Abstract
Guanine-rich DNA and RNA sequences can fold into higher-order structures known as G-quadruplexes (or G4-DNA and G4-RNA, respectively). The prevalence of the G4 landscapes in the human genome, transcriptome and ncRNAome (non-coding RNA), collectively known as G4ome, is strongly suggestive of biological relevance at multiple levels (gene expression, replication). Small-molecules can be used to track G4s in living cells for the functional characterization of G4s in both normal and disease-associated changes in cell biology. Here, we describe biotinylated biomimetic ligands referred to as BioTASQ and their use as molecular tools that allow for isolating G4s through affinity pull-down protocols. We demonstrate the general applicability of the method by purifying biologically relevant G4s from nucleic acid mixtures in vitro and from human cells through the G4RP-RT-qPCR protocol. Overall, the results presented here represent a step towards the optimization of G4-RNAs identification, a key step in studying G4s in cell biology and human diseases.
INTRODUCTION
Guanine-rich (G-rich) DNA and RNA sequences have recently earned a solid reputation as key genetic regulators, owing to their capacity to fold into higher-order, four-stranded structures known as G-quadruplexes (or G4). (1). Recent advances in massively parallel sequencing (2) have helped gain insights into both the prevalence and functional relevance of G4s in the human genome and transcriptome. Quadruplex-specific small-molecules (or G4-ligands) (3) were pivotal for these next-generation sequencing (NGS) investigations, allowing for promoting and/or stabilizing quadruplex structures, therefore making them detectable during the NGS analyses. Pyridostatin (PDS) has been particularly instrumental: (4) PDS was used in G4-seq to cause polymerase stalling at ligand-stabilized DNA G4 sites which leads to altered nucleobase calling used as a convenient NGS readout (the Phred quality score). (5) This allowed for the identification of over 716 000 potentially DNA quadruplex-forming sequences (QFS) in the human genome. Using G4 ChIP-seq, 10 000 G4 sites were detected, a reduction in number of detected G4s from previous estimate, likely due to the intrinsic topological constraints of the chromatin environment. (6,7) PDS was also used in rG4-seq since PDS-stabilized RNA G4s likewise trigger reverse transcriptase stalling (RTS), leading to the report of a RNA G4 landscape comprising 13 400 QFS (in polyA-enriched RNA, corresponding to 5 800 genes) in the human transcriptome. (8) Chemically modified PDS derivatives were useful as well, allowing for the first isolation of quadruplexes from purified human DNA (biotinylated PDS) (9) and the first visualization of G4s in human cells (alkynylated PDS, known as PDS-α). (10) It is thus indisputable that selective and chemically versatile ligands are invaluable chemical biology tools for providing a deeper understanding of the complex cell biology of DNA/RNA quadruplexes.
Over the past years, template-assembled synthetic G-quartet (TASQ)-based G4 ligands have proven successful in targeting quadruplexes via an actively selective, bioinspired ligand/target association: (11) for instance, twice-as-smart ligands such as PyroTASQ allowed for visualizing G4s in vitro (12) while NaphthoTASQ (or N-TASQ) allowed for tracking both DNA and RNA G4s in fixed cells (13) and RNA G4s in living cells. (14) More recently, we reported on a biotinylated TASQ named BioTASQ (here referred to as BioTASQ v.1) that was used for developing the G4RP-seq protocol (G4-RNA–specific precipitation-sequencing). (15) As further discussed below, G4RP-seq is a NGS technique that can be used for studying RNA G4-landscapes transcriptome-wide, evaluating the quantitative changes upon G4-ligand treatment and uniquely highlighting a strong correlation between high G4RP signals and the number of G4s in RNA, notably long non-coding RNA (lncRNA) (16). Collectively, results obtained with PDS, TASQ and others such as the pyridodicarboxamide (PDC) 360A derivatives (17) (e.g. the fluorescent dye PDC-M-TO, (18) photo-cross-linking PDC-XL (19) and affinity-tagged PDC-biotin (20)) strongly advocate for the strategic relevance of multifunctional G4-ligands, which precisely illustrate the critical importance of G4s in cell biology.
Here, we continued our efforts to develop multifunctional TASQ ligands. We designed a new BioTASQ prototype (referred to as BioTASQ v.2) and performed in-depth study of both BioTASQ probes (v.1 and v.2) as G4-specific affinity reagents. We showed that both BioTASQs allow for capturing G4s in vitro and from human cells extracts, confirming the reliability and versatility of the G4RP protocol. We evaluated their efficiency via two different pull-down protocols, highlighting the need to exercise caution in selecting biotin linkages, whose nature (length and flexibility of the linker) could drastically influence the nucleic acid-interacting properties of the resulting conjugates. Globally, we provide here new insights into the mechanistic basis for the efficiency of biotinylated TASQs as G4-specific baits for the isolation and identification of G4-RNAs in human transcriptome, expand the portfolio of molecular devices to study G4s in vivo and further improve our understandings of the G4ome.
MATERIALS AND METHODS
Two-step pull-down protocol
The in vitro quadruplex capture experiments were performed in 250 μl final volume as follows: BioTASQ (0, 20 and 50 μM) was mixed with 5′-fluorescently labeled Myc oligonucleotide (F-Myc, FAM-d[5′GAG3TG4AG3TG4A2G3′]) (1 μM) in Caco.K buffer, comprised of 10 mM lithium cacodylate buffer (pH 7.2) plus 10 mM KCl/90 mM LiCl for 2 h at 25°C. The mixtures were thus taken up for the first fluorescence analysis (that is, control experiments without beads, which will be used for normalization, see below), being distributed in 3 wells (80 μl each) of a 96-well plate, using a ClarioStar® machine (BMG Labtech) equipped with FAM filters (λex = 492 nm; λem = 516 nm). The streptavidin MagneSphere® beads (Promega) were washed three times with Caco.K buffer: 200 μl of the commercial solution of beads (1 mg.ml−1) were washed three times with 200 μl of Caco.K. After the original solutions being reconstituted (as 250-μl mixtures), 25 μl (corresponding to 10% of the final volume, 25 μg) of MagneSphere® beads were added to the BioTASQ/F-Myc mixtures; after 1-h incubation at 25°C, beads were immobilized (attracted by a magnet) (21) and the supernatant was taken up for the second fluorescence analysis (that is, experiments with beads), after being distributed in three wells (80 μl each) of a 96-well plate. Data analysis: raw FAM emission (FE) values were collected as triplicates, averaged (with SD) and treated as followed: (a) to evaluate the impact of the beads, results obtained with solutions of F-Myc alone and (F-Myc+beads) were compared and showed as normalized FAM emission (NFE) values, using NFE = [FE/FEF-Myc] × 100 (by definition, NFEF-Myc = 100%); (b) to evaluate the BioTASQ efficiency, (F-Myc+BioTASQ) and (F-Myc+BioTASQ+beads) were compared (at both 20 and 50 μM BioTASQ, with both BioTASQ v.1 and v.2) and showed as NFE values, using NFE = [FE/FE(F-Myc+BioTASQ)]x100 (by definition, NFE(F-Myc+BioTASQ) = 100%, for both BioTASQs, at both concentrations). Next, the solid residue was resuspended in 240 μl of TBS 1X buffer, heated for 8 min at 90°C (under gentle stirring 800 rpm), centrifuged for 2 min (8 900 rpm) and the supernatant was taken up for the third fluorescent analysis (i.e. after elution from the beads), after being distributed in three wells (80 μl each) of a 96-well plate. Data analysis: raw FE values were collected as triplicates, averaged (with SD) and treated as followed: results obtained with solutions of (F-Myc+beads) and (F-Myc+beads+BioTASQ) were compared (at both 20 and 50 μM BioTASQ, with both BioTASQ v.1 and v.2) and showed as NFE values, using NFE = [FE/FE(F-Myc+beads)] (by definition, NFE(F-Myc+beads) = 1).
One-step pull-down protocol
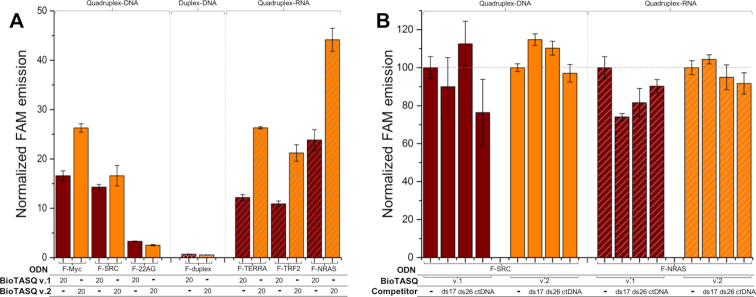
The streptavidin MagneSphere® beads (Promega) were washed three times with Caco.K buffer as indicated above. BioTASQ (20 μM) was mixed with various 5′-labeled oligonucleotides (F-ON, 1 μM, here: F-Myc, F-SRC, F-22AG, F-duplex, F-TERRA, F-TRF2 and F-NRAS) and MagneSphere® beads (25 μg) in Caco.K buffer (250 μl final volume) and stirred for 16 h at 25°C. The beads were immobilized (attracted by a magnet) and the supernatant was removed (21). The solid residue was resuspended in 240 μl of TBS 1X buffer, heated for 8 min at 90°C (under gentle stirring 800 rpm) and then centrifuged for 2 min (8 900 rpm). The supernatant was taken up for analysis (the beads being immobilized by a magnet), after being distributed in 3 wells (80 μl each) of a 96-well plate, using a ClarioStar® machine (BMG Labtech) equipped with FAM filters (λex = 492 nm; λem = 516 nm). Data were analyzed as above: raw FE values were collected as triplicates, averaged (with SD) and treated as followed: results obtained with solutions of (F-ON+beads) and (F-ON+beads+BioTASQ) were compared (with both BioTASQ v.1 and v.2) and showed as NFE values, using NFE = [FE/FE(F-ON+beads)] (by definition, NFE(F-ON+beads) = 1; for sake of clarity, NFE(F-ON+beads) is not shown in Figure 4A). For competitive pull-down experiments: results obtained with solutions of (F-ON+beads+BioTASQ) and (F-ON+beads+BioTASQ+competitor) were compared (with ds17, ds26 and CT-DNA, with both BioTASQ v.1 and v.2) and showed as NFE values, using NFE = [FE/FE(F-ON+beads+BioTASQ)]x100 (by definition, NFE(F-ON+beads+BioTASQ) = 100%, for both F-SRC and F-NRAS, with both BioTASQs).
Figure 4.
(A) One-step pull-down results (FAM intensity of F-ON after release from the beads) for experiments carried out with 1 μM of G4-DNA (F-Myc, F-SRC and F-22AG, plain bars), ds-DNA (F-duplex, cross-hatched bars) or G4-RNA (F-TERRA, F-TRF2 and F-NRAS, diagonally hatched bars) and 20 μM of both BioTASQ (v.1 and v.2, brown and orange bars, respectively). (B) Competitive pull-down experiments carried out with 1 μM of G4-DNA (F-SRC) or G4-RNA (F-NRAS), 20 μM of both BioTASQ (v.1 and v.2) in absence or presence of competitive ds-DNA (either 20 μM of ds17 or ds26, or 100 μM of calf thymus DNA (ctDNA)).
G4RP-RT-qPCR protocol
MCF7 cells were seeded at 3.5 × 105 cells per 10-cm dish before treatment with either vehicle (PBS) or BRACO-19 5 μg/ml (LD15) for 72 h. Cells were then crosslinked using 1% formaldehyde/PBS for 5 min at 25°C and the crosslink was then quenched with 0.125 M glycine for 5 min. Cells were scraped and resuspended in G4RP buffer (150 mM KCl, 25 mM Tris pH7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, RNase inhibitor (Roche), homebrew protease inhibitor cocktail). Cells were then sonicated using Covaris m220 Ultrasonicator using default settings at 10% duty for 2 min. The sonicated fractions were then incubated with 100 μM BioTASQ v.1 or BioTASQ v.2 (or 100 μM biotin for controls) overnight at 4°C. 5% of the sonicate was collected as input control. 10 μg of MagneSphere® beads (Promega) were added and the extract was incubated for 2 h at 4°C. Magnetic beads were then washed four times in G4RP buffer for 5 min. The beads were then incubated at 70°C for 1 h to release captured G4-forming targets from the beads as unfolded sequences (through the reversal of the formaldehyde crosslink) for RT-qPCR quantification purposes. TRIZOL was then used to extract the RNA from the beads using manufacturer's instructions.
The primer sets used for RT-qPCR are:
| mRNA | Forward | Reverse |
|---|---|---|
| VEGFA | 5′CCTTGCCTTGCTGCTCTACC3′ | 5′AGATGTCCACCAGGGTCTCG3′ |
| NRAS | 5′ATGACTGAGTACAAACTGGTGGT3′ | 5′CATGTATTGGTCTCTCATGGCAC3′ |
| HPRT1 | 5′ACCAGTCAACAGGGGACATAA3′ | 5′CTTCGTGGGGTCCTTTTCACC3′ |
| MALAT1 | 5′AAAGCAAGGTCTCCCCACAAG3′ | 5′GGTCTGTGCTAGATCAAAAGGCA3′ |
| XIST | 5′GGTCTGTGCTAGATCAAAAGGCA3′ | 5′AGCTCCTCGGACAGCTGTAA3′ |
| RPPH1 | 5′GAGCTGAGTGCGTCCTGTC3′ | 5′TCAGGGAGAGCCCTGTTAGG3′ |
Extracted RNA was reverse transcribed with Superscript III (Thermos) and random hexamer primers were used according to the manufacturer's protocol to generate cDNA. cDNAs were quantified for target RNAs using 2× SYBR green mix (Bimake) and specific primer sets with three technical replicates in each assay. C(t) values of pull-down samples were normalized to the input control. Three biological replicates were used for all RT-qPCR-based quantifications. Exon-spanning primers for quantifying mRNA levels were derived from Primerbank (see the Supplementary data).
RESULTS
The design of two different BioTASQs
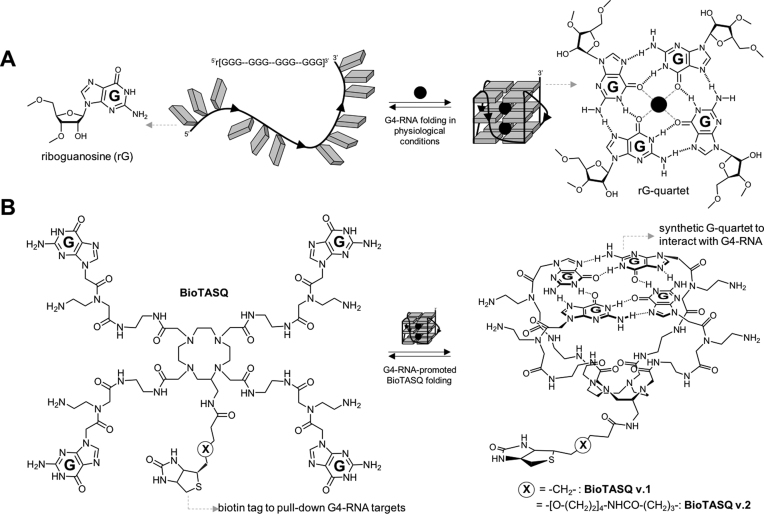
The BioTASQ derivatives belong to the family of biomimetic G4-ligands that uniquely interact with G4s through a biomimetic, like-likes-like recognition between the synthetic G-quartet of the ligand and the native G-quartet of the G4 targets (Figure 1A) (11,12,22). The different prototypes of TASQ studied so far display high affinity for both DNA/RNA G4s and more importantly an excellent selectivity over other nucleic acid structures, making them well-suited to cell-based investigations (13,14,23). We recently studied a biotinylated TASQ named BioTASQ (referred to as BioTASQ v.1 here) that allows for purifying (pull-down protocol) and identifying (G4RP-seq technology) transcriptomic G4-RNA targets from human cells (15). We wondered whether our initial molecular design was optimal, particularly regarding the position of the biotin tag as compared to the template of TASQ, which allows for the intramolecular G-quartet formation. With BioTASQ v.1, we initially selected a short, 7-membered linker (Figure 1B, Supplementary Figures S1, S2), with the hope that it is long enough not to affect or impede TASQ/G4 association, notably upon streptavidin interaction. First in vitro pull-down results showed that BioTASQ v.1 was an efficient affinity reagent, with possible room for improvement. We thus decided to synthesize a novel prototype that exhibits a longer, 23-membered linker (BioTASQ v.2, Figure 1B), with the hope of discarding the possible steric clash that may occur upon streptavidin interaction, which may be responsible for TASQ/G4 dissociation and target release (Supplementary Figure S2). As detailed in the Supporting Information, the synthesis of BioTASQ v.2 was directly inspired by the methodology developed for BioTASQ v.1, which relied on a C-functionalized DOTA derivative we previously described (24), making this chemical access fast and fairly efficient (Supplementary Figures S3–S9).
Figure 1.
(A) Schematic representation of a guanine-rich RNA sequence (guanines as grey squares) under its random-coil (left) and folded, that is, quadruplex structures (G4, right). Detailed structures of guanine (left) and G-quartet (right). (B) Structure of both BioTASQ v.1 and v.2 (left) displaying a biotin affinity tag and schematic representation of its closed, G4-affinic conformation (right), in which the intramolecular G-quartet is formed.
The first, two-step BioTASQ-based G4-affinity pull-down protocol
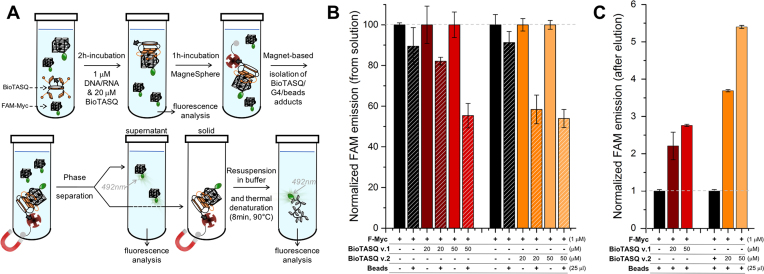
To assess whether BioTASQ v.1 and v.2 efficiently pull-down quadruplexes from solutions in vitro, we employed fluorescein (FAM or F)-labeled nucleic acids given that fluorescence measurements allow for convenient analyses and a high detection sensitivity. The first pull-down protocol was attempted according to published literature (9), which allows for quantifying the capture efficiency by the depletion of FAM fluorescence from the solution (Figure 2). We first used F-Myc (FAM-d[5′GAG3TG4AG3TG4A2G3′]), a well-known G4-forming sequence found in the promoter region of the Myc gene (25). F-Myc (1 μM) was incubated with increasing amounts of BioTASQ (0, 20 and 50 μM) for 2h at 25°C. The fluorescence of these solutions was measured (λex = 492 nm, λem = 516 nm) to be used as controls (vide infra). Then, the streptavidin-coated magnetic beads (25 μg, 10% of the final volume) were added and incubated for 1 h at 25°C. The beads were precipitated, the supernatant collected and subjected to a second fluorescence analysis (Figure 2B): this allowed for quantification of the nucleic acid capture in a differential manner, that is, by comparison with the fluorescence before (controls, fixed at 100%) and after addition of and incubation with the beads (the pull-down experiment per se, whose fluorescence emission is expressed (in %) relative to ad hoc controls). It is worth noting that these results have value insofar as they are compared and normalized to relevant controls, the normalization being performed here in pairs, one for each concentration (0, 20 and 50 μM) and each BioTASQ (v.1 and v.2), obtained in strictly identical experimental conditions (see Materials and Methods). We next quantified the amount of F-Myc immobilized on the beads: to this end, the beads were re-suspended in TBS 1X buffer and the bead/BioTASQ/nucleic acids assemblies dissociated by thermal treatment (8 min at 90°C). After separation from the beads, the fluorescence of the resulting solution was measured to quantify the nucleic acid capture in a direct manner (Figure 2C). Data were compared to control experiments performed without BioTASQ (normalized to 1, see Materials and Methods) to take into account possible unspecific binding of F-Myc on the beads.
Figure 2.
(A) Schematic representation of the in vitro pull-down protocol. (B,C) Results of the two-step pull-down experiments quantified through the fluorescence analysis of the supernatant (B) and of the precipitate after phase separation and release from the beads (C) of experiments carried out with F-Myc (1 μM), in absence (black bars) or presence of BioTASQ (20–50 μM, colored bars).
Our data demonstrated that both BioTASQ efficiently capture G4-DNA from solution, in a concentration-dependent manner. However, the overall efficiency of the process was restricted to 46% at best (Figure 2B: 18–45% and 42–46% decrease of the fluorescence as compared to controls (normalized to 100%) for BioTASQ v.1 and v.2, respectively), in a manner that was not dependent on the used G4-forming oligonucleotides since comparable results were obtained with 2 other G4s (Supplementary Figure S10), i.e. the DNA F-SRC (FAM-d[5′G3AG3AG3CTG53′]) and the RNA F-TERRA (FAM-r[5′AG3(U2AG3)33′]), two sequences found in the SRC gene (10) and telomeric transcripts, (26) respectively. The efficiency of the BioTASQ capture of G4s in solution (up to 45 and 46% for BioTASQ v.1 and v.2, respectively) was not consistent with the excess of ligand as compared to the DNA (20- to 50-fold). This could be attributed to two potential reasons: the non-covalent, that is, reversible TASQ/G4 association that poorly withstands the streptavidin capture step or the disruption of the TASQ/G4 interaction by the introduction of biotin tags. The small difference between BioTASQ v.1 and v.2 efficiencies suggests that it was not due to the aforementioned target release due to streptavidin/biotin association. The results collected after the release of F-Myc from the beads by a short thermal denaturation step (8 min at 90°C, Figure 2C) also provided dose-response results but confirmed the modest efficiency of the overall process. Indeed, as compared to controls (that is, the fluorescence of samples without BioTASQ, normalized to 1), the fluorescence of BioTASQ-enriched fractions increased, but only in a modest manner (2.2–2.8 and 3.7–5.4-fold increase of the fluorescence as compared to controls for BioTASQ v.1 and v.2, respectively). This again pointed to the need for optimizing the pull-down protocol. In particular, we reasoned that the biotin tag may alter the efficiency of the BioTASQs as G4-ligands per se and decided to investigate this through in vitro affinity assays.
Assessing the interaction between BioTASQ and G4s
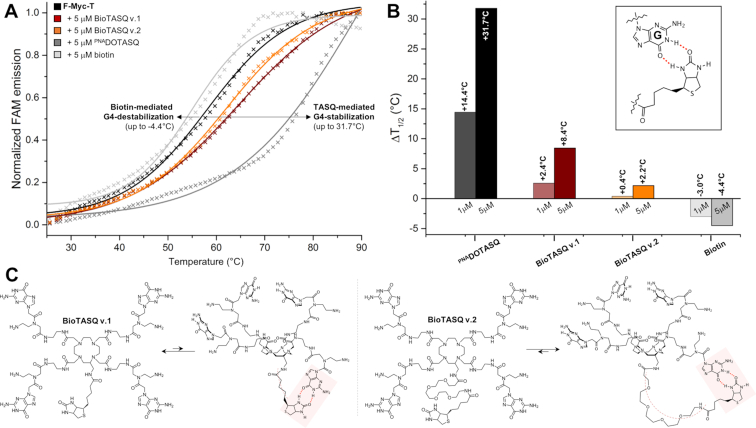
The apparent affinity of each new G4-ligand is quantified via fluorescence resonance energy transfer (FRET)-melting experiments. (27) Both BioTASQs were thus assayed against the dual-labeled F-Myc-T (FAM-d[5′GAG3TG4AG3TG4A2G3′]-TAMRA), in a dose-response manner (experiments were performed with 1 and 5 μM ligand versus 0.2 μM DNA, i.e. 5 and 25 mol. equiv. ligand). Results seen in Figures 3 were surprising: first, the quadruplex-stabilizing capacity of both BioTASQs was strongly decreased as compared to the parent PNADOTASQ (11,28) (ΔT1/2 = 2.4 and 0.4°C for BioTASQ v.1 and v.2, respectively versus 14.4°C for PNADOTASQ at 1 μM ligand concentration); second, the BioTASQ with the longest biotin appendage (BioTASQ v.2) was even less efficient than BioTASQ v.1 (with ΔT1/2 up to 8.4 and 2.2°C for BioTASQ v.1 and v.2, respectively). These results clearly demonstrated that the introduction of the biotin tag negatively impacted the quadruplex-stabilizing properties of BioTASQs, reminiscently of what was observed with PDS, (9) in a manner that was connected with the length and flexibility of the biotin linker (the shorter, the better). To further investigate this, we performed control FRET-melting experiments with biotin (Figures 3). Quite unexpectedly, we found that biotin destabilized F-Myc-T (with ΔT1/2 up to –4.4°C at 5 μM ligand concentration). The origins of this interaction remain at present unclear; we postulate that biotin may interact with guanines through the creation of hydrogen bonds (H-bonds, insert Figure 3B) given that it comprises an ureido ring (fused to a tetrahydrothiophene ring) that is known to target GC base-pair via H-bond-directed G-interaction. (29) In our conditions, the biotin/guanine association took place with quadruplexes when the biotin was used alone (Figure 3A,B) and most certainly occurred intramolecularly within the BioTASQ structure (Figure 3C): this could explain why the ligand with the longer linker (BioTASQ v.2) was less effective in stabilizing quadruplexes than BioTASQ v.1 since the long linker provides more flexibility and thus, makes the intramolecular biotin/guanine association sterically easier and more stable. We thus decided to modify the in vitro pull-down protocol according to this new data.
Figure 3.
(A) FRET-melting results obtained with F-myc-T (0.2 μM, black line) and ligands (5 μM), either BioTASQ v.1 (brown line), BioTASQ v.2 (orange line), PNADOTASQ (dark grey line) or biotin (grey line). (B) Summary of ΔT1/2 values obtained with F-myc-T (0.2 μM) and increasing amounts of ligands (1 and 5 μM), either BioTASQ v.1, BioTASQ v.2, PNADOTASQ or biotin; inset: a schematic representation of the possible biotin/guanine association. (C) Schematic representation of the biotin/guanine interactions that could take place intramolecularly within the structure of BioTASQ v.1 (left panel) or BioTASQ v.2 (right panel).
The second, one-step BioTASQ-based G4-affinity pull-down protocol
We reasoned that a way to improve the efficiency of the pull-down protocol could be to hijack biotin from guanine poisoning: to this end, we decided to directly co-incubate the oligonucleotides, BioTASQ and the magnetic beads in order to favor the streptavidin/biotin association at the very beginning of the experiment, thus making this novel approach a one-step protocol. To assess the validity of this strategy, we used 3 G4-DNAs, which correspond to sequences found in Myc and SRC genes (F-Myc and F-SRC) and in the human telomeric region (F-22AG, FAM-d[5′AG3(T2AG3)33′]), (30) 3 G4-RNAs that correspond to sequences found in the mRNA of TRF2 and NRAS genes (F-TRF2 and F-NRAS, FAM-r[5′CG3AG3CG4AG3C3′] and FAM-r[5′G3AG4CG3UCUG33′], respectively) (31) and in the human telomeric transcripts (F-TERRA), along with 1 duplex used as control (F-duplex, FAM-d[5′(TA)2GC(TA)2T6(TA)2GC(TA)23′]). The labeled oligonucleotides (1 μM) were incubated with both BioTASQ (20 μM) and the streptavidin-coated magnetic beads (25 μg). After an overnight incubation at 25°C, the beads were precipitated, supernatant removed and beads re-suspended in TBS 1X buffer before thermal denaturation (8 min at 90°C). After separation from the beads, fluorescence of the solution was monitored to quantify the nucleic acid capture.
Results seen in Figure 4A highlighted the better overall efficiency of the one-step strategy as compared to the two-step protocol: the FAM fluorescence increased 26-fold (as compared to controls, that is, the fluorescence of samples without BioTASQ treatment, normalized to 1 as above) with G4-DNA (versus 5.4-fold for the two-step protocol, Figure 2) and 44-fold with G4-RNA. Significant discrepancies were nonetheless noticeable: first, the capture of G4-RNA (from 10.9- to 44.1-fold) was more efficient than that of G4-DNA (from 2.5- to 26.3-fold). This could be attributed to the unique topology of the G4-RNA (parallel or type I quadruplex), (32) which makes their external G-quartet more accessible to BioTASQ, that is, more suited to interact efficiently with the sterically demanding TASQ ligand. This assumption was further substantiated by the modest results obtained with the highly polymorphic F-22AG (3.3- and 2.5-fold enhancement with BioTASQ v.1 and v.2, respectively), which was mainly found as a hybrid-type (or type II) quadruplex under the condition of our assay. Second, the efficiency of BioTASQ v.2 (from 2.5- to 44.1-fold enhancement) was better than that of BioTASQ v.1 (from 3.3- to 23.8-fold enhancement), which was not in line with the FRET-melting results (with ΔT1/2 up to 8.4 and 2.2°C for BioTASQ v.1 and v.2, respectively, Figure 3), therefore supporting the relevance of the biotin hijacking strategy. Finally, none of the BioTASQ interacted with F-duplex (0.6- and 0.7-fold enhancement, Figure 4): this indicated that while the introduction of the biotin tag strongly affects the G4-stabilizing properties of the TASQ, it does not alter their specificity toward G4s.
This was further investigated via competitive pull-down experiments, performed with 1 μM labeled G4s (1 G4-DNA, F-SRC and 1 G4-RNA, F-NRAS) in the presence of an excess of unlabeled duplex competitors, including the synthetic duplexes ds17 (d[5′C2AGT2C(GTA)2AC33′]/d[5′G3T(TAC)2GA2CTG23′], 20 μM) and ds26 (the self-complementary sequence d[5′CA2TCG2ATCGA2T2CGATC2GAT2G3′], 20 μM) or DNA extracts (calf thymus DNA, or ctDNA, 100 μM). Results seen in Figure 4B demonstrated that i- both BioTASQs can efficiently capture G4s in the presence of duplex competitors (76–114% efficiency with G4-DNA and 74–104% with G4-RNA as compared to control experiments carried out without competitor), and ii- that BioTASQ v.2 performed better in the duplex competition, with an efficiency comprised between 97–114% with F-SRC and 91–104% with F-NRAS (versus 76–112% with F-SRC and 75–90% with F-NRAS for BioTASQ v.1). Collectively, these results validated the strategic relevance of the one-step pull-down protocol and demonstrated that both BioTASQs are promising molecular tools to purify G4s for cell-based investigations.
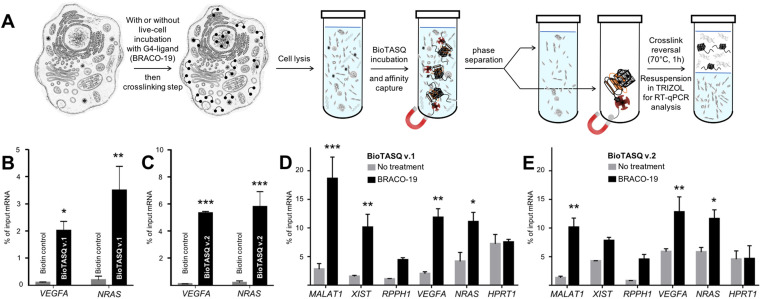
Using both BioTASQs in G4RP-RT-qPCR protocols
We then decided to implement both BioTASQs in the G4RP protocol that allows for both isolating/identifying RNA quadruplexes from human cells and assessing the changes in the G4ome induced by live-cell treatment with G4-ligands. (15) As schematically depicted in Figure 5A, MCF7 human breast cancer cells were first cross-linked with formaldehyde to structurally immobilize transiently folded quadruplexes; harvested cells were then sonicated briefly to release cellular content, and cell lysates were incubated with either BioTASQ v.1 or v.2 overnight before affinity purification with magnetic streptavidin beads. The reverse transcription quantitative polymerase chain reaction (RT-qPCR) method was employed to analyze BioTASQ-enriched fractions, alleviating the need of removing DNA from cell extracts given that RT-qPCR quantifications were done using RNA-specific primers. As seen in Figures 5B,C, results collected through the G4RP-RT-qPCR protocol showed that non-specific binding was negligible (biotin used as control) while demonstrating the enrichment of two firmly established G4-rich mRNAs, i.e. VEGFA (Vascular endothelial growth factor A) (33) and NRAS (neuroblastoma rat sarcoma viral oncogene). (34) Of note, BioTASQ v.2 was again found to be more efficient than BioTASQ v.1 in the G4RP-RT-qPCR protocol (59- and 25-fold improvement as compared to biotin for VEGFA and NRAS, respectively, versus 22- and 16-fold for BioTASQ v.1), in line with its overall better performances in in vitro pull-down assays.
Figure 5.
(A) Schematic representation of the G4RP-RT-qPCR protocol. (B,C) G4RP signals of biotin control versus BioTASQ v.1 (B) and BioTASQ v.2 (C) through RT-qPCR quantification of VEGFA and NRAS mRNA levels in untreated MCF7 cells. Three biological replicates were used for the quantifications. (D,E) G4RP signals obtained with BioTASQ v.1 (D) and BioTASQ v.2 (E) quantified with RT-qPCR measurements of MALAT1, XIST, RPPH1, VEGFA, NRAS and HPRT1 RNA levels in untreated (grey bars) or BRACO-19-treated MCF7 cells (black bars). Values are normalized to their individual input control used for the BioTASQ pull-down. Two-way ANOVA was performed against No Rx control. *P < 0.05, **P < .01, ***P < 0.001. Experiments were conducted with three biological replicates. Error bars represent standard error of the mean (SEM).
Next, we focused on MCF7 cells treated with BRACO-19. (35) We previously demonstrated that treatment with G4-ligands significantly increased G4RP signals of G4-harboring RNAs, (15) most notably of long non-coding RNA (lncRNA) (16,36) such as MALAT1 (metastasis associated lung adenocarcinoma transcript 1), (37) XIST (X-inactive specific transcript) (38) and RPPH1 (ribonuclease P RNA component H1). (39) We thus evaluated here the BRACO-19-induced modifications of G4RP signals using the two BioTASQs. As seen in Figures 5D,E, both BioTASQs can specifically enrich for G4-containing RNA sequences including lncRNA (MALAT1, XIST and RPPH1), mRNA (VEGFA and NRAS), while no changes was observed for HPRT1 (hypoxanthine phosphoribosyltransferase 1), a housekeeping gene transcript that was selected as an unstructured RNA control. Trends were similar in both conditions, given that i- the enrichments obtained with both BioTASQs were higher for transcripts from BRACO-19-treated cells (black bars) than for untreated cells (gray bars), with the notable exception of the control HPRT1 (control); and ii- the order of efficiencies for lncRNA is MALAT1 > XIST > RPPH1 (with 18.6, 10.1 and 4.4% of input RNA for BioTASQ v.1, and 10.1,7.8 and 4.5% for BioTASQ v.2), and for protein coding RNA is VEGFA > NRAS (with 11.8 and 11.1 for BioTASQ v.1, and 12.8 and 11.6% for BioTASQ v.2). Some discrepancies between the two BioTASQs were nonetheless noted: first, the lower overall enrichment of BioTASQ v.1 in untreated cells (up to 3.5% of input RNA) as compared to BioTASQ v.2 (up to 5.5%), which could originate in a better accessibility of the biotin bait once BioTASQ v.2 is embedded in the RNA/protein (RNP) complexes; second, the more sensitive BRACO-19-induced changes with BioTASQ v.1 (up to 9.5-fold enrichment as compared to untreated cells) than with BioTASQ v.2 (up to 4.3-fold enrichment), since G4 ligand-treatments may result in G4 targets being more accessible (thus decreasing the BioTASQ v.1 versus v.2 differences, with the notable exception of MALAT1 here). Altogether, these results highlight that the choice of a suitable BioTASQ molecule for performing G4RP experiments must be done according to the purpose of the experiments, that is, using BioTASQ v.2 for assessing the prevalence of less accessible G4s in cells, and BioTASQ v.1 for evaluating the G4-ligand induced changes in the human transcriptome. They also confirm that both BioTASQs are efficient devices for the isolation and identification of G4-containing RNAs from human cells as well as for gaining precious insights into the G4ome in vivo.
DISCUSSION
The ability of G-rich DNA/RNA sequences to adopt G4 secondary structures in vitro has been thoroughly documented over the past decades through a wide range of biophysical and biochemical techniques (40). To do so in living cells represents a bigger challenge, owing to the many–and yet poorly understood–cellular mechanisms the cell evolved to resolve G4 structures efficiently (41). Small-molecular probes uniquely offer the possibility of interacting with and isolating G4 nucleic acid targets from functional cells and identifying these purified targets via next-generation sequencing technologies (1,7,42). Here, we report on two biotinylated biomimetic quadruplex probes (BioTASQ v.1 and v.2) and their use as affinity reagents to purify quadruplexes in vitro and in vivo through G4-selective precipitation approaches. The efficiency of these new molecular probes was studied through their interactions with different G4-forming sequences (both biologically relevant DNA (Myc, SRC, 22AG) and RNA sequences (TRF2, NRAS, TERRA)), and different pull-down protocols (one- and two-step protocols, in both comparative and competitive manners), optimized to the specifications of biotinylated G4-ligands (in response to the unexpected guanine-interacting properties of the biotin). The demonstration that both BioTASQs are efficient to assess the prevalence of G4-RNA in the human transcriptome (focusing on both coding (VEGFA and NRAS) and non-coding RNAs (MALAT1, XIST and RPPH1)) and that the quantitative changes in their abundance in response to treatments with G4-ligands (here, BRACO-19) can be easily monitored via the G4RP-RT-qPCR technology lends further credence to the use of biotinylated TASQ as highly specific molecular bait for quadruplexes. We anticipate the use of these new-generation molecular tools will contribute to the continued identification of cellular DNA/RNA quadruplexes as key regulators of gene expression in normal and disease-associated changes in cell biology and as promising targets for innovative treatments of genetic dysregulations and associated diseases, i.e. cancers and neurodegenerative diseases.
Supplementary Material
ACKNOWLEDGEMENTS
D.M. is grateful to the Lycée and BTS Le Castel (for A.R.).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche [ANR-17-CE17–0010-01]; European Research Council [H2020-MSCA-IF-2016-750368]; Pharmacoimagerie & agents théranostiques project supported by the Université de Bourgogne and Conseil Régional de Bourgogne (PARI) and the European Union (PO FEDER-FSE Bourgogne 2014/2020 programs). Funding for open access charge: Agence Nationale de la Recherche [ANR-17-CE17–0010-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hänsel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279. [DOI] [PubMed] [Google Scholar]
- 2. Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., Waterston R.H.. DNA sequencing at 40: past, present and future. Nature. 2017; 550:345. [DOI] [PubMed] [Google Scholar]
- 3. Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017; 1:0041. [Google Scholar]
- 4. Rodriguez R., Mueller S., Yeoman J.A., Trentesaux C., Riou J.-F., Balasubramanian S.. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008; 130:15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 6. Hänsel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 7. Hänsel-Hertsch R., Spiegel J., Marsico G., Tannahill D., Balasubramanian S.. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018; 13:551. [DOI] [PubMed] [Google Scholar]
- 8. Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S.. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016; 13:841–844. [DOI] [PubMed] [Google Scholar]
- 9. Mueller S., Kumari S., Rodriguez R., Balasubramanian S.. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2010; 2:1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodriguez R., Miller K.M., Forment J.V., Bradshaw C.R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S.P.. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012; 8:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haudecoeur R., Stefan L., Denat F., Monchaud D.. A Model of Smart G-Quadruplex Ligand. J. Am. Chem. Soc. 2013; 135:550–553. [DOI] [PubMed] [Google Scholar]
- 12. Laguerre A., Stefan L., Larrouy M., Genest D., Novotna J., Pirrotta M., Monchaud D.. A twice-as-smart synthetic G-quartet: PyroTASQ is both a smart quadruplex ligand and a smart probe. J. Am. Chem. Soc. 2014; 136:12406–12414. [DOI] [PubMed] [Google Scholar]
- 13. Laguerre A., Wong J.M.Y., Monchaud D.. Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic mechanism. Sci. Rep. 2016; 6:32141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laguerre A., Hukezalie K., Winckler P., Katranji F., Chanteloup G., Pirrotta M., Perrier-Cornet J.-M., Wong J.M., Monchaud D.. Visualization of RNA-quadruplexes in live cells. J. Am. Chem. Soc. 2015; 137:8521–8525. [DOI] [PubMed] [Google Scholar]
- 15. Yang S.Y., Lejault P, Chevrier S., Boidot R., Robertson A. G., Wong J.M.Y., Monchaud D.. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018; 9:4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qian X., Zhao J., Yeung P. Y., Zhang Q. C., Kwok C. K.. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochim. Sci. 2018; 44:33–52. [DOI] [PubMed] [Google Scholar]
- 17. Pennarun G., Granotier C., Gauthier L.R., Gomez D., Hoffschir F., Mandine E., Riou J.-F., Mergny J.-L., Mailliet P., Boussin F.D.. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005; 24:2917–2928. [DOI] [PubMed] [Google Scholar]
- 18. Yang P., De Cian A., Teulade‐Fichou M.P., Mergny J.L., Monchaud D.. Engineering Bisquinolinium/Thiazole orange conjugates for fluorescent sensing of G‐Quadruplex DNA. Angew. Chem. Int. Ed. 2009; 48:2188–2191. [DOI] [PubMed] [Google Scholar]
- 19. Verga D., Hamon F., Poyer F., Bombard S., Teulade‐Fichou M.P.. Photo‐Cross‐Linking probes for trapping G‐Quadruplex DNA. Angew. Chem. Int. Ed. 2014; 126:1012–1016. [DOI] [PubMed] [Google Scholar]
- 20. de la Faverie A.R., Hamon F., Di Primo C., Largy E., Dausse E., Delaurière L., Landras-Guetta C., Toulmé J.-J., Teulade-Fichou M.-P., Mergny J.-L.. Nucleic acids targeted to drugs: SELEX against a quadruplex ligand. Biochimie. 2011; 93:1357–1367. [DOI] [PubMed] [Google Scholar]
- 21. A very fast (<30 s) centrifugation can be performed to help magnet immobilization. Though this step is not recommended by the MagneSphere® provider, it improves experiment reproducibility. [Google Scholar]
- 22. Stefan L., Guedin A., Amrane S., Smith N., Denat F., Mergny J.-L., Monchaud D.. DOTASQ as a prototype of nature-inspired G-quadruplex ligand. Chem. Commun. 2011; 47:4992–4994. [DOI] [PubMed] [Google Scholar]
- 23. Yang S.Y., Amor S., Laguerre A., Wong J.M., Monchaud D.. Real-time and quantitative fluorescent live-cell imaging with quadruplex-specific red-edge probe (G4-REP). Biochim. Biophys. Acta. 2017; 1861:1312–1320. [DOI] [PubMed] [Google Scholar]
- 24. Laguerre A., Levillain M., Stefan L., Haudecoeur R., Katranji F., Pirrotta M., Monchaud D.. Synthetic G-Quartets as versatile nanotools for the luminescent detection of G-Quadruplexes. Chimia. 2015; 69:530–536. [DOI] [PubMed] [Google Scholar]
- 25. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Azzalin C.M., Lingner J.. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015; 25:29–36. [DOI] [PubMed] [Google Scholar]
- 27. De Rache A., Mergny J.-L.. Assessment of selectivity of G-quadruplex ligands via an optimised FRET melting assay. Biochimie. 2015; 115:194–202. [DOI] [PubMed] [Google Scholar]
- 28. Haudecoeur R., Stefan L., Monchaud D.. Multitasking Water-Soluble synthetic G-Quartets: From preferential RNA-Quadruplex interaction to biocatalytic activity. Chem. Eur. J. 2013; 19:12739–12747. [DOI] [PubMed] [Google Scholar]
- 29. Malnuit V., Duca M., Benhida R.. Targeting DNA base pair mismatch with artificial nucleobases. Advances and perspectives in triple helix strategy. Org. Biomol. Chem. 2011; 9:326–336. [DOI] [PubMed] [Google Scholar]
- 30. Xu Y. Chemistry in human telomere biology: structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011; 40:2719–2740. [DOI] [PubMed] [Google Scholar]
- 31. Bugaut A., Balasubramanian S.. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012; 40:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karsisiotis A.I., Hessari N.M.A., Novellino E., Spada G.P., Randazzo A., da Silva M.W.. Topological characterization of nucleic acid G-Quadruplexes by UV absorption and circular dichroism. Angew. Chem., Int. Ed. 2011; 50:10645–10648. [DOI] [PubMed] [Google Scholar]
- 33. Ferrara N. Vascular endothelial growth factor. Eur. J. Cancer. 1996; 32:2413–2422. [DOI] [PubMed] [Google Scholar]
- 34. Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003; 3:11. [DOI] [PubMed] [Google Scholar]
- 35. Moore M.J.B., Schultes C.M., Cuesta J., Cuenca F., Gunaratnam M., Tanious F.A., Wilson W.D., Neidle S.. Trisubstituted acridines as G-quadruplex telomere targeting agents. Effects of extensions of the 3,6-and 9-side chains on quadruplex binding, telomerase activity, and cell proliferation. J. Med. Chem. 2006; 49:582–599. [DOI] [PubMed] [Google Scholar]
- 36. Morris K.V., Mattick J.S.. The rise of regulatory RNA. Nat. Rev. Genet. 2014; 15:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma K.-X., Wang H.-J., Li X.-R., Li T., Su G., Yang P., Wu J.-W.. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumor Biol. 2015; 36:3355–3359. [DOI] [PubMed] [Google Scholar]
- 38. Gilbert S.L., Pehrson J.R., Sharp P.A.. XIST RNA associates with specific regions of the inactive X chromatin. J. Biol. Chem. 2000; 275:36491–36494. [DOI] [PubMed] [Google Scholar]
- 39. Cai Y., Sun Z., Jia H., Luo H., Ye X., Wu Q., Xiong Y., Zhang W., Wan J.. Rpph1 upregulates CDC42 expression and promotes hippocampal neuron dendritic spine formation by competing with miR-330-5p. Front. Mol. Neurosci. 2017; 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murat P., Singh Y., Defrancq E.. Methods for investigating G-quadruplex DNA/ligand interactions. Chem. Soc. Rev. 2011; 40:5293–5307. [DOI] [PubMed] [Google Scholar]
- 41. Tian T., Chen Y.-Q., Wang S.-R., Zhou X.. G-Quadruplex: a regulator of gene expression and its chemical targeting. Chem. 2018; 4:1314–1344. [Google Scholar]
- 42. Kwok C.K., Marsico G., Balasubramanian S.. Detecting RNA G-Quadruplexes (rG4s) in the Transcriptome. Cold Spring Harbor Perspect. Biol. 2018; 10:a032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.