Abstract
The DNA in mitochondria contributes essential components of the organelle’s energy producing machinery that is essential for life. In 1971, many mitochondrial DNA molecules were found to have a third strand of DNA that maps to a region containing critical regulatory elements for transcription and replication. Forty-five years later, a third strand of RNA in the same region has been reported. This mitochondrial R-loop is present on thousands of copies of mitochondrial DNA per cell making it potentially the most abundant R-loop in nature. Here, I assess the discovery of the mitochondrial R-loop, discuss why it remained unrecognized for almost half a century and propose for it central roles in the replication, organization and expression of mitochondrial DNA, which if compromised can lead to disease states.
R-LOOPS: FRIENDS AS WELL AS FOES
The versatility of RNA is so extensive that it has been proposed as the starting point for the whole of biology (1). RNA naturally hybridizes to complementary DNA sequences, and in the case of duplex DNA this results in the formation of triple stranded structures termed R-loops. Concerns about RNA hybridizing to DNA inappropriately and causing DNA damage (2–4) have tended to obscure the positive roles of RNA interactions with DNA. The idea that RNA purposefully interacts with DNA has a long history (5), and the subject has been enjoying a renaissance in recent years with the recognition that R-loops regulate and participate in most aspects of nuclear DNA metabolism (2,6–13). There is burgeoning evidence that nowhere is the fate of DNA more heavily tied to interactions with its RNA products than in the mitochondria.
MITOCHONDRIAL DNA ORGANIZATION, EXPRESSION AND REPLICATION
Mitochondria are descendants’ of free-living prokaryotes, whose assimilation to form the eukaryotic cell catalyzed the explosion of multicellular life on earth (14). The key faculty of mitochondria is their ability to produce copious amounts of energy from food, via respiration, a process that depends on a vestigial piece of DNA in the organelle. Consequently, aberrant or insufficient mitochondrial DNA causes cell and tissue dysfunction that manifests in a wide range of human diseases (15). Mitochondrial DNA (mtDNA) of mammals is typically circular and its two strands are denoted heavy (H) and light (L) based on their different nucleotide compositions. Important regulatory elements, which function as origins of replication, the replication terminus and transcriptional promoters, are concentrated in only substantial non-coding, or control, region of the molecule (Figure 1) (16–20). Thirty-seven genes are compressed into the other 15 kb of the mtDNA and are expressed as polycistronic RNAs of both strands, which are processed and modified to yield mature transfer, messenger and ribosomal RNAs (21).
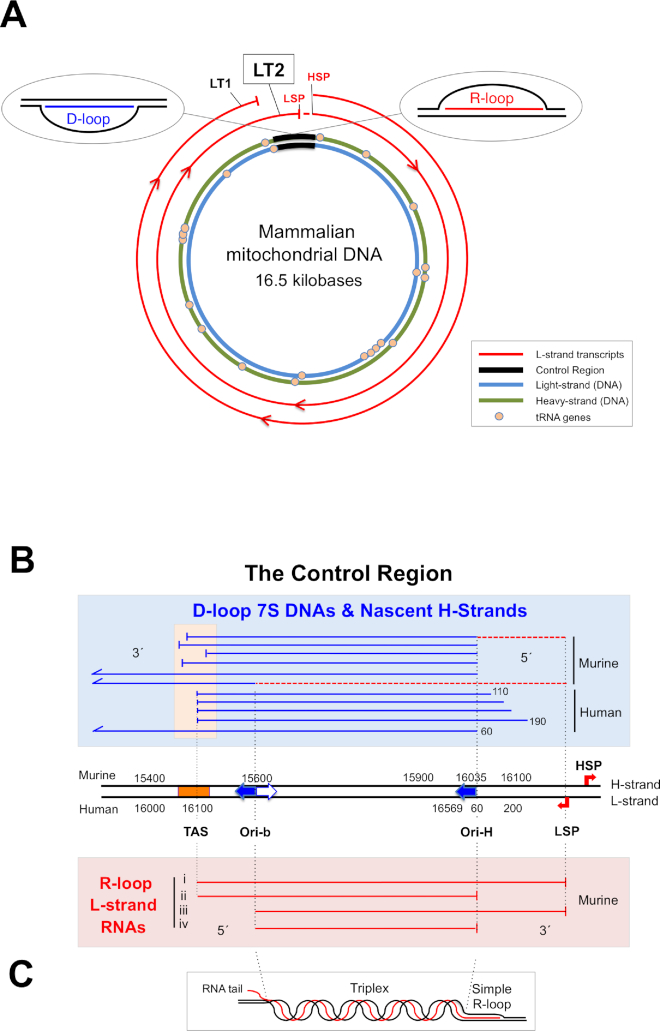
Figure 1.
Mammalian mitochondrial DNA and the D-loop. (A) A map of duplex mammalian mtDNA, encircled by two polycistronic RNAs (L-strand transcripts 1 and 2: LT1 and LT2) originating from the heavy strand promoter (HSP). LT1 is processed to produce all the RNAs for aerobic energy production encoded by the heavy (H) strand; further synthesis to the light strand promoter (LSP) yields LT2, which includes RNA spanning much of the major non-coding or control region. Processing of LT2 would yield RNA(s) corresponding to those associated with highly abundant R-loops (inset, right and detailed in panel B). The control region is also the location of the displacement loop or D-loop (inset, left). (B) An expanded view of the control region. Ori-H is the canonical origin of unidirectional replication initiated from a primer starting at LSP; another prominent RNA–DNA transition point maps to nucleotide position (np) 15 620 of murine mtDNA (39), which is similar to the map position of a bidirectional origin in human mtDNA (20), both of which are designated Ori-b. In mice, the RNA–DNA transition point for replication, defined as Ori-H, is identical to the 5′ ends of 7S DNAs that form the D-loop (np 16 035). However, in human cells nascent DNA strands start at approximately np 60, placing Ori-H downstream of the 5′ ends of 7S DNA (46). Many of the 3′ ends of the Light-strand control region RNAs, LC-RNAs, are close to Ori-H, none extends beyond LSP, none starts before TAS, and many 5′ ends are concentrated around Ori-b. Note that the 3′ ends of LC-RNAs terminating at LSP coincide with the start of the 7S DNA primers, two examples of which are represented as broken red lines as they are ordinarily barely detectable at steady-state (39). TAS—termination-associated sequence, a predicted stem–loop structure (47) located near the 3′ end of the D-loop. Nucleotide numbering for mouse and human mtDNA is according to the reference sequences (NC_012920 and NC_005089). (C) An illustration of a possible arrangement of a mitochondrial R-loop. Other more complex structures for LC-RNA involving G-quadruplexes and cruciform structures (termination intermediates) are also possible.
Although the major chromosomes of prokaryotes and eukaryotes are replicated via concurrent synthesis of the two strands of DNA, the predominant mechanism of DNA replication in mammalian mitochondria involves a prolonged interval between the start of leading and lagging strand DNA synthesis, i.e. strand-asynchronous replication (22–26). The RNAs produced via transcription from the heavy strand promoter (HSP) have been proposed to play a critical role in strand-asynchronous replication (Figure 2A) (27). In this transcript-dependent, or bootlace, mechanism the template for lagging strand DNA synthesis is hybridized to complementary mitochondrial RNAs prior to lagging-strand DNA synthesis. There is also evidence that widely dispersed RNA/DNA hybrids are involved in mtDNA replication in other metazoans, albeit not as extensively as in mammals (28–30). On the other hand, the earlier strand-displacement mechanism of mammalian mtDNA replication proposes that protein, rather than RNA, coats the lagging-strand template (Figure 2B) (31,32). An analysis of the mtDNA-binding pattern of mtSSB in HeLa cell mitochondria was consistent with this model (32); however, the replication intermediates may have lost regions of RNA/DNA hybrid during the mitochondrial isolation process (27). Moreover, a recent study indicates that mtSSB, unlike its prokaryotic homologs, does not form nucleoprotein tracts in vitro (33), suggesting the protein would leave substantial gaps on long segments of single-stranded DNA (Figure 2B). Taking all these observations into account, mtSSB could play an important role in filling any gaps between the RNA/DNA hybrids (Figure 2C). The proposed bootlace mechanism raises a number of questions, in particular how are the RNAs hybridized to the lagging strand template and later removed to permit second strand DNA synthesis? Both RNA hybridization to the lagging-strand template and its subsequent removal could be mediated by the mitochondrial RNA granules that handle multiple aspects of mtRNA metabolism (34,35), and which include two key mtDNA maintenance and replication factors (Twinkle DNA helicase and mtSSB) (36), as well as enzymes involved in processing aberrant R-loops (37,38).
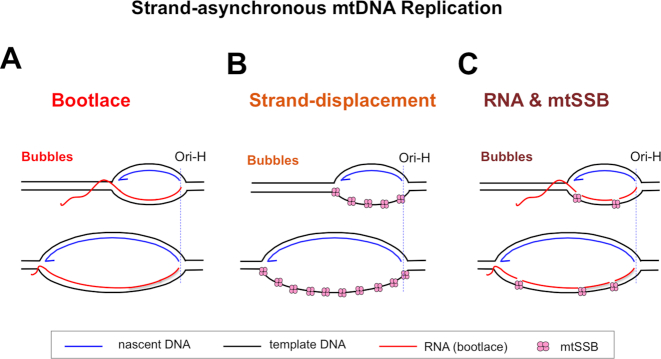
Figure 2.
Proposed models of mammalian mtDNA replication. Two alternative mechanisms of strand-asynchronous replication have been proposed: (A) the bootlace mechanism that relies on mature transcripts. (B) The strand-displacement model that depends on protein (mtSSB) to coat the displaced lagging-strand template. (C) Models A and B can be merged into one in which transcripts are hybridized to the lagging strand template with mtSSB occupying any gaps between RNAs. Ori-H—origin of heavy-strand DNA replication.
In all versions of strand-asynchronous mtDNA replication, RNA primes mtDNA replication at specific sites, and after the initiation of DNA synthesis the primers are processed rapidly by ribonuclease H1 (RNase H1) (39). In mammals, the primers for leading strand DNA synthesis in the control region start at the light-strand promoter (LSP) and the RNA transitions to DNA at one of two positions, Ori-H (origin of heavy strand DNA replication) or Ori-b (origin of bidirectional replication), in the control region (39) (Figure 1B).
THE MITOCHONDRIAL D-LOOP
In the early 1970s, many molecules of mammalian mtDNA were found to contain a triple stranded region, or displacement (D-) loop spanning half a kilobase or more of the control region (Figure 1A, inset, left) (40–42). The third strand, 7S DNA [The letter S denotes the sedimentation rate in velocity gradients in units of Svedbergs], ends at or close to a sequence proposed to act as a termination signal for D-loop synthesis, called TAS (Figure 1B) (43). Between 1% and 65% of mtDNA molecules contain a D-loop in mammals (44). The frequent synthesis of a piece of DNA across such a pivotal region of the mitochondrial genome strongly implies an important role for the D-loop in mtDNA metabolism.
Many discussions of the D-loop suggest that 7S DNAs represent paused, or stalled, replication intermediates; however, this is difficult to reconcile with the recent finding that nascent H-strands associated with strand-asynchronous replication (i.e. Ori-H) (45,46) differ from the 5′ ends of 7S DNAs in human cells (17,18,47,48) (Figure 1B). Nevertheless, the formation of the D-loop could be a quite separate, precursor step in the initiation of DNA replication, as detailed later in the article. Moreover, the D-loop form of mtDNA has been implicated in protein recruitment and mtDNA organization (49,50), as well as being proposed to have a role in the termination of replication (51,52).
R-LOOP DISCOVERY
The RNA/DNA hybrids associated with replicating mtDNA were not detected for many years because they are readily lost during isolation (23,53); however, inter-strand cross-linking can preserve them (27). One species greatly enhanced by cross-linking of the mtDNA was a small bubble-like structure with a similar mass and location to the D-loop. Unlike D-loops, the novel small bubbles proved refractory to restriction digestion at multiple sites in the control region, and contained a DNase-resistant, RNase H-sensitive component of the opposite strand to 7S DNA, all of which suggests they are R-loops containing a third strand of pure RNA (45). Reverse-transcription, cDNA synthesis and cloning yielded sequences spanning much of the control region, which we termed LC-RNA, as the RNA is L-strand in sequence and maps to the Control region of the mtDNA (Figure 1) (45). The 5′ ends of LC-RNA map predominantly to TAS and Ori-b, whereas the 3′ ends map close to Ori-H or LSP (Figure 1B). It is not yet known how the LC-RNAs hybridized to mtDNA are arranged in vivo, parts might form triplexes and tails, as well as simple R-loops (Figure 1C).
Collectively, the R-loops containing LC-RNA are highly abundant, accounting for 15% of all mtDNAs in mouse liver samples, without cross-linking, and over 50% in human fibroblasts with cross-linking (45). Although other R-loops are found in mtDNA, notably involving the confusingly named 7S RNA (which represents the RNA primer for synthesis of 7S DNA (45,54)), they are present at very low abundance compared with the LC-RNA (39,45). Therefore, for convenience, we refer to the structure formed by LC-RNA together with mtDNA as ‘the’ R-loop, using the definite article. The discovery that long non-coding RNAs form persistent R-loops in the control region of mammalian mtDNA (45) warrants a thorough assessment of their potential roles in mtDNA maintenance and expression both in normal and disease states.
The difficulty of preserving the mitochondrial R-loop also raises the question: are we underestimating R-loops in the nucleus? This question is all the more pertinent in that part of the difficulty of preserving RNA/DNA hybrids in mitochondria is attributable to contaminating activities from other cell compartments, most likely the nucleus, as more highly purified mitochondria have better preserved RNA–DNA hybrids than crude mitochondrial preparations (23). Thus notwithstanding the fact that R-loops have been documented for millions of base pairs (55), refined extraction procedures for nuclear DNA, using conditions where RNase H or RNase H-like activities are inhibited, could reveal new or more abundant R-loops in the nucleus. Reciprocally methods used for characterizing R-loops in nuclear DNA could aid the further characterization of mitochondrial R-loops (55,56).
R-LOOP AND D-LOOP GENESIS
The location and span of the mitochondrial R-loop coupled with prior knowledge of mtDNA metabolism offer possible insights into its production. The RNAs of the R-loop are L-strand and map to the control region (Figure 1B and (45)) and so the most straightforward way of synthesizing LC-RNA is via a near full-genome-length polycistronic transcript arising from the heavy strand promoter. At a minimum, the HSP-derived, L-strand transcript must encompass 95% of the mitochondrial genome to produce all the gene products for respiration encoded by the H-strand of mtDNA (LT1, Figure 1A). Continued synthesis across the control region to the light-strand promoter would generate a polycistronic transcript, LT2, whose final portion constitutes LC-RNA; hence, RNA processing of LT2 at TAS could give rise to the RNA component of the R-loops (Figures 1A, B and 3A).
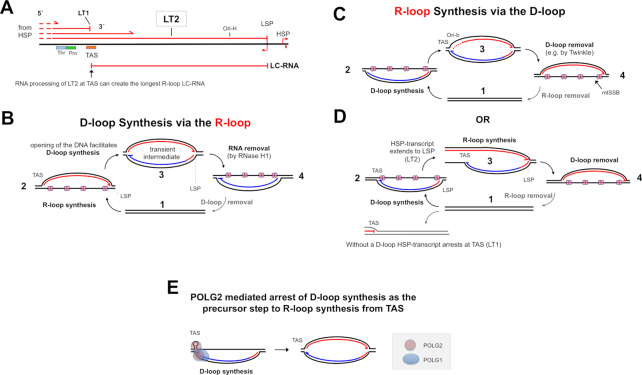
Figure 3.
R-loop and D-loop formation. (A) R-loops can derive from a part of a polycistronic RNA initiating at the heavy strand promoter of mammalian mtDNA, if transcription does not always terminate at the termination-associated sequence(s) TAS (60). (B) The longest LC-RNAs end at the light-strand promoter, LSP; hence, strand-switching of POLRMT at this site could initiate primer synthesis for 7S DNA. Moreover, extensive triplex DNA/RNA/DNA (see Figure 1C) could help to favor initiation from LSP, while protecting the R-loop from RNase H1 degradation. Synthesis of the D-loop would inevitably disrupt the triplexes, and make them vulnerable to RNase H1. (C) Opposite to panel (B), D-loop synthesis precedes R-loop synthesis. Secondary structure and trans-acting factors (e.g. panel E) at TAS or Ori-b could favor these as start sites for POLRMT, possibly in conjunction with triplexes preventing POLRMT loading elsewhere. (D) D-loop synthesis could allow transcription from HSP to extend beyond TAS. (E) Coupling of the termination of D-loop synthesis and the initiation of R-loop synthesis by POLG. 7S DNA synthesis by mitochondrial DNA polymerase gamma (1 copy of POLG1 and 2 of POLG2) could be arrested by POLG2 binding to the predicted stem–loop at TAS, followed by the recruitment of POLRMT to enable synthesis of LC-RNA. RNA, Red lines; template DNA, black lines, nascent DNA, blue lines. LSP, light strand promoter; Thr, tRNA threonine gene; Pro, tRNA proline gene; Ori-b and Ori-H, origins of replication. Twinkle, mitochondrial DNA helicase; RNase H1, Ribonuclease H1; mtSSB, mitochondrial single-stranded DNA binding protein.
As LC-RNAs are complementary to the 7S DNAs of the D-loops, R-loops and D-loops may be co-regulated or interdependent, and either could be the progenitor of the other (Figure 3B–D). The longest LC-RNAs (TAS to LSP (Figure 1B)) create an R-loop with a potential template corresponding to 7S DNA plus its primer. Hence, R-loops of LC-RNA could, by exposing LSP, promote the initiation of transcription for D-loop synthesis (Figure 3B), aided by the mitochondrial transcription factor, TFB2M, which is known to recruit POLRMT at this site (57).
Alternatively, D-loops may be the precursors of R-loops—a simple triple-stranded DNA form of the D-loop might facilitate LC-RNA synthesis (Figure 3C), and secondary structure or other features of TAS and Ori-b, in combination with partial triplex formation (Figure 1C), could ensure that the initiation of LC-RNA synthesis in the control region is site-specific. D-loops could instead allow transcription to extend from TAS to LSP, thereby yielding LT2 (Figure 3D); i.e. in the absence of a D-loop, L-strand transcripts might always terminate at TAS (yielding LT1). However, if D-loops adopt a triplex structure then this could cause transcription to arrest at TAS, preventing the synthesis of LT2. Hence, the form of the D-loop rather than its presence or absence could determine the balance of LT1 and LT2 transcripts, which could be regulated by interacting proteins that alter triplex DNA stability. As mentioned above and discussed further below, the different R-loop lengths and forms (Figure 1C) could perform different functions; accordingly, more than one of the models (Figure 3B–D) can apply.
Defining the mechanisms behind the R-loop length variation would be helpful but is not straightforward, as it can arise from a combination of alternative initiation, termination and RNA-processing sites. Thus, the 3′ ends mapping to Ori-H and LSP may reflect either alternate transcription termination sites or post-transcriptional RNA processing events. LC-RNAs starting at TAS and Ori-b could arise directly from the initiation of RNA synthesis in the control region (e.g. Figure 3C) or via transcript processing. Labeling of terminal 5′ triphosphates with a capping enzyme (58) can identify LC-RNAs initiated in the control region, whereas RNAs derived from polycistronic transcripts (i.e. LT2) would not be labeled by any such treatment. This approach might also clarify whether the increase in LC-RNAs in two previous studies (59,60) was the result of extended transcription (to create LT2 in the terminology of this article) (Figures 1 and 3A), or if it resulted instead from the initiation of transcription in the control region of the mtDNA (Figure 3C).
THE PROTEIN MACHINERY OF THE ‘LOOPS’
The models of D-loop and R-loop formation outlined above (Figure 3) reconcile readily with the properties and activities of known factors in the mitochondria, none more so than the dedicated mitochondrial RNA polymerase, POLRMT. It is POLRMT that produces the polycistronic mitochondrial transcripts (61), and so will be essential to generate LC-RNA via LT2 (Figure 1). POLRMT is also presumed to generate the primers for D-loop synthesis starting at the light-strand promoter (Figure 1), and it is the obvious candidate for R-loop synthesis on D-loops (Figure 3C). Specifically, POLRMT might initiate R-loop synthesis from the stem–loop at TAS, analogous to it priming second-strand DNA synthesis at Ori-L (62). TAS has other potential roles: it could mediate transcription termination (yielding LT1 rather than LT2) (60), perhaps in concert with a member of the mTERF family (63,64), or an RNA processing factor (such as GSRF1 (65), or RNase Z (66)). Alternatively, TAS could engage other proteins, such as POLG2 (the accessory subunit of mitochondrial DNA polymerase, POLG) to promote LT2 synthesis. POLG2 is known to prefer D-loops to circular and linear duplex DNA (49), and to bind to single-stranded DNA (67), and its recognized homology to amino-acyl tRNA synthetases (68) could enable it to recognize the stem–loop structure of TAS. POLG2 binding to TAS could also arrest 7S DNA synthesis by POLG holoenzyme and promote R-loop synthesis in the opposite direction, giving it a pivotal role in R-loop synthesis via a D-loop (Figure 3E).
Although the non-cooperative binding of mtSSB, to single-stranded DNA (33), suggests it cannot fully coat the displaced strand of the mitochondrial R-loops, it could nevertheless help to stabilize them, in the manner of another single-stranded DNA binding protein, AtNDX (69). Therefore, a contribution of mtSSB to mtDNA replication (70) may be R-loop, as well as D-loop, maintenance. Ribonuclease H1, or RNase H1, is another anticipated key player in the metabolism of the mitochondrial R-loop, as it degrades RNA only when it is hybridized to DNA. RNase H1 is established to process primers involved in mtDNA replication (39) and the RNA primer (7S RNA) for D-loop synthesis (45). The R-loop is a larger target located in the same region of the mtDNA. Hence, RNase H1 can remove the D-loop primer and the R-loop to generate a D-loop (Figure 3B), with final processing of the 7S DNA by mitochondrial endonuclease MGME1 (71,72) or Flap endonuclease 1 (FEN1). Less obviously, RNase H1 might also stabilize mitochondrial R-loops, as it contains a distinct RNA/DNA hybrid binding domain (73). Thus, in conditions where the catalytic activity of RNase H1 is repressed (e.g. via oxidation (74), or perhaps ligand binding or post-translational modification), the enzyme could potentially bind to, but not degrade, the mitochondrial R-loop. However, activation of the catalytic function of RNase H1 would signal the immediate destruction of the mitochondrial R-loops (not in a triplex conformation), which conceivably is what happens during all but the most delicate mtDNA isolation procedures.
The difficulty of preserving mitochondrial R-loops could explain why some nucleic acid binding proteins in mitochondria fail to co-purify with mtDNA. For example, the mitochondrial isoform of FEN1, FENMIT, preferentially binds to R-loops in vitro, but little of it was associated with isolated mitochondrial nucleoprotein complexes (75). FENMIT might play a role in the initiation of mtDNA replication through interaction with the long H-strand primer, known as 7S RNA, as earlier proposed (75). However, the R-loop is another potential substrate that could explain the need for this FEN1 variant that appears to be exclusive to mitochondria.
THE MITOCHONDRIAL R-LOOP AND STRAND-ASYNCHRONOUS REPLICATION
Having outlined the ways in which the R-loops might be generated, the critical question arising is what are its functional roles in mtDNA metabolism? One predicted function of LC-RNA predated its discovery. LC-RNA is integral to the model of mtDNA replication that involves the incorporation of RNA on the lagging strand: transcript-dependent replication (TDR), or the ‘bootlace’ mechanism, although this was not made explicit in the original report (27). Strand-asynchronous replication begins in the control region with the synthesis of a long primer from the light-strand promoter to the origin (Ori-H or Ori-b), where the transition to DNA synthesis occurs (39); hence, if the incorporation of transcripts is obligatory for strand-asynchronous replication then the first RNA element will be a L-strand RNA corresponding to the control region, i.e. LC-RNA-i or LC-RNA-ii (Figure 1B). Moreover, the generation of LC-RNA via transcription from the heavy strand promoter offers an attractive regulatory system for such a mechanism of mtDNA replication. Extending transcription into the control region, i.e. to synthesize LT2 could permit replication (Figure 4), whereas transcriptional arrest at TAS would deprive the mtDNA of the first ‘bootlace’, and could thereby prevent replication initiating in the control region. In the one context that has been studied to date where LC-RNAs hybridized to mtDNA are greatly depressed there is a switch to strand-coupled replication (46), which is consistent with the idea that LC-RNA is required for the initiation of strand-asynchronous mtDNA replication.
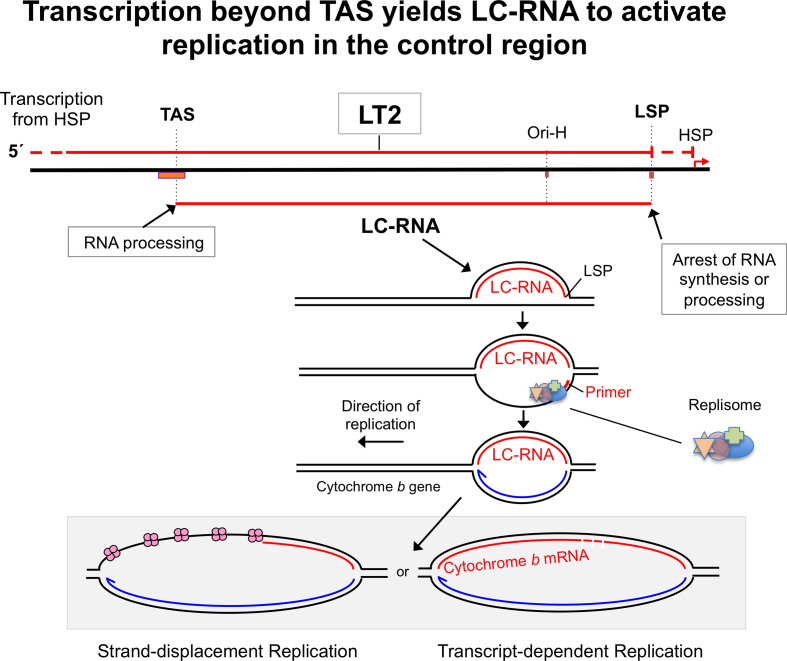
Figure 4.
A proposed role of the mitochondrial R-loop in strand-asynchronous replication. (A) According to Transcript-dependent Replication, mature transcripts hybridize to the lagging strand prior to second strand DNA synthesis (27). Because this mechanism of DNA replication starts in the Control Region (with the synthesis of a primer from LSP and transition to DNA synthesis at Ori-H, the first transcript required is a LC-RNA. LC-RNA dependent initiation of mtDNA replication is also compatible with the strand-displacement model, as illustrated.
If LC-RNA synthesis is dependent on the presence of a D-loop (Figure 3D) and LC-RNA is a requirement of strand-asynchronous replication (Figure 4), then the D-loop is itself essential for this mechanism of replication. However, in this model 7S DNA does not contribute to the nascent strands of DNA.
Separation (unwinding or melting) of the two strands of DNA is a critical step in the initiation of DNA replication in diverse systems, which is typically protein-mediated. R-loop formation achieves the same result, with the caveat that only one strand is rendered single-stranded. Thus, the mitochondrial R-loop might initiate strand-asynchronous mtDNA replication, by exposing LSP—the primer start site for nascent H-strand DNA synthesis (Figure 4). In this way, the R-loop potentially offers a much simpler system for initiating DNA replication than operated in the nucleus (76), as it permits only one replisome to load (Figure 4). Thus, the mitochondrial R-loop can explain why (strand-asynchronous) DNA replication initiates in the control region and is unidirectional (22,46). Moreover, because mtDNA replication is unidirectional on circular molecules, both initiation and termination occur in the control region, and so LC-RNA could contribute to completing the replication cycle.
Evolutionary conservation often provides clues as to function; however, there are no data as to the occurrence of a long non-coding RNA forming a R-loop in mtDNA outside of mammals. Nevertheless, D-loops have been identified in a range of vertebrates and invertebrates (40,77–78), and so assuming D-loops and R-loops are inter-dependent, or functionally complementary, R-loops are expected to be present in a broad range of animals, and an initial screen of cross-linked mtDNA could readily support or refute this prediction. Including insects in such an analysis could be informative, as they appear to lack a D-loop form of mtDNA (79), and so may also lack R-loops. However, the mitochondrial R-loop might be dispensible for the initiation of replication in insects because the control region of mtDNA has an exceptionally high A:T content (79), which facilitates strand separation (i.e. melting at the origin of replication).
Once formed, the R-loop could inhibit strand separation at proximal sites and thereby prevent transcription from HSP, in the manner of long non-coding RNAs found in the nucleus (69,80). Hence, in the model where LC-RNA is needed for the initiation of replication, inhibiting transcription from HSP with LC-RNA would prevent transcription and replication occurring on the same molecule, thereby avoiding collisions between the two machineries and the formation of unwanted R-loops on actively replicating mtDNAs. Likewise, if the R-loop increases supercoiling in the rest of the molecule, it will make DNA melting for the initiation of replication outside the control region thermodynamically less favorable and could thereby actively inhibit strand-coupled mtDNA replication elsewhere (46). Thus, the presence or absence of mitochondrial R-loops could determine the choice of replication mechanism.
MITOCHONDRIAL DNA SEGREGATION AND MEMBRANE ATTACHMENT
The close proximity of mtDNA to the inner mitochondrial membrane (IMM) in electron microscope images led Nass to propose that mtDNA is attached to the membrane in 1969 (81), and evidence that this specifically involves the control region, dates back to 1977 (82). Located in the control region, D-loops and R-loops are credible candidates for playing one or more roles in attaching mtDNA to the IMM. Although the role of the loops could be transient, serving to recruit proteins for particular tasks, including membrane-attachment, they could also play a structural role. The case is strongest for the R-loops, as mitochondria retain numerous features of their prokaryotic ancestors, and in bacteria RNA has long been implicated in DNA organization (5). Moreover, in the contexts we have studied, R-loops outnumber D-loops (45), and given the problems of preserving RNA/DNA hybrids of mtDNA, especially LC-RNA, the true proportion may be still higher. It is therefore conceivable that every mtDNA molecule (or nucleoid) has either a D-loop or a R-loop on a (semi-) permanent basis.
Membrane attachment is far more than a ‘clothes-hanger’ for mtDNA; by localizing the DNA to rigid cholesterol-rich microdomains (83), it can provide traction to facilitate the processes of replication, transcription, RNA processing and segregation, as well as the assembly of the mitochondrial ribosome at the nucleoid (84–86). Thus, any role of R-loops and D-loops in mtDNA membrane-attachment offers the possibility of them impacting almost every facet of mtDNA metabolism.
Putative mtDNA segregation intermediates map to the control region and are disrupted by the repression of POLG2 or ATAD3 (49,50). Both these proteins display a strong preference for binding to DNA molecules with a D-loop, and this preference is expected to extend to R-loops. Hence, R-loops may play a pivotal role in mtDNA segregation. The mitochondrial R-loop is a potential target of RNase H1, and a pathological variant of the enzyme has been shown to cause mtDNA aggregation and R-loop instability, with the inference that the mutant enzyme impairs mtDNA segregation because it has greater than usual access to R-loops (45).
CONCLUDING REMARKS
Discovery of the R-loop almost half a century after the D-loop reinforces the status of mammalian mtDNA as a model of efficiency: high gene density, and a single substantial non-coding region packed with features required for its maintenance, expression, organization and propagation. The firm predictions are that the R-loops of LC-RNA play a central role in mtDNA metabolism, being required for, and regulating, the unique mechanism of strand-asynchronous replication in mitochondria, as well as optimizing mtDNA organization and expression. The most pressing questions concern the relationship between the varieties of D-loops and R-loops and their specific binding factors, as addressing these can clarify the roles of the loops in mtDNA metabolism; and improved protocols to isolate mitochondrial nucleoids with the mitochondrial R-loop intact would help considerably to achieve this aim. Finally, the disturbances to mtDNA organization in cells and solid tissues with mutant RNase H1 and the concomitant effect on mitochondrial R-loops (45) suggest that this new feature of mtDNA will prove to be relevant to a range of mitochondrial disorders, especially as mtDNA aggregation is increasingly recognized in these diseases (45,87–91). Therefore, the mitochondrial R-loop should be investigated whenever mtDNA perturbation is apparent or suspected.
ACKNOWLEDGEMENTS
The author thanks his many colleagues and collaborators down the years, especially Takehiro Yasukawa for his exceptional ability to isolate mtDNAs with substantially intact R-loops; and Howy Jacobs and Antonella Spinazzola for helpful discussions, encouragement and a critical reading of the manuscript.
FUNDING
Carlos III Program of Health [Pl17/00380]; Department of Health, País Vasco [2018111043 and 2018222031]. Funding for open access charge: Medical Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bernhardt H.S. The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others)(a). Biol. Direct. 2012; 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wahba L., Koshland D.. The Rs of biology: R-loops and the regulation of regulators. Mol. Cell. 2013; 50:611–612. [DOI] [PubMed] [Google Scholar]
- 3. Potenski C.J., Klein H.L.. R we there yet? R-loop hazards to finishing the journey. Mol. Cell. 2011; 44:848–850. [DOI] [PubMed] [Google Scholar]
- 4. Sollier J., Cimprich K.A.. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015; 25:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pettijohn D.E., Hecht R.. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harbor Symp. Quant. Biol. 1974; 38:31–41. [DOI] [PubMed] [Google Scholar]
- 6. Briggs E., Hamilton G., Crouch K., Lapsley C., McCulloch R.. Genome-wide mapping reveals conserved and diverged R-loop activities in the unusual genetic landscape of the African trypanosome genome. Nucleic Acids Res. 2018; 46:11789–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guil S., Esteller M.. Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 2012; 19:1068–1075. [DOI] [PubMed] [Google Scholar]
- 8. Kuznetsov V.A., Bondarenko V., Wongsurawat T., Yenamandra S.P., Jenjaroenpun P.. Toward predictive R-loop computational biology: genome-scale prediction of R-loops reveals their association with complex promoter structures, G-quadruplexes and transcriptionally active enhancers. Nucleic Acids Res. 2018; 46:7566–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quinn J.J., Chang H.Y.. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016; 17:47–62. [DOI] [PubMed] [Google Scholar]
- 10. Wiedemann E.M., Peycheva M., Pavri R.. DNA replication origins in immunoglobulin switch regions regulate class switch recombination in an R-Loop-Dependent manner. Cell Rep. 2016; 17:2927–2942. [DOI] [PubMed] [Google Scholar]
- 11. Lombrana R., Almeida R., Alvarez A., Gomez M.. R-loops and initiation of DNA replication in human cells: a missing link. Front. Genet. 2015; 6:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keskin H., Shen Y., Huang F., Patel M., Yang T., Ashley K., Mazin A.V., Storici F.. Transcript-RNA-templated DNA recombination and repair. Nature. 2014; 515:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costantino L., Koshland D.. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 2015; 34:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lane N., Martin W.. The energetics of genome complexity. Nature. 2010; 467:929–934. [DOI] [PubMed] [Google Scholar]
- 15. Area-Gomez E., Schon E.A.. Mitochondrial genetics and disease. J. Child Neurol. 2014; 29:1208–1215. [DOI] [PubMed] [Google Scholar]
- 16. Chang D.D., Clayton D.A.. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crews S., Ojala D., Posakony J., Nishiguchi J., Attardi G.. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979; 277:192–198. [DOI] [PubMed] [Google Scholar]
- 18. Fish J., Raule N., Attardi G.. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science. 2004; 306:2098–2101. [DOI] [PubMed] [Google Scholar]
- 19. Montoya J., Christianson T., Levens D., Rabinowitz M., Attardi G.. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:7195–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yasukawa T., Yang M.Y., Jacobs H.T., Holt I.J.. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 2005; 18:651–662. [DOI] [PubMed] [Google Scholar]
- 21. D'Souza A.R., Minczuk M.. Mitochondrial transcription and translation: overview. Essays Biochem. 2018; 62:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kasamatsu H., Vinograd J.. Unidirectionality of replication in mouse mitochondrial DNA. Nat. New Biol. 1973; 241:103–105. [DOI] [PubMed] [Google Scholar]
- 23. Yang M.Y., Bowmaker M., Reyes A., Vergani L., Angeli P., Gringeri E., Jacobs H.T., Holt I.J.. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002; 111:495–505. [DOI] [PubMed] [Google Scholar]
- 24. Brown T.A., Cecconi C., Tkachuk A.N., Bustamante C., Clayton D.A.. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Develop. 2005; 19:2466–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yasukawa T., Reyes A., Cluett T.J., Yang M.Y., Bowmaker M., Jacobs H.T., Holt I.J.. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006; 25:5358–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phillips A.F., Millet A.R., Tigano M., Dubois S.M., Crimmins H., Babin L., Charpentier M., Piganeau M., Brunet E., Sfeir A.. Single-Molecule analysis of mtDNA replication uncovers the basis of the common deletion. Mol. Cell. 2017; 65:527–538. [DOI] [PubMed] [Google Scholar]
- 27. Reyes A., Kazak L., Wood S.R., Yasukawa T., Jacobs H.T., Holt I.J.. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 2013; 41:5837–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joers P., Lewis S.C., Fukuoh A., Parhiala M., Ellila S., Holt I.J., Jacobs H.T.. Mitochondrial transcription terminator family members mTTF and mTerf5 have opposing roles in coordination of mtDNA synthesis. PLoS Genet. 2013; 9:e1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reyes A., Yang M.Y., Bowmaker M., Holt I.J.. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J. Biol. Chem. 2005; 280:3242–3250. [DOI] [PubMed] [Google Scholar]
- 30. Ciesielski G.L., Nadalutti C.A., Oliveira M.T., Jacobs H.T., Griffith J.D., Kaguni L.S.. Structural rearrangements in the mitochondrial genome of Drosophila melanogaster induced by elevated levels of the replicative DNA helicase. Nucleic Acids Res. 2018; 46:3034–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clayton D.A. Replication of animal mitochondrial DNA. Cell. 1982; 28:693–705. [DOI] [PubMed] [Google Scholar]
- 32. Miralles Fuste J., Shi Y., Wanrooij S., Zhu X., Jemt E., Persson O., Sabouri N., Gustafsson C.M., Falkenberg M.. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLos Genet. 2014; 10:e1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaur P., Longley M.J., Pan H., Wang H., Copeland W.C.. Single-molecule DREEM imaging reveals DNA wrapping around human mitochondrial single-stranded DNA binding protein. Nucleic Acids Res. 2018; 46:11287–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jourdain A.A., Boehm E., Maundrell K., Martinou J.C.. Mitochondrial RNA granules: Compartmentalizing mitochondrial gene expression. J. Cell Biol. 2016; 212:611–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Antonicka H., Shoubridge E.A.. Mitochondrial RNA Granules Are Centers for Posttranscriptional RNA Processing and Ribosome Biogenesis. Cell Rep. 2015; 10:920–932. [DOI] [PubMed] [Google Scholar]
- 36. Hensen F., Potter A., van, Esveld S.L., Tarres-Sole A., Chakraborty A., Sola M., Spelbrink J.N.. Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res. 2019; doi:10.1093/nar/gkz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Silva S., Camino L.P., Aguilera A.. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:11024–11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pietras Z., Wojcik M.A., Borowski L.S., Szewczyk M., Kulinski T.M., Cysewski D., Stepien P.P., Dziembowski A., Szczesny R.J.. Controlling the mitochondrial antisense - role of the SUV3-PNPase complex and its co-factor GRSF1 in mitochondrial RNA surveillance. Mol. Cell Oncol. 2018; 5:e1516452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holmes J.B., Akman G., Wood S.R., Sakhuja K., Cerritelli S.M., Moss C., Bowmaker M.R., Jacobs H.T., Crouch R.J., Holt I.J.. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:9334–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arnberg A., van Bruggen E.F., Borst P.. The presence of DNA molecules with a displacement loop in standard mitochondrial DNA preparations. Biochim. Biophys. Acta. 1971; 246:353–357. [DOI] [PubMed] [Google Scholar]
- 41. Kasamatsu H., Robberson D.L., Vinograd J.. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc. Natl. Acad. Sci. U.S.A. 1971; 68:2252–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walberg M.W., Clayton D.A.. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981; 9:5411–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown G.G., Gadaleta G., Pepe G., Saccone C., Sbisa E.. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. J. Mol. Biol. 1986; 192:503–511. [DOI] [PubMed] [Google Scholar]
- 44. Annex B.H., Williams R.S.. Mitochondrial DNA structure and expression in specialized subtypes of mammalian striated muscle. Mol. Cell Biol. 1990; 10:5671–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Akman G., Desai R., Bailey L.J., Yasukawa T., Dalla Rosa I., Durigon R., Holmes J.B., Moss C.F., Mennuni M., Houlden H. et al.. Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E4276–E4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cluett T.J., Akman G., Reyes A., Kazak L., Mitchell A., Wood S.R., Spinazzola A., Spelbrink J.N., Holt I.J.. Transcript availability dictates the balance between strand-asynchronous and strand-coupled mitochondrial DNA replication. Nucleic Acids Res. 2018; 46:10771–10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doda J.N., Wright C.T., Clayton D.A.. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:6116–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kang D., Miyako K., Kai Y., Irie T., Takeshige K.. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 1997; 272:15275–15279. [DOI] [PubMed] [Google Scholar]
- 49. Di Re M., Sembongi H., He J., Reyes A., Yasukawa T., Martinsson P., Bailey L.J., Goffart S., Boyd-Kirkup J.D., Wong T.S. et al.. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009; 37:5701–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He J., Mao C.C., Reyes A., Sembongi H., Di Re M., Granycome C., Clippingdale A.B., Fearnley I.M., Harbour M., Robinson A.J. et al.. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007; 176:141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bowmaker M., Yang M.Y., Yasukawa T., Reyes A., Jacobs H.T., Huberman J.A., Holt I.J.. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 2003; 278:50961–50969. [DOI] [PubMed] [Google Scholar]
- 52. Nicholls T.J., Minczuk M.. In D-loop: 40 years of mitochondrial 7S DNA. Expt. Gerontol. 2014; 56:175–181. [DOI] [PubMed] [Google Scholar]
- 53. Pohjoismaki J.L., Holmes J.B., Wood S.R., Yang M.Y., Yasukawa T., Reyes A., Bailey L.J., Cluett T.J., Goffart S., Willcox S. et al.. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010; 397:1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang D.D., Hauswirth W.W., Clayton D.A.. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985; 4:1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nadel J., Athanasiadou R., Lemetre C., Wijetunga N.A., P O.B., Sato H., Zhang Z., Jeddeloh J., Montagna C., Golden A. et al.. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin. 2015; 8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu Z., Zhang Q.C., Lee B., Flynn R.A., Smith M.A., Robinson J.T., Davidovich C., Gooding A.R., Goodrich K.J., Mattick J.S. et al.. RNA duplex map in living cells reveals Higher-Order transcriptome structure. Cell. 2016; 165:1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Posse V., Gustafsson C.M.. Human mitochondrial transcription factor B2 is required for promoter melting during initiation of transcription. J. Biol. Chem. 2017; 292:2637–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shuman S. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli. J. Biol. Chem. 1990; 265:11960–11966. [PubMed] [Google Scholar]
- 59. Freyer C., Park C.B., Ekstrand M.I., Shi Y., Khvorostova J., Wibom R., Falkenberg M., Gustafsson C.M., Larsson N.G.. Maintenance of respiratory chain function in mouse hearts with severely impaired mtDNA transcription. Nucleic Acids Res. 2010; 38:6577–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jemt E., Persson O., Shi Y., Mehmedovic M., Uhler J.P., Davila Lopez M., Freyer C., Gustafsson C.M., Samuelsson T., Falkenberg M.. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015; 43:9262–9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fernandez-Silva P., Enriquez J.A., Montoya J.. Replication and transcription of mammalian mitochondrial DNA. Expt. Physiol. 2003; 88:41–56. [DOI] [PubMed] [Google Scholar]
- 62. Fuste J.M., Wanrooij S., Jemt E., Granycome C.E., Cluett T.J., Shi Y., Atanassova N., Holt I.J., Gustafsson C.M., Falkenberg M.. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010; 37:67–78. [DOI] [PubMed] [Google Scholar]
- 63. Linder T., Park C.B., Asin-Cayuela J., Pellegrini M., Larsson N.G., Falkenberg M., Samuelsson T., Gustafsson C.M.. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005; 48:265–269. [DOI] [PubMed] [Google Scholar]
- 64. Terzioglu M., Ruzzenente B., Harmel J., Mourier A., Jemt E., Lopez M.D., Kukat C., Stewart J.B., Wibom R., Meharg C. et al.. MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 2013; 17:618–626. [DOI] [PubMed] [Google Scholar]
- 65. Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Martinou J.C.. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013; 17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brzezniak L.K., Bijata M., Szczesny R.J., Stepien P.P.. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011; 8:616–626. [DOI] [PubMed] [Google Scholar]
- 67. Carrodeguas J.A., Pinz K.G., Bogenhagen D.F.. DNA binding properties of human pol gammaB. J. Biol. Chem. 2002; 277:50008–50014. [DOI] [PubMed] [Google Scholar]
- 68. Fan L., Sanschagrin P.C., Kaguni L.S., Kuhn L.A.. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9527–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sun Q., Csorba T., Skourti-Stathaki K., Proudfoot N.J., Dean C.. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013; 340:619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ruhanen H., Borrie S., Szabadkai G., Tyynismaa H., Jones A.W., Kang D., Taanman J.W., Yasukawa T.. Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochim. Biophys. Acta. 2010; 1803:931–939. [DOI] [PubMed] [Google Scholar]
- 71. Kornblum C., Nicholls T.J., Haack T.B., Scholer S., Peeva V., Danhauser K., Hallmann K., Zsurka G., Rorbach J., Iuso A. et al.. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 2013; 45:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Uhler J.P., Thorn C., Nicholls T.J., Matic S., Milenkovic D., Gustafsson C.M., Falkenberg M.. MGME1 processes flaps into ligatable nicks in concert with DNA polymerase gamma during mtDNA replication. Nucleic Acids Res. 2016; 44:5861–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nowotny M., Cerritelli S.M., Ghirlando R., Gaidamakov S.A., Crouch R.J., Yang W.. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 2008; 27:1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lima W.F., Wu H., Nichols J.G., Manalili S.M., Drader J.J., Hofstadler S.A., Crooke S.T.. Human RNase H1 activity is regulated by a unique redox switch formed between adjacent cysteines. J. Biol. Chem. 2003; 278:14906–14912. [DOI] [PubMed] [Google Scholar]
- 75. Kazak L., Reyes A., He J., Wood S.R., Brea-Calvo G., Holen T.T., Holt I.J.. A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS One. 2013; 8:e62340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yeeles J.T., Deegan T.D., Janska A., Early A., Diffley J.F.. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015; 519:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hallberg R.L. Mitochondrial DNA in Xenopus laevis oocytes. I. Displacement loop occurrence. Dev. Biol. 1974; 38:346–355. [DOI] [PubMed] [Google Scholar]
- 78. Jacobs H.T., Herbert E.R., Rankine J.. Sea urchin egg mitochondrial DNA contains a short displacement loop (D-loop) in the replication origin region. Nucleic Acids Res. 1989; 17:8949–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lewis D.L., Farr C.L., Farquhar A.L., Kaguni L.S.. Sequence, organization, and evolution of the A+T region of Drosophila melanogaster mitochondrial DNA. Mol. Biol. Evol. 1994; 11:523–538. [DOI] [PubMed] [Google Scholar]
- 80. Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A.. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007; 445:666–670. [DOI] [PubMed] [Google Scholar]
- 81. Nass M.M. Mitochondrial DNA. I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J. Mol. Biol. 1969; 42:521–528. [DOI] [PubMed] [Google Scholar]
- 82. Albring M., Griffith J., Attardi G.. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:1348–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gerhold J.M., Cansiz-Arda S., Lohmus M., Engberg O., Reyes A., van Rennes H., Sanz A., Holt I.J., Cooper H.M., Spelbrink J.N.. Human mitochondrial DNA-protein complexes attach to a cholesterol-rich membrane structure. Sci. Rep. 2015; 5:15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bogenhagen D.F., Martin D.W., Koller A.. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 2014; 19:618–629. [DOI] [PubMed] [Google Scholar]
- 85. Dalla Rosa I., Durigon R., Pearce S.F., Rorbach J., Hirst E.M., Vidoni S., Reyes A., Brea-Calvo G., Minczuk M., Woellhaf M.W. et al.. MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res. 2014; 42:8500–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. He J., Cooper H.M., Reyes A., Di Re M., Kazak L., Wood S.R., Mao C.C., Fearnley I.M., Walker J.E., Holt I.J.. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012; 40:6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Almalki A., Alston C.L., Parker A., Simonic I., Mehta S.G., He L., Reza M., Oliveira J.M., Lightowlers R.N., McFarland R. et al.. Mutation of the human mitochondrial phenylalanine-tRNA synthetase causes infantile-onset epilepsy and cytochrome c oxidase deficiency. Biochim. Biophys. Acta. 2014; 1842:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bonnen P.E., Yarham J.W., Besse A., Wu P., Faqeih E.A., Al-Asmari A.M., Saleh M.A., Eyaid W., Hadeel A., He L. et al.. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am. J. Hum. Genet. 2013; 93:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Desai R., Frazier A.E., Durigon R., Patel H., Jones A.W., Dalla Rosa I., Lake N.J., Compton A.G., Mountford H.S., Tucker E.J. et al.. ATAD3 gene cluster deletions cause cerebellar dysfunction associated with altered mitochondrial DNA and cholesterol metabolism. Brain. 2017; 140:1595–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Durigon R., Mitchell A.L., Jones A.W., Manole A., Mennuni M., Hirst E.M., Houlden H., Maragni G., Lattante S., Doronzio P.N. et al.. LETM1 couples mitochondrial DNA metabolism and nutrient preference. EMBO Mol. Med. 2018; 10:e8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nicholls T.J., Nadalutti C.A., Motori E., Sommerville E.W., Gorman G.S., Basu S., Hoberg E., Turnbull D.M., Chinnery P.F., Larsson N.G. et al.. Topoisomerase 3alpha is required for decatenation and segregation of human mtDNA. Mol. Cell. 2018; 69:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]