Abstract
BACKGROUND: Hepatitis B virus (HBV) is one of the most prominent risk factors for hepatocellular carcinoma (HCC) development and virus-mediated cases represents more than 80% of HCC in East Asia, where it is endemic. Currently, the HBV status of pathological HCC is not fully clarified, especially by comparison to nontumorous tissues. Lack of clinicopathological animal models of HCC impedes clinical application of antiviral treatment in the field. MATERIALS AND METHODS: A cohort sample of 14 HCC and corresponding stroma tissues were analyzed for pathological patterns of HBV antigens using immunohistochemistry; 10 fresh primary tumor tissues were inoculated into NOD/SCID mice and risk factors for patient-derived xenograft (PDX) model were identified by the univariate F test. Consistency of HBV features and cellular biomarkers between patient tissues and tumor grafts were examined. Results: In HCC, HBV surface antigen (HBsAg) was mainly absent. Only 9.9% of samples showed HBsAg positivity in the tumor tissue that was limited to benign hepatocytes. In contrast, HBV core antigen (HBcAg) exhibited positive staining in all HCC tissues, located mainly in the cytoplasm of tumor cells. Of 14 HCC cases, three were diagnosed as occult infection of HBV based on HBcAg expression. The successful rate for the PDX model was 20% (2/10). Tumor lesions on hepatic lobes of V and VI, severe liver dysfunction and higher CA125 showed p-values of 0.01, 0.035, and 0.01, respectively. HBsAg absence in original tumors of #6 and 8 patients were faithfully reproduced by engraftments. Mixed distribution of HBcAg in cellular compartments of original tumor cells was also observed in mice. ki67 was dramatically increased in tumor grafts. Conclusion: We delineated pathological HBV profiles of HCC specimens and perilesional areas, which provided evidence for virus-based therapy in the future. PDX mice may phenocopy virological and cellular features of patient tissues, which is novel in the virus-related hepatocarcinogenesis field.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most prevalent tumor type and the third leading cause of cancer-related deaths worldwide. More than 250,000 new cases of HCC occur annually [1], [2]. Half of cases are thought to occur in China, rendering HCC the top priority in China's anticancer campaign [3], [4]. Major risk factors for HCC include infection with hepatitis virus B or C, alcoholic liver disease, and probably nonalcoholic fatty liver disease [5], [6], [7]. In Asia and Africa, where HBV is endemic, 60% of HCC is associated with HBV and 20% is related to HCV [8].
Many studies of HCC derived from gene expression profiling focus mainly on the host and little on the virus [9], [11]. HCC patients are often analyzed as a single group regardless of the etiological factor involved, and the clinical, virological, and histologic features studied are often limited or missing [9], [11]. Limited information is available on the gene expression profiles of surrounding nontumorous tissue.
Antiviral treatment represents the cornerstone endpoint of all our current therapeutic attempts. The inhibition of viral replication by nucleos(t)ide analogues (NAs) has been shown to achieve the elimination of chronic HBV-induced necroinflammatory activity and progressive fibrotic liver processes in the vast majority of patients, in turn reducing the risk of HCC. However, those data are controversial [12], [13], [14], [15]. A recent retrospective study involving 4568 HBV chronic hepatitis patients indicated that all male patients and female patients aged above 50 years were still at risk of HCC even after HBsAg seroclearance [16]. The authors urgently requested evaluating drugs by parallel systems on standardized HBV-related HCC models, which recapitulate the many forms of this disease.
The current models for HBV-induced HCC mainly comprise transgenic mice expressing specific fragments of the HBV genome [17], [18], [19] These models demonstrate a primary function of viral hepatitis genes in initiating or promoting liver carcinogenesis. However, two features differ from clinical settings; cirrhosis and inflammation are absent in the preneoplastic stages [17], [20]. In addition, mice livers do not permit the formation and persistence of covalently closed circular DNA (cccDNA) of HBV. [21] Another category of model is based on recombinant HBV plasmids in transgenic mice for chronic liver disease [22], [23]. However, whether a patient-derived xenograft (PDX) model can also be applied in virus research is unknown [24], [25], [26]. In the study, we analyzed the clinicopathological pattern of HBV in a unique cohort of HCC samples and matched stroma tissues. Pieces of solid primary tumors maintained as tissue structures were collected by surgical procedures and inoculated into nonobese diabetic severe combined immunodeficiency (NOD/SCID) mice. Both viral expression and cellular biomarkers in clinical HCC tissues were faithfully reproduced by tumor engraftments in mice. This study may suggest a possible application of PDX model in the tumor virology field.
Material and Methods
Ethics Statement
The clinical section of the research had been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). Written informed consent was obtained from all participants, and all samples were anonymized. All participants were adults.
The animal experiments were complied with the ARRIVE guidelines. All animal care and use protocols were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China.
HCC Tissue Acquisition and Processing
tlsb=-0.09pt?>HCC tissues obtained from surgery were washed three times in cold PBS solution and transferred to laboratory in DMEM (Gibco) containing antibiotics (penicillin 250 U/mL, streptomycin 250 mg/mL). Specimen transfer and procedures were under 4 °C or on ice. All procedures from tissue acquisition to tumor implantation were finished in 1 hour. Tumor lesions from surrounding normal tissues were dissected as thoroughly as possible to remove residual hemorrhagic necrosis, fat tissue, and fibrosis. Single or multiple lesions were collected depending on individual specimens. Harvested tumors were divided into pieces and fresh-frozen in liquid nitrogen, fixed in 4% paraformaldehyde, and embedded in paraffin, or cryoprotected consecutively with 5%, 15%, and 30% sucrose prior to embedding in Tissue-Tek OCT embedding medium (Sakura) [27].
Tumor Implantation
A minimum of three mice were used per experimental group. We implanted mixed fragments of fresh tumor (~20 mm3) in Matrigel (growth factors reduced, BD Company), subcutaneously into two sides of flanks of 4-week-old male NOD-SCID mice. Mice (NOD CB17-Prkdcscid/NcrCrl) were from Tengxin Company (Chongqing, China). Tumor growth was measured weekly using calipers. Tumor volumes were calculated using the formula ½ × length × (width) [2]. After 10–12 weeks, visible tumors were detected. When tumors reached 0.5–2.0 cm3, mice were euthanized and killed, and graft tissues were separated as previously described and analyzed for histology and gene expression, or frozen for later use.
Histology Assays
Staining used standard microtome sectioning, deparaffinization, and hydration. To deparaffinize and rehydrate slide sections, xylene and ethanol from 100% to 80% ethanol were used. Hematoxylin was used for staining. Harris hematoxylin with glacial acetic acid was applied for 5 minutes with rinsing with deionized water and acid ethanol. Eosin phloxine was used for transient staining, with 95% and 100% of ethanol and xylene were used for dehydration. Permount mounting medium was used to cover all tissue with drying overnight in a hood. Original magnifications were ×100 and ×400.
Immunohistochemistry Assays
All immunostaining was on paraffin-embedded tissues. Tissue blocks were sectioned, mounted on microscope slides, and heated at 56 °C overnight. Paraffin was removed with xylene and tissues were serially rehydrated through descending ethanol concentrations to water. For immunohistochemistry, antigen retrieval was performed by boiling samples in a microwave oven using 10 mmol/L sodium citrate buffer (pH 6.0). Slides were washed twice in PBS and once in PBS-1% Tween-20 for 15 minutes. Tissue specimens were blocked for 1 hour with PBS containing 3% bovine serum albumin. Slides were incubated with specific primary antibodies at 4 °C overnight: α-HBsAg (Abcam, ab18797) and α-HBcAg at 1:200 (Abcam, ab115992); α-Collagen I (Abcam, ab34710) and α-SMA at 1:100 (Abcam, ab5694), or α-ki67 at 1:50 (Santa Cruz Biotechnology, sc23900). Slides were stained subsequently with corresponding secondary antibodies (goat anti-mouse at 1:2000 and goat anti-rabbit at 1:5000) conjugated to HRP (Thermal Fisher, 62–6520 and 31,460) for 1 hour at room temperature. After color development with diaminobenzidine, sections were counter-stained with hematoxylin. Entire tumor sections within glass slides were evaluated under low and high power magnification (×100 and ×400 objectives).
Statistics
The successful rate of PDX models was calculated and log-rank tests were performed for statistical univariate analysis of prognostic variables. Twenty clinicopathological variables for clinical features, laboratory data, operative findings, and pathological features were analyzed. All parameters were dichotomized based on the most significant parametric factors to examine their influence on PDX model development, except for three ranges of HBsAg titers. In all cases, a P value lower than 0.05 was considered statistically significant.
Results
Characteristics of Patients
We studied 14 patients with HBV-associated HCC. HCC tissues with their peritumor stroma controls from surgical resection were collected from July to November 2017. Of the samples, 10 HCC tissues from (#5 to #14) were injected into the lateral abdomens of NOD/SCID mice. And clinical, serological, and pathological features including grade and size of tumor for those 10 tissues were provided in Supplementary Table 1.
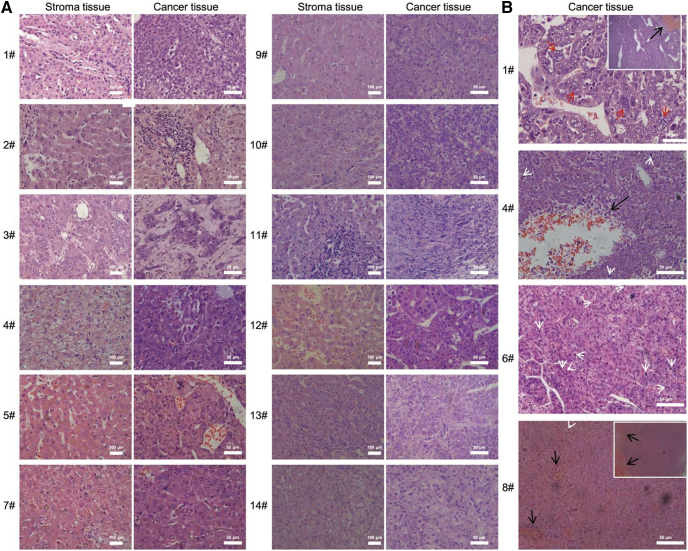
All 14 samples exhibited histological morphology of HCC from well, moderate to poorly differentiated types. Tissues #2, 5, 7, 9, and 12 had well differentiation with polygonal tumor cells that resembled hepatocytes, 2- to 3-cell-thick hepatocellular plates/cords and/or degraded balloon-like cells with enlarged nuclei and prominent nucleoli (Fig. 1A). As the most easily recognizable form of moderate differentiation, tissues #3, 10, 11, 13, and 14 showed a trabecular growth pattern with dilated sinusoids and untypical cell proliferation. A solid growth pattern of samples #3, 11, and 14 with large tumor nodules separated by thick fibrous septa may have represented moderately to poorly differentiated types (Fig. 1A). As with poorly differentiated type, tissues #1, 4, 6, and 8 contained small and large necrosis foci due to thrombi in tumor vessels accompanied by microvessel invasion (Fig. 1B). Other hallmark morphology included giant tumor cells and atypical mitoses indicated and undifferentiated tumor cells (Fig. 1B).
Fig. 1.
Histological morphology and differentiation subtypes were analyzed for pair-matched HCC tissues and corresponding stromas. (A) Typical features of well-differentiated (#2, 5, 7, and 9), middle-grade moderately differentiated (#3, 10, 11, 12, 13, and 14), and grade III poorly differentiated cases (#1, 4, 6, and 8), individually presented. (B) Hematoxylin–eosin staining for four HCC grade III tissues; ×100 fields indicate hemorrhagic necrosis, black arrows. In patient #1, anaplastic tumor giant cells are indicated with red arrows; in patients #4 and 6, with white arrows for mitoses. Original magnifications were ×100 and ×400. Scale bars, 100 μm and 50 μm.
Differential HBV Expression Between HCC Tumors and Peritumor Tissues
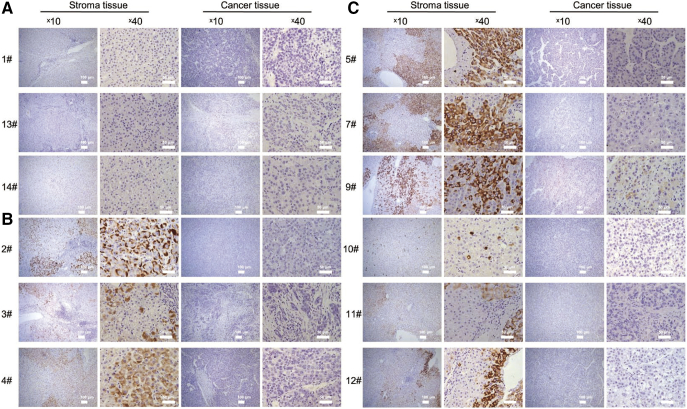
Access to a unique series of pair-matched liver specimens provided us with the opportunity to delineate the intrahepatic pattern of HBsAg and HBcAg in pathological livers for HCC diagnosis from virological aspects. Among the 14 samples series, #1, 13, and 14 were HBsAg seronegative. The absence of HBsAg was also identified in patient liver tissues (Fig. 2A). For the remaining 11 samples, HBsAg was decreased in HCC tissues compared with stroma controls. Immunostaining was positive in 1 of 11 (9.1%) tumor tissues and was positive for 90.9% of nontumor specimens (Figs. 2, B and C, and 5A). In continuous fields of normal and malignant hepatocytes, we observed exclusiveness of HBcAg in tumor nodules (Fig. S1). HBsAg was located with patterns of spotty, diffuse, membranous and cytoplasmic scattering, even in an individual patient (Fig. S2).
Fig. 2.
HBsAg was downregulated in HCC tissues compared to tumor periphery. (A) Cases #1, 13, and 14 with negative signals from serological assays and viral DNA showed no HBsAg in liver tissues. (B, C) HBsAg positive HCC patients had different expression patterns in nontumor and tumor tissue. Distinctive features by cytoplasmic spots (#2 and 10), diffuse (#2, 3, 4, 5, 7, 9, 11, and 10) and membranous (#2, 5, 7, 9, and 12). Scale bars, 100 μm and 50 μm.
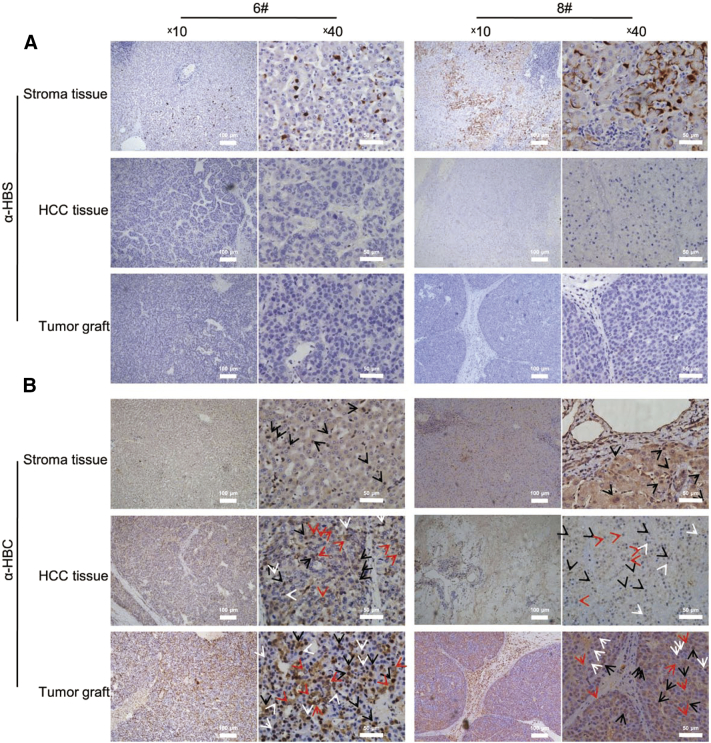
Fig. 5.
HBV viral markers in engraftments were generally consistent with original tissues. (A) HBsAg was nearly absent in HCC tissues compared to adjacent livers. In normal liver tissues, #6 exhibited cytoplasmic spots and #8 showed an additional membranous type. (B) HBcAg exhibited distinctive patterns in HCC tissues that was faithfully recapitulated and increased in engraftments. In normal liver tissues, #6 and 8 were cases of expressing HBcAg in whole hepatocytes. HBcAg decreased in HCC tissues and exhibited low level of nuclear expression. The mixed pattern of cytoplasmic, nuclear and whole cell distribution of HBcAg in original tissues was retained and increased in PDX grafts. Arrows were: black, representative hepatocytes with whole cell HBcAg expression; red, alternative cytoplasmic; white nuclear localization. Scale bars, 100 μm and 50 μm.
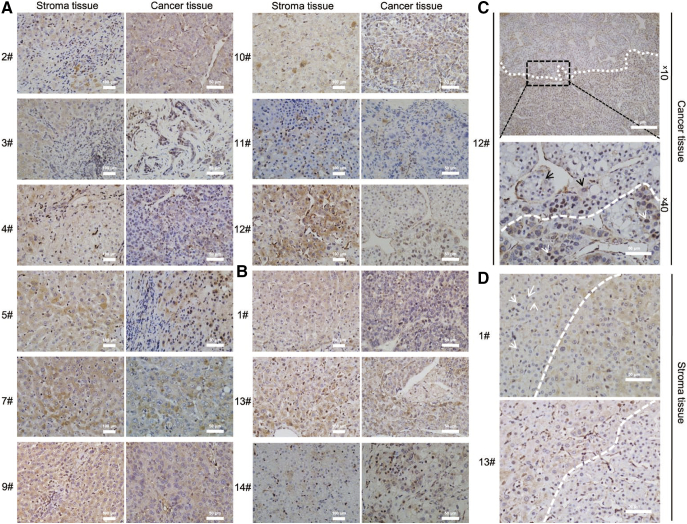
The tendency for decreased HBcAg in tumor tissues was not significant compared to HBsAg. Although all samples were HBcAg positive, levels were lower in tumor tissues compared to corresponding stromas (Figs. 3A, 5B). HBsAg negative samples #1, 13, and 14 showed strong positivity for intrahepatic HBcAg with signals sustained even when the primary antibody was diluted from 1:200 to 1:400 (Figs. 3B and S3), indicating the presence of occult HBV infection. In addition to quantitative differences, locations differed between nontumorous tissues and tumors. HBcAg was localized mainly in the cytoplasm of malignant hepatocytes, except for solid growth types of #3, 11, and 14, which had distribution throughout the cell (Fig. 3, A and B). The results were confirmed by a representative boundary field of hyperplasia tumor cells and benign nodules in sample 12. We observed an exclusive distribution of cytoplasmic HBcAg in tumor cells, characterized by hyperplasia clusters of enlarged and irregular nuclear (Fig. 3C). A distinctive pattern of HBcAg location was observed in different types of cells in normal liver tissue. Regenerative cells with enlarged and light-staining nuclei, had a propensity to have cytoplasmic HBcAg (Fig. 3D).
Fig. 3.
HBcAg expressed at lower levels in HCC tumors than stroma tissues, but positive in each case and limited to cytoplasmic tumor cells. (A) HBcAg was decreased in HCC tissues compared to stroma tissues. Except for #3, 5, and 14, HBcAg was mainly distributed in the tumor cell cytoplasm. (B) Three HBsAg-negative tissues showed strong positivity for HBcAg, indicating HBV occult infection. HBcAg was expressed extensively throughout normal liver and HCC tissues. (C) Cytoplasm of HBcAg in tumor cells had a boundary region of benign neoplasm and nuclear atypia tumor cells. Crossed pattern of sporadic cytoplasmic (black arrows) or nuclear (white arrows) HBcAg in each region. The boundary was indicated by white dotted line. (D) Distinctive pattern of HBcAg in perilesional normal liver tissues. HBcAg was expressed in the nucleus in normal hepatocytes and cytoplasmic in proliferating regenerative cells. Boundaries were highlighted. Sporadic nuclear expression in tissue #1 is indicated by white arrows. Scale bars, 100 μm and 50 μm.
Risk Factors for HCC-Associated PDX Model
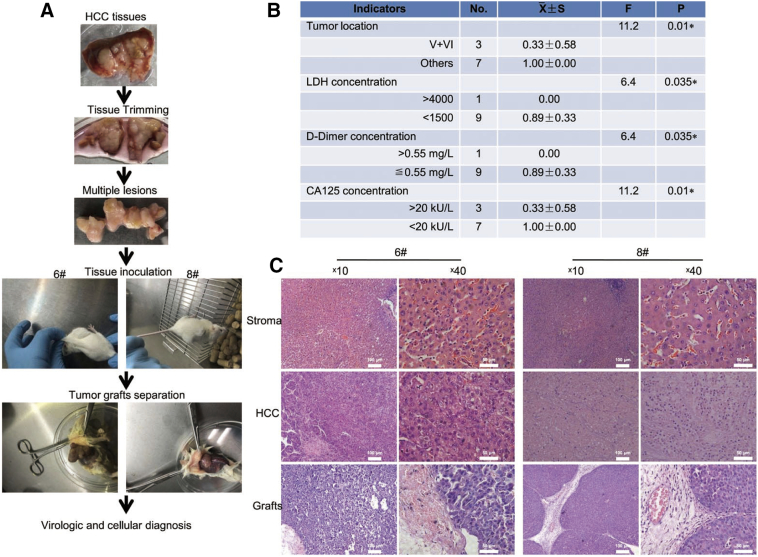
We transplanted 10 freshly isolated primary HCC tissues obtained immediately after surgery from #5 and 14 patients, into the lateral abdomens of male NOD-SCID mice. Multiple lesions from individual patients were collected and thoroughly trimmed before inoculation (Fig. 4A). Tumors grew to an average of more than 0.5 cm3 after 10–12 weeks in two of 10 samples (20%). Tumors proliferated substantially in the last 2 weeks. To determine if successful engraftment correlated with clinical characteristics of HCC, univariate F tests were performed. Tumors at hepatic V and VI fragment (P = .01), LDH (P = .035), D-dimer (P = .035), and CA125 (P = .01) were independent risk factors (Fig. 4B). Intrahepatic metastasis, hemorrhagic necrosis, and higher levels of CA19–9, glutathione transpeptidase were regional risk factors (Table S1).
Fig. 4.
PDX model developed by inoculation of fresh primary HCC tissues into NOD/SCID mice. (A) Flowchart of experimental procedures. (B) Risk factors identified by univariate F test. Of 20 variables, indicators of tumor location in the V and VI regions, impaired liver function, and CA125 showed significant p values. (C) Detailed histological morphology of large amounts of blood vessels (#6 and 8), thick trabecular (#6) and collagen in (#8) were reproduced in tumor grafts. Scale bars, 100 μm and 50 μm.
We demonstrated that histological morphology of parental HCC tissues was authentically reproduced by mice engraftments, however with a slight modification (Fig. 4C). Instead of typical clear cell type in patient 8, tumor grafts exhibited a thin trabecular morphology (Fig. 4C). Similar to primary tumors, engraftments exhibited distinctive mitosis and undifferentiated tumor cells in the field (Fig. 4C).
PDX Tumor Grafts Recapitulate HBV Behavior in Human Tissues
We analyzed if HBV was expressed in engraftments consistent with parental tissues. In samples #6 and 8, HBsAg was expressed at low levels in cytoplasmic spots or diffuse forms in liver tissues and was not detected in tumor tissues. The phenotype of the original tissue was faithfully reproduced in graft tissues (Fig. 5A). For HBcAg, expression was increased in tumor cells of engraftments (Fig. 5B). In a large fraction of engraftment cells, HBcAg was expressed extensively, in contrast to the mainly cytoplasmic location in original cells (Fig. 5B).
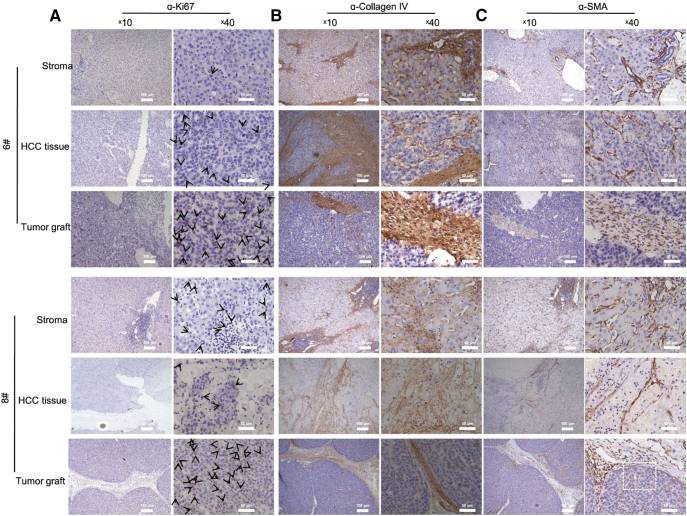
Several cellular markers were analyzed. Few cells were identified as ki67 positive in HCC tissues #6 and 8, and we observed that ki67 was increased in tumor grafts (Fig. 6A). Bridging of necrosis and fibrosis areas characterized by collagen IV in parental tissues was reproduced by tumor cells in engraftment (Fig. 6B). The specific distribution of SMA in sinusoid endothelial cells surrounding proliferative neoplasms or hemorrhagic necrosis changed to be mainly concentrated in infiltrated spindle cells in fibrotic brunch in engraftments and tumor cells (Fig. 6C). We found that PDX mice authentically recapitulated the clinicopathological properties of patient tissues, but potentially evolved towards an aggressive phenotype with increased ki67 and HBcAg expression.
Fig. 6.
Cellular markers ki67, collagen IV, and SMA were generally reproduced in mice with a preferential selection of tumor cells. (A) Proliferative cellular marker ki67 significantly increased in mice engraftments. ki67 expressed at low level in tumor tissues #6 and 8. The staining of engraftment tissues was strong and extensive. Black arrows, ki67 positive cells. (B) Fibrosis structure represented by collagen IV positivity was significant in tumor tissue of two cases and reproduced in mice. (C) SMA expressed distinctively in tumor grafts differing from patient tissues. In mice, SMA expression concentrated in fibrotic structures without the multiple scattered SMA positive pericytes or fibroblasts in patient tissues. Tumor cells in engraftments expressed cytoplasmic SMA that was absent from patient tissues. Black arrows and box, representative tumor cells. Scale bars, 100 μm and 50 μm.
Discussion
Epidemiological studies demonstrate that the relative risk of HCC among HBV carriers is 10-fold higher than among noncarriers, which necessitates further investigation of the molecular mechanism of viral-mediated hepatocarcinogenesis [5], [6], [7], [8]. We took advantage of a unique collection of liver specimens from patients who underwent partial hepatectomy for HBV-associated HCC to analyze virological features under pathological condition. Furthermore, we firstly attempt to address if PDX mice could be a candidate model for virus-induced disease.
A major goal of our study was to delineate gene expression patterns of viral biomarkers in HCC. We noticed decreased HBsAg expression in tumors by compared with perilesional areas. The phenomenon had long been confirmed to chronic type B hepatitis patients; however if it is true for tumor patients were largely unknown. [28], [29], [30] Only tumor tissue of patient #9 was positive for HBsAg. Expression was restricted to normal hepatocytes and was absent in neighboring neoplastic nodules. Previous studies reported that HBsAg positivity ranges from 72%–80% in samples of perilesional tissues vs. 15%–23% in HCC tissues [31], [32], [33]. Concordant with that, decreased HBsAg in HCC samples in our study raised doubts about the clinical applications of treating HBV-related HCC patients with autologous T cells engineered to recognize HBsAg [34]. Similarly, HBcAg expression was decreased in tumor tissues, but to a relatively lower degree, and its cytoplasmic location was distinctive. Decreased or even fully abrogated expression of HBV antigens may result from HBV DNA integration into the host genome in 85%–90% of patients [35]. Integration often leads to disruption of viral proteins and expression of HBV-host fusion constructs. In HCC, genes coding for viral proteins are often methylated (mostly viral core and surface genes) or mutated (mostly HBx gene), reducing the chance of presenting regular HBV sequences in MHCs [36]. Previous reports indicated that in patients with chronic active liver disease, the expression of nuclear HBcAg decreased significantly with concomitant increase in cytoplasmic membranous HBcAg expression [33], [38], [39], which was highly coincided by HCC samples in our study, and raised a role of HBcAg in tumor progression.
Although HBsAg was negative in serum and tissue samples from patients #1, 13, and 14, samples showed intrahepatic HBV occult infections by strong HBcAg-positivity in liver tissues, highlighting further diagnosis by combined viral markers in liver tissue [40].
Of viral models of HBV-related HCC in standardized mice, transgenic mice harboring HBx, pre S, or truncated S were developed. However, intact Dane particles in these models do not represent natural HBV infection [17], [18], [19], [20]. PDX models mostly retain the principal histologic and genetic characteristics of their donor tumor and remain stable across passages [24], [25], [26], [27]. Few studies have properly addressed the efficiency of methods for tumor virology studies [24], [25], [26]. The take rate in our study was 20% (2/10), slightly lower than previous studies [19], [27], which may have resulted from a limited number of high grade malignant tumors in our study. The risk factors identified by univariate analysis were a high level of LDH, D-dimer, and CA125, emphasizing the role of liver dysfunction in HCC progression. Of note, poorly differentiated HCC tissues of #6 and 8 were successfully taken up and had distinctive mitosis. Thus, prescreening by histological and serological risk factors before tissue implantation is important for implementing real-time PDX data for personalized cancer treatment.
Although primary tumors are generally thought to be faithfully represented by PDX models [24], [25], [26], [27], assessing if they retain virological characteristics throughout propagation is important. Limited studies reported that a dotty pattern of HBsAg is correlated with HCC and active viral replication [28], [30]. Samples #6 and 8 both exhibited the pattern. The special pattern of whole cell expression of HBcAg in tissues #6 and 8 were recapitulated in two tumor grafts with stronger positivity. The pathological meaning for that was still need to be clarified. [28], [29], [30], [33], [38] For cellular markers, PDX models authentically mimicked the fibrosis structure of samples #6 and 8, but with a different pattern of SMA. SMA was mainly located in the cytoplasm of tumor cells in mice. We further identified that active proliferation indicated by ki67 stain was significantly increased when compared to original tissues, which implied that the process of generating a PDX model resulted in the selection of tumors that engraft and propagate in mice [41]. No antibodies had cross reactions with mouse species. These results may suggest an evolutionary progression of extracellular matrix remodeling during PDX model development, but needs further investigation.
The model was novel and appealing; however, we are still in collection of HCC samples for developing PDX, which may lead to more accurate identification of risk factors. And deep sequencing profiles to construct a virus-associated landscape for the disease will be fulfilled in our future research. Here, we concluded that accurate delineation of viral status in HCC is a critical prerequisite for HBV-based therapy. And we firstly developed an alternative model by PDX mice, which represent physiological characteristics of clinical HCC.
Declarations
Ethics Approval and Consent to Participate
All procedures performed in studies involving human participants were approved by the institutional review board of Army Medical University (Chongqing, China), and informed consent was obtained from all patients. All procedures were in accordance with the Helsinki Declaration. The animal care and experimental procedures were conducted in accordance with the regulations of the Animal Care and Use Committee of Army Medical University (Chongqing, China).
The following are the supplementary data related to this article.
HBcAg was exclusively expressed in tumor nodules but not in normal hepatocytes in 9# tissue.
Various patterns of HBsAg expression were identified in an individual sample, which were spotty, diffuse, membranous and cytoplasmic scattering types.
Sero-negative HBsAg of samples #13, and 14 still showed strong positivity for intrahepatic HBcAg when lower dilution of the primary antibody was used, with #9 and 13 as positive controls.
Risk factors were statistically analyzed for HCC-PDX model development. Clinical, serological, and pathological features including grade and size of tumor for those 10 tissues were provided and univariate F tests were performed.
Consent for Publication
Not applicable.
Availability of Data and Material
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare that they have no competing interests.
Funding
This study was funded grants from the Natural Science Foundation (81373966) to X.W. and China Postdoctoral Science Foundation (2017M613439) to J.L.
Authors' Contributions
XW wrote the manuscript and generated the figures; JL, SYC, ZZ, DHT, and XDL contributed to the acquisition of clinical and experimental data; JL and XW contributed to editing the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Shiming Yang for sample collection.
Contributor Information
Jiao Liu, Email: liujiao861119@hotmail.com.
Siyuan Chen, Email: csy383117770@163.com.
Zhe Zou, Email: 522429449@qq.com.
Dehong Tan, Email: tdh12345678@163.com.
Xiangde Liu, Email: liuxd@medmail.com.cn.
Xing Wang, Email: xwang@ips.ac.cn.
References
- 1.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Hernaez R, El-Serag HB. Hepatocellular carcinoma surveillance: The road ahead. Hepatology. 2017;65(3):771–773. doi: 10.1002/hep.28983. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273. doi: 10.1053/j.gastro.2011.12.061. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13(2):123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 6.Papatheodoridis GV, Chan HL, Hansen BE, Janssen HL, Lampertico P. Risk of hepatocellular carcinoma in chronic hepatitis B: assessment and modification with current antiviral therapy. J Hepatol. 2015;62(4):956–967. doi: 10.1016/j.jhep.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25(27):3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 8.Global Burden of Disease Cancer C, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4(11):1553–1568. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15(11):653–667. doi: 10.1038/nrc4017. [DOI] [PubMed] [Google Scholar]
- 11.Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7 doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(2):370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Menendez-Arias L, Alvarez M, Pacheco B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr Opin Virol. 2014;8:1–9. doi: 10.1016/j.coviro.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Wu C-Y, Chen Y-J, Ho HJ, Hsu Y-C, Kuo KN, Wu M-S. Association Between Nucleoside Analogues and Risk of Hepatitis B Virus–Related Hepatocellular Carcinoma Recurrence Following Liver Resection. JAMA. 2012;308(18):1906. doi: 10.1001/2012.jama.11975. [DOI] [PubMed] [Google Scholar]
- 15.Kurokawa M, Hiramatsu N, Oze T, Yakushijin T, Miyazaki M, Hosui A. Long-term effect of lamivudine treatment on the incidence of hepatocellular carcinoma in patients with hepatitis B virus infection. J Gastroenterol. 2012;47(5):577–585. doi: 10.1007/s00535-011-0522-7. [DOI] [PubMed] [Google Scholar]
- 16.Yip TC, Chan HL, Wong VW, Tse YK, Lam KL, Wong GL. Impact of age and gender on risk of hepatocellular carcinoma after hepatitis B surface antigen seroclearance. J Hepatol. 2017;67(5):902–908. doi: 10.1016/j.jhep.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Tang ZY, Hou JX. Hepatocellular carcinoma: insight from animal models. Nat Rev Gastroenterol Hepatol. 2011;9(1):32–43. doi: 10.1038/nrgastro.2011.196. [DOI] [PubMed] [Google Scholar]
- 18.Daniel HD, Torbenson M. Transgenic hepatitis B: a new model of HBV infection. Sci Rep. 2017;7(1):2610. doi: 10.1038/s41598-017-02862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15(5):311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 20.Zhou SJ, Deng YL, Liang HF, Jaoude JC, Liu FY. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017;24(9):1577–1587. doi: 10.1038/cdd.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riviere L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63(5):1093–1102. doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Zhu Y, Shao D, Chang H, Zhang X, Zhou D. Recombinant covalently closed circular DNA of hepatitis B virus induces long-term viral persistence with chronic hepatitis in a mouse model. Hepatology. 2018;67(1):56–70. doi: 10.1002/hep.29406. [DOI] [PubMed] [Google Scholar]
- 23.Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25(3):335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 24.Bondarenko G, Ugolkov A, Rohan S, Kulesza P, Dubrovskyi O, Gursel D. Patient-derived tumor xenografts are susceptible to formation of human lymphocytic tumors. Neoplasia. 2015;17(9):735–741. doi: 10.1016/j.neo.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Facompre ND, Sahu V, Montone KT, Harmeyer KM, Nakagawa H, Rustgi AK. Barriers to generating PDX models of HPV-related head and neck cancer. Laryngoscope. 2017;127(12):2777–2783. doi: 10.1002/lary.26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieter SM, Giessler KM, Kriegsmann M, Dubash TD, Mohrmann L, Schulz ER. Patient-derived xenografts of gastrointestinal cancers are susceptible to rapid and delayed B-lymphoproliferation. Int J Cancer. 2017;140(6):1356–1363. doi: 10.1002/ijc.30561. [DOI] [PubMed] [Google Scholar]
- 27.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ML, Gerber MA, Thung SN, Thornton JC, Chung WK. Morphometric study of hepatocytes containing hepatitis B surface antigen. Am J Pathol. 1984;114(2):217–221. [PMC free article] [PubMed] [Google Scholar]
- 29.Chu CM, Liaw YF. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology. 1987;92(1):220–225. doi: 10.1016/0016-5085(87)90863-8. [DOI] [PubMed] [Google Scholar]
- 30.Kondo Y, Ninomiya M, Kakazu E, Kimura O, Shimosegawa T. Hepatitis B surface antigen could contribute to the immunopathogenesis of hepatitis B virus infection. ISRN Gastroenterol. 2013;2013 doi: 10.1155/2013/935295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringelhan M, O'Connor T, Protzer U, Heikenwalder M. The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J Pathol. 2015;235(2):355–367. doi: 10.1002/path.4434. [DOI] [PubMed] [Google Scholar]
- 32.Fan C, Li M, Gan Y, Chen T, Sun Y, Lu J. A simple AGED score for risk classification of primary liver cancer: development and validation with long-term prospective HBsAg-positive cohorts in Qidong, China. Gut. 2019;68(5):948–949. doi: 10.1136/gutjnl-2018-316525. [DOI] [PubMed] [Google Scholar]
- 33.Uzun Y, Bozkaya H, Erden E, Cinar K, Idilman R, Yurdaydin C. Hepatitis B core antigen expression pattern reflects the response to anti-viral treatment. J Gastroenterol Hepatol. 2006;21(6):977–981. doi: 10.1111/j.1440-1746.2006.04263.x. [DOI] [PubMed] [Google Scholar]
- 34.Qasim W, Brunetto M, Gehring AJ, Xue SA, Schurich A, Khakpoor A. Immunotherapy of HCC metastases with autologous T cell receptor redirected T cells, targeting HBsAg in a liver transplant patient. J Hepatol. 2015;62(2):486–491. doi: 10.1016/j.jhep.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44(7):765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez AF, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009;19(3):438–451. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu CM, Yeh CT, Sheen IS, Liaw YF. Subcellular localization of hepatitis B core antigen in relation to hepatocyte regeneration in chronic hepatitis B. Gastroenterology. 1995;109(6):1926–1932. doi: 10.1016/0016-5085(95)90760-2. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Lu W, Zheng Y, Wang W, Bai L, Chen L. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest. 2016;126(3):1079–1092. doi: 10.1172/JCI83339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollicino T, Raimondo G. Occult hepatitis B infection. J Hepatol. 2014;61(3):688–689. doi: 10.1016/j.jhep.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 41.Byrne AT, Alferez DG, Amant F, Annibali D, Arribas J, Biankin AV. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17(4):254–268. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HBcAg was exclusively expressed in tumor nodules but not in normal hepatocytes in 9# tissue.
Various patterns of HBsAg expression were identified in an individual sample, which were spotty, diffuse, membranous and cytoplasmic scattering types.
Sero-negative HBsAg of samples #13, and 14 still showed strong positivity for intrahepatic HBcAg when lower dilution of the primary antibody was used, with #9 and 13 as positive controls.
Risk factors were statistically analyzed for HCC-PDX model development. Clinical, serological, and pathological features including grade and size of tumor for those 10 tissues were provided and univariate F tests were performed.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.