called with MuTect2. The identified variants were annotated with ANNOVAR. We only considered variants that passed the standard MuTect filters, and we excluded common SNPs with minor allele frequency of >0.01 as recorded in dbsnp138, the NHLBI exome sequencing project, 1000 Genomes. We also excluded variants in non-coding regions, synonymous variants, and variants present in highly repetitive regions. IPA (ingenuity pathway analysis) was used for pathway enrichment analysis.
Statistical Analysis
Statistical analysis was performed using IBM SPSS Statistics 22.0 (SPSS, Chicago, IL) and GraphPad Prism 5 (GraphPad, San Diego, CA). Demographic and clinicopathologic characteristics, as well as mutation burden and frequency, were compared using Fisher's exact test for categorical variables, and a Student's t test was used for continuous variables. The distribution of somatic mutation burdens in the tumor stages was analyzed with Kruskal-Wallis Test (Nonparametric One-way ANOVA) and Dunn's Multiple Comparison Test. A two-tailed P < .05 was considered statistically significant.
Results
Demographic and Clinicopathologic Characteristics of Samples
In the test cohort (Table 1), median age was 60 (range 51–69) and 57(range 31–85) years for diabetic and non-diabetic PDAC patients, respectively. There was no significant difference in demographic and clinicopathologic characteristics between groups (P > .05). Likewise, no statistically significant differences were observed in the validation group (Table 2). Among a total of 32 patients in the test cohort, 12 were diagnosed as Nx in TNM staging, the lymph nodes of whom cannot be evaluated in this way. These data thus were not presented in Table 1.
Detection of Somatic Mutations in Test Cohort of PDACS With or Without Diabetes
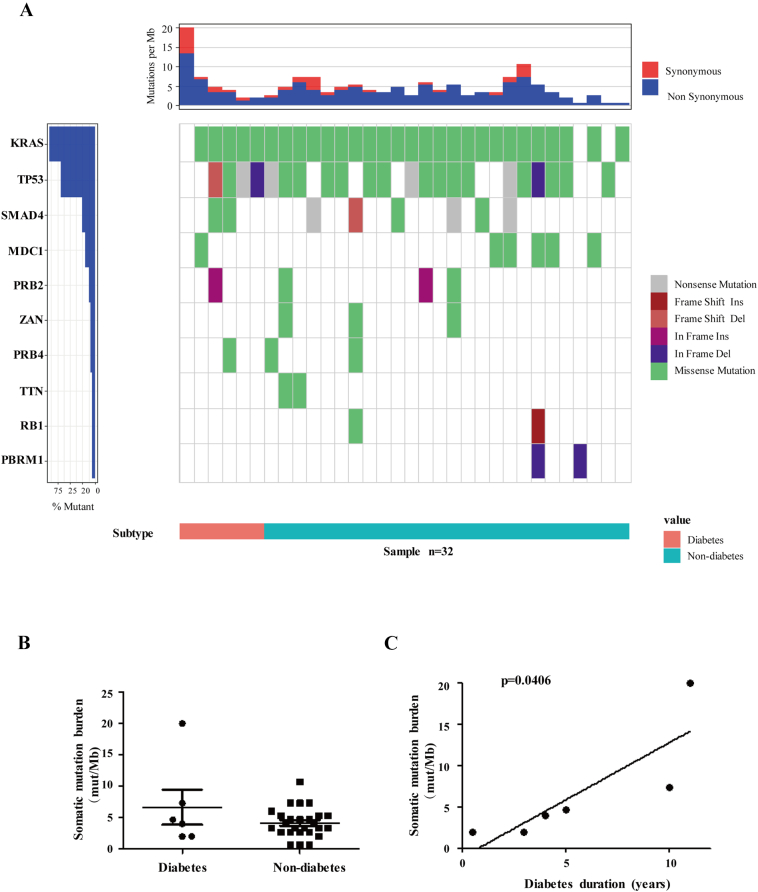
We characterized 32 PDAC cases with a 551 gene panel, six of which were with diabetes. Totally, 345 somatic mutations in 153 genes were detected, including 249 SNVs and 96 indels. As shown in Figure 1A, commonly mutated genes that characterize PDAC (KRAS 90.63% vs. 65.4%, TP53 68.75% vs. 59.8%, SMAD4 25% vs 20.7%) were reaffirmed in our result with a higher frequency as reported by the Cancer Genome Atlas (TCGA) Research Network (http://www.cbioportal.org/study?id=paad_tcga_pan_can_atlas_2018#summary). Interestingly, CDKN2A mutation, as a famous hotspot mutation in PDAC, was not detected in most of the cases. Instead, we identified mutations in MDC1 (mediator of DNA damage checkpoint 1), PRB2, and PRB4 genes in more than four cases, accounting for over 10% of 32 subjects, which were not reported by TCGA. Overall, the targeted capture data from our Biotecan PanCancer Panoramic Detection (BTC-PCPD) panel were consistent with the TCGA findings.
Figure 1.
Mutational signatures and tumor mutation burden derived from BTC-PCPD targeted sequencing data of test cohort. (A) Frequency and types of mutations in top 10 genes identified by targeted sequencing. Different mutations and subtypes are colored in the middle panel. Frequency of mutations is shown on left (%) with dark blue color. Pink bar represents diabetes, while light blue bar stands for non-diabetes at the bottom. Top panel shows the somatic mutation burden. (B) Scatter diagram depicting the somatic mutation burdens (mutations/Mb) in the diabetic and non-diabetic groups. (C) The association between tumor mutation burden (mutations/Mb) and diabetes duration (years) in six diabetic PDACs.
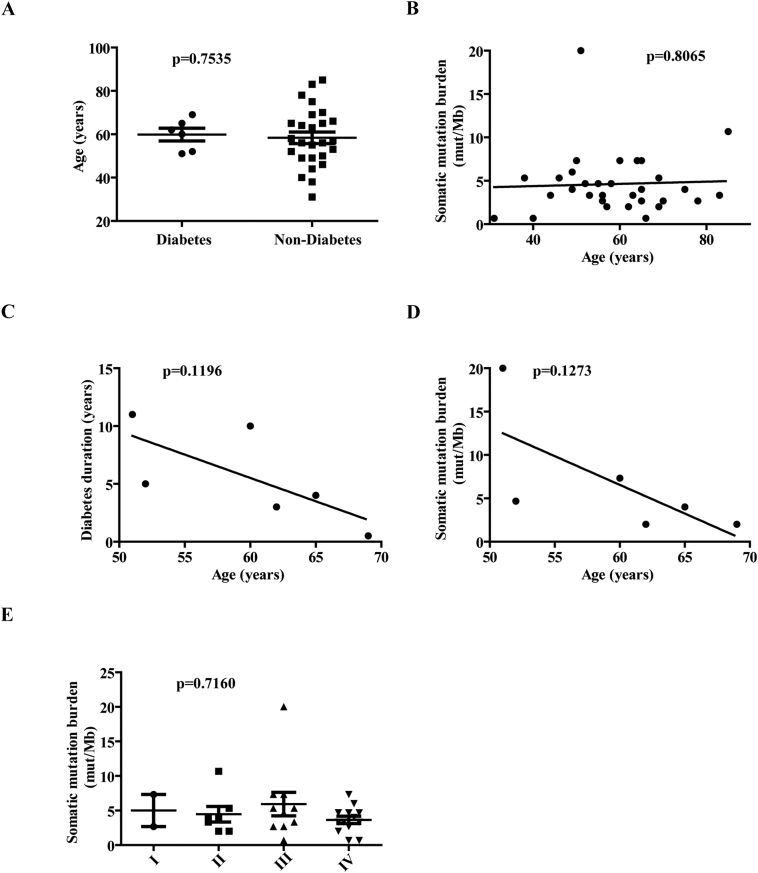
For 26 non-diabetic primary tumors, their median somatic mutation burden showed 3.665 (range 0.67–10.67) (mutations/MB), while for the six diabetic tumors, the number was 4.335 (range 2–20). Unfortunately, no significant differences of somatic mutation burden between PDAC patients with or without diabetes were identified in this cohort (Figure 1B), which might be attributed to the limited size. Nevertheless, a positive association between somatic mutation burden and diabetes duration was identified in the test cohort of diabetic PDAC patients, suggesting the negative contribution of diabetes to genomic stability (Figure 1C). Considering the higher mutation frequency in long-term diabetes could be a consequence of older age, we also explored the distribution of age in diabetic and non-diabetic group, and the association between age and somatic mutation burden in the overall test cohort and diabetic group, separately (Supplementary Figure S1, A–D). Actually, no significant difference or association was found, suggesting that the higher mutation frequency in long-term diabetes is not due to old age in this study. Although patients with stage III PDAC had the highest mean tumor mutation burden, Kruskal-Wallis and Dunn's multiple comparison test showed that there was no significant difference among tumor stages (P = .7160), indicating that tumor mutation burden is probably not related to tumor stage in the test cohort (Supplementary Figure S1E).
Supplementary Figure S1.
Distribution of age, and the association between age and somatic mutation burden in the test cohort. (A) Scatter diagram depicting the distribution of age (years) in the diabetic and non-diabetic groups. (B, D) The association between tumor mutation burden (mutations/Mb) and age (years) in test cohort (B) and six diabetic PDACs (D). (C) The association between diabetes duration and age (years) in six diabetic PDACs. (E) Scatter diagram depicting the somatic mutation burdens (mutations/Mb) in the tumor stages.
Frequently Mutated Pathways in PDACs With or Without Diabetes
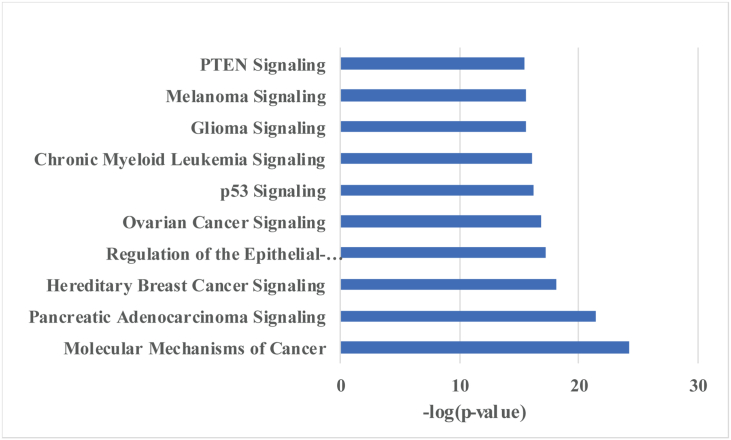
All somatic mutations were further analyzed using Ingenuity Pathway Analysis software. The result of this analysis is summarized in Figure 2 and shows that the mutated genes are most representative to the p53, PTEN and several cancer-related signaling pathways.
Figure 2.
Key pathways analysis of the test cohort by IPA.
Analysis of Somatic Mutation Differences Between Two Groups
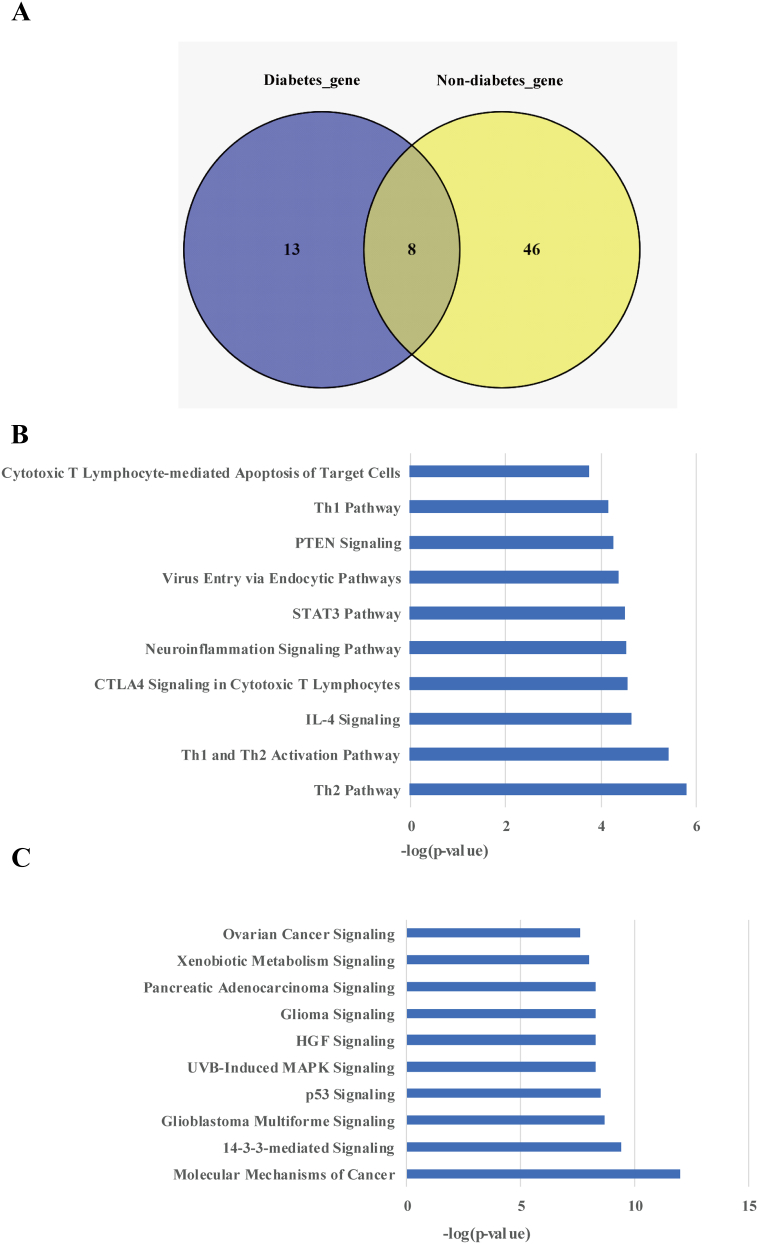
To further understand the differential molecular signatures, we next compared somatic mutations between diabetic and non-diabetic groups, and found eight mutated genes were overlapped (Figure 3A). In addition, 13 mutated genes were only identified in diabetic PDACs, while 46 mutated genes were specifically present in non-diabetic group. Pathway enrichment analysis with IPA demonstrated that the 13 genes related to diabetic PDACs were prevailingly distributed in immune-related pathways like Th2 Pathway, Th1 and Th2 Activation Pathway, IL-4 Signaling, etc. (Figure 3B). Comparatively, the other 46 genes specifically present in non-diabetic group were more enriched in pathways associated with Molecular Mechanisms of Cancer, 14–3-3-mediated signaling, and p53 Signaling (Figure 3C).
Figure 3.
Analysis of the mutated genes and related pathways in diabetic and non-diabetic groups. (A) Venn diagram of genes with somatic mutations in diabetic and non-diabetic groups. Key pathways analysis of the (B) diabetic and (C) non-diabetic PDACs in test cohort by IPA.
Validation Cohort Analysis of Gene-Panel Targeted Sequencing
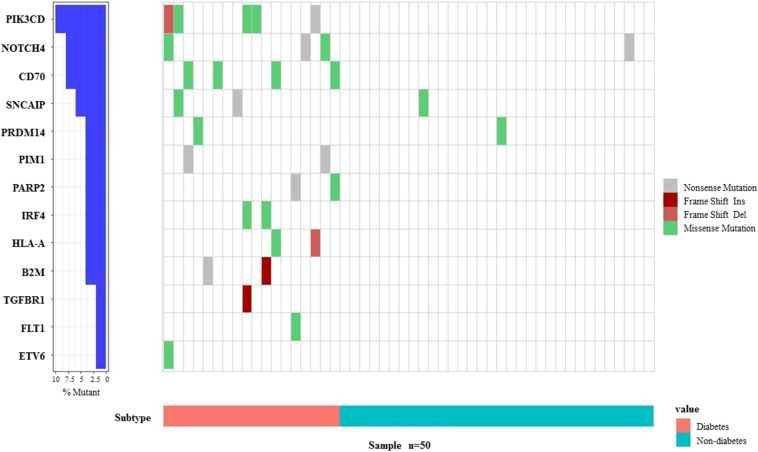
To verify the differences of somatic mutations between two groups, we further tested the 13 genes that specifically exist in the diabetic group in another independent cohort of 50 PDAC patients. Among them, 18 cases were with diabetes, and 32 cases were non-diabetic PDACs. In the diabetic group, there was at least one gene mutation in 16 cases (16/18), while in the non-diabetic group, only three cases had mutations (3/32). The extremely significant difference (P = 2.6964E-08) between them suggests the specificity of these genes to distinguish the diabetic PDAC patients from the non-diabetic cases (Figure 4).
Figure 4.
Validation of mutations in the 13 genes detected in diabetic PDACs within an independent cohort (n = 50). Colors depict different types of mutations as described in.
Discussion
Increasing epidemiological studies have shown that the prevalence of diabetes among PDAC patients is much higher than that among other malignancies since these two diseases originate from the same organ and share a set of canonical risk factors such as age, obesity, and a family history of diabetes [9]. In a study with 512 newly diagnosed pancreatic cancer (PanCa) cases and 933 controls of similar age, diabetes was reported to be more prevalent (47% vs 7%; P < .001) in PanCa. Among PanCa with diabetes in this cohort, 74% cases are new onset (<2-year duration), suggesting that new-onset diabetes is probably caused by tumor and could be used as a potential biomarker for the diagnosis of PDAC [10], [11]. Despite the close relationship between PDAC and T2DM, the molecular mechanism to address this connection is still obscure. Here we offer data regarding genomic patterns of key genes related to PDAC patients with or without diabetes.
Considering the high cost and uncertain meaning of mutation profiling like whole exome sequencing (WES) or whole genome sequencing (WGS) in clinical application, we developed and implemented BTC-PCPD, a hybridization capture–based NGS panel to detect all protein-coding mutations, selected copy number alterations (CNAs), and promoter mutations in 521 (and, more recently, 551) cancer-associated genes. With this method, we sequenced 32 PDAC cases with or without diabetes and compared the data with those from untreated primary tumors characterized by TCGA. Although a higher frequency of the major driver genes like KRAS, TP53, and SMAD4 were also identified in our test cohort, to our surprise, CDKN2A was not among the top 10 mutated genes and present in only one case (3.125%,1/32), which could be resulted from the limited sample size or population differences and need further investigation. Demographically, there are more untreated patients with advanced cancer (Stage III + IV, 71.9%) in our test cohort, which differs substantially from TCGA cohort characterized in primary, untreated cases (stage III + IV, 5.4%). Intriguingly, we also identified mutations in MDC1, PRB2, and PRB4 genes in more than four cases, which were not reported in TCGA data. As a DNA damage response (DDR) mediator, MDC1 is known to maintain genomic stability. Besides, its expression is negatively associated with perineural and lymph node invasion and metastasis of PDAC under regulation of Sox9 and oncogenic Kras [12]. PRB2, and PRB4 mutations have been detected in several types of cancers [13], [14], [15], but their functions in tumor development and metastasis need to be further explored.
Mechanically, the abnormal carbohydrate and lipid metabolism, high levels of circulating insulin, as well as chronic inflammation resulted from Type 2 diabetes, could lead to the alteration of energy sensing pathways such as the AMP-activated protein kinase (AMPK), mechanistic (mammalian) target of rapamycin (mTOR), sirtuins, and autophagy signaling pathways, which might increase the risk of gene mutation and cancer genesis by decreasing genetic stability and DNA mismatch repair [16], [17], [18], [19]. Consistently, we observed slightly increased somatic mutation burden in diabetic PDAC patients compared with the non-diabetic patients. A positive association between somatic mutation burden and diabetes duration was also found in the diabetic group. In the six diabetic cases, only 1 is new onset (6-month duration) and the somatic mutation burden of this 69-year-old female is 2 (mutations/Mb) with stage IIB. For the remaining five long-standing (3- to 11-year duration) DM patients, the somatic mutation burden increases slowly with time among patients with diabetes for ≤10 years. Since there was no significant association between diabetes duration and age of patients, our observation of the positive link between somatic mutation burden and diabetes duration in the diabetic PDACs provides new evidence for the contribution of long-standing diabetes to PDAC development.
Specifically, 13 genes (PIK3CD, SNCAIP, IRF4, HLA-A, NOTCH4, PIM1, ETV6, B2M, CD70, PRDM14, TGFBR1, FLT1, and PARP2) stand out in the diabetic PDAC group, which are predominately present in immune-related pathways. The pathophysiological changes of T2D are mainly characterized by β-cell dysfunction, insulin resistance, and chronic inflammation [20]. As expected, we observed mutations of the 13 genes were enriched in STAT3 and activation of pro-inflammatory Th1 pathways. Surprisingly, the IPA result also showed Interleukin (IL)-4 and activation of Th2 signaling pathways. IL-4, the major product of Th2 cells, is also a principle inducer of Th2. This strong positive feedback also exists in other Th differentiations like Th1 or Th17 [21]. The Th2 cytokines, especially IL-4 and IL-13, mediate immune responses and the immune microenvironment under normal physiological conditions, as well as in cancer. They are important in humoral immunity and involved in multiple biological effects including cell survival and adhesion, tumor proliferation as well as metastasis. Besides, they also play a role in modulating the immune system for tumor growth and cancer stem cells [22]. The mutated genes enriched in activation of Th2 and IL-4 signaling pathways, might be a result of T2D, which further contributed to tumorigenesis and development of PDAC. But it needs more evidence to prove whether this is specific for diabetic PDACs.
Overall, we depicted a part of the unique genomic picture of diabetic PDACs in this study, however, the significance was more or less limited by the sample size. Additionally, of all the six diabetic PDAC patients in test cohort, only 1 belongs to new onset diabetes (≤2-year duration), and the remaining five cases are long-standing (≥3-year duration). Therefore, more new-onset diabetes patients should be recruited and compared with the long-standing cases, which could be improved in future studies.
The following is the supplementary data related to this article.
Acknowledgement
This study was supported by the National Natural Science Foundation of China (81672830), the Shanghai Municipal Education Commission(2017-01-07-00-07-E00012); National Natural Science Foundation of China(81372482); Major Projects of Special Development Funds in Zhangjiang National Independent Innovation Demonstration Zone, Shanghai (ZJ2017-ZD-012)
Contributor Information
Kaixuan Wang, Email: wangkaixuan224007@163.com.
Duowu Zou, Email: zdw_pi@163.com.
Gang Jin, Email: Drjingang@126.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson DA, Hruban RH. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 3.Federation ID. 8th edn. 2017. IDF Diabetes Atlas. [Google Scholar]
- 4.Andersen DK, Korc M, Petersen GM, Eibl G, Li D, Rickels MR, Chari ST, Abbruzzese JL. Diabetes, Pancreatogenic Diabetes, and Pancreatic Cancer. Diabetes. 2017;66(5):1103–1110. doi: 10.2337/db16-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 6.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas L, McCarthy DJ. The genetic architecture of type 2 diabetes. Nature. 2016;536(7614):41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aggarwal G, Kamada P, Chari ST. Prevalence of diabetes mellitus in pancreatic cancer compared to common cancers. Pancreas. 2013;42(2):198–201. doi: 10.1097/MPA.0b013e3182592c96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pannala R, Leirness JB, Bamlet WR, Basu A, Petersen GM, Chari ST. Prevalence and clinical profile of pancreatic cancer-associated diabetes mellitus. Gastroenterology. 2008;134(4):981–987. doi: 10.1053/j.gastro.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma A, Kandlakunta H, Nagpal SJS, Feng Z, Hoos W, Petersen GM, Chari ST. Model to Determine Risk of Pancreatic Cancer in Patients With New-Onset Diabetes. Gastroenterology. 2018;155(3):730–739. doi: 10.1053/j.gastro.2018.05.023. (e3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H, Qin Y, Ji S, Ling J, Fu J, Zhuang Z, Fan X, Song L, Yu X, Chiao PJ. SOX9 activity is induced by oncogenic Kras to affect MDC1 and MCMs expression in pancreatic cancer. Oncogene. 2018;37(7):912–923. doi: 10.1038/onc.2017.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu P, Wu H, Tang Y, Luo S, Fang X, Xie C, He J, Zhao S, Wang X, Xu J. Whole-exome sequencing reveals novel mutations and epigenetic regulation in hypopharyngeal carcinoma. Oncotarget. 2017;8(49):85326–85340. doi: 10.18632/oncotarget.19674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Huang JY, Chen YN, Yuan F, Zhang H, Yan FH, Wang MJ, Wang G, Su M, Lu G. Whole genome and transcriptome sequencing of matched primary and peritoneal metastatic gastric carcinoma. Sci Rep. 2015;5:13750. doi: 10.1038/srep13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan X, Gao H, Wang F, Feng J, Bai J, Zhao P, Cao L, Gui S, Gong L, Zhang Y. Whole-exome sequencing identifies variants in invasive pituitary adenomas. Oncol Lett. 2016;12(4):2319–2328. doi: 10.3892/ol.2016.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer. 2018;18(12):744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 17.Jeon SM. Regulation and function of AMPK in physiology and diseases. Exp Mol Med. 2016;48(7):e245. doi: 10.1038/emm.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19(2):121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tornovsky-Babeay S, Dadon D, Ziv O, Tzipilevich E, Kadosh T, Schyr-Ben Haroush R, Hija A, Stolovich-Rain M, Furth-Lavi J, Granot Z. Type 2 diabetes and congenital hyperinsulinism cause DNA double-strand breaks and p53 activity in beta cells. Cell Metab. 2014;19(1):109–121. doi: 10.1016/j.cmet.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 20.DeFronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ, Hu FB, Kahn CR, Raz I, Shulman GI. Type 2 diabetes mellitus. Nat Rev Dis Primers. 2015;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 21.Paul WE. History of interleukin-4. Cytokine. 2015;75(1):3–7. doi: 10.1016/j.cyto.2015.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki A, Leland P, Joshi BH, Puri RK. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine. 2015;75(1):79–88. doi: 10.1016/j.cyto.2015.05.026. [DOI] [PubMed] [Google Scholar]