Abstract
Bacillus subtilis is recognized as a safe and reliable human and animal probiotic and is associated with bioactivities such as production of vitamin and immune stimulation. Additionally, it has great potential to be used as an alternative to antimicrobial drugs, which is significant in the context of antibiotic abuse in food animal production. In this study, we isolated one strain of B. subtilis, named WS-1, from apparently healthy pigs growing with sick cohorts on one Escherichia coli endemic commercial pig farm in Guangdong, China. WS-1 can strongly inhibit the growth of pathogenic E. coli in vitro. The B. subtilis strain WS-1 showed typical Bacillus characteristics by endospore staining, biochemical test, enzyme activity analysis, and 16S rRNA sequence analysis. Genomic analysis showed that the B. subtilis strain WS-1 shares 100% genomic synteny with B. subtilis with a size of 4,088,167 bp. Importantly, inoculation of newborn piglets with 1.5 × 1010 CFU of B. subtilis strain WS-1 by oral feeding was able to clearly inhibit diarrhea (p < 0.05) and death (p < 0.05) caused by pathogenic E. coli in piglets. Furthermore, histopathological results showed that the WS-1 strain could protect small intestine from lesions caused by E. coli infection. Collectively, these findings suggest that the probiotic B. subtilis strain WS-1 acts as a potential biocontrol agent protecting pigs from pathogenic E. coli infection.
Importance: In this work, one B. subtilis strain (WS-1) was successfully isolated from apparently healthy pigs growing with sick cohorts on one E. coli endemic commercial pig farm in Guangdong, China. The B. subtilis strain WS-1 was identified to inhibit the growth of pathogenic E. coli both in vitro and in vivo, indicating its potential application in protecting newborn piglets from diarrhea caused by E. coli infections. The isolation and characterization will help better understand this bacterium, and the strain WS-1 can be further explored as an alternative to antimicrobial drugs to protect human and animal health.
Keywords: Bacillus subtilis, genomic analysis, biocontrol agent, newborn piglets, Escherichia coli
Introduction
Bacillus was first reported by Christian Gottfried Ehrenberg in 1835 (Sella et al., 2015). Since then, Bacillus has been found in a wide variety of organisms, such as pigs, and environments, such as ponds and soil (Borsodi et al., 2011; Gu et al., 2015; Wei et al., 2016). Bacillus is a common bacterium and is Gram-positive, rod-shaped with a size range of 0.3~22 μm × 1.2~7.0 μm, spore-forming, and aerobic-to-facultative (Cowan, 1974). A circular colony with rough, opaque, fuzzy white or slightly yellow, and jagged edges was observed by culturing Bacillus on nutrient agar (Lu et al., 2018). The genus Bacillus was initially proposed by Cohn in 1872 (Harwood, 1989), since then the genus has been expanded with many novel strains. In general, Bacillus can be classified into three categories based on the morphology of the spore: (1) seven species including Bacillus subtilis in the first genus with ovular- or pillar-shaped spores and with sporocyte that is not significantly expanded; (2) nine species including Bacillus circulans in the second genus with oval spore and enlarged sporocyte; and (3) Bacillus sphaericus in the third genus with a circular spore and enlarged sporocyte (Ruth et al., 1973).
Bacillus is widely used in industry, agriculture, and the medical field since a variety of functional Bacillus strains are extensively used in the production of industrial enzymes, bioinsecticides, antibiotics, and other products (Schallmey et al., 2004). B. subtilis is one of the most commonly functional strains. Currently, several studies have confirmed that B. subtilis has great application value in animal husbandry. It was reported that pig feed with B. subtilis natto could significantly improve meat quality and flavor (Sheng et al., 2016). Liu et al. (2017) also demonstrated that dietary corn bran fermented by B. subtilis MA139 could decrease gut cellulolytic bacteria and microbiota diversity in finishing pigs. In addition, B. subtilis as delivery vectors is also used to develop vaccines against TGEV in pigs (Mou et al., 2016). Additionally, more functions such as the antimicrobial activity of B. subtilis have been identified with the discovery and isolation of new strains (Sato et al., 2001; Phelan et al., 2013; Piewngam et al., 2018; Bartolini et al., 2019; Swartzendruber et al., 2019). For example, it has been found by Bartolini et al. (2019) that the stress-responsive alternative sigma factor (SigB) of B. subtilis enhanced antifungal proficiency by increasing the synthesis of lipopeptide surfactin. In addition, Piewngam proved that fengycin-producing Bacillus could inhibit Staphylococcus aureus colonization in mice (Piewngam et al., 2018).
The accurate identification of a new bacterium is an essential part of studying its function. The traditional techniques to identify a new bacterium are mainly based on the use of selective media by observing bacterial colony characteristics and morphology and by biochemical tests (Lu et al., 2018). Although these methods can preliminarily identify the type of the new strain, the detailed information such as genus, genomic composition, and functional proteins remains unclear. After the conserved bacterial genomic regions are amplified and sequenced, the genus of the bacteria will be accurately identified by homology analysis with the sequences from GenBank (Wellinghausen et al., 2009). These methods contribute to rapid and accurate identification of new bacterial strains.
Despite various applications of B. subtilis in many fields, detailed information underlying resistance against intestinal infections in the pig remains unclear. In this study, we isolated one strain of B. subtilis WS-1 from apparently healthy pigs raised with sick cohorts on one Escherichia coli endemic commercial pig farm in Guangdong, China and analyzed its genome. In addition, B. subtilis WS-1 was identified to inhibit the growth of pathogenic E. coli in vitro and in vivo, which implied that the isolate of B. subtilis WS-1 could have potential usage in the future.
Materials and Methods
Bacterial Strain and Experimental Newborn Piglets
E. coli strain 4–1, belonging to serogroup O149:K88 and containing LT and ST genes, was isolated from the same commercial pig farms as B. subtilis WS-1 and was confirmed to be highly pathogenic to the newborn piglets. Four-day-old crossbred (Duroc × Landrace × Big White) healthy conventional female newborn piglets without diarrheic symptoms were procured from Wen’s Foodstuffs Group Co, Ltd. (Guangdong, China). All piglets were fed a mixture of skim milk powder (Inner Mongolia Yili Industrial Group Co., Ltd., China) with warm water. The animal study was approved by the Institutional Animal Care and Use Committee of the Sun Yat-sen University (Guangdong, China), and animals were treated in accordance with the regulations and guidelines of this committee.
Reagents and Culture Medium
The Tryptic Soy Broth (TSB) medium and Tryptic Soy Agar (TSA) medium were purchased from Becton, Dickinson and Company (USA). The Oxford cup and Gram Stain Kit were purchased from Guangzhou Heyue Biotechnology Co., Ltd. (China). Endospore Stain Kit was purchased from Solarbio Company (China). TIANamp Bacteria DNA Kit was purchased from TIANGEN Biotech Co., Ltd. (Beijing, China). The Premix Taq™ (LA Taq™ Version 2.0 plus dye) and pMD19-T were purchased from Takara (Dalian, China).
Bacteria Isolation and Inoculum Preparation
Fresh pig feces (10 g) were collected from apparently healthy pigs growing with diarrheic cohorts on one E. coli endemic commercial pig farm in Guangdong, China. Prior to isolation, pig feces were mixed with 90 μl sterile 1 × phosphate buffer saline (PBS; pH 7.4), then incubated in an orbital shaker incubator (Shanghai Bluepard Instruments Co., Ltd., China) at room temperature with a shaking speed of 180 rpm for 30 min, then serial diluted up to 10−7 with a sterile distilled water. Isolation of bacteria from this mixture was done with a serial dilution technique in TSA medium (BD, USA). Bacteria were purified by repeated streaking and single colony culture at 37°C for 17–24 h. A total of 35 unknown bacterial isolates were recovered in TSB medium (BD, USA). Exponential phase growing cultures were washed twice with sterile 1 × PBS and maintained at 4°C until use.
Screening of the Antimicrobial Isolates Using the Oxford Cup Method
The Oxford cup method was performed as previously described with some modifications (Bian et al., 2016). Briefly, the culture fluid of E. coli strain 4–1 was mixed with the TSA medium at 50°C and poured into a bacterial culture dish. The Oxford cups containing 80 μl (7.2 × 108 CFU/ml) culture fluid of 35 unknown bacteria strains were affixed to the uniform coating on the medium, placed at 37°C for 18–24 h, and the inhibitory rings were observed and measured with a Vernier caliper (Guangzhou Heyue Biotechnology Co., Ltd., China). The Oxford cups containing culture fluid without bacteria were used as control.
Identification of the No. 1 Unknown Bacterial Strain
(1) The No. 1 unknown bacterial strain was examined for being a member of the family Bacillus by means of endospore staining according to the manufacturer’s instruction (Solarbio Company, China), Gram staining (Lu et al., 2018), and biochemical trait test (Cowan, 1974) and further by enzyme activity (Cowan, 1974). (2) Molecular identification of the Bacillus isolate by PCR and total DNA of the No. 1 unknown bacterial strain were prepared according to the manufacturer’s instruction (TIANGEN Company, China). The universal bacterial primers for the 16S rRNA gene of Bacillus (sense: 5′-AGAGTTGATCCTGGCTAAG-3′; antisense: 5′-GGTTACCTTGTTACGACTT-3′) were designed with reference to a previous publication (Lu et al., 2018) and were synthesized by Sangon Company (Shanghai, China). The PCR was performed in a volume of 50 μl containing 1 μl of DNA, 25 μl Premix Taq™ (LA Taq™ Version 2.0 plus dye), upstream and downstream primer (50 μmol/L) each at 1 μl and 22 μl ddH2O. The thermal cycling parameters were as follows: 94°C for 5 min; 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 15 min. The positive PCR products were cloned into the pMD19-T (TaKaRa, Dalian) and sequenced by Sangon Company (Shanghai, China). Sequence alignments of 16S rRNA of different Bacillus species were performed using the DNAStar Lasergene 7.0. A genome homology analysis and phylogenetic trees were constructed by using the maximum likelihood method with MEGA 5 software1 based on the 16S rRNA nucleotide sequences of 14 Bacillus strains from different countries.
Genome Sequencing and Bioinformatics Analysis
After we successfully identified the No. 1 unknown strain as a member of B. subtilis, which was thereafter named as B. subtilis WS-1, the complete genome was sequenced by the PacBio platform (Single Molecule, Real-Time technology) (Magigene, Guangdong) as previously described with some modifications (Cameron et al., 2015; Tanizawa et al., 2015; Carrión et al., 2018). Briefly, the bacterial genomic DNA was extracted as described above. DNA integrity and purity were monitored on 1% agarose gels, and the concentration and purity of DNA were measured using Qubit 2.0 (Thermo Fisher Scientific Waltham, USA) and Nanodrop one (Thermo Fisher Scientific Waltham, USA) at the same time. Then, the qualified genomic DNA was fragmented with G-tubes (Covaris) and end-repaired to prepare SMRTbell DNA template libraries (with a fragment size of >10 kb selected by bluepippin system) according to the manufacturer’s specification (PacBio, Menlo Park, USA). Library quality was detected by Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY, USA), and average fragment size was estimated on an Agilent 4,200 (Agilent, Santa Clara, CA, USA). SMRT sequencing was performed on the Pacific Biosciences RSII sequencer (PacBio, Menlo Park, USA) according to standard protocols. The low-quality reads were filtered by the SMRT 2.3.0 and assembled to generate one contig without gaps after sequencing. For the genome component prediction, the whole genome sequence was performed by Gene Marks for coding gene prediction (Besemer et al., 2001), diamond and BLAST for gene annotation, and PHAST for pre-phage prediction (Zhou et al., 2011). Related functional proteins were analyzed by BLAST. The genome overview was created by Circos (Krzywinski et al., 2009) to show the annotation information. Genomic synteny was analyzed by MUMmer software (Delcher et al., 2002) based on the alignment results with B. subtilis (GenBank no: AL009126.3).
Measurement of Bacterial Growth
To determine the growth rate of B. subtilis WS-1 (3.4 × 104), the strain was grown in TSB medium at 37°C for 48 h with agitation, and the CFU was determined at 0, 6, 12, 18, 24, 30, 36, 42, and 48 h.
Experimental Infection With the E. coli 4–1 Strain After B. subtilis Strain WS-1 Treatment in Conventional Newborn Piglets
Twelve newborn piglets were randomly divided into two groups (6 piglets/group) and were housed in two separate rooms. Prior to inoculation, newborn piglets were confirmed negative for the major porcine enteric viruses (PDCoV, PEDV, TGEV, and PRoV) by testing the rectal swabs collected from the newborn piglets on day 1 as previously described (Xu et al., 2018). On day 0, newborn piglets in group 1 were orally inoculated with 5 ml/day of TSB medium for 3 days. Newborn piglets in group 2 were orally inoculated with 5 ml/day of TSB medium containing a total of 5 × 109 CFU of the B. subtilis strain WS-1 (1 ml of medium contained 1 × 109 CFU of B. subtilis strain WS-1) for 3 days. Afterward, all piglets were orally challenged with 5 ml TSB medium containing 1 × 1010 CFU of the E. coli 4–1 strain. The piglets were observed daily for clinical signs of diarrhea and lethargy. One piglet from each group was necropsied at 3 days post challenge (d.p.c). At necropsy, the fresh duodenum, jejunum, and ileum were collected and fixed by 10% formalin for histopathology analysis. In addition, the mortality rate of newborn piglets in different treatment groups was recorded daily from day 1 to day 6 after challenge for protection rate analysis.
Histological Staining
Histological staining was performed as previously described (Xu et al., 2018). Briefly, tissue samples of the duodenum, jejunum, and ileum of the piglets from the B. subtilis strain WS-1 treatment and control groups were routinely fixed in 10% formalin for 36 h at room temperature and then dehydrated in graded ethanol, embedded in paraffin, cut in 5-μm sections, and mounted onto glass slides. After the sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E), the slides were examined and analyzed with conventional light microscopy (Nikon, Japan).
Statistical Analysis
Statistical comparisons were performed using GraphPad Prism software. The significance of the differences between the treatment group and control in the inhibitory rings, diarrhea rate, and survival rate was determined by ANOVA and Mann-Whitney accordingly.
Results
Ten Unknown Bacterial Strains Isolated From the Feces of Apparently Healthy Pigs Inhibit the Growth of Pathogenic E. coli in vitro
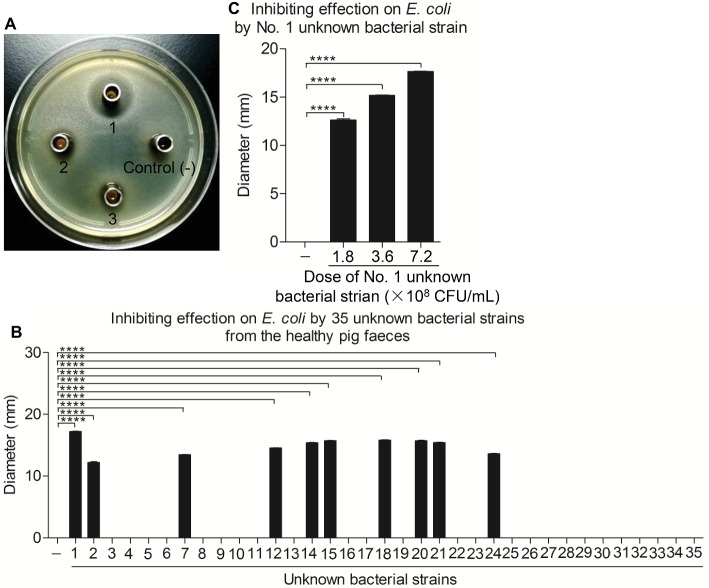
Some apparently healthy piglets without any symptoms of diarrhea were found on one E. coli endemic commercial pig farm in Guangdong, China. To determine the protective agent against the diarrhea outbreak in this pig farm, fresh feces from one healthy pig were collected and used to isolate the bacteria. Of the 10 g of feces examined, 35 unknown bacterial strains were isolated. Furthermore, we found that 10 unknown bacterial strains could markedly inhibit the growth of E. coli by the Oxford cup method (Figures 1A,B). Of them, the No. 1 unknown bacterial strain had the best antibacterial effect in a dose-dependent manner (Figures 1B,C).
Figure 1.
Ten unknown bacterial strains from healthy pig feces significantly inhibited the growth of E. coli in vitro. (A,B) Inhibiting effect on E. coli of total 35 unknown bacterial strains isolated from the healthy pig feces. E. coli strain 4–1 was maintained on bouillon agar medium (108 CFU/ml); the Oxford cup with 80 μl single bacterial culture fluid was placed in the middle. After 24 h, the inhibitory ring was observed and measured with ruler. (C) The No. 1 unknown bacterial strain significantly inhibited the growth of E. coli. E. coli strain 4–1 was maintained on bouillon agar medium (108 CFU/ml); the Oxford cups with 80 μl of No. 1 unknown bacterial culture fluid containing different amounts of 1.8 × 108, 3.6 × 108, and 7.2 × 108 CFU/ml were placed in the middle. The inhibitory rings were observed and measured with a ruler. Data are representative of three independent experiments. Data are represented as mean ± SD, n = 3. ****p < 0.0001.
A Strain of B. subtilis Was Identified
The No. 1 unknown bacterial strain was identified as Bacillus by morphological and biochemical examinations. The isolate displayed the morphology of Bacillus by visual and microscopic observations and was shaped as Gram-positive (data not shown) Bacillus, which could form spores (Figure 2). A biochemical test was further employed to analyze the characteristic of this bacterium. The results clearly showed that the strain was up to 99% in consistency with the standard of Bacillus (Table 1). These results indicated that the No. 1 unknown bacterial strain was a strain of Bacillus. To determine the species of Bacillus of this strain, we further analyzed the 16S rRNA by sequencing the PCR-amplified product. As shown in Figure 3,2 the No. 1 unknown bacterial strain shared 99% identity with B. subtilis based on the sequence of 16S rRNA. Phylogenetic analysis showed that the No. 1 unknown bacterial strain was clustered into a clade with B. subtilis B4 from Hubei, China. Taken together, the No. 1 unknown bacterial strain belongs to B. subtilis and was named WS-1.
Figure 2.
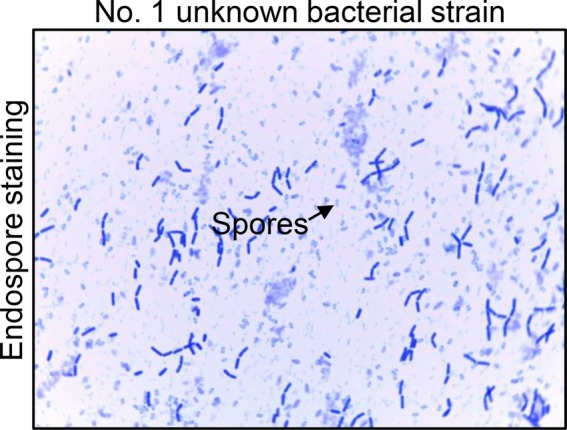
The endospore staining of No. 1 unknown bacterial strain. Endospore staining to analyze the characteristic of No. 1 unknown bacterial strain. The arrows show the forming spores of this unknown bacterium.
Table 1.
Biochemical traits of the Bacillus isolate (B. subtilis strain WS-1).
| Test | Result | Test | Result | Test | Result |
|---|---|---|---|---|---|
| Glycerin | + | Inositol | + | Inulin | + |
| Erythritol | − | Mannitol | + | Melezitose | − |
| D-arabinose | − | Sorbitol | + | Raffinose | + |
| L-arabinose | + | α-methyl-D mannoside | − | Starch | + |
| Ribose | + | α-methyl-D glucoside | + | Glycogen | + |
| D-xylose | + | N-acetyl-glucosamine | − | Xylitol | − |
| L-xylose | − | Amygdalin | + | Gentian disaccharide | − |
| Amoxicol | − | Arbutin | + | D-Turanose | + |
| β-methyl-D-xyloside | − | Esculin | + | D-lyxose | − |
| Galactose | − | 2-Hydroxybenzyl alcohol | + | D-Tagatose | − |
| Glucose | + | Cellobiose | + | D-arabinitol | − |
| fructose | + | Maltose | + | L-arabinitol | − |
| Mannose | + | Lactose | − | Gluconate | − |
| Sorbose | − | Melibiose | + | 2-keto-gluconate | − |
| L-Rhamnose | − | Sucrose | + | 5-keto-gluconate | − |
| Dulcitol | − | Trehalose | + |
Figure 3.
Phylogenetic tree constructed on the basis of the 16S rRNA gene from B. subtilis strain WS-1 and 13 B. subtilis strains from different countries. The dendrogram was constructed using the neighbor joining method in the MEGA software package, version 5. Reference sequences obtained from GenBank are indicated by strain name. The scale bar represents 0.01 nucleotide substitutions per site.
Complete Genome Sequencing and Analysis of B. subtilis Strain WS-1
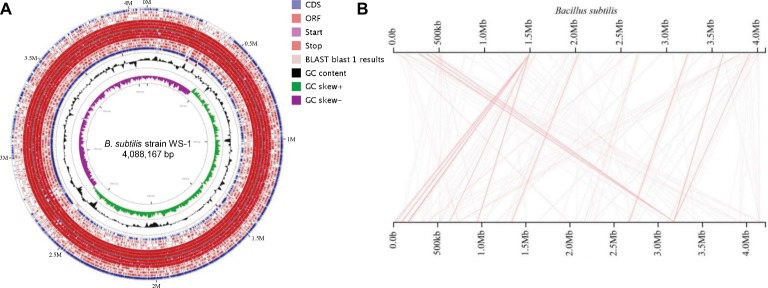
The complete genome of B. subtilis strain WS-1 was acquired and uploaded onto GenBank (No. CP024921). The genome of B. subtilis strain WS-1 has a size of 4,088,167 bp (Figure 4A), with G + C content being 43.8%. Some genes encoded by this strain of Bacillus have protease, lipase, and amylase activity (Table 2), indicating that the WS-1 strain might have antimicrobial activity. In addition, approximately 89% of nucleotides were predicted as protein-coding regions, and 86.8% (3,704) of the open reading frames were annotated with known proteins. Some putative proteins were predicted to be associated with antimicrobial or probiotic activity by the Swiss-Prot database (Table 3). We also confirmed that the WS-1 strain contained sporulation genes like CgeA, CotB, and CotZ (data not shown). These data confirmed that the WS-1 strain shares 100% genomic synteny with B. subtilis (Figure 4B).
Figure 4.
Genome visualization and genomic synteny of the B. subtilis strain WS-1. (A) Genome visualization of B. subtilis strain WS-1. The genome of B. subtilis strain WS-1 was sequenced and analyzed using the PacBio platform. (B) Genomic synteny of B. subtilis strain WS-1 was analyzed based on the alignment results with B. subtilis.
Table 2.
Enzyme activity analysis of B. subtilis strain WS-1.
| Variety | Nucleotide position* | Enzyme activity (U/ml) |
|---|---|---|
| Protease | (−) 210183-211151, 684509-686197, 725622-727559, 758838-761258, 1122374-1122967, 2696411-2697742, 2813385-2814671, 2814668-2815948, 2829137-2830327, 2847680-2848948, 2882897-2884300, 2884317-2884862, 3384242-3385801, 3415417-3418101, 3467502-3468782, 3906905-3907594; (+) 516616-517818, 1045409-1046851, 1283746-1285122, 1364459-1365088, 1450962-1451639, 1783051-1784313, 1784465-1786123, 1786304-1788628, 1804131-1804667, 1812254-1813120, 1895220-1895846, 2095165-2095638, 2134016-2135539, 2187051-2188331, 2300552-2301154, 2306573-2307229, 2439721-2441121, 2710268-2711596, 3031354-3032919, 3135226-3137355, 3185998-3186957, 3466390-3467463, 3498168-3498779, 3899889-3900581 |
40.1 |
| Lipase | (−) 163484-164155, 3661219-3661836, 4066571-4067212; (+) 2415760-2416569 |
28.6 |
| Amylase | (−) 126705-128684 | 158.2 |
Nucleotide position is numbered based on B. subtilis WS-1 strain (CP024921).
(+): Positive-sense strand; (−): negative-sense strand.
Table 3.
Functional proteins prediction of B. subtilis strain WS-1.
| Name | Nucleotide position* | Number |
|---|---|---|
| Polyketides | (−) 2714457-2722088, 2722103-2738647, 2738637-2751425, 2751441-2754173, 2754134-2764267, 2764255-2765067, 2765212-2765610, 2765607-2780180, 2780225-2780974, 2781014-2781793, 2781781-2783043, 2783044-2784291, 2784579-2786513, 2786501-276881, 278678-2787852, 2788356-2789222, 2790462-2791079; (+) 3033214-3034185 |
18 |
| Lipopeptides | (−) 20315-21268, 90550-91134, 132322-133203, 147031-147990, 164284-165411, 600839-601852, 962123-963040, 1243143-1243943, 1249990-1250721, 1428128-1429255, 1570007-1571287, 1865158-1865979, 2955757-2956074, 2956062-2956208, 3207178-3208827, 3350410-3352047, 3463183-3464229,3586153-3586992, 3608704-3609702, 3784560-3785786, 3824696-3825895, 4012394-4013515, 4063703-4065709 (+) 44837-45643, 87329-88180, 267938-269866, 272995-273948, 608703-609497, 650251-651399, 683398-684237, 740201-741358, 1074976-1075785, 1113395-1114648, 1160684-1161949, 1207777-1208697, 1264758-1265702, 1306601-1307425, 1318172-1318681, 1318687-1319430, 1523167-1524108, 1581821-1583335, 1660893-1661702, 1661692-1662528, 1726112-1727431, 1965356-1966111, 2058797-2059528, 2125691-2126728, 2214635-2215402, 2415760-2416569, 2437636-2438457, 2457696-2458565, 2676968-2677603, 3500643-3501521, 3651495-3652505, 3745050-3746072 |
55 |
| Flagella | (−) 1138642-1139100, 2861243-2862133, 2862130-2863230, 2863230-2865263, 2865296-2866378, 2866378-2867157, 2867165-2867410, 2867449-2868114, 2868107-2868766, 2869169-2870305, 2870295-2871293, 2871327-2871749, 2871746-2871961, 2872001-2872762, 2872817-2873239, 2873236-2874699, 2875337-2875780, 2877096-2877857, 2877841-2878857, 2878870-2880480, 2880526-2880852, 2880857-2881312, 2881309-2881704, 2892718-2892999; (+) 921972-922808, 922854-923651, 1029455-1030963, 1030974-1031870, 1032513-1032944, 1034454-1034783, 1034801-1034947, 1034977-1036296, 1036318-1036719, 1625025-1625846, 1625836-1626564, 3137648-3138550, 3138522-3139307 |
37 |
| Exopolysaccharide biosynthesis | (+) 556979-558043, 1143948-1144784, 1149168-1150202, 1152318-1152968, 1153006-1154139, 1154136-1155086 | 6 |
| Enolase | (−) 3144704-3145948; (+) 3395921-3397036 | 2 |
Nucleotide position is numbered based on B. subtilis WS-1 strain (CP024921).
(+): Positive-sense strand; (−): negative-sense strand.
The Growth Rate of B. subtilis Strain WS-1 in vitro
To determine the growth rate in vitro, we inoculated B. subtilis strain WS-1 in TSB medium and detected the living bacterial count at corresponding time points. As shown in Figure 5, the B. subtilis strain WS-1 grew exponentially post inoculation and reached a plateau 12 h post inoculation, which lasted for at least 36 h, indicating that B. subtilis strain WS-1 could adapt to the TSB medium in vitro.
Figure 5.
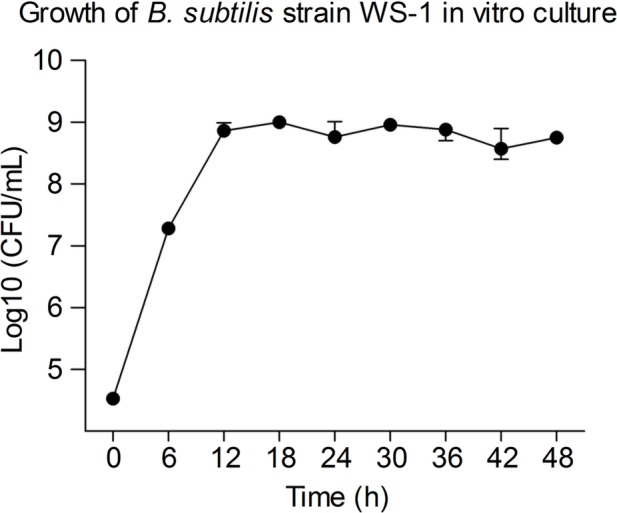
Measurement of bacterial growth. The growth curves of B. subtilis strain WS-1. Bacteria were grown in bouillon fluid medium at 37°C for 48 h with agitation, and the Colony Forming Unit (CFU) was determined at 0, 6, 12, 18, 24, 30, 36, 42, and 48 h. The data are representative of three independent experiments. Data are represented as mean ± SD, n = 3.
B. subtilis Strain WS-1 Inhibits Diarrhea and Death Caused by E. coli in Newborn Piglets
Since B. subtilis strain WS-1 was confirmed to significantly inhibit the growth of E. coli in vitro, we attempted to determine whether WS-1 has the same effect in vivo. We experimentally infected newborn piglets with E. coli strain 4–1 after B. subtilis strain WS-1 treatment. As expected, the newborn piglets pre-inoculated with TSB medium containing B. subtilis strain WS-1 via oral feeding showed a lower diarrhea rate (16.7%) at the first day. By contrast, all newborn piglets (100%) pre-inoculated with TSB medium without B. subtilis strain WS-1 via oral feeding showed acute and severe watery diarrhea in the first 2 days (Figure 6A), indicating that B. subtilis strain WS-1 might serve as a probiotic in newborn piglets. Importantly, two piglets died that pre-inoculated with TSB medium without B. subtilis strain WS-1 via oral feeding group at 2 d.p.c., and no piglets died in the B. subtilis strain WS-1 treatment group during the study (Figure 6B). Taken together, these results demonstrated that B. subtilis strain WS-1 works as a probiotic to inhibit E. coli infection in vivo.
Figure 6.
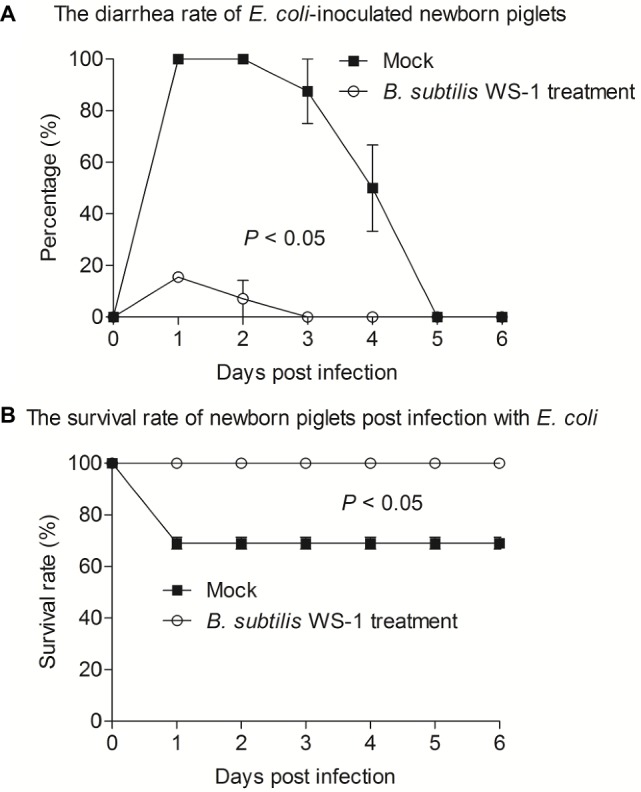
B. subtilis strain WS-1 inhibited diarrhea and death caused by E. coli in newborn piglets. Newborn piglets were first orally fed with TSB medium or TSB medium containing B. subtilis strain WS-1. At 3 days, all piglets were orally challenged with E. coli strain 4–1. The diarrhea rate (A) and survival rate (B) of newborn piglets post-challenge with E. coli between the control group and the B. subtilis strain WS-1 treatment group were recorded daily from the first day to the sixth day after challenge. The data are representative of two independent experiments. Data are represented as mean ± SD, n = 6 or n = 7.
Histopathological Results of Newborn Piglets Infected With E. coli After B. subtilis Strain WS-1 Treatment
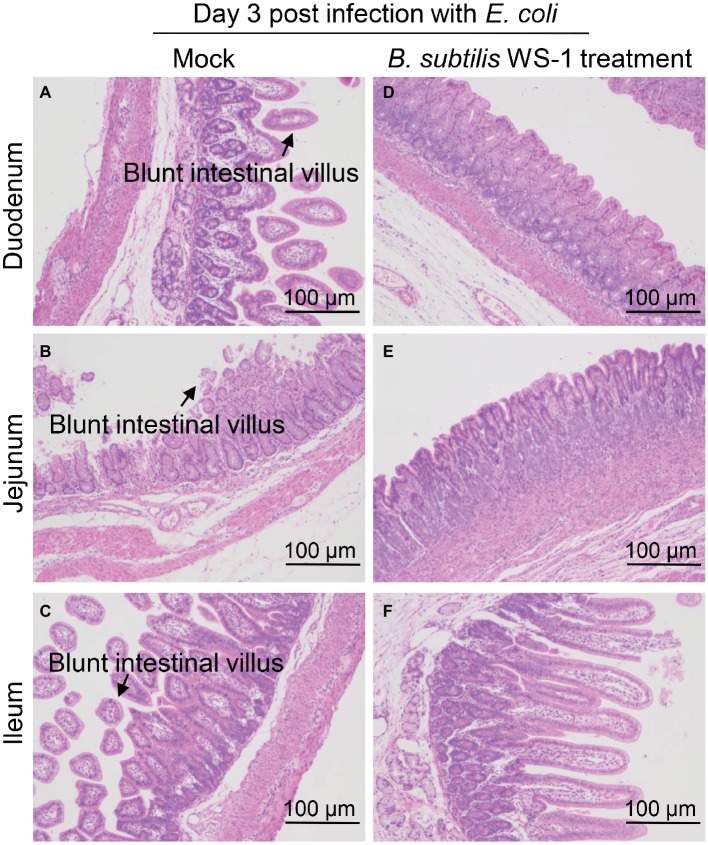
To determine the histological changes in the intestine of newborn piglets infected with E. coli after B. subtilis strain WS-1 treatment, piglets were necropsied at 3 d.p.c. As shown in Figure 7, blunt intestinal villus was observed in the duodenum, jejunum, and ileum in the TSB medium only group, while the villus in the B. subtilis strain WS-1 treatment group remained intact (Figures 7A–F).
Figure 7.
Intestinal changes in newborn piglets post-challenge with E. coli of the control group and the B. subtilis strain WS-1 treatment group. (A–C) Hematoxylin and eosin (H&E)-stained intestinal tissue section (blunt intestinal villus was indicated by arrows) of a control piglet at 3 d.p.c. (D–F) H&E-stained intestinal tissue section of a B. subtilis strain WS-1 treatment piglet at 3 d.p.c. The data are representative of two independent experiments. Data are represented as mean ± SD, n = 1 or n = 2.
Discussion
Since the first report of B. subtilis by Christian Gottfried Ehrenberg in 1835 (Sella et al., 2015), this ancient bacteria had been widely detected and isolated in many organisms and environments (Borsodi et al., 2011; Gu et al., 2015; Wei et al., 2016). Many subsequent studies demonstrated that B. subtilis had a variety of functions. Although B. subtilis was widely used in the field of fermentation and livestock and poultry breeding (Nguyen et al., 2015; Liu et al., 2017; Amorim et al., 2019), currently limited information is available regarding the protection of B. subtilis against intestinal diseases in newborn piglets. In the present study, we reported an isolate of B. subtilis from apparently healthy pigs in a pathogenic E. coli endemic commercial farm with severe diarrhea symptoms in piglets, which could inhibit the growth of E. coli in vitro and protect piglets from diarrhea and death caused by pathogenic E. coli in newborn piglets.
Diarrhea in piglets is mainly caused by viruses and bacteria such as Porcine epidemic diarrhea virus (PEDV) and E. coli (Vlasova et al., 2014; Garcia-Menino et al., 2018), which results in significant economic losses to the pig industry. Occasionally, some apparently healthy pigs without symptoms of diarrhea were found commingling with E. coli diarrheic piglets. To determine the protective agent in these pigs, 35 unknown bacterial strains from fresh feces of one healthy pig were isolated and 10 unknown bacterial strains were determined to markedly inhibit the growth of E. coli and Salmonella typhi (data not shown) by the Oxford cup method (Figure 1), indicating that these unknown strains might be the key to protect from diarrhea in these pigs. Of them, the No. 1 unknown bacterial strain had the best antibacterial effect. After analysis through conventional bacterial identification methods, the No. 1 unknown bacterial strain was identified as Gram-positive bacteria of Bacillus, having protease, lipase, and amylase activity, which might be utilized to generate antimicrobial substances such as bioactive peptides (Daroit et al., 2012) and chloramphenicol esters (Dong et al., 2017) to inhibit bacterial biofilm formation (Vaikundamoorthy et al., 2018). Bosshard et al. (2003) demonstrated that 95–99% similarity for 16S rRNA gene sequencing between two bacteria hints toward a similar species, while >99% indicates the same bacterium. In the study, the No. 1 unknown bacterial strain was identified as a member of B. subtilis by the analysis of 16S rRNA gene sequencing and therefore named as B. subtilis WS-1. To further characterize the isolate, the complete genome of B. subtilis strain WS-1 was sequenced and analyzed. The WS-1 strain shares 100% genomic synteny with B. subtilis and encoded multiple functional proteins like lipopeptides, which might be associated with the antibacterial activity of the WS-1 strain.
Bacterial resistance is becoming more common with the abuse of antibiotics over the past years, and it urgently demands an alternative to antimicrobial drugs. Of note, several groups have confirmed the protective role of Bacillus strains as probiotics in vivo (Wu et al., 2014; Ramesh et al., 2015). Collectively, these previous results confirmed that Bacillus strains could antagonize Vibrio parahaemolyticus and Aeromonas hydrophila. Swine enteric colibacillosis affects all sectors of the pig industry and all cycles of production. Compared with other age groups, newborn piglets are more vulnerable to swine enteric colibacillosis. We hypothesized that B. subtilis strain WS-1 was also capable of antagonizing colibacillosis in newborn piglets based on its probiotic effect in vitro. Therefore, we experimentally infected newborn piglets with the pathogenic E. coli after B. subtilis strain WS-1 treatment. As expected, B. subtilis strain WS-1 was able to work against diarrhea and death caused by E. coli in newborn piglets (Figure 6), indicating that the isolated B. subtilis strain WS-1 also had a probiotic effect in vivo and might be used as an alternative of antimicrobial drugs. In addition, we also found that none of the WS-1 strain-inoculated newborn piglets showed any clinical signs (data not shown) before E. coli infection, proving that B. subtilis strain WS-1 is safe for pigs. Furthermore, no evident gross lesions were observed in the intestinal tract of the B. subtilis strain WS-1 treatment piglets at necropsy at 3 d.p.c. (data not shown). Similarly, no microscopic lesions were observed in the small intestine in the B. subtilis strain WS-1 treatment group after E. coli infection (Figure 7), which furthered our understandings of the role of the B. subtilis as a probiotic. It was reported that several lipopeptides such surfactin, secreted by B. subtilis, can confer strong antipathogenic effects and thus benefit the host by balancing the intestinal microbiome (Zhou et al., 2018). Polyketide from the seaweed-associated heterotrophic bacterium B. subtilis MTCC 10403 has potential antibacterial activities against clinically important pathogens (Chakraborty et al., 2017). In addition, some studies concluded that some proteins involved in aggregation, such as flagella, might be associated with probiotic effects (Kleta et al., 2014). We determined that some genes in the WS-1 genome encode these proteins (Table 3). Whether the interference effect of WS-1 on E. coli infection in newborn piglets was associated with these putative proteins needs to be further explored. Nevertheless, there are still several important questions that need to be addressed. For example, what is the exact underlying mechanism of B. subtilis strain WS-1 inhibiting enteric diseases caused by E. coli in newborn piglets? Can B. subtilis strain WS-1 resist enteric diseases caused by viruses like PEDV in pigs? Does B. subtilis strain WS-1 have other applications apart from protecting the intestinal tract? Elucidation of these questions will elevate our understandings of the function of B. subtilis strain WS-1 and may help extend its application in many fields.
In summary, we isolated a field strain of B. subtilis from apparently healthy pigs growing with sick cohorts on one E. coli endemic commercial pig farm in Guangdong, China. Remarkably, inoculation of newborn piglets with 1.5 × 1010 CFU of WS-1 by oral feeding could prevent diarrhea and death caused by pathogenic E. coli in piglets. Collectively, these findings suggest that B. subtilis strain WS-1 has great potential to be explored as biocontrol agent protecting piglets from enteric diseases in pigs.
Ethics Statement
The animal study was supervised by the Institutional Animal Care and Use Committee of Sun Yat-sen University (IACUC-2018-000178), and animals were used in accordance with regulations and guidelines of this committee.
Author Contributions
YC and YD conceived and designed the experiments. YD, ZX, GY, WL, and DY performed the experiments. YD and ZX analyzed the data. YC, QZ, JL, LC, YZ, and CX contributed the reagents, materials, and analysis tools. ZX wrote the manuscript. YC checked and finalized the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Key Research and Development Program, China (2016YFD0500101) and the Foundation Research Project of Jiangsu Province, China (BK20181034).
References
- Amorim C., Silverio S. C., Goncalves R. F. S., Pinheiro A. C., Silva S., Coelho E., et al. (2019). Downscale fermentation for xylooligosaccharides production by recombinant Bacillus subtilis 3610. Carbohydr. Polym. 205, 176–183. 10.1016/j.carbpol.2018.09.088, PMID: [DOI] [PubMed] [Google Scholar]
- Bartolini M., Cogliati S., Vileta D., Bauman C., Ramirez W., Grau R. (2019). The stress-responsive alternative sigma factor SigB plays a positive role in the antifungal proficiency of Bacillus subtilis. Appl. Environ. Microbiol. 85. 10.1128/aem.00178-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J., Lomsadze A., Borodovsky M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. 10.1093/nar/29.12.2607, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X., Evivie S. E., Muhammad Z., Luo G. W., Liang H. Z., Wang N. N., et al. (2016). In vitro assessment of the antimicrobial potentials of Lactobacillus helveticus strains isolated from traditional cheese in Sinkiang China against food-borne pathogens. Food Funct. 7, 789–797. 10.1039/C5FO01041A, PMID: [DOI] [PubMed] [Google Scholar]
- Borsodi A. K., Pollak B., Keki Z., Rusznyak A., Kovacs A. L., Sproer C., et al. (2011). Bacillus alkalisediminis sp. nov., an alkaliphilic and moderately halophilic bacterium isolated from sediment of extremely shallow soda ponds. Int. J. Syst. Evol. Microbiol. 61, 1880–1886. 10.1099/ijs.0.019489-0, PMID: [DOI] [PubMed] [Google Scholar]
- Bosshard P. P., Abels S., Zbinden R., Bottger E. C., Altwegg M. (2003). Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory (an 18-month evaluation). J. Clin. Microbiol. 41, 4134–4140. 10.1128/JCM.41.9.4134-4140.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D. R., Jiang J. H., Hassan K. A., Elbourne L. D., Tuck K. L., Paulsen I. T., et al. (2015). Insights on virulence from the complete genome of Staphylococcus capitis. Front. Microbiol. 6:980. 10.3389/fmicb.2015.00980, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión V. J., Cordovez V., Tyc O., Etalo D. W., De B. D., De J. V., et al. (2018). Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 12, 2307–2321. 10.1038/s41396-018-0186-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K., Thilakan B., Raola V. K. (2017). Antimicrobial polyketide furanoterpenoids from seaweed-associated heterotrophic bacterium Bacillus subtilis MTCC 10403. Phytochemistry 142, 112–125. 10.1016/j.phytochem.2017.06.019, PMID: [DOI] [PubMed] [Google Scholar]
- Cowan S. T. (1974). Bergey’s manual of determinative bacteriology. (USA: American Journal Public Health and the Nations Health; ). [Google Scholar]
- Daroit D. J., Corrêa A. P. F., Canales M. M., Coelho J. G., Hidalgo M. E., Tichota D. M., et al. (2012). Physicochemical properties and biological activities of ovine caseinate hydrolysates. Dairy Sci. Technol. 92, 335–351. 10.1007/s13594-012-0068-3 [DOI] [Google Scholar]
- Delcher A. L., Phillippy A., Carlton J., Salzberg S. L. (2002). Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 1, 2478–2483. 10.1093/nar/30.11.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Li L., Lin L., He D., Chen J., Wei W., et al. (2017). Transesterification synthesis of chloramphenicol esters with the lipase from Bacillus amyloliquefaciens. Molecules 22, 1–11. 10.3390/molecules22091523, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Menino I., Garcia V., Mora A., Diaz-Jimenez D., Flament-Simon S. C., Alonso M. P., et al. (2018). Swine enteric colibacillosis in Spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front. Microbiol. 9:2659. 10.3389/fmicb.2018.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. B., Zhao L. N., Wu Y., Li S. C., Sun J. R., Huang J. F., et al. (2015). Potential probiotic attributes of a new strain of Bacillus coagulans CGMCC 9951 isolated from healthy piglet feces. World J. Microbiol. Biotechnol. 31, 851–863. 10.1007/s11274-015-1838-x, PMID: [DOI] [PubMed] [Google Scholar]
- Harwood C. R. (1989). Introduction to the biotechnology of Bacillus. (New York: Plenum Press; ), 1–4. [Google Scholar]
- Kleta S., Nordhoff M., Tedin K., Wieler L. H., Kolenda R., Oswald S., et al. (2014). Role of F1C fimbriae, flagella, and secreted bacterial components in the inhibitory effect of probiotic Escherichia coli Nissle 1917 on atypical enteropathogenic E. coli infection. Infect. Immun. 82, 1801–1812. 10.1128/IAI.01431-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. 10.1101/gr.092759.109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Zhao J., Guo P., Lu W., Geng Z., Levesque C. L., et al. (2017). Dietary corn bran fermented by Bacillus subtilis MA139 decreased gut cellulolytic bacteria and microbiota diversity in finishing pigs. Front. Cell. Infect. Microbiol. 7:526. 10.3389/fcimb.2017.00526, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Guo W., Liu C. (2018). Isolation, identification and characterization of novel Bacillus subtilis. J. Vet. Med. Sci. 80, 427–433. 10.1292/jvms.16-0572, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou C., Zhu L., Xing X., Lin J., Yang Q. (2016). Immune responses induced by recombinant Bacillus subtilis expressing the spike protein of transmissible gastroenteritis virus in pigs. Antivir. Res. 131, 74–84. 10.1016/j.antiviral.2016.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Nguyen A. T., Nguyen D. V., Tran M. T., Nguyen L. T., Nguyen A. H., Phan T. N. (2015). Isolation and characterization of Bacillus subtilis CH16 strain from chicken gastrointestinal tracts for use as a feed supplement to promote weight gain in broilers. Lett. Appl. Microbiol. 60, 580–588. 10.1111/lam.12411, PMID: [DOI] [PubMed] [Google Scholar]
- Phelan R. W., Barret M., Cotter P. D., O’connor P. M., Chen R., Morrissey J. P., et al. (2013). Subtilomycin: a new lantibiotic from Bacillus subtilis strain MMA7 isolated from the marine sponge Haliclona simulans. Mar. Drugs 11, 1878–1898. 10.3390/md11061878, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piewngam P., Zheng Y., Nguyen T. H., Dickey S. W., Joo H. S., Villaruz A. E., et al. (2018). Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562, 532–537. 10.1038/s41586-018-0616-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh D., Vinothkanna A., Rai A. K., Vignesh V. S. (2015). Isolation of potential probiotic Bacillus spp. and assessment of their subcellular components to induce immune responses in Labeo rohita against Aeromonas hydrophila. Fish Shellfish Immunol. 45, 268–276. 10.1016/j.fsi.2015.04.018, PMID: [DOI] [PubMed] [Google Scholar]
- Ruth E. G., Haynes W. C., Hor-Nay Pang C. (1973). The genus Bacillus. (USA: U.S. Department of Agriculture; ). [Google Scholar]
- Sato T., Yamada Y., Ohtani Y., Mitsui N., Murasawa H., Araki S. (2001). Production of menaquinone (vitamin K2)-7 by Bacillus subtilis. J. Biosci. Bioeng. 91, 16–20. 10.1016/S1389-1723(01)80104-3, PMID: [DOI] [PubMed] [Google Scholar]
- Schallmey M., Singh A., Ward O. P. (2004). Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50, 1–17. 10.1139/w03-076, PMID: [DOI] [PubMed] [Google Scholar]
- Sella S. R., Vandenberghe L. P., Soccol C. R. (2015). Bacillus atrophaeus: main characteristics and biotechnological applications - a review. Crit. Rev. Biotechnol. 35, 533–545. 10.3109/07388551.2014.922915 [DOI] [PubMed] [Google Scholar]
- Sheng Q. K., Zhou K. F., Hu H. M., Zhao H. B., Zhang Y., Ying W. (2016). Effect of Bacillus subtilis natto on meat quality and skatole content in TOPIGS pigs. Asian-Australas. J. Anim. Sci. 29, 716–721. 10.5713/ajas.15.0478, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzendruber J. A., Incrocci R. W., Wolf S. A., Jung A., Knight K. L. (2019). Bacillus subtilis exopolysaccharide prevents allergic eosinophilia. Allergy 74, 819–821. 10.1111/all.13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa Y., Tohno M., Kaminuma E., Nakamura Y., Arita M. (2015). Complete genome sequence and analysis of Lactobacillus hokkaidonensis LOOC260(T), a psychrotrophic lactic acid bacterium isolated from silage. BMC Genomics 16:240. 10.1186/s12864-015-1435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaikundamoorthy R., Rajendran R., Selvaraju A., Moorthy K., Perumal S. (2018). Development of thermostable amylase enzyme from Bacillus cereus for potential antibiofilm activity. Bioorg. Chem. 77, 494–506. 10.1016/j.bioorg.2018.02.014, PMID: [DOI] [PubMed] [Google Scholar]
- Vlasova A. N., Marthaler D., Wang Q., Culhane M. R., Rossow K. D., Rovira A., et al. (2014). Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 20, 1620–1628. 10.3201/eid2010.140491, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Xin D., Xin Y., Zhang H., Wang T., Zhang J. (2016). Bacillus depressus sp. nov., isolated from soil of a sunflower field. Antonie Leeuwenhoek 109, 13–20. 10.1007/s10482-015-0605-1, PMID: [DOI] [PubMed] [Google Scholar]
- Wellinghausen N., Kochem A. J., Disque C., Muhl H., Gebert S., Winter J., et al. (2009). Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J. Clin. Microbiol. 47, 2759–2765. 10.1128/JCM.00567-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Sun L. B., Li C. B., Li Z. Z., Zhang Z., Wen X. B., et al. (2014). Enhancement of the immune response and protection against Vibrio parahaemolyticus by indigenous probiotic Bacillus strains in mud crab (Scylla paramamosain). Fish Shellfish Immunol. 41, 156–162. 10.1016/j.fsi.2014.08.027, PMID: [DOI] [PubMed] [Google Scholar]
- Xu Z., Zhong H., Zhou Q., Du Y., Chen L., Zhang Y., et al. (2018). A highly pathogenic strain of porcine deltacoronavirus caused watery diarrhea in newborn piglets. Virol. Sin. 33, 131–141. 10.1007/s12250-018-0003-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J., Wishart D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39, W347–W352. 10.1093/nar/gkr485, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Liu F., Zhang X., Zhou X., Zhong Z., Su H., et al. (2018). Cellulose-dependent expression and antibacterial characteristics of surfactin from Bacillus subtilis HH2 isolated from the giant panda. PLoS One 13:e0191991. 10.1371/journal.pone.0209902, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]