Abstract
Background:
Prostate cancer patients who have a detectable prostate-specific antigen (PSA) postprostatectomy may harbor pre-existing metastatic disease. To our knowledge, none of the commercially available genomic biomarkers have been investigated in such men.
Objective:
To evaluate if a 22-gene genomic classifier can independently predict development of metastasis in men with PSA persistence postoperatively.
Design, setting, and participants:
A multi-institutional study of 477 men who underwent radical prostatectomy (RP) between 1990 and 2015 from three academic centers. Patients were categorized as detectable PSA (n = 150) or undetectable (n = 327) based on post-RP PSA nadir ≥0.1 ng/ml.
Outcome measurements and statisitical analysis:
Cumulative incidence curves for metastasis were constructed using Fine-Gray competing risks analysis. Penalized Cox univariable and multivariable (MVA) proportional hazards models were performed to evaluate the association of the genomic classifier with metastasis.
Results and limitations:
The median follow-up for censored patients was 57 mo. The median time from RP to first postoperative PSA was 1.4 mo. Detectable PSA patients were more likely to have higher adverse pathologic features compared with undetectable PSA patients. On MVA, only genomic high-risk (hazard ratio [HR]: 5.95, 95% confidence interval [CI]: 2.02–19.41, p = 0.001), detectable PSA (HR: 4.26, 95% CI: 1.16–21.8, p = 0.03), and lymph node invasion (HR: 12.2, 95% CI: 2.46–70.7, p = 0.003) remained prognostic factors for metastasis. Among detectable PSA patients, the 5-yr metastasis rate was 0.90% for genomic low/intermediate and 18% for genomic high risk (p < 0.001). Genomic high risk remained independently prognostic on MVA (HR: 5.61, 95% CI: 1.48–22.7, p = 0.01) among detectable PSA patients. C-index for Cancer of the Prostate Risk Assessment Postsurgical score, Gandaglia nomogram, and the genomic classifier plus either Cancer of the Prostate Risk Assessment Postsurgical score or Gandaglia were 0.69, 0.68, and 0.82 or 0.81, respectively. Sample size was a limitation.
Conclusions:
Despite patients with a detectable PSA harboring significantly higher rates of aggressive clinicopathologic features, Decipher independently predicts for metastasis. Prospective validation of these findings is warranted and will be collected as part of the ongoing randomized trial NRG GU-002.
Patient summary:
Decipher independently predicted metastasis for patients with detectable prostate-specific antigen after prostatectomy.
Keywords: Prostate cancer, Genomic classifier, Decipher, Detectable PSA, Persistent PSA, Prostatectomy, Biomarker, PSA
1. Introduction
Prostate cancer is now the third most common malignancy in men in the USA, with an estimated 160 000 new cases diagnosed annually in 2017 [1]. Radical prostatectomy (RP) is a common form of radical treatment for localized prostate cancer [2]. However, men with high-risk features, including high-grade group, stage T3–4, lymph node (LN) invasion, or positive margins have a >40% chance of recurrence 5–10-yr postoperatively [3]. A subset of these patients will even have immediate evidence of persistent disease and their prostate-specific antigen (PSA) will never become undetectable after surgery. A detectable PSA after RP is a poor prognostic factor, and is often associated with more advanced disease and aggressive clinical course [4]. Functionally, it indicates either persistent local disease or potentially pre-existent metastatic disease. Therefore, treatment of these patients today often is both local (post-RP radiotherapy) and systemic (androgen deprivation therapy [ADT]) [5].
Multiple genomic classifiers have recently been developed and are commercially available to help prognosticate outcomes for men with prostate cancer [6,7]. Decipher, a 22-gene RNA-based genomic classifier, utilizes tissue from the RP specimens to help determine a patients risk of metastasis, independent of clinicopathologic variables [8,9]. However, none of the previous 40+ studies testing Decipher, including the recent individual patient meta-analysis of Decipher, included or specifically examined the genomic classifier in men with persistently detectable PSAs post-RP [10–14]. Given that a subset of men with detectable PSAs postoperatively likely already harbor metastatic disease, it is unclear if a tissue-based biomarker could help in this patient population.
Herein, we conducted the first study to examine the performance of a commonly used prognostic genomic classifier in men with persistently detectable PSAs post-RP to determine if it can independently add prognostic benefit to predict for metastases.
2. Materials and methods
2.1. Study cohort
Institutional Review Board approval was obtained from the participating institutions prior to initiating the current study [15,16]. Patients were included from three centers: MD Anderson Cancer Center, Durham VA Hospital, and Thomas Jefferson University. Patients were required to have undergone RP, sufficient tissue for genomic analysis, and serial PSA measurements post-RP to document undetectable versus persistently detectable PSAs postoperatively. Noedajuvant ADT was not allowed, and only 1% (N = 4) had ADT within 3 mo post-RP. A total of 477 men, who underwent RP between 1990 and 2015, met study inclusion criteria and formed the study cohort. None of the patients in the study received neoadjuvant treatment prior to RP. Patients whose PSA level fails to fall to undetectable levels (<0.1 ng/ml) within approximately 8 wk after RP were categorized as detectable PSA group.
2.2. Decipher score
Expression analysis of 22 biomarkers that comprise the commercial Decipher test were extracted and analyzed as previously described [8]. Analyses of the Decipher score were performed using two methods: (1) a continuous score between 0 and 1, and (2) previously established cutpoint scores of 0.45 and 0.60 to categorize patients into low-, intermediate-, and high-risk categories [10].
2.3. Covariables and endpoints
Analyzed variables included whether patients had a detectable or undetectable PSA post-RP (defined by a post-RP PSA within 8 wk of surgery), International Society of Urological Pathology (ISUP) grade (Gleason grade), pathologic T-stage, LN invasion, surgical margin status, Cancer of the Prostate Risk Assessment Postsurgical (CAPRA-S) risk group, preoperative PSA, use of radiotherapy post-RP and prior to developing metastatic disease, and Decipher score (continuous or categorical). Given the years of the study cohort [17], adjuvant radiotherapy was defined as those who received radiotherapy <6 mo post-RP and who had PSA levels <0.5 ng/ml, and salvage radiotherapy were all other patients. Patients with a detectable PSA post-RP who received radiotherapy were classified as salvage radiotherapy. Given that men with detectable PSAs postoperatively already had evidence of biochemical failure, rates of distant metastasis were used as the primary endpoint for the study. Time to distant metastasis was calculated using a landmark analysis from time of 8-wk post-RP until radiographic evidence by computed tomography or bone scan of either bone, viscera or LN metastasis, or last follow-up.
2.4. Statistical analysis
To compare patient characteristics among detectable and undetectable PSA patients, Wilcoxon rank sum test and Fisher’s exact test were used for continuous and categorical variables, respectively. Cumulative incidence curves to estimate distant metastasis were constructed using Fine-Gray competing risks analysis for deaths from other causes [18]. Cox univariable (UVA) and multivariable (MVA) proportional hazards models with Firth’s penalized likelihood method were used on both the overall cohort (patients with detectable and undetectable PSAs post-RP) and solely within patients with a detectable post-RP PSA to determine predictors of distant metastasis [19]. Firth’s penalized likelihood method was used for identification of the prognostic risk factors to ensure the robustness of the analyses and avoid overestimation of the resulting estimates [19]. Variables of categorical Decipher, whether patients had a detectable or undetectable PSA post-RP, ISUP grade, pathologic T-stage, LN invasion (LNI), surgical margin status, and preoperative PSA were included in the UVA and MVA models on the overall cohort. The same variables except whether patients with detectable PSA and LNI (only 2 PSA-detectable patients with LNI) were included in the models on patients with detectable PSA. Further, we performed a sensitivity analysis including institution as a stratification and observed no significant differences (data not shown). Then, we performaed another sensitivity analysis including Decipher, CAPRA-S, and whether patients had a detectable PSA or post-RP PSA on overall cohort, and including Decipher, CAPRA-S, and with/without post-RP PSA on patients with detectable PSA. Survival c-indices of continuous Decipher alone, CAPRA-S, clinical model (preoperative PSA, RP grade groups, surgical margins, and pathological stage), Gandaglia 2017 nomogram [20] and their combination with categorical Decipher were calculated at 5-yr post-RP. Time-dependent c-indices were used to measure the discrimination of the risk factors [21]. C-index of the combined models was estimated by subjecting the model to bootstrapping with 500 resamples to correct for optimism. Extension of the decision curve analysis to survival data with 10-fold cross validation to correct for overfitting was employed to evaluate the net benefit of Decipher, CAPRA-S, clinical model, Gandaglia 2017 nomogram, and their combination with Decipher across clinically relevant threshold probabilities [22]. All statistical tests were two-sided using a 5% significance level and analyses were performed in R version 3.3 (R Foundation, Vienna, Austria). In this manuscript, we adhered to the European Urology’s guidelines for reporting of statistics [23].
3. Results
The median follow-up for the censored patients was 57 mo (Q1–3, 31–90 mo), and the median follow-up for censored patients with a detectable PSA post-RP (n = 150) was 77 mo (Q1–3, 46–126 mo). The median age was 60 yr, 21% were African-American and 73% were Caucasian, 26% were CAPRA-S high risk, the median pre-RP PSA was 6.4 ng/ml, the median post-RP PSA was 0.1 ng/ml, the median time from RP to when post-RP PSA was measured is 1.4 mo (Q1–3, 1.4–1.6 mo), the majority of patients (79%) had grade group 2 or 3 cancers, 46% had extraprostatic extension, 21% had seminal vesicle invasion, 48% had positive margins, and 8% had LNI (Table 1). In the entire cohort, 53% received post-RP radiotherapy prior to developing metastasis, and of the detectable PSA patients, 95% received post-RP radiotherapy. Ten out of 150 patients with detectable PSA and six out of 327 undetectable PSA patients developed metastases during the study follow-up period.
Table 1 –
Demographic and clinical characteristics among undetectable and detectable prostate-specific antigen (PSA) patients and overall cohort
| Variable | Undetectable PSA | Detectable PSA | p value |
|---|---|---|---|
| No. patients | 327 | 150 | |
| Patient age (yr) | 0. 3 | ||
| Median (Q1, Q3) | 60 (55, 64) | 61 (56, 65) | |
| Race, n (%) | <0.001a | ||
| African American | 46 (14) | 55 (37) | |
| Caucasian | 258 (79) | 92 (61) | |
| Other | 19 (5.8) | 3 (2.0) | |
| Unknown | 4 (1.2) | ||
| Pre-operative PSA (ng/ml) | <0.001a | ||
| Median (Q1, Q3) | 6.1 (4.6, 9) | 7.3 (5.1, 12) | |
| Unknown | 4 (2.7%) | ||
| RP grade groups, n (%) | <0.001a | ||
| Group 1 | 13 (4.0) | 19 (13) | |
| Group 2 | 161 (49) | 60 (40) | |
| Group 3 | 115 (35) | 41 (27) | |
| Group 4 | 16 (4.9) | 18 (12) | |
| Group 5 | 20 (6.1) | 12 (8.0) | |
| Unknown | 2 (0.61) | ||
| Extraprostatic extension, n (%) | 133 (41) | 86 (57) | <0.001a |
| Unknown | 2 (1.3) | ||
| Seminal vesicle invasion, n (%) | 50 (15) | 49 (33) | <0.001a |
| Unknown | 2 (0.61) | 1 (0.67) | |
| Lymph node invasion, n (%) | 36 (11) | 2 (1.3) | <0.001a |
| Unknown | 1 (0.31) | ||
| Positive surgical margins, n (%) | 110 (34) | 119 (79) | <0.001a |
| Pathological stage, n (%) | <0.001a | ||
| T2 | 182 (56) | 49 (33) | |
| T3a | 91 (28) | 44 (29) | |
| T3b | 50 (15) | 49 (33) | |
| T4 | 2 (0.61) | 6 (4.0) | |
| Unknown | 2 (0.61) | 2 (1.3) | |
| CAPRA-S risk, n (%) | <0.001a | ||
| Low | 109 (33) | 14 (9.3) | |
| Intermediate | 142 (43) | 61 (41) | |
| High | 55 (17) | 66 (44) | |
| Unknown | 21 (6.4) | 9 (6) | |
| Post-operative PSA (ng/ml) | <0.001a | ||
| Median (Q1, Q3) | — | 0.2 (0.1, 0.5) | |
| Time from RP to post-operative PSA (mo) | NA | ||
| Median (Q1, Q3) | 1.4 (1.4, 1.4) | 2.5 (1.8, 3.9) | |
| Follow-up time (mo) | NA | ||
| Median (Q1, Q3) | 45 (27, 73) | 77 (46, 126) | |
| RT before mets, n (%) | 109 (33) | 142 (95) | <0.001a |
| Unknown | 1 (0.67) | ||
| ADT before mets, n (%) | 23 (7.0) | 38 (25) | <0.001a |
| Unknown | 1 (0.31) | 1 (0.67) | |
| Decipher risk group, n (%) | 0.12 | ||
| Low | 142 (43) | 79 (53) | |
| Intermediate | 93 (28) | 40 (27) | |
| High | 92 (28) | 31 (21) |
ADT = androgen deprivation therapy; CAPRA-S = Cancer of the Prostate Risk Assessment Postsurgical Score; mets = metastasis; RP = radical prostatectomy; RT = radiotherapy.
To compare patient characteristics among detectable PSA and undetectable patients, Wilcoxon rank sum test, and Fisher’s exact test were used for continuous and categorical variables, respectively.
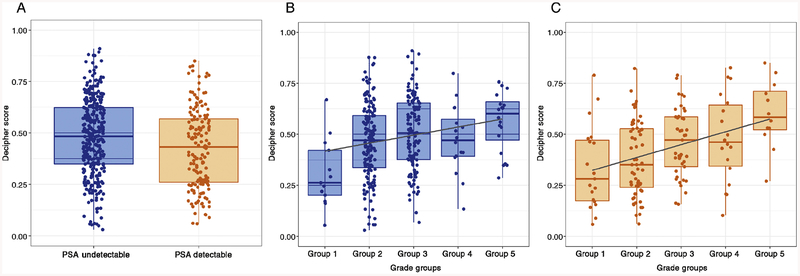
Detectable PSA patients were significantly more likely than undetectable PSA patients to be African-American, harbor higher clinical risk features such as higher pre-RP PSA, post-RP PSA, extraprostatic extension, seminal vesicle invasion, positive margins, had received post-RP radiotherapy, and had received post-RP ADT (p < 0.001). Decipher scores correlated strongly within ISUP grade group in detectable and undetectable PSA patients (Figs. 1B–C).
Fig. 1 –
(A) Boxplot of Decipher score among undetectable prostate-specific antigen (PSA) and detectable PSA patients. (B) Boxplot of Decipher score versus radical prostatectomy grade groups among undetectable PSA patients. (C) Boxplot of Decipher score versus radical prostatectomy grade groups among detectable PSA patients.
Within the entire cohort, univariable analysis demonstrated that a higher Decipher score, grade group 4–5, higher pathologic T-stage, and LNI were all significant predictors of metastasis (Table 2). Decipher intermediate risk was not significantly different from Decipher low risk and were thus grouped together for the MVA. On MVA, the only independent predictors of metastases were Decipher high risk (hazard ratio [HR]: 5.95, 95% confidence interval [CI]: 2.02–19.4, p = 0.001), detectable PSA (HR: 4.26, 95% CI: 1.16–21.8, p = 0.03), and LNI (HR: 12.2, 95% CI: 2.46–70.7, p = 0.003). Gleason score, margin status, and T-stage were no longer significant (Table 2). In a second MVA model incorporating CAPRA-S into the model, Decipher score remained significant (p = 0.001; Supplementary Table 3). Additionally, an interaction analysis was performed, and there was no significant interaction effect of Decipher with patients with or without detectable PSA (p = 0.6) or by institution.
Table 2 –
Univariable (UVA) and multivariable (MVA) of Decipher adjusting for detectable prostate-specific antigen (PSA) and clinical data for prediction of metastasis for the overall cohort
| Variables | Category | UVA | MVA | ||
|---|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | ||
| Decipher risk (2 categories) | Low/intermediate | Ref | |||
| High | 7.71 (2.88–23.3) | <0.001 | 5.95 (2.02–19.4) | 0.001 | |
| Pre-operative PSAa | 1.55 (0.95–2.43) | 0.08 | 1.24 (0.79–1.92) | 0.4 | |
| Group 1 & 2 | Ref | ||||
| RP grade groups | Group 3 | 3.04 (0.92–11.0) | 0.07 | 1.57 (0.45–5.95) | 0.5 |
| Group 4 & 5 | 5.1 (1.55–18.4) | 0.008 | 1.53 (0.42–6.06) | 0.5 | |
| Positive surgical margins | 3.76 (1.42–9.93) | 0.009 | 1.22 (0.4–4.05) | 0.7 | |
| Lymph node invasion | 9.74 (2.85–27.9) | <0.001 | 12.18 (2.46–70.7) | 0.003 | |
| T2 | Ref | ||||
| Pathological stage | T3a | 7.93 (1.73–75.19) | 0.006 | 4.74 (0.96–46.3) | 0.056 |
| T3b & T4 | 11.5 (2.59–108.4) | <0.001 | 4.6 (0.95–44.9) | 0.059 | |
| Detectable PSA | 2.36 (0.89–6.72) | 0.08 | 4.26 (1.16–21.8) | 0.03 | |
CI = confidence interval; HR = hazard ratio; Ref = reference; RP = radical prostatectomy.
Preoperative PSA values were log2 transformed.
Within only the detectable PSA cohort, UVA analysis demonstrated that Decipher score, higher grade group, and preoperative PSA were statistically significant predictors of metastasis (Table 3). On MVA, the only independent predictor of metastases was Decipher high risk (HR: 5.61, 95% CI: 1.48–22.7, p = 0.01). Grade group, pre-RP PSA, and T-stage were no longer significant. In a second MVA model incorporating CAPRA-S into the model in only PSA-detectable patients, Decipher score remained significant (p = 0.002; Supplementary Table 4). Furthermore, the c-index for Decipher (0.86) alone without the use of any information from clinicopathologic features was higher than CAPRA-S (0.69), which is a composition of six clinicopathologic factors, and the Gandaglia nomogram (0.68), which is composed of five clinicopathologic factors (Supplementary Table 5). The combination of Decipher and CAPRA-S or Decipher with the Gandaglia nomogram both resulted in a c-index for 5-yr metastasis of 0.83 and 0.81. In the decision curve analyses (Supplementary Fig. 2), clinical model, CAPRA-S, or Gandaglia all resulted in higher net benefit with combination of Decipher.
Table 3 –
Univariable (UVA) and multivariable (MVA) of Decipher adjusting for clinical data for prediction metastasis among detectable prostate-specific antigen (PSA) patients
| Variables | Category | UVA | MVA | ||
|---|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | ||
| Decipher risk (2 categories) | Low/intermediate | Ref | |||
| High | 7.23 (2.17–26.3) | 0.002 | 5.61 (1.48–22.7) | 0.01 | |
| Pre-operative PSAa | 2.26 (1.35–3.69) | 0.003 | 1.62 (0.98–2.74) | 0.062 | |
| Group 1 & 2 | Ref | ||||
| RP grade groups | Group 3 | 3.96 (0.88–22.7) | 0.07 | 2.69 (0.52–18.7) | 0.2 |
| Group 4 & 5 | 5.49 (1.21–31.6) | 0.028 | 1.78 (0.28–12.8) | 0.5 | |
| Positive surgical margins | 0.71 (0.19–3.75) | 0.6 | 0.85 (0.19–5.04) | 0.8 | |
| T2 | Ref | ||||
| Pathological stage | T3a | 3.4 (0.63–33.9) | 0.2 | 1.57 (0.21–18.2) | 0.7 |
| T3b & T4 | 4 (0.80–39.1) | 0.10 | 1.27 (0.18–14.6) | 0.8 | |
CI = confidence interval; HR = hazard ratio; RP = radical prostatectomy; Ref = reference.
Preoperative PSA values were log2 transformed. Since there were only two patients with lymph node invasion, it was not included in the MVA model
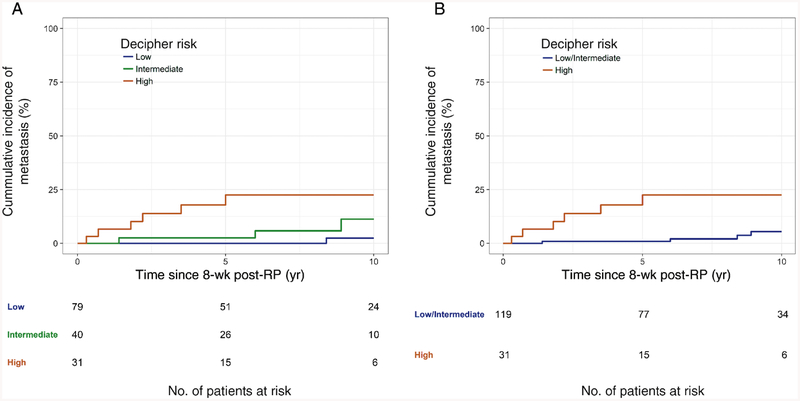
Figure 2 demonstrates the cumulative incidence curves for metastases with death as a competing risk in patients with a detectable PSA post-RP. Consistent with the UVA results (Table 3), there were no significant differences between Decipher low- and Decipher intermediate-risk patients. However, Decipher high patients had significantly higher 5-yr rates of metastases compared with Decipher low/intermediate-risk patients (23% vs 0.90%, p < 0.001). Ten-yr metastases rates were also significantly higher in Decipher high patients (23% vs 5.4%).
Fig. 2 –
Cumulative incidence curves of metastasis for (A) detectable prostate-specific antigen patients with Decipher low-(<0.45), intermediate-(0.45–0.60), and high-risk (>0.60) (B) detectable prostate-specific antigen patients with Decipher low/intermediate (≤0.60) and high risk (>0.60). RP = radical prostatectomy.
4. Discussion
In the first study to examine the use of the 22-gene genomic classifier, Decipher, in men with persistently detectable PSAs post-RP, we demonstrate that Decipher independently predicts for metastasis when correcting for grade group, T-stage, margin status, LNI, and receipt of post-RP radiotherapy. Furthermore, despite patients with a detectable PSA having relatively aggressive clinicopathologic risk features (44% were CAPRA-S high risk, 67% had T3–T4 disease, 79% had positive margins, and 1% had LNI), Decipher reclassified 79% with a low/intermediate risk of metastasis (1% metastasis rate at 5-yr for Decipher low/intermediate risk).
Multiple genomic classifiers have been widely studied in a variety of clinicopathologic subgroups and have demonstrated a consistent prognostic ability to predict for the specific tests outcome of interest [12,24–28]. For example, a recent meta-analysis of individual patient data from Decipher studies on over 850 patients, demonstrated that the prognostic biomarker was independently associated with metastasis across race, treatment, and clinicopathologic characteristics [12]. However, no studies to date to the authors’ knowledge, including the recent meta-analysis, have examined the use of any tissue-based genomic biomarker to predict outcomes in men who have persistently detectable PSAs post-RP.
It is plausible that a biomarker from the prostate itself may not accurately discriminate who has persistent local versus metastatic disease, and thus limiting the utility of these commercial tests in this subset of men. Given that men with persistently detectable PSAs also often harbor additional aggressive clinicopathologic features, pre-existing micrometastatic disease is common. However, our results interestingly demonstrate that genomic testing on the primary specimen not only is able to independently predict for the development of distant metastasis, it was the only prognostic factor that remained significant on MVA in men with persistently detectable PSAs post-RP. Thus, Decipher appears to be able to capture the biology of the disease process (indolent vs aggressive) regardless of the potential location of the persistent disease process.
Consistent with the literature, we demonstrate that having a persistently detectable PSA is one of the worst prognostic factors associated with the development of metastasis [4]. We demonstrated on MVA, the presence of a persistently detectable PSA post-RP is associated with a four-fold increase in the risk of development of metastasis. This population in general is at a much higher risk of metastasis compared with men that reach an undetectable PSA post-RP, and despite 95% of patients receiving post-RP radiotherapy, many patients (7.0%) developed metastasis during the follow-up period. For this reason, treatment intensification is warranted. However, it will be critical for multi-modal therapy trials in men with persistently detectable PSAs to stratify by more than merely clinicopathologic features, as our analyses demonstrate Decipher high patients have a five-fold increased rate of metastases compared with Decipher low/intermediate. This is important not only for balancing treatment arms through stratification, but also for powering the study given that Decipher high patients had a 23% 5-yr metastasis rate compared with 1.0% for low/intermediate-risk patients.
A recently opened NRG randomized phase 2 trial, with a potential lead in to phase 3 trial, is testing if the addition of docetaxel to post-RP radiotherapy with ADT will improve outcomes in men with persistently detectable PSAs post-RP. This trial will be one of, if not the first, cooperative group trial to utilize Decipher as a stratification variable. Based on the present work, we believe that stratifying by Decipher low/intermediate versus high risk would be of value, and NRG GU-002 ( NCT03070886) would provide prospective validation for the present study. Importantly, nearly all men with detectable PSAs in our study received salvage radiotherapy, so the primary utility of Decipher in these men would be to guide systemic treatment (eg, ADT and/or chemotherapy).
Limitations of our work exist. First, the retrospective methodology is subject to bias. Second, the sample size of men with detectable PSAs post-RP may have limited the power to detect differences in various prognostic variables. Third, although the present study was comprised from three institutions, our results need validation from independent cohorts. Lastly, although men with a low/intermediate risk Decipher score had a very low rate of metastasis at 5 yr, this information should not be used to guide omission of post-RP radiotherapy as 95% of patients with detectable PSAs post-RP received salvage radiotherapy.
5. Conclusions
Despite patients with a detectable PSA harboring significantly higher rates of aggressive clinicopathologic features, Decipher independently was able to identify men at low and high risk for developing metastasis. Prospective validation of these findings is warranted and will be collected as part of the ongoing randomized trial NRG GU-002 ( NCT03070886).
Supplementary Material
Funding/Support and role of the sponsor:
GenomeDx Biosciences assisted with the design and conduct of the study, collection of the data, management of the data, analysis, and iterpretation of the data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures:
Daniel E. Spratt certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Nguyen has consulted for Medivation, GenomeDx, Ferring, Nanobiotix, and Dendreon. Nguyen has received research funding from Astellas. Feng has consulted for Medivation and GenomeDx. Davis has received research grant from GenomeDx Biosciences. Haddad, Yousefi, Dai, and Davicioni are employees of GenomeDx Biosciences. Spratt was supported by the Prostate Cancer Foundation Young Investigator Award.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Cooperberg MR, Carroll PR. Trends in management for patients with localized prostate cancer, 1990–2013. JAMA 2015;314:80–2. [DOI] [PubMed] [Google Scholar]
- [3].MSKCC. Prostate cancer nomograms. https://www.mskcc.org/nomograms/prostate/preop.
- [4].Wiegel T, Bartkowiak D, Bottke D, et al. Prostate-specific antigen persistence after radical prostatectomy as a predictive factor of clinical relapse-free survival and overall survival: 10-year data of the ARO 96–02 trial. Int J Radiat Oncol Biol Phys 2015;91:288–94. [DOI] [PubMed] [Google Scholar]
- [5].Mohler JL, Armstrong AJ, Bahnson RR, et al. Prostate cancer, Version 1. 2016. J Natl Compr Canc Netw 2016;14:19–30. [DOI] [PubMed] [Google Scholar]
- [6].Ross AE, D’Amico AV, Freedland SJ. Which, when and why? Rational use of tissue-based molecular testing in localized prostate cancer. Prostate Cancer Prostatic Dis 2016;19:1–6. [DOI] [PubMed] [Google Scholar]
- [7].Colicchia M, Morlacco A, Cheville JC, Karnes RJ. Genomic tests to guide prostate cancer management following diagnosis. Expert Rev Mol Diagn 2017;17:367–77. [DOI] [PubMed] [Google Scholar]
- [8].Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 2013;8:e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Den RB, Santiago-Jimenez M, Alter J, et al. Decipher correlation patterns post prostatectomy: initial experience from 2 342 prospective patients. Prostate Cancer Prostatic Dis 2016;19:374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ross AE, Johnson MH, Yousefi K, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol 2016;69:157–65. [DOI] [PubMed] [Google Scholar]
- [11].Glass AG, Leo MC, Haddad Z, et al. Validation of a genomic classifier for predicting post-prostatectomy recurrence in a community based health care setting. J Urol 2016;195:1748–53. [DOI] [PubMed] [Google Scholar]
- [12].Spratt DE, Yousefi K, Deheshi S, et al. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol 2017;35:1991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karnes RJ, Choeurng V, Ross AE, et al. Validation of a genomic risk classifier to predict prostate cancer-specific mortality in men with adverse pathologic features. Eur Urol. In press 10.1016/j.eururo.2017.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Klein EA, Santiago-Jiménez M, Yousefi K, et al. Molecular analysis of low grade prostate cancer using a genomic classifier of metastatic potential. J Urol 2017;197:122–8. [DOI] [PubMed] [Google Scholar]
- [15].Den RB, Yousefi K, Trabulsi EJ, et al. Genomic classifier identifies men with adverse pathology after radical prostatectomy who benefit from adjuvant radiation therapy. J Clin Oncol 2015;33:944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Freedland SJ, Choeurng V, Howard L, et al. Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol 2016;70:588–96. [DOI] [PubMed] [Google Scholar]
- [17].Thompson IM, Tangen CM, Paradelo J, et al. Adjuvant radiotherapy for pathological T3N0M0 prostate cancer significantly reduces risk of metastases and improves survival: long-term followup of a randomized clinical trial. J Urol 2009;181:956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 1999;94:496–509. [Google Scholar]
- [19].Firth D Bias reduction of maximum likelihood estimates. Biometrika 1993;80:27–38. [Google Scholar]
- [20].Gandaglia G, Boorjian SA, Parker WP, et al. Impact of postoperative radiotherapy in men with persistently elevated prostate-specific antigen after radical prostatectomy for prostate cancer: a long-term survival analysis. Eur Urol 2017;72:910–7. [DOI] [PubMed] [Google Scholar]
- [21].Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337–44. [DOI] [PubMed] [Google Scholar]
- [22].Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 2008;8:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vickers AJ, Sjoberg DD. Guidelines for reporting of statistics in European urology. Eur Urol 2015;67:181–7. [DOI] [PubMed] [Google Scholar]
- [24].Klein E, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol 2014;66:440–60. [DOI] [PubMed] [Google Scholar]
- [25].Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol 2013;31:1428–34. [DOI] [PubMed] [Google Scholar]
- [26].Klein EA, Haddad Z, Yousefi K, et al. Decipher genomic classifier measured on prostate biopsy predicts metastasis risk. Urology 2016;90:148–52. [DOI] [PubMed] [Google Scholar]
- [27].Freedland SJ, Choeurng V, Howard L, et al. Utilization of a genomic classifier for prediction of metastasis following salvage radiation therapy after radical prostatectomy. Eur Urol 2016;70:588–96. [DOI] [PubMed] [Google Scholar]
- [28].Nguyen PL, Haddad Z, Ross AE, et al. Ability of a genomic classifier to predict metastasis and prostate cancer-specific mortality after radiation or surgery based on needle biopsy specimens. Eur Urol 2017;72:845–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.