Abstract
Aberrant splicing in exon 11 of the LMNA gene causes the premature aging disorder Hutchinson-Gilford Progeria Syndrome. A de novo C1824T mutation activates an internal alternative 5′ splice site, resulting in formation of the disease-causing progerin protein. The underlying mechanism for this 5′ splice site selection is unknown. Here, we have applied a combination of targeted mutational analysis in a cell-based system and structural mapping by SHAPE-MaP to comprehensively probe the contributions of primary sequence, secondary RNA structure and linear splice site position in determining in vivo mechanisms of splice site choice in LMNA. While splice site choice is in part defined by sequence complementarity to U1 snRNA, we identify RNA secondary structural elements near the alternative 5′ splice sites and show that splice site choice is significantly influenced by the structural context of the available splice sites. Furthermore, relative positioning of the competing sites within the primary sequence of the pre-mRNA is a predictor of 5′ splice site usage, with the distal position favored over the proximal, regardless of sequence composition. Together, these results demonstrate that 5′ splice site selection in LMNA is determined by an intricate interplay among RNA sequence, secondary structure and splice site position.
INTRODUCTION
Our understanding of how RNA impacts diverse cellular processes has grown considerably in recent years. A long appreciated, but still relatively poorly understood, regulatory mechanism of RNA function is alternative pre-mRNA splicing (1), which enables generation of multiple mature mRNAs from the same gene (2). While alternative splicing is a highly effective mechanism to generate proteomic diversity in higher eukaryotes (3), the complex nature of 5′ and 3′ splice site (SS) selection by the major and minor spliceosomes makes splicing outcome susceptible to mutations within the splice site regions (4). It is estimated that 10% of all disease-causing mutations impact either a 3′ or 5′ SS (5) and consequentially result in exon skipping, intron retention, or alternative splice site activation.
A typical 5′ SS consists of three nucleotides at the end of the exon (–3 to –1) and six nucleotides at the beginning of the adjacent intron (+1 to +6). The strength of a 5′ SS, and thus its likelihood of use, has conventionally been strongly related to its potential for base pairing to the 5′ end of the U1 small nuclear RNA (snRNA) (6). However, the permissible sequence to serve as functional 5′ SS is context-dependent and can vary between different splicing events (7). In addition to primary sequence, several factors have been shown to be involved in the regulation of splice site usage. Prominent among these are the Ser/Arg-rich (SR) proteins (8–12) and heterogeneous nuclear ribonucleoproteins (hnRNP) (13). In addition, features inherent to the RNA itself have been implicated in 5′ SS choice, including the relative positions of candidate splice sites in the RNA primary sequence (14–17), as well as local RNA secondary structure, which can suppress 5′ SS usage by limiting accessibility to the spliceosome machinery (18–20). All these factors add multiple layers to the control of splicing regulation and must be taken into account, especially for splicing events which involve competition between multiple 5′ splice sites.
Hutchinson–Gilford Progeria Syndrome (HGPS) is a prominent example of disease-related alternative 5′ SS usage. This rare premature aging disease is caused by a de novo point mutation (C1824T) in exon 11 of the LMNA gene (21,22). The mutation activates an alternative exonic 5′ SS, which in turn results in an internal 150 nucleotide deletion in the mRNA (Δ150) and synthesis of the disease-causing progerin protein (23). While the mutation increases complementarity of the exonic alternative 5′ SS for the 5′ end of U1 snRNA and in this way likely contributes to its preferential use in HGPS (24), sequence alone is not sufficient to explain the dramatic increase in alternative 5′ SS usage in mutant LMNA since analysis by sequence-based splice site scoring algorithms (e.g. MaxENT (25)) demonstrates that the alternative 5′ SS score is lower than the nearly perfect score of the normal 5′ SS, even when augmented by the presence of the point mutation (24). Based on these considerations, it is clear that non-sequence related factors must contribute to LMNA 5′ SS choice.
Here, we perform a structure-function analysis to comprehensively explore the underlying mechanisms for 5′ SS selection in exon 11 of the normal and HGPS-causing LMNA isoform. We used systematic mutational analysis in a cell-based system and SHAPE-MaP to gain detailed information on the local and global features of LMNA RNA. We find that 5′ SS choice in exon 11 of LMNA is determined by an intricate interplay of primary sequence, RNA secondary structure, distal regulatory elements and splice site position.
MATERIALS AND METHODS
Cell culture and transfection conditions
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% FBS (Sigma), 2 mM l-glutamine, 1 mM sodium pyruvate, non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2. Cells were seeded in six-well plates at a density of 350 000 cells/well and incubated overnight. PEI was used for transient transfection of cells with 1 μg of plasmid DNA. Following transfection, cells were cultured for an additional 72 h.
CRL 1474 normal human fibroblast cells were used for antisense oligonucleotide (ASO) experiments. Cells were cultured in Minimum Essential Medium (Life Technologies) supplemented with 15% FBS (Sigma), 2 mM l-glutamine, 1 mM sodium pyruvate, non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2. Cells were seeded in 60 mm plates at a density of 200 000 cells/plate and incubated overnight. The next day, cells were transfected with the appropriate ASO using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Following transfection, cells were cultured for an additional 72 h. ASO1919 was purchased from Integrated DNA Technologies and contains 2′-O-methyl RNA bases with a phosphorothioate backbone. ASO074 and ASOscrambled (SCR) were obtained from Ionis Pharmaceuticals and contain a 2′-O-methoxy-ethyl ribose modification and a phosphorothioate backbone. ASO sequences are listed in Table 1.
Table 1.
Antisense oligonucleotides sequence
| Name | Sequence |
|---|---|
| ASO 1919 | 5′-AGGAGCGGGUGACCAGAU-3′ |
| ASO 074 | 5′-AGTGACCGTGACACTGGA-3′ |
| ASO SCR | 5′-ATGGCTGGCAGAGCCACTCA-3′ |
Plasmids
LMNA-GFP (WT in this study) and C1824U LMNA-GFP (MUT in this study) minigene splicing reporters were constructed as previously described (26,27). A schematic representation of the minigene reporter is shown in Supplementary Figure S1. Although not pictured, dsRed is also included in the minigene construct and is used as a loading control. Systematic mutational analysis was performed using the Q5 Site-Directed Mutagenesis Kit (New England BioLabs) based on the manufacturer's instructions. Primers for site directed mutagenesis (SDM) are listed in Table 2.
Table 2.
Site directed mutagenesis primers
| Name | Forward | Reverse |
|---|---|---|
| 2X NORM | 5′-AGCCCAGGTGAGTGGACCCATC-3′ | 5′-CCTGAGCCGCTGGCAGAT-3′ |
| MUT/2X-ALT | 5′-AACCCAGGTGGGTTGTCTCTGCTTTG-3′ | 5′-CGGGGGCTGGAGTTGCCC-3′ |
| WT/2X-ALT | 5′-ACCCAGGTGGGCTGTCTCTGCTTTTG-3′ | 5′-TCGGGGGCTGGAGTTGCC-3′ |
| SWAP | 5′-AGCCCAGGTGAGTGGACCCATC-3′ | 5′-CCTGAGCCGCTGGCAGAT-3′ |
| WT/NORM-ALT MUT | 5′-CCCAGGTGGGTTGTCTCTGCTTTTG-3′ | 5′-TTCGGGGGCTGGAGTTGC-3′ |
| WT/Alt-SS-Open-1 | 5′-AATCTGGTCAGGGGCTCCTAACTCCTGGGCAACTCCAGC-3′ | 5′-GTCCCCGAGGCTGCCACC-3′ |
| WT/Alt-SS-Open-2 | 5′-TGGCCTAACTCCTGGGCAACTCCAG-3′ | 5′-CCCTGACCAGATTGTCCCCGAGGC-3′ |
| WT/1928–1930 C→U | 5′-AATCTGGTCATTTGCTCCTAACTCCTGGGCAACTCCAGC-3′ | 5′-GTCCCCGAGGCTGCCACC-3′ |
| WT/Norm-SS-Closed-1 | 5′-TCCTGCAGGCAACTCCCTGGTCATC-3′ | 5′-TTTGGAGACAAAAGCAGAG-3′ |
| WT/Norm-SS-Closed-2 | 5′-ACTCACCTGGTCATCGAGGGGTA-3′ | 5′-TGTCTCTGCAGGATTTGGAGACAAAAG-3′ |
| WT/ALT +3G→C | 5′-GAGCCCAGGTCGGCGGACCCA-3′ | 5′-CTGAGCCGCTGGCAGATGC-3′ |
| WT/ALT +3G→A | 5′-GAGCCCAGGTAGGCGGACCCA-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
| WT/ALT +3G→T | 5′-GAGCCCAGGTTGGCGGACCCA-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
| WT/ALT +4G→C | 5′-AGCCCAGGTGCGCGGACCCAT-3′ | 5′-CCTGAGCCGCTGGCAGATG-3′ |
| WT/ALT +4G→A | 5′-AGCCCAGGTGAGCGGACCCAT-3′ | 5′-CCTGAGCCGCTGGCAGATG-3′ |
| WT/ALT +4G→T | 5′-AGCCCAGGTGTGCGGACCCAT-3′ | 5′-CCTGAGCCGCTGGCAGATG-3′ |
| WT/ALT +34GG→AA | 5′-GAGCCCAGGTAAGCGGACCCATC-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
| MUT/ALT −3C→A | 5′-CTCAGGAGCCAAGGTGGGTGG-3′ | 5′-CCGCTGGCAGATGCCTTG-3′ |
| MUT/ALT −3C→T | 5′-CTCAGGAGCCTAGGTGGGTGG-3′ | 5′-CCGCTGGCAGATGCCTTG-3′ |
| MUT/ALT −3C→G | 5′-CTCAGGAGCCGAGGTGGGTGG-3′ | 5′-CCGCTGGCAGATGCCTTG-3′ |
| MUT/ALT −2A→T | 5′-TCAGGAGCCCTGGTGGGTGGA-3′ | 5′-GCCGCTGGCAGATGCCTT-3′ |
| MUT/ALT −2A→C | 5′-TCAGGAGCCCCGGTGGGTGGAC-3′ | 5′-GCCGCTGGCAGATGCCTT-3′ |
| MUT/ALT −2A→G | 5′-TCAGGAGCCCGGGTGGGTGGA-3′ | 5′-GCCGCTGGCAGATGCCTTG-3′ |
| MUT/ALT +3G→C | 5′-GAGCCCAGGTCGGTGGACCCA-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
| MUT/ALT +3G→A | 5′-GAGCCCAGGTAGGTGGACCCA-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
| MUT/ALT +3G→T | 5′-GAGCCCAGGTTGGTGGACCCA-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
| MUT/ALT +4G→C | 5′-AGCCCAGGTGCGTGGACCCATC-3′ | 5′-CCTGAGCCGCTGGCAGAT-3′ |
| MUT/ALT +4G→A | 5′-AGCCCAGGTGAGTGGACCCATC-3′ | 5′-CCTGAGCCGCTGGCAGAT-3′ |
| MUT/ALT +4G→T | 5′-AGCCCAGGTGTGTGGACCCATC-3′ | 5′-CCTGAGCCGCTGGCAGAT-3′ |
| MUT/ALT +34GG→TC | 5′-GAGCCCAGGTTCGTGGACCCATC-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
| MUT/ALT +34GG→CA | 5′-GAGCCCAGGTCAGTGGACCCATC-3′ | 5′-CTGAGCCGCTGGCAGATG-3′ |
The WT/NORM—ALT MUT, WT/1927A→G, WT/1927A→U, WT/Alt-SS-Open-1, WT/1928-1930C→U, WT/Norm-SS-Closed-1, WT/Norm-SS-Closed-2, WT/Norm-SS-Sil, ALT +6A, ALT +6G, WT/ALT +3G→C, WT/ALT +3G→A, WT/ALT +3G→T, WT/ALT +4G→C, WT/ALT +4G→A, WT/ALT +4G→T, and WT/ALT +34GG→AA mutants were all generated from the WT minigene construct. The MUT/1928-1930C→G, 2X-NORM, MUT/2X-ALT, MUT/Alt-SS-Closed, MUT/Norm-SS-Open, MUT/ALT -3C→A, MUT/ALT -3C→T, MUT/ALT -3C→G, MUT/ALT -2A→T, MUT/ALT -2A→C, MUT/ALT -2A→G, MUT/ALT +3G→C, MUT/ALT +3G→A, MUT/ALT +3G→T, MUT/ALT +4G→C, MUT/ALT +4G→T, MUT/ALT +34GG→TC, and MUT/ALT +34GG→CA mutants were all generated from the MUT minigene construct. The MUT/ALT +4G→A, is the same mutation as 2X-NORM. The WT/Alt-SS-Open-2 mutant was generated from the WT/Alt-SS-Open-1 construct. The WT/2X-ALT mutant was generated via two rounds of SDM. In the first round, the MUT/2X-ALT forward and reverse primers were used to make an A→G substitution at position 1972 of the WT construct. In the second round, the end product of the first round was used as the starting material and the WT/2X-ALT forward and reverse primers were used to introduce a T→C substitution at position 1974. The SWAP mutant was generated from the MUT/2X-ALT construct. The MUT/Alt-Closed-Norm-Open mutant was generated from the MUT/Alt-SS-Closed construct.
Secondary structure mapping
Secondary structure models were obtained by SHAPE-MaP structure probing of in vitro transcribed segments of WT and C1824U LMNA RNA. Transcription templates containing five 3′-terminal nt of exon 9, all of exons 10 and 11, and all but 2 nt of intron 11/12 were amplified by PCR from the LMNA-GFP and C1824U LMNA-GFP expression vectors described above. The two 687 nt RNAs were transcribed using a MEGAshortscript T7 transcription kit (Thermo Fisher Scientific), purified using a MEGAclear Transcription Cleanup Kit (Thermo Fisher Scientific), and folded by thermal denaturation at 95°C for 5 min in 10 mM Tris–HCl, pH 8.0, rapid cooling on ice, and equilibration at 37°C for 20 min after supplementing with 80 mM KCl, 5 mM MgCl2. Reactivity of Exon 11 nucleotides to 1-methyl-7-nitroisatoic anhydride (1M7) was assessed by SHAPE-MaP using a variation of the standard protocol (28,29), as follows.
As appropriate, RNA in negative control (−), modification reaction (+) and denatured control (dc) reactions were probed with the 1M7 acylation reagent under standard conditions. In separate reactions, the purified WT(−), WT(+), WT(dc), C1824U(−), C1824U(+), C1824U(dc) RNAs were then reverse transcribed from a primer hybridized to nt 2139–2158 under conditions favoring mutagenesis instead of premature termination of reverse transcription, opposite acylated sites (28,29). Due to the lengths of the RNA segments being analyzed, the Exon 11 sequences were divided into two overlapping zones (Z1: nt 913–1885, Z2: 1832–2047), and the respective zones in each of the six cDNA libraries amplified in separate 20-cycle PCR1 reactions (WT(−), Z1; WT(−), Z2, etc.; 12 reactions total) using appropriate primer sets containing partial Illumina adapter sequences. One-tenth of each PCR1 amplicons library was then reamplified in a 10-cycle PCR2 reaction using indexed primers suitable for library multiplexing and that added the remaining Illumina adapter sequences. PCR2 amplicon libraries were purified from a 2% non-denaturing agarose gel, quantified using a KAPA Library Quantitation Kit, Illumina Platform, and sequenced on an Illumina MiniSeq using a Mid Output Kit (paired end, 2 × 150 nt).
Sequencing output was trimmed using a custom Python script to remove portions of each sequence read containing primer hybridization sites and merge the respective forward and reverse Z1 and Z2 reads into individual FASTQ files (WT(–), WT(+), WT(dc), C1824U(–), C1824U(+) and C1824U(dc)). These files were processed using ShapeMapper software (28,29) to produce 1M7 reactivity profiles that, together with the respective RNA sequences, were inputted into the RNA secondary structure prediction program RNAStructure (30) to generate structure models. Both sequence (.seq) and reactivity profile (.shape) files used for this purpose are provided as Supplementary Data. Images of the RNA structure models of lowest energy were refined using VARNA (31).
RNA isolation and RT-PCR analysis
RNA was isolated from cell culture samples at 72 h post transfection using the RNeasy Plus Micro Kit (Qiagen). 1 μg of each RNA sample was reverse-transcribed to cDNA using the iScript cDNA synthesis kit (Bio-Rad). RT-PCR was performed on the cDNA samples using the Quick-Load Taq 2X master mix (New England BioLabs). The primers used for RT-PCR are provided in Table 3. DsRed expression was used as an internal loading control for the minigene experiments and TATA-binding protein (TBP) expression was used as a loading control for the ASO experiments. The RT-PCR protocol consisted of 30 s at 95°C, 30 cycles of 15 s at 95°C, 20 s at 55°C and 30 s at 68°C, followed by a final extension of 5 min at 68°C. PCR products were resolved on a 2% agarose gel and imaged using a ChemiDoc MP Imaging System (Bio-Rad). Quantification of mRNA isoforms was performed using Image Lab 5.2.1 software (Bio-Rad). Data are displayed as mean expression values ± SEM from at least three independent experiments.
Table 3.
RT-PCR and qPCR primers
| Gene | Forward | Reverse |
|---|---|---|
| LMNA (minigene) | 5′-AAGAAGTGGCCATGCGCAAGC-3′ | 5′-GGTGAACAGCTCCTCGCCCTT-3′ |
| DsRed | 5′-AGGACGGCTGCTTCATCTAC-3′ | 5′-CCCATGGTCTTCTTCTGCAT-3′ |
| LMNA (total endogenous) | 5′-GTGTGGAAGGCACAGAACAC-3′ | 5′-ACATGATGCTGCAGTTCTGG-3′ |
| TBP | 5′-AACAACAGCCTGCCACCTTA-3′ | 5′-GCCATAAGGCATCATTGGAC-3′ |
| qPCR Primers | ||
| LMNA (Δ150 isoform) | 5′-GCGTCAGGAGCCCTGAGC-3′ | 5′-GACGCAGGAAGCCTCCAC-3′ |
| LMNA (WT isoform) | 5′-CTGGTCACCCGCTCCTAC-3′ | 5′-ACATGATGCTGCAGTTCTGG-3′ |
| TBP | 5′-AACAACAGCCTGCCACCTTA-3′ | 5′-GCCATAAGGCATCATTGGAC-3′ |
| DsRed | 5′-AGGACGGCTGCTTCATCTAC-3′ | 5′-CCCATGGTCTTCTTCTGCAT-3′ |
Quantitative PCR analysis
RNA was isolated from cell culture samples at 72 h post transfection using the RNeasy Plus Micro Kit (Qiagen). 1 μg of each RNA sample was reverse-transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR was performed on cDNA samples using the iQ SYBR Green Supermix (Bio-Rad) on a CFX96 Real Time PCR System (Bio-Rad) to measure LMNA and Δ150 expression levels. Primers used for qPCR are provided in Table 3. The qPCR protocol consisted of 3 min at 95°C and 40 cycles of 20 s at 95°C and 30 s at 60°C. Reactions were performed in triplicate. Analysis was performed using the Bio-Rad CFX Maestro Software. LMNA and Δ150 expression levels were normalized to expression of TATA-binding protein (TBP). Data are displayed as mean expression values ± SEM from a single experiment (n = 3).
Splice site strength calculation
The strength of each 5′ splice site sequence was calculated using MaxEntScan: http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html to obtain scores based on the Maximum Entropy Model (MAXENT), Maximum Dependence Decomposition Model (MDD), Markov Model (MM) or Weight Matrix Model (WMM). The splice site score model was calculated using: http://rulai.cshl.edu/new_alt_exon_db2/HTML/score.html.
RESULTS
Sequence-independent preferential use of the distal 5′ SS in LMNA
Exon 11 of LMNA pre-mRNA contains an internal alternative 5′ SS located 150 nt upstream to its normal 5′ SS (Figure 1A). Predominant usage of the normal 5′ SS produces the wild-type LMNA mRNA isoform, whereas in the premature aging disorder HGPS the alternative 5′ SS is strongly activated by a point mutation (C1824T) in position +6 of the alternative 5′ SS (+6C>T) to produce the Δ150 mRNA isoform and consequently the HGPS disease-causing protein progerin (23) (Figure 1A).
Figure 1.
Positional preferences in splice site selection. (A) Schematic representation of the LMNA exon 11 splicing event. Wild-type and mutant (HGPS) splicing events are represented by dotted black lines. Grey box in exon 11 represents the 150 nt removed in the LMNA C1824T mutant. (B) Splice site strength calculation for the normal 5′ SS, unmutated alternative 5′ SS (WT) and the mutated alternative 5′SS (MUT) using the Maximum Entropy Model (MAXENT), Maximum Dependence Decomposition Model (MDD), Markov Model (MM) or Weight Matrix Model (WMM) and Splice site score. (C) Design of mutant minigene constructs to compare the effect of splice site sequences at the proximal and distal locations relative to intron 11/12. Splice sites with the same color have the same sequence. Blue: WT, alternative splice site; yellow: WT, normal splice site; green: mutant, alternative splice site. (D, F) RT-PCR analysis for LMNA/Δ150 expression of each mutant. 293T cells were transfected with 1 μg of the indicated plasmids. RNA was harvested at 72 h post transfection. DsRed expression served as an internal loading control. (E, G) Quantification of mRNA isoforms expressed as relative usage of the normal vs. alternative 5′ SS. Data are expressed as mean ± s.d. from three independent experiments. WT: LMNA wild-type minigene construct; MUT: LMNA C1824U mutant minigene construct; 2X-NORM: minigene construct with normal splice site sequence at both proximal and distal splice site positions; MUT/2X-ALT: minigene construct with C1824U mutant alternative splice site sequence at both proximal and distal splice site positions; WT/2X-ALT: minigene construct with un-mutated alternative splice sequence at both proximal and distal splice positions; SWAP: minigene construct with C1824U mutant alternative splice site sequence at the proximal splice site and normal splice site sequence at the distal splice site; WT/NORM-ALT MUT: minigene construct with C1824U mutant alternative splice site sequence at the proximal splice site and unmutated alternative splice site sequence at the distal splice site; NC: negative control (PCR reaction mix without cDNA template).
Application of several computational tools to calculate splice site strength indicates that the splice site scores for the normal 5′ SS are consistently higher than the scores for the mutated alternative site (24) (Figure 1B). Yet, the mutant alternative site is strongly dominant in HGPS, demonstrating that sequence-based splice site strength alone is not the only parameter driving selection of competing 5′ SS in LMNA exon 11. The discrepancy between splice site strength and its usage prompted us to probe the mechanism for the predominant usage of the mutated and unmutated alternative 5′ SS in LMNA. To do so, we used systematic mutational analysis in a well-established LMNA minigene system which faithfully reflects endogenous LMNA splicing (26,27) consisting of exons 9–11, intron 11/12 and exon 12 (see Supplementary Figure S1; Materials and Methods). Usage of the two competing 5′ SS was measured by RT-PCR with the long amplicon (LMNA) reflecting splicing at the normal 5′ SS and the short amplicon (Δ150) reflecting use of the exonic alternative 5′ SS (Supplementary Figure S1; Materials and Methods). To validate the system and to demonstrate that the unmutated alternative 5′ SS can function as a splice site, we mutated the normal 5′ SS sequence to a non-5′ SS sequence (16). As expected, inactivation of the normal 5′ SS sequence redirects the splicing machinery to the unmutated alternative 5′ SS demonstrating that the unmutated alternative 5′ SS can act as a functional splice site (Supplementary Figure S2).
In addition to splice site sequence, relative position is known to contribute to 5′ SS choice of competing sites (18). To first directly assess the contribution of location to selection of distal (alternative) or proximal (normal) sites, we generated a series of mutants containing normal 5′ SS, mutated (1824T) alternative 5′ SS, or unmutated (1824C) alternative 5′ SS sequences in both the proximal and distal 5′ SS positions (Figure 1C). Additionally, we created a mutant in which the sequence of the normal 5′ SS was swapped with that of the mutated alternative 5′ SS (Figure 1C). We found that when the proximal and distal splice site positions in exon 11 of LMNA RNA have identical sequence compositions, the distal position is always predominantly used as indicated by greater relative abundance of the Δ150 product in all mutants (Figure 1D and E). The preference for alternative 5′ SS usage was even more pronounced for the SWAP mutant, likely because both sequence and position favor its use (Figure 1D and E). To pinpoint the interplay of location and sequence, we mutated the normal (proximal) 5′ SS sequence to have the mutated alternative sequence, making the proximal site weaker compared to the normal 5′ SS sequence (Figure 1C). In this case, the proximal site is still dominant over the distal site, probably due to sequence advantage. However, the distal site usage is higher compared to normal 5′ SS sequence (Figure 1F and G). We conclude that relative positioning of competing 5′ SS is a critical determinant of splice site choice in LMNA exon 11.
SHAPE-MaP analysis of LMNA RNA secondary structure
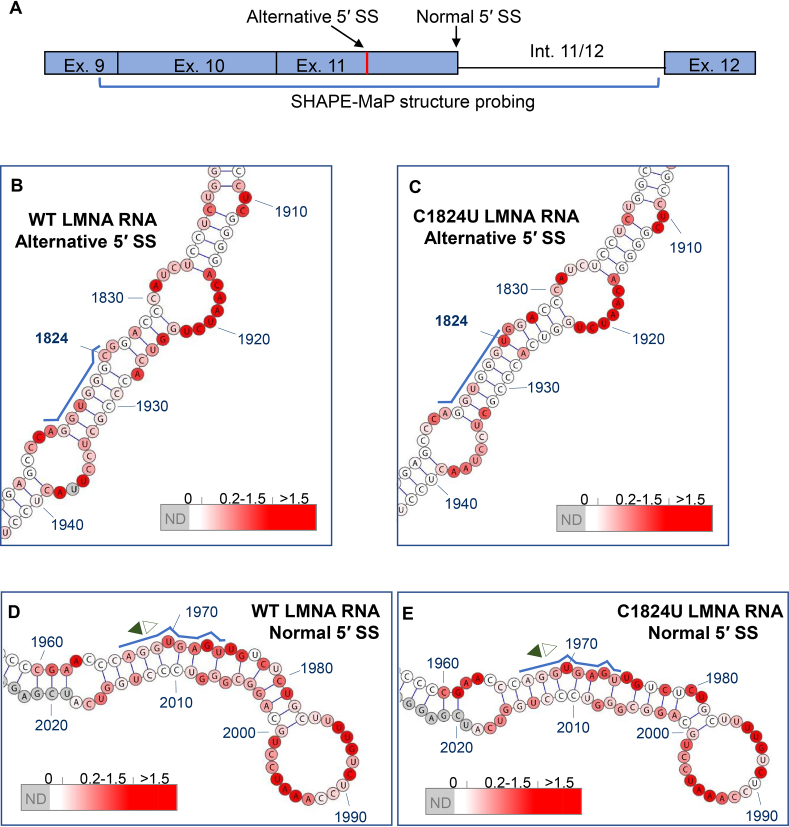
Given the preferential position of the distal site, it is unclear what mechanisms inhibit selection of the unmutated alternative 5′ SS. To investigate a potential role of RNA secondary structure in differential utilization of these two competing 5′ SS, we used SHAPE-MaP to determine the secondary structures of WT and C1824U LMNA RNA constructs. For SHAPE-MaP analysis, transcription templates containing five 3′-terminal nt of exon 9, all of exons 10 and 11, and all but two nt of intron 11/12 were amplified by PCR from the LMNA-GFP and C1824U LMNA-GFP vectors (26) (Figure 2A). The two 687 nt RNAs were in vitro transcribed, purified and folded, and reactivity of Exon 11 nucleotides to 1-methyl-7-nitroisatoic anhydride (1M7) was assessed by SHAPE-MaP using a variation of the standard protocol (28,29) (see Materials and Methods).
Figure 2.
Secondary structures of wild-type (WT) and C1824U mutant LMNA RNA. RNA constructs probed by SHAPE-MaP were 687 nt in length and comprised the five 3′-terminal nt of exon 9, all of exons 10 and 11, and all but two nt of intron 11/12. Ribonucleotides are numbered in accordance with standard numbering of the human LMNA gene, and splice junctions are indicated by opposite-facing pairs of arrows (filled, toward exon; open, toward intron). Nucleotides for which no SHAPE-MaP data was obtained are shaded grey, while a red-to-white color gradient is used to indicate nucleotides that are more or less reactive to the 1M7 probing reagent, respectively. Within the insets, the alternative and normal 5′ SS sequences are highlighted with blue lines. (A) Schematic indicating the SHAPE-MaP structure probing region of LMNA. (B) Sub-motif of WT LMNA RNA containing the alternative 5′ SS. (C) Sub-motif of C1824U mutant LMNA RNA containing the alternative 5′ SS. (D) Sub-motif of WT LMNA RNA containing the normal 5′ SS. (E) Sub-motif of C1824U mutant LMNA RNA containing the normal 5′ SS. Reactivity values obtained for depicted nucleotides in all structure figures are color coded as follows: white (0.0–0.2), white-red linear gradient (0.2–1.5), red (>1.5) and grey (not determined).
SHAPE-MaP analysis demonstrates that the WT LMNA RNA is highly structured and consists of a series of interrupted stem–loop motifs arranged around a six-way junction (Supplementary Figure S3). The first and last of these stem–loop elements, principally comprising exon 10 and intron 11/12, respectively, are branched. A short 5′-to-3′ base pairing interaction is also predicted, suggesting that the fragment may form a discrete, self-contained structure in the context of the larger LMNA RNA (Supplementary Figure S3). The alternative exonic 5′ SS and the normal 5′ SS are contained in adjacent 142 nt and 80 nt hairpins, respectively, each interrupted by a series of bulges and internal loops (Figure 2B and D). The former motif is stabilized by nine consecutive base pairs between the junction and the first internal loop, while the corresponding segment of the latter is destabilized by a bulge and a mismatched pair. Importantly, positions +1 to +6 of the alternative 5′ SS are predicted to be completely base paired, including four G–C pairs, three of which are consecutive (Figure 2B and C). In contrast, although three guanines in the normal 5′ SS are also predicted to pair with cytosines, these G–C pairs are interrupted at position U1970, which represents one of two unpaired nucleotides in the normal 5′ SS core (Figure 2D).
The structure of the C1824U mutant LMNA RNA closely resembles that of the WT LMNA RNA, indicating that the disease-causing mutation does not affect global LMNA RNA folding (compare Figure 2B with C, D with E; Supplementary Figure S3). Base pairing between U1824 and A1927 is predicted in the mutant RNA, although the impact of this on even local duplex stability is likely to be minimal. Imperfect pairing and high reactivity values within the distal regions of the normal 5′ SS hairpin produce slight differences in predicted local structures between the WT and HGPS mutant (compare Figure 2D with E), suggesting that these nucleotides may be single stranded in alternative RNA conformations. These SHAPE-Map data show that the alternative 5′ SS nucleotides are sequestered in base pairing interactions within a long, stable RNA helix and that the normal 5′ SS exists in a less structured environment enabling it to sample alternative base pairing partners, including the U1-snRNA. In addition, the effect of the disease-causing C1824U mutation on global LMNA RNA structure is negligible but the HGPS mutation results in a subtle local alteration of the alternative 5′ SS in exon 11
Local structure around the HGPS point mutation is not sufficient to account for splice site activation
Although the SHAPE-MaP data suggest that the effects of the C1824U mutation on overall LMNA RNA structure are minimal, it is conceivable that the induced structural changes contribute to the mutated alternative 5′ SS selection. To test this possibility, we introduced the more closed structure of the HGPS mutant into the WT background by generating mutants WT/1927A→G and WT/1927A→U, the latter of which re-created base pairing in the WT background of similar strength as in the mutant (Supplementary Figure S4). Neither of these mutants induced usage of the alternative 5′ SS (Supplementary Figure S4). Conversely, introducing the more open structure, as present in the WT, into the region of the alternative 5′ SS in the mutant background (MUT/1928-1930C→G) resulted in a slight increase in Δ150 expression and concomitant decrease in LMNA isoform expression (Supplementary Figure S4). Overall, these results demonstrate that the local structural changes in the HGPS mutant around the point mutation are not sufficient to account for the dramatic increase in usage of the alternative 5′ SS.
Alternative 5′ SS usage is inhibited by base pairing with downstream elements
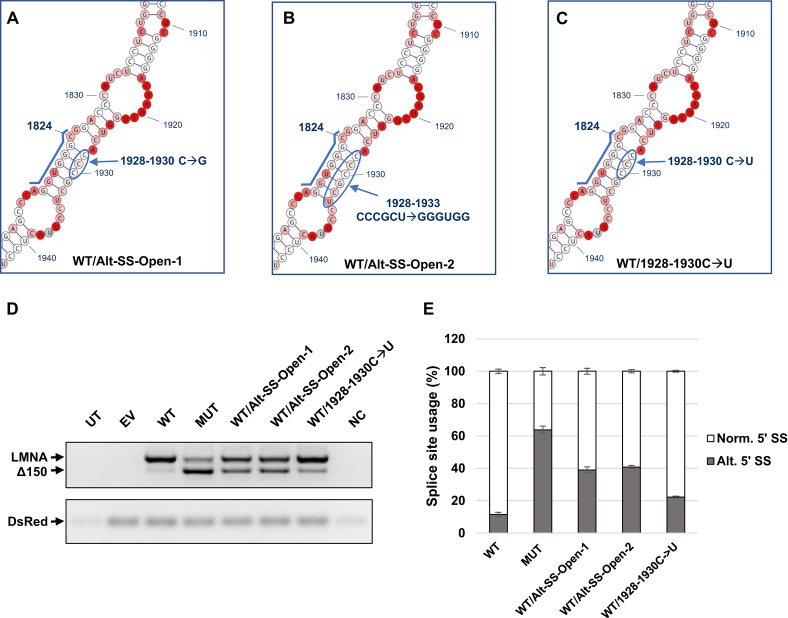
The SHAPE-MaP data indicated that the alternative 5′ SS structure is stable while the normal 5′ SS structure may be more accessible and thus more reactive (Figure 2). We set out to determine whether disrupting the alternative site hairpin in the WT background would increase utilization of this site. Indeed, introducing 1928–1930 triple C→G mutations into the WT background (Figure 3A) resulted in a pronounced increase in the amount of Δ150 product compared to WT (Figure 3D and E), demonstrating that freeing the WT alternative 5′ SS from intrastrand base pairing substantially increases its usage. Disrupting the hairpin even further by mutating nucleotides 1928–1933 (Figure 3B) resulted in an even greater increase in alternative 5′ SS usage relative to WT (Figure 3D and E). This finding is in line with the almost complete suppression of aberrant LMNA RNA splicing observed in healthy individuals unaffected by HGPS (21).
Figure 3.
Splice site inhibition by downstream complementary sequence prevents usage of alternative splice site. (A–C) Design of the WT/Alt-SS-Open-1, WT/Alt-SS-Open-2, and WT/1928-1930C→U mutants generated from the WT minigene construct. The alternative 5′ SS sequence is highlighted by a blue line. (D) RT-PCR analysis for LMNA/Δ150 expression levels of each mutant. 293T cells were transfected with 1 μg of the indicated plasmids. RNA was harvested at 72 hours post transfection. DsRed expression served as an internal loading control. (E) Quantification of mRNA isoforms expressed as relative usage of the normal vs. alternative 5′ SS. Data are expressed as mean ± s.d. from three independent experiments. UT: untransfected; EV: empty vector; WT: LMNA wild-type minigene construct; MUT: LMNA C1824U mutant minigene construct; NC: negative control (PCR reaction mix without cDNA template).
Given these findings, we sought to confirm that the shift in splicing observed upon mutation of this downstream region was due to disruption of structural elements and not due to interference with sequence-based elements such as splicing factor binding sites. A triple C→U mutation (Figure 3C), which maintains the secondary structure at the alternative 5′ SS through wobble base pairing while changing the RNA sequence, resulted in a splicing pattern similar to the WT, with only a slight increase in alternative 5′ SS usage likely due to the weaker bonding strengths of the G-U versus G-C base pairs (Figure 3D and E).
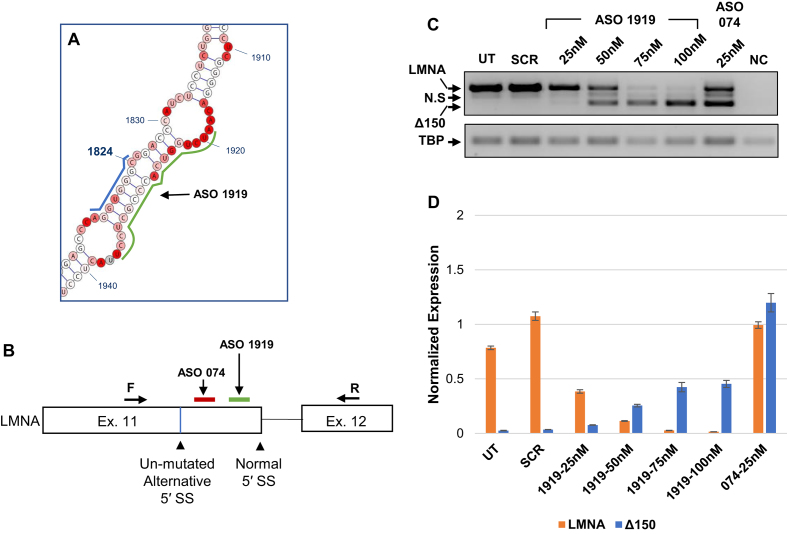
To validate that the effect of structural stability of the alternative 5′ SS observed in the minigene system reflects splicing outcome of endogenous LMNA, we designed an antisense oligonucleotide (ASO1919) to hybridize to nucleotides 1919–1936 in the LMNA transcript and thus prevent intra-strand base pairing with the alternative 5′ SS (Figure 4A and B, green). As a control, we used ASO074 which hybridizes to nucleotides 1851–1868 in LMNA and has previously been shown to increase production of the Δ150 isoform (32) (Figure 4B, red). In CRL 1474 normal human fibroblasts transfected with varying concentrations of ASOs, we observe a dose-dependent shift from predominant expression of the LMNA isoform to the Δ150 isoform at higher concentrations of ASO1919, but not in the presence of the scrambled ASO (Figure 4C and D). Taken together, these results suggest a mechanism by which alternative 5′ SS usage is suppressed by base paring with an RNA element ∼100 nt downstream of the 5′ SS.
Figure 4.
Increased usage of the endogenous alternative splice site in CRL 1474 cells following ASO treatment. (A) Diagram indicating the target location of ASO1919 on the LMNA WT structure model, highlighted by a green line. The alternative 5′ SS sequence is highlighted by a blue line. (B) Diagram indicating the target location of ASO074 (red) and ASO1919 (green) in the context of the splicing cassette. The locations of the forward and reverse primers used to measure endogenous expression of LMNA/Δ150 are indicated. (C) RT-PCR analysis for LMNA/Δ150 expression levels of each treatment group. CRL 1474 cells were transfected with 25–100nM of ASO1919, 25nM of ASO074, or 25nM of scrambled ASOSCR control. RNA was harvested at 72 hours post transfection. TBP expression served as a loading control. (D) Quantitative PCR results for LMNA and Δ150 expression levels for the same samples tested in panel C. Expression levels are normalized to expression of TBP. Data are expressed as mean ± SEM. N.S: nonspecific PCR product. ASO: antisense oligonucleotide; UT: untransfected; SCR: scrambled ASO; NC: negative control (PCR reaction mix without cDNA template).
Crosstalk between the 5′ SS affects splicing outcome
Given our observation of the importance of secondary structure in inhibition of the alternative exonic 5′ SS, we sought to determine whether usage of the normal 5′ SS is similarly affected by structural context. We generated two mutants from the WT construct in which the structural stability of the normal 5′ SS was increased by an intermediate or more substantive degree, respectively, through introduction of complementary base pairs (Figure 5A and B). Both mutants led to increased usage of the alternative 5′ SS while usage of the normal 5′ SS was concomitantly reduced (Figure 5C and D). In addition, stabilization of this region resulted in complete usage of the alternative 5′ SS (see WT/Norm-SS-Closed-2 mutant). Together, these findings support the notion that the structural stability of the normal 5′ SS, like that of the alternative 5′ SS, impacts splicing outcome.
Figure 5.
Increased structural stability around the normal splice site induces usage of the alternative splice site without the C1824U point mutation. (A, B) Design of the WT/Norm-SS-Closed-1 and WT/Norm-SS-Closed-2 mutants, generated from the WT minigene construct. The normal 5′ SS sequence is highlighted by a blue line. Ins. A: Sites at which adenines are inserted. (C) RT-PCR analysis for LMNA/Δ150 expression levels of each mutant. 293T cells were transfected with 1 μg of the indicated plasmids. RNA was harvested at 72 h post transfection. DsRed expression served as an internal loading control. (D) Quantification of mRNA isoforms expressed as relative usage of the normal versus alternative 5′ SS. Data are expressed as mean ± s.d. from three independent experiments. UT: untransfected; EV: empty vector; WT: LMNA wild-type minigene construct; MUT: LMNA C1824U mutant minigene construct; NC: negative control (PCR reaction mix without cDNA template).
The HGPS point mutation confers a dominant effect on 5′ SS choice
In part due to its potential relevance to designing HGPS therapeutics, we next asked whether altering the structural stability of the alternative and normal 5′ SS may re-direct splicing in the MUT minigene construct towards expression of the WT LMNA isoform. To do so, we designed three C1824U mutant variants to either further stabilize the stem containing the alternative site, or alternatively to destabilize the stem containing the normal site or to combine these two mutations (Supplementary Figure S5A and B). In all three mutants, regardless of the structural stability of the available 5′ splice sites, the alternative site is always favored (Supplementary Figure S5C and D). We thus conclude that the HGPS point mutation confers a dominant effect on 5′ SS choice in LMNA.
Alternative 5′ SS selection and U1 snRNP pairing are correlated in LMNA exon 11
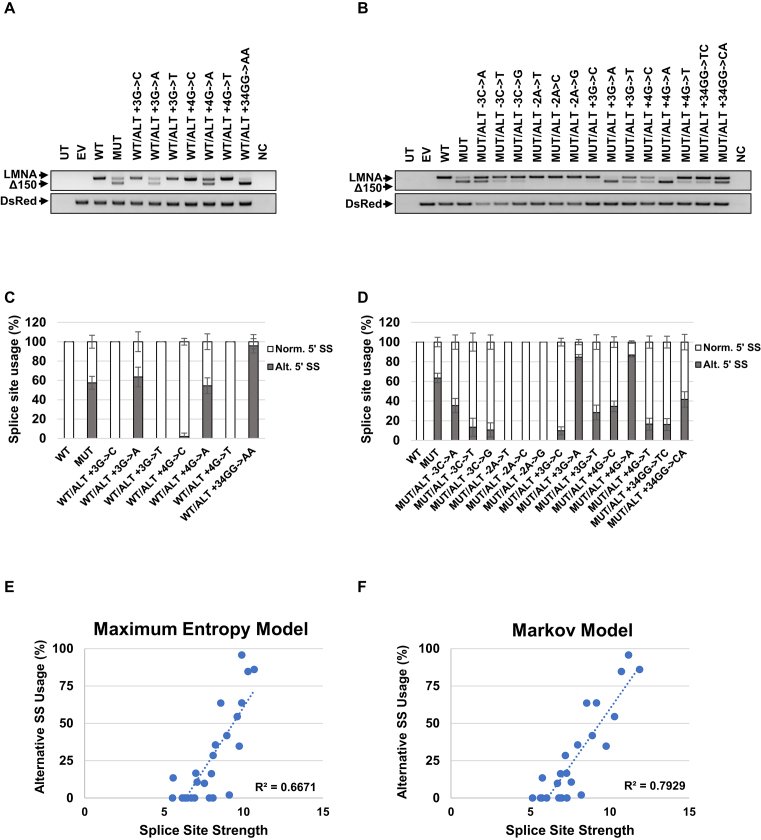
Given the strong effect of the HGPS point mutation on splice site choice, we sought to determine the role of 5′ SS strength in selection of the alternative 5′ SS. We conducted a systematic analysis by mutating the +3, +4 and +6 positions of the alternative 5′ SS and also the –2 and –3 positions in the background of the +6C>T mutation. Mutation of the +6 position into G or A did not induce usage of the alternative splice site (Supplementary Figure S6). In the WT background, G→A mutations at the +3 or +4 positions induced selection of the alternative splice site, similar to the +6 C→T mutation (Figure 6A and C). Notably, changing G→A at both +3 and +4 positions (+34GG→AA) shifted the selection completely towards the alternative splice site (Figure 6A and C). Similarly, in the +6 C→T background, mutations at the +3 or +4 positions induced even greater preference for the alternative splice site. Note that the +3 G→A mutation has the same sequence as 2X-NORM used above (Figure 6B and D; Figure 1).
Figure 6.
Alternative 5′ SS selection and splice site strength are correlated in LMNA exon 11. (A, B) Usage of the alternative 5′ SS measured by RT-PCR for LMNA/Δ150 expression levels of each mutant in the background of (A) WT or (B) the +6C→T mutation. 293T cells were transfected with 1μg of the plasmids. RNA was harvested at 72 h post transfection. DsRed expression served as an internal loading control. (C, D) Quantification of mRNA isoforms expressed as relative usage of the normal versus alternative 5′ SS in in the background of (C) WT or (D) +6C→T mutation. Data are expressed as mean ± s.d. from three independent experiments. (E, F) Graphs represent splice site usage relative to splice site strength. Splice site strength was calculated using (E) Maximum Entropy Model or (F) Markov Model. UT: untransfected; EV: empty vector; WT: LMNA wild-type minigene construct; MUT: LMNA C1824U mutant minigene construct; NC: negative control (PCR reaction mix without cDNA template).
To finally probe the effect of the +6 C→T mutation, we compared the experimentally measured splicing ratio with the strength of the 5′ SS determined using several complementary computational methods (Supplementary Figure S7). Mutations in the +6 positions of the alternative 5′ SS to A (ALT +6A) or G (ALT +6G) are predicted to have similar 5′ SS strength as the WT (ALT +6C) and indeed did not induce selection of the alternative 5′ SS (Supplementary Figure S6). Mutations at position +3 and +4 in the WT background (+6C), on the other hand, are predicted to have high scores and were effective in changing the splicing ratio (Figure 6A, C, E and F). Likewise, mutations in the +6T background displayed a similar trend (Figure 6B, E and F). Maximum Entropy and Markov models (Figure 6E and F) as well as the Maximum Dependence Decomposition and the Weight Matrix Models (supplementary Figure S8A and B) demonstrated correlation between SS strength and selection. Based on these results we conclude that the sequence advantage introduced by the HGPS mutation significantly contributes to splice site selection in LMNA pre-mRNA.
DISCUSSION
Sequence analysis alone frequently fails to accurately predict experimentally determined splicing outcomes and splice site usage (18), strongly suggesting that other factors contribute to splice site choice. One such case is the alternative splicing of the LMNA RNA in the premature aging disorder HGPS. Here, we have used a combination of SHAPE-MaP RNA structural analysis and targeted mutagenesis to investigate the factors that contribute to 5′ SS choice in exon 11 of LMNA. In particular, we sought to probe the role of RNA structure on splice site choice. We identify several contributors to splice site selection in LMNA and our data reveal that 5′ SS selection is governed by an intricate interplay amongst RNA linear position, secondary structure, and primary RNA sequence.
First, we identified relative positioning of competing 5′ SSs in the primary RNA sequence as a key determinant of splice site choice in LMNA. We observed a prominent advantage of the distal 5′ SS over the proximal 5′ SS when the two positions are composed of identical sequences, regardless of the sequence of the splice site. Interestingly, this behavior in LMNA differs from that reported for β-globin where the proximal 5′ SS selection is favored (14–17). A possible explanation for this difference is the uncharacteristic length of LMNA exon 11 (270 nt) in addition to relatively short flanking introns (744 and 322 nt). Usage of the alternative 5′ SS produces an exon of 120 nt which is closer to average exon length and may therefore be favored. Furthermore, it is likely that the effect of positioning of the splice sites is also influenced by the action of local binding proteins. U1 snRNA binding potential has been shown to be a key determinant when competing 5′ SS are in close proximity (33). In LMNA, the relatively large separation in primary sequence between the competing 5′ SS may include exonic elements that can be recognize by members of the SR proteins which are known to interfere with splicing outcome (34). While it is likely that some of the observed effects of RNA elements on splice site choice are mediated by RNA-binding proteins, cursory inspection of CLIP data does not reveal any obvious candidate proteins and full RNA–protein interaction analysis of wild type and mutant LMNA will be required to address this issue.
To probe potential contributions of secondary structure to 5′ SS selection we used SHAPE-MaP analysis. Interestingly, only minor local differences were found between the wild-type and mutant forms of LMNA pre-mRNA which undergo distinct alternative splicing fates and result in dramatic cellular and organismal consequences due to differential 5′ SS usage, suggesting that other factors, such as relative positioning or primary sequence, govern the shift in splicing in mutant LMNA. To probe the accuracy of our mechanistic model, a systemic mutational study was carried out to validate the SHAPE-MaP data. As observed in a previously published structure of LMNA (24), the normal 5′ SS forms an unstable structure and accordingly is likely more accessible. In contrast to the earlier study, we find that the alternative 5′ SS region is highly structured in both unmutated and mutated background and the mutation has neglectable effects on the local structure of the alternative 5′ SS. Most notably, we find that structural features of LMNA RNA identified by SHAPE-MaP are important determinants of 5′ SS selection in vivo. Specifically, we find that a complementary region ∼100 nt downstream of the alternative exonic 5′ SS serves to sterically hinder its accessibility to the splicing machinery. Disruption of base pairing between this region and the alternative 5′ SS facilitates its selection, demonstrating the inhibitory function of the downstream element. This finding is supported by preferential usage of the alternative 5′ SS lacking the HGPS point mutation upon inactivation of the normal 5′ SS. Importantly, we also find that increasing the rigidity of the local structure at the normal 5′ SS diminishes its selection and promotes usage of the competing site. These results demonstrate that secondary RNA structure, in addition to primary sequence, is functionally important for alternative splicing of LMNA. This interpretation is in line with conclusions of previous studies which provide evidence for the influence of secondary structures on 5′ SS suppression of other RNAs, suggesting that this may be an general mechanism of alternative splicing regulation (19,35,36).
The variation between our structural model and a previously published model most likely reflects the fact that the RNA constructs previously probed are markedly shorter than those utilized here (329 nt vs. 687 nt), precluding detection of long-range interactions that we identify and which appear to influence the overall structure of the RNA. Our combined use of SHAPE-MaP and RNAStructure software is also advantageous compared to the earlier RNAse-based mapping methods in that the 1M7 reagent used in SHAPE-MaP can acylate and probe all four nucleotides, eliminating problems with steric clashes that can sometimes alter enzymatic cleavage profiles. In addition, in contrast to nuclease-based mapping, 1M7 reactivity values can be incorporated as soft, pseudo-energy constraints into the RNAStructure folding algorithm, from which unbiased models that account for all of the probing data can be generated (37). The observed close correspondence between structural information derived in an in vitro assay on an RNA fragment and in vivo mutational analysis of a minigene is reassuring and gives high confidence that our conclusions regarding structural features are valid in the context of the intact RNA in living cells. Having said that, confounding effects of co-transcriptional folding and of RNA binding proteins on the RNA structure can not be excluded. In addition, it is also likely that the LMNA RNA exists in multiple co-existing structures in vivo.
We find a strong correlation between the strength of a splice site and its selection when analyzed using several computational approaches to calculate splice site strength. This relationship partially explains the dramatic shift in splicing observed in the C→T mutation found in HGPS. However, while 5′ splice site selection clearly correlates with 5′ splice site strength in the context of single splice sites (18), these rules may not be valid when more than one splice site is present such as in LMNA. Our results illustrate that with the C→T mutation in LMNA not only primary sequence but also relative linear position must be taken into account for splicing outcome.
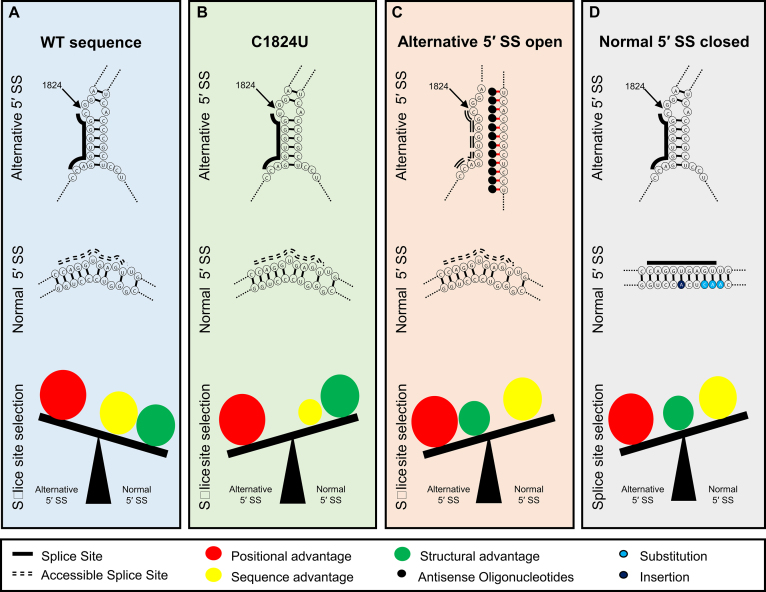
Given the disease relevance of the aberrant splicing event examined here, our findings have implications for HGPS. Based on our observations, it seems likely that a therapeutic strategy aimed at switching 5′ SS selection from the alternative 5′ SS to the normal 5′ SS by stabilizing the alternative splice site and making the normal splice site more open will not be successful due to the strong effect of the alternative 5′ SS location along the transcript, as observed here. Our data indicate that in WT LMNA RNA, the positional advantage of the alternative 5′ SS is not sufficient to outcompete the structural and sequence-based advantages favoring the normal 5′ SS (Figure 7). However, the disease-causing mutation sufficiently enhances the strength of the alternative 5′ SS, so that, combined with its positional advantage, it outweighs the sequence-based advantage of the normal 5′ SS (Figure 7). Experimentally, this effect is mimicked by disrupting the structure by reducing the stability of the alternative 5′ SS or by increasing the stability of the normal 5′ SS, tipping the balance towards usage of the alternative 5′ SS (Figure 7). An RNA-based therapeutic strategy for HGPS should thus focus on effectively blocking the alternative splice site or on promoting degradation of the spliced progerin RNA.
Figure 7.
A model for 5′ SS selection in exon 11 of LMNA. (A) In the wild-type sequence, usage of the alternative 5′ SS in hindered by stable base pairing with the downstream sequence. Thus, it is not as accessible as the normal 5′ SS which forms an unstable structure. While the positional advantage favors the alternative 5′ SS, this advantage is outweighed by the sequence and structure which favor selection of the normal 5′ SS. (B) In the case of the disease-causing C1824U mutation, the sequence advantage for the normal 5′ SS is not as potent as in the wild-type sequence. However, the positional advantage of the alternative 5′ SS shifts the balance to favor its selection. (C) ASO treatment interfering with the inhibitory structural elements of the alternative splice site confers the structural advantage to the alternative 5′ SS. Combined with the positional advantage, the alternative 5′ SS is selected even though the sequence favors the normal 5′ SS. (D) Reducing reactivity of the normal 5′ SS, by making the region more structurally stable, eliminates the structural advantage of the normal 5′ SS. The alternative 5′ SS is selected due to the positional advantage over the sequence advantage of the normal 5′ SS.
In summary, the data provided here offers insight into the mechanisms of alternative 5′ splice site selection of LMNA in the premature aging disorder HGPS. Our results suggest that the preference for the normal splice site in the wild-type gene is largely determined by the sequence composition at the splice sites and structural features of the RNA, while the sequence and the linear position of the splice sites is sufficient to account for the activation of the cryptic splice site. While this study has clarified the influence of factors that are intrinsic to the RNA such as sequence, structure and position in 5′ SS selection of exon 11 of LMNA, there are unanswered questions regarding the transition from 5′ SS recognition to selection and whether a threshold of 5′ SS/U1 binding strength is required for splice site commitment. Furthermore, the influence of other splicing factors and the implications of RNA polymerase II transcription kinetics on this splicing event require further characterization. Regardless, our observations highlight the involvement of multiple factors in 5′ SS selection of a prominent disease-causing splicing event. More broadly, they suggest that consideration of parameters other than sequence, particularly secondary RNA structure, is warranted when predicting 5′ SS usage.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and Center for Cancer Research [TM: 1-ZIA BC010309-19, SLG: 1-ZIA BC010493-15]. Funding for open access charge: National Institutes of Health [1-ZIABC010309-19].
Conflict of interest statement. None declared.
REFERENCES
- 1. Keren H., Lev-Maor G., Ast G.. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010; 11:345–355. [DOI] [PubMed] [Google Scholar]
- 2. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 3. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scotti M.M., Swanson M.S.. RNA mis-splicing in disease. Nat. Rev. Genet. 2016; 17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krawczak M., Thomas N.S., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N.. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007; 28:150–158. [DOI] [PubMed] [Google Scholar]
- 6. Zhuang Y., Weiner A.M.. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986; 46:827–835. [DOI] [PubMed] [Google Scholar]
- 7. Wong M.S., Kinney J.B., Krainer A.R.. Quantitative activity profile and context dependence of all human 5′ splice sites. Mol. Cell. 2018; 71:1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ge H., Manley J.L.. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990; 62:25–34. [DOI] [PubMed] [Google Scholar]
- 9. Ge H., Zuo P., Manley J.L.. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991; 66:373–382. [DOI] [PubMed] [Google Scholar]
- 10. Tarn W.Y., Steitz J.A.. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994; 8:2704–2717. [DOI] [PubMed] [Google Scholar]
- 11. Zahler A.M., Roth M.B.. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:2642–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eperon I.C., Ireland D.C., Smith R.A., Mayeda A., Krainer A.R.. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993; 12:3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han S.P., Tang Y.H., Smith R.. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010; 430:379–392. [DOI] [PubMed] [Google Scholar]
- 14. Reed R., Maniatis T.. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986; 46:681–690. [DOI] [PubMed] [Google Scholar]
- 15. Cunningham S.A., Else A.J., Potter B.V., Eperon I.C.. Influences of separation and adjacent sequences on the use of alternative 5′ splice sites. J. Mol. Biol. 1991; 217:265–281. [DOI] [PubMed] [Google Scholar]
- 16. Yu Y., Maroney P.A., Denker J.A., Zhang X.H., Dybkov O., Luhrmann R., Jankowsky E., Chasin L.A., Nilsen T.W.. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008; 135:1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hicks M.J., Mueller W.F., Shepard P.J., Hertel K.J.. Competing upstream 5′ splice sites enhance the rate of proximal splicing. Mol. Cell Biol. 2010; 30:1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roca X., Krainer A.R., Eperon I.C.. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 2013; 27:129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin Y., Yang Y., Zhang P.. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol. 2011; 8:450–457. [DOI] [PubMed] [Google Scholar]
- 20. Warf M.B., Berglund J.A.. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010; 35:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P. et al.. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003; 423:293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M. et al.. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003; 300:2055. [DOI] [PubMed] [Google Scholar]
- 23. Gordon L.B., Rothman F.G., Lopez-Otin C., Misteli T.. Progeria: a paradigm for translational medicine. Cell. 2014; 156:400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopez-Mejia I.C., Vautrot V., De Toledo M., Behm-Ansmant I., Bourgeois C.F., Navarro C.L., Osorio F.G., Freije J.M., Stevenin J., De Sandre-Giovannoli A. et al.. A conserved splicing mechanism of the LMNA gene controls premature aging. Hum. Mol. Genet. 2011; 20:4540–4555. [DOI] [PubMed] [Google Scholar]
- 25. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 26. Scaffidi P., Misteli T.. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 2005; 11:440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scaffidi P., Misteli T.. Lamin A-dependent nuclear defects in human aging. Science. 2006; 312:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Siegfried N.A., Busan S., Rice G.M., Nelson J.A., Weeks K.M.. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods. 2014; 11:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smola M.J., Rice G.M., Busan S., Siegfried N.A., Weeks K.M.. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 2015; 10:1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu Z.Z., Mathews D.H.. Secondary structure prediction of single sequences using RNAstructure. Methods Mol. Biol. 2016; 1490:15–34. [DOI] [PubMed] [Google Scholar]
- 31. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fong L.G., Vickers T.A., Farber E.A., Choi C., Yun U.J., Hu Y., Yang S.H., Coffinier C., Lee R., Yin L. et al.. Activating the synthesis of progerin, the mutant prelamin A in Hutchinson-Gilford progeria syndrome, with antisense oligonucleotides. Hum. Mol. Genet. 2009; 18:2462–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roca X., Sachidanandam R., Krainer A.R.. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005; 11:683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ibrahim E.C., Schaal T.D., Hertel K.J., Reed R., Maniatis T.. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5002–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eperon L.P., Estibeiro J.P., Eperon I.C.. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986; 324:280–282. [DOI] [PubMed] [Google Scholar]
- 36. Donahue C.P., Muratore C., Wu J.Y., Kosik K.S., Wolfe M.S.. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J. Biol. Chem. 2006; 281:23302–23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mathews D.H. RNA secondary structure analysis using RNAstructure. Curr. Protoc. Bioinformatics. 2006; doi:10.1002/0471250953.bi1206s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.