Abstract
Association of the initiation factor eIF4E with the mRNA cap structure is a key step for translation. Trypanosomatids present six eIF4E homologues, showing a low conservation and also differing significantly from the IF4Es of multicellular eukaryotes. On the mRNA side, while in most eukaryotes the mRNA contains cap-0 (7-methyl-GTP), the trypanosomatid mRNA features a cap-4, which is formed by a cap-0, followed by the AACU sequence containing 2′-O-ribose methylations and base methylations on nucleotides 1 and 4. The studies on eIF4E-cap-4 interaction have been hindered by the difficulty to synthesize this rather elaborated cap-4 sequence. To overcome this problem, we applied a liquid-phase oligonucleotide synthesis strategy and describe for the first time the crystal structure of a trypanosomatid eIF4E (T. cruzi EIF4E5) in complex with cap-4.
The TcEIF4E5-cap-4 structure allowed a detailed description of the binding mechanism, revealing the interaction mode for the AACU sequence, with the bases packed in a parallel stacking conformation and involved, together with the methyl groups, in hydrophobic contacts with the protein. This binding mechanism evidences a distinct cap interaction mode in comparison with previously described eIF4E structures and may account for the difference of TcEIF4E5-cap-4 dissociation constant in comparison with other eIF4E homologues.
INTRODUCTION
Translation initiation in eukaryotes is a complex and highly regulated process dependent on the action of trans acting factors to assemble the large (60S) and small (40S) ribosomal subunits with an initiator methionyl-tRNA positioned over the start codon of the mRNA. Initiation begins with mRNA recruitment, through the association of the so-called eIF4F complex with the cap structure at the 5′ end and binding of the poly(A)-binding protein (PABP) to the 3′ end of the mRNA. The eIF4F complex comprises a scaffold and regulatory subunit, eIF4G, which interacts with the cap-binding factor eIF4E and the RNA helicase eIF4A. The eIF4G subunit interacts also with PABP forming the activated mRNP complex (reviewed in 1). Thus, the mRNA cap recognition by eIF4E is a key step for translation and any dysregulation of cap-binding proteins leads to numerous physiological consequences. Due to its critical role in cancer cell proliferation, eIF4E has also been pointed out as a potential therapeutic target (reviewed in 2).
The first three-dimensional structure described for the eukaryotic translation initiation factor eIF4E (3) revealed a curved-shape α/β protein with the cap binding site located on the concave basal surface of the protein, and the nucleotide base sandwiched between two tryptophan residues. In the past years, X-ray crystal structures and NMR solution structures of eIF4E from several organisms, including human, were determined in complex with cap analogues, peptides derived from eIF4G as well as the inhibitory protein 4E-BP. These structural studies, together with functional assays, provided details on the cap binding mechanism (4–6), on the interaction of eIF4G and 4E-BP at the convex side of eIF4E and on the motifs modulating these interactions to regulate translation initiation (7–11).
In addition to the canonical eukaryotic translation initiation factor 4E (also called eIF4E1), two other classes of eIF4E have been identified and were then called eIF4E2, or 4E homologous proteins (4EHP), and eIF4E3 (12,13). Both are characterized by substitutions of conserved tryptophan residues, including one of the tryptophans involved in the m7G base sandwich responsible for cap binding. Structural and functional characterization of human eIF4E2 (14) and mouse eIF4E3 (15) showed that, despite residue substitutions, the cap binding pocket is preserved, and that its binding activity is functionally relevant. However, in comparison with the canonical eIF4E, eIF4Es from classes 2 and 3 both feature a much lower affinity for m7GTP cap. Accordingly, class 2 and 3 members of eIF4E have been implicated in distinct physiological roles, such as translation of specific mRNA subsets or translational repression (reviewed in 16,17).
Kinetoplastida, a class of early-diverging eukaryotes, which includes pathogenic Trypanosoma and Leishmania species, display key differences in their translation machinery and interaction patterns. The first major difference involves the mRNA 5′-cap structure. While in most eukaryotes the mRNAs feature a cap-0 component, the trypanosomatid mRNAs have a cap-4. This cap-4 is composed of cap-0, followed by a hypermodified AACU sequence with 2′-O-ribose methylations on all residues, and a base methylation on nucleotides 1 and 4 (18). Moreover, trypanosomatid genomes encode for six divergent homologues of eIF4E (EIF4E1 to EIF4E6), with a low degree of conservation among themselves (sequence identity below 22%, except for EIF4E5 and EIF4E6, with ∼30% of sequence identity), and significantly different from their counterparts from multicellular eukaryotes. In addition, five homologues of eIF4G and two or three of PABP are found in Trypanosoma and Leishmania species. Interaction between these homologues to form eIF4F-like and activated mRNP complexes has been shown to be species-specific (reviewed in 19,20).
It is reasonable to assume that these divergent trypanosomatid EIF4E homologues have undergone structural changes to enable them to interact with the unusual cap-4. Phylogenetic analysis revealed that Trypanosomatids EIF4E cluster in three groups, E1–E2, E3–E4, E5–E6, none of them belonging to the metazoan classes 1, 2 and 3 of eIF4E proteins (21,22). Studies of trypanosomatid EIF4Es have mostly focused on Trypanosoma brucei and Leishmania species. EIF4E3 and EIF4E4, which were described as the most abundant, are essential for cell survival and can form canonical eIF4F complexes implicated in translation (23,24). On the other hand, although limited functional data are available for EIF4E from groups 1 and 3, studies have shown that these are unlikely to be involved in general translation (reviewed in 20). Group 3 members are not essential in T. brucei. However, knockdown of TbEIF4E5 and TbEIF4E6 caused, respectively, loss of cell motility and defects in motility and instability of flagellar attachment (25,26). TbEIF4E5 was found in two distinct complexes. One including TbEIF4G1, a 47.5-kDa protein containing conserved RNA-binding domains and either the regulatory protein 14–3–3 II or a 117.5-kDa protein with guanylyltransferase and methyltransferase domains. The second complex was formed by TbEIF4E5, TbEIF4G2, a 17.9-kDa hypothetical protein and both 14–3–3 variants I and II (25). TbEIF4E6, was found in a complex with the TbEIF4G5 homologue and a hypothetical protein of 70.3 kDa, referred to as TbG5-IP, which contains a P-loop NTPase domain (26). These phenotypes and interaction patterns suggest possible roles for these factors in specific cellular pathways.
In vitro interaction assays have shown that Leishmania and T. brucei EIF4E homologues have different affinities for cap-4 (25–27). Again, these differences are probably related to their function either in translation or in regulatory processes. Moreover, such differences in affinity, suggest a distinct binding mode to the mRNA cap-4 for each of these EIF4E homologues. Further information on trypanosomatid EIF4E-cap-4 interactions are dependent on structure determination of EIF4E-cap-4 complexes. So far, the three-dimensional structure of L. major EIF4E1 has been the only reported for a trypanosmatid EIF4E factor and it was described in its apo form (28). This may be partly explained by the fact that trypanosomatid EIF4E homologues are not easily amenable to structural studies. To overcome this problem, we screened T. cruzi EIF4E homologues for crystallization and obtained useful crystals for TbEIF4E5. Equally important, the unavailability of mRNA caps in milligram amount greatly hampers any biological and structural studies of these cap-binding proteins. Accordingly, with the key help of a large-scale chemical synthesis, we could prepare a cap-1 dinucleoside derivative in milligram amount as well as the hypermodified cap-4 in similar amount. With these cap derivatives, we have then obtained the crystal structures of T. cruzi EIF4E5 in complex with m7GTP, cap-1 and cap-4. These structures provide unprecedented details on the interaction of an EIF4E homologue with the mRNA cap-4, specific to trypanosomatids. Their analysis combined with biophysical assays revealed significant differences in comparison with IF4E homologues of other eukaryotes. The unique features mediating T. cruzi EIF4E5-cap-4 interaction thus provide a molecular basis for its distinct mRNA affinity, which may account for the different functions of trypanosomatid homologues in translation mechanisms.
MATERIALS AND METHODS
Chemical synthesis of cap-1 and cap-4
General methods
Chemicals and solvents were purchased from Acros Organics or Sigma-Aldrich and were used without further purification. Nucleosides were purchased from Carbosynth. Standard RNA phosphoramidites and reagents for RNA synthesis were purchased from Sigma-Aldrich or Glen Research. Chemical reactions were monitored by thin layer chromatography (TLC) on pre-coated Merck silica gel plates (60 F254/0.2 mm), by LC-MS (Agilent 1260 series, Quadrupole 6130) or by HPLC (Agilent 1200 series) using a C18 reverse phase column. Purification by flash chromatography was performed using Merck silica gel 60 (0.063–0.200 mm). Analytical HPLC was carried out on an Agilent 1200 series system equipped with a diode-array detector using a C18 reverse phase column (Kromasil, 5 μm, 100 Å, 4.6 × 150 mm) at a flow rate of 1 ml.min−1 and a linear gradient of acetonitrile in 10 mM triethylammonium acetate buffer (TEAA) over 20 min. Purification by HPLC was carried out on an Agilent 1100 Series system using a C18 reverse phase column (Kromasil, 5 μm, 100 Å, 10 × 250 mm) at a flow rate of 4.0 ml min−1, and an isocratic or linear gradient of acetonitrile in 10 mM TEAA over 20 min. Yields refer to chromatographically and spectroscopically pure (>95%) compounds. 1H NMR, 13C NMR and 31P NMR spectra were recorded on a Bruker Advance 400 system operating at 400.13, 100.62 and 168.98 MHz, respectively. Chemical shifts are given in ppm (δ) relative to the residual solvent peak, coupling constants (J) are reported in Hertz and standard abbreviations are used. Assignment of 1H and 13C signals was performed by analysis of the correlated homonuclear 1H,1H-COSY and heteronuclear 1H,13C-HMBC, 1H,13C-HSQC spectra. High-resolution mass spectra were recorded on a Waters Q-TOF micro MS instrument under electrospray ionization in positive ionization mode using a mobile phase of acetonitrile/water with 0.1% formic acid, or negative ionization mode using a mobile phase of acetonitrile/water with 10 mM ammonium acetate. MALDI-MS analysis was performed on a Bruker UltrafleXtreme MALDI TOF/TOF in positive mode. Samples were dissolved in pure dichloromethane (1 mg/ml). An aliquot (1 μl) was deposited onto the Maldi plate and the matrix solution (1 μl, 10 mg/ml of 2,5-dihydroxybenzoic acid in 1:1 acetonitrile/water + 0.1% TFA) was immediately added. All mass spectra were externally calibrated from 700 to 3500 Da using the peptide calibration standard from Bruker Starter kit.
General procedures for RNA assembly
Coupling and oxidation reaction
A solution of benzylthio-1H-tetrazole (BMT) (12.5 eq) in a mixture of dry acetonitrile (25 ml, 0.5 mmol) and dry dichloromethane (37.5 ml, 0.5 mmol) was added dropwise during 20 min to a premixed solution of amidite monomer (2.5–3.5 eq) and RNAx-ACSS (0.5 mmol) in dry dichloromethane (37.5 ml) containing 4 Å molecular sieves (0.2 g) kept at 35°C. Reaction completion was confirmed by monitoring substrate consumption using TLC. After stirring for 1 h at room temperature, 2-butanone peroxide solution (BPO, 50 ml of 0.67% Luperox® in dichloromethane) was added dropwise, and the mixture was stirred at room temperature for 5 min. The resulting solution was diluted with methanol and concentrated under vacuum. After addition of methanol and filtration, the product DMT-RNAx+1-ACSS was obtained as a white solid.
Deprotection
To DMT-RNAx+1-ACSS in dichloromethane (53 ml, 0.5 mmol) was added 5% DCA in dichloromethane (17 ml). After stirring for 15 min at room temperature, the reaction mixture was diluted with methanol (38 ml) and concentrated under vacuum. The residue was taken up in methanol, and the precipitate was filtrated off, rinsed with methanol and dried to give RNAx+1-ACSS as a white solid.
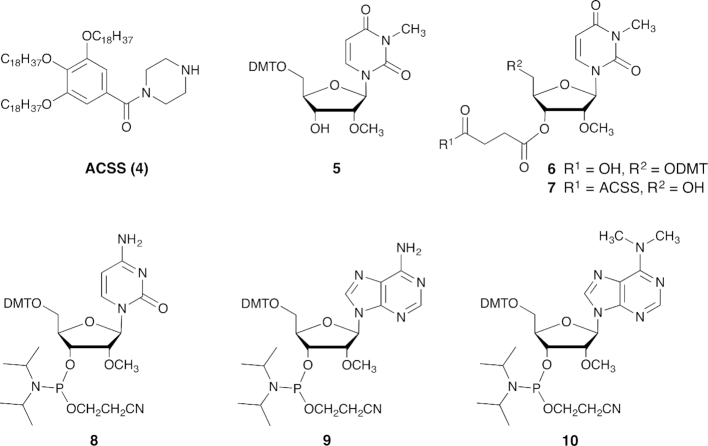
5′-O-Dimethoxytrityl-N3,2′-O-dimethyluridine (5)
Compound 5 was prepared in two steps in 71% overall yield from commercially available 2′-O-methyluridine by N-3 methylation with methyl iodide (29) followed by 5′-O-dimethoxytritylation. 1H and 13C NMR spectra are in accordance with the literature (30).
5′-O-Dimethoxytrityl-3′-O-succinyl-N3,2′-O-dimethyluridine (6)
A mixture of compound 5 (0.50 g, 0.87 mmol), anhydrous succinic anhydride (0.13 g, 1.31 mmol), 4-dimethylaminopyridine (0.05 g, 0.44 mmol) and triethylamine (0.12 ml, 0.87 mmol) in dichloromethane (5 ml) was stirred for 2 h at room temperature. After addition of dichloromethane, the organic layer was washed with 2% citric acid, water and then dried with anhydrous sodium sulfate and concentrated under vacuum to give compound 6 (0.55 g, 88%). 1H NMR (400 MHz, DMSO-d6): δ 2.46–2.60 (m, 4H, CH2), 3.17 (s, 3H, CH3N), 3.26–3.40 (m, 5H, CH3O, H-5′, H-5″), 3.75 (s, 6H, CH3O), 4.13–4.18 (m, 1H, H-4′), 4.20 (t, J = 4.9 Hz, 1H, H-2′), 5.26 (t, J = 5.6 Hz, 1H, H-3′), 5.56 (d, J = 8.0 Hz, 1H, H-5), 5.86 (d, J = 4.4 Hz, 1H, H-1′), 6.87–6.96 (m, 4H, H-arom), 7.20–7.28 (m, 5H, H-arom), 7.29–7.40 (m, 4H, H-arom), 7.78 (d, J = 8.0 Hz, 1H, H-6 pyrimidine); 13C NMR (100 MHz, DMSO-d6): δ 27.7, 29.2, 39.4, 39.6, 39.8, 40.0, 40.3, 40.5, 40.7, 55.3, 55.5, 58.7, 62.8, 70.5, 74.0, 80.7, 80.9, 86.6, 88.8, 101.5, 113.8, 127.3, 128.1, 128.4, 130.2, 135.5, 135.7, 139.2, 145.0, 151.1, 158.7, 162.4, 172.0, 173.7; HRMS (ESI-TOF) m/z: calcd for [C36H38N2O11+Na]+ 697.2373, found: 697.2395.
3′-O-[4-oxo-4-(4-(3,4,5-tris(octadecyloxy)benzoyl)piperazin-1-yl)]-succinyl-N3,2′-O-dimethyluridine (7, namely RNA1-ACSS)
To compound 6 (0.74 g, 0.72 mmol) in dichloromethane (5 ml) were added ACSS 4 (0.50 g, 0.74 mmol), O-benzotriazole-N,N,N’,N’-tetramethyl-uronium-hexafluoro-phosphate (HBTU) (0.67 g, 2.39 mmol), 1-hydroxybenzotriazole (HOBT) (0.32 g, 2.13 mmol) and N,N-diisopropylethylamine (DIEA) (0.38 ml, 2.16 mmol). After stirring for 1 h at room temperature, the reaction mixture was diluted with methanol (10 ml), concentrated and the precipitate was filtrated off, rinsed with methanol and dried under vacuum. The precipitate was taken up in dichloromethane (40 ml) and a solution of 5% DCA in dichloromethane (12 ml) was added. After stirring for 1 h, the reaction mixture was diluted with methanol (25 ml) and concentrated under vacuum. The precipitate was filtrated off, rinsed with methanol and dried to give RNA1-ACSS (7) as a white solid (0.61 g, 0.45 mmol, 63%). 1H NMR (400 MHz, CDCl3): δ 0.90 (t, J = 6.7 Hz, 9H, OCH2CH2CH2(CH2)14CH3), 1.19–1.41 (m, 84H, OCH2CH2CH2(CH2)14CH3), 1.42–1.55 (m, 6H, OCH2CH2CH2(CH2)14CH3), 1.72–1.92 (m, 6H, OCH2CH2CH2(CH2)14CH3), 2.6–2.85 (m, 4H, CH2 succinyl), 3.35 (s, 3H, NCH3), 3.48 (s, 3H, OCH3), 3.51–3.77 (m, 8H, CH2 piperazine), 3.78–3.87 (m, 1H, H-5′), 3.92–4.04 (m, 7H, H-5″ and OCH2CH2CH2(CH2)14CH3), 4.22–4.29 (m, 2H, H-2′ and H-4′), 5.31 (t, J = 4.7 Hz, 1H, H-3′), 5.78 (d, J = 4.3 Hz, 1H, H-1′), 5.81 (d, J = 8.2 Hz, 1H, H-5), 6.61 (s, 2H, H arom), 7.71 (d, J = 8.2 Hz, 1H, H-6); 13C NMR (100 MHz, CDCl3): δ 14.0, 22.6, 26.1, 27.6, 28.0, 29.1, 29.3, 29.3, 29.4, 29.5, 29.6, 29.7, 30.3, 31.9, 58.9, 61.4, 69.4, 70.5, 73.5, 76.7, 77.0, 77.2, 77.3, 81.2, 82.9, 90.7, 102.0, 105.9, 129.7, 139.0, 139.7, 151.1, 153.3, 162.6, 169.7, 170.7, 172.7; Maldi-TOF-MS: m/z calcd for [C80H140N4O12+Na]+ 1372.036, found: 1372.079.
RNA2-ACSS
Coupling of RNA1-ACSS (7) (0.54 g, 0.40 mmol) and 5′-O-dimethoxytrityl-2′-O-methyl-N-acetyl-cytidine-3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (8) (0.80 g, 2.5 eq) following the general procedures afforded RNA2-ACSS (0.64 g, 0.36 mmol, 94%). Maldi-TOF-MS: m/z calcd for [C95H159N8O20P+Na]+ 1786.131, found 1786.142.
RNA3-ACSS
Coupling of RNA2-ACSS (0.64 g, 0.36 mmol) and 5′-O-dimethoxytrityl-2′-O-methyl-N-benzoyl-adenosine-3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (9) (0.80 g, 2.5 eq) following the general procedures afforded RNA3-ACSS (0.73 g, 0.32 mmol, 89%). Maldi-TOF-MS: m/z calcd for [C116H180N14O27P2+Na]+ 2286.251, found 2286.263.
RNA4-ACSS
Coupling of RNA3-ACSS (0.72 g, 0.32 mmol) and 5′-O-dimethoxytrityl-6-N,6-N,2′-O-trimethyl-adenosine-3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (10) (see Supplementary Information) (0.72 g, 2.8 eq) following the general procedures afforded RNA4-ACSS (0.74 g, 0.27 mmol, 84%). Maldi-TOF-MS: m/z calcd for [C132H201N20O33P3+Na]+ 2710.377, found: 2710.402.
DMT-P-RNA4-ACSS
Coupling of RNA4-ACSS (0.31 g, 0.115 mmol) and 2-[2-(4,4′-dimethoxytrityloxy)ethylsulfonyl]ethyl-(2-cyanoethyl)-(N,N-diisopropyl)-phosphorami-dite (3.5 eq) following the general procedure A afforded DMT-P-RNA4-ACSS (0.37 g, 0.114 mmol, 99%). Maldi-TOF-MS: m/z calcd for [C160H231N21O41P4S-DMT+Na]+ 2979.390, found: 2979.701.
5′-Phosphorylated tetramer (2)
Cleavage of RNA from the support and base/phosphate deprotection were performed by treatment of DMT-P-RNA4-ACSS (42.6 mg, 0.015 mmol) with 28% aqueous ammonia in ethanol (1.0 ml, 3:1 v/v) for 90 min at 80°C. After lyophilization, the residue was taken up twice in water, centrifuged and the supernatant was filtered and purified by reverse-phase HPLC (5–20% linear gradient of ACN in 10 mM TEAA buffer over 20 min) to give P-RNA4 (2) as 2.8 equiv triethylammonium salt (12.4 mg, 0.009 mmol, 61%). 1H NMR (400 MHz, D2O): δ 1.26 (t, J = 7.5 Hz, 24H, CH3 NEt3), 3.13–3.36 (m, 26H, OCH3 and CH2 NEt3), 3.49 (s, 3H, OCH3), 3.54 (s, 3H, OCH3), 3.66 (s, 3H, OCH3), 3.70 (s, 3H, OCH3), 3.74–4.92 (m, 20H, H-2′, H-3′, H-4′, H-5′ and H-5″), 5.68 (br s, 1H, H-1′), 5.70 (d, J = 7.7 Hz, 1H, H-5 rU), 5.84 (d, J = 8.1 Hz, 1H, H-5 rC), 5.95–6.00 (m, 2H, H-1′), 6.11 (d, J = 2.3 Hz, 1H, H-1′), 7.69 (d, J = 7.7 Hz, 1H, H-6 rU), 7.83 (d, J = 8.1 Hz, 1H, H-6 rC), 7.89 (s, 1H, H-2 purine), 8.13 (s, 2H, H-8 purine), 8.38 (s, 1H, H-2 purine); 31P NMR (169 MHz, D2O): δ -0.23, -0.10, 0.03, 1.51; Maldi-TOF-MS: m/z calcd for [C45H63N15O28P4+H]+ 1386.299, found: 1386.339.
P 2-Imidazolide-N7-methylguanosine 5′-O-diphosphate (3)
A mixture of triethylammonium salt of 7-methylguanosine 5′-diphosphate (157 mg, 0.115 mmol) [synthesized from GDP by reaction of methyliodide in DMSO according to reported procedures (29), imidazole (155 mg, 2.28 mmol), 2,2′-dithiodipyridine (128 mg, 0.90 mmol), triethylamine (157 μl) and triphenylphosphine (240 mg, 0.92 mmol) in dimethylformamide (1.6 ml) was stirred overnight at room temperature. A solution of sodium perchlorate (100 mg, 0.80 mmol) in anhydrous acetone (4 ml) was then added at 4°C. The precipitate recovered by centrifugation was washed with four portions of cold acetone, and dried to give Im(m7GDP) (9) as sodium salt (50.9 mg, 92 μmol, 80%). NMR spectra are in accordance to literature data (30). HRMS (ESI-TOF): m/z calcd for [C13H20N5O7P-H]− 388.1022, found: 388.1016.
Trypa cap-4 (1)
A mixture of the triethylammonium salt of 2 (12.9 mg, 7.74 μmol), the sodium salt of Im(m7GDP) 3 (27.0 mg, 49 μmol) and anhydrous ZnCl2 (14 mg, 105 μmol) in dimethylformamide (1.2 ml) was stirred overnight at room temperature. The reaction mixture was then poured into 1.8 M aqueous solution of EDTA (10 ml) and pH was adjusted to 6.5 with 1M Na2CO3. Preparative HPLC purification (10–17% linear gradient of acetonitrile in 10 mM TEAA over 14 min) gave trypa cap-4 (1) as triethylammonium salt (5.9 mg, 2.95 μmol, 40%). To tetramer 1 dissolved in water (80 μl) was added slowly at 4°C a cooled solution of sodium perchlorate (40 mg) in anhydrous acetone (800 μl). The precipitate was recovered by centrifugation, washed with four portions of cold acetone and then dried (5.15 mg, 2.66 μmol, 34%). 31P NMR (169 MHz, D2O): δ –0.23, –0.10, 0.03, 1.49; HRMS (ESI-TOF): m/z calcd for [C56H79N20O38 P6–3H]3− 607.0992, found: 607.1012.
Protein Expression and Purification
The nucleotide sequences encoding the full-length and a truncated construct comprising residues 1 to 189 of TcEIF4E5 (EKG01513.1) were synthesized by Biomatik, after codon optimization for Escherichia coli expression system, and cloned into the pET28a vector (Novagen). In this vector, the recombinant proteins are expressed in fusion with a N-terminal His-tag. E. coli BL21 Star (DE3) cells were transformed with the expression vector and incubated at 37°C in LB medium containing the selection antibiotics. When the cultures reached OD600 ∼0.6, the temperature was reduced to 18°C and protein expression was induced with 0.25 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) for ∼16 h. Cells from 1 l culture were harvested by centrifugation at 6000 × g for 15 min, suspended and lysed in 20 ml of buffer A (50 mM Tris–HCl pH 8, 200 mM NaCl, 20 mM imidazole, 1 mM de DTT) using a microfluidizer (Microfluidics™ 110L series 0300).
The soluble fractions were isolated by centrifugation at 40 000 × g for 30 minutes at 4°C. The extract was loaded onto a 1 ml His-Trap HP column (GE Healthcare Life Sciences) equilibrated with buffer A. TcEIF4E5 was eluted with a 10 column volumes (CV) linear gradient from 0 to 15% buffer B (buffer A + 500 mM imidazole), followed by a 15 CV linear gradient from 15 to 100% buffer B. TcEIF4E5-ΔC was eluted with a 10 column volumes (CV) linear gradient from 0 to 10% buffer B, followed by a 15 CV linear gradient from 10 to 100% buffer B. Fractions containing the target protein were pooled, concentrated to a final volume of 2 ml and loaded onto a Superdex 75 16/60 (GE Healthcare Life Sciences) equilibrated with 20 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM DTT. TcEIF4E5 and TcEIF4E5-ΔC purified by this procedure were used in microscale thermophoresis assays.
Purification of TcEIF4E5 for crystallization assays followed a slightly different protocol. pDEST17 expression vector (Invitrogen) containing the gene encoding TcEIF4E5 sequence was kindly provided by the laboratory of Dr. Fabiola Holetz (Instituto Carlos Chagas, FIOCRUZ, Brazil) and used to transform E. coli BL21 (DE3) ΔSlyD_pRARE2. Protein expression and affinity chromatography followed the same procedures described above. After Ni+2 affinity chromatography, the fractions containing the target protein were pooled, dialyzed against 50 mM Tris–HCl pH 8, 20 mM NaCl, 2 mM DTT and loaded onto a 5 ml Hi-Trap SP HP column (GE Healthcare Life Sciences). TcEIF4E5 was eluted with a 20 CV linear gradient from 20 mM to 1 M NaCl. The sample was concentrated to a final volume of 2 ml and loaded onto a Superdex 75 16/60 (GE Healthcare Life Sciences) equilibrated with 20 mM Tris–HCl pH 8, 150 mM NaCl, 2 mM DTT. The eluted protein was concentrated to ∼5 mg/ml for crystallization assays.
Expression of selenomethionine-labeled (SeMet) TcEIF4E5 was performed based on the metabolic inhibition method (31). Briefly, E. coli BL21 Star (DE3) cells transformed with the expression vector were incubated at 37°C, 200 rpm in M9 minimal medium supplemented with the selection antibiotic. When the culture reached OD600 ∼0.6, it was supplemented with 10 ml of a sterile mixture containing lysine, phenylalanine and threonine (10 mg/ml), isoleucine, leucine and valine (5 mg/ml) and selenomethionine (6 mg/ml) diluted into M9 medium without glucose and thiamine. The temperature was reduced to 18°C and after 30 min of incubation, expression was induced with 0.25 mM IPTG for approximately 16 h. Cells were harvested by centrifugation and TcEIF4E5-SeMet was purified using the same protocol applied to non-labelled TcEIF4E5.
Crystallization, data collection and processing
TcEIF4E5 was incubated with m7GTP (Jena Bioscience), cap-1 and cap-4 at a molar ratio of 1:1.5 (protein:ligand). Protein samples in the presence and absence of ligands were submitted to crystallization trials by sitting drop vapor-diffusion method using commercial screens. Initial hits were obtained using TcEIF4E5 after incubation with cap analogues but not in the absence of ligands. Optimization of the initial crystallization conditions was performed by varying precipitant and protein concentrations. Best crystals were obtained by hanging drop vapor diffusion at 18°C by mixing the TcEIF4E5 protein at 5 mg/ml (0.2 mM) in 20 mM Tris–HCl pH 8, 150 mM NaCl, 2 mM DTT with crystallization buffer containing 100 mM BIS–TRIS pH 6 or 6.5, 200 mM Lithium sulfate, 25–30% PEG 3350. TcEIF4E5-SeMet at 5 mg/ml in 20 mM Tris–HCl pH 8, 150 mM NaCl, 2 mM DTT was crystallized in buffer containing 20 mM ammonium sulfate, 10 mM HEPES pH 7.5, 25% PEG 3350.
Before data collection, the crystals were cryo-protected with 20% (v/v) glycerol added to the mother liquor prior to flash cooling in liquid nitrogen. X-ray diffraction data from TcEIF4E5-m7GTP crystals were collected at the PROXIMA 1 beam line of the Synchrotron SOLEIL using a PILATUS 6M detector (Dectris). Diffraction data from TcEIF4E5-cap1 and TcEIF4E5-cap4 crystals were collected at the same beam line using an EIGER X 16M detector (Dectris). X-ray diffraction data from TcEIF4E5-SeMet crystals were collected at the PROXIMA 2 beam line of the Synchrotron SOLEIL using an EIGER X 9M detector (Dectris) at the Se-absorption wedge wavelength (0.97830 Å). Diffraction data were processed with XDS package (32). Data statistics are presented in Table 1 and Supplementary Table S1.
Table 1.
Crystallographic data and refinement statistics
| Crystal | TcIF4E5-m7GTP | TcIF4E5-cap1 | TcIF4E5-cap4 |
|---|---|---|---|
| Data statistics | |||
| Source | SOLElL-PX1 | SOLElL-PX1 | SOLElL-PX1 |
| Space group | P3121 | P3121 | P3121 |
Unit cell ( ) ) |
a = b = 100.72, c = 40.63 | a = b = 102.82 c = 42.50 | a = b = 102.52, c = 41.11 |
Resolution (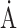 ) ) |
50–2.10 (2.23–2.10) | 50–2.70 (2.86–2.70) | 50–2.20 (2.33–2.20) |
| Number of observations | 204 339 (32 204) | 149 068 (23 611) | 262 446 (41 327) |
| Number of unique reflections | 13 943 (2205) | 7339 (1176) | 12 900 (2040) |
| Completeness (%) | 99.2 (98.2) | 100.0 (99.9) | 100.0 (99.9) |
| Redundancy | 14.6 (14.6) | 20.3 (20.1) | 20.3 (20.3) |
| Rmeas (%) | 6.9 (262.6) | 6.6 (262.6) | 6.4 (204.7) |
| I/σ | 19.9 (1.05) | 24.1 (0.98) | 23.7 (1.01) |
CC 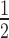
|
100.0 (58.1) | 100.0 (54.4) | 100.0 (58.7) |
| Refinement statistics | |||
| R work/Rfree (%) | 17.8/19.8 | 19.4/25.2 | 20.7/22.6 |
RMSD bond length(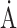 )/angle (°) )/angle (°) |
0.01/1.09 | 0.01/1.19 | 0.01/1.15 |
Mean B value (overall, 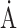 2) 2) |
84.7 | 136.6 | 93.3 |
| Protein atoms | 1414 | 1447 | 1452 |
| Heterogen atoms | 38 | 59 | 130 |
| Number of water molecules | 72 | 23 | 68 |
| Ramachandran plot | |||
| Favored (%) | 98.3 | 96.1 | 98.3 |
| Outliers (%) | 0 | 0 | 0 |
Values in parentheses are for the highest resolution shell.
Structure determination and refinement
The structure of TcEIF4E5 was determined by single-wavelength anomalous diffraction using diffraction data from TcEIF4E5-SeMet. The heavy atoms’ positions were determined using the program SHELXD (33), the initial phases were calculated using PHASER (34) and density modification was performed using PARROT (35). The low quality of the experimental electron density map hindered the construction of an initial model, either manually with COOT (36) or automatically using the program BUCCANEER (37). Therefore, we used MOLREP (38) to fit a homology model generated using the Phyre2 server (39) into the electron density map. After model edition using COOT to keep only selected fragments which were in good accordance with the experimental electron density map, we used this partial model together with the heavy atoms’ positions to recalculate phases using PHASER, followed by density modification with PARROT. The new electron density map allowed improvement of the model construction using COOT. At this point, we used the higher resolution data set of the TcEIF4E5-m7GTP crystal in order to procced with model building and structure refinement. TcEIF4E5-m7GTP refined structure was used as model for TcEIF4E5-cap1 and TcEIF4E5-cap4 structure determination by molecular replacement. Refinement of the structures were performed alternating cycles of BUSTER (40) with visual inspection and manual rebuilding using COOT. The geometrical restraints for cap-1 and cap-4 molecules were generated using the Grade Web Server (http://grade.globalphasing.org) (41). Model validation was performed using the MolProbity Web Server (http://molprobity.biochem.duke.edu/) (42).
Microscale thermophoresis
TcEIF4E5 and TcEIF4E5-ΔC were labeled using the Monolith NT Protein Labeling Kit RED-Tris-NTA (NanoTemper Technologies) according to the supplier instructions. The concentration of labeled proteins was kept constant at 12.5 nM. The unlabeled binding partners (m7GTP or cap-4) were titrated in 1:1 dilutions with the highest final concentration of 2.5 mM. The assays were performed in triplicate in buffer containing 20 mM Tris–HCl pH 8, 100 mM NaCl, 2 mM DTT and 0.05% Tween-20. Measurements were performed in treated capillaries (NanoTemper Technologies) on a Monolith NT.115 Pico system (Nano Temper Technologies) using 20% LED and Medium IR-laser power. Data analysis were performed using MO.affinity analysis v.2.2.7 software (NanoTemper Technologies).
RESULTS
Large scale synthesis of cap-4
An efficient and robust synthesis protocol for synthetic route to trypanosome cap-4 (m7Gpppm36,6,2’Apm2’Apm2’Cpm23,2’U) (compound 1, Figure 1) was developed by taking advantage of recent developments in liquid-phase oligonucleotide synthesis (LPOS) which combine the beneficial aspects of both conventional solid-phase and solution-phase chemistry (43). We have also produced the cap-1 analogue (m7Gpppm36,6,2’A) for comparative structural studies.
Figure 1.
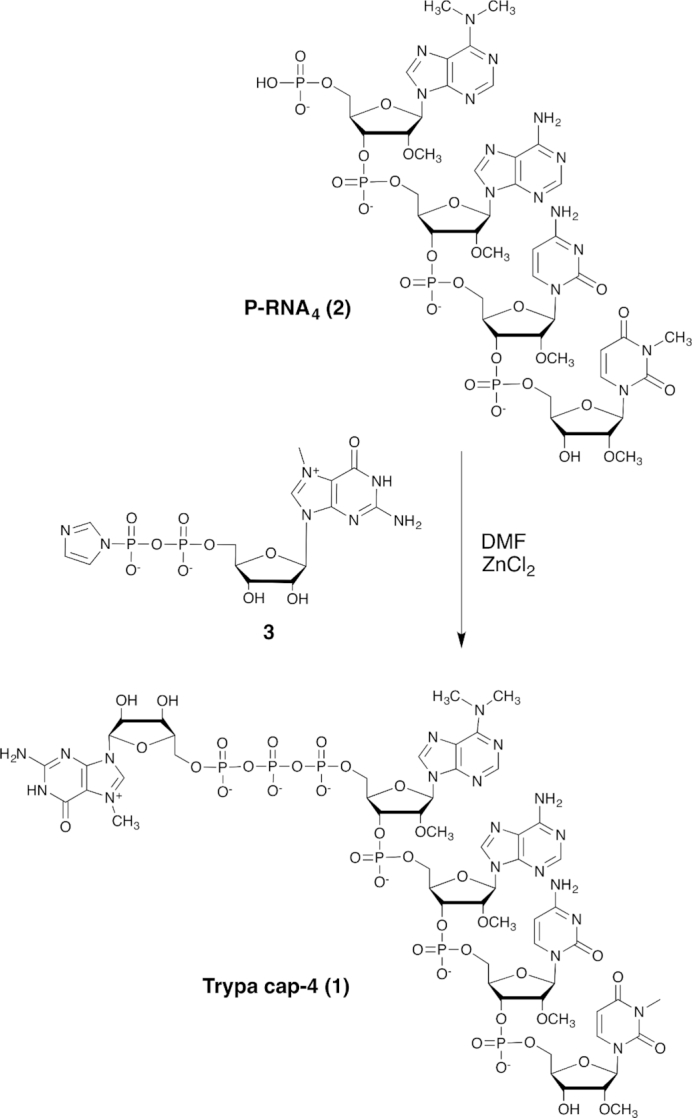
Capping step of 5′-phosphorylated tetramer 2 with imidazolide 3 in the presence of ZnCl2 as catalyst.
As illustrated in Figure 1, the key step in trypa cap-4 synthesis is the formation of the triphosphate linkage by coupling the phosphorylated tetramer P-RNA4 (2) with the m7GDP imidazolide (3) in the presence of ZnCl2 as catalyst (44). The 5′-phosphorylated tetramer (2, P-RNA4) was prepared by liquid-phase synthesis on an alkyl-chain-soluble support (ACSS, 4) using phosphoramidite chemistry. The soluble support 4 was obtained in 5 steps and 56% overall yield from commercially available methyl gallate (43). Two of the four RNA building blocks (5 and 10, Figure 2) are not commercially available. The detailed procedures for their synthesis and analytical data can be found in the Supplementary Data. The first modified nucleoside, 5′-O-dimethoxytrityl-N3,2′-O-dimethyluridine (5), was obtained in two steps from 2′-O-methyluridine by N-3 methylation with methyl iodide (29), followed by 5′-O-dimethoxytritylation. The fourth nucleoside phosphoramidite 10 was obtained in six steps from 6-chloropurine riboside using slight changes in established procedures (30). Briefly, amination of 6-chloropurine riboside with dimethylamine was achieved by microwave irradiation at 95°C for 30 min. 3′-O-Methylation was performed with methyl iodide in DMSO after selective protection of the 3′ and 5′-hydroxyl groups using the Markiewicz's reagent. Deprotection of silyl ethers, followed by 5′-O-dimethoxytritylation and 3′-O-phosphitylation according to classical protocols, afforded the phosphoramidite 10 in an overall yield of 25% in six steps (Supplementary Scheme S1).
Figure 2.
Building blocks 4–10 used for RNA assembly.
RNA synthesis started with attachment of the nucleoside 5 to the soluble support via a succinate linkage. The resulting anchored nucleoside 7 was then submitted to phosphoramidite coupling cycle using the building blocks 8, 9 and 10 under optimized conditions (43). The last step of the synthesis consisted of the 5′-phosphorylation of the tetramer still anchored to the support by using the commercially available phophitylating reagent (2-[2-(4,4′-dimethoxytrityloxy)ethylsulfonyl]ethyl-(2-cyanoethyl)-(N,N-diisopropyl)phosphoramidite). Removal of all protecting groups and cleavage from the support were achieved by conventional ammonolysis. The fully deprotected tetramer 5′-monophosphate (1) was isolated after reverse phase HPLC purification in high yield and purity. Finally, the phosphorylated tetramer 2 was coupled to 7-methylguanosine 5′-diphosphate imidazolide (3) in the presence of anhydrous zinc chloride as catalyst (30,44). Trypa cap-4 (1) was isolated as sodium salt in 34% yield. The chemical structure and purity of 1 were confirmed by mass spectrometry and HPLC analysis. Cap-1 (m7Gpppm36,6,2’A, 17) was obtained by coupling N6,N6,2′-O-trimethyladenosine 5′-monophosphate (16) and phosphoimidazolide 3 (Supplementary Scheme S2).
The crystal structure of T. cruzi EIF4E5 in complex with cap analogs and cap-4 binding mechanism
Initial attempts to determine TcEIF4E5 structure by molecular replacement methods failed, although several homologue structures are available in the Protein Data Bank. Therefore, selenomethionine-labelled (SeMet) TcEIF4E5 was produced and experimental phases were obtained by single wavelength anomalous diffraction. As TcEIF4E5-SeMet crystals showed poor diffraction, the partial model obtained from experimental electron density maps was used as an initial model to complete building and refinement against a higher resolution native data set (see Materials and Methods).
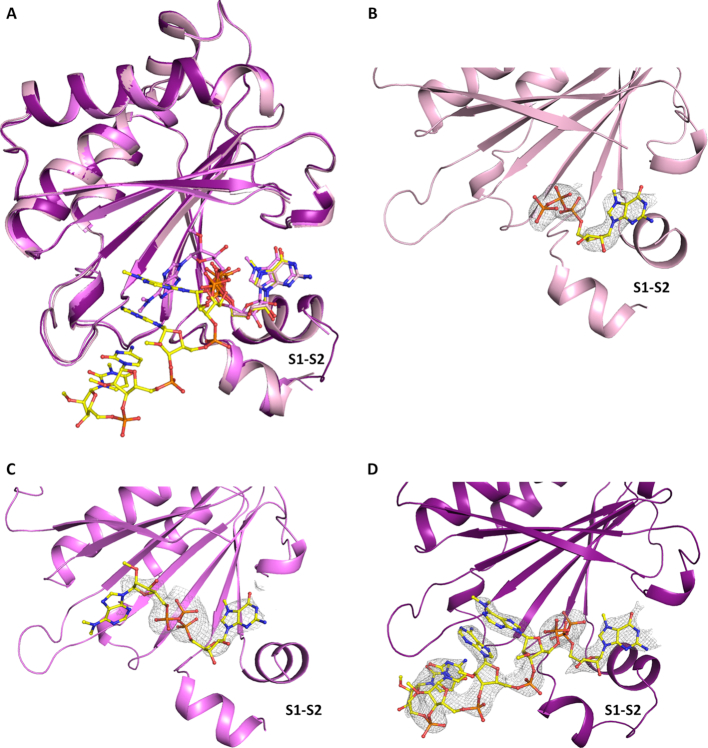
The TcEIF4E5 structure in complex with 7-methyl-GTP (m7GTP) was refined at 2.10 Å resolution to final Rfactor/Rfree of 17.8%/19.8%. The TcEIF4E5 structures in complex with cap-1 and cap-4 were refined at 2.70 and 2.20 Å resolution to final Rfactor/Rfree of 19.4%/25.2% and Rfactor/Rfree of 20.7%/22.6%, respectively (Table 1). The models comprise residues 4–186 with the polypeptide chain clearly defined by the electron density. The exception is the region connecting β1–β2 strands, which participates in the cap binding and is better defined in TcEIF4E5-cap-4 structure. Residues 30 to 32 could not be modeled in TcEIF4E5-m7GTP complex whereas residues 30–31 are lacking in TcEIF4E5-cap-1 model (Figure 3). Superposition of TcEIF4E5 structures resulted in RMSD values of 0.37 Å. TcEIF4E5 conserves the overall structure of the eIF4E family, a curved-shape α/β protein with the cap binding site located on the concave surface of the protein (Figure 3A). The cap analogues were well defined by the electron density maps, except for the base moiety of cap-1 second nucleotide (Figure 3B–D). The second ribose and base moieties of cap-1 adopt a distinct conformation compared with cap-4, probably due to flexibility of the cap-1 analogue in the TcIF4E5-cap-1 complex (discussion below).
Figure 3.
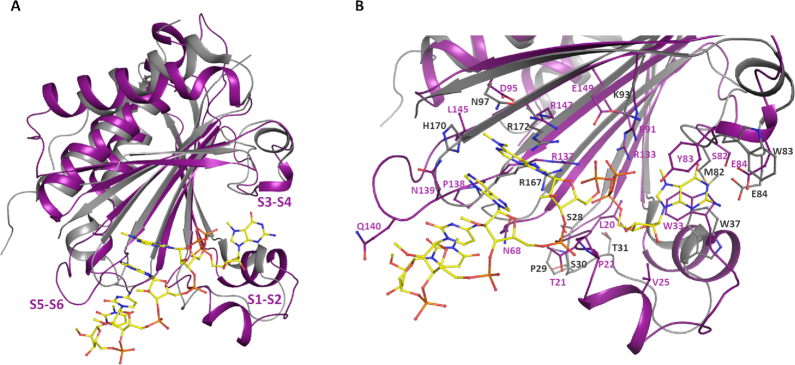
Three-dimensional structure of T. cruzi EIF4E5. (A) Overall superposition of TcEIF4E5 in complex with m7GTP, cap-1 and cap-4 (light pink, magenta and purple, respectively). (B–D) Electron density maps corresponding to the cap analogues. The ligands m7GTP (B), cap-1 (C) and cap-4 (D) are clearly defined by the electron density, except for the base moiety of cap-1 second nucleotide. 2Fo – Fc electron density maps are represented in gray and contoured at 1.0σ. The ligands are represented in sticks. The region connecting β1–β2 strands (S1–S2) is indicated.
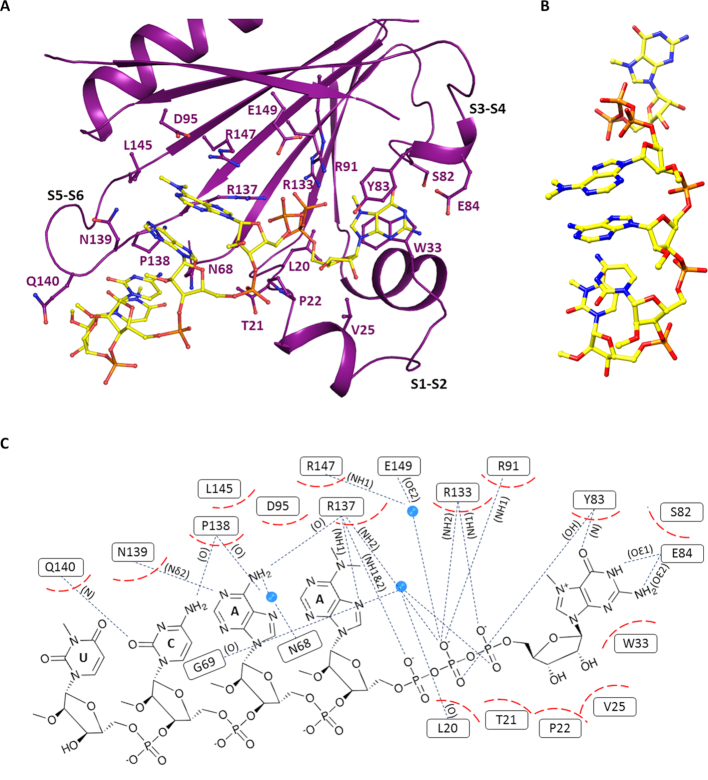
The structure of the TcEIF4E5-cap-4 complex revealed an extensive protein-ligand interaction surface. Binding of cap-4 is mediated by hydrophobic interactions as well as hydrogen bonds (Figure 4). TcEIF4E5 conserves the classical stacking interaction with the m7G moiety, with W33 and Y83 (at equivalent positions of human eIF4E1 W56 and W102) forming the m7G base sandwich. The highly conserved adjacent glutamic acid (E84) forms two hydrogen bonds with N1 and N2 of the guanine base and Y83 main chain-amide forms a H-bond with the guanine C-6 carbonyl group. The arginine residues R91, R133, R137 are involved in hydrogen bonds with the phosphate groups of m7GTP. In addition, the m7GTP ribose moiety is involved in hydrophobic interactions with L20, P22, V25 and main chain of T21, located in the region connecting β1–β2 strands. Interestingly, the cap-4 AACU bases pack together in a parallel stacking conformation (Figure 4B), oriented towards the protein, leaving the phosphate groups exposed to the solvent. Besides stacking between the bases, the first methylated adenine is involved in additional hydrophobic interactions with the aliphatic moieties of residues D95, L145 and R147. The second adenine base is involved in hydrophobic interactions with N139, in addition to two hydrogen bonds with R137 main chain-carbonyl and N139 side chain. P138 main chain-carbonyl and Q140 main chain-amide form H-bonds with the cytosine base, whereas the last nucleotide (U) is only involved in hydrophobic interactions. Figure 4C shows cap-4-TcEIF4E5 interactions in detail. Methylation of cap-4 bases seems to be determinant of its structural arrangement in the TcEIF4E5-cap-4 complex. Notably, the N6 and N3 methyl groups of adenine and uracil are involved in hydrophobic interactions with TcEIF4E5 residues, besides having a role in stacking of AACU bases. The 2′-O-methyl groups of cytidine and uridine also participate in hydrophobic interactions involving the TcEIF4E5 loop connecting β5–β6 strands, whereas the first two adenosine ribose methyl groups are exposed to the solvent.
Figure 4.
T. cruzi EIF4E5 cap recognition site. (A) Representation of the cap-4 binding site. Residues involved in the interaction with cap-4 are shown in sticks and labeled. The regions connecting β1–β2, β3–β4 and β5–β6 strands (S1–S2, S3–S4 and S5–S6) are indicated. (B) Three-dimensional structure of cap-4 evidencing the AACU base stacking. (C) Schematic representation showing the cap-4-TcEIF4E5 interactions in detail. Hydrophobic interactions are represented in dashed red and hydrogen bonds in blue dotted lines. The atoms involved in the hydrogen bonds are indicated in parentheses. Water molecules are represented as blue spheres.
Unique molecular features mediate T. cruzi EIF4E5-cap-4 interaction
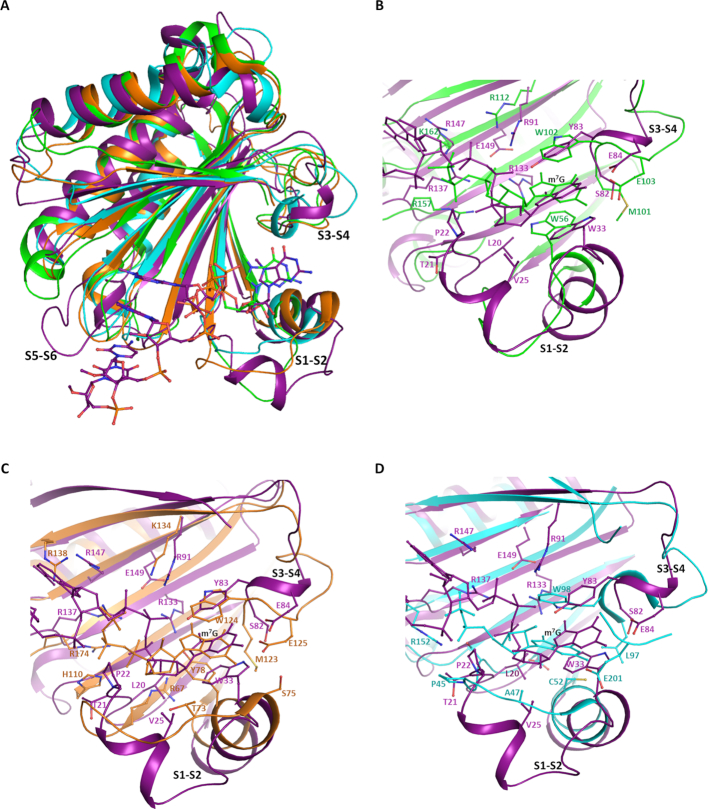
Superposition of the TcEIF4E5 structure (cap-4 complex) with mammalian eIF4E family members resulted in RMSD varying from 1.90 to 2.77 Å (146 and 157 C-alpha atoms aligned with human eIF4E1 and eIF4E2 (PDB: 1IPC and PDB: 2JGB) respectively, 157 C-alpha atoms aligned with mouse eIF4E3 (PDB: 4B6U)). The main structural differences of TcEIF4E5 compared with eIF4E family members reside in the regions connecting β1–β2, β3–β4 and β5–β6 strands (Figure 5A). The first one, also called S1-S2 loop, comprises one of the residues that forms the m7G sandwich (W33 in TcIF4E5, W56 in human eIF4E1) and is described to undergo structural rearrangement upon cap binding (14). The corresponding region in TcEIF4E5 is longer, comprising 19 residues, in contrast to a 7-residues loop present in human eIF4E1. In TcEIF4E5, two short α-helices are formed, neighboring residue W33. Human eIF4E2 and mouse eIF4E3 also present one helix in S1-S2 (14,15), however their β1–β2 connections are significantly shorter compared with TcEIF4E5 (Figure 5). The second residue forming the m7G sandwich (Y83 in TcIF4E5, W102 in human eIF4E1) is located in the region connecting β3–β4 strands (S3–S4 loop), which also shows a conformational variation among the structures. Finally, the region connecting β5-β6 strands is longer in TcEIF4E5, encompassing 9 residues, in contrast to a 4-residues loop in the other counterparts (Figure 5A). Interestingly, the TcEIF4E5 extended S5–S6 loop creates an additional molecular surface that mediates the interaction with the cap-4 AACU bases (Figure 4A).
Figure 5.
Structural comparison of T. cruzi EIF4E5 with other eukaryotic IF4E factors. (A) Overall superposition: TcEIF4E5 is shown in purple, human eIF4E1 and eIF4E2 (HsIF4E1, PDB 1IPC; HsIF4E2, PDB 2JGB) are represented in green and orange, respectively. Mouse eIF4E3 (MmIF4E3, PDB: 4B6U) is represented in cyan. TcEIF4E5-bound cap-4 and HsIF4E1-bound m7GTP are shown in sticks. The regions connecting β1–β2, β3–β4 and β5–β6 strands (S1–S2, S3–S4 and S5–S6) are indicated. (B, C and D) Detailed comparison between the m7GTP recognition sites. (B) TcEIF4E5-cap-4 (purple) versus HsIF4E1-m7GTP (green), (C) TcEIF4E5-cap-4 (purple) versus HsIF4E2-m7GTP (orange), (D) TcEIF4E5-cap-4 (purple) versus MmIF4E3-m7GTP (cyan). Cap-4 is represented in purple and m7GTP follows the color of the counterpart structure. The m7G group position is indicated. Residues involved in the interaction with cap analogues are shown in sticks and labeled, following the color of the protein structure.
A detailed comparison of the residues involved in TcEIF4E5 cap recognition reveals further differences from other eIF4E family members. Although substitution of the tryptophan residue located at the flexible S1–S2 region for other aromatic or non-aromatic residue has been previously described, this is the first eIF4E homologue structure showing a substitution of the tryptophan equivalent to human W102 located at the S3–S4 loop (TcEIF4E5 Y83) (Figure 5B–D). In addition, an adjacent methionine (human M101) also involved in hydrophobic interactions with the guanine base is replaced by a serine (S82) in TcEIF4E5. The correct positioning of less bulky residues Y83 and S82 to interact with the m7G moiety in TcEIF4E5 was most likely made possible by the flexibility of S3–S4 loop.
Interestingly, TcEIF4E5 W33, located at the beginning of the S1–S2 second short helix, interacts with the cap-4 guanine base in a distinct orientation, with the pyrrole ring stacking with the pyrimidine moiety of the m7G (Figure 5B). The extended β1-β2 connection observed in TcEIF4E5 creates additional surface for cap interaction with an additional short helix, placing residues V25, P22, L20 in position to participate in hydrophobic interactions with the first ribose moiety. The implication of aliphatic and backbone residues of the S1–S2 loop to form the cap pocket is also observed in mouse eIF4E3 (Figure 5D). However, in the case of mouse eIF4E3 this additional hydrophobic surface is shorter and proposed to compensate the lack of the S1–S2 aromatic residue to form the m7G sandwich (15). Similar to human eIF4E1 and eIF4E2, basic side chains are involved in TcEIF4E5 interactions with the m7GTP phosphate groups (Figure 5B, C). However, in striking contrast with other eukaryotic counterparts, very few water molecules are present in the cap-4 pocket (Supplementary Figure S1). In the TcEIF4E5-cap-4 complex there is only three water molecules participating in the cap-binding interface, two of them mediating phosphate interactions with the protein side chains (Figure 4B). In the TcEIF4E5-m7GTP complex, seven water molecules are involved in the hydration of the cap pocket, four of them participating in phosphates interaction (see Discussion).
Comparison of T. cruzi EIF4E5 and Leishmania EIF4E1 structures provides insights into cap-4 binding specificities of trypanosomatids EIF4E homologues
Superposition of the TcEIF4E5 and L. major EIF4E1 (PDB 5wb5) structures resulted in an RMSD of 2.06 Å for 157 C-alpha atoms aligned. Significant differences were observed in the cap-binding pocket of these two trypanosomatid homologues (Figure 6). The LmEIF4E1 structure was determined in its apo form showing tryptophans W37 and W83, that are expected to sandwich the cap m7G-moiety in a flipped conformation (28). The LmEIF4E1 S1–S2 loop is shorter than in TcEIF4E5 (9 residues instead of 19), but a structural rearrangement upon cap-binding could bring a proline containing segment (28-SPST-31) in position to form a hydrophobic interacting surface with the cap ribose moiety, similar to that observed in TcEIF4E5 (Figure 6B). Basic side chains in position to interact with the m7GTP phosphates (K93, R167, R172) are also present in LmEIF4E1 (Figure 6B). However, the S5–S6 loop that forms the surface of interaction with the cap-4 AACU bases in TcEIF4E5 has a short configuration in LmEIF4E1 similarly to the other eukaryotic counterparts.
Figure 6.
Structural comparison of T. cruzi EIF4E5-cap-4 complex and L. major EIF4E1 in the apo state. (A) Overall superposition: TcEIF4E5 is shown in purple and LmEIF4E1 (PDB 5WB5) is shown in gray. (B) Detailed comparison of the cap recognition sites. Cap-4 is represented in yellow. TcEIF4E5 residues involved in the interaction with cap-4 and corresponding residues in LmEIF4E1 are shown in sticks and labeled, following the color of the protein structure. The regions connecting β1-β2, β3-β4 and β5-β6 strands (S1–S2, S3–S4 and S5–S6) are indicated.
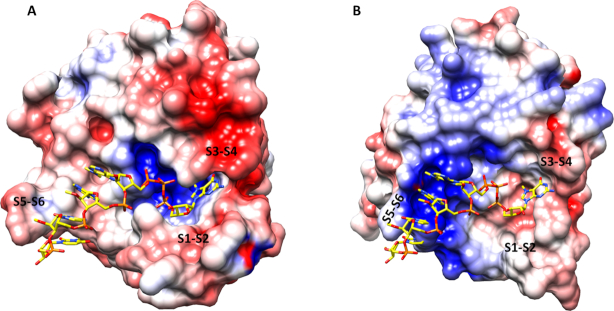
Analysis of the TcEIF4E5 electrostatic potential surface evidences the positive charge mediating the interaction of the m7GTP phosphate groups while the remaining interacting surface is mostly neutral or lightly acidic (Figure 7A). This surface pattern most likely dictates the cap-4 interacting conformation, with the stacked bases towards the protein. In TcIF4E5, we observe a cavity that accommodates the m7GTP formed by the regions connecting strands β1–β2 and β3–β4, whereas the remaining cap-4 residues follow an open surface of the protein towards the long β5-β6 loop. Interestingly, the L. major EIF4E1 electrostatic potential surface strongly indicates a distinct cap-4 binding mechanism (Figure 7B). Four basic side chains present in the LmEIF4E1 S5–S6 loop (166-KRKYH-170) confer a positive potential to the corresponding surface. In contrast to TcEIF4E5, LmEIF4E1-cap-4 interaction would most likely occur with the phosphate groups of the AACU nucleotides oriented towards the protein, leaving the base moieties more exposed. Differences in EIF4E molecular mechanisms for cap binding can account for distinct mRNA affinities and are consistent with the proposal that trypanosomatid EIF4E homologues play distinct roles in translation.
Figure 7.
Electrostatic potential surface of (A) T. cruzi EIF4E5 and (B) L. major EIF4E1 (PDB 5WB5). The boundaries for potential contour map visualization are ± 5 kT/e. The cap-4 molecule (shown in sticks) is positioned on LmEIF4E1, based on the superposition of TcEIF4E5-cap-4 structure. The regions corresponding to the connections between β1–β2, β3–β4 and β5–β6 strands (S1–S2, S3–S4 and S5–S6) are indicated. Electrostatic potential was calculated using the Adaptive Poisson–Boltzmann Solver through the software Chimera (45).
T.cruzi EIF4E5 shows low affinity to cap analogues in vitro which is abolished by deletion of the C-terminal region
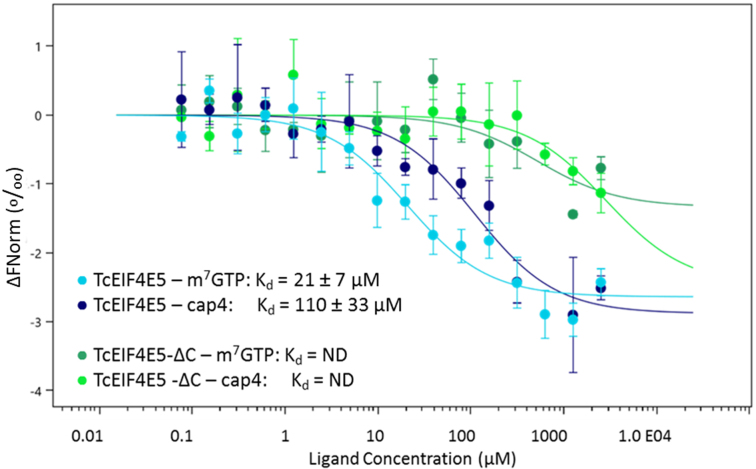
Affinity of TcEIF4E5 to m7GTP and cap-4 were investigated by microscale thermophoresis. Since the role of eIF4E C-terminal region in cap binding has been previously described in other organisms (4,5), a truncated TcEIF4E5 construction, lacking C-terminal residues 190 to 200 (TcEIF4E5-ΔC) was also used in the assays. Our results showed that TcEIF4E5 has low affinity to m7GTP and cap-4 (Kd = 21 ± 7 and 110 ± 33 μM, respectively) and interaction was abolished by deletion of the C-terminal region (Figure 8). TcEIF4E5 crystallographic models lack residues 187–200 even in the presence of cap-1 or cap-4. Therefore, although deletion of TcEIF4E5 residues 190–200 abolished cap binding, our structural analysis indicates that the role of C-terminal region is not in stabilizing the cap interaction.
Figure 8.
Microscale thermophoresis analysis of T. cruzi EIF4E5 binding to mRNA cap analogues. TcEIF4E5 presented low affinity to m7GTP and cap-4 (light and dark blue, Kd = 21 ± 7 and 110 ± 33 μM, respectively). Deletion of the C-terminal region abolishes the interaction (TcEIF4E5-ΔC, dark and light green).
The crystal structure of human eIF4E in complex with m7GpppA showed that the C-terminal loop is stabilized by interaction with the adenine nucleoside, while in the m7GTP complex the C-terminal remains disordered (4). In the crystal structure of parasitic Schistosoma mansoni eIF4E in complex with m7GpppA or m7GpppG, the C-terminal is also ordered, although a distinct orientation of the second base is observed, compared with the mammalian structure (5). In TcEIF4E5, cap-1 and cap-4 follow different orientations (Figure 9). Electron density was missing for cap-1 second base, indicating a flexibility for this nucleotide (Figure 3C). Cap-4 conformation is stabilized by the presence of the additional nucleotides, due to their interactions with the protein besides the stacking arrangement of the bases. Interestingly, four out of the eleven residues lacking in the TcEIF4E5-ΔC construction, and also absent from TcEIF4E5 crystal structure, are lysines. We hypothesize whether a flexible and basic C-terminal arm could function as a carrier, favoring the recruitment of the capped mRNA, before its positioning in the interaction pocket.
Figure 9.
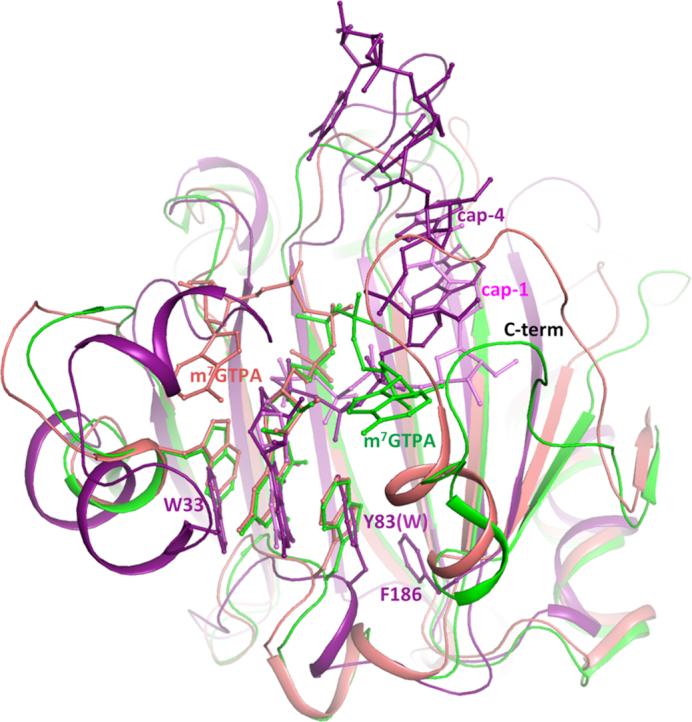
Structural superposition of TcEIF4E5-cap-4 (purple), human eIF4E1 (green) and Schistosoma mansoni eIF4E1 (salmon) in complex with m7GTPA (PDB: 1IPB and 3HXG, respectively). The ligands are represented in sticks and identified by the positions of the second base. Cap-1 (pink) is superimposed. The residues forming the m7G sandwich are represented in sticks and numbered according to TcEIF4E5 sequence. Human and Schistosoma eIF4E1 C-terminal regions, structured upon binding of a dinucleotide, are indicated. TcEIF4E5 model lacks C-terminal residues 187–200. The last residue built in TcEIF4E5 model (F186) is shown in sticks.
DISCUSSION
The interaction mode between EIF4E homologues and the mRNA cap in trypanosomatids is of special interest since the unique and extensive covalent modifications of cap-4 could be at the source of trypanosomatid-specific interaction mechanisms. The cap-4 structure includes multiple chemical modifications on the RNA body: the 2′-O-methylation on the first four ribose moieties and unique base methylations on the first adenine and the fourth uracil (m7Gpppm36,6,2’Apm2’Apm2’Cpm23,2’U) (compound 1, Figure 1). Access to mRNA cap structures remains a major bottleneck for structural studies of cap-binding proteins. Known synthetic routes to 5′-capped RNA are based on chemical or enzymatic preparation, as well as on a combination of both (46). A multi-step synthesis of trypanosome cap-4 has been previously reported on a 5–10 μmol scale by using RNA solid-support synthesis and phosphoramidite chemistry (30). However, a ten-fold excess of phosphoramidite monomers is usually required to guarantee a high efficiency for each coupling step leading to the target oligomer in good overall yield. Scaling up the cap synthesis with this approach (i.e. solid support synthesis) would have required the preparation of rather large amounts of the corresponding phosphoramidite derivatives. In this work, these limitations were overcome by designing a new synthetic strategy providing large amounts of highly pure cap-4, suitable for structural and interaction analysis. Based on liquid-phase oligonucleotide synthesis (LPOS), it combines the beneficial aspects of conventional solid-phase and solution-phase chemistry (43). Our approach thus significantly reduced the amount of phosphoramidite building blocks required for RNA assembly, and consequently reduced the cost and time for the preparation of large quantity of cap-4.
T. cruzi EIF4E5 belongs to the so-called group 3 of kinetoplastid EIF4E homologues, which also includes EIF4E6. The functions of the extended EIF4E family in these organisms are poorly understood. The current knowledge, mostly obtained from studies in T. brucei and L. major, indicates that EIF4E3 and EIF4E4 form typical translation promoting eIF4F complexes and are the trypanosomatid homologues involved in general translation (reviewed in 20). T. brucei EIF4E5 is cytosolic and was shown to bind cap analogs in vitro. Knockdown of EIF4E5 in T. brucei resulted in loss of cell motility, suggesting a direct or indirect role in regulation of specific cellular pathways. In addition, the association of TbEIF4E5 with two distinct multiprotein complexes containing either the TbEIF4G1 or TbEIF4G2 homologues raised new questions about EIF4E5 function in trypanosomatids (25).
Structural comparisons of T. cruzi EIF4E5 with eIF4E family members achieved in this work pointed out significant differences in the cap recognition pocket. In addition to residue substitutions, long insertions in the TcEIF4E5 polypeptide chain, notably in the regions connecting β1-β2 and β5-β6 strands, are modifying the molecular interaction surface. Analysis of the electrostatic potential surfaces revealed a neutral to acidic surface, which is a unique feature of TcEIF4E5, providing the interaction path for the cap-4 AACU nucleotides (Figure 7A). Moreover, the cap-4 base moieties and methyl groups turned out to be involved in a number of hydrophobic interactions with the protein residues. In contrast, the eukaryotic counterparts, including Leishmania EIF4E1, feature an extended basic surface, that would be favorable to accommodate the ribonucleotide chain through an interaction with the phosphate groups (Figure 7B and Supplementary Figure S2). Structurally conserved waters were previously shown to participate in the cap binding interface. Fourteen water molecules are partially conserved in eIF4E structures in complex with cap analogues (47). In contrast, TcEIF4E5-m7GTP and TcEIF4E5-cap-4 structures present, respectively, seven and three waters in the cap pocket (Supplementary Figure S1). The reduced number of water molecules mediating TcEIF4E5-cap-4 interaction is a further distinct feature, which is most likely associated with its specific binding mechanism.
The differences in cap binding mode could be related to the role of the eIF4F-like complexes involving trypanosomatid homologues of EIF4E group 3. Sequence alignment of EIF4E5 and EIF4E6 homologues from T. cruzi, T. brucei and L. major (Supplementary Figure S3) pointed out a conservation of the extended loop connecting β5–β6 strands, which is implicated, in TcEIF4E5, in cap-4 AACU interaction through the base moieties. However, the region connecting β1–β2 strands, which includes one of the aromatic residues that participates in the m7G stacking, is shorter in EIF4E6 when compared with EIF4E5 homologues (Supplementary Figure S3), indicating that, in EIF4E6, a distinct structural arrangement of the S1–S2 loop is likely to take place upon cap binding.
The crystal structures of T. cruzi EIF4E5 in complex with m7GTP, cap-1 and cap-4 demonstrate the flexibility of the polypeptide chain in the extended region connecting β1-β2 strands. Such flexibility was previously seen in the course of a structural comparison of human eIF4E2 in apo and m7GTP bound states (14). The TcEIF4E5 S1–S2 region forms the base of the cavity which accommodates the m7G nucleoside. Interestingly, electron density corresponding to S1–S2 residues is poor in the m7GTP and cap-1 containing structures but is better defined in the cap-4 bound structure. The electron density map corresponding to the ligand is also of a better quality in the cap-4 containing structure (Figure 3). This density improvement indicates that the interactions involving N1–N4 nucleotides stabilize the cap-4 conformation and also the m7GTP recognition pocket. In addition, our results suggest an indirect involvement of the flexible TcEIF4E5 C-terminal in the cap binding mechanism. We speculate whether the basic C-terminal arm could function as a carrier for the capped mRNA, through interaction with the phosphate groups.
Our biophysical analysis revealed a very low affinity of TcEIF4E5 for cap-4, which might be related to its specific interaction mode, engaging the cap-4 N1-N4 base moieties. Eukaryotic IF4E2 and IF4E3 were shown to bind the m7GTP cap with affinities lower than the canonical eIF4E1 (14,15). The affinity of TcEIF4E5 for cap-4 is approximately 10-fold lower than the affinity of human eIF4E2 and mouse eIF4E3 for m7GTP and 2000-fold lower than the affinity of canonical eIF4E1 for m7GTP.
Studies on cap binding affinity of trypanosomatid EIF4E homologues have reported intriguing differences between homologues and species. Among L. major homologues, the highest binding affinity for m7GTP and cap-4 was observed for EIF4E4 (equivalent dissociation constants of approximately 1.3 μM). L. major EIF4E1 showed preferential binding to methylated cap-4 (dissociation constants of 6.3 and 4 μM for m7GTP and cap-4, respectively). On the other hand, a clear reduction in the binding affinity for cap-4 was observed for EIF4E3 (dissociation constants of 3.1 μM and 21.7 μM for m7GTP and cap-4, respectively) (27). T. brucei EIF4E5 was reported to bind slightly better to m7GTP than to cap-4 (dissociation constants of 1.5 and 1.8 μM, respectively) (25). Surprisingly, our interaction assays revealed the lowest affinity to cap analogues described for trypanosomatid homologues so far. We have confirmed our results using different batches of purified recombinant protein. Differences in binding constant values are expected when distinct experimental methods are applied. However, a difference of approximately two orders of magnitude as observed for TcEIF4E5-cap-4 binding most likely indicates a very weak interaction, which is intrinsic to T. cruzi EIF4E5 homologue. Previous studies demonstrated that the interaction of eIF4E with the mRNA 5′-cap is enhanced by eIF4G (48). Moreover, in plants, association of PABP with the eIF4F complex was shown to increase the affinity of eIF4F for the cap structure (49). We hypothesize whether TcEIF4E5 affinity for cap-4 would be enhanced within the context of the translational machinery, or by the binding of an EIF4G factor. We also speculate about the influence of additional nucleotides of the spliced leader over the interaction of the mRNA with EIF4E in trypanosomatids. Further structural and interaction studies are necessary to elucidate the spectrum of molecular mechanisms for capped mRNA binding to the different trypanosomatid EIF4E homologues.
Biological data on T. cruzi EIF4E homologues are not available. Preliminary information on the interaction pattern between T. cruzi initiation factor homologues involved in the formation of eIF4F-like complexes indicates that TcEIF4E5 interacts with EIF4G1, EIF4G2 and EIF4G5 proteins (Dr. Fabiola Holetz, personal communication). TcEIF4E5 conserves the key residues that are implicated in the canonical 4E-4G and 4E-4EIP interactions, however the role of these interactions and the function of TcEIF4E5 in translation or other RNA metabolism pathway remain to be determined. The structural data presented in this work provide key starting points for the design of new experiments to investigate T. cruzi EIF4E5 function and cap-4 interaction mechanism with the remaining EIF4E homologues from trypanosomatids.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 6O80, 6O7Z and 6O7Y for TcEIF4E5-m7GTP, TcEIF4E5-cap-1 and TcEIF4E5-cap-4 complexes, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the Platform for Protein Purification and Characterization of the FIOCRUZ Technical Platform program. The authors are grateful to Synchrotron SOLEIL for beam time awarded at PROXIMA1 and PROXIMA2a beam lines (Proposal 20160778) and all the support they benefited as SOLEIL’s users. We thank Jean-Sebastien Vartanian-Grimaldi for technical help during the preparation of ACSS. Yves L. Janin is also acknowledged for his help in proof-reading this manuscript. Electrostatic potential analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundação Araucária [05/2016]; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES Financing code 001); Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq, NITZ 312195/2015-0 BGG 304027/2015-4 and LWR fellowship]; Fundação Oswaldo Cruz. The authors also acknowledge financial support from Institut Pasteur and Centre National de la Recherche Scientifique (CNRS). Funding for open access charge: Fundação Oswaldo Cruz.
Conflict of interest statement. None declared.
REFERENCES
- 1. Aitken C.E., Lorsch J.R.. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012; 19:568–576. [DOI] [PubMed] [Google Scholar]
- 2. Ramon Y., Cajal S., Castellvi J., Hümmer S., Peg V., Pelletier J., Sonenberg N.. Beyond molecular tumor heterogeneity: protein synthesis takes control. Oncogene. 2018; 37:2490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K.. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997; 89:951–961. [DOI] [PubMed] [Google Scholar]
- 4. Tomoo K., Shen X., Okabe K., Nozoe Y., Fukuhara S., Morino S., Ishida T., Taniguchi T., Hasegawa H., Terashima A. et al.. Crystal structures of 7-methylguanosine 5′-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem. J. 2002; 362:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu W., Zhao R., McFarland C., Kieft J., Niedzwiecka A., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., Jones D.N., Davis R.E.. Structural insights into parasite eIF4E binding specificity for m7G and m2,2,7G mRNA caps. J. Biol. Chem. 2009; 284:31336–31349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu W., Jankowska-Anyszka M., Piecyk K., Dickson L., Wallace A., Niedzwiecka A., Stepinski J., Stolarski R., Darzynkiewicz E., Kieft J. et al.. Structural basis for nematode eIF4E binding an m(2,2,7)G-Cap and its implications for translation initiation. Nucleic Acids Res. 2011; 39:8820–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K.. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 1999; 3:707–716. [DOI] [PubMed] [Google Scholar]
- 8. Gross J.D., Moerke N.J., von der Haar T., Lugovskoy A.A., Sachs A.B., McCarthy J.E., Wagner G.. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003; 115:739–750. [DOI] [PubMed] [Google Scholar]
- 9. Umenaga Y., Paku K.S., In Y., Ishida T., Tomoo K.. Identification and function of the second eIF4E-binding region in N-terminal domain of eIF4G: comparison with eIF4E-binding protein. Biochem. Biophys. Res. Commun. 2011; 414:462–467. [DOI] [PubMed] [Google Scholar]
- 10. Grüner S., Peter D., Weber R., Wohlbold L., Chung M.Y., Weichenrieder O., Valkov E., Igreja C., Izaurralde E.. The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol. Cell. 2016; 64:467–479. [DOI] [PubMed] [Google Scholar]
- 11. Grüner S., Weber R., Peter D., Chung M.Y., Igreja C., Valkov E., Izaurralde E.. Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast. Nucleic Acids Res. 2018; 46:6893–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joshi B., Cameron A., Jagus R.. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004; 271:2189–2203. [DOI] [PubMed] [Google Scholar]
- 13. Joshi B., Lee K., Maeder D.L., Jagus R.. Phylogenetic analysis of eIF4E-family members. BMC Evol. Biol. 2005; 5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosettani P., Knapp S., Vismara M.-G., Rusconi L., Cameron A.D.. Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J. Mol. Biol. 2007; 368:691–705. [DOI] [PubMed] [Google Scholar]
- 15. Osborne M.J., Volpon L., Kornblatt J.A., Culjkovic-Kraljacic B., Baguet A., Borden K.L.B.. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhoads R.E. eIF4E: new family members, new binding partners, new roles. J. Biol. Chem. 2009; 284:16711–16715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernández G., Proud C.G., Preiss T., Parsyan A.. On the Diversification of the Translation Apparatus across Eukaryotes. Comp. Funct. Genomics. 2012; 2012:256848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bangs J.D., Crain P.F., Hashizume T., McCloskey J.A., Boothroyd J.C.. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 1992; 267:9805–9815. [PubMed] [Google Scholar]
- 19. Zinoviev A., Shapira M.. Evolutionary conservation and diversification of the translation initiation apparatus in trypanosomatids. Comp. Funct. Genomics. 2012; 2012:813718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freire E.R., Sturm N.R., Campbell D.A., de Melo Neto O.P.. The role of cytoplasmic mRNA cap-binding protein complexes in Trypanosoma brucei and other trypanosomatids. Pathogens. 2017; 6:E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jagus R., Bachvaroff T.R., Joshi B., Place A.R.. Diversity of eukaryotic translational initiation factor eIF4E in protists. Comp. Funct. Genomics. 2012; 2012:134839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bannerman B.P., Kramer S., Dorrell R.G., Carrington M.. Multispecies reconstructions uncover widespread conservation, and lineage-specific elaborations in eukaryotic mRNA metabolism. PLoS One. 2018; 13:e0192633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dhalia R., Reis C.R., Freire E.R., Rocha P.O., Katz R., Muniz J.R., Standart N., de Melo Neto O.P.. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 2005; 140:23–41. [DOI] [PubMed] [Google Scholar]
- 24. Freire E.R., Dhalia R., Moura D.M., da Costa Lima T.D., Lima R.P., Reis C.R., Hughes K., Figueiredo R.C., Standart N., Carrington M. et al.. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol. Biochem. Parasitol. 2011; 176:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freire E.R., Vashisht A.A., Malvezzi A.M., Zuberek J., Langousis G., Saada E.A., Nascimento J.F., Moura D., Darzynkiewicz E., Hill K. et al.. eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA. 2014; 20:1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Freire E.R., Malvezzi A.M., Vashisht A.A., Zuberek J., Saada E.A., Langousis G., Nascimento J.D., Moura D., Darzynkiewicz E., Hill K. et al.. Trypanosoma brucei translation initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot. Cell. 2014; 13:896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoffe Y., Zuberek J., Lerer A., Lewdorowicz M., Stepinski J., Altmann M., Darzynkiewicz E., Shapira M.. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot. Cell. 2006; 5:1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meleppattu S., Arthanari H., Zinoviev A., Boeszoermenyi A., Wagner G., Shapira M., Léger-Abraham M.. Structural basis for LeishIF4E-1 modulation by an interacting protein in the human parasite Leishmania major. Nucleic Acids Res. 2018; 46:3791–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamamoto I., Kimura T., Tateoka Y., Watanabe K., Ho I.K.. N-substituted oxopyrimidines and nucleosides: structure-activity relationship for hypnotic activity as central nervous system depressant. J. Med. Chem. 1987; 30:2227–2231. [DOI] [PubMed] [Google Scholar]
- 30. Lewdorowicz M., Yoffe Y., Zuberek J., Jemielity J., Stepinski J., Kierzek R., Stolarski R., Shapira M., Darzynkiewicz E.. Chemical synthesis and binding activity of the trypanosomatid cap-4 structure. RNA. 2004; 10:1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Duyne G.D., Standaert R.F., Karplus P.A., Schreiber S.L., Clardy J.. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993; 229:105–124. [DOI] [PubMed] [Google Scholar]
- 32. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheldrick G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cowtan K. Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 37. Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:1002–1011. [DOI] [PubMed] [Google Scholar]
- 38. Vagin A., Teplyakov A.. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:22–25. [DOI] [PubMed] [Google Scholar]
- 39. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O.S., Vonrhein C., Womack T.O.. BUSTER version 2.10.2. 2017; Cambridge: Global Phasing Ltd. [Google Scholar]
- 41. Smart O.S., Womack T.O., Sharff A., Flensburg C., Keller P., Paciorek W., Vonrhein C., Bricogne G.. Grade, version 1.2.13. 2011; Cambridge: Global Phasing Ltd. [Google Scholar]
- 42. Chen V.B., Arendall W.B 3rd., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim S., Matsumoto M., Chiba K.. Liquid-phase RNA synthesis by using alkyl-chain-soluble support. Chem. Eur. J. 2013; 19:8615–8620. [DOI] [PubMed] [Google Scholar]
- 44. Kadokura M., Wada T., Urashima C., Sekine M.. Efficient synthesis of γ-methyl-capped guanosine 5′-triphosphate as a 5′-terminal unique structure of U6 RNA via a new triphosphate bond formation involving activation of methyl phosphorimidazolidate using ZnCl2 as a catalyst in DMF under anhydrous conditions. Tetrahedron Lett. 1997; 38:8359–8362. [Google Scholar]
- 45. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 46. Muttach F., Muthmann N., Rentmeister A.. Synthetic mRNA capping. Beilstein J. Org. Chem. 2013; 13:2819–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lama D., Pradhan M.R., Brown C.J., Eapen R.S., Joseph T.L., Kwoh C.K., Lane D.P., Verma C.S.. Water-bridge mediates recognition of mRNA cap in eIF4E. Structure. 2017; 25:188–194. [DOI] [PubMed] [Google Scholar]
- 48. Haghighat A., Sonenberg N.. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 1997; 272:21677–21680. [DOI] [PubMed] [Google Scholar]
- 49. Wei C.C., Balasta M.L., Ren J., Goss D.J.. Wheat germ poly(A)binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry. 1998; 37:1910–1916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 6O80, 6O7Z and 6O7Y for TcEIF4E5-m7GTP, TcEIF4E5-cap-1 and TcEIF4E5-cap-4 complexes, respectively.