Abstract
Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight disease that limits the rice production globally. The bacterium secretes effector proteins directly into plant cells through a type III secretion system (T3SS). Here, we examined the role of a conserved XopR T3SS-effector in the suppression of host basal defense response. Phylogenetic and sequence analysis showed that XopR is well conserved within Xoo strains but shares varying degree of similarity among the other Xanthomonas species. The expression of XopR was shown to be regulated by hrpX, a key regulator of hrp cluster. For functional analysis we employed two mutant strains of Xoo, one lacks xopR gene and other lacks hrpX gene (making the strain defective in T3SS). Programmed cell death (PCD) events was examined both in rice and tobacco leaves through trypan blue staining method. In XopR expressing tobacco leaves the PCD induction was compromised. We observed higher PCD on rice leaves inoculated with Xoo mutants lacking either xopR or functional T3SS as compared to wild type. Contrary, when xopR gene was complemented in mutated strain the PCD was suppressed which clearly suggests that XopR acts as suppressor of the PCD mediated defense response. The EYFP::XopR fusion protein was shown to be localized to the plasma membrane of Nicotiana benthamiana and onion epidermal cells. Altogether our study leads to the understanding that XopR T3SS-effector is essential for Xoo to suppress PCD, primarily to support the in planta colonization of Xoo during blight pathogenesis.
Keywords: Xanthomonas oryzae, T3SS, Phylogenetic analysis, PCD, EYFP
Introduction
Rice is an important staple food crop grown in various agro-climatic regions all over world including India. Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight that potentially threatens the rice production. Xanthomonas for its pathogenicity relies on the effector proteins that are secreted through the type III secretion system (T3SS) (Leyns et al. 1984; Tampakaki et al. 2004). Effectors are classified into two categories, one is TALE (transcription activation like effectors) of the avrBs3/pthA family and the other is Xop (Xanthomonas outer protein) effectors (Buttner and He 2009; Long et al. 2018; Medina et al. 2018; Mudgett 2005). Xop T3SS-effectors are known to play a vital role in disease induction. Pathogens use Xop T3SS-effectors to suppress PAMP (pathogen-associated molecular patterns) triggered immunity (PTI) in host plants (Bartetzko et al. 2009; Zhang et al. 2015). Genomic analysis revealed nine “core T3SS-effectors” present in all Xanthomonas species (Hajri et al. 2009; Jalan et al. 2011; Moreira et al. 2010; Potnis et al. 2011). The core T3SS-effectors are suggested to have a critical role in Xanthomonas pathology. Thus, in depth studies on these core T3SS-effectors may lead to strategies for disease mitigation (Dangl et al. 2013; Potnis et al. 2011).
Plants counteract the pathogen’s invasion by activating various immune responses including PCD (programmed cell death), callose deposition, ROS production that often leads to effector-triggered immunity (ETI) (Spoel and Dong 2012). However, T3SS-effectors mediated plant immune system is of complex nature. The matching T3SS-effectors of the pathogen against the host receptors actually determine the outcome of a plant–pathogen interaction. Xoo strains from Japan, Philippines and Korea are well characterized for their effector profiles as well as for their possible role in blight development. Mutational analysis suggested that XopZ is crucial for blight induction in Philippine Xoo strain PX099A. Further, its transient expression in Nicotiana benthamiana also reduced callose deposition and cell-wall based defense response (Song and Yang 2010). In Japan strain (MAFF311018), XopR was reported to suppress PAMP-induced defense responses in Arabidopsis thaliana and a XopR deletion mutant showed reduced pathogenicity in rice plants (Akimoto-Tomiyama et al. 2012). All these reports indicated that T3SS-effectors are indispensable for pathogen during pathogenesis. However, sufficient information pertaining to the role of effectors is yet to be generated for Xoo strain from India, one of the major rice eating region in the Asian continent. A virulent Xoo strain (race 4) from India was reported to possess 21 Xop effectors (Mondal et al. 2014). Subsequent studies on these effectors revealed that T3SS-effector-like XopR plays a crucial role during blight induction. XopR was shown to be essential for in planta bacterial growth and virulence as well as for the suppression of plant defense responses including callose deposition and ROS production (Verma et al. 2018). This study aims to further characterize XopR from Indian Xoo strain with reference to its phylogenetic relationship with other Xanthomonas species with the purpose to verify its subcellular localization and more importantly, its role in regulating PCD mediated rice immunity during blight disease development.
Materials and methods
Bacterial culture, media and transformation
All bacterial strains and plasmids used in this study are given in Table 1. Escherichia coli and Agrobacterium tumefaciens were grown in Luria broth. Xoo strain ITCCBB0002 was grown on either NB (1% peptone and 0.5% beef extract) or PSB (1% peptone, 1% sucrose, 0.1% Na-glutamate) medium. In NA and PSA plates, 2% w/v agar was used for solidification. Antibiotics including rifampicin (100 µg mL−1), ampicillin (100 μg mL−1), kanamycin (50 µg mL−1) and spectinomycin (50 µg mL−1) were used when required. E. coli DH5α cells were transformed by heat shock method while electroporation was used for Xoo and A. tumefaciens.
Table 1.
List of plasmid vectors, constructs, bacterial strains used
| Plasmid or strains | Properties | Source |
|---|---|---|
| Escherichia coli DH5α | Broad range host for cloning purpose | Stratagene, USA |
| X. oryzae pv. oryzae ITCCBB0002 Rif100(Xoo-Rif100) | A virulent Xoo strain (pathotyped as race 4) from North India having resistance to rifampicin | Available in lab |
| Agrobacterium tumefaciens strain EHA105 | Resistance to rifampicin | Available in lab |
| pEZRK-LCY | Resistance to kanamycin | Gifted from Professor MB Mudgett lab, Stanford University, USA |
| EYFP::XopR | EYFP-tagged XopR in pEZRK-LCY vector | Developed in this study |
Plant stages and growing condition
N. benthamiana was directly seeded in pots filled with soilrite. The rice seedlings after germination were transplanted into pots filled with garden soil (clay loam mixed with dry farm yard manure @ 3:1 ratio). The pots were kept in glasshouse maintaining optimum temperature (25 °C for N. benthamiana and 28 °C for rice) and relative humidity (~ 80%). The plants were monitored throughout its growing period and watered regularly whenever required. For infiltration, 4–6 weeks old rice seedlings and 6–8 weeks old N. benthamiana were used.
Isolation of bacterial culture
The wild Xoo strain used throughout the present study was having rifampicin resistance as natural phenotypic marker (referred to as Xoo-Rif100). For long-term storage, cell suspension of Xoo-Rif100 in 50% glycerol was kept at − 80 °C deep freezer. Prior to each experiment, the strain was grown on PSA (after autoclaving) plates supplemented with rifampicin.
Construction of mutants and complement strains
The mutants, Xoo ΔhrpX (lacking hrpX, a regulator of T3SS genes), Xoo ΔxopR (lacking xopR, a T3SS-effector) and a complemented strain for xopR were constructed following a PCR based double crossing over-mediated homologous recombination strategy as described previously (Verma et al. 2018).
Sequence and phylogenetic analysis of XopR homologs
The XopR amino acid sequences from diverse strains of Xanthomonas were downloaded from NCBI GenBank. These sequences were aligned using MUSCLE algorithm and the evolutionary history was inferred using neighbor-joining method with 1000 bootstrap replicates in Mega 6 software (Tamura et al. 2013). The pair-wise sequence comparison was performed by sequence demarcation tool (SDT) (Muhire et al. 2014).
Studying the hrp-dependent regulation of xopR gene
To investigate the hrp-dependent regulation of xopR gene, two different media, XOM2 (inducing) and NB (non-inducing) were used. The wild Xoo and Xoo ΔhrpX were grown in 5 mL NB medium (OD600nm 0.2). The culture was divided into two fractions. In one fraction, the cells were harvested through centrifugation and the resultant pellet was washed twice with XOM2. XOM2 is a synthetic medium used to induce the expression of hrp genes (Tsuge et al. 2002). The harvested cells were allowed to grow on XOM2 for 12 h. The other fraction was concurrently grown on NB medium. From both the growth, cells were harvested for RNA extraction. Total bacterial RNA was pulled down using kit (Fermentus Co., USA). The DNA contamination in the RNA (1 μg) samples was removed with DNase I (Invitrogen, CA, USA) and the cDNA was synthesized using Verso cDNA synthesis kit (Thermo Scientific Inc., USA). The hrp-dependent xopR regulation was examined by reverse-transcription PCR (RT-PCR) using specific primers, XopR-F (5′-TTGTCGTTGGTGATCTCGCTC-3′) and XopR-R (5′-CCGTTCTCCATTGAGTCTCCG-3′). The 16S ribosomal RNA expression was used as internal control using 16S rRNA-F (5′AGAGTTTGATCCTGGCTCA 3′) and 16S rRNA-R (5′AAGGAGGTGATCCAGCCGC 3′) primers.
Subcellular localization of EYFP::XopR fusion protein
To monitor the subcellular localization, the xopR gene sequence was cloned in C-terminal fusion with the coding sequence of enhanced yellow fluorescence protein (eyfp). The full-length xopR gene with integrated restriction sites was PCR amplified using specific primers (EYFP_XopR_F, 5′GAATTCATGCGCACGAATTTTCTTCCG3′ and EYFP_XopR_ R, 5′TCTAGATTATCGGTAACCGTTCTCCA3′). The PCR amplicon was digested with EcoR1 and Xba1 restriction enzymes and further cloned into pEZRK-LCY vector in fusion with EYFP and downstream of cauliflower mosaic virus (CaMV) 35S promoter. The xopR construct (eyfp::xopR) and empty pEZRK-LCY vector were transformed into electrocompetent A. tumefaciens (strain EHA105) cells. The transformed cells were overnight grown at 28 °C and the culture was pelleted down by centrifugation. The pellet was resuspended in the induction buffer (10 mM MgCl2 and 10 mM MES, pH 5.6, supplemented with 0.5 mM acetosyringone) to an optical density of 0.5 at 600 nm and further incubated at 28 °C for 2 h with gentle shaking. The cell suspension was finally infiltrated into N. benthamiana leaves using a needleless syringe. After 3 days post-infiltration, leaves were visualized under confocal microscope (Leica Microsystems) for the expression and accumulation of EYFP::XopR fusion protein in plant cells. The leaf samples were also stained with 6-diamidino-2-phenylindole (DAPI) (5 mg/mL, Sigma, USA) to visualize the nucleus.
The XopR localization was also examined in living onion epidermal cells using a modified method as described by Xu et al. (2014). Before agro-infiltration, the onion bulbs were pre-conditioned under dark for 72 h at 28 °C. After infiltration, the selected 2 mm infiltrated areas were seen under confocal microscopy. The samples were stained with DAPI to examine the nucleus and were kept under darkness for 20 min before observation. The EYFP-tagged XopR (yellow) field was superimposed with the DAPI (blue)-field. The emission and excitation for DAPI-stained samples were 461 nm and 368 nm, respectively; and emission and excitation for EYFP-tagged, samples were 568 nm and 514 nm, respectively.
Role of XopR in PCD
The role of XopR in PCD suppression was investigated by trypan blue staining in both rice and N. benthamiana. Trypan blue staining was performed as described by Van Wees (2008). The rice leaves were infiltrated using a needleless syringe at different points with Xoo suspension (108 CFU mL−1) of wild (Xoo), Xoo ΔhrpX, Xoo ΔxopR and complement (Xoo ΔxopR + xopR) strains. In another experiment, EYFP::XopR and EYFP were transiently expressed in N. benthamiana as described earlier. From both the experiments, the whole leaves were harvested for trypan blue staining (at 7 dpi in case of rice and at 3 dpi in case of tobacco). The staining solution composed of 67% ethanol, 8.3% lactic acid, 8.3% phenol, 8.3% glycerol and 2.5 mg mL−1 trypan blue. The leaves were submerged in 50 mL of staining solution and boiled for 5 min. Leaves were kept incubated in the same solution for overnight at room temperature. After staining leaves were transferred to the destaining solution (67% ethanol, 8.3% lactic acid, 8.3% phenol, 8.3% glycerol) and kept overnight at room temperature. Leaves were dried briefly after washing in ethanol before taking photographs.
Quantification of cell death through measuring ion leakage
To assess cell death, we measured ion leakage in rice leaves on the infiltrated areas. Three leaf disks (1 cm2) were sampled from infiltrated region at 72 hpi. The disks were gently rinsed and then immersed in deionized water at shaking (80 rpm) under continuous light for 4 h. After incubation, the conductivity of bathing water (in which the leaf disks were immersed) was measured with the help of conductivity meter (Labman Scientific Instruments Pvt. Ltd., India). The conductivity data are mean of three replications.
Results
Sequence and phylogenetic analysis of XopR
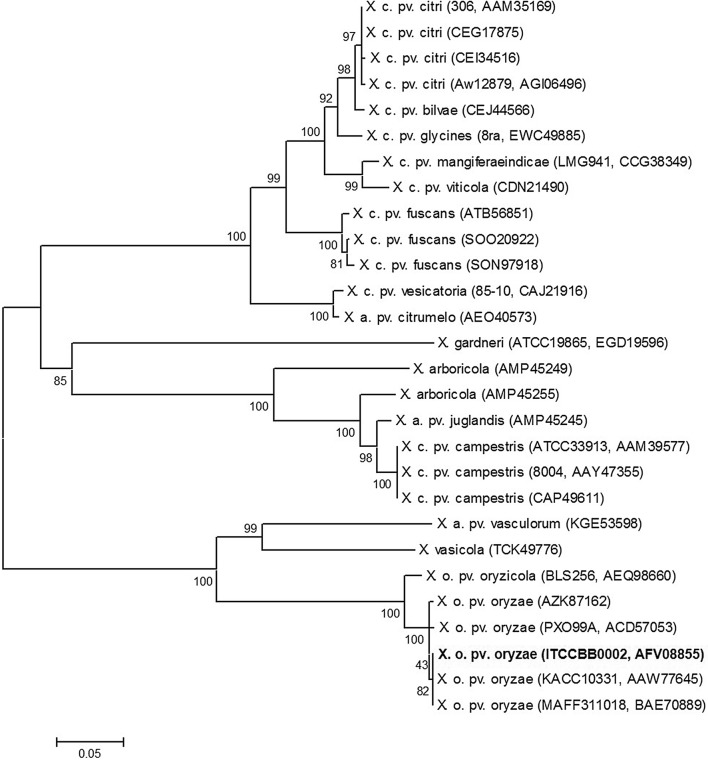
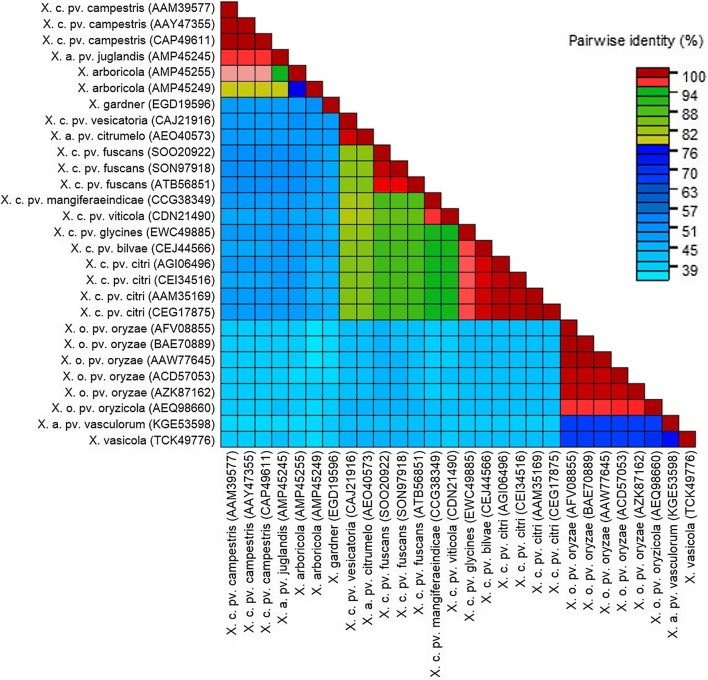
Phylogenetic analysis shows that the XopR is well conserved among Xoo species. The XopR from Indian strain (ITCCBB0002) clusters with various XopR sequences of Xoo reported from Japan (MAFF311018), Korea (KACC10331) and Philippines (PX099A) but is distantly related to other species in Xanthomonas genus (Fig. 1). SDT analysis shows that XopR of Indian Xoo strain is 97–100% similar to other XopR sequences of different Xoo strains whereas it shows 36–67% sequence identity to other Xanthomonas XopR sequences. It is closely related to X. axonopodis pv. vasculorum and shares 67% sequence identity and is least similar (36%) to X. arboricola (Fig. 2).
Fig. 1.
Phylogenetic tree of XopR. The bootstrap values are shown at nodes. The abbreviation of various Xanthomonas species taken for analysis are depicted with their locus tag and Gen-Bank accession number in parenthesis
Fig. 2.
A colored similarity matrix of XopR generated through sequence demarcation tool with various Xanthomonas XopR sequences taken into analysis
hrp-dependent regulation of xopR gene
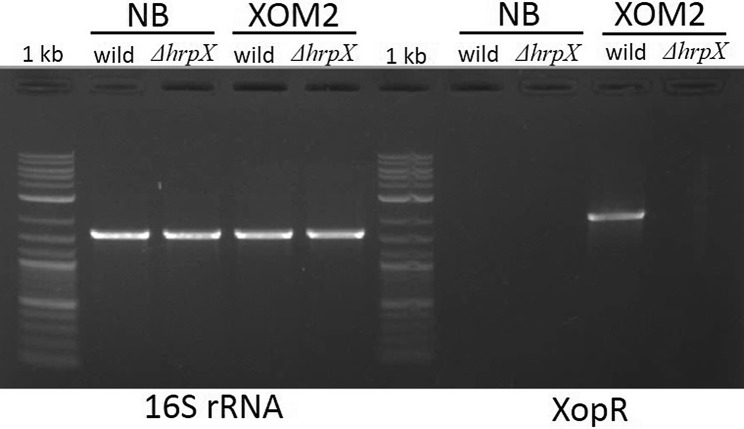
We verified the hrp-dependent regulation of xopR gene using two different media. The hrp-inducing medium XOM2 supported the induction of xopR gene of Xoo wild (Fig. 3) as we could not detect any amplification of xopR in NB used as non-hrp-inducing medium. Further, no amplification of xopR gene was observed in hrpX-deficient mutant, irrespective of medium used indicating the hrp-dependent regulation of xopR gene. The amplification of 16S rRNA, the housekeeping gene, was observed in both wild and Xoo ΔhrpX, irrespective of the media used.
Fig. 3.
Induced hrp-dependent regulation of xopR. RT-PCR based expression of xopR in wild but not in hrpX mutant grown in Xanthomonas hrp-inducing medium (XOM2) medium. No expression of xopR was observed from the wild as well as in hrpX mutant grown in NB medium, used as a reference check. However, the housekeeping gene 16S rRNA used as control was found to be expressed in both, mutant as well as wild under both the media conditions
EYFP::XopR fusion protein localizes to the plasma membrane
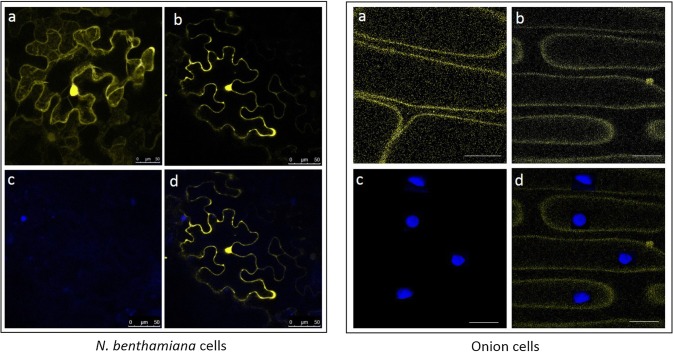
The subcellular localization of XopR was visualized through transiently expressing EYFP::XopR protein in N. benthamiana. The XopR is localized to the plasma membrane as confocal microscopy revealed the YFP fluorescence only on the membrane, not on the nucleus (Fig. 4). Low level of non-specific (both on nuclear and plasma membrane) fluorescence was detected on leaves infiltrated with empty vector expressing only EYFP.
Fig. 4.
Subcellular localization of transiently expressed EYFP::XopR fusion protein in Nicotiana benthamiana (left panel), and in onion (right panel). a Empty vector (without XopR) showing non-specific localization. b XopR::EYFP fusion protein showing localization only on cytoplasmic membrane; c DAPI-stained nuclei, d superimposed images of (b) and (c). A. tumefaciens EHA 105, which mediated T-DNA transfer of XopR::EYFP was infiltrated at 1.0 OD600 nm. Samples were observed by confocal microscopy 3 days after infiltration. EYFP fluorescence was shown in yellow; DAPI-stained nuclei was shown in blue. The emission and excitation for DAPI-stained samples were 461 nm and 368 nm, respectively; and for EYFP-tagged samples were 568 nm and 514 nm, respectively. Scale bar 25 µm
The XopR localization was further validated in uninucleated living onion cells. In consistence with the previous observation, yellow fluorescence was detected into the onion plasma membrane. In contrary, no fluorescence was observed on the nuclei, as the nucleus took blue stain of the DAPI (Fig. 4). This study clearly demonstrates that XopR is localized to the plasma membrane of the plants to exert its putative function during the suppression of rice defense.
XopR is essential for suppression of PCD
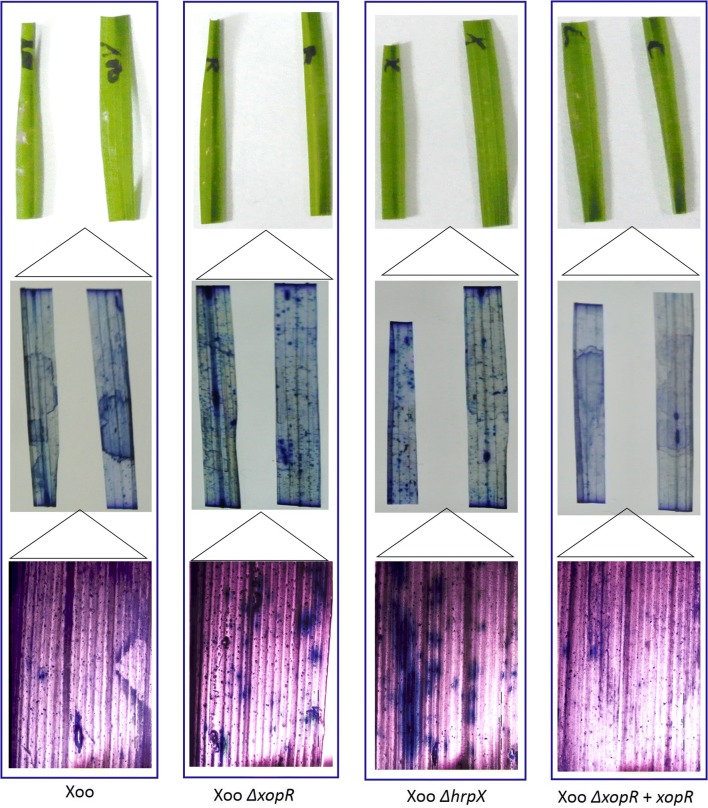
We investigated whether XopR play any role in PCD suppression during blight pathogenesis. Trypan blue that specifically stains dead cells (but not absorbed by living cells having an intact plasma membrane) was used to distinguish the dead cells from living one. We observed that N. benthamiana cells expressing only EYFP took dark blue color compared to the cells expressing the EYFP::XopR (Fig. 5), suggesting the role of XopR as PCD suppressor. Further, we examined the PCD suppression activity of XopR in rice leaves infiltrated with wild Xoo, Xoo ΔxopR, Xoo ΔhrpX and complement (Xoo ΔxopR + xopR) strains. The rice leaves inoculated with Xoo ΔxopR as well as Xoo ΔhrpX took dark blue stain compared to that of wild Xoo suggesting the PCD suppression activity by these two mutants lacking either XopR or T3SS regulator. The mutant Xoo ΔhrpX thus showed non-suppressive action on PCD events due to lacking any of the essential T3SS-effectors (Fig. 6). The role of XopR on the suppression of PCD was further verified by the no or less appearance of blue area in and around the infiltrated region with xopR-complemented strain (Fig. 6). This result suggests that XopR suppresses the PCD of necrosis induced onset during Xoo:rice interaction. This further corroborates that the suppression of PCD was mediated through T3SS-effectors including XopR of Xoo.
Fig. 5.
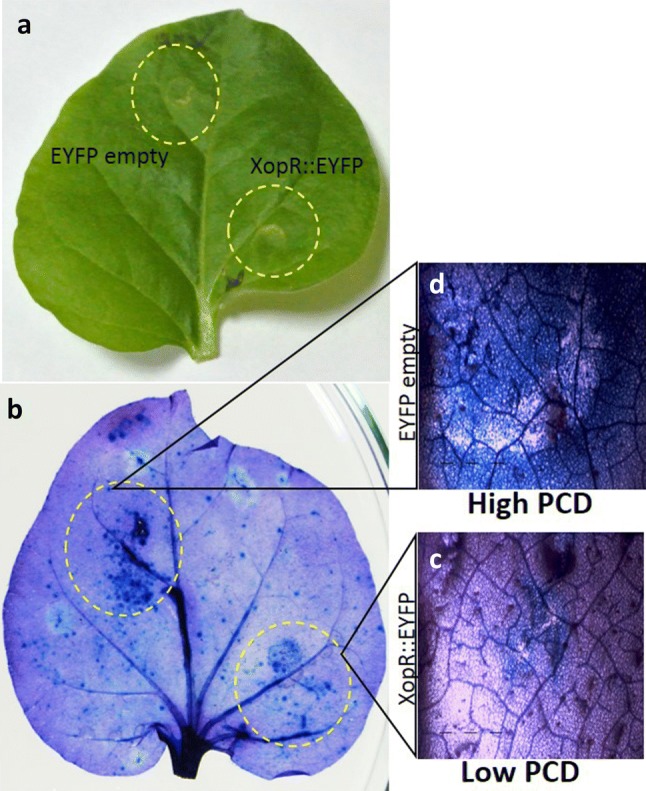
Transiently expressed XopR compromises the PCD induction on Nicotiana benthamiana. Agrobacterium tumefaciens strains bearing EYFP or EYFP::XopR fusion protein vector was inoculated into leaves (OD600 = 1.0). Leaves were harvested at 3 dpi and photographed before (a) and after staining with trypan blue to indicate regions of PCD (b). Magnified microscopic view of the agro-infiltrated regions of leaves showing lesser dark blue patches, indicative of reduced cell death, in case of EYFP::XopR (c) as compared to empty vector control (d)
Fig. 6.
XopR suppresses the PCD in rice during infection. Leaves were inoculated (OD600 = 1.0) using needleless syringe independently with Xoo wild, Xoo ΔxopR, Xoo ΔhrpX, Xoo ΔxopR + xopR. The inoculated leaves were excised after 7 dpi, photographed before (top panel in each row) and after staining with trypan blue (middle panel in each row) to indicate regions of PCD. Magnified microscopic view of the infiltrated regions of the respective leaves (bottom panel in each row). Cell death is directly proportional to the blue colored deposits in the infiltrated region. The leaves infiltrated with Xoo wild as well as complemented strains showing lesser dark blue patches, indicative of reduced cell death, compared to the leaves infiltrated with either Xoo ΔxopR or Xoo ΔhrpX
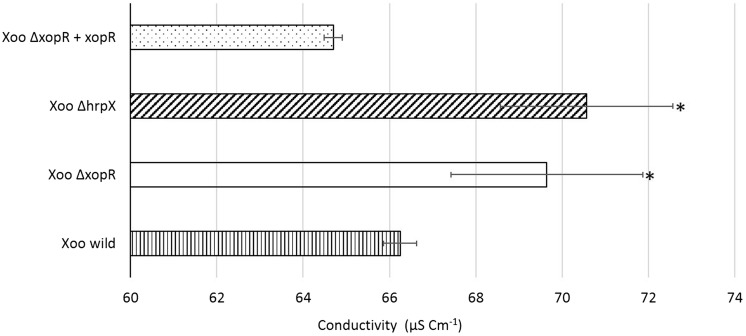
We quantitatively measured PCD indirectly by estimating the ion leakage in rice leaves. As a result of membrane damage, the immediate events followed by PCD were enhanced ion leakage in dying cells. The conductivity was significantly high in samples infiltrated with Xoo ΔxopR, in comparison to Xoo wild justifying the role of XopR in the suppression of PCD (Fig. 7). Also, the conductivity of leaves inoculated with Xoo ΔhrpX was observed to be highest suggesting the role of T3SS-effectors in PCD. The results were in accordance to those in trypan blue PCD assay. The experiment was repeated twice and every time the same trend was observed.
Fig. 7.
Quantification of PCD based on mean conductivity of the electrolytic ion leakage. Mean conductivity of the bathing water used for soaking rice leaf disks (three independent infiltrated regions) was determined. Error bar represents the standard error for three independent observations; Asterisk indicates significant difference (P < 0.05) relative to Xoo wild control
Discussion
Bacterial blight being one of the major biotic threats reduces substantial rice productivity globally (Mondal et al. 2014). The causal bacterium, Xoo like other phytopathogenic Xanthomonads employs T3SS-effectors to overcome the rice immune responses. Xop T3SS-effectors play a significant role in governing virulence to the pathogen and suppression of plant immune response. Several Xop T3SS-effectors were documented as virulence factors in phytopathogenic Xanthomonads including X. axonopodis pv. citri infecting citrus, X. oryzae pv. oryzae infecting rice, X. campestris pv. vesicatoria infecting tomato, X. perforans infecting pepper, X. campestris pv. campestris infecting crucifers (Ryan et al. 2011). However, few studies have been reported concerning the functions of Xop effectors in disease development (Kim et al. 2009; Song and Yang 2010). Earlier we conducted a thorough search for T3SS-effectors in a virulent Indian Xoo strain ITCCBB0002 (pathotyped as race 4); and found that race 4 contained 21 Xop effectors (Mondal et al. 2014). Our continued investigation on the effectors of Xoo race 4 revealed that core effectors like XopF, XopR play important role during blight pathogenesis in rice (Mondal et al. 2016). We experimentally verified the importance of XopR for blight disease development by Xoo and demonstrated its critical role as suppressor of PTI- and ROS-mediated rice defense response during pathogenesis (Verma et al. 2018). In this study, we further investigated its phylogenetic relationship with homologs from other Xanthomonas species and also tracked its subcellular localization and contribution to the modulation of PCD mediated immune events.
Phylogenetic and sequence comparison revealed that XopR is well conserved among Xoo strains from Asia and other continents. However, XopR is distantly related to Xanthomonads infecting other than rice. SDT analysis demonstrated that besides Xoo, XopR showed high similarity (67%) with X. axonopodis pv. vasculorum while it showed least similarity with X. arboricola. The phylogeny based information has great relevance in understanding the structural identity of XopR protein and thereby predicting its putative functions in suppression of plant defense including PCD. The Indian strain showed highest homology with XopR from Japanese Xoo strain, the only strain so far for which the role of XopR in PCD was earlier demonstrated (Akimoto-Tomiyama et al. 2012). In this study, we conclusively confirmed that XopR from the Indian strain also possesses the PCD suppressive role.
We demonstrated the PCD-suppressive role of XopR using trypan blue staining method. Rice leaves inoculated with Xoo ΔxopR or Xoo ΔhrpX caused lesser suppression of PCD as indicated by larger blue stained area compared to Xoo wild. The leaves inoculated with xopR complement strain suppressed PCD comparable to wild Xoo demonstrating the PCD suppressor activity of XopR. Further, in N. benthamiana, the induction of cell death was compromised in plants expressing XopR compared to mock. In addition, we quantitatively determined the cell death through ion leakage experiment and observed higher conductivity in samples infiltrated with Xoo ΔxopR than Xoo wild. This confirms that XopR suppresses PCD during infection. It suggests that Xoo utilizes XopR to suppress PCD that ultimately favors Xoo for its multiplication and spread to the host plants during blight disease development. Similar results of the T3SS-effector mediated suppression of PCD were detected in tomato by XopD of Xcv (Kim et al. 2008) and in pepper by XopJ of Xcv (Ustun et al. 2013). The role of XopR in suppressing MAMP-induced immune responses was well documented in experimental host, A. thaliana (Akimoto-Tomiyama et al. 2012). This study further supports that XopR acts as immune suppressor, particularly cell death associated immune responses in its natural host, rice during blight pathogenesis.
We studied the subcellular localization of EYFP::XopR fusion protein upon transient expression onto two experimental hosts, N. benthamiana and onion epidermal cells. In both the plants, we observed that XopR is localized to the plasma membrane. We understood that XopR suppresses immune responses like PCD and possibly it is required to be localized to the plasma membrane to execute its function. Similar to our results, the T3SS effectors like XopR of Japanese and XopF of Indian Xoo strains, XopJ of X. euvesicatoria, XopN and XopL of X. axonopodis pv. punicae were previously reported to be localized to the plasma membrane indicating their possible roles to suppress cell-wall based defense responses (Kumar et al. 2016; Mondal et al. 2016; Soni and Mondal 2018; Ustun et al. 2013).
The present investigation thus brought a further detailed account on various aspects like sequence analysis, phylogenetic relationship, subcellular localization and mode of action of XopR, a core member of T3SS-effectors of Indian Xoo strain. XopR was demonstrated to localize to the plasma membrane, the predicted site to execute its function during rice::Xoo interaction. Further, we conclusively proved its role as suppressor of PCD, an immune event in plant, apparently to support the Xoo for its sufficient multiplication and spread in the invaded tissues. This insight leads to the understanding that XopR T3SS-effector is essential for Xoo to suppress PCD and thereby it supports the in planta colonization of Xoo during blight pathogenesis.
Acknowledgements
The authors gratefully acknowledge the help from Dr. MB Mudgett, Professor, Department of Biology, Stanford University, USA for providing the constructs and valuable suggestions during plasmid constructs development. The authors acknowledge the support of Dr. Madhvi Soni during formatting of the manuscript. The authors also thank the Head, Division of Plant Pathology, ICAR-Indian Agricultural Research Institute, New Delhi, India for necessary support during the investigation.
Funding
The study was funded by Department of Biotechnology, Government of India (Grant Number BT/PR14870/AGR/02/767/2010).
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Akimoto-Tomiyama C, Furutani A, Tsuge S, Washington EJ, Nishizawa Y, Minami E, Ochiai H. XopR, a type III effector secreted by Xanthomonas oryzae pv. oryzae, suppresses microbe-associated molecular pattern triggered immunity in Arabidopsis thaliana. Mol Plant Microbe Interact. 2012;25:505–514. doi: 10.1094/MPMI-06-11-0167. [DOI] [PubMed] [Google Scholar]
- Bartetzko V, Sonnewald S, Vogel F, Hartner K, Stadler R, Hammes UZ, Börnke F. The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall-associated defense responses. Mol Plant Microbe Interact. 2009;22:655–664. doi: 10.1094/MPMI-22-6-0655. [DOI] [PubMed] [Google Scholar]
- Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–1664. doi: 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajri A, Brin C, Hunault G, Lardeux F, Lemaire C, Manceau C, Boureau T, Poussier S. A “repertoire for repertoire” hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One. 2009;4:e6632. doi: 10.1371/journal.pone.0006632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalan N, Aritua V, Kumar D, Yu F, Jones JB, Graham JH, Setubal JC, Wang N. Comparative genomic analysis of Xanthomonas axonopodis pv. citrumelo F1, which causes citrus bacterial spot disease, and related strains provides insights into virulence and host specificity. J Bacteriol. 2011;193:6342–6357. doi: 10.1128/JB.05777-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Hotson A, Keegan M, Schmelz EA, Mudgett MB. XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell. 2008;20:1915–1929. doi: 10.1105/tpc.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, McLane H. XanthomonasT3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato a typical receptor-like kinase and TFT1. Plant Cell. 2009;21:1305–1323. doi: 10.1105/tpc.108.063123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Soni M, Mondal KK. XopN-T3SS effector of Xanthomonasaxonopodis pv. punicae localizes to the plasma membrane and modulates ROS accumulation events during blight pathogenesis in pomegranate. Microbiol Res. 2016;193:111–120. doi: 10.1016/j.micres.2016. [DOI] [PubMed] [Google Scholar]
- Leyns F, De Cleene M, Swings JG, De Ley J. The host range of the genus Xanthomonas. Bot Rev. 1984;50:308–356. doi: 10.1007/BF02862635. [DOI] [Google Scholar]
- Long J, Song C, Yan F, Zhou J, Zhou H, Yang B. Non-TAL effectors from Xanthomonas oryzae pv. oryzae suppress peptidoglycan-triggered MAPK activation in rice. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.01857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina CA, Reyes PA, Trujillo CA, Gonzalez JL, Bejarano DA, Montenegro NA, Jacobs JM, Joe A, Restrepo S, Alfano JR, Bernal A. The role of type III effectors from Xanthomonas axonopodis pv. manihotis in virulence and suppression of plant immunity. Mol Plant Pathol. 2018;19(3):593–606. doi: 10.1111/mpp.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal KK, Meena BR, Junaid A, Verma G, Mani C, Majumder D, Manju Khicher, Kumar S, Banik S. Pathotyping and genetic screening of type III effectors in Indian strains of Xanthomonas oryzae pv. oryzae causing bacterial leaf blight of rice. Physiol Mol Plant Pathol. 2014;86:98–106. doi: 10.1016/j.pmpp.2014.03.005. [DOI] [Google Scholar]
- Mondal KK, Verma G, Junaid A, Mani C. Rice pathogen Xanthomonas oryzae pv. oryzae employs inducible hrp-dependent XopF type III effector protein for its growth, pathogenicity and for suppression of PTI response to induce blight disease. Eur J Plant Pathol. 2016;144(2):311–323. doi: 10.1007/s10658-015-0768-7. [DOI] [Google Scholar]
- Moreira LM, Almeida NF, Potnis N, Digiampietri LA, Adi SS, Bortolossi JC, da Silva AC, da Silva AM, de Moraes FE, de Oliveira JC, de Souza RF. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics. 2010;1:238. doi: 10.1186/1471-2164-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 2014;9(9):e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potnis N, Krasileva K, Chow V, Almeida NF, Patil PB, Ryan RP, Sharlach M, Behlau F, Dow JM, Momol MT, White FF. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics. 2011;12:146. doi: 10.1186/1471-2164-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Vorhölter FJ, Potnis N, Jones JB, Van Sluys MA, Bogdanove AJ, Dow JM. Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat Rev Microbiol. 2011;9:344. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- Song C, Yang B. Mutagenesis of 18 type III effectors reveals virulence function of XopZ PXO99 in Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 2010;23:893–902. doi: 10.1094/MPMI-23-7-0893. [DOI] [PubMed] [Google Scholar]
- Soni M, Mondal KK. Xanthomonasaxonopodis pv. punicae employs XopL effector to suppress pomegranate immunity. J Integr Plant Biol. 2018;60:341–357. doi: 10.1111/jipb.12615. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Tampakaki AP, Fadouloglou VE, Gazi AD, Panopoulos NJ, Kokkinidis M. Conserved features of type III secretion. Cell Microbiol. 2004;6(9):805–816. doi: 10.1111/j.1462-5822.2004.00432.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge S, Furutani A, Fukunaka R, Takashi OKU, Tsuno K, Ochiai H, Inoue Y, Hisatoshi KAKU, Yasuyuki KUBO. Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J Gen Plant Pathol. 2002;68:363–371. doi: 10.1007/PL00013104. [DOI] [Google Scholar]
- Ustun S, Bartetzko V, Bornke F. The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog. 2013;9:e1003427. doi: 10.1371/journal.ppat.1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees S. Phenotypic analysis of Arabidopsis mutants: trypan blue stain for fungi, oomycetes and dead plant cells. Cold Spring Harb Protoc. 2008 doi: 10.1101/pdb.prot4982. [DOI] [PubMed] [Google Scholar]
- Verma G, Sharma M, Mondal KK. XopR TTSS-effector regulates in planta growth, virulence of Indian strain of Xanthomonas oryzae pv. oryzae via suppressing reactive oxygen species production and cell wall-associated rice immune responses during blight induction. Funct Plant Biol. 2018;45:561–574. doi: 10.1071/FP17147. [DOI] [PubMed] [Google Scholar]
- Xu K, Huang X, Wu M, Wang Y, Chang Y, Liu K, Zhang J, Zhang Y, Zhang F, Yi L, Li T. A rapid, highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS One. 2014;9:e83556. doi: 10.1371/journal.pone.0083556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yin Z, White F. TAL effectors and the executor R genes. Front Plant Sci. 2015;6:641. doi: 10.3389/fpls.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]