Abstract
Prokaryotic Argonaute proteins (pAgos) constitute a diverse group of endonucleases of which some mediate host defense by utilizing small interfering DNA guides (siDNA) to cleave complementary invading DNA. This activity can be repurposed for programmable DNA cleavage. However, currently characterized DNA-cleaving pAgos require elevated temperatures (≥65°C) for their activity, making them less suitable for applications that require moderate temperatures, such as genome editing. Here, we report the functional and structural characterization of the siDNA-guided DNA-targeting pAgo from the mesophilic bacterium Clostridium butyricum (CbAgo). CbAgo displays a preference for siDNAs that have a deoxyadenosine at the 5′-end and thymidines at nucleotides 2–4. Furthermore, CbAgo mediates DNA-guided DNA cleavage of AT-rich double stranded DNA at moderate temperatures (37°C). This study demonstrates that certain pAgos are capable of programmable DNA cleavage at moderate temperatures and thereby expands the scope of the potential pAgo–based applications.
INTRODUCTION
Eukaryotic Argonaute proteins (eAgos) play a key role in RNA interference (RNAi) processes (1–3). As the core of the multiprotein RNA-induced silencing complex (RISC), eAgos bind small non-coding RNA molecules as guides to direct the RISC complex towards complementary RNA targets (3–5). Reflecting their physiological function, variation among eAgos is observed with respect to the presence or absence of a catalytic site, and to their potential to interact with other proteins (6). Depending on the eAgo and on the sequence complementarity between guide and target RNA, eAgo-guide complexes either catalyze endonucleolytic cleavage of the target RNA (7) or indirectly silence the target RNA by repressing its translation and promoting its degradation through recruitment of additional silencing factors (8). Independent of the mechanism, eAgo-mediated RNA binding generally results in sequence-specific silencing of gene expression. As such, eAgos can coordinate various cellular processes by regulating intracellular RNA levels.
Prokaryotes also encode Argonaute proteins (pAgos) (9,10). Various pAgos share a high degree of structural homology with eAgos as both pAgos and eAgos adopt the same four domain (N-PAZ-MID-PIWI) architecture (9–12). Despite their structural homology, several recently characterized pAgos have distinct functional roles and different guide and/or target preferences compared to eAgos. For example, several pAgos have been implicated in host defense by directly targeting DNA instead of RNA (13–16). One of the best characterized mechanisms that pAgos utilize is DNA-guided DNA interference, which is demonstrated for pAgos from Thermus thermophilus (TtAgo), Pyrococcus furiosus (PfAgo) and Methanocaldococcus jannaschii (MjAgo) (13–15,17–20). These pAgos use 5′-end phosphorylated small interfering DNAs (siDNAs) for recognition and successive cleavage of complementary DNA targets. This mechanism enables both TtAgo and PfAgo to mediate host defense against invading nucleic acids. Prokaryotes lack homologs of eukaryotic enzymes that are involved in guide biogenesis (21). Instead, both TtAgo and MjAgo—besides the canonical siDNA-dependent target cleavage termed ‘slicing’—exhibit an alternative nuclease activity termed ‘chopping’ (14,17). Chopping facilitates autonomous generation of small DNA fragments from dsDNA substrates. Subsequently, these DNA fragments generated during chopping can serve as siDNAs for canonical slicing (14,17).
TtAgo and PfAgo can be programmed with short synthetic siDNA which allows them to target and cleave dsDNA sequences of choice in vitro (13,15). This activity has enabled the repurposing of PfAgo as an universal restriction endonuclease for in vitro molecular cloning (22). In addition, a diagnostic TtAgo-based application termed NAVIGATER (Nucleic Acid enrichment Via DNA Guided Argonaute from T. thermophilus) was developed which enables enhanced detection of rare nucleic acids with single nucleotide precision (23). In analogy with the now commonly used CRISPR-Cas9 and CRISPR-Cas12a enzymes (24–26), it has also been suggested that pAgos could be repurposed as next-generation genome editing tools (27). However, due to the thermophilic nature (optimum activity temperature ≥ 65°C) and low levels of endonuclease activity at the relevant temperatures (20–37°C), it is unlikely that the well-studied TtAgo, PfAgo and MjAgo are suitable for genome editing. The quest for a pAgo that can cleave dsDNA at moderate temperatures has resulted in the characterization of the Argonaute protein from Natronobacterium gregory (NgAgo), which was claimed to be the first pAgo suitable for genome editing purposes (28). However, the study reporting this application has been retracted after a series of reproducibility issues (28–30). Instead, it has been suggested that NgAgo targets RNA rather than DNA (31).
Although considerable efforts have been made to elucidate the mechanisms and biological roles of pAgos, efforts have mainly focused on pAgo variants from (hyper)thermophiles. This has left a large group of mesophilic pAgos unexplored. We here report the characterization of the Argonaute protein from the mesophilic bacterium Clostridium butyricum (CbAgo). We demonstrate that CbAgo can utilize siDNA guides to cleave both ssDNA and dsDNA targets at moderate temperatures (37°C). In addition, we have elucidated the macromolecular structure of CbAgo in complex with a siDNA guide and complementary ssDNA target in a catalytically competent state. CbAgo displays an unusual preference for siDNAs with a deoxyadenosine at the 5′-end and thymidines at nucleotides 2–4. The programmable DNA endonuclease activity of CbAgo provides a foundation for the development of pAgo-based applications at moderate temperatures.
MATERIALS AND METHODS
Plasmid construction
The CbAgo gene was codon harmonized for Escherichia coli Bl21 (DE3) and inserted into a pET-His6 MBP TEV cloning vector (obtained from the UC Berkeley MacroLab, Addgene #29656) using ligation independent cloning (LIC) using oligonucleotides oDS067 and oDS068 (Supplementary Table S1) to generate a protein expression construct that encodes the CbAgo polypeptide sequence fused to an N-terminal tag comprising a hexahistidine sequence, a maltose binding protein (MBP) and a Tobacco Etch Virus (TEV) protease cleavage site.
Generation of the double mutant
CbAgo double mutant (D541A, D611A) was generated using an adapted Quick Directed Mutagenesis Kit instruction manual (Stratagene). The primers were designed using the web-based program primerX (http://bioinformatics.org/primerx).
CbAgo expression and purification
The CbAgo WT and DM proteins were expressed in E. coli Bl21(DE3) Rosetta™ 2 (Novagen). Cultures were grown at 37°C in LB medium containing 50 μg ml–1 kanamycin and 34 μg ml–1 chloramphenicol until an OD600 nm of 0.7 was reached. CbAgo expression was induced by addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. During the expression cells were incubated at 18°C for 16 h with continuous shaking. Cells were harvested by centrifugation and lysed by sonication (Bandelin, Sonopuls. 30% power, 1 s on/2 s off for 5 min) in lysis buffer containing 20 mM Tris–HCl pH 7.5, 250 mM NaCl, 5 mM imidazole, supplemented with a EDTA free protease inhibitor cocktail tablet (Roche). The soluble fraction of the lysate was loaded on a nickel column (HisTrap Hp, GE Healthcare). The column was extensively washed with wash buffer containing 20 mM Tris–HCl pH 7.5, 250 mM NaCl and 30 mM imidazole. Bound protein was eluted by increasing the concentration of imidazole in the wash buffer to 250 mM. The eluted protein was dialysed at 4°C overnight against 20 mM HEPES pH 7.5, 250 mM KCl and 1 mM dithiothreitol (DTT) in the presence of 1 mg TEV protease (32) to cleave of the His6-MBP tag. Next, the cleaved protein was diluted in 20 mM HEPES pH 7.5 to lower the final salt concentration to 125 mM KCl. The diluted protein was applied to a heparin column (HiTrap Heparin HP, GE Healthcare), washed with 20 mM HEPES pH 7.5, 125 mM KCl and eluted with a linear gradient of 0.125–2 M KCl. Next, the eluted protein was loaded onto a size exclusion column (Superdex 200 16/600 column, GE Healthcare) and eluted with 20 mM HEPES pH 7.5, 500 mM KCl and 1 mM DTT. Purified CbAgo protein was diluted in size exclusion buffer to a final concentration of 5 μM. Aliquots were flash frozen in liquid nitrogen and stored at −80°C.
Co-purification nucleic acids
To 500 pmol of purified CbAgo in SEC buffer CaCl2 and proteinase K (Ambion) were added to final concentrations of 5 mM CaCl2 and 250 μg/ml proteinase K. The sample was incubated for 4 h at 65°C. The nucleic acids were separated from the organic fraction by adding Roti phenol/chloroform/isoamyl alcohol pH 7.5–8.0 in a 1:1 ratio. The top layer was isolated and nucleic acids were precipitated using ethanol precipitation by adding 99% ethanol in a 1:2 ratio supplied with 0.5% linear polymerized acrylamide as a carrier. This mixture was incubated overnight at −20°C and centrifuged in a table centrifuge at 16 000 g for 30 min. Next, the nucleic acids pellet was washed with 70% ethanol and solved in 50 μl MilliQ water. The purified nucleic acids were treated with either 100 μg/ml RNase A (Thermo), 2 units DNase I (NEB) or both for 1 h at 37°C and resolved on a denaturing urea polyacrylamide gel (15%) and stained with SYBR gold.
Single stranded activity assays
Unless stated otherwise 5 pmol of each CbAgo, siDNA and target were mixed in a ratio of 1:1:1, in 2× reaction buffer containing 20 mM Tris–HCl pH 7.5 supplemented with 500 μM MnCl2. The target was added after the CbAgo and siDNA had been incubated for 15 min at 37°C. Then the complete reaction mixture was incubated for 1 h at 37°C. The reaction was terminated by adding 2× RNA loading dye (95% formamide, 0.025% bromophenol blue, 5 mM ETDA) and heating it for 5 min at 95°C. After this, the samples were resolved on a 20% denaturing (7 M urea) polyacrylamide gel. The gel was stained with SYBR gold nucleic acid stain (Invitrogen) and imaged using a G:BOX Chemi imager (Syngene).
Double stranded activity assay
In two half reactions 12.5 pmol of CbAgo was loaded with either 12.5 pmol of forward or reverse siDNA in reaction buffer containing 10 mM Tris–HCl pH 7.5, 10 μg/ml BSA and 250 μM MnCl2. The half reactions were incubated for 15 min at 37°C. Next, both half reactions were mixed together and 120 ng target plasmid was added after which the mixture was incubated for 1 h of 37°C. After the incubation the target plasmid was purified from the mixture using a DNA clean and concentrate kit (DNA Clean & Concentrator™-5, Zymogen) via the supplied protocol. The purified plasmid was subsequently cut using either EcoRI-HF (NEB) or SapI-HF (NEB) in Cutsmart buffer (NEB) for 30 min at 37°C. A 6× DNA loading dye (NEB) was added to the plasmid sample prior to resolving it on a 0.7% agarose gel stained with SYBR gold (Invitrogen). In the double stranded assays in which linear plasmid was targeted, 300 ng pUC IDT, linearized by EcoRI, was added to both half reactions and incubated for 1 h of 37°C. The reaction products were resolved on a 0.7% agarose gel.
Crystallization
To reconstitute the CbAgo DM–siDNA–target DNA complex, siDNA and target DNA were pre-mixed at a 1:1 ratio, heated to 95°C, and slowly cooled to room temperature. The formed dsDNA duplex (0.5 M) was mixed with CbAgo DM in SEC buffer at a 1:1:4 ratio (CbAgo DM:duplex DNA), and MgCl2 was added to a final concentration of 5 mM. The sample was incubated for 15 min at 20°C to allow complex formation. The complex was crystallized at 20°C using the hanging drop vapour diffusion method by mixing equal volumes of complex and reservoir solution. Initial crystals were obtained at a CbAgo DM concentration of 5 mg/ml with a reservoir solution consisting of 4 M sodium formate. Data was collected from crystals grown obtained using a complex concentration of 4.3 mg/ml and reservoir solution containing 3.8 M Sodium Formate and 5 mM NiCl2 at 20°C. For cryoprotection, crystals were transferred to a drop of reservoir solution and flash-cooled in liquid nitrogen.
X-ray diffraction data were measured at beamline X06DA (PXIII) of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). Data were indexed, integrated and scaled using AutoPROC (33). Crystals of the CbAgo–siDNA–target DNA complex diffracted to a resolution of 3.55 Å and belonged to space group P63 2 2, with one copy of the complex in the asymmetric unit. The structure was solved by molecular replacement using Phaser-MR (34). As search model, the structure of TtAgo in complex with guide and target DNA strands (PDB: 5GQ9) was used after removing loops and truncating amino acid side chains. Phases obtained using the initial molecular replacement solution were improved by density modification using phenix.resolve (35) and phenix.morph_model (36). The atomic model was built manually in Coot (37) and refined using phenix.refine (37). The final binary complex model contains CbAgo residues 1–463 and 466–748, guide DNA residues 1–16, and target DNA residues (−18)–(−1).
Structure analysis
Core root means square deviations (rmsd) of structure alignments were calculated using Coot SSM superpose (38). Hydrogen bonding interactions were analysed using PDBePISA (39), while other interactions with the Mg2+ ion were determined manually in Coot (37). Figures were generated using PyMOL (Schrödinger).
Single-molecule experimental set-up
Single-molecule fluorescence FRET measurements were performed with a prism-type total internal reflection fluorescence microscope. Cy3 and Cy5 molecules were excited with 532 and 637 nm wavelength, respectively. Resulting Cy3 and Cy5 fluorescence signal was collected through a 60× water immersion objective (UplanSApo, Olympus) with an inverted microscope (IX73, Olympus) and split by a dichroic mirror (635dcxr, Chroma). Scattered laser light was blocked out by a triple notch filter (NF01-488/532/635, Semrock). The Cy3 and Cy5 signals were recorded using a EM-CCD camera (iXon Ultra, DU-897U-CS0-#BV, Andor Technology) with exposure time 0.1 s. All single-molecule experiments were done at room temperature (22 ± 2°C).
Fluorescent DNA and RNA preparation
The RNAs with amine-modification (amino-modifier C6-U phosphoramidite, 10-3039, Glen Research) were purchased from STPharm (South Korea) and DNAs with amine-modification (internal amino modifier iAmMC6T) Ella biotech (Germany). The guide and target strands were labeled with donor (Cy3) and acceptor (Cy5), respectively, using the NHS-ester form of Cy dyes (GE Healthcare). 1 μl of 1 mM of DNA/RNA dissolved in MilliQ H2O was added to 5 μl labeling buffer of (freshly prepared) sodium tetraborate (380 mg/10ml, pH 8.5). 1 μl of 20 mM dye (1 mg in 56 μl DMSO) was added and incubated overnight at room temperature in the dark, followed by washing and ethanol precipitation. The labeling efficiency was ∼100%.
Single-molecule sample preparation
A microfluidic chamber was incubated with 20 μl Streptavidin (0.1 mg/ml, Sigma) for 30 s. Unbound Streptavidin was washed with 100 μl of buffer T50 (10 mM Tris–HCl pH 8.0, 50 mM NaCl buffer). The 50 μl of 50 pM acceptor-labeled target construct were introduced into the chamber and incubated for 1 min. Unbound labeled constructs were washed with 100 μl of buffer T50. The CbAgo binary complex was formed by incubating 10 nM purified CbAgo with 1 nM of donor-labeled guide in a buffer containing 50 mM Tris–HCl pH 8.0 (Ambion), 1 mM MnCl2 and 100 mM NaCl (Ambion) at 37°C for 20 min. For binding rate (kon) measurements, the binary complex was introduced into the fluidics chamber using syringe during the measurement. The experiments were performed at the room temperature (23 ± 1°C).
For fluorescence Guide Loading Experiments before immobilizing CbAgo on the single-molecule surface, 1 μl of 5 μM His-tagged apo-CbAgo was incubated with 1 μl of 1 μg/ml biotinylated anti-6x His antibody (Abcam) for 10 min. Afterward, the mixture was diluted 500× in T50 and 50 μl were loaded in the microfluidic channel for 30 s incubation, followed by washing with 100 μl of T50 buffer. Cy3-labeled ssDNA (0.1 nM) was applied to the microfluidic chamber in imaging buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM MnCl2, 1 mM Trolox ((±)-6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid), supplemented with an oxygen-scavenging system (0.5 mg/ml glucose oxidase (Sigma), 85 mg/ml catalase (Merck) and 0.8% (v/v) glucose (Sigma)).
Single-molecule data acquisition and analysis
CCD images of time resolution 0.1 or 0.3 s were recorded, and time traces were extracted from the CCD image series using IDL (ITT Visual Information Solution). Co-localization between Cy3 and Cy5 signals was carried out with a custom-made mapping algorithm written in IDL. The extracted time traces were processed using Matlab (MathWorks) and Origin (Origin Lab).
The binding rate (kon) was determined by first measuring the time between when CbAgo binary complex was introduced to a microfluidic chamber and when the first CbAgo-guide docked to a target; and then fitting the time distribution with a single-exponential growth curve, 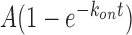 . The dissociation rate was estimated by measuring the dwell time of a binding event. A dwell time distribution was fitted by single-exponential decay curve (
. The dissociation rate was estimated by measuring the dwell time of a binding event. A dwell time distribution was fitted by single-exponential decay curve (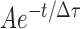 ).
).
Fluorescence competition experiments
MBP-tagged CbAgo was immobilized on the quartz surface using an anti-MBP antibody. An equimolar mixture of let7 DNA guide (Cy3 labeled) and let7 RNA guide (Cy5 labeled) in imaging buffer was introduced to the microfluidic chamber. After 5 min, 10 snapshots of independent fields of view with simultaneous illumination were collected to estimate the amount of guide molecules bound to protein. Movies were taken for 200 s (2000 frames) at continuous illumination of Cy3 and Cy5 molecules to determine the dwell times of the binding events. Dwell times were binned in a histogram and fitted with a single exponential decay curve.
FRET targeting experiments of ATTT and AAAA guide target combinations
100 pM of target construct annealed with biotin handle were flushed in the microfluidic chamber. After incubation of 1 min, the microfluidic chamber was rinsed with 100 μl T50 buffer. 10 nM of apo-CbAgo was loaded with 1 nM of ATTT seed DNA guide or with AAAA seed DNA guide at 37°C for 30 min in imaging buffer after which the mixture is introduced inside the microfluidic chamber. Movies of 200 s were taken at continuous illumination of the Cy3 signal. Site specific protein target interactions were identified as FRET signals and were further analyzed.
RESULTS
CbAgo mediates siDNA-guided ssDNA cleavage
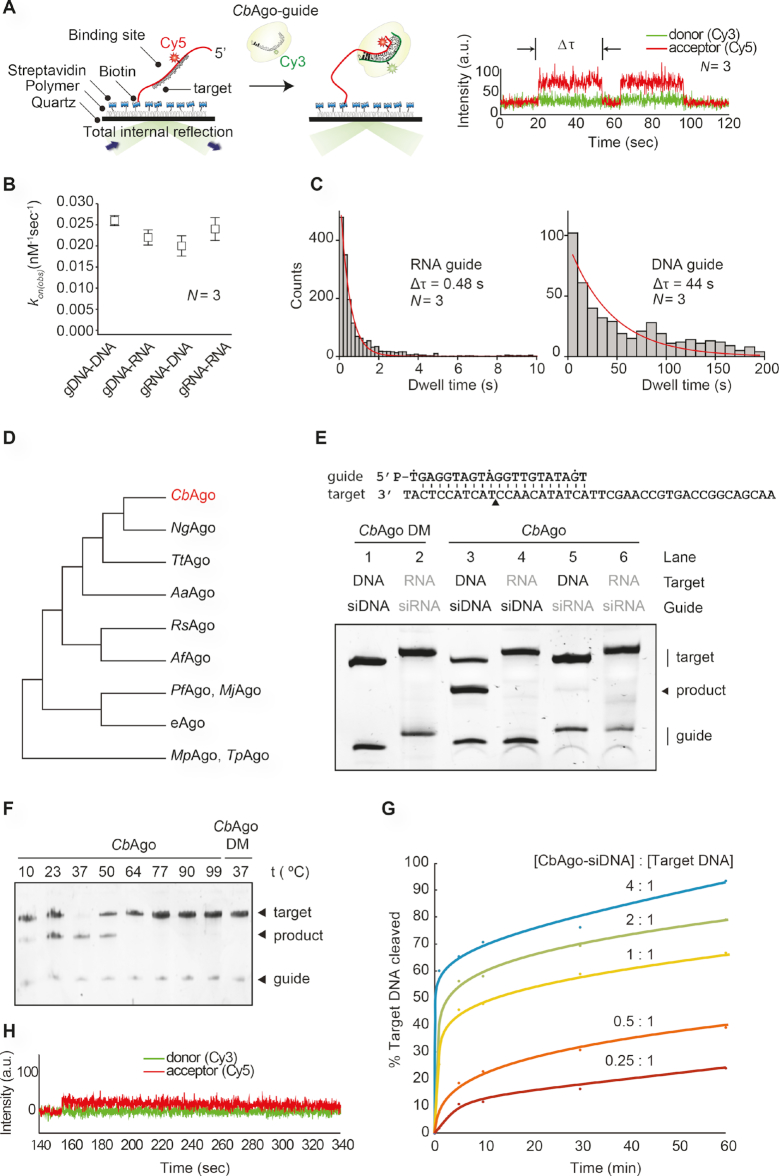
CbAgo was successfully expressed in E. coli from a codon-optimized gene using a T7-based pET expression system and purified (Supplementary Figure S1A). To determine the guide and target binding characteristics of CbAgo, we performed single-molecule experiments using Förster resonance energy transfer (FRET). We immobilized either Cy5-labeled single stranded RNA or DNA targets (FRET acceptor) on a polymer-coated quartz surface (Figure 1A). Next, we introduced CbAgo in complex with either a Cy3-labeled siRNA or siDNA guide (FRET donor) and recorded the interactions. Strikingly, CbAgo could utilize both siRNAs and siDNAs to bind DNA or RNA targets (Figure 1B). To test which guide is preferentially bound by CbAgo we performed a competition assay in which CbAgo was immobilized into the microfluidic chamber, and an equimolar mixture of siDNA and siRNAs was introduced. While only short-lived interactions (average dwell time: 0.48 s) were observed for siRNA, siDNA was strongly bound (average dwell time: 44 s) by CbAgo (Figure 1C). This results suggests that CbAgo utilizes siDNA rather than siRNA as a guide.
Figure 1.
CbAgo exhibits DNA-guided DNA endonuclease activity at 37°C. (A) Left: Overview of the single molecule assay to determine the binding characteristics of CbAgo. Right: FRET diagram of a CbAgo–siDNA complex that has three complementary base pairs (2–4nt) to the DNA target. Indicated is the dwell time (Δτ). (B) Comparison of the binding rates (kon) of CbAgo in complex with siDNA or siRNA to bind DNA or RNA targets. The rates are similar for each nucleic acid type guide and target. N is the number of base paired nucleotides. (C) Dwell time histograms showing CbAgo preferentially binds siDNAs in siDNA-siRNA competition experiments. (D) Schematic phylogenetic tree of characterized pAgos. (E) CbAgo exhibits DNA-guided DNA endonuclease activity. Upper panel: Sequence of the synthetic let7 miRNA-based siDNA guide and target DNA sequences that were used for the in vitro cleavage assays. Lower panel: CbAgo, guides and targets were mixed in a 1:1:1 molar ratio and incubated for 1 h at 37°C. Catalytic mutant CbAgoDM was used as a control. Cleavage products were analysed by denaturing polyacrylamide electrophoresis. (F) CbAgo displays highest activity at 37°C. CbAgo and siDNA were mixed and pre-incubated at various temperatures for 10 min. Next, target DNA was added and the sample was incubated for 1 h at the same temperature. CbAgoDM was used as a control. Cleavage products were analysed by denaturing polyacrylamide electrophoresis. (G) Quantified data of a CbAgo-mediated siDNA-guided ssDNA cleavage turnover experiment using 5 pmol target DNA and increasing concentrations of CbAgo-siDNA (1.25–20 pmol). (H) FRET diagram showing that a cleavage compatible CbAgo-siDNA remains bound to a fully complementary target DNA (N = 21) during the entire the measurement (340 seconds).
CbAgo is phylogenetically closest related to the clade of halobacterial pAgos, among which also pAgo from Natronobacterium gregoryi (NgAgo) can be found (Figure 1D and Supplementary Figure S2). A multiple sequence alignment of CbAgo with other pAgos (Supplementary Figure S1B) suggests that CbAgo contains the conserved DEDX catalytic residues (where X can be a D, H or N) which are essential for nuclease activity in ‘slicing’ Agos (40). In the case of CbAgo, this concerns residues D541, E577, D611 and D727.
To confirm whether CbAgo indeed is an active nuclease, we performed in vitro activity assays in which CbAgo was loaded with either synthetic siDNAs or siRNAs (21 nucleotides in length). Next the complexes were incubated at 37°C with 45-nucleotide complementary single stranded RNA or DNA target oligonucleotides. While no activity was found in any of the combinations in which siRNAs or target RNAs were used, CbAgo was able to cleave target DNAs in a siDNA-dependent manner (Figure 1E). In agreement with the predicted DEDD catalytic site (Supplementary Figure S1B), alanine substitutions of two of aspartic acids (D541A, D611A) in the expected catalytic tetrad abolished the nuclease activity, demonstrating that the observed siDNA-guided ssDNA endonucleolytic activity was indeed catalyzed by the DEDD catalytic site. To further investigate the full temperature range at which CbAgo is active, we performed additional cleavage assays at temperatures ranging from 10 to 95°C. While CbAgo displayed the highest activity at its physiologically relevant temperature (37°C), CbAgo also catalyzed siDNA-guided target DNA cleavage at temperatures as low as 10°C and as high as 50°C (Figure 1F).
When CbAgo–siDNA complexes and target ssDNA substrates (45 nt) were mixed in equimolar amounts, cleavage of the target DNA was not complete after one hour incubation (Figure 1E). Therefore, we investigated the substrate turnover kinetics of CbAgo by monitoring the cleavage assays in a time course using variable CbAgo:siDNA:target DNA ratios (Figure 1G). A rapid burst of activity was observed during the first minute, likely indicating the first target binding and cleavage event. This stage was followed by a slow steady state, suggesting that under these conditions the CbAgo–siDNA complex slowly dissociates from the cleaved target DNA product before being able to bind and cleave a new target DNA strand. The cleavage kinetics were confirmed using single-molecule assays which demonstrated that the CbAgo-siDNA complex remains bound to the DNA target (N = 21) for several minutes (Figure 1H), which prevents CbAgo–siDNA complexes from binding and cleaving new DNA targets. Thus, while CbAgo functions as a multi-turnover nuclease enzyme, its steady-state rate is limited by product release.
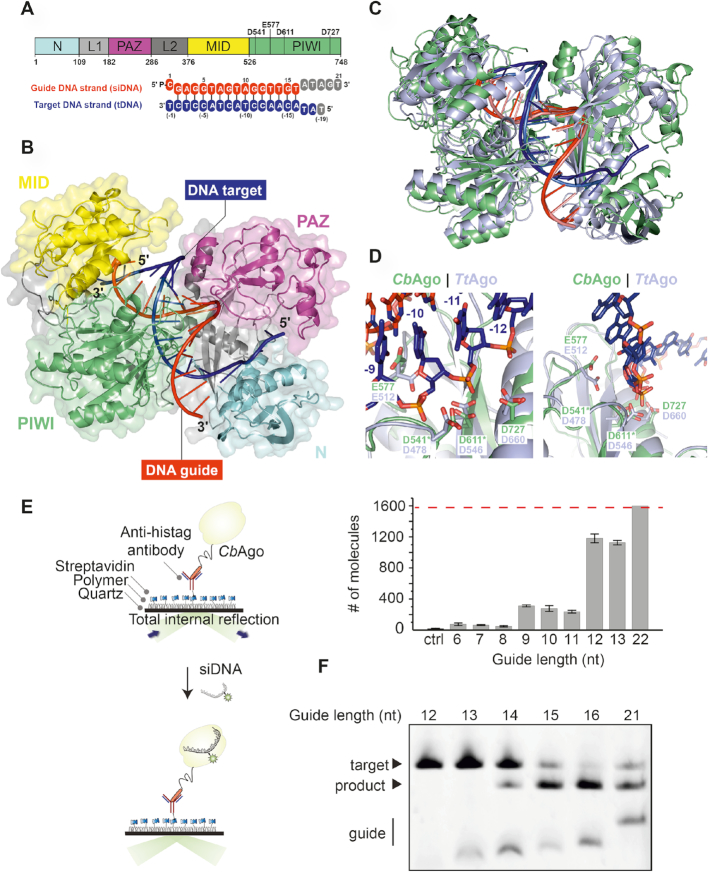
Structure of CbAgo in the cleavage-competent conformation
To investigate the molecular architecture of CbAgo in light of its biochemical activity, we crystallized CbAgoDM in complex with both a 21 nt siDNA and a 19 nt DNA target, and solved the structure of the complex at 3.54 Å resolution (Figure 2 and Supplementary Table S1). Like other Agos, CbAgo adopts a bilobed conformation in which one of the lobes comprises the N-terminal, linker L1, and PAZ domains, which are linked by linker L2 to the other lobe comprising the MID and PIWI domains. Nucleotides 2–16 of the siDNA constitute a 15 base-pairs A-form-like duplex with the target DNA (Figure 2A). The 5′-terminal nucleotide of the siDNA is anchored in the MID domain pocket, where the 5′-phosphate group of the siDNA makes numerous interactions with MID domain residues and the C-terminal carboxyl group of CbAgo (Supplementary Figure S3), as observed for in structures of TtAgo and MjAgo bound to siDNAs (12,19,41). To test whether the interactions with the 5′-phosphate group of the siDNA are important for CbAgo activity, we performed target DNA cleavage assays in which we used siDNAs with a 5′ phosphate or a 5′ hydroxyl group (Supplementary Figure S4). As observed for other pAgos (42,43), CbAgo is able to utilize both siDNAs for target DNA cleavage, but it cleaves target DNA much more efficiently when the siDNA contains a 5′-phosphate group. This is in agreement with the siDNA–protein interactions predicted form the crystal structure. Furthermore, the backbone phosphates of the siDNA seed segment appear to form hydrogen-bonding and ionic interactions with specific residues in the MID, PIWI and L1 domains (Supplementary Figure S3). At the distal end of the siDNA-target DNA duplex, the N-domain residue His35 caps the duplex by stacking onto the last base pair. After this point, the remaining 3′-terminal nucleotides of the siDNA are disordered, while the target DNA bends away from the duplex and enters the cleft between the N-terminal and PAZ domains. In agreement with other ternary pAgo complexes (18,44,45), the PAZ domain pocket, which normally binds the 3′ end of the guide in a binary Ago-guide complex, is empty.
Figure 2.
Structure of CbAgo in complex with a siDNA and a DNA target. (A) Upper panel: Schematic diagram of the domain organization of CbAgo. L1 and L2 are linker domains. Lower panel: Sequences of the siDNA (red) and target DNA (blue). Nucleotides that are unordered in the structure are coloured grey. See also Supplementary Table S1. (B) Overall structure of the CbAgo-siDNA-target DNA complex. Domains are coloured according to the colour scheme in panel A. (C) Structural alignment of CbAgo (green) and TtAgo (light purple; PDB: 4NCB). Core Root Mean Square Deviation of 3.0 Å over 563 residues. (D) Close-up views of the aligned DDED catalytic sites of CbAgo (green) and TtAgo (light purple; PDB: 4NCB). Modelled side chains of D541 and D611 in CbAgo are indicated with green asterisks. The glutamate finger of both pAgos (E512 in TtAgo or E577 in CbAgo) are inserted into the catalytic site. The scissile phosphate between nucleotide −10 and −11 of the target DNA strand (blue) is indicated with a black asterisk in the left panel. (E) Total internal reflection microscopy (TIRM) was used to determine the minimal length for siDNA to be bound by CbAgo. Left panel: Graphical overview of the TIRM method. Right panel: Histogram with TIRM results demonstrated that synthetic siDNAs of at least 12 nt in length are efficiently bound by CbAgo. The red line indicates the total number of countable molecules within the microscope image. The raw microscope images are given in Supplementary Figure S5. (F) CbAgo mediates target DNA cleavage with siDNAs as short as 14 nucleotides. CbAgo was incubated with siDNA and target DNA in a 1:1:1 ratio. Cleavage products were analyzed by denaturing polyacrylamide electrophoresis.
CbAgo is phylogenetically closely related to TtAgo (Figure 1D). However, CbAgo is 63 amino acids (9.2%) longer than TtAgo (748 amino acids versus 685 amino acids) and CbAgo and TtAgo share only 23% sequence identity. Superposition of the CbAgo complex structure with the structure of TtAgo bound to a siDNA and DNA target (PDB: 4NCB) (Figure 2C) reveals that the macromolecular architecture and conformation of these TtAgo and CbAgo structures are highly similar (Core root mean square deviation of 3.0 Å over 563 residues). In the TtAgo structure, which is thought to represent a catalytically competent state, a ‘glutamate finger’ side chain (Glu512TtAgo) is inserted into the catalytic site completing the catalytic DDED tetrad (44). Similarly, the corresponding residue in CbAgo (Glu577) is located within a flexible loop and is positioned near the other catalytic residues (Figure 2D; Asp541, Asp611 and Asp727). All pAgos and eAgos characterized to date cleave the target strand in between nucleotide 10 and 11 of the target strand. In line with the consensus, the catalytic residues of CbAgo perfectly align with the scissile phosphate linking these nucleotides in our structure (Figure 2D). This observation implies that this structure represents the cleavage competent conformation of CbAgo.
Only 15 siDNA–target DNA base pairs are formed in the complex, which suggests that additional siDNA-target DNA binding is not essential for target DNA cleavage. To determine the minimum siDNA length that CbAgo requires for target binding, we performed single-molecule fluorescence assays. First, CbAgo was immobilized on a surface and next it was incubated with 5′-phosphorylated Cy3-labeled siDNAs (Figure 2E). These assays demonstrate that CbAgo can bind siDNAs with a minimal length of 12 nucleotides. Next, we determined the minimum siDNA length for CbAgo-siDNA mediated target DNA cleavage (Figure 2F). In line with the observation that the CbAgo adopts a cleavage-competent confirmation when only 14 base pairs are formed, CbAgo can cleave target DNAs when programmed with siDNAs as short as 14 nt (forming 13 siDNA-target DNA base pairs) under the tested conditions. This resembles the activity of PfAgo, MjAgo and MpAgo, which require siDNAs with a minimal length of 15 nt to catalyze target DNA cleavage (14,15,43). Only TtAgo has been reported to mediate target DNA cleavage with siDNAs as short as 9 nt (12).
CbAgo associates with plasmid-derived siDNAs in vivo
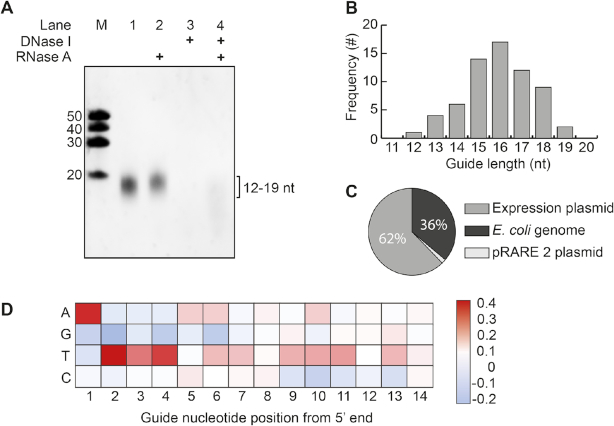
It has previously been demonstrated that certain pAgos co-purify with their guides and/or targets during heterologous expression in Escherichia coli (13,16). To determine whether CbAgo also acquires siDNAs during expression, we isolated and analyzed the nucleic acid fraction that co-purified with CbAgo. Denaturing polyacrylamide gel electrophoresis revealed that CbAgo co-purified with small nucleotides with a length of ∼12–19 nucleotides (Figure 3A). These nucleic acids were susceptible to DNase I but not to RNase A treatment, indicating that CbAgo acquires 12–19 nucleotide long siDNAs in vivo, which fits with its observed binding and cleavage activities in vitro (Figures 1 and 2).
Figure 3.
CbAgo associates with small plasmid derived siDNA in vivo. (A) Nucleic acids that co-purified with CbAgo were treated with either RNAse A, DNAse I or both, and were analyzed by denaturing polyacrylamide gel electrophoresis. (B) Histogram displaying the length of DNA co-purified with CbAgo as determined by sequencing. (C) Sequenced nucleic acids that co-purified with CbAgo are mostly complementary to the CbAgo expression plasmid. (D) Heat map showing the base preference of the co-purified nucleic acids at each position. The red squares indicate bases that were more often found compared to a random distribution (25%); blue squares indicate bases that were less frequently found.
We cloned and sequenced the siDNAs that co-purified with CbAgo to determine their exact length and sequence. The majority of the siDNAs had a length of 16 nucleotides and are complementary to the plasmid used for expression of CbAgo (Figure 3B and C). Likewise the siRNAs and siDNAs that co-purify with respectively Rhodobacter sphaeroides (RsAgo) and TtAgo are also mostly complementary to their expression plasmids (13,16). As both TtAgo and RsAgo have been demonstrated to interfere with plasmid DNA, this suggests that also CbAgo might play a role in protecting its host against invading DNA. However, no significant reduction of plasmid content could be detected during or upon expression of CbAgo in E. coli (Supplementary Figure S6). We also investigated whether CbAgo co-purified with nucleic acids that were enriched for certain motifs. Sequence analysis revealed that most siDNAs co-purified with CbAgo contain a deoxyadenosine at their 5′ ends (Figure 3D). In addition, we observed an enrichment of thymidine nucleotides in the three positions directly downstream of the siDNA 5′ end (nt 2–4) (Figure 3D).
The sequence of the siDNA affects CbAgo activity
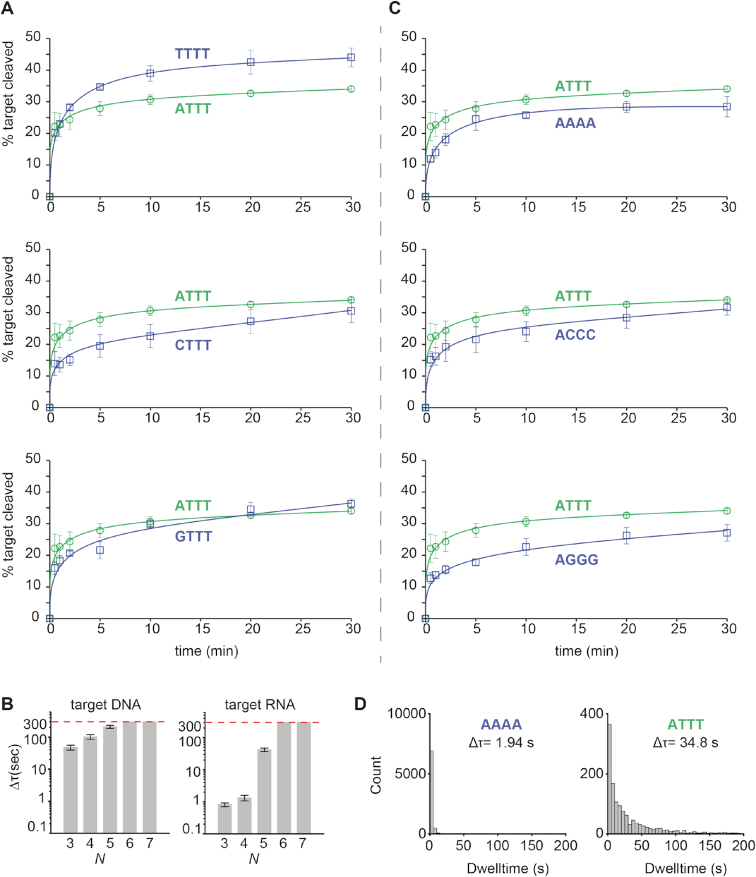
To investigate if the 5′-terminal nucleotide of the siDNA affects the activity of CbAgo, we performed cleavage assays. CbAgo was loaded with siDNA guides with varied nucleotides at position 1 (g1N) and incubated with complementary target DNAs (Figure 4A). Surprisingly, the highest cleavage rates were observed when CbAgo was loaded with siDNAs containing a 5′-T, followed by siDNAs containing 5′-A. CbAgo bound 5′-G or 5′-C siDNAs displayed slightly lower initial cleavage rates. Also for other pAgos the g1N preference observed in vivo is not reflected in the in vitro activities; TtAgo (which preferentially co-purifies with g1C siDNAs) as well as PfAgo and MpAgo (of which the in vivo g1N preferences are unknown) demonstrate no clear preference for a specific g1N during in vitro cleavage reactions (13,17,43). Instead, the preference of TtAgo for 5′-C siDNAs is determined by specific recognition of a guanosine nucleotide in the corresponding position t(–1) in the target DNA (17). Indeed, TtAgo structures and models have revealed base-specific interactions with target strand guanine, while base-specific interactions with the 5′-terminal cytidine in the siDNA are less obvious (17). Similarly, we predict no obvious base-specific interactions with the 5′-terminal cytidine in the structure of the CbAgo complex (Supplementary Figure S7). When we investigated potential base-specific interactions with the base at the opposing target strand t(-1) position, we observed that the t(-1) thymine base is not placed in the t(-1) binding pocket as has been observed in TtAgo, RsAgo and hAGO2 (17,46,47). Instead, the thymine bases is flipped and stacks on Phe557 that also caps the siDNA-target DNA duplex (Supplementary Figure S7). At present, we are unable to rationalize the preferential co-purification of 5′-adenosine siDNAs with CbAgo.
Figure 4.
The siDNA sequence affects CbAgo activity. (A) CbAgo has no strong 5′-end nucleotide preference. CbAgo was incubated with siDNA with varied 5′-end and incubated with complementary DNA targets. Cleavage products were analysed by denaturing polyacrylamide electrophoresis and quantified. Graphs display the amount of target DNA cleaved. Error bars indicate the standard variation of three independent experiments. (B) Histograms displaying dwell time of CbAgoDM-siDNA complexes binding either DNA or RNA targets with a varied sequence complementarity (N = number of complementary nucleotides between the siDNA and the target, starting at nt 2. Thus N 3 = nt 2–4) The photobleaching limit is reached where the signal is deactivated (300s). (C) CbAgo preferentially utilizes siDNAs with thymidines at position 2–4. CbAgo-siDNA complexes with siDNA in which nt 2–4 varied were incubated with complementary DNA targets. Cleavage products were analysed by denaturing polyacrylamide electrophoresis and quantified. Graphs display the amount of target DNA cleaved. Error bars indicate the standard variation of three independent experiments. (D) Histograms displaying dwell time of CbAgoDM in complex with a 5′-ATTT siDNA or 5′-AAAA siDNA binding to a target DNA. Interactions of CbAgo are on average ∼18-fold longer with the siDNA containing a 5′-ATTT motif compared to interactions with siDNAs containing a 5′-AAAA motif.
In order to characterize the seed segment of CbAgo, and to test whether the seed length changes depending on the nature of the guide and the target (i.e. DNA versus RNA), we performed additional single-molecule binding assays. The length of seed was determined based on the minimal number of complementary nucleotide pairs between guide and target that were required to achieve a stable binding event. When only nt 2–4 of the siDNA are complementary to the DNA and RNA targets, CbAgo–siDNA complexes bound to the DNA target with an average dwell time 58-fold longer compared to RNA target-binding (Figure 4B). When nt 2–7 of the siDNA were complementary to the target, the CbAgo–siDNA complex stably bound to both to target DNA and RNA beyond our observation time of 300 s. This suggests CbAgo prefers DNA targets above RNA targets and that the seed segment of the siDNAs bound by CbAgo comprises nt 2–7.
Next, we set out to investigate whether CbAgo displays a preference for siDNAs with thymidines at nt 2–4 in vitro, similar to the observed sequence preference for siDNAs that co-purified with CbAgo in vivo. CbAgo was incubated with siDNAs in which nt 2–4 were varied and complementary target DNAs were added. In contrast to the 5′-base preference, the preference for thymidines in the nt 2–4 segment that we observed in vivo is also reflected in vitro: CbAgo displays the highest target cleavage rates when programmed with siDNAs containing thymidines at nt 2–4 (Figure 4C). To confirm these findings, we performed single-molecule assays in which we compared the target binding properties of CbAgo-siDNA complexes containing siDNAs with either thymidines or a deoxyadenosines at position 2–4. These assays demonstrate that the dwell time of CbAgo loaded with siDNA containing thymidines at nt 2–4 on a target was 18-fold longer compared to CbAgo loaded with siDNA containing deoxyadenosines at nt 2–4 (Figure 4D). Combined, these data indicate that CbAgo displays a preference for siDNAs containing thymidines at position 2–4.
A pair of CbAgo–siDNA complexes can cleave double stranded DNA
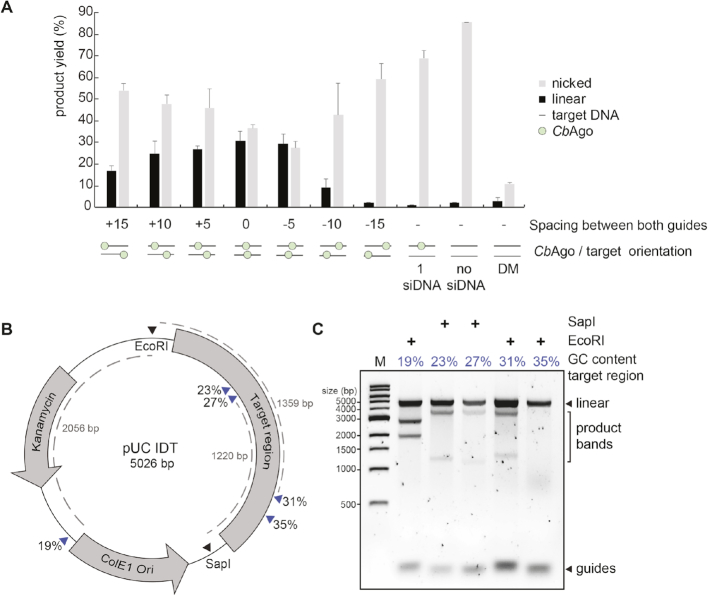
Thermophilic pAgos have successfully been used to generate double stranded DNA breaks in plasmid DNA (13,15). As each pAgo–siDNA complex targets and cleaves a single strand of DNA only, two individual pAgo–siDNA complexes are required for dsDNA cleavage, each targeting another strand of the target dsDNA. Although all pAgos characterized so far appear to lack the ability to actively unwind or displace a dsDNA duplex substrate, it has been proposed that, at least in vitro, thermophilic pAgos rely on elevated temperatures (>65°C) to facilitate local melting of the dsDNA targets to target each strand of the DNA individually. However, CbAgo is derived from a mesophilic organism and we therefore hypothesize that it is able to mediate protection against invading DNA at moderate temperatures (37°C). To test if CbAgo can indeed cleave dsDNA targets at 37°C, we incubated apo-CbAgo and pre-assembled CbAgo-siDNA complexes with a target plasmid. Previous studies showed that the ‘chopping’ activity of siDNA-free apo-TtAgo and apo-MjAgo can result in plasmid linearization or degradation, respectively (14,17). We observed that apo-CbAgo converted the plasmid substrate from a supercoiled to open-circular state, possibly by nicking one of the strands, but did not observe significant linearization or degradation of the plasmid DNA (Figure 5A). When the plasmid was targeted by CbAgo loaded with a single siDNA, we also observed loss of supercoiling (Figure 5A). As this activity was not observed with nuclease-deficient CbAgoDM, we conclude that apo–CbAgo and CbAgo–siDNA complexes are generating nicks in dsDNA plasmid targets with their DEDD catalytic site. When using two CbAgo–siDNA complexes, each targeting one strand of the plasmid, we observed that a fraction of the target plasmid DNA becomes linearized (Figure 5A). This implies that CbAgo-siDNA complex-mediated nicking of each of the target plasmid DNA strands resulted in the generation of a double stranded DNA break. Next, we investigated if the spacing between the two siDNAs affects the ability of CbAgo to cleave the plasmid. The most efficient plasmid linearization was achieved when the siDNAs were orientated exactly or almost opposite to each other (Figure 5A).
Figure 5.
Double stranded plasmid DNA cleavage by CbAgo. (A) Two CbAgo–siDNA complexes can generate double stranded DNA breaks in plasmid DNA. CbAgo–siDNA complexes were pre-assembled and incubated with target plasmid DNA. Cleavage products were analysed by agarose gel electrophoresis (Supplementary Figure S8B) and quantified. The spacing between both CbAgo–siDNA target sites affects the linearization efficiency (nucleotide spacing between the predicted cleavage sites: +15 nt, +10 nt, +5 nt, 0 nt, −5 nt, −10 nt, −15 nt, a single siDNA, no siDNA). With 0 nt spacing, both CbAgo–siDNA complexes are exactly on top of each other. (B) Schematic overview of the pUC IDT target plasmid. Blue arrows indicate target sites while percentages indicate the GC-content of the 100 bp segments in which these target sites are located. (C) Pre-assembled CbAgo-siDNA complexes targeting various pUC IDT segments were incubated with pUC IDT. Cleavage products were incubated with EcoRI or SapI and were further analysed by agarose gel electrophoresis. The GC-content of the segments in which the target sites were located are indicated by the percentage (in blue).
Finally, we investigated whether the GC-content of the target DNA plays a role during DNA targeting by CbAgo. For TtAgo, it has been observed that AT-rich DNA is cleaved more efficiently than GC-rich DNA (17), possibly because AT-rich DNA is more prone to unwinding. To test if such preference also exists for CbAgo, we designed a target plasmid containing 16 gene fragments of 100 bp complementary to sequences from the human genome, with an increasing GC content (Figure 5B). The target plasmid was digested with a restriction enzyme (SapI or EcoRI) either before or after the incubation with two CbAgo–siDNA complexes (Figure 5B and Supplementary Figure S8A). Interestingly, CbAgo could cleave the negatively supercoiled plasmid, but not the linearized plasmid, suggesting that, like TtAgo, CbAgo relies on the negative supercoiled state of the target plasmid to facilitate local DNA melting before targeting and cleavage can take place (13). Furthermore, CbAgo–siDNA complexes were only able to generate dsDNA breaks in gene fragments with a GC-content of 31% or lower (Figure 5C). We hypothesize that, at moderate temperatures, the supercoiled state of the target plasmid allows for local DNA unwinding especially in AT-rich DNA regions. Taken together, this suggests that, like thermophilic pAgos, CbAgo relies on unwinding of the dsDNA before targeting and cleavage can take place.
DISCUSSION
Several prokaryotic Argonaute proteins have been demonstrated to protect their host against invading nucleic acids, such as plasmid DNA (13,15,16). Similar to TtAgo and RsAgo, CbAgo co-purifies with guides which are preferentially acquired from the plasmid used for its heterologous expression in E. coli. In addition, CbAgo mediates programmable DNA-guided DNA cleavage in vitro. This suggests that, similar to the phylogenetically related TtAgo, also CbAgo can interfere with plasmid DNA via DNA-guided DNA interference.
Sequencing of the nucleic acids that co-purified with CbAgo revealed that CbAgo preferentially associates with siDNAs with a 5′-ATTT-3′ sequence at their 5′ end. It was previously shown that RNA guides utilized by eAgos can be divided into functional segments. These segments are (from 5′ to 3′) the anchor nucleotide (nt 1), the seed (nt 2–8) and sub-seed segments (nt 2–4/2–5), the central (nt 9–12), the 3′ supplementary (nt 13–16) and the tail (nt 17–21) segments (48,49). Extending this knowledge to the siDNAs that co-purified with CbAgo, CbAgo preferentially associates with siDNAs that have a 5′-terminal adenosine anchor (nt 1) and a thymidine-rich sub-seed. In RNAi pathways, the preference for a specific 5′-terminal nucleotide is important for guide RNA loading into a subset of eAgos (50–52). Similarly, several pAgos including RsAgo, TtAgo, and now CbAgo also preferentially associate with specific 5′-terminal nucleotides in vivo (13,16). However, for both CbAgo and TtAgo, there is no clear preference for siDNAs with that specific 5′-base during cleavage assays in vitro. Rather than having a functional importance, the preference of pAgos for a specific nucleotide at the siDNA 5′ end might be a consequence of siDNA generation and/or loading, as has been demonstrated for TtAgo (17). Several studies on human Ago2 have described the importance of the sub-seed segment (nt 2–4/2–5) in its RNA guides (48,53,54). For hAgo2, a complete match between the guide RNA sub-seed segment and the target RNA triggers a conformational change that first exposes the remainder of the seed (nt 5–8/6–8), and eventually the rest of the guide. This facilitates progressive base paring between the guide RNA and the target (55). However, a specific nucleotide preference in the sub-seed segment, as we have observed for CbAgo, has not been described for any other Argonaute protein. The preference for the T-rich sub-seed is not only observed in the in vivo acquired siDNAs, but also plays a clear role during target binding and cleavage assays in vitro. This may reflect a structural preference for these thymidines in the cleft of the PIWI domain. We have not been able to obtain diffracting crystals of CbAgo in complex with siDNAs that have a 5′-ATTT-3′ sequence at the 5′-end. Future research will thus be necessary to determine the structural basis the apparent preference for these nucleotides at these positions. We hypothesize that this bias might reflect the mesophilic nature of CbAgo, which might have better access to AT-rich dsDNA fragments, both for siDNA acquisition and for target cleavage.
Several DNA-targeting pAgos have been repurposed for a range of molecular applications among which a cloning, recombineering and nucleic acid-detection method (22,23,56,57). Additionally, the potential repurposing of pAgos for genome editing applications has previously been discussed (27). However, all characterized DNA-cleaving pAgos to date originate from thermophilic prokaryotes and are solely active at elevated temperatures, which limits the potential repurposing of pAgos for applications that require moderate temperatures, such as genome editing. The biochemical characterization of CbAgo reported herein is the first example of a pAgo that catalyzes siDNA-guided dsDNA cleavage at 37°C, indicating that the pool of mesophilic pAgos contains candidates that—in theory—can be utilized for potential applications that require moderate temperatures, such as genome editing. If CbAgo or other mesophilic pAgos could be harnessed for genome editing, they will have certain advantages over the currently well-established genome editing tools CRISPR-Cas9 and CRISPR-Cas12a; While CRISPR-based genome editing tools can be programmed with a guide RNA to target DNA sequences of choice, target DNA cleavage additionally requires the presence of a protospacer adjacent motif (PAM) next to the targeted sequence (5′-NGG-3′ for Cas9 and 5′-TTTV-3′ for Cas12a) (58). This limits the possible target sites of Cas9 and Cas12a. In contrast, pAgos do not require a PAM for DNA targeting, which would make them much more versatile tools compared to CRISPR-associated nucleases. However, PAM binding by Cas9 and Cas12a also promotes unwinding of dsDNA targets (59–61) which subsequently facilitates strand displacement by the RNA guide, and eventually R-Loop formation. The absence of such mechanism in pAgos might explain their limited nuclease activity on dsDNA targets.
Here, we have demonstrated that CbAgo does not strictly rely on other proteins when targeting AT-rich dsDNA sequences in vitro. As such, this study provides a foundation for future efforts to improve double stranded DNA target accessibility of pAgos and to facilitate the further development of pAgo-based applications at moderate temperatures.
DATA AVAILABILITY
Atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB) with the accession number PDB: 6QZK.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Meitian Wang, Vincent Olieric and Takashi Tomizaki at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) for assistance with X-ray diffraction measurements.
Authors contributions: J.W.H. and J.v.d.O. conceived the project and designed the biochemical experiments, which were performed by J.H. and J.K. Single-molecule experiments were designed by S.C., T.J.C. and C.J. and performed by S.C. and T.J.C. X-ray crystallographic analysis was designed and performed by D.C.S. under the supervision of M.J., J.W.H., D.C.S., C.H., M.J., C.J. and J.v.d.O. wrote the manuscript. All authors read and approved the manuscript.
Notes
Present address: Stanley D. Chandradoss, Oxford Nanoimaging Ltd, Oxford, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Meitian Wang, Vincent Olieric and Takashi Tomizaki at the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) for assistance with X-ray diffraction measurements; Netherlands Organization of Scientific Research (NWO) [ECHO grant 711013002 and NWO-TOP grant 714.015.001 to J.v.d.O.]; Swiss National Science Foundation (SNSF) Project Grant [SNSF 31003A_149393 to to M.J.]; European Molecular Biology Organization (EMBO) [ALTF 179-2015 and aALTF 509-2017 to to D.C.S.]; M.J. is International Research Scholar of the Howard Hughes Medical Institute and Vallee Scholar of the Bert L & NKuggie Vallee Foundation; Vidi [864.14.002 to C.J.] of the Netherlands Organization for Scientific research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ketting R.F. MicroRNA biogenesis and function: an overview. Adv. Exp. Med. Biol. 2010; 700:1–14. [PubMed] [Google Scholar]
- 2. Joshua-Tor L., Hannon G.J.. Ancestral roles of small RNAs: an ago-centric perspective. Cold Spring Harb. Perspect. Biol. 2011; 3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 4. Pratt A.J., MacRae I.J.. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009; 284:17897–17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuhn C.D., Joshua-Tor L.. Eukaryotic Argonautes come into focus. Trends Biochem. Sci. 2013; 38:263–271. [DOI] [PubMed] [Google Scholar]
- 7. Hutvagner G., Simard M.J.. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008; 9:22–32. [DOI] [PubMed] [Google Scholar]
- 8. Ketting R.F. The many faces of RNAi. Dev. Cell. 2011; 20:148–161. [DOI] [PubMed] [Google Scholar]
- 9. Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V.. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F.R.F., Koonin E. V., Patel D.J., Van Der Oost J.. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song J., Smith S.K., Hannon G.J., Joshua-Tor L.. Crystal structure of argonaute and its implications for RISC slicer activity. Science. 2004; 305:1434–1437. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J.J. et al.. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stöckl S., Tinnefeld P., Schneider S., Klingl A. et al.. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017; 2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., Van Der Oost J.. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Olovnikov I., Chan K., Sachidanandam R., Newman D., Aravin A.. Bacterial Argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013; 51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D., Zhu Y., Wang Y., Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D. et al.. Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell. 2017; 65:985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zander A., Holzmeister P., Klose D., Tinnefeld P., Grohmann D.. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol. 2014; 11:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S.. Structural and mechanistic insights into the DNA-guided DNA endonuclease activity of an archaeal Argonaute. Nat. Microbiol. 2017; 17035:1–7. [DOI] [PubMed] [Google Scholar]
- 20. Swarts D.C., Koehorst J.J., Westra E.R., Schaap P.J., Van Der Oost J.. Effects of argonaute on gene expression in Thermus thermophilus. PLoS One. 2015; 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shabalina S.A., Koonin E.V.. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008; 23:578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Enghiad B., Zhao H.. Programmable DNA-guided artificial restriction enzymes. ACS Synth. Biol. 2017; 6:752–757. [DOI] [PubMed] [Google Scholar]
- 23. Song J., Hegge J.W., Mauk M.G., Chen J., Bhagwat N., Till J.E., Azink L.T., Peng J., Sen M., Mays J. et al.. Highly specific enrichment of rare nucleic acids using Thermus thermophilus Argonaute. 2019; bioRxiv doi: 10.1101/491738, 15 January 2018, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- 24. Savić N., Schwank G.. Advances in therapeutic CRISPR/Cas9 genome editing. Transl. Res. 2016; 168:15–21. [DOI] [PubMed] [Google Scholar]
- 25. Fellmann C., Gowen B.G., Lin P.C., Doudna J.A., Corn J.E.. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017; 16:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knott G.J., Doudna J.A.. CRISPR-Cas guides the future of genetic engineering. Science. 2018; 361:866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hegge J.W., Swarts D.C., van der Oost J.. Prokaryotic Argonaute proteins: novel genome-editing tools?. Nat. Rev. Microbiol. 2018; 16:5–11. [DOI] [PubMed] [Google Scholar]
- 28. Gao F., Shen X.Z., Jiang F., Wu Y., Han C.. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat. Biotech. 2016; 34:768–772. [DOI] [PubMed] [Google Scholar]
- 29. Lee S.H., Turchiano G., Ata H., Nowsheen S., Romito M., Lou Z., Ryu S.-M., Ekker S.C., Cathomen T., Kim J.-S.. Failure to detect DNA-guided genome editing using Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 2017; 35:17–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cyranoski D. Replications, ridicule and a recluse: the controversy over NgAgo gene-editing intensifies. Nature. 2016; 536:136–137. [DOI] [PubMed] [Google Scholar]
- 31. Ye S., Bae T., Kim K., Habib O., Lee S.H., Kim Y.Y, Lee K.-I., Kim S., Kim J.-S.. DNA-dependent RNA cleavage by the Natronobacterium gregoryi. 2017; bioRxiv doi: 10.1101/101923, 20 January 2017, preprint: not peer reviewed. [DOI]
- 32. Tropea J.E., Cherry S., Waugh D.S.. Doyle SA. Expression and purification of soluble His6-Tagged TEV protease. High Throughput Protein Expression and Purification: Methods and Protocols. 2009; Totowa, NJ: Humana Press; 297–307. [DOI] [PubMed] [Google Scholar]
- 33. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G.. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; 67:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 2004; 11:49–52. [DOI] [PubMed] [Google Scholar]
- 36. Terwilliger T.C., Read R.J., Adams P.D., Brunger A.T., Afonine P.V., Hung L.-W.. Model morphing and sequence assignment after molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013; 69:2244–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 39. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 40. Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J.. Structure of yeast Argonaute with guide RNA. Nature. 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J.. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008; 456:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Willkomm S., Makarova K., Grohmann D.. DNA-silencing by prokaryotic Argonaute proteins adds a new layer of defence against invading nucleic acids. FEMS Microbiol. Rev. 2018; 42:376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schirle N.T., Sheu-Gruttadauria J., Chandradoss S.D., Joo C., MacRae I.J.. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife. 2015; 4:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu Y., Esyunina D., Olovnikov I., Teplova M., Kulbachinskiy A., Aravin A.A., Patel D.J.. Accommodation of helical imperfections in R hodobacter sphaeroides Argonaute ternary complexes with guide RNA and target DNA. Cell Rep. 2018; 24:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides Its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Frank F., Sonenberg N., Nagar B.. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010; 465:818–822. [DOI] [PubMed] [Google Scholar]
- 51. Frank F., Hauver J., Sonenberg N., Nagar B.. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012; 31:3588–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T. et al.. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006; 442:203–207. [DOI] [PubMed] [Google Scholar]
- 53. Chandradoss S.D., Schirle N.T., Szczepaniak M., Macrae I.J., Joo C.. A dynamic search process underlies MicroRNA targeting. Cell. 2015; 162:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V.. Single-Molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015; 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klein M., Chandradoss S.D., Depken M., Joo C.. Why Argonaute is needed to make microRNA target search fast and reliable. Semin. Cell Dev. Biol. 2017; 65:20–28. [DOI] [PubMed] [Google Scholar]
- 56. Lapinaite A., Doudna J.A., Cate J.. Programmable RNA recognition using a CRISPR-associated Argonaute. PLoS One. 2018; 115:3368–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xie C., Fu L., Jin Z., Han L., Zhang A., Jin M., Tu Z., Xiang Y.. The prokaryotic Argonaute proteins enhance homology sequence-directed recombination in bacteria. Nucleic Acids Res. 2019; 47:3568–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Swarts D.C., Jinek M.. Cas9 versus Cas12a/Cpf1: Structure–function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA. 2018; 9:1–19. [DOI] [PubMed] [Google Scholar]
- 59. Anders C., Niewoehner O., Duerst A., Jinek M.. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014; 513:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yamano T., Zetsche B., Ishitani R., Zhang F., Nishimasu H., Nureki O.. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Mol. Cell. 2017; 67:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Swarts D.C., van der Oost J., Jinek M.. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell. 2017; 66:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB) with the accession number PDB: 6QZK.